Highlights
-
•
We designated an alternative to qtPCR for the identification of SARS-Cov-2.
-
•
The method was a PCR with 5-end fluorescent primers and capillary electrophoresis.
-
•
Two viral fragments amplified in several SARS-Cov-2 positive and negative specimens.
-
•
This approach would increase the testing capacity of diagnostic labs.
Keywords: COVID19, SARS-Cov-2, Viral genome detection, Quantitative PCR, Capillary electrophoresis
Abstract
Due to the huge demand for SARS-Cov-2 determination,alternatives to the standard qtPCRtestsare potentially useful for increasing the number of samples screened. Our aim was to develop a direct fluorescent PCR capillary-electrophoresis detection of the viral genome. We validated this approach on several SARS-Cov-2 positive and negative samples.We isolated the naso-pharingealRNA from 20 positive and 10 negative samples. The cDNA was synthesised and two fragments of the SARS-Cov-2 were amplified. One of the primers for each pair was 5´-end fluorochrome labelled. The amplifications were subjected to capillary electrophoresis in ABI3130 sequencers to visualize the fluorescent peaks.The two SARS-Cov-2 fragments were successfully amplified in the positive samples, while the negative samples did not render fluorescent peaks. In conclusion, we describe and alternative method to identify the SARS-Cov-2 genome that could be scaled to the analysis of approximately 100 samples in less than 5 h. By combining a standard PCR with capillary electrophoresis our approach would overcome the limits imposed to many labs by the qtPCR and increase the testing capacity.
PCR tests for theaccurate and fastSARS-CoV-2 testing are crucial for the identification of carriers and control the spreading of COVID19. Most of these tests rely on the isolation and retro-transcription of the viral RNA from nasal, pharyngeal or pulmonary specimens followed by real-time quantitative PCR (qtPCR) with primers/probes designated from the SARS-Cov-2 sequence (Hong et al., 2020; Pfefferle et al., 2020; Nörz et al., 2020; Zhen et al., 2020). Most of the labs used commercial kits that were subject to supply restrictions in case of huge global demand. This bottle-neck for massive screening of the new coronavirus and reinforced the necessity of developing new technical approaches to overcome these barriers. Fluorescent capillary electrophoresis (FCE) of PCR-fragments amplified with 5´-fluoresecent primers has been previously used to detect human infection disease (Li et al., 2019 ).
FCE has several advantages compared to other techniques such as visualization of PCR fragments on agarose or polyacrilamide gels. It requires lower amounts of the target nucleic acid, and the whole process is automated and consume less time and resources. Moreover, the absence/presence of a PCR-fragment is interpreted by a software that measures the fluorescent intensity, that is proportional to the amount of the target sequence in the sample. FCE has also advantages relative to the qtPCR, such as the rapid and sensitive detection of dozens of different PCR fragments in single runs (Lian and Zhao, 2016). During the first months of the COVID19 pandemic we faced a demand for SARS-Cov-2 testing that exceeded our qtPCR capacity. Nowadays, the COVID 19 pandemic scenario is being increasing over the world. Furthermore, a second wave of the infection in countries that have already dropped down the epidemic curve it is a real possibility. For this reason we worked in the design and validation of an alternative method that could take advantage of capillary sequencers, available in many labs.
We describe a technical approach to SARS-Cov-2 testing by amplifying fragments of the viral genome with 5´-fluorescent primers followed by capillary electrophoresis in an ABI3130xl equipment. This method would permit the analysis of 96 samples in approximately five hours.Our method was validated with nasal-swap samples from 30 individuals, 20 positive and 10 negative for SARS-Cov-2 with a standard qtPCR. The total RNA was isolated with MagNa Pure-96 System (Roche) in a final vol of 100 μL. A total of 10 μL of the RNA were retrotranscribed (RT) with the RetroScript™ kit (Invitrogene) in a final vol of 20 μL. To check for quality of the cDNA synthesis we amplified a fragment of the human beta-actin gene (ACTB) and two fragments of the SARS-Cov-2. The ACTB transcript was also confirmed in all the samples by a qtPCR (Supplementary Fig. 1). The 20 positive samples amplified the two SARS-Cov-2 fragments, that were not detected in the 10 negative samples (Fig. 1 ). The viral fragments from the 20 positive samples were Sanger sequenced with BigDye chemistry in an ABI3130xl equipment (Supplementary figures 2, 3). The SARS-Cov-2 primers were designated from the reference sequence in the NCBI Virus (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/) (Wang et al., 2020).We identified three patients who were heterozygous for 3037 T/C and 8782 T/C, while 17 were 3037 T and 8782 C. The two are nucleotide changes reported from viral isolates worldwide, and 8782 C is in disequilibrium with ORF8: C251 T (p.S84 L in the spike protein) (Yin, 2020; Chang et al., 2020).
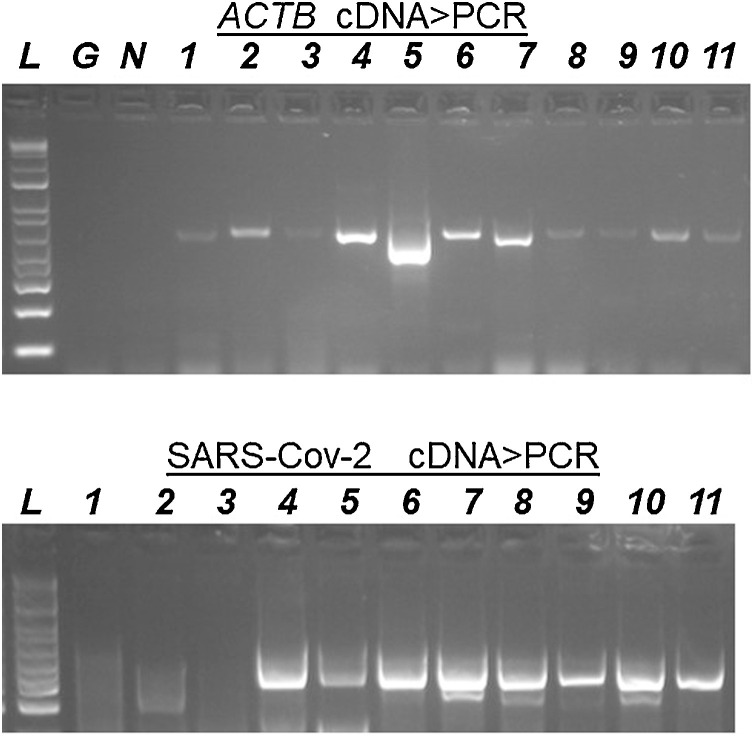
Fig. 1.
Upper: PCR fragments (agarose gels) for the human ACTB gene in cDNAs from nasal-samples (lanes 1–11). G and N, lack of amplification in the tubes containing genomic DNA and water (negative controls). Lower: PCR of a SARS-Cov-2 genome in cDNAs from viral positives (4–11) and negatives (1–3). These fragments were Sanger sequenced (Supplementary figures).
Briefly, two μL of the RT reactions were subjected to 32 PCR cycles with primer-pairs specific for either ACTB or the viral genome in a final vol of 30 μL. 10 μL of the PCRs were electrophoresed on 3% agarose gels to visualize the presence of the corresponding fragments.
L=1 kb ladder, size marker.
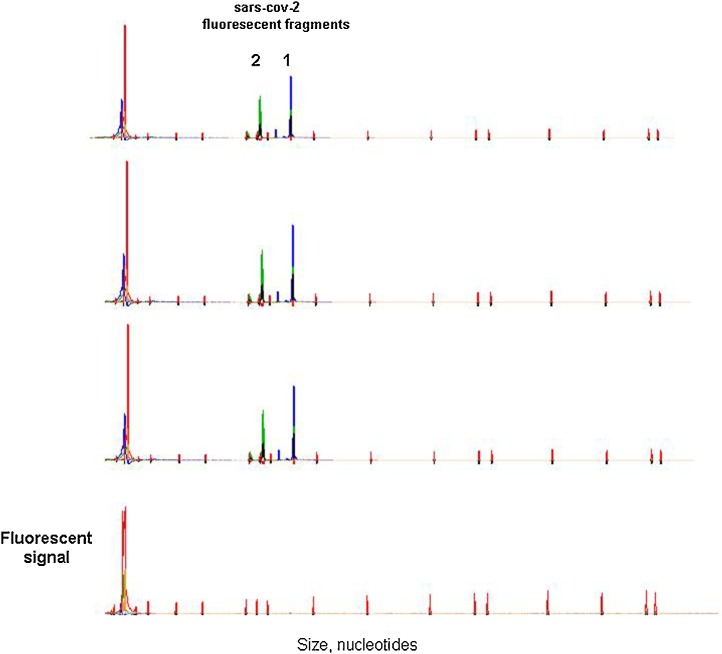
Based on the observed viral sequences we designated two primer pairs to amplify fragments of approximately 120 bp. To avoid the chance of false negatives the primers were specific for non-polymorphic sites in the 20 positive samples. The forward primers were 5´-end labelled with the 5-FAM or HEX fluorochromes. The samples were subjected to capillary electrophoresis in an ABI3130xl and analysed with the Gene-Mapper software. The two fluorescent peaks were present in the SARS-Cov-2 samples, and absent in the negative specimens (Fig. 2 ).
Fig. 2.
Capillary electrophoresis of the fluorescent PCR fragments from three SARS-Cov-2 positive cDNAs (three upper lines) and a negative sample (lower line). The two peaks corresponded to fragment 1 (180 bp) and fragment 2 (162 bp).
2 μL of the cDNAs were subjected to PCR with the two primer pairs in the same tube (30 μL final volume; 32 cycles of 95 °C-30 s, 62 °C-30 s, 72 °C-30 s). The primers for fragment 1 were 5-FAM- 5´CTTCACACTCAAAGGCGGTGCACC (forward) and 5´GCGAACTCATTTACTTCTGTACCGAGTTC (fragment size = 180 bp). The primers for fragment 2 were HEX- 5´GCTG TCAAATTACAGAATAATGAGCTTAGTCC (forward) and 5´CGGATAACA GTGCAAGTACAAACCTACC (fragment size = 162 bp).10 μL of the PCRs were mixed with 20 μL of deionised formamide and 0.5 μL of the fragment size-marker (250-ROX) in 96-well plates.
The total amplification time of our protocol takes about 90 min, and for 96-well plates the electrophoresis should be complete in approximately 3 h. Thus, the analysis of 96 samples could be done in < 5 h, compared to approximately 3 h for the analysis of the same amount of samples by qtPCR. The cDNA and PCR steps could be performed simultaneously with one-step RT and PCR kits with a reduction of the required time.
The protocol here described for the identification of SARS-Cov-2 is flexible in terms of the viral genome fragments prone to amplification, and could permit the simultaneous detection of multiple fragments (multiplex PCR). Moreover, compared to qtPCR the fluorescent PCR is not limited for the “compatibility” between the Taqman probe and the flanking PCR primers. An almost unlimited combination of primers to amplify the viral genome is thus possible (Nagel et al., 2009 ). Also, because the fluorescent primers are purchased on demand for thousand of reactions and the other are common reactive with many suppliers there is not a foreseeable bottle-neck for the analysis of large amounts of samples.
In conclusion, we present an alternative method for the detection of the SARS-Cov-2 genome in biological samples. Our approach was based on capillary electrophoresis of fluorescent PCRs, and permits the screening of 96 samples in less than 5 h. The method was validated with 20 virus positive and 10 virus negative samples, and would require a validation by each lab before its application. Our aim was not to “substitute” the gold qtPCR technique, but to develop an alternative to facilitate the analysis when the demand for COVID19 testing exceeds the capacity of the qfPCR of any lab that can implement the FCE technique.
Contributorship
All the authors contributed to this work by recruiting the cohorts or performing the genetic and statistical analysis.
Declaration of Competing Interest
None of the authors have competing interests related to this work.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2020.113937.
Contributor Information
Juan Gómez, Email: juan.gomezde@sespa.es.
Eliecer Coto, Email: eliecer.coto@sespa.es.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Chang T.J., Yang D.M., Wang M.L., Liang K.H., Tsai P.H., Chiou S.H., Lin T.H., Wang C.T. Genomic analysis and comparative multiple sequence of SARS-CoV2. J. Chin. Med. Assoc. 2020;83:537–543. doi: 10.1097/JCMA.0000000000000335. [DOI] [PubMed] [Google Scholar]
- Hong K.H., Lee S.W., Kim T.S., Huh H.J., Lee J., Kim S.Y., Park J.S., Kim G.J., Sung H., Roh K.H., Kim J.S., Kim H.S., Lee S.T., Seong M.W., Ryoo N., Lee H., Kwon K.C., Yoo C.K. Guidelines for laboratory diagnosis of coronavirus disease 2019 (COVID-19) in Korea. Ann. Lab. Med. 2020;40:351–360. doi: 10.3343/alm.2020.40.5.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Chen B., Zhang S., Li X., Chang J., Tang Y., Wu Y., Lu X. Rapid detection of respiratory pathogens for community-acquired pneumonia by capillary electrophoresis-based multiplex PCR. SLAS Technol. 2019;24:105–116. doi: 10.1177/2472630318787452. [DOI] [PubMed] [Google Scholar]
- Lian D.S., Zhao S.J. Capillary electrophoresis based on nucleic acid detection for diagnosing human infectious disease. Clin. Chem. Lab. Med. 2016;54:707–738. doi: 10.1515/cclm-2015-0096. [DOI] [PubMed] [Google Scholar]
- Nagel M.A., Gilden D., Shade T., Gao B., Cohrs R.J. Rapid and sensitive detection of 68 unique varicella zoster virusgene transcripts in five multiplex reverse transcription polymerasechain reactions. J. Virol. Methods. 2009;157:62–68. doi: 10.1016/j.jviromet.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nörz D., Fischer N., Schultze A., Kluge S., Mayer-Runge U., Aepfelbacher M., Pfefferle S., Lütgehetmann M. Clinical evaluation of a SARS-CoV-2 RT-PCR assay on a fully automated system for rapid on-demand testing in the hospital setting. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferle S., Reucher S., Nörz D., Lütgehetmann M. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.9.2000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liu Z., Chen Z., Huang X., Xu M., He T., Zhang Z. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J. Med. Virol. 2020 doi: 10.1002/jmv.25762. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C. Genotyping coronavirus SARS-CoV-2: methods and implications. Genomics. 2020;20:30318–30319. doi: 10.1016/j.ygeno.2020.04.016. pii: S0888-7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen W., Manji R., Smith E., Berry G.J. Comparison of four molecular in vitro diagnostic assays for the detection of SARS-CoV-2 in nasopharyngeal specimens. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00743-20. pii: JCM.00743-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.