Abstract
COVID-19, which is caused by the emerging human coronavirus SARS-CoV-2, has become a global pandemic that poses a serious threat to human health. To date, no vaccines or specific antiviral drugs have been approved for the treatment of this disease in clinic. Herein, therapeutic antibodies for SARS-CoV-2 were obtained from hyperimmune equine plasma. First, a recombinant SARS-CoV-2 spike protein receptor-binding domain (RBD) was obtained in gram-level quantities through high-cell density fermentation of Chinese hamster ovary cells. Then, the binding of the RBD to the SARS-CoV-2 receptor, human angiotensin-converting enzyme 2, was verified by several biochemical methods. The efficacy of the RBD in triggering antibody response in vivo was subsequently tested in both mice and equines, and the results showed that the RBD triggered high-titer neutralizing antibody production in vivo. Immunoglobulin F(ab’)2 fragments were prepared from equine antisera via removal of the Fc region from the immunoglobulins. Finally, a neutralization test with live virus demonstrated that RBD-specific F(ab’)2 inhibited SARS-CoV-2 with an EC50 of 0.07 μg/ml and an EC80 of 0.18 μg/ml, showing a potent inhibitory effect on SARS-CoV-2. These results highlight RBD-specific equine immunoglobulin F(ab’)2 fragment as a candidate for the treatment of SARS-CoV-2.
Keywords: SARS-CoV-2, Neutralizing antibody, Receptor-binding domain, Immunoglobulin fragment, COVID-19
Highlights
-
•
SARS-CoV-2 RBD triggered strong antibody responses both in mice and equines.
-
•
High neutralizing-titer antisera were obtained by immunizing equine with CHO cell-expressed RBD protein.
-
•
RBD-specific immunoglobulin F(ab’)2 fragments potently neutralized SARS-CoV-2 in vitro.
1. Introduction
In recent years, emerging or reemerging viruses, including severe acute respiratory syndrome coronavirus (SARS-CoV), Ebola virus, Lassa virus, Zika virus, H1N1 influenza virus, Middle East respiratory syndrome-related coronavirus (MERS-CoV) and others, have challenged the global biosafety system and attracted substantial attention worldwide. SARS-CoV-2, identified at the beginning of 2020 (Zhou et al., 2020), causes coronavirus disease 2019 (COVID-19), has become a global pandemic now. Unfortunately, no vaccines or drugs have been approved for clinical use in the treatment of this disease, although some are already in clinical trials (Zhang and Wang, 2020; Zhang et al., 2020), such as chloroquine and remdesivir (Gao et al., 2020; Wang et al., 2020). The epidemic situation urgently calls for effective, specific, and quickly accessible drugs (Jiang et al., 2020).
Neutralizing antibodies (nAbs) play important roles in antiviral therapy (Corti et al., 2016; Hastie et al., 2017; Sapparapu et al., 2016) because they effectively inhibit viral entry through mechanisms such as prevention of viral attachment or membrane fusion. Polyclonal antibodies, such as those in convalescent plasma from recovered patients, are commonly produced as emergency treatments for emerging infectious diseases (Ankcorn et al., 2019; Brown et al., 2018; Ruggiero et al., 1986; Shen et al., 2020). However, the lack of blood sources and the risk of bloodborne diseases have typically impeded the widespread clinical application of convalescent plasma (Tiberghien et al., 2020). Antisera produced by large animals, such as equines, after active immunization can be used as alternatives to convalescent plasma (Lu et al., 2005; Pan et al., 2020; Wang et al., 2019). They are effective in treating complex and intractable diseases, especially snakebites and highly pathogenic infectious diseases (Ratanabanangkoon et al., 2016; Wang et al., 2019).
SARS-CoV-2 has been reported to use angiotensin-converting enzyme 2 (ACE2) to enter host cells (Hoffmann et al., 2020), uses the same receptor as SARS-CoV (Song et al., 2018). The amino acid sequence identity between the SARS-CoV-2 and SARS-CoV spike (S) proteins is approximately 76% (Zheng and Song, 2020). The S protein consists of S1 and S2; S1 is responsible for receptor attachment, and S2 is responsible for membrane fusion (Walls et al., 2020). The receptor-binding domain (RBD) of SARS-CoV S1 can potently induce nAb production in vivo (Du et al., 2009). Thus, the SARS-CoV-2 RBD may be a good immunogen for induction of nAb production in vivo.
Based on the above, we tried to produce equine antiserum against SARS-CoV-2 RBD and explore its potential in anti-SARS-CoV-2 in the current study. Therefore, SARS-CoV-2 RBD was expressed in mammalian cells, and its antigenicity and efficacy were tested in both mice and equines. Through traditional systemic immunization, the RBD elicited high-titer nAbs in equines. The immunoglobulins were purified from hyperimmune equine plasma, immunoglobulin F(ab’)2 fragments were acquired via removal of the Fc regions from the immunoglobulins, and the efficacy of the F(ab’)2 fragments was evaluated through a set of in vitro neutralization tests with live virus. The results reported here confirm the efficacy of the RBD in triggering nAb production in vivo and highlight RBD-specific F(ab’)2 fragments as a therapeutic option for the treatment of COVID-19.
2. Materials and methods
2.1. Cells lines and viruses
Vero-E6 and HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM, Gibco, NY, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, NY, USA) at 37 °C with 5% CO2. ExpiCHO-S cells were cultured in ExpiCHO Expression Medium (Gibco, NY, USA) in an incubator at 37 °C under 8% CO2 with shaking at 125 rpm/min.
Live SARS-CoV-2 (nCoV-2019BetaCoV/Wuhan/WIV04/2019) was obtained from the National Virus Resource, Wuhan Institute of Virology, Chinese Academy of Sciences, and handled in a BSL-3 laboratory. SARS-CoV-2 was passaged in Vero-E6 cells.
2.2. Protein expression and purification
The sequence of the SARS-CoV-2 RBD gene (amino acids 319–541) was synthesized by GenScript Co., Ltd., and cloned into the pCAGGS eukaryotic expression plasmid to obtain pCAGGS-Signal peptide-RBD-(Thrombin site)-Fc. The plasmid was purified from E. coli (DH5α) with an endotoxin-free plasmid extraction kit (Invitrogen, CA, USA) and transfected into ExpiCHO-S cells using an ExpiFectamine Transfection Kit (Gibco, NY, USA). Cell supernatants containing RBD-Fc were collected 8 days later and filtered with a 0.22-μm membrane before being subjected to affinity chromatography through Protein A agarose. The eluted fraction that contained RBD-Fc was collected for further analysis. The RBD without an Fc tag was obtained from RBD-Fc by removing the Fc region by thrombin digestion. The RBD proteins were collected from the flow-through faction, and affinity chromatography against Protein A was repeatedly conducted to completely remove the residual Fc regions.
2.3. Reducing SDS-PAGE
The prepared RBD-Fc and RBD proteins were analyzed by SDS-PAGE with Coomassie Brilliant Blue staining. To verify the sizes and purities of these proteins, reducing SDS-PAGE was employed. The reduced samples were prepared by mixing 2 μg of the proteins with loading buffer, adding 2-β-mercaptoethanol, and then boiling the mixtures in water for 10 min. The samples were concentrated via 4% SDS-PAGE and separated via 10% SDS-PAGE. Finally, the SDS-PAGE gels were stained with Coomassie Brilliant Blue. After decolorization, images of the gels were captured with a ChemiDoc MP Imaging system (Bio-Rad, CA, USA).
2.4. Flow cytometry
The binding of the RBD to human ACE2 was detected by flow cytometry as described elsewhere (Tai et al., 2020). HeLa cells were seeded in 6-well plates overnight. PcDNA3.1-human ACE2 was transfected into HeLa cells with Lipofectamine 2000 (Invitrogen), and PcDNA3.1 was used as a control. Twenty-four hours later, the cells were scraped off and washed with PBS. The RBD was labeled with biotin and dialyzed with PBS to remove the residual biotin before use. The biotin-labeled RBD was added to ACE2-overexpressing cells or mock-treated cells at a final concentration of 10 μg/ml. After incubation at 4 °C for 30 min, the cells were washed with PBS. Then, PE-Cy7-conjugated streptavidin (BD Biosciences, CA, USA) was added to the cells. After incubation at 4 °C for 15 min, the cells were washed and resuspended in PBS before detection by flow cytometry (BD, CA, USA).
2.5. Receptor blockade assay
The inhibition of SARS-CoV-2 entry by the RBD protein was carried out as previously described, with some modifications (Tai et al., 2016). Vero-E6 cells were seeded in 48-well plates at a density of 5 × 104 cells/well overnight. After the culture medium was removed, RBD protein diluted in 2% FBS-DMEM was incubated with cells at concentrations of 20, 6.6 and 2.2 μg/ml at 37 °C for 1 h, and PBS was used as a control. Then, the proteins were removed, and the cells were washed twice with PBS. SARS-CoV-2 in 100 μl of 2% FBS-DMEM was added to each well at a multiplicity of infection (MOI) of 0.05. After incubation at 37 °C for 1 h, the supernatant was completely removed, and the cells were washed twice with PBS before fresh 2% FBS-DMEM was added. Twenty-four hours later, the cell supernatants were collected for viral copy number detection. Infection by SARS-CoV-2 was calculated as a percentage of that in the control group.
2.6. Mouse immunization and sampling
Female BALB/c mice aged 6–8 weeks (n = 6) were housed in specific pathogen-free animal care facilities. According to a homogeneous prime-boost-boost protocol, immunization was performed three times in total with two-week intervals. In detail, 25 μg of RBD protein in a volume of 100 μl of PBS was mixed with 100 μl of Freund's complete adjuvant (Sigma-Aldrich, St. Louis, MO) for priming or with 100 μl of Freund's incomplete adjuvant (Sigma-Aldrich) for boosting. A total of 200 μl of the mixture was subcutaneously injected into each mouse. Blood samples were collected from the ophthalmic vein 10 days after each immunization.
2.7. Equine immunization and sampling
Ten healthy equines aged 6–10 years were housed under standard breeding conditions after quarantine inspection. Before immunization, 3 mg of RBD protein in 3 ml of PBS was mixed with an equal volume of Freund's complete adjuvant (Sigma-Aldrich) for priming, 6 mg of RBD protein in 3 ml of PBS was mixed with an equal volume of Freund's incomplete adjuvant (Sigma-Aldrich) for the first boost, and 12 mg of RBD protein in 3 ml of PBS was mixed with an equal volume of Freund's incomplete adjuvant (Sigma-Aldrich) for the second boost. The equines received intramuscular injections on day 0, 12, and 22. Serum samples were collected from the jugular vein on day 7, 19, and 27 to monitor variations in antibody responses. A large amount of plasma was collected after the third boost, and each collection was conducted 7 days after the boost with 12 mg of RBD.
2.8. Enzyme-linked immunosorbent assay (ELISA)
The recombinant RBD protein was diluted to 2 μg/ml and coated onto 96-well plates at a volume of 100 μl/well overnight at 4 °C. The liquid was aspirated, and the plates were washed three times with 0.1% Tween 20-PBS and then blocked with 2% nonfat milk at 37 °C for 1 h. Gradient-diluted murine or equine serum was added to each well, and the plates were incubated at room temperature for 1 h. PBS and irrelevant sera were used as controls. Then, the liquid was aspirated, and the plates were washed five times with 0.05% Tween 20-PBS and incubated with HRP-conjugated secondary antibodies at room temperature for 1 h. After the plates were washed five times as described above, 3,3′,5,5′-tetramethylbenzidine was added, and the chromogenic reaction was terminated by the addition of H2SO4 approximately 10 min later. Finally, the absorbance was measured at 450 nm with a microplate reader (Tecan, Switzerland), and samples with values greater than twice those of the controls were considered positive.
2.9. Virus neutralization test
Vero-E6 cells were seeded in 48-well plates at a density of 5 × 104 cells/well overnight. Murine or equine sera and immunoglobulins or F(ab’)2 fragments were first diluted in 100 μl of 2% FBS-DMEM and incubated with 5 μl of SARS-CoV-2 (MOI = 0.05) at 37 °C for 1 h. Then, the media were aspirated from the cells, and 100 μl of antiserum-virus or antibody-virus mixture was added to the cells. After incubation at 37 °C for 1 h, the supernatants were completely removed, and the cells were washed with PBS and supplemented with fresh 10% FBS-DMEM. After 24 h, the cell supernatants were collected and subjected to viral RNA isolation, and the cells were stored for indirect immunofluorescence analysis. The viral genome copy numbers were determined by RT-qPCR with primers targeting the S gene.
2.10. Indirect immunofluorescence assay (IFA)
Cell plates were collected after the virus neutralization test. The cells were washed with PBS, fixed with 4% paraformaldehyde, and permeabilized with Triton X-100. Then, the cells were blocked with 2% nonfat milk at room temperature for 1 h, washed with PBS and incubated with rabbit anti-NP antibodies at room temperature for 2 h. The cells were washed again before incubation with Alexa Fluor 488-conjugated goat anti-rabbit antibodies at room temperature for 1 h. Finally, the cells were washed and stained with 4,6-diamino-2-phenylindole (DAPI) for 10 min at room temperature. Images were captured with a fluorescence microscope (Olympus, Japan).
2.11. Preparation of immunoglobulins and F(ab’)2 fragments
When 50% neutralization titer (NT50) of the equine antisera exceeded the value of 10000, the plasma was collected with a plasma collection machine 7 days after each boost. Briefly, the plasma was first diluted with phosphate buffer (pH 7.0) at a ratio of 1:4 and then subjected to affinity chromatography against Protein A. The immunoglobulins eluted by glycine (1 M, pH 3.0) were dialyzed with phosphate buffer (pH 7.0). For preparation of the F(ab’)2 fragments, the plasma was first diluted with double-distilled water at a ratio of 1:4. Before pepsin (Sigma-Aldrich, USA) was added, the pH was adjusted to 3.0 with HCl and the temperature was adjusted to 30 °C. The incubation was continued for 1.5 h with stirring. The pepsin was inactivated by increasing the temperature, and gradient concentrations of ammonium sulfate were added in proper order; each addition was followed by a filtration step. Finally, the supernatant was run through a Protein A column to remove the residual immunoglobulins, and the F(ab’)2 fragments were collected from the flow-through fraction.
2.12. Biomolecular interaction analysis (BIA)
The affinities of the immunoglobulins and F(ab’)2 fragments for the RBD were monitored by biolayer interferometry (BLI) using an Octet-Red 96 device (Pall ForteBio LLC, CA, USA) according to previously described protocols (Pan et al., 2018). Briefly, the RBD was biotinylated by incubation with biotin at a molar ratio of 1:3 at room temperature for 0.5 h. The residual biotin was removed by dialysis with PBS. The biotinylated RBD was loaded onto streptavidin biosensors (ForteBio, CA, USA) at a concentration of 10 μg/ml until saturation, and the immunoglobulins or F(ab’)2 fragments were then loaded. The kinetics of association (K on) and dissociation (K dis) were measured, and the data were processed with an Octet data analysis system.
2.13. Purification of RBD-specific immunoglobulins and F(ab’)2 fragments
For purification of RBD-specific immunoglobulins and F(ab’)2 fragments, RBD protein expressed in CHO cells and prepared as described above was coupled to preactivated resin (PabPurSulfolink Beads, SMART Life Sciences, Changzhou, China) through an amino reaction (Pan et al., 2020). The RBD-coupled resin was then used to purify the RBD-specific immunoglobulins or F(ab’)2 fragments from the total immunoglobulins or F(ab’)2 fragments. In brief, the total immunoglobulins or F(ab’)2 fragments were diluted in 20 mM phosphate buffer (pH 8.0) and repeatedly run through the RBD-coupled resin to bind to the RBD. Then, the resin was adequately washed with 20 mM phosphate buffer (pH 8.0) before glycine (1 M, pH 3.0) was added to elute the strongly bound immunoglobulins or F(ab’)2 fragments. The eluted components, that is, the RBD-specific immunoglobulins and F(ab’)2 fragments, were dialyzed with PBS to remove glycine and stored in PBS until use.
2.14. Ethics statements
All animal experiments were performed in strict accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals in China, and the protocols were approved by the Laboratory Animal Care and Use Committee of Wuhan Institute of Virology, Chinese Academy of Sciences (Wuhan, China).
2.15. Data analysis
The data were analyzed using GraphPad Prism 8.0 software (CA, USA) and are presented as the means±SEMs based on at least three independent experiments.
3. Results
3.1. Designation, preparation, and characterization of the SARS-CoV-2 RBD
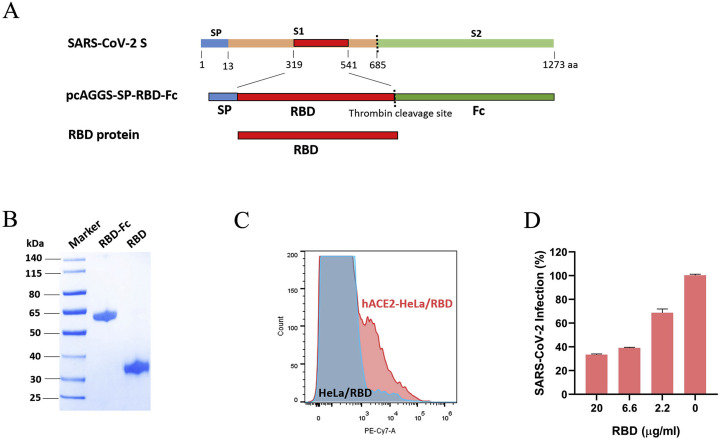
In order to effectively elicit nAbs production without triggering the production of irrelevant antibodies in vivo, we selected the RBD as the immunogen rather than the full-length S protein, inactivated SARS-CoV-2 whole virus or virus-like particles. The SARS-CoV-2 S gene was obtained by de novo synthesis with codon optimization. An RBD expression plasmid was constructed as described in Fig. 1 A. To obtain large amounts of the RBD protein, the plasmid was transfected into CHO cells, and high-cell density fermentation was conducted for 8 days. The RBD-Fc proteins secreted into the medium were purified by affinity chromatography against Protein A. Considering that the Fc tag can induce unexpected antibody production in vivo, we completely removed the Fc region via thrombin digestion and repeatedly purified the products against Protein A to remove the residual Fc regions. Through this method, the RBD protein was obtained in gram-level quantities.
Fig. 1.
Preparation and characterization of the RBD. A The SARS-CoV-2 S protein contains a signal peptide; a receptor-binding subunit, S1; and a fusion subunit, S2. The RBD was predicted to include amino acids 319–541 of the S protein. For construction of the RBD expression plasmid, the RBD sequence was inserted after an efficient signal peptide and was followed by a thrombin site. Fc was linked as a purification tag. The RBD construct was expressed by CHO cells, purified from cell culture supernatants through affinity chromatography against Protein A, and digested with thrombin. B The RBD-Fc and RBD proteins were detected by reducing SDS-PAGE with Coomassie Brilliant Blue staining. C The binding of the RBD to human ACE2 was detected by flow cytometry. HeLa cells were transfected with a human ACE2 plasmid for 24 h. The cells were incubated with biotin-labeled RBD protein and stained with fluorescent antibodies. Binding of the RBD to ACE2-HeLa cells was indicated by the presence of additional peaks beyond those observed for mock-treated HeLa cells. D Vero-E6 cells were pretreated with RBD protein at the indicated concentrations and then infected with SARS-CoV-2 at an MOI = 0.05. The inhibition by the RBD was calculated compared with the inhibition in mock-treated cells.
To validate the RBD and RBD-Fc proteins produced in our study, we characterized these proteins based on their sizes, purities, and capacities to bind the SARS-CoV-2 receptor, human ACE2. In addition, we conducted a series of verification experiments involving dichroism spectroscopy, capillary isoelectric-focused electrophoresis and Western blot analysis, as shown in the supplemental figures (Fig s1, s2 and s3). As indicated in Fig. 1B, single bands were observed in the lanes containing the RBD and RBD-Fc proteins, as detected by reducing SDS-PAGE with Coomassie Bright Blue staining; the results showed that the recombinant proteins exhibited high purities of over 92%. Then, the binding of the RBD and RBD-Fc proteins to human ACE2 was assessed by flow cytometry, which revealed that both proteins bound to HeLa cells transiently transfected with a human ACE2 plasmid (Fig. 1C and Fig s4A). In contrast, these proteins rarely bound to HeLa cells without ACE2 overexpression. Furthermore, in a cellular receptor blockade assay, the RBD and RBD-Fc proteins both inhibited the entry of SARS-CoV-2 in a dose-dependent manner (Fig. 1D and Fig s4B). The effective binding of the RBD to ACE2 demonstrated the structural validity of the RBD protein prepared in our study and confirmed its suitability for further research.
3.2. Antigenicity and efficacy of the SARS-CoV-2 RBD in mice
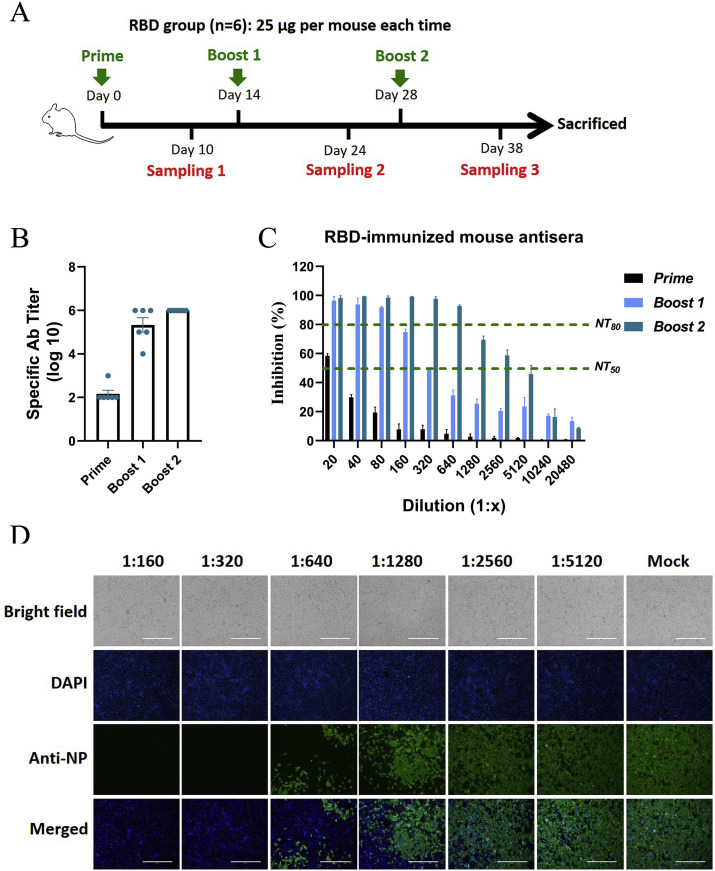
The effectiveness of the RBD in triggering an antibody response in vivo was first tested in mice. According to the traditional immunization scheme described in the Methods section, each mouse was immunized with 25 μg of protein in Freund's adjuvant via subcutaneous injections. As shown in Fig. 2 A, the mice were immunized three times in total with two-week intervals, and serum samples were collected ten days after each immunization to monitor the antibody response during the immunization process.
Fig. 2.
Antigenicity and efficacy of the RBD in mice. A Scheme of mouse immunization. Each mouse was immunized with 25 μg of RBD protein each time. A timeline of immunization and sampling is shown, and the detailed immunization methods are described in the Methods section. B The titers of specific antibodies in sera from RBD-immunized mice were detected by antigen-capture ELISAs. C The NT50s and NT80s of sera from RBD-immunized mice were detected by a set of neutralization tests with live SARS-CoV-2 in Vero-E6 cells. The serum dilutions ranged from 1:20 to 1:20480, as shown in the figures. The NT50 and NT80 values are marked with green lines. D The inhibitory effects of the sera collected from RBD-immunized mice after the third immunization were detected by IFA. Sera diluted 1:160 to 1:5120 were used in the neutralization tests, and SARS-CoV-2 infection of Vero-E6 cells was detected with antibodies against NP. The cell nuclei were stained with DAPI. Brightfield and merged images are also shown.
Then, the titers of RBD-specific antibodies were measured by antigen-capture ELISAs. As shown in Fig. 2B, the RBD-specific antibody titers were significantly elevated by the second immunization and reached 106 after the third immunization, showing that the RBD effectively elicited the production of specific antibodies in mice. The neutralization test revealed that sera collected from the RBD-immunized mice after the second immunization inhibited SARS-CoV-2 by 50% at a dilution of 1:320, while sera collected after the third immunization inhibited SARS-CoV-2 by 50% at a dilution of over 1:2560, showing that the effects of immunization were time-dependent (Fig. 2C). In addition, the 80% neutralization titers (NT80s) of the sera collected from the RBD-immunized mice after the second and third immunizations exceeded 80 and 640, respectively. The inhibition of SARS-CoV-2 by the sera collected from the RBD-immunized mice after the third immunization was also confirmed by indirect IFA. Infection of Vero-E6 cells by SARS-CoV-2 was sharply reduced with decreasing serum dilutions, as shown in Fig. 2D, showing a trend similar to that of the results in Fig. 2C. Similar specific antibody and nAb responses were also observed in the RBD-Fc-immunized mice, and the corresponding results are shown in Fig s5. These results collectively demonstrated that the RBD could be used as an immunogen to trigger nAb production in vivo.
3.3. Equine immunization and determination of the neutralizing potency of immunoglobulin F(ab’)2 fragments
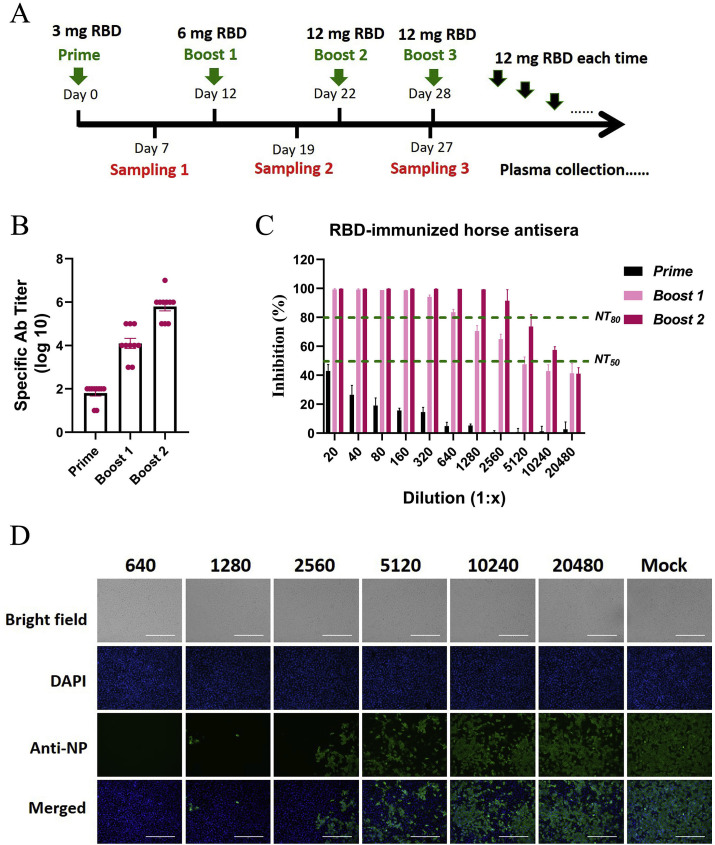
Based on the results described above, the RBD was used as an immunogen to prepare equine antisera. As described in the Methods section, ten equines were immunized with the RBD protein in complete Freund's adjuvant for the first immunization and with the RBD protein in incomplete Freund's adjuvant for subsequent immunizations. All immunizations were delivered via intramuscular injection. The amount of RBD protein was doubled over the first three immunizations, ranging from 3 mg to 12 mg for each equine, and was fixed at 12 mg for each equine for boosting before each plasma collection (Fig. 3 A). The serum samples were routinely adopted after each immunization and were used to monitor the antibody response.
Fig. 3.
Equine immunization strategy and characterization of antisera against SARS-CoV-2. A Scheme of equine immunization. The SARS-CoV-2 RBD was used as the immunogen. Equines were vaccinated with 3 mg of RBD protein in Freund's complete adjuvant for the first immunization via intramuscular injection and then boosted with 6 mg of RBD protein and 12 mg of RBD protein in Freund's incomplete adjuvant for the second and third immunizations. Venous blood was collected 7 days after each immunization to monitor the antibody response. Before each plasma collection, 12 mg of RBD protein was injected with a standard procedure. B The titers of specific antibodies in the equine sera after each immunization were examined by RBD-capture ELISAs. C The NT50s and NT80s of sera from RBD-immunized equines were examined in neutralization tests with live SARS-CoV-2 in Vero-E6 cells. The serum dilutions ranged from 1:20 to 1:20480, as shown in the figure. The NT50 and NT80 values are marked with green lines. D The inhibitory effects of the sera collected from RBD-immunized equines after the third immunization were detected by IFA. Sera diluted 1:640 to 1:20480 were used in the neutralization test, and SARS-CoV-2 infection of Vero-E6 cells was detected with antibodies against NP. The cell nuclei were stained with DAPI. Brightfield and merged images are also shown.
The titer of the RBD-specific antibodies was 102 after the first immunization, ~104 after the second immunization, and ~106 after the third immunization, increased with the number of immunizations (Fig. 3B). In addition, the NT50s of the sera after the second and third immunizations were approximately 5120 and over 10240, respectively, and the NT80s were 640 and over 2560, respectively, indicating potent neutralization of SARS-CoV-2 (Fig. 3C). Correspondingly, fewer than 20% of Vero-E6 cells were infected by SARS-CoV-2 following treatment with sera collected after the third immunization at a dilution of 1:2560, and the infection rate was reduced to less than 50% following treatment with sera at a dilution of 1:10240 (Fig. 3D). Through the immunization strategy described in this study, high-titer neutralizing sera were obtained from equines.
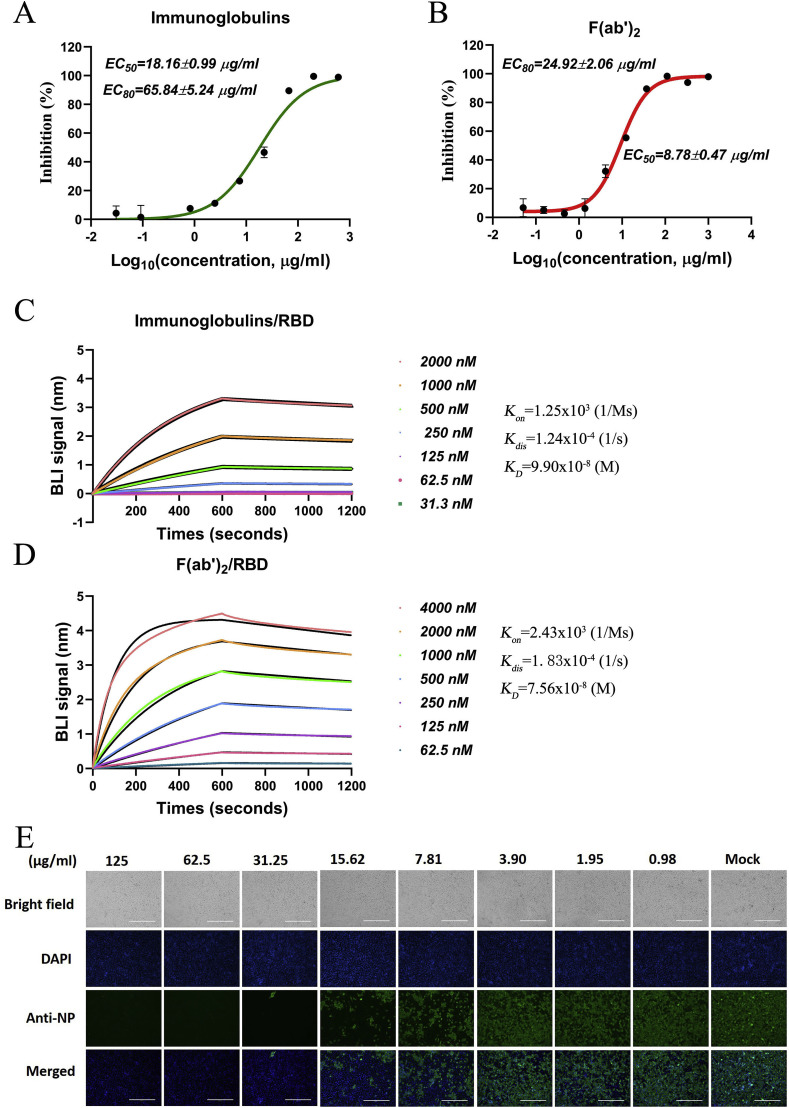
3.4. Characterization of immunoglobulins and F(ab’)2 fragments in vitro
Subsequently, immunoglobulins were purified from the antisera through affinity chromatography against Protein A. In addition, F(ab’)2 fragments were purified from the equine antisera via removal of the Fc regions from the immunoglobulins by digestion with pepsin at 37 °C according to a conventional process, as described in the Methods section. A set of neutralization tests showed that the immunoglobulins neutralized SARS-CoV-2 with a half-maximal effective concentration (EC50) of 18.16 μg/ml and a concentration eliciting 80% of the maximal effect (EC80) of 65.84 μg/ml (Fig. 4 A). The F(ab’)2 fragments were found to inhibit SARS-CoV-2 with an EC50 of 8.78 μg/ml and an EC80 of 24.92 μg/ml (Fig. 4B). Correspondingly, the kinetics with which the immunoglobulins and F(ab’)2 fragments bound to and dissociated from the recombinant RBD were determined by BIA. The K D of total immunoglobulin binding to the RBD was calculated as 99.0 nM (Fig. 4C), and that of total F(ab’)2 fragment binding to the RBD was calculated as 75.6 nM (Fig. 4D). Furthermore, the SARS-CoV-2-neutralizing ability of the total F(ab’)2 fragments was detected by IFA. As a result, SARS-CoV-2 was over 90% inhibited by treatment with the F(ab’)2 fragments at 31.15 μg/ml and over 50% inhibited by treatment with the F(ab’)2 fragments at 7.81 μg/ml. The inhibition of SARS-CoV-2 was observed to occur in a clearly dose-dependent manner, as shown in Fig. 4E.
Fig. 4.
Neutralization of SARS-CoV-2 by immunoglobulins and F(ab’)2fragments and their binding to the RBD. A and B The inhibitory effects of total immunoglobulins and F(ab’)2 fragments on SARS-CoV-2 were examined by neutralization tests, and both the EC50 and EC80 are shown. C and D The affinities of the immunoglobulins and F(ab’)2 fragments for the SARS-CoV-2 RBD were detected by BLI, and the kinetics (Kon and Kdis) were investigated with an Octet data analysis system. The binding curves were obtained by passing the F(ab’)2 fragments over biotinylated RBD protein immobilized on a streptavidin biosensor surface. The kinetics values (KD, M) were calculated by subtraction from the baseline and fitting of the association and dissociation responses to a 1:1 Langmuir binding model. E SARS-CoV-2 infection of Vero-E6 cells after F(ab’)2 fragment treatment was detected by IFA against NP. The working concentrations of the F(ab’)2 fragments ranged from 125 to 0.98 μg/ml.
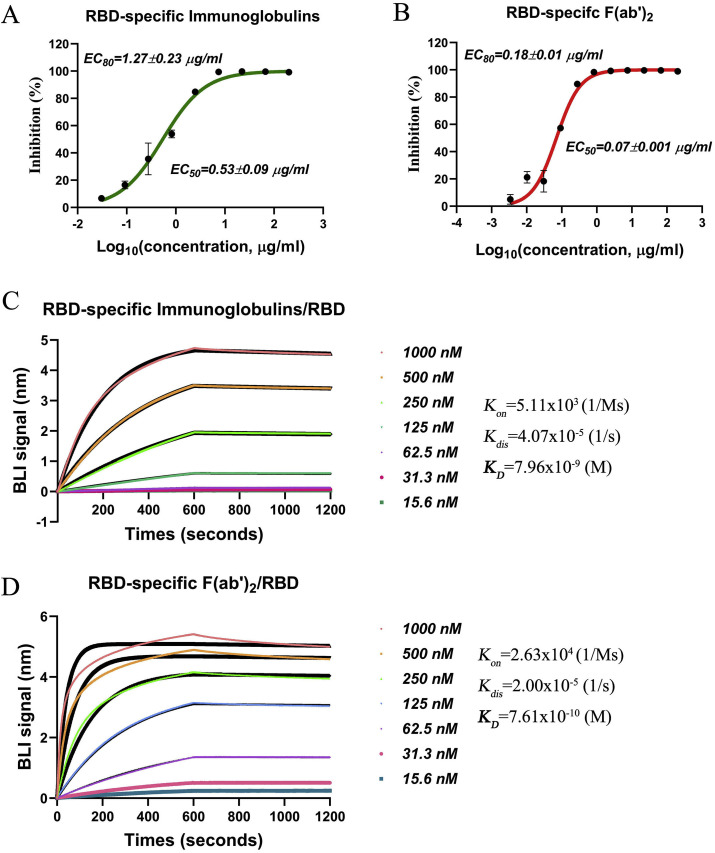
To further improve the neutralizing activity of the F(ab’)2 fragments and to confirm the target of these antibodies, the immunoglobulins and F(ab’)2 fragments that bound strongly to the RBD were purified from the total immunoglobulins and F(ab’)2 fragments by affinity chromatography against the RBD. The neutralizing activities of the RBD-specific immunoglobulins and F(ab’)2 fragments were tested by the same method used for the total immunoglobulins and F(ab’)2 fragments. As expected, the RBD-specific immunoglobulins neutralized SARS-CoV-2 with an EC50 of 0.53 μg/ml and an EC80 of 1.27 μg/ml (Fig. 5 A), and the RBD-specific F(ab’)2 fragments neutralized SARS-CoV-2 with an EC50 of 0.07 μg/ml and an EC80 of 0.18 μg/ml (Fig. 5B), showing strong inhibitory effects against SARS-CoV-2. Correspondingly, the K D of the RBD-specific immunoglobulins to the RBD was determined to be 7.96 nM through BIA (Fig. 5C). Additionally, the high affinity of the RBD-specific F(ab’)2 fragments for the RBD was reflected by a K D of 0.76 nM (Fig. 5D). The affinities of the RBD-specific immunoglobulins and F(ab’)2 fragments were further evaluated through RBD-specific purification, which suggested that the immunoglobulins and F(ab’)2 fragments that bound the RBD with high affinity were successfully selected from the antisera. The positive correlation between the SARS-CoV-2-neutralizing activity and the affinity for the RBD demonstrated that the antibodies produced in our study (the immunoglobulins and F(ab’)2 fragments) indeed targeted the RBD to work.
Fig. 5.
Neutralization of SARS-CoV-2 by RBD-specific immunoglobulins and F(ab’)2fragments and their binding to the RBD. A and B The inhibitory effects of RBD-specific immunoglobulins and F(ab’)2 fragments on SARS-CoV-2 were examined by neutralization tests, and the EC50 and EC80 are shown in figure. C and D The affinities of RBD-specific immunoglobulins and F(ab’)2 for recombinant RBD protein were detected by BLI. The binding curves were obtained by passing the RBD-specific immunoglobulins or F(ab’)2 fragments over biotinylated RBD protein immobilized on a streptavidin biosensor surface.
In addition, an in vivo safety evaluation was performed. No obvious toxic reactions were observed in rhesus monkeys after intravenous injection with a single dose of 762.5 mg of the F(ab’)2 fragments (data not shown). Furthermore, sera collected from mice 8 h after administration of a 10-mg dose of F(ab’)2 fragments via tail intravenous injection inhibited the entry of SARS-CoV-2 pseudovirus with an NT50 of over 750 (Fig s6). Together, these results suggest that the F(ab’)2 fragment is a therapeutic option for the treatment of SARS-CoV-2 infection.
4. Discussion
COVID-19 is an emerging infectious disease caused by a new member of the coronavirus family. SARS-CoV-2 became a serious threat to public health worldwide within a short period of time during the outbreak. Although the biotechnology and pharmaceutical fields have advanced rapidly in the 21st century, responses to public health emergencies remain slow. Due to close contact between humans and wild animals such as pangolins and bats, infectious diseases caused by coronaviruses such as SARS-CoV, MERS-CoV, and SARS-CoV-2 continuously pose threats to humans. Because the pace at which specific drugs are developed is not fast enough to address emerging threats, development of broad-spectrum drugs and vaccines is particularly important (Totura and Bavari, 2019). Notably, antiviral serum obtained from hyperimmune equine plasma has long been used for the treatment of life-threatening viral diseases (Bal et al., 2015). In the current study, we found that equine immunoglobulin F(ab’)2 fragments have the potential to provide protection against COVID-19.
Clinical evidence has shown that the latent period of COVID-19 is short (approximately 5 days to 2 weeks), and most patients appear to recover within a short time with no persistent or latent infection. It can reasonably be hypothesized that nAbs may play important roles in preventing SARS-CoV-2 infection (Lauer et al., 2020). Shen et al. recently reported that administration of convalescent plasma that contains nAbs can improve the clinical status of COVID-19 patients (Shen et al., 2020). Convalescent plasma from patients has been an important material for the treatment of infectious diseases (Wong and Lee, 2020). Such plasma has been used to treat severe acute respiratory syndrome (SARS) (Cheng et al., 2005; Zhang et al., 2005), H5N1 influenza infection (Rockman et al., 2017; Zhou et al., 2007), Ebola infection (Kraft et al., 2015), and other viral infections, but its use carries the risk of infection with bloodborne diseases, and the supply of convalescent plasmas is insufficient.
Seventeen years ago, we prepared an inactivated SARS-CoV whole-virus vaccine. Tests on mice demonstrated that the inactivated vaccine induced the production of antibodies specific to SARS-CoV, and an in vitro neutralization test also proved that the induced antibodies could neutralize SARS-CoV (Tang et al., 2004). A major problem that impeded further study regarding the induction of nAb production by inactivated SARS-CoV whole virus is that some nAbs may enhance coronavirus infection (Luo et al., 2018). Indeed, several studies have demonstrated the occurrence of antibody-dependent enhancement (ADE) of SARS coronavirus infection (Jaume et al., 2012; Yip et al., 2014). Additionally, ADE has been found in studies on human immunodeficiency virus (HIV), simian immunodeficiency virus (SIV) and Dengue virus (de Alwis et al., 2014; Jolly and Weiss, 2000; Montefiori et al., 1990). Thus, RBD could induce the production of highly effective nAbs with greater safety than the whole virus, the SARS-CoV-2 S protein RBD was selected as the immunogen in the current study.
As the RBD-specific equine F(ab’)2 fragments are relatively specific antibodies, they have unique advantages over monoclonal antibodies and human immunoglobulins after natural infection. These F(ab’)2 fragments specifically target the RBD and prevent the binding of the virus to its receptor, ACE2, avoiding the risk associated with the Fc region and further avoiding ADE. In addition, these F(ab’)2 fragments bind to multiple epitopes of the SARS-CoV-2 RBD, suggesting that they can neutralize a broad spectrum of human or bat SARS-related coronaviruses and bat SARS-like coronaviruses. Moreover, equines can be immunized multiple times, high titers of antibodies are expected to be obtained from hyperimmune equine plasma for the treatment of SARS-CoV-2.
In this study, large amounts of equine antiserum were prepared via immunization of equines with the SARS-CoV-2 RBD. Before using the RBD protein as the immunogen, we verified its conformation in receptor binding experiments and tested its antigenicity in mice. Through a strategy involving increasing immunogen doses, high-titer equine antisera were obtained, and F(ab’)2 fragments were manufactured at a good manufacturing practice (GMP) facility to be processed for clinical study. Notably, monoclonal antibodies against SARS-CoV, such as CR3022, have been found to potently bind to SARS-CoV-2, but the neutralization of SARS-CoV-2 has not been verified (ter Meulen et al., 2006). Other nAbs against SARS-CoV have been proven to weakly bind to the SARS-CoV-2 RBD (Tian et al., 2020). The F(ab’)2 fragments prepared in our study have the potential to exhibit cross neutralization on SARS-CoV to some extent since they contain antibodies that bind multiple epitopes of the SARS-CoV-2 RBD. In addition, the amino acid sequence identity between the SARS-CoV-2 and SARS-CoV RBDs is 73%, and these two proteins use the same receptor, ACE2, to enter host cells.
An F(ab’)2 fragment is a kind of immunoglobulin fragment that is prepared via removal of the Fc region from immunoglobulin and retention of the active fragment, this process significantly reduces the side effects in the body. Although equine F(ab’)2 fragments are heterologous proteins to the human immune system, equine serum proteins and Fc-related proteins can be removed to a considerable extent using modern industrial techniques to reduce adverse effects. Additionally, single-dose administration can prevent the reduced efficacy associated with repeated administration. Finally, the process of Fc removal eliminates the major concern of ADE in the context of coronavirus infection. These advantages enable antibody-based drugs, such as our equine F(ab’)2 fragments, to be candidates for the treatment of COVID-19.
5. Conclusions
In summary, we have successfully obtained therapeutic antibodies from hyperimmune equine plasma. Equine immunoglobulin F(ab’)2 fragments against the SARS-CoV-2 RBD is highlighted as a potential therapeutic option for the treatment of COVID-19.
Authors’ contributions
X.-Y. P. drafted the manuscript, processed the data, and performed parts of the mouse and horse experiments. P.-F. Z., J. Z., L.-J. F. and J. S. prepared the immunogen. Y. W., W.-J. S., X.-M. J. and Y. S. performed the neutralization tests and viral detection. T.-J. F. and X.-Y. S. vaccinated the horses and prepared the F(ab’)2 fragments. S.-J. Z. and R. G. provided the plasmids. G.-F. X. and Z. C. revised the manuscript and supervised the project process.
Declaration of competing interest
The authors declare no competing financial interests.
Acknowledgments
This study was financially supported by grants from the National Ministry of Science and Technology Emergency Project (2020YFC0841400) and the Hubei Provincial Department of Science and Technology Emergency Project (2020FCA003). This study was also supported by the Wuhan National Biosafety Laboratory of the Chinese Academy of Sciences.
We would like to thank Ding Gao and Juan Min at the Centre for Instrumental Analysis and Metrology, Wuhan Institute of Virology, Chinese Academy of Sciences, for providing technical assistance. We also thank the P3 laboratory staff at the Wuhan Institute of Virology, Chinese Academy of Sciences.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2020.104868.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ankcorn M., Gallacher J., Ijaz S. Convalescent plasma therapy for persistent hepatitis E virus infection. J. Hepatol. 2019;71:434–438. doi: 10.1016/j.jhep.2019.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal C., Herbreteau C.H., Buchy P. Safety, potential efficacy, and pharmacokinetics of specific polyclonal immunoglobulin F(ab')(2) fragments against avian influenza A (H5N1) in healthy volunteers: a single-centre, randomised, double-blind, placebo-controlled, phase 1 study. Lancet Infect. Dis. 2015;15:285–292. doi: 10.1016/S1473-3099(14)71072-2. [DOI] [PubMed] [Google Scholar]
- Brown J.F., Dye J.M., Tozay S. Anti-ebola virus antibody levels in convalescent plasma and viral load after plasma infusion in patients with Ebola virus disease. J. Infect. Dis. 2018;218:555–562. doi: 10.1093/infdis/jiy199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Wong R., Soo Y.O. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur. J. Clin. Microbiol. Infect. Dis. 2005;24:44–46. doi: 10.1007/s10096-004-1271-9. official publication of the European Society of Clinical Microbiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Misasi J., Mulangu S. Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science. 2016;351:1339–1342. doi: 10.1126/science.aad5224. [DOI] [PubMed] [Google Scholar]
- de Alwis R., Williams K.L., Schmid M.A. Dengue viruses are enhanced by distinct populations of serotype cross-reactive antibodies in human immune sera. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Zhao G., Chan C.C. Recombinant receptor-binding domain of SARS-CoV spike protein expressed in mammalian, insect and E. coli cells elicits potent neutralizing antibody and protective immunity. Virology. 2009;393:144–150. doi: 10.1016/j.virol.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- Hastie K.M., Zandonatti M.A., Kleinfelter L.M. Structural basis for antibody-mediated neutralization of Lassa virus. Science. 2017;356:923–928. doi: 10.1126/science.aam7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaume M., Yip M.S., Kam Y.W. SARS CoV subunit vaccine: antibody-mediated neutralisation and enhancement. Hong Kong Med. J. = Xianggang yi xue za zhi. 2012;18(Suppl. 2):31–36. [PubMed] [Google Scholar]
- Jiang S., Du L., Shi Z. An emerging coronavirus causing pneumonia outbreak in Wuhan, China: calling for developing therapeutic and prophylactic strategies. Emerg. Microb. Infect. 2020;9:275–277. doi: 10.1080/22221751.2020.1723441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly P.E., Weiss H.L. Neutralization and enhancement of HIV-1 infection by sera from HIV-1 infected individuals who progress to disease at different rates. Virology. 2000;273:52–59. doi: 10.1006/viro.2000.0401. [DOI] [PubMed] [Google Scholar]
- Kraft C.S., Hewlett A.L., Koepsell S. The use of TKM-100802 and convalescent plasma in 2 patients with Ebola virus disease in the United States. Clin. Infect. Dis. 2015;61:496–502. doi: 10.1093/cid/civ334. an official publication of the Infectious Diseases Society of America. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer S.A., Grantz K.H., Bi Q. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J.H., Guo Z.M., Han W.Y. Preparation and development of equine hyperimmune globulin F(ab')2 against severe acute respiratory syndrome coronavirus. Acta Pharmacol. Sin. 2005;26:1479–1484. doi: 10.1111/j.1745-7254.2005.00210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo F., Liao F.L., Wang H. Evaluation of antibody-dependent enhancement of SARS-CoV infection in rhesus macaques immunized with an inactivated SARS-CoV vaccine. Virol. Sin. 2018;33:201–204. doi: 10.1007/s12250-018-0009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori D.C., Robinson W.E., Jr., Hirsch V.M. Antibody-dependent enhancement of simian immunodeficiency virus (SIV) infection in vitro by plasma from SIV-infected rhesus macaques. J. Virol. 1990;64:113–119. doi: 10.1128/jvi.64.1.113-119.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Wu Y., Wang W. Novel neutralizing monoclonal antibodies against Junin virus. Antivir. Res. 2018;156:21–28. doi: 10.1016/j.antiviral.2018.06.002. [DOI] [PubMed] [Google Scholar]
- Pan X., Wu Y., Wang W. Development of horse neutralizing immunoglobulin and immunoglobulin fragments against Junin virus. Antivir. Res. 2020;174 doi: 10.1016/j.antiviral.2019.104666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratanabanangkoon K., Tan K.Y., Eursakun S. A simple and novel strategy for the production of a pan-specific antiserum against elapid snakes of asia. PLoS Neglected Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman S., Lowther S., Camuglia S. Intravenous immunoglobulin protects against severe pandemic influenza infection. EBioMedicine. 2017;19:119–127. doi: 10.1016/j.ebiom.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero H.A., Perez Isquierdo F., Milani H.A. Treatment of Argentine hemorrhagic fever with convalescent's plasma. 4433 cases. Presse Med. 1986;15:2239–2242. [PubMed] [Google Scholar]
- Sapparapu G., Fernandez E., Kose N. Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice. Nature. 2016;540:443–447. doi: 10.1038/nature20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Wang Z., Zhao F. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Gui M., Wang X. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., He L., Zhang X. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., Zhao G., Sun S. A recombinant receptor-binding domain of MERS-CoV in trimeric form protects human dipeptidyl peptidase 4 (hDPP4) transgenic mice from MERS-CoV infection. Virology. 2016;499:375–382. doi: 10.1016/j.virol.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L., Zhu Q., Qin E. Inactivated SARS-CoV vaccine prepared from whole virus induces a high level of neutralizing antibodies in BALB/c mice. DNA Cell Biol. 2004;23:391–394. doi: 10.1089/104454904323145272. [DOI] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microb. Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiberghien P., de Lambalerie X., Morel P. Collecting and evaluating convalescent plasma for COVID-19 treatment: why and how. Vox Sang. 2020 doi: 10.1111/vox.12926. [DOI] [PubMed] [Google Scholar]
- Totura A.L., Bavari S. Broad-spectrum coronavirus antiviral drug discovery. Expet Opin. Drug Discov. 2019;14:397–412. doi: 10.1080/17460441.2019.1581171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wong G., Zhu W. Equine-Origin immunoglobulin fragments protect nonhuman primates from Ebola virus disease. J. Virol. 2019;93 doi: 10.1128/JVI.01548-18. e01548-01518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H.K., Lee C.K. Pivotal role of convalescent plasma in managing emerging infectious diseases. Vox Sang. 2020;10(1111):12927. doi: 10.1111/vox.12927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip M.S., Leung N.H., Cheung C.Y. Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. Virol. J. 2014;11:82. doi: 10.1186/1743-422X-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Wang Y. Clinical trial analysis of 2019-nCoV therapy registered in China. J. Med. Virol. 2020;92:540–545. doi: 10.1002/jmv.25733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Wang Y., Qi C. Clinical trial analysis of 2019-nCoV therapy registered in China. J. Med. Virol. 2020;92:540–545. doi: 10.1002/jmv.25733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Xie Y.W., Hong J. Purification of severe acute respiratory syndrome hyperimmune globulins for intravenous injection from convalescent plasma. Transfusion. 2005;45:1160–1164. doi: 10.1111/j.1537-2995.2005.00179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Song L. Novel antibody epitopes dominate the antigenicity of spike glycoprotein in SARS-CoV-2 compared to SARS-CoV. Cell. Mol. Immunol. 2020;17:536–538. doi: 10.1038/s41423-020-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Zhong N., Guan Y. Treatment with convalescent plasma for influenza A (H5N1) infection. N. Engl. J. Med. 2007;357:1450–1451. doi: 10.1056/NEJMc070359. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.