Abstract
Coronavirus Disease 2019 (COVID-19), caused by the novel coronavirus, has spread rapidly across China. Consequently, there is an urgent need to sort and develop novel agents for the prevention and treatment of viral infections. A rapid structure-based virtual screening is used for the evaluation of current commercial drugs, with structures of human angiotensin converting enzyme II (ACE2), and viral main protease, spike, envelope, membrane and nucleocapsid proteins. Our results reveal that the reported drugs Arbidol, Chloroquine and Remdesivir may hinder the entry and release of virions through the bindings with ACE2, spike and envelope proteins. Due to the similar binding patterns, NHC (β-d-N4-hydroxycytidine) and Triazavirin are also in prospects for clinical use. Main protease (3CLpro) is likely to be a feasible target of drug design. The screening results to target 3CL-pro reveal that Mitoguazone, Metformin, Biguanide Hydrochloride, Gallic acid, Caffeic acid, Sulfaguanidine and Acetylcysteine seem be possible inhibitors and have potential application in the clinical therapy of COVID-19.
Keywords: 87.14.E-, 87.15.A-, 87.15.B-, 87.15.ap
Coronavirus disease 2019 (COVID-19) is an acute respiratory infection that is caused by the 2019 novel coronavirus (SARS-CoV-2), which has spread rapidly across China and has become a global health challenge confronting the entire international community.[1] However, the agents that are available to treat COVID-19 cannot be used for prevention and they have serious side-effects, such as diarrhea, emesis and hyperlipidemia.[2–4] Furthermore, their mechanisms of action are unclear, and this has hindered the individualized strategies of prevention and treatment. Therefore, there is an urgent need to sort and develop novel agents, and efforts at improving the optimistic outcome have been directed more recently at clinical therapies of COVID-19.[5–7]
The complete viral genome analysis has revealed that SARS-CoV-2 belongs to the β-coronavirus genus, and its gene sequence is most closely related (89.1% nucleotide similarity) to that of coronavirus derived from Rhinolophus sinicus, in contrast the homology of SARS-CoV-2 with SARS-CoV (MERS-CoV) is ∼70(40)%.[8, 9] In addition, the open reading frame (ORF1a), which encodes the replicase complex, accounts for about 2/3 of the total length of SARS-CoV-2 genome. The other 1/3 encodes spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins.[9] The sequence homologies of the five proteins between SARS-CoV-2 and SARS-CoV (or MERS-CoV) are relatively low, especially the considerable genetics distance of the spike (S) protein.[9, 10] During the preparation of this paper, cryo-electron microscopy (cryo-EM) and biolayer interferometry experiments evidenced that the binding affinity of SARS-CoV-2 spike protein and human ACE2 is ∼20 times higher than the case of SARS-CoV with ACE2, and antibodies that work against SARS-CoV may not work against SARS-CoV-2.[10] Thus, numerous efforts have been made to explore specific therapeutic agents of COVID-19, including their explicit mechanisms.[10–12]
Nowadays, standard structure-based virtual screening has been routinely implemented in drug discovery to quickly prioritize potential compounds for in vitro activity tests. However, a significant practical problem is encountered when we account for intrinsic receptor flexibility, which leads to considerable computational costs. In our previous works, we applied an ensemble-based screening method to determine the binding profiles of ligands with flexible receptors, with advantages in the discovery of novel efficacious agents and cost-effectiveness.[13, 14] With this in mind, a rapid structure-based virtual screening strategy was used to identify compounds as therapeutic agents of COVID-19 by utilizing crystal structures of human ACE2 (accession code: 1R42[15]) and SARS-CoV-2 main protease (3CLpro, accession code: 6LU7), and homology modeling structures of viral spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins.
The coordinates of viral spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins were constructed by the MODELER module,[16] with the templates of 5X58(6NB6), 2MM4, 1BUC and 1SSK. Each homology modeling structure is partially in accordance with the respective templates, with the amino acid sequence similarities of 91.1, 70.7, 31.1 and 31.0%, respectively (Fig. S1 in Supplemental Material). Note that the constructed spike (S) structure is in a manner consistent with the latest x-ray and cryo-EM results, with the RMSD values being 1.7 and 1.8 Å (Fig. S2). Virtual screening was performed using the cDocker algorithm[17] and CHARMm force field,[18] which has shown to have many advantages in the development of novel antiviral drugs.[13, 19] During the screening processes, each binding site sphere was assigned with a sphere of 10.0 Å. The optimal orientations of ligands within proteins were probed on the basis of interactions with binding residues and geometrical matching qualities,[14, 20–22] and then the selected docked complexes were energy-minimized using the conjugate gradient (CG) method, and further refined by 100.0-ns explicit solvent molecular dynamics (MD) simulations using the AMBER16 package.[23] All values of binding free energies () were calculated in averages over 200 snapshots, which were evenly extracted from the 60–100 ns MD trajectories. Details of this simulation were published previously.[14, 22]
Primarily, 10 agents with the in vitro cellular activities (Arbidol, (R)-Chloroquine, (S)-Chloroquine, Darunavir, Lopinavir, Remdesivir, Ritonavir, Ribavirin, Triazavirine and β-d-N4-hydroxycytidine (NHC)), were, respectively, docked to the envelope, spike, main protease (3CLpro), membrane, nucleocapsid and human ACE2 structures. Note that interaction energy ( ) refers to the receptor-ligand interaction energy, and total energy ( ) includes and internal ligand strain energy. These were derived from the cDocker module, consistent with the previous works.[14, 22] It was found that Arbidol, Chloroquine, Remdesivir, NHC and Triazavirin have relatively good binding affinities, and envelope, ACE2, spike and 3CLpro are more likely target proteins for drug design (Table 1). For example, the envelope-Remdesivir, ACE2-Arbidol, spike-(S)-Chloroquine and 3CLPro-Remdesivir complexes are well-behaved during the 100-ns MD simulations (Figs. S3 and S4), while the spike-Ribavirin and membrane-Ribavirin complexes represent obvious structural fluctuates and thermodynamic instabilities. In particular, Ribavirin moves far from the binding pocket of membrane protein over the 100-ns MD simulation (Fig. S5). This motion indicated that though the interactions ( ) between agents and membrane/nucleocapsid structures look relatively good, yet their docked complexes ( ) might find it difficult to maintain stability (e.g., membrane-Ribavirin complex) (Table 1). Recent in vitro cell experiments have confirmed that Arbidol, Chloroquine, and Remdesivir can effectively inhibit the infection of SARS-CoV-2, and the treatments in combination with necessary supportive cares could significantly improve the pneumonia-related symptoms.[24] Arbidol effectively inhibits SARS-CoV-2 at 10–30 μM, with the suppression of cytopathic effect. On Vero E6 cells, half maximal effective concentration (EC50) value of Chloroquine (antimalarial drug) equals 1.13 μM, and selection index (SI) >88. Remdesivir (GS-5734) is a nucleoside analogue and is currently in phase III clinical trials for COVID-19, with the EC50 value of 0.77 and SI . NHC is also a nucleotide analogue, and its effect is similar to that of Remdesivir. Triazavirin could protect influenza virus infected mice and inhibit the accumulation of virions.
Table 1.
Total (Etotal) and interaction energies (Eint) of docked complexes. Energy is in units of kcal/mol, derived from cDocker module.
| Protein | Compound | E total | E int |
|---|---|---|---|
| Envelope | Ritonavir | −37.43 | −51.44 |
| Lopinavir | −32.79 | −43.87 | |
| Arbidol | −18.67 | −38.62 | |
| Remdesivir | −15.74 | −52.20 | |
| (S)-Chloroquine | −11.90 | −32.82 | |
| (R)-Chloroquine | −11.53 | −28.51 | |
| NHC | −2.04 | −30.79 | |
| Darunavir | −1.15 | −36.71 | |
| Triazavirine | −11.68 | −23.69 | |
| ACE2 | Arbidol | −16.65 | −29.81 |
| (S)-Chloroquine | −13.68 | −30.84 | |
| (R)-Chloroquine | −11.91 | −27.44 | |
| NHC | −6.59 | −35.24 | |
| Ribavirin | 13.03 | −30.22 | |
| Triazavirine | 13.77 | −24.35 | |
| Spike | (S)-Chloroquine | −17.11 | −36.02 |
| (R)-Chloroquine | −16.89 | −34.12 | |
| NHC | −8.46 | −38.22 | |
| Triazavirine | 6.05 | −29.35 | |
| Ribavirin | 8.64 | 34.02 | |
| 3CLpro | Ritonavir | −71.80 | −67.97 |
| Lopinavir | −56.81 | −56.89 | |
| Darunavir | −31.39 | −70.68 | |
| Remdesivir | −6.70 | −59.86 | |
| Triazavirine | 6.69 | −29.07 | |
| Membrane | NHC | 5.01 | −30.36 |
| Triazavirine | 7.11 | −28.83 | |
| Ribavirin | 13.11 | −34.42 | |
| (R)-Chloroquine | 53.06 | 1.32 | |
| (S)-Chloroquine | 64.59 | 8.58 | |
| Nucleocapsid | NHC | 2.56 | −35.84 |
| Triazavirine | 7.02 | 30.89 | |
| (S)-Chloroquine | 7.85 | −26.75 | |
| (R)-Chloroquine | 11.78 | −23.96 | |
| Ribavirin | 16.72 | −32.68 |
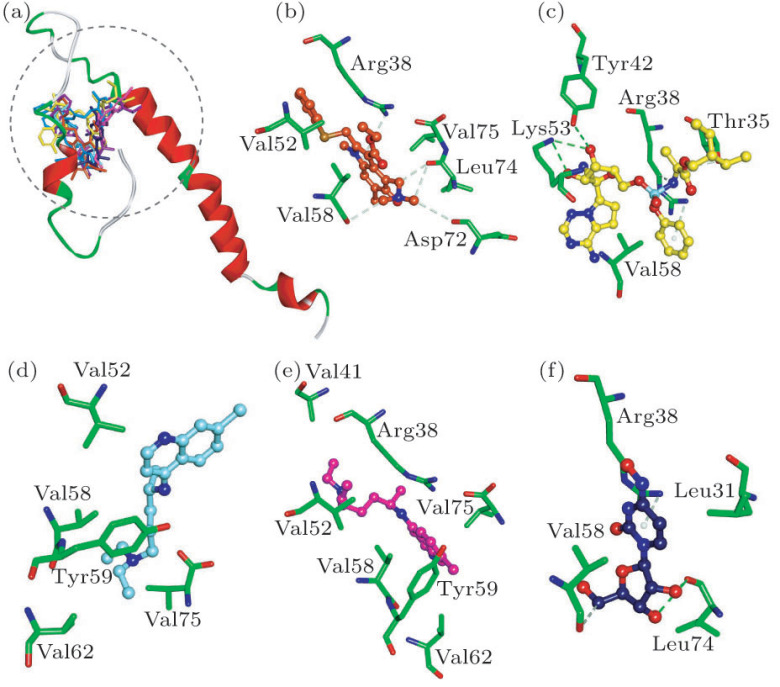
Envelope protein plays an important role in the assembly and release of SARS-CoV-2 virion. According to our results, Arbidol, (S)-Chloroquine, (R)-Chloroquine, Remdesivir, NHC and Triazavirin can bind to envelope protein. The HIV-1 protease inhibitors Ritonavir and Lopinavir interact with envelope protein in a similar manner, with the interaction energies () values of −51.44 and −43.87 kcal/mol (Table 1). While, the interaction energies () of Arbidol, Remdesivir and NHC with envelope protein are −38.62, −52.20 and −30.79 kcal/mol (Table 1), and they possess the H-bonding interactions with residues Arg38 and Val58 (Fig. 1). It is worth noting that (S)-/(R)-Chloroquine bind with envelope protein by the hydrophobic interactions with residues Val52, Val58, Tyr59, Val62 and Val75 (Figs. 1(d) and 1(e)).
Fig. 1.
(a) Compounds superposed in envelope structure and views of the binding modes of (b) Arbidol, (c) Remdesivir, (d) (S)-Chloroquine, (e) (R)-Chloroquine and (f) NHC with the active-site residues. Key residues are represented by stick models. Compounds are represented by ball and stick models. The O, N, C, S, P atoms are colored in red, blue, green, dark yellow and Cambridge blue. The important H-bonding (or electrostatic) interactions are labeled in the green dotted lines.
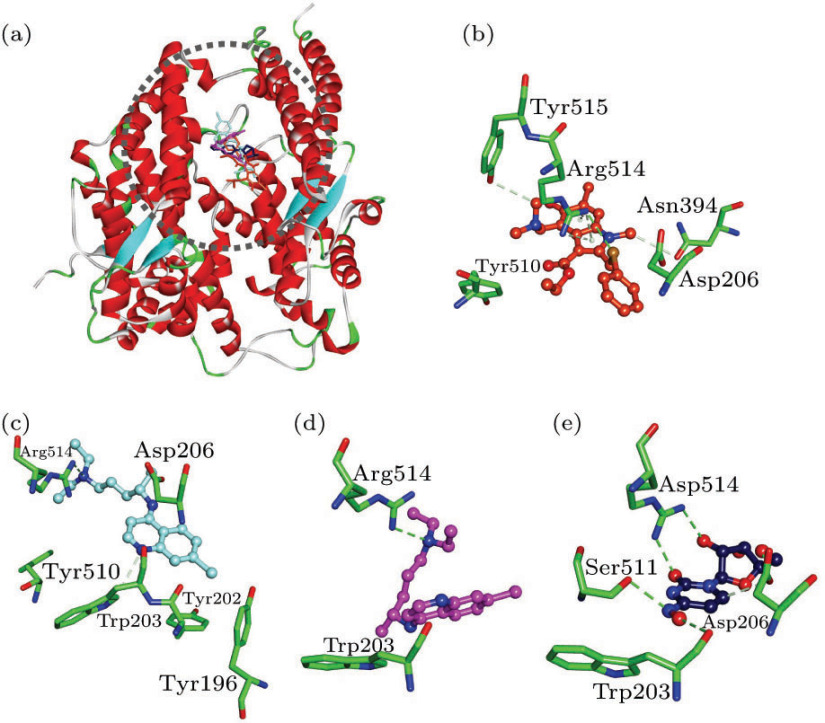
The receptor-binding region (RBD) of SARS-CoV-2 spike protein has high binding affinity with human ACE2, and this motion is responsible for the recognition between virions and host cells, and subsequent membrane fusion.[10] Our results revealed that Arbidol, (S)-Chloroquine, (R)-Chloroquine and NHC can bind with ACE2, with the interaction energies () values of −29.81, −30.84, −27.44 and −35.24 kcal/mol, respectively (Table 1). Among them, Arbidol has the H-bonding interactions with residues Asn394, Arg514 and Tyr515, electrostatic interaction with residue Asp206, and the hydrophobic interaction with residue Tyr510 (Fig. 2(b)). There are H-bonding interactions involving (S)-/(R)-Chloroquine with residue Arg514. In addition, (S)-Chloroquine also forms the hydrophobic interactions with three TYR amino acids (Figs. 2(c) and 2(d)). NHC has the H-bonding interactions with residues Tpr203, Asp206, Ser511 and Arg514, respectively (Fig. 2(e)). Taken together, the residues Asp206 and Arg514 are important in the binding processes of ligands with ACE2.[10]
Fig. 2.
(a) Compounds superposed in ACE2 structure and views of the binding modes of (b) Arbidol, (c) (S)-Chloroquine, (d) (R)-Chloroquine and (e) NHC with the active-site residues.
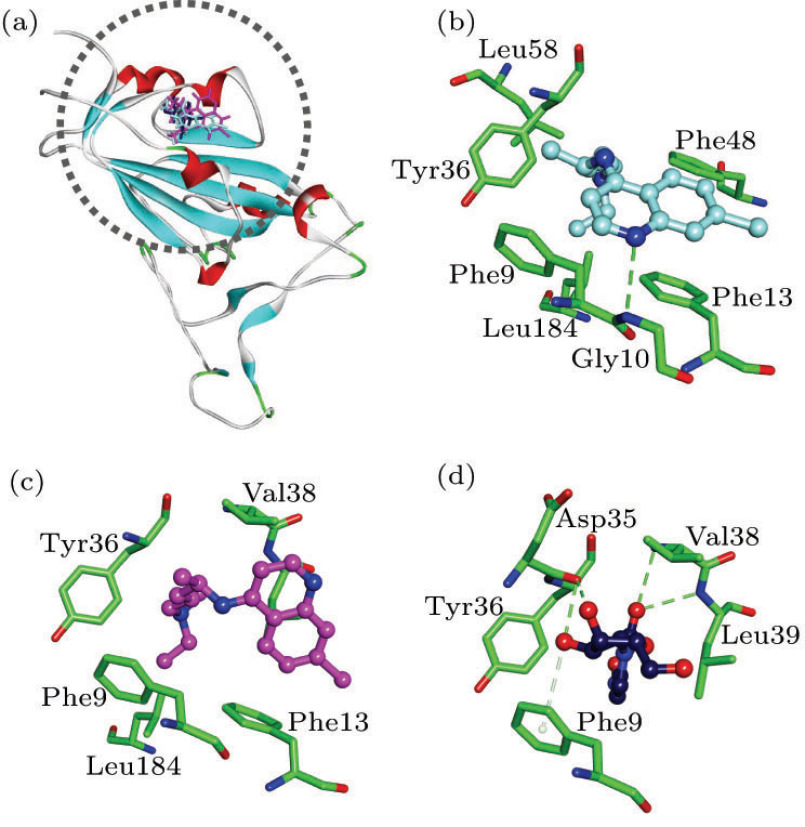
Biophysical results revealed that antibodies of spike protein against other coronaviruses should not work against the case of SARS-CoV-2, and small-molecule drugs may prove to be a better approach.[10–12] The interaction energies () of (S)-Chloroquine, (R)-Chloroquine and NHC with spike protein are −36.02, −34.12 and −38.22 kcal/mol, respectively (Table 1). (S)-Chloroquine has the H-bonding interaction with residue Gly10. The two isomers of Chloroquine both have the hydrophobic interactions with residues Phe9, Phe13, Tyr36 and Leu184 (Figs. 3(b) and 3(c)). NHC possesses the H-bonding interactions with residues Asp35 and Val38, as well as the hydrophobic interactions with residues Phe9, Tyr36 and Leu39 (Fig. 3(d)).
Fig. 3.
(a) Compounds superposed in spike structure and view of the binding modes of (b) (S)-Chloroquine, (c) (R)-Chloroquine and (d) NHC with the active-site residues.
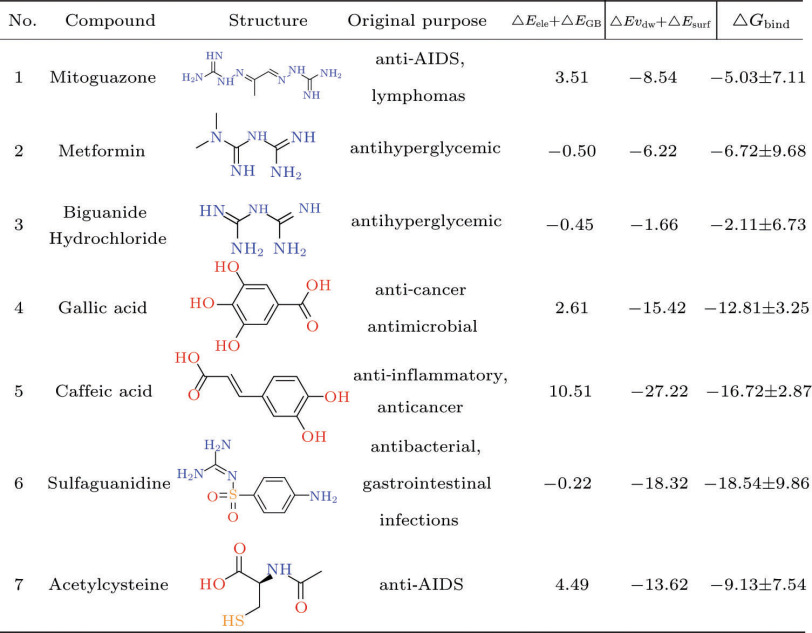
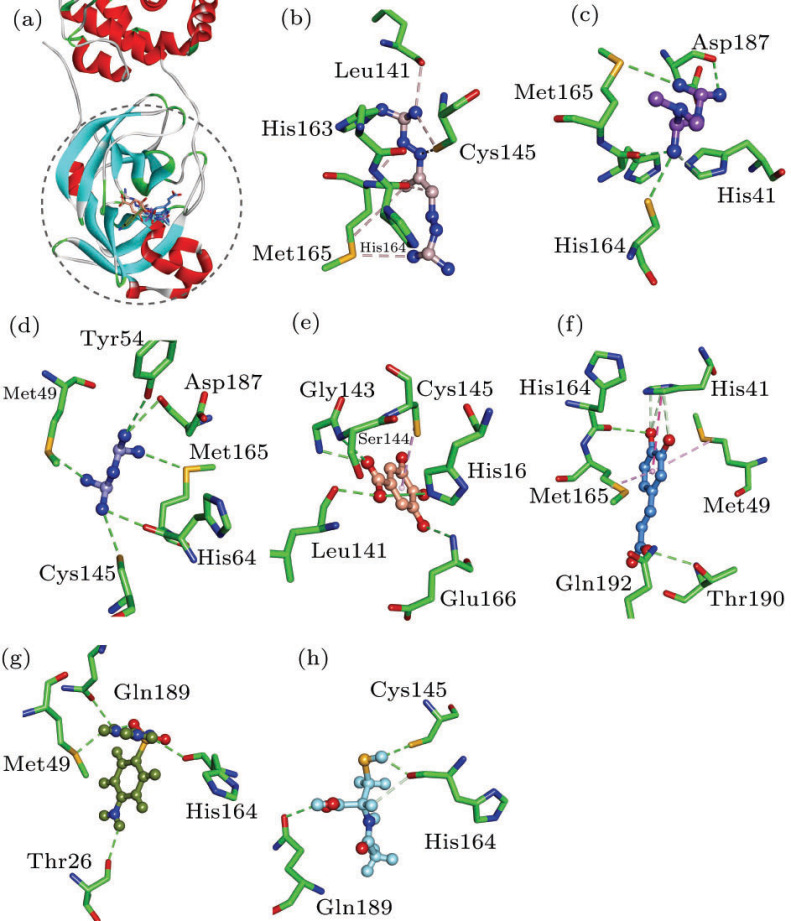
This analysis of structure and energy shows that the main protease (3CLpro) of SARS-CoV-2 should be a rational target for the drug development, with a relatively explicit and conservative structure. While the high-resolution crystal structure (accession code: 6LU7) determined by the group of Professor Zihe Rao was used in the rapid structure-based screening with approved drug library of ZINC database.[25] In accordance with our results, seven commercial drugs (Mitoguazone, Metformin, Biguanide Hydrochloride, Gallic acid, Caffeic acid, Sulfaguanidine and Acetylcysteine) should have therapeutic potentials in infections of SARS-CoV-2, with the values of −5.03, −6.72, −2.11, −12.81, −16.72, −18.54 and −9.13 kcal/mol, respectively (Fig. 5). Van der Waals components () primarily drive the binding processes, with the contributions over 60% of , which is consistent with previous simulation results of antiviral drugs.[13, 19] In contrast to reported agents Remdesivir, Arbidol and Chloroquine, the seven agents seem to induce more favorable bindings and possible inhibition of 3CLpro (Fig. 4 and Fig. S6). For instance, the seven sorted agents generally have the H-bonding interactions with residues Met49, Cys145, His164 and Gln189 of 3CLpro (Fig. 4), and all the docked complexes represent relatively good thermodynamic stabilities (Fig. S6). What is more interesting is that some of them are known to be used for the antiviral applications. Mitoguazone (MGBG), which is a guanidino-containing compound with the similar structure of spermidine, inhibits the key enzymes of S-adenosylmethionine decarboxylase pathway or polyamine biosynthesis pathway. MGBG has been widely used in the treatment of AIDS. Metformin and Biguanide Hydrochloride are hydrophilic and metabolically stable drugs, with minimal passive membrane permeability. Metformin is now used as an oral hypoglycemic agent.[26] Gallic acid has many potential therapeutic properties including anticancer and antimicrobial properties.[27] Caffeic acid is an anti-inflammatory antioxidant, and has shown significant efficacy as an inhibitor of the JAK2/STAT3 pathway in the cancer cell lines.[28] Sulfaguanidine is a very useful antibacterial drug that is not absorbed from the gastrointestinal tract. In addition, it does not enter the bloodstream, and even very young children may be given in fairly large doses.[29] Acetylcysteine is an antioxidant with thiol group, and could improve the experimental or clinical toxicity of ischemia-reperfusion syndrome in the heart, kidney, lung and liver. Moreover, acetylcysteine could inhibit inflammatory stimulation and HIV replication.[30] On basis of the steric and hydrophobicity/hydrophilicity characteristics of 3CLpro, the charged groups (e.g., guanidino/carboxylate group) might contribute considerably to the ligand bindings, and benefit the inhibition of protease activities.
Fig. 5.
Top hits of approved drug library against 3CLpro. Energy is in units of kcal/mol, calculated by the MM-GBSA method.
Fig. 4.
(a) Compounds superposed in 3CLpro structure and views of the binding modes of (b) Mitoguazone, (c) Metformin, (d) Biguanide Hydrochloride, (e) Gallic acid, (f) Caffeic acid, (g) Sulfaguanidine and (h) Acetylcysteine with the active-site residues.
In summary, we have performed a rapid structure-based virtual screening of the available information of protein structures and approved drug library. Molecular modeling, docking and molecular dynamic simulations are used to reveal the inhibiting mechanisms of reported drugs (e.g., Arbidol, Chloroquine and Remdesivir). These drugs may hinder the entry and release of virions through the bindings with human ACE2, and viral spike and envelope proteins. In addition, NHC and Triazavirin present the potential of clinical application. Main protease (3CLpro) is a kind of protease related to the virus replication, and should be a feasible target for rational drug design. Based on the mechanism of action, Mitoguazone, Metformin, Biguanide Hydrochloride, Gallic acid, Caffeic acid, Sulfaguanidine and Acetylcysteine should be potential inhibitors of 3CLpro, and their guanidino/carboxylate groups are helpful for the binding processes. In terms of low toxicity and druggability, they also seem to be drugs that can be used in clinical studies.
The authors wish to thank Professor Zihe Rao for supplying the crystal structure of main protease.
Footnotes
Supported by the National Natural Science Foundation of China (Grant Nos. 11774279 and 11774280), the Fundamental Research Funds for the Central Universities of China (Grant Nos. xjj2017029 and xzy032020038), and the Natural Science Basic Research Plan in Shaanxi Province of China (Grant No. 2019JQ-603).
References
- [1].The L. Lancet. 2020;395:311. doi: 10.1016/S0140-6736(20)30186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sheahan T P, Sims A C, Leist S R, Schäfer A, Won J, Brown A J, Montgomery S A, Hogg A, Babusis D, Clarke M O, Spahn J E, Bauer L, Sellers S, Porter D, Feng J Y, Cihlar T, Jordan R, Denison M R, Baric R S. Nat. Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Liu Y, Wei Y, Xia J A, Yu T, Zhang X, Zhang L. Lancet. 2020;395:507. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung K S M, Lau E H Y, Wong J Y, Xing X, Xiang N, Wu Y, Li C, Chen X, Li D, Liu T, Zhao J, Li M, Tu W, Chen C, Jin L, Wang Q, Zhou S, Guan X, Wang R, Wang X, Luo Y, Liu Y, Ren R, Shao G, Li H, Tao Z, Yang Y, Jin L, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam T T Y, Wu J T K, Cowling B J, Yang B, Leung G M, Feng Z. New Engl. J. Med. (in press) 2020.
- [5].Zumla A, Chan J F W, Azhar E I, Hui D S C, Yuen K Y. Nat. Rev. Drug Discov. 2016;15:327. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Holshue M L, DeBolt C, Lindquist S, Lofy K H, Wiesman J, Tong Y, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, Diaz G, Cohn A, Fox L, Patel A, Gerber S I, Kim L, Tong S, Lu X, Lindstrom S, Pallansch M A, Weldon W C, Biggs H M, Uyeki T M, Pillai S K. New Engl. J. Med. (in press) 2020. Washington State -nCo V C I T.
- [7].Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Lancet. 2020;395:497. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhou P, Yang X L, Wang X G, Hu B, Zhang L, Si H R, Zhu Y, Li B, Huang C L, Cheng Z, Chen H D, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Nature. 2020;579:270. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wu F, Zhao S, Yu B, Chen Y M, Wang W, Song Z G, Tian J H, Pei Y Y, Yuan M L, Zhang Y L, Dai F H, Liu Y, Wang Q M, Zheng J J, Xu L, Holmes E C, Zhang Y Z. Nature. 2020;579:270. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wrapp D, Wang N, Corbett K S, Goldsmith J A, Hsieh C L, Abiona O, Graham B S, McLellan J S. Science. 2020;367:1260. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Xu X, Chen P, Wang J, Feng J, Zhou H, Li X, Zhong W, Hao P. Sci. Chin.-Life Sci. 2020;63:457. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wan Y, Shang J, Graham R, Baric R S, Li F. 2020. pii: JVI.00127.
- [13].Yang Z W, Hao D X, Che Y Z, Yang J H, Zhang L, Zhang S L. Chin. Phys. B. 2018;27:018704. doi: 10.1088/1674-1056/27/1/018704. [DOI] [Google Scholar]
- [14].Xia J W, Yang L, Dong L, Niu M J, Zhang S L, Yang Z W, Wumaier G, Li Y, Wei X M, Gong Y, Zhu N, Li S Q. Front. Pharmacol. 2018;9:134. doi: 10.3389/fphar.2018.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Towler P, Staker B, Prasad S G, Menon S, Tang J, Parsons T, Ryan D, Fisher M, Williams D, Dales N A, Patane M A, Pantoliano M W. J. Biol. Chem. 2004;279:17996. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Accelrys Disc. Studio. 2011;3.1 Available online at http://accelrys.com. [Google Scholar]
- [17].Wu G, Robertson D H, Brooks C L, Vieth M. J. Comput. Chem. 2003;24:1549. doi: 10.1002/jcc.10306. [DOI] [PubMed] [Google Scholar]
- [18].Brooks B R, Bruccoleri R E, Olafson B D, States D J, Swaminathan S, Karplus M. J. Comput. Chem. 1983;4:187. doi: 10.1002/jcc.540040211. [DOI] [Google Scholar]
- [19].Yang Z W, Li Q Y, Yang G. Future Med. Chem. 2016;8:2245. doi: 10.4155/fmc-2016-0176. [DOI] [PubMed] [Google Scholar]
- [20].Yang Z W, Yang G, Zhou L J. J. Comput.-Aid. Mol. 2013;27:935. doi: 10.1007/s10822-013-9691-1. [DOI] [PubMed] [Google Scholar]
- [21].Yang Z W, Cao Y, Hao D X, Yuan X H, Zhang L, Zhang S L. J. Biomol. Struct. Dyn. 2018;36:2567. doi: 10.1080/07391102.2017.1363661. [DOI] [PubMed] [Google Scholar]
- [22].Li Z, Chen S, Gao C, Yang Z, Shih K C, Kochovski Z, Yang G, Gou L, Nieh S L, Jiang M, Zhang L, Chen G. J. Am. Chem. Soc. 2019;141:19448. doi: 10.1021/jacs.9b10505. [DOI] [PubMed] [Google Scholar]
- [23].Chen H Y, Fu W T, Wang Z, Wang X W, Lei T L, Zhu F, Li D, Chang S, Xu L, Hou T J. Acs Chem. Neurosci. 2019;10:677. doi: 10.1021/acschemneuro.8b00489. [DOI] [PubMed] [Google Scholar]
- [24].Wang Z, Chen X, Lu Y, Chen F, Zhang W. Biosci. Trends. 2020;14:64. doi: 10.5582/bst.2020.01030. [DOI] [PubMed] [Google Scholar]
- [25].Irwin J J, Sterling T, Mysinger M M, Bolstad E S, Coleman R G. J. Chem. Inf. Model. 2012;52:1757. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gong L, Goswami S, Giacomini K M, Altman R B, Klein T E. Pharmacogenet. Genom. 2012;22:820. doi: 10.1097/FPC.0b013e3283559b22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ow Y Y, Stupans I. Curr. Drug Metab. 2003;4:241. doi: 10.2174/1389200033489479. [DOI] [PubMed] [Google Scholar]
- [28].Priebe W, Fokt I, Szymanski S, Madden T, Myers J, Conrad C. Orally bioavailable caffeic acid related anticancer drugs. 2014;US8779151:B2. [Google Scholar]
- [29].Jones R D C, Matthews B A, Rhodes C T. Adv. Colloid. Interfac. 2006;59:518. [Google Scholar]
- [30].Roederer M, Staal F J T, Ela S W, Herzenberg L A, Herzenberg L A. Pharmacology. 1993;46:121. doi: 10.1159/000139037. [DOI] [PubMed] [Google Scholar]