Abstract
Heterogeneous transcriptional start site usage by HIV-1 produces 5’-capped RNAs beginning with one, two, or three 5’-guanosines (Cap1G, Cap2G, Cap3G, respectively) that are either selected for packaging as genomes (Cap1G) or retained in cells as translatable mRNAs (Cap2G/Cap3G). To understand how 5’-guanosine number influences fate, we probed the structures of capped HIV-1 leader RNAs by 2H-edited NMR. The Cap1G transcript adopts a dimeric multi-hairpin structure that sequesters the cap, inhibits interactions with translation initiation factor eIF4E, and resists decapping. The Cap2G/Cap3G transcripts adopt an alternate structure with an elongated central helix, exposed splice donor residues, and an accessible cap. Extensive remodeling, achieved at the energetic cost of a G-C base pair, explains how a single 5’ guanosine modifies the function of a ~9 kilobase HIV-1 transcript.
One Sentence Summary
HIV-1 transcripts that differ by as few as one or two 5’-guanosines adopt distinct structures that modulate RNA function and fate.
All viral constituents required for HIV-1 replication are encoded within a single integrated proviral DNA and expressed using a single promoter (1). Diversification of transcript function is achieved primarily by splicing, which produces mRNAs encoding the viral envelope and accessory proteins, and by regulated frame shifting during translation of unspliced transcripts to produce the Gag and Gag-Pol polyproteins. Some unspliced transcripts do not function as mRNAs but are instead selected for packaging into assembling virions as progeny genomes (gRNA). Genomes are packaged as dimers (2–4), a requirement for strand-transfer mediated recombination during reverse transcription (5). Dimerization, packaging, and other RNA dependent functions required for viral replication are mediated by conserved elements within the HIV-1 5’-leader (1, 6–8), and there is considerable evidence that transcript structure and function are established by the dimerization state of the leader (2–4). Although dimerization could be modulated by a riboswitch-like mechanism (8–10), recent studies indicate that dimerization and function are instead controlled at the level of transcription by heterogeneous start site usage (11, 12).
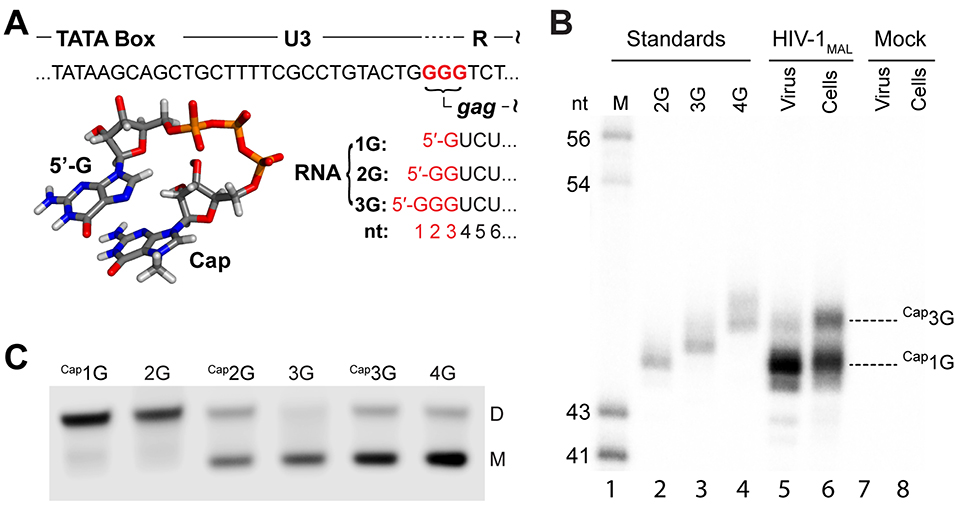
The HIV-1 promoter contains three sequential guanosines that can function as the transcription initiation site (U3-R junction; Fig. 1A). Cells infected with the laboratory adapted NL4–3 strain of HIV-1 (subtype B; HIV-1NL4–3) utilize all three start sites to express transcripts containing one, two, or three 5’-guanosines (1G, 2G, 3G, respectively). Most of these RNAs (~ 90%) are transcribed with 1G or 3G 5’-ends (11, 12), consistent with a predominant “twinning” transcription initiation mechanism (12). Like eukaryotic mRNAs, HIV-1 transcripts are co-transcriptionally capped by a 5’-5’ triphosphate linked 7-methylguanosine (Fig. 1A) (13–16). Capping is important for RNA splicing, nuclear export, translation, and metabolic stability (17). 5’-Capped 1G transcripts (CaplG) preferentially form dimers in vitro (12) and are selectively packaged into assembling virions in infection assays (11, 12) whereas those containing two or three guanosines (Cap2G/Cap3G) preferentially form monomers and are retained in cells and enriched on polysomes (12).
Fig. 1.
Heterogenous transcriptional start site usage modulates HIV-1MAL RNA dimerization and function. (A) Three guanines (red) can serve as alternative transcription start sites. Transcripts are co-transcriptionally capped by 7-methylguanosine. (B) 5’ ends of cellular and virion RNAs expressed from transiently transfected HIV-1 MAL-GPP-pA analyzed by RNase protection. Lane 1: molecular size standards. Lanes 2–4: HIV-1MAL RNA standards: 2G, 3G and 4G protected products serve as mobility standards for Cap1G, Cap2G, and Cap3G ends, correspondingly. Lanes 5 and 6: protected fragments from RNA samples harvested from 293T cells transfected with the HIV-1 derivative MAL-GPP-pA (lane 6) and from virus produced by these cells (lane 5). Lanes 7 and 8: protected fragments from RNA samples harvested from mock transfected 293T cells (lane 8) and from media produced by these cells (lane 7). (C) Full length leader RNAs (L371) that begin with a single capped guanosine or two non-capped guanosines favor the dimer, whereas RNAs with additional 5’ G or cap favor the monomer.
To understand how transcriptional addition of as few as one or two 5’-guanosines modulates RNA dimerization and fate, we probed the structures of Cap1G, Cap2G, and Cap3G HIV-1 leader RNAs by 2H-edited NMR and examined their abilities to interact with cellular proteins important for RNA processing and metabolic stability. Studies focused on the MAL strain of HIV-1 that is widely distributed among humans (M group subtype A; HIV-1MAL) (18). The HIV-1MAL leader contains a dimer-promoting GUGCAC palindrome and adopts a monomer-dimer equilibrium insensitive to the presence of the cognate NC protein (19) (fig. S1). Cultured 293T cells were transiently transfected with an HIV-1 vector containing the first 368 nucleotides of HIV-1MAL followed by the NL4–3 strain gag/pol sequence(MAL-GPP-pA) and 5’-end sequences of cellular and virion-associated RNA transcripts were determined by RNase protection assays (12). Of the three potential 5’-ends encoded by the provirus, only CaplG and Cap3G RNAs were detected (Fig. 1A,B). CaplG transcripts were enriched in virions produced from MAL-GPP-pA transfected cells (> 95%) whereas Cap3G transcripts were retained in cells during virus replication (Fig. 1B). As observed for HIV-1NL4–3 RNAs, in vitro transcribed HIV-1MAL Cap1G 5’-leader RNAs (Cap1G-L) preferentially formed dimers in vitro under physiological-like conditions (PI buffer =10 mM phosphate, 1 mM Mg2+, 122 mM K+, pH 7.4) whereas Cap3G-L (and Cap2G-L) preferentially formed monomers (Fig. 1C). These findings confirm that HIV-1 subtypes A and B encode 5’-leaders with similar start-site dependent dimerization propensities and gRNA versus mRNA control.
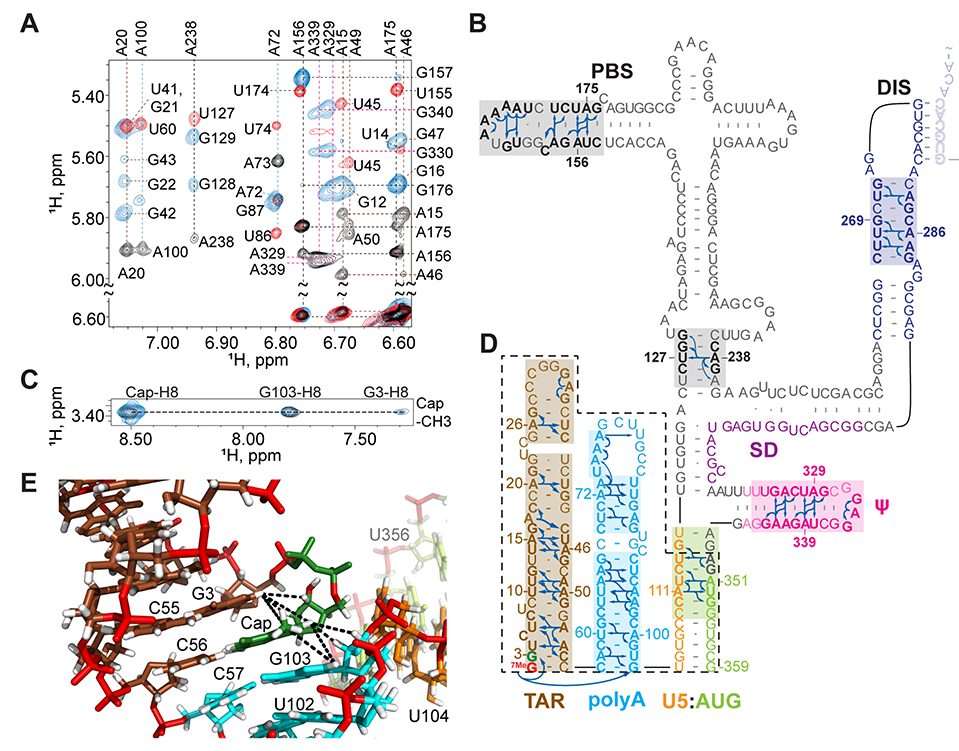
The secondary structure of the dimeric HIV-1MAL Cap1G leader (capped residue G3 through G359, [Cap1G-L359]2; 232 kDa) was probed by 2H-edited NMR. Sequential and long-range adenosine-H2 detected nuclear Overhauser effects (NOEs) that are diagnostic of RNA secondary structure (20) and can be employed for larger RNAs (8, 21) were detected for leader constructs prepared with the following nucleotide-specific 2H-labeling schemes (superscripts denote sites of protonation, all other sites deuterated; e.g., A2r = adenosines protonated at C2 and ribose carbons): A2r, A2rGr, A2rUr, A2Gr, and A2Ur. NMR assignments were corroborated by comparisons with spectra obtained for fragment RNAs (fig. S2) and by database 1H NMR chemical shift analyses (22). Adenosine-H2 NOEs were assigned for stretches of residues within most of the expected secondary structures of [Cap1G-L359]2, including those in the transcriptional activation (TAR), primer binding (PBS), dimerization (DIS), packaging (Ψ), and cleavage and polyadenylation site (polyA) elements (Fig. 2A,B) (8, 21). Well resolved NOEs for A111 and A351 confirmed the presence of the U5:AUG helix that pairs upstream sequences with those flanking the gag start codon (Fig. 2B and fig. S3) (23).
Fig. 2.
NMR and structural findings for the dimeric Cap1G form of the HIV-1MAL leader. (A) Portions of 2D NOE spectra for 2H-labeled [Cap1G-L359]2 samples (A2r, black; A2Ur, red; A2Gr, blue). (B) Assigned A-H2 NOEs and deduced secondary structure; discrete functional elements differentiated by color and intermolcular “kissing” interactions denoted by shaded residues. (C) Portions of 2D NOE spectra showing similarities of Cap-CEf to Cap-H8, G3, and G108 NOEs observed for G8-[Cap1G-L359]2 (blue) and truncated leader fragment Cap1G-LTPUA (D) (dashed black lines). (E) Portion of the Cap1G-LTPUA NMR structure showing Cap NOEs (dashed lines) indicative of end-to-end stacking of the TAR (brown) and polyA (cyan) helices.
NOEs between the Cap methyl group and protons of G3 and G103 were detected for the intact dimeric leader, suggesting that the Cap is sandwiched between these residues (Fig. 2C). Similar spectra were obtained for a truncated portion of the leader comprising the TAR and polyA hairpins and the U5:AUG helix (CaP1G-LTPUA) (Fig. 2C,D; fig S4). The improved sensitivity and spectral resolution obtainable for the smaller Cap1G-LTPUA construct (42 kDa) was sufficient for 3D structural studies (Table S1 and fig. S4). Residues G103, Cap, and G3 are sequentially stacked, as are residues C55, C56 and C57, leading to an overall end-to-end stacking of the TAR and polyA hairpins (Fig. 2E and fig. S4). The structure juxtaposes the Cap and C56 bases in a manner consistent with Cap:C56 base pairing.
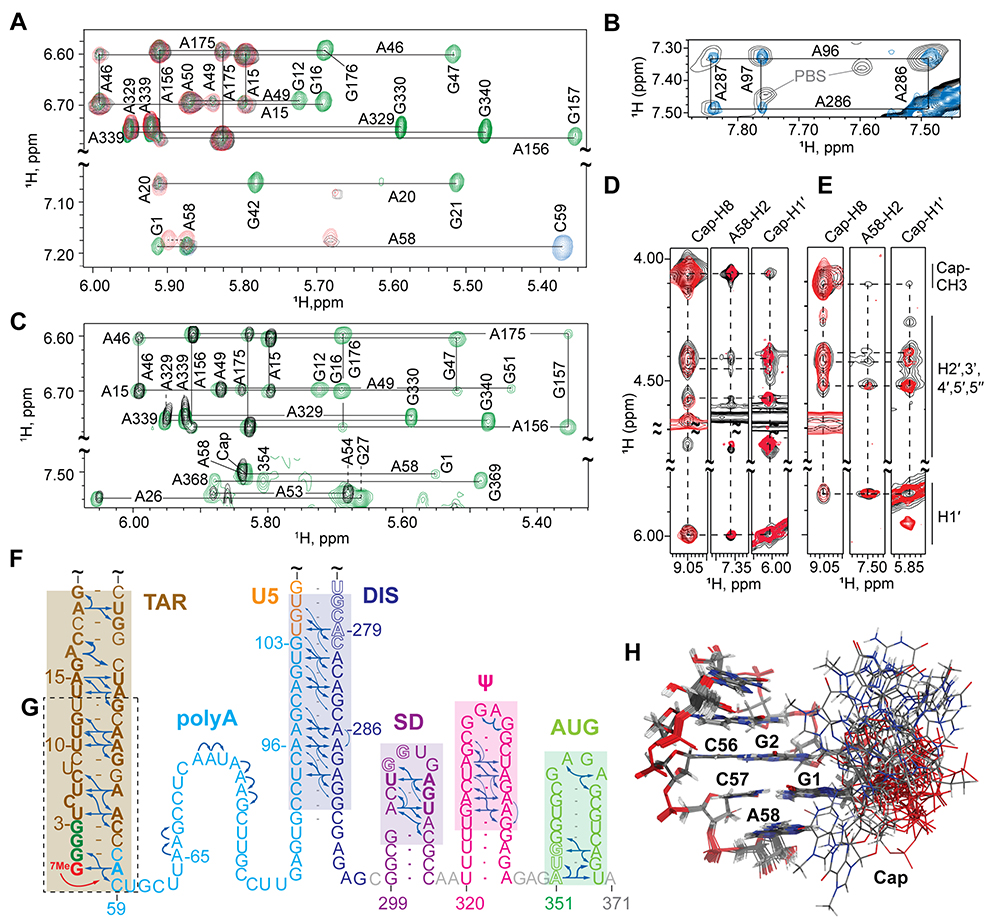
NMR studies were also conducted with monomeric 2G and 3G leader RNAs. NOESY spectra obtained for non-capped 3G-L371 (Fig. 3A) and capped Cap2G-L371 and Cap3G-L371 (Fig. 3B–E) RNAs exhibited similar cross peak patterns indicative of a common structure, with residues of AUG forming a hairpin rather than the U5:AUG helix observed in the dimer (8). NOE patterns for TAR, Ψ, and a portion of PBS were similar to those observed for [Cap1G-L359]2 indicating that these substructures exist in both the monomeric and dimeric forms of the leader (figs. S5 and S6). Long-range A58-H2 NOEs to G1 (but not G103) ribose protons were observed (confirmed using a sample in which only the G1 guanosine and adenosines were protonated; Fig. 3A), indicating that the lower portion of the polyA hairpin was remodeled (12). In addition, signals diagnostic of the DIS hairpin in the [Cap1G-L359]2 RNA were absent in the 3G-L371 NOESY spectra. 1H NMR chemical shifts of the H2 and H8 protons of adenosines A65, A66, A72, A73, and A75-A77 were also significantly different from those observed for [Cap1G-L359]2, and none of these adenosines exhibited long-range NOEs (fig. S6A). However, adenosine residues in the downstream portion of polyA exhibited long-range NOEs and chemical shifts indicative of base pairing with residues of DIS (Fig. 3B). Similar NMR results were obtained for Cap3G-L371 (Fig. 3C,E) and Cap2G-L371 (Fig. 3D). The capped RNAs exhibited additional NOEs between A58-H2 and the Cap methyl and ribose protons (Fig. 3D,E). The NMR data are consistent with a secondary structure that is substantially remodeled relative to that of the dimeric [Cap1G-L359]2 leader, with residues of TAR, SD, Ψ, and AUG adopting independently folded hairpin structures (Fig. 3F) and residues of polyA, U5 and DIS forming an elongated helix (Fig. 3F).
Fig. 3.
NMR and structural findings for the monomeric Cap3G, Cap2G, and 3G forms of the HIV-1MAL leader. (A-E) Portions of 2D NOE spectra used to make secondary structure assignments shown in (F). (A) Non-capped 3G-L371 spectra (AH, black; A2rGr, green; A2rCr, blue; G1HA2r, red); G1 is the only protonated guanosine in the G1HA2r-labeled sample, enabling unambiguous assignment of A58-H2 NOEs to G1. (B) NOE spectra for A2_Cap3G-L371 (black) showing A-H2 to A-H2 NOEs of the extended polyA-DIS helix matches a non-capped analog lacking the PBS loop ( A2rGr-4G-L37l−ΔPBS, blue; see fig. S5). (C) Cap3G-L371 spectra (AH, black; A2rGr, green) showing that the Cap is in close proximity to A58. (D) Comparison of 2D NOE spectra for A2_Cap2G-L371 (black) and TAR fragment Cap2G-TARm (G) (red), showing NOEs between the Cap and A58. (E) Similar Cap-to-A58 NOEs were observed for Cap3G-L371 (black) and a Cap3G-TARm RNA (red). (H) Portion of the NMR structure of Cap3G-TARm showing the disordered cap residue.
No sequential NOEs between the Cap and G1 residues were detected in spectra obtained for the intact Cap3G-L371 or Cap2G-L371 leader RNAs. Spectra with improved sensitivity and resolution were obtained for constructs corresponding to the lower portion of the capped TAR hairpin (Cap2G-TARm in Fig. 3D, and Cap3G-TARm in Fig. 3E,G). NMR chemical shifts and NOE patterns were similar to those observed for analogous residues in Cap3G-L371 and indicate that the Cap residue does not stack with G1 or G2 and is disordered (Fig. 3H and fig. S7).
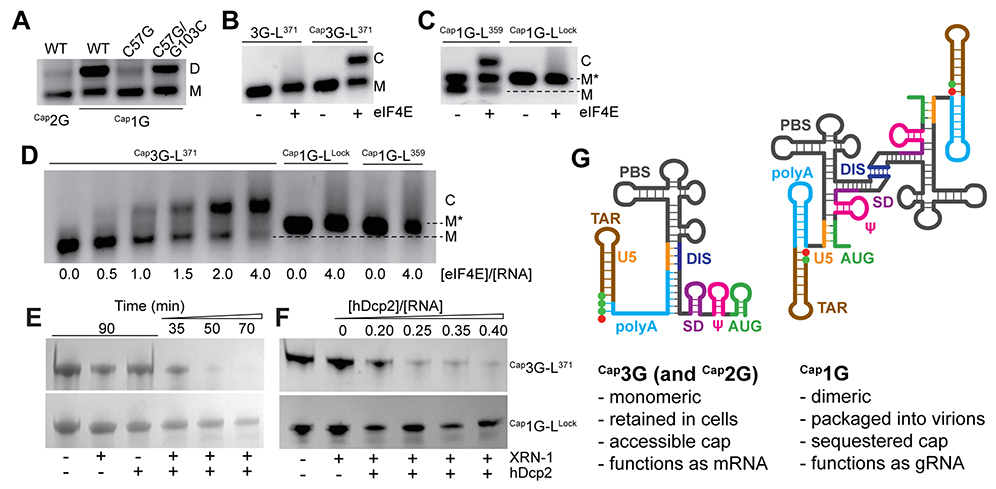
The NMR data suggest that structural remodeling of the capped 2G/3G RNAs relative to the capped 1G leader is a consequence of a single additional base pair (Cap:C57 in Cap2G-L or G1:C57 in Cap3G-L). Consistent with this hypothesis, substitution of C57 by G in Cap1G-L371 to ablate the base pair at the terminus of the polyA helix (C57:G103) (Fig. 2B) shifted the monomer-dimer equilibrium to the monomer (Fig. 4A), and compensatory substitution of G103 to C reverted the equilibrium toward dimer (Fig. 4A). This indicates that structural remodeling is achieved at an energetic cost equivalent to a single G-C base pair (~ 3–5 kcal/mol) (24). The compensatory mutant did not fully recapitulate the dimerization properties of the WT sequence, suggesting that the C57:G103 base pair, which is conserved in 99% of deposited sequences reporting full-length 5’UTRs (see SI materials), is important both for stabilizing the dimeric form of the Cap1G transcripts and enabling remodeling via C57:Cap or C57:G3 base pairing in the Cap2G and Cap3G transcripts, respectively.
Fig. 4.
Influence of 5’-guanosine number on RNA function. (A) Disruption of a single base pair (C57-G103) by C57 to G mutagenesis disrupts Cap1G-L371 dimerization. Compensatory G103C substitution substantially restores dimerization. (B) eIF4E binds Cap3G-L371 (C denotes the eIF4E:RNA complex) but not the non-capped RNA. (C) At low ionic strength, eIF4E binds the M conformer of Cap1G-L359, butnotM* or the Cap1G-LLock construct. (D) Similar results were obtained in PI buffer. (E-F) The 5’-RNA exonuclease (XRN-1) and decapping enzyme (hDcp2) are independently unable to degrade Cap1G- or Cap3G-leader RNAs. In the presence of both enzymes, The Cap1G-LLock resists degradation over time (E) and with increasing hDcp2 (F) compared to the cap-exposed Cap3G-L371 leader. (G) Mechanism for transcriptional control of HIV-1 RNA function. Capped RNAs containing two or three 5’-guanosines adopt a monomeric structure that exposes the cap and enables RNA processing and metabolism, whereas those with a single capped G adopt a cap-sequestered conformation that promotes dimerization and packaging.
Because the cap is exposed in monomeric Cap2G/Cap3G leader RNAs and sequestered between the TAR and polyA helices in the dimeric Cap1G leader, we examined the abilities of these RNAs to interact with two cellular cap binding proteins: the eukaryotic translation initiation factor eIF4E and the human decapping enzyme hDcp2. eIF4E initiates recruitment and assembly of the eukaryotic translation machinery (25), and cap recognition and removal by hDcp2 is required for 5’-exonucleolytic mRNA turnover (26). Native agarose gel shift experiments revealed that eIF4E binds the Cap2G and Cap3G leader RNAs with affinities (Kd ~ 0.7 μM) similar to that of a single capped guanosine (Kd = 1.44 μM) (25) (results for non-capped 3G-L371 and Cap3G-L371 shown in Fig. 4B). Cap1G-L titrations were also conducted under non-physiological low ionic strength conditions that favor the monomer (10 mM NaCl, no Mg+2). Under these conditions, Cap1G-L359 adopts two monomeric conformations that are resolvable on tris borate (TB) gels, M and M* (Fig. 4C and fig. S8). The M conformer exhibits gel mobility similar to that of the cap-exposed Cap3G-L monomer whereas M* exhibits mobility of a cap-sequestered CaplG-L359 mutant engineered to form a monomer while retaining the secondary structure of the dimer (DIS residues A273-A281 mutated to GAGA to prevent dimerization (27); Cap1G-LLock; Fig. 4C and figs. S2 and S8). Titration of CaplG-L359 with eIF4E resulted in a mobility shift for the cap-exposed M conformer, but not for the cap-sequestered M* species, even at excess (two-fold) molar ratios of eIF4E (Fig. 4C). Cap1G-LLock was likewise unable to bind eIF4E (Fig. 4C). Differential eIF4E binding between Cap3G-L371 and Cap1G-L was also observed in PI buffers (Fig. 4D). The capped 1G leader also exhibited reduced sensitivity to hDcp2-dependent 5’-exonuclease digestion compared to the capped 3G leader RNA (Fig. 4, E and F). These findings indicate that cap-binding proteins important for mRNA translation and processing bind efficiently to monomeric, cap-exposed forms of the leader but not to the cap-sequestered Cap1G dimer.
Our findings support a structure-based mechanism for diversification of HIV-1 transcript function by heterogeneous transcriptional start site usage (Fig. 4G). Analogous to riboswitches, which undergo structural remodeling and functional activation upon binding of small exogenous ligands (28), the structure and function of HIV-1 transcripts are controlled by transcriptional addition of one or two 5’ guanosines. Transcripts that begin with a single 5’-capped guanosine adopt a dimeric branched multi-helical structure that promotes dimerization and exposes Gag binding sites while simultaneously sequestering the 5’-cap, the major splice-donor site, and the translational start site. Cap sequestration is likely to inhibit both translation and splicing as both processes depend on initial interactions with cap binding proteins (29), and may also inhibit decapping dependent 5’-exonuclease-dependent degradation of the gRNA during cytoplasmic transport and particle assembly. Subgenomic flaviviral RNAs are similarly protected from exonuclease digestion by structural sequestration of 5’-nucleotides (30). HIV-1 transcripts that contain additional 5’ guanosines adopt an alternate structure that inhibits dimerization (12), sequesters Gag binding sites (8), and exposes the cap, the major splice donor site, the gag start codon, and unstructured residues immediately downstream of the TAR hairpin. Cap exposure enables eIF4E binding, and the unstructured polyA residues immediately downstream of the TAR hairpin could facilitate eIF4E-dependent association of additional factors required for splicing and translation (31). A genome-wide study of mammalian promoter architecture by cap analysis of gene expression revealed that twinned transcriptional start sites comprise a significant subset of mammalian promoters (32, 33). Start-site dependent modulation of transcript structure and cap exposure could serve as a general mechanism for expanding cellular RNA function.
Supplementary Material
ACKNOWLEDGMENTS
We thank HHMI staff at UMBC for technical assistance, Cleo Burnett at Michigan for help with manuscript preparation, and Remco Sprangers (University of Regensburg, Germany) for helpful suggestions.
Funding: This research was supported by research grants from the National Institutes of Health (NIAID 8R01 AI150498 to M.F.S. and A.T., NIAID U54 AI150470 to AT. and D.A.C.). J.D.B. was supported by NIH predoctoral fellowship F31 GM123803; M.L., K.Singh, M.O., T.R., and G.G. were supported by a NIGMS grant for enhancing minority access to research careers (MARC U*STAR 2T34 GM008663); and M.O. and T.R. were supported by an HHMI undergraduate education grant; A.S.I., M.L., K.Singh, M.O., T.R., and F.G.G. were supported by the Meyerhoff Scholars Program at UMBC
Footnotes
Competing interests: Authors declare no competing interests.
Data and materials availability: Depositions for Cap1G-LTPUA and Cap3G-TARm structures include atomic coordinates (PDB ID 6VU1 and 6VVJ, respectively) and NMR chemical shifts and restraints for structure calculations (BMRB ID 30723 and 30724, respectively).
References and Notes
- 1.Coffin JM, Hughes SH, Varmus HE, Retroviruses. (Cold Spring Harbor Laboratory Press, Plainview, N.Y., 1997). [PubMed] [Google Scholar]
- 2.D’Souza V, Summers MF, How retroviruses select their genomes. Nature Reviews Microbiology 3, 643–655 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Lu K, Heng X, Summers MF, Structural determinants and mechanism of HIV-1 genome packaging. J. Mol. Biol. 410, 609–633 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuzembayeva M, Dilley K, Sardo L, Hu W-S, Life of psi: how full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology 454–455, 362–370 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Onafuwa-Nuga A, Telesnitsky A, The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol Mol Biol Rev 73, 451–480 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lever AM, HIV-1 RNA packaging. Adv Pharmacol 55, 1–32 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Abbink TEM, Ooms M, Haasnoot PCJ, Berkhout B, The HIV-1 Leader RNA Conformational Switch Regulates RNA Dimerization but Does Not Regulate mRNA Translation†. Biochemistry 44, 9058–9066 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Lu K et al. , NMR detection of structures in the HIV-1 5’-leader RNA that regulate genome packaging. Science 344, 242–245 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darlix J-L, Gabus C, Nugeyre M-T, Clavel F, Barre-Sinoussi F, Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J. Mol. Biol. 216, 689–699 (1990). [DOI] [PubMed] [Google Scholar]
- 10.Ooms M, Huthoff H, Russell R, Liang C, Berkhout B, A riboswitch regulates RNA dimerization and packaging in human immunodeficiency virus type 1 virions. J. Virol. 78,10814–10819(2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masuda T et al. , Fate of HIV-1 cDNA intermediates during reverse transcription is dictated by transcription initiation site of virus genomic RNA. Sci Rep 5, 17680 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kharytonchyk S et al. , Transcriptional start site heterogeneity modulates the structure and function of the HIV-1 genome. Proc Natl Acad Sci USA 113, 13378–133832016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiu YL, Coronel E, Ho CK, Shuman S, Rana TM, HIV-1 Tat protein interacts with mammalian capping enzyme and stimulates capping of TAR RNA. J Biol Chem 276, 12959–12966 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Zhou M et al. , The Tat/TAR-dependent phosphorylation of RNA polymerase II Cterminal domain stimulates cotranscriptional capping of HIV-1 mRNA. Proc Natl Acad Sci USA 100, 12666–12671 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menees TM, Muller B, Krausslich HG, The major 5’ end of HIV type 1 RNA corresponds to G456. AIDS Res Hum Retroviruses 23, 1042–1048 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Sharma A, Yilmaz A, Marsh K, Cochrane A, Boris-Lawrie K, Thriving under stress: selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoSPathog 8, el002612 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramanathan A, Robb GB, Chan SH, mRNA capping: biological functions and applications. Nucleic Acids Res 44, 7511–7526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alizon M, Wain-Hobson S, Gluckman J-C, Sonigo P, Genetic variability of the AIDS virus: nucleotide sequence analysis of two isolates from African patients. Cell, 63–74 (1986). [DOI] [PubMed] [Google Scholar]
- 19.Tran T et al. , Conserved determinants of lentiviral genome dimerization. Retrovirolosy 12, 83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wüthrich K, NMR of Proteins and Nucleic Acids. (John Wiley & Sons, New York, 1986). [Google Scholar]
- 21.Heng X et al. , Identification of a minimal region of the HIV-1 5’-leader required for RNA dimerization, NC binding, and packaging. JMolBiol 417, 224–239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown JD, Summers MF, Johnson BA, Prediction of hydrogen and carbon chemical shifts from RNA using database mining and support vector regression. Journal of biomolecular NMR 63, 39–52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abbink TEM, Berkhout B, A novel long distance base-pairing interaction in Human Immunodeficiency Virus Type 1 RNA occludes the Gag start codon. J. Biol. Chem. 278, 11601–11611 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Xia et ah T, Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry 37, 14719–14735. (1998). [DOI] [PubMed] [Google Scholar]
- 25.Gingras AC, Raught B, Sonenberg N, eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68, 913–963 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Jiao X, Carr-Schmid A, Kiledjian M, The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci USA 99, 12663–12668 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keane etal SC., Structure of the HIV-1 RNA packaging signal. Science 348, 917–921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serganov A, Nudler E, A decade of riboswitches. Cell 152, 17–24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis JD, Izaurralde E, The role of the cap structure in RNA processing and nuclear export. Eur J Biochem 247, 461–469 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Akiyama BM et al. , Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science 354, 1148–1152 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boeras I et al. , The basal translation rate of authentic HIV-1 RNA is regulated by 5’UTR nt-pairings at junction of R and U5. Sci Rep 7, 6902 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carninci P et al. , Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38, 626–635 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Frith MC et al. , A code for transcription initiation in mammalian genomes. Genome Res 18, 1–12 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De la Pena M, Kyrieleis OJ, Cusack S, Structural insights into the mechanism and evolution of the vaccinia virus mRNA cap N7 methyl-transferase. EMBO J 26, 4913–4925. (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.https://www.hiv.lanl.gov/.
- 36.Thompson JD, Higgins DG, Gibson TJ, CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ, Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norris M, Fetler B, Marchant J, Johnson BA, NMRFx Processor: a cross-platform NMR data processing program. Journal of biomolecular NMR, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson BA, Blevins RA, NMRview: a Computer Program for the Visualization and Analysis of NMR Data. J. Biomol. NMR 4, 603–614 (1994). [DOI] [PubMed] [Google Scholar]
- 40.Barton S, Heng X, Johnson BA, Summers MF, Database proton NMR chemical shifts for RNA signal assignment and validation. Journal of biomolecular NMR 55, 33–46 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchant J, Summers MF, Johnson BA, Assigning NMR spectra of RNA, peptides and small organic molecules using molecular network visualization software. Journal of biomolecular NMR 73, 525–529 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tolbert BS et al. , Major groove width variations in RNA structures determined by NMR and impact of 13C residual chemical shift anisotropy and 1H-13C residual dipolar coupling on refinement. J. Biomol. NMR 47, 205–219 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Case DA et al. , The Amber biomolecular simulation programs. J. Computat. Chem. 26, 1668–1688 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yildirim I, Stern HA, Tubbs JD, Kennedy SD, Turner DH, Benchmarking AMBER force fields for RNA: comparisons to NMR spectra for single-stranded r(GACC) are improved by revised chi torsions. J Phys Chem B 115, 9261–9270 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zgarbova M et al. , Refinement of the Cornell et al. Nucleic Acids Force Field Based on Reference Quantum Chemical Calculations of Glycosidic Torsion Profiles. J Chem Theory Comput 7, 2886–2902 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mongan J, Simmerling C, McCammon JA, Case DA, Onufriev A, Generalized born model with a simple, robust molecular volume correction. J. Chem. Theory Comput. 3, 156–169(2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aduri R et al. , AMBER Force Field Parameters for the Naturally Occurring Modified Nucleosides in RNA. J Chem Theory Comput 3, 1464–1475 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Meagher KL, Redman LT, Carlson HA, Development of polyphosphate parameters for use with the AMBER force field. J Comput Chem 24, 1016–1025 (2003). [DOI] [PubMed] [Google Scholar]
- 49.DeLano WL, "The PyMOL molecular graphics system," (DeLano Scientific, San Carlos, CA, 2002). [Google Scholar]
- 50.Zuker M, Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31, 3406–3415 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reuter JS, Mathews DH, RNAstructure: software for RNA secondary structure prediction and analysis. BMC bioinformatics 11, 129 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marquet R, Paillart J-C, Skripkin E, Ehresmann C, Ehresmann B, Dimerization of human immunodeficiency virus type 1 RNA involves sequences located upstream of the splice donor site. Nucl. Acids Res. 22, 145–151 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davis IW et al. , MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35, W375–383 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen VB et al. , MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.