Abstract
PURPOSE
Human herpesvirus 6B (HHV-6B) DNA is frequently detected in bronchoalveolar lavage fluid (BALF) from immunocompromised subjects with lower respiratory tract disease (LRTD). Whether HHV-6B is a pulmonary pathogen is unclear.
METHODS
We tested BALF for HHV-6B DNA using polymerase chain reaction in allogeneic hematopoietic cell transplantation (HCT) recipients who underwent a BAL for evaluation of LRTD from 1992 to 2015. We used multivariable proportional hazards models to evaluate the association of HHV-6B+ BALF with overall mortality, death from respiratory failure, and the effect of anti–HHV-6B antivirals on these outcomes. We used branched-chain RNA in situ hybridization to detect HHV-6 messenger RNA (U41 and U57 transcripts) in lung tissue.
RESULTS
We detected HHV-6B+ BALF from 147 of 553 (27%) individuals. Subjects with HHV-6B+ BALF, with or without copathogens, had significantly increased risk of overall mortality (adjusted hazard ratio [aHR], 2.18; 95% CI, 1.41-3.39) and death from respiratory failure (aHR, 2.50; 95% CI, 1.56-4.01) compared with subjects with HHV-6B- BALF. Subjects with HHV-6B+ BALF who received antivirals within 3 days pre-BAL had an approximately 1 log10 lower median HHV-6B BALF viral load, as well as a lower risk of overall mortality (aHR, 0.42; 95% CI, 0.16-1.10), compared with subjects with HHV-6B+ BALF not receiving antivirals. We detected intraparenchymal HHV-6 gene expression by RNA in situ hybridization in lung tissue in all three tested subjects with HHV-6B+ BALF and sufficient tissue RNA preservation.
CONCLUSION
These data provide evidence that HHV-6B detection in BALF is associated with higher mortality in allogeneic hematopoietic cell transplantation recipients with LRTD. Definitive evidence of causation will require a randomized prevention or treatment trial.
INTRODUCTION
Lower respiratory tract disease (LRTD) occurs frequently in the early period after allogeneic hematopoietic cell transplantation and is an important contributor to morbidity and mortality.1-3 Despite advances in diagnostic methods, causative pathogens are often not identified, and approximately 2% to 8% of hematopoietic cell transplantation (HCT) recipients with LRTD are diagnosed with idiopathic pneumonia syndrome (IPS).4 Current diagnostic algorithms may miss treatable pathogens such as human herpesvirus 6B (HHV-6B),5 a β-herpesvirus closely related to the well-established pulmonary pathogen human cytomegalovirus (CMV).6
HHV-6B infects more than 95% of individuals during early childhood and establishes latency in a broad range of cells, including bronchial glands.7,8 HHV-6B DNA is detectable in blood in approximately 40% of allogeneic HCT recipients within 100 days and is the most frequent infectious cause of encephalitis after HCT.9 HHV-6B was first demonstrated as a possible cause of post-HCT LRTD in 1991.10 Subsequent studies with small sample sizes and heterogeneous patient populations, and that used variable sample types and testing strategies frequently detected HHV-6B DNA in respiratory samples but demonstrated inconsistent associations with LRTD.11-15 There is no consensus whether HHV-6B is a pulmonary pathogen, as reflected by a lack of testing and treatment guidelines.2,16 Importantly, several therapies have activity against HHV-6B.17
In this study, we addressed limitations of prior publications by using established strategies to define pathogen-disease associations in the molecular era18 and a rigorous study design to determine whether HHV-6B is a pulmonary pathogen in allogeneic HCT recipients with LRTD. Our approach included testing for HHV-6B DNA in bronchoalveolar lavage fluid (BALF) from subjects with LRTD, analyzing the dose-response association of HHV-6B+ BALF with mortality and death from respiratory failure, and determining the association of anti–HHV-6B antivirals with HHV-6B viral load and outcomes. We compared these findings to those for CMV DNA detection in BALF in the same cohort. Finally, we performed in situ hybridization for HHV-6 gene expression in lung tissue.
METHODS
Subjects
The study cohort consisted of patients who received allogeneic HCT between 1992 and 2015 at Fred Hutchinson Cancer Research Center (FHCRC) and who had stored BALF from clinical evaluation of LRTD within 100 days after HCT. LRTD was defined as abnormal radiographic findings with lower respiratory tract symptoms (ie, cough, shortness of breath, or decreased oxygen saturation). Leftover BALF and blood were stored at −80°C at the time of collection. Subjects were excluded if the HCT donor or recipient had inherited chromosomally integrated HHV-6 (iciHHV-6; Data Supplement).19 The study was approved by the FHCRC Institutional Review Board.
Transplantation and Supportive Care Practices
HCT regimens, supportive care, and post-HCT complications varied over time.3,6 Subjects in whom LRTD developed underwent a bronchoalveolar lavage (BAL) using standard procedures based on international consensus guidelines.20 Subjects were not routinely tested or treated for HHV-6B in blood or BALF during the study period. Antiviral treatment with agents active against HHV-6B (namely, ganciclovir, foscarnet, or cidofovir) was primarily for CMV.6
Microbiologic Testing
We retrospectively tested BALF and the closest available plasma sample within 14 days before or after BAL for HHV-6B and HHV-6A DNA using real-time quantitative polymerase chain reaction (qPCR) assays, as previously described.21 A threshold of at least 50 copies/mL was considered positive. We also tested BALF for CMV DNA.6 We performed qPCR for the human β-globin gene from BALF to confirm cellularity and the quality of DNA extraction.6,12 Additional microbiologic testing is detailed in the Data Supplement.
LRTD Categorization
We newly categorized the cause of LRTD according to consensus guidelines on the basis of review of clinical, microbiologic, histopathologic, and radiographic records (Data Supplement).2,22,23
Outcome Measures
Primary outcome measures were overall mortality or death from respiratory failure within 100 days after the BAL. We defined death from respiratory failure as any death caused exclusively or predominantly by respiratory failure.
Lung Tissue Detection of HHV-6B DNA and RNA by Droplet Digital PCR and In Situ Hybridization
We obtained formalin-fixed paraffin-embedded lung-tissue specimens from subjects with HHV-6B DNA detection in BALF (HHV-6B+ BALF). We used droplet digital PCR and qPCR to determine the absolute number of HHV-6 DNA copies per cell and the HHV-6 species (Data Supplement).19 We used RNAscope branched chain RNA in situ hybridization24 (RISH; ACDBio, Newark, CA) to stain tissue for early (U41 transcript) and late (U57 transcript) HHV-6 messenger RNA transcripts (Data Supplement).25
Statistical Analysis
We abstracted clinical data from medical records and databases and were blinded to research HHV-6B test results. Descriptive statistics were used to detail the prevalence of HHV-6B+ BALF and the HHV-6B viral load over time; results were compared with the Wilcoxon rank-sum or Kruskal-Wallis tests as appropriate. Spearman rank correlation was used to test correlations between HHV-6B viral load and BALF cellularity.
We calculated the cumulative incidence of overall mortality and death from respiratory failure (treating death for other reasons as a competing risk event) among subjects alive at day 60 post-HCT to graphically display associations with HHV-6B+ BALF in the preceding 60 days. We next performed multivariable Cox proportional hazards regression to examine the association of HHV-6B+ BALF, the primary predictor of interest, with outcomes of overall mortality or death from respiratory failure within 100 days after the last BAL. We modeled HHV-6B+ BALF as (1) positive versus negative and (2) quartiles of viral load. To evaluate the association of anti−HHV-6B antivirals with overall mortality and death from respiratory failure, we created separate Cox models incorporating a composite variable for HHV-6B+ BALF and antiviral therapy to illustrate the varying degrees of association depending on whether each factor was present singly or in combination. For subjects with more than one BAL, time-dependent variables were updated with each additional BAL. We used the same approach to test the association of BALF CMV DNA detection with overall mortality among subjects in whom the donor or recipient was CMV seropositive. We selected potential confounding variables a priori. Variables with P < 0.1 in univariable models were considered for multivariable models and excluded if their exclusion did not change the hazard ratios (HRs) for HHV-6B–related variables by more than 10%. No adjustments were made for multiple comparisons. We used SAS, version 9.4 (TS1M3) for Windows (SAS Institute, Cary, NC) for analyses. Additional details are in the Data Supplement.
RESULTS
Subjects
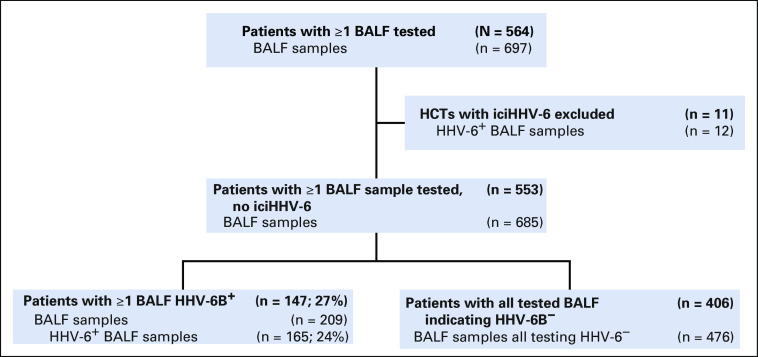
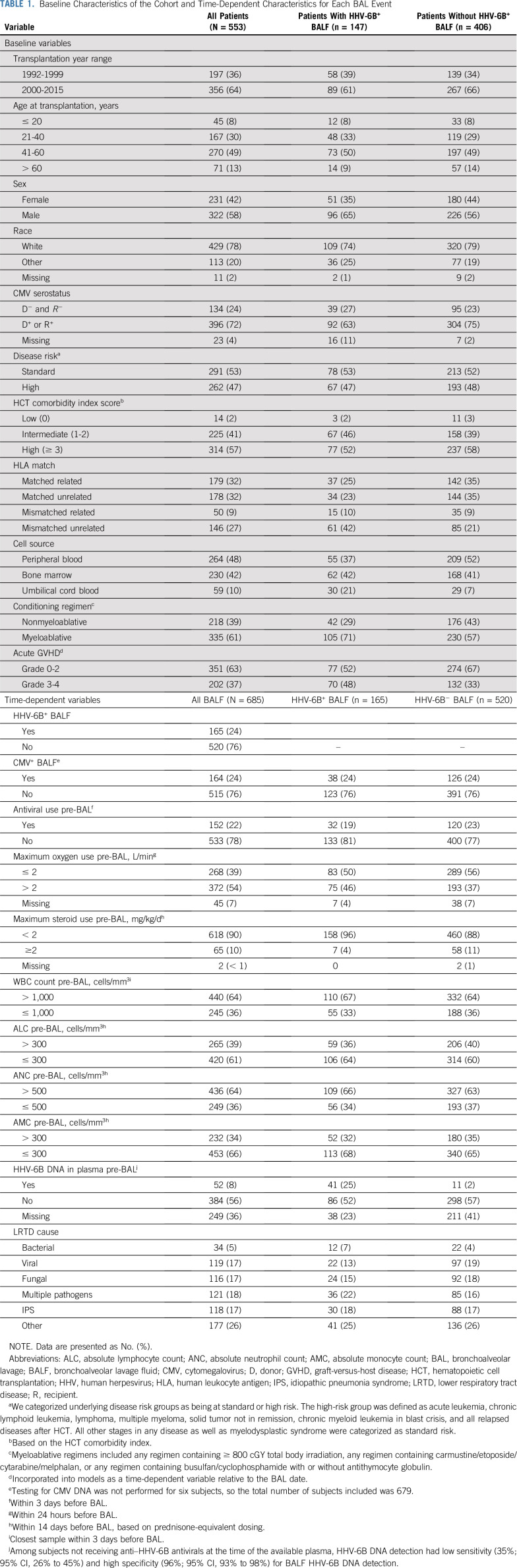
During the study period, 6,126 patients received an allogeneic HCT at FHCRC and 1,391 (23%) had a BAL within 100 days after HCT. We identified 564 patients (41%) with 697 BALF samples that were available for testing (Fig 1). Eleven recipients with 12 BALF samples had iciHHV-6 (species A or B) and were excluded. Characteristics of the 553 included patients are described in Table 1. The characteristics of excluded patients were largely representative of this cohort (Data Supplement).
FIG 1.
CONSORT diagram. BALF, bronchoalveolar lavage fluid; HHV, human herpesvirus; iciHHV-6, inherited chromosomally integrated HHV-6.
TABLE 1.
Baseline Characteristics of the Cohort and Time-Dependent Characteristics for Each BAL Event
HHV-6B Detection in BALF
We detected HHV-6B DNA in 165 of 685 BALF samples (24%) from 147 of 553 patients (27%) with LRTD. The distribution of LRTD categories between days 0 to 100 after HCT is depicted in the Data Supplement. Among those with HHV-6B+ BALF, the median day of the first HHV-6B+ BALF occurred 29 (interquartile range [IQR], 20-64]) days after HCT. The median HHV-6B BALF viral load was 3.3 log10 copies/mL (IQR, 2.5-4.1 copies/mL). We did not detect HHV-6A DNA in any BALF.
The majority of HHV-6B+ BALF and the highest HHV-6B BALF viral loads occurred between days 11 and 40 after HCT (Fig 2A and 2B). There were relatively similar proportions of HHV-6B+ BALF and HHV-6B BALF viral loads across strata defined by LRTD category (Fig 2C and 2D). Higher proportions of HHV-6B+ BALF occurred in patients with diffuse abnormalities with a mixed pattern than other radiographic patterns (Data Supplement). Median HHV-6B BALF viral loads were higher in patients with diffuse ground glass opacities or diffuse mixed changes compared with focal changes or diffuse nodules (Data Supplement).
FIG 2.
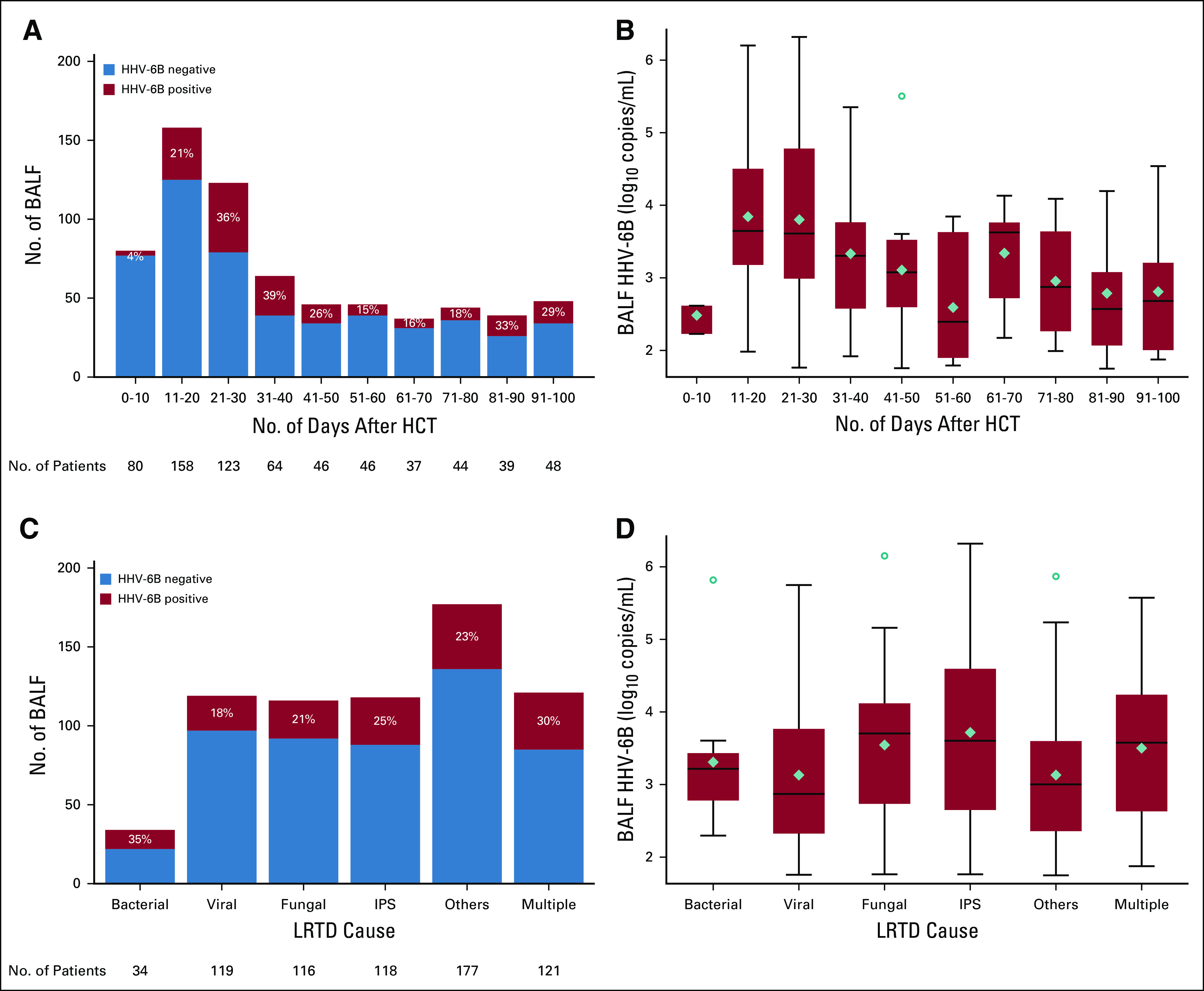
The distribution of human herpesvirus (HHV)-6B+ bronchoalveolar lavage fluid (BALF) detection and viral load after allogeneic hematopoietic cell transplantation (HCT). (A) A histogram of the number of BALF samples (N = 685) and the proportion of HHV-6B+ BALF (n = 165) over time after allogeneic HCT. (B) A box-and-whisker plot of the HHV-6B viral load over time among patients with HHV-6B+ BALF. (C) A histogram of the number of BALF samples and the proportion of HHV-6B+ BALF samples stratified by cause of lower respiratory tract disease (LRTD). (D) A box-and-whisker plot of the HHV-6B viral load stratified by cause of LRTD among patients with HHV-6B+ BALF. (B, D) The boxes represent the interquartile range, the horizontal lines and diamonds within the boxes represent the median and mean, respectively, and the upper and lower whiskers extend to the third and first quartiles ± 1.5 times the interquartile range, respectively. Circles represent data points that fall outside these parameters. IPS, idiopathic pneumonia syndrome.
We performed additional analyses to test whether HHV-6B+ BALF was due to lower respiratory tract infection versus contamination from blood. We demonstrated that patients with diffuse alveolar hemorrhage and HHV-6B+ BALF had a higher median HHV-6B viral load in BALF than in plasma obtained within a median of 1 day of the BALF (IQR, 1-3 days), and the plasma was often negative (Data Supplement). Similar findings were demonstrated in patients without diffuse alveolar hemorrhage (Data Supplement). Next, we demonstrated no meaningful correlation of BALF total β-globin or WBC count with HHV-6B BALF viral load (Data Supplement). In comparison, there was a correlation between BALF β-globin and HHV-6 viral load in BALF from patients with iciHHV-6, as expected due to latent HHV-6 DNA in every nucleated recipient cell (Data Supplement). These findings support HHV-6B replication within the lung as opposed to contamination from the blood.
HHV-6B+ BALF and Risk for Overall Mortality and Death From Respiratory Failure
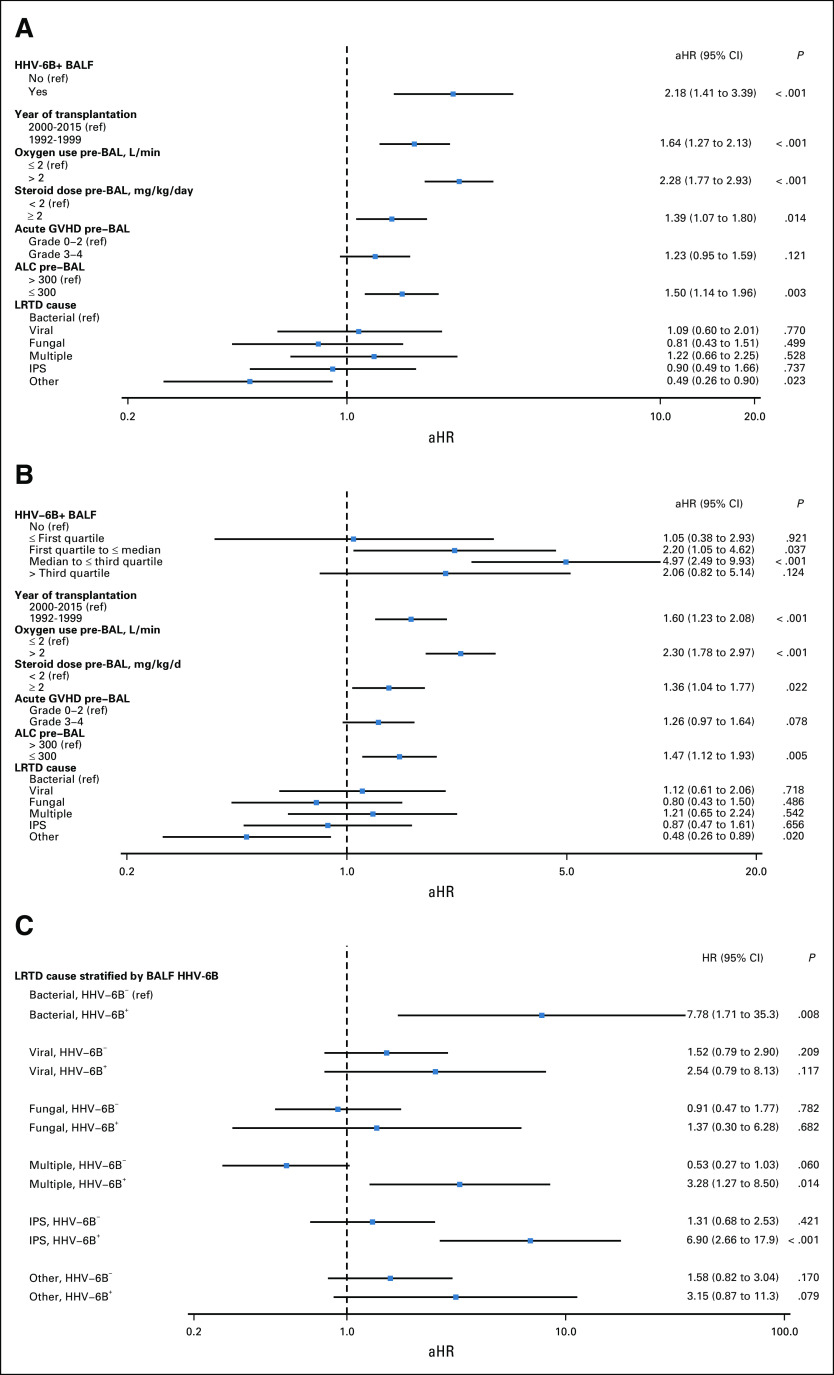
There were 269 deaths (49%) due to any cause and 219 deaths (40%) from respiratory failure within 100 days of the last BAL per patient. Three patients died on the day of the BAL and were excluded. Among day 60 survivors, those who had a preceding HHV-6B+ BALF had increased overall mortality and death from respiratory failure compared with those without HHV-6B+ BALF (Fig 3). In adjusted Cox models incorporating the entire cohort, patients with HHV-6B+ BALF had a higher risk of overall mortality (adjusted hazard ratio [aHR], 2.18; 95% CI, 1.41 to 3.39; Fig 4A). Patients with HHV-6B BALF viral loads greater than the lower quartile (2.41 log10 copies/mL) had a higher risk for overall mortality (Fig 4B). In univariate analysis, there was increased risk for mortality in every LRTD category when HHV-6B was detected in BALF as a copathogen (Fig 4C). Similar findings were demonstrated for death from respiratory failure (Data Supplement). These findings were recapitulated in subgroup analyses among patients receiving a HCT on or after 2006 (Data Supplement).
FIG 3.
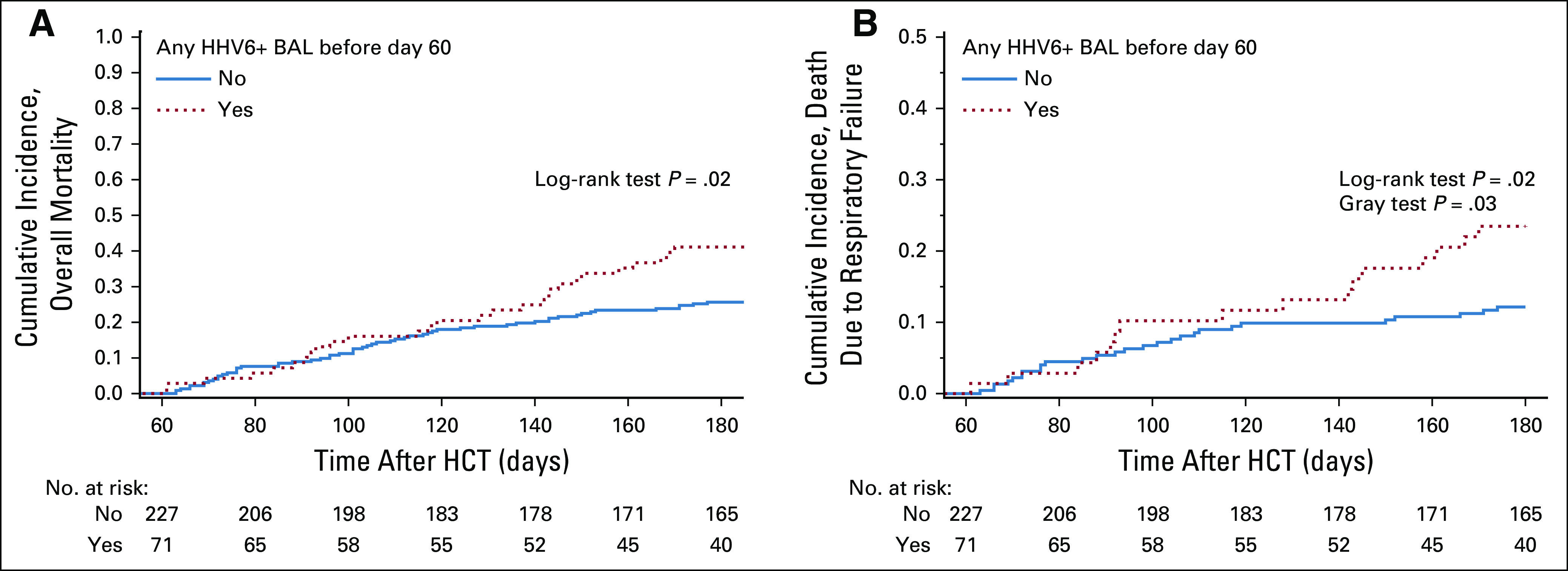
Cumulative incidence curves of overall mortality and death from respiratory failure among subjects alive at day 60 after hematopoietic cell transplantation (HCT). The graphs are stratified by whether a subject had human herpesvirus (HHV)-6B detected in a BALF sample between day 0 and day 60 after HCT. Only subjects who had a BAL for evaluation of lower respiratory tract disease after HCT were included in this study. (A) Cumulative incidence curve of overall mortality. (B) Cumulative incidence curve of deaths due to respiratory failure. Deaths due to other causes were treated as competing risk events.
FIG 4.
Forest plots of risk factors for overall mortality within 100 days after the BAL. (A) Model 1 demonstrates the results of an adjusted model in which the human herpesvirus (HHV)-6B+ BALF variable was incorporated as positive or negative. (B) Model 2 demonstrates the results of an adjusted model in which the HHV-6B+ BALF variable was incorporated in quartiles of viral load. (C) Model 3 demonstrates the results of a univariate analysis of LRTD categories stratified by HHV-6B+ BALF; bacterial pneumonia without HHV-6B detection in the BALF is the referent (ref) for all other categories. The variables shown in the figure were all the variables included in the final models. aHR, adjusted hazard ratio; ALC, absolute lymphocyte count; BAL, bronchoalveolar lavage; BALF, bronchoalveolar lavage fluid; GVHD, graft-versus-host disease; HCT, hematopoietic cell transplantation; IPS, idiopathic pneumonia syndrome; LRTD, lower respiratory tract disease.
HHV-6B BALF Viral Load and Mortality Among Subjects Receiving Antivirals
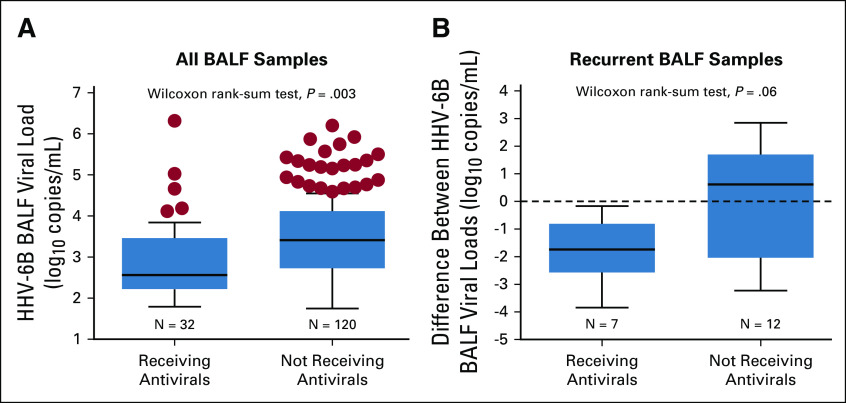
Antivirals active against HHV-6B were administered to 152 patients within 3 days before BAL at a median of 33 days after HCT (IQR, 23-52 days). Antivirals primarily consisted of ganciclovir or foscarnet and were indicated for CMV in 86% of patients. Patients who received antivirals within 3 days before HHV-6B+ BALF had lower median HHV-6B viral loads (2.6 log10 copies/mL) compared with those not receiving anti–HHV-6B antivirals (3.4 log10 copies/mL; Fig 5A). Among patients with HHV-6B+ BALF who had a subsequent BAL within 28 days, there was a greater decrease in the HHV-6B BALF viral load in patients who received anti–HHV-6B antivirals in between BALs compared with those who did not (Fig 5B; Data Supplement).
FIG 5.
Median HHV-6B BALF viral loads stratified by the receipt of antivirals before the BAL. (A) Among subjects with HHV-6B+ BALF, individuals who received anti–HHV-6B antivirals within 3 days before the BAL had significantly lower median HHV-6B BALF viral loads (2.6 log10 copies/mL) compared with individuals not receiving anti–HHV-6B antivirals (3.4 log10 copies/mL) . (B) Among seven subjects and 12 subjects who did and did not receive anti–HHV-6B antivirals in between subsequent BALs, respectively, there was a greater decrease in the HHV-6B BALF viral load in the subjects receiving antivirals. Only subsequent BALF samples within 28 days of the first HHV-6B+ BALF were considered. The boxes represent the interquartile range, the horizontal lines within the boxes represent the median, and the upper and lower whiskers extend to the third and first quartiles ± 1.5 times the interquartile range, respectively. Circles represent data points that fall outside these parameters.
Patients with HHV-6B+ BALF who received antivirals within 3 days before the BAL had a lower risk for overall mortality compared with patients with HHV-6B+ BALF not receiving antivirals (aHR, 0.42; 95% CI, 0.16 to 1.10; Data Supplement). This approximated the HR in patients without HHV-6B+ BALF who did or did not receive antivirals (Data Supplement). Findings were similar for death from respiratory failure (Data Supplement).
CMV+ BALF and Risk for Overall Mortality
We tested BALF for CMV DNA as a comparison with this related and established pulmonary pathogen. Detection of CMV DNA in BALF was similar in HHV-6B+ and HHV-6B− BALF (Table 1). CMV+ BALF was similarly associated with an increased risk of overall mortality (aHR, 1.78; 95% CI, 1.01 to 3.15; Data Supplement).
Lung Tissue Detection of HHV-6B DNA and RNA by Droplet Digital PCR and RISH
We identified six patients with HHV-6B+ BALF who had available formalin-fixed paraffin-embedded lung tissue from within 3 weeks of the BAL. HHV-6B was identified by PCR in all samples. We performed RISH for HHV-6 messenger RNA in tissue from three patients whose samples met prespecified quality metrics (Data Supplement). Scattered intraparenchymal cells stained with HHV-6 U41 and/or U57 probes in all three lung samples (Fig 6). One patient with clinically diagnosed IPS had the highest viral detection; virologic and clinical findings are detailed in the Data Supplement.
FIG 6.
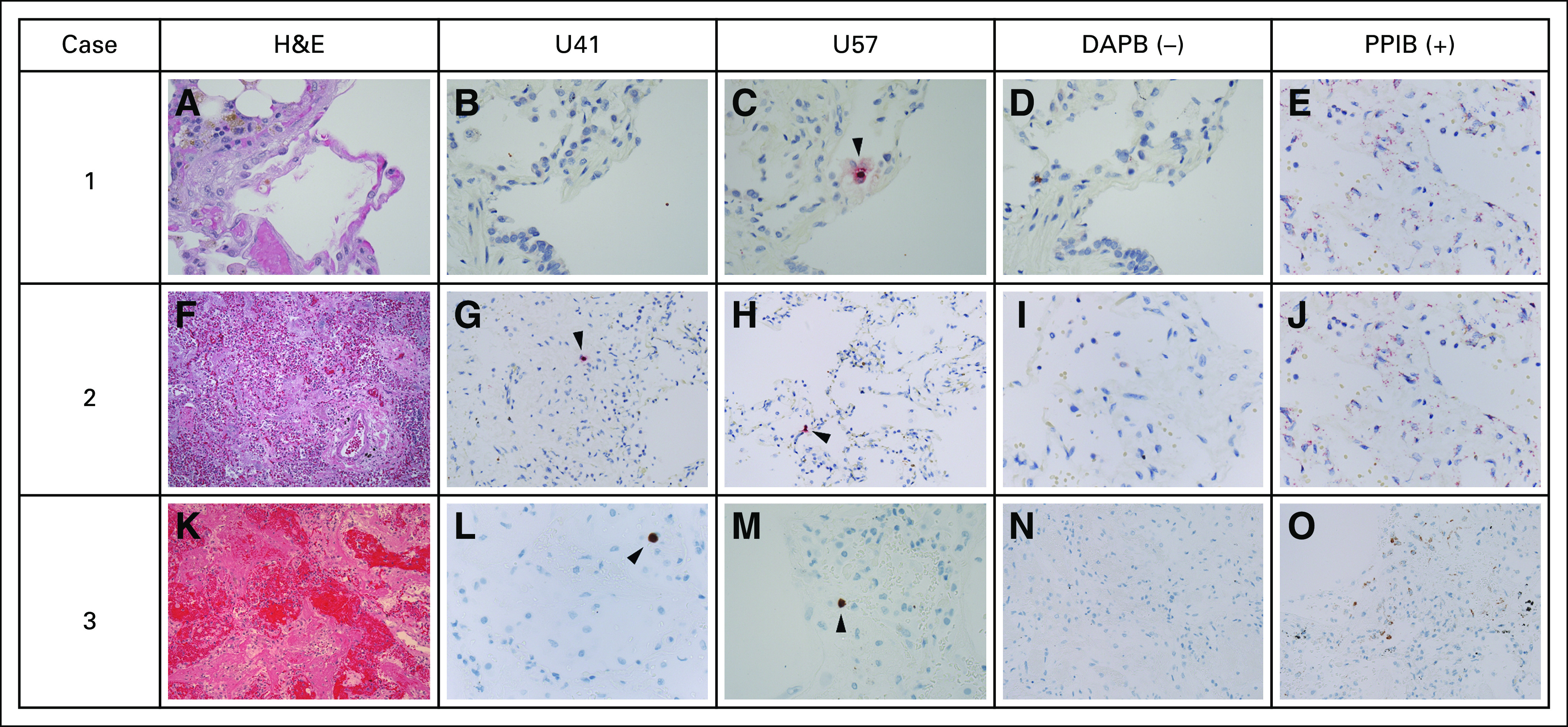
RNAscope branched-chain RNA in situ hybridization (RISH) for HHV-6 messenger RNA transcripts (U41 and U57 transcripts) in formalin-fixed paraffin-embedded lung-tissue specimens from subjects with HHV-6B+ BALF. (A-O) The images under the columns labeled “U41” and “U57” correspond to HHV-6 RISH targeting HHV-6 early (U41) and late (U57) messenger RNA transcripts expressed during lytic viral infection. The images under the column labeled “DAPB” represent the negative control, which is a cocktail of probes targeting bacterial genes. The images under the column labeled “PPIB” represent the positive control, which is a cocktail of normally weakly expressed human control genes. Each row represents a subject with HHV-6B+ BALF sample collected within 10 days of the tissue-sample collection date. Only tissue samples that met prespecified quality of RNA preservation based on PPIB staining were included (Data Supplement). (A) In case 1, the subject was diagnosed with bacterial and fungal pneumonia. The hematoxylin and eosin (H&E) stain demonstrated a thin alveolar wall, some congested capillaries and hemosiderin pigment, but no overt viral cytopathic changes. (B) RISH (with red chromogen staining) for U41 was negative . (C) RISH for U57 demonstrated rare positive cells. In case 2, the subject was diagnosed with bacterial and fungal pneumonia. (F) The H&E stain demonstrated diffuse alveolar damage and organizing pneumonia with significant vascular congestion but no overt viral cytopathic changes. RISH (with red chromogen staining) for (G) U41 and (H) U57 demonstrated rare positive cells. In case 3, the subject was diagnosed with idiopathic pneumonia syndrome and diffuse alveolar hemorrhage. (K) The H&E stain demonstrated marked destruction of normal lung parenchyma with hemorrhage and diffuse alveolar damage. There was a comparatively mild inflammatory infiltrate without overt viral cytopathic changes. RISH (with brown chromogen staining) for (L) U41 and (M) U57 demonstrated rare positive cells. (C, G, H, L, M) The rare positive cells highlighted by the arrowheads are intraparenchymal, often seen within alveolar walls, and, therefore, favored to represent pneumocytes or lymphocytes. (D, I, N) The negative control stains for each case demonstrated minimal to no background staining. (E, J, O) The positive control stains for each case demonstrated good staining to indicate preservation of RNA in the samples.
DISCUSSION
We found that allogeneic HCT recipients with LRTD and HHV-6B+ BALF had an increased risk for overall mortality and death from respiratory failure compared with those without HHV-6B detection in BALF. Anti–HHV-6B antiviral use was associated with decreased HHV-6B BALF viral load and lower mortality risk. Analogous findings were demonstrated in analyses of CMV+ BALF. Lung tissue from three patients with HHV-6B+ BALF demonstrated HHV-6 gene expression in intraparenchymal cells.
Ascribing a causal role to HHV-6B in human diseases on the basis of DNA detection is challenging in the context of observational studies because of the high prevalence of HHV-6B and its ability to establish latency in many cell types.7 Despite these challenges, we used multiple strategies to approach causal inference based on the Bradford Hill epidemiologic criteria26 and an adaptation of these criteria that consider sequence-based identification of pathogens.18 We present data that fulfill the majority of these criteria in support of HHV-6B as a pulmonary pathogen.
The primary finding of this study is a strong association between HHV-6B+ BALF and increased risk for mortality. Furthermore, we demonstrated a dose-dependent response between HHV-6B BALF viral load and mortality. Low-level HHV-6B detection in BALF was not associated with increased mortality risk and may be due to asymptomatic shedding, similar to that described for CMV.6 Although the increased risk for mortality did not reach statistical significance in patients in the highest viral-load quartile, the confidence intervals were wide, and interpretation is limited by fewer events in this exploratory stratified analysis. A viral load threshold may be more important, as demonstrated for CMV.6 We also demonstrated that patients with HHV-6B+ BALF who received antivirals within 3 days before the BAL, typically for CMV, had a lower risk for overall mortality and death from respiratory failure that approximated the risk in patients with HHV-6B- BALF. Although the associations did not reach statistical significance in adjusted models, the power of this analysis was limited by the low number of patients with HHV-6B+ BALF who received antivirals before BAL. Thus, these results should be interpreted cautiously. Nonetheless, the findings are notable, given that HCT recipients receiving antiviral treatment for CMV tend to have higher mortality.27 Together, the data demonstrate strong associations of HHV-6B detection with mortality and a dose-response relationship, supporting the plausibility of HHV-6B as a pulmonary pathogen.
We performed a variety of analyses to demonstrate HHV-6B replication within the lungs as opposed to detection of latently infected cells or viral DNA from blood. Findings that supported HHV-6B replication in the lungs included differences in HHV-6B BALF detection over time after HCT, lack of correlation between HHV-6B BALF viral load and blood contamination, and lower HHV-6B BALF viral load among recipients of anti–HHV-6B antivirals. The similar distribution of HHV-6B BALF detection and viral loads across LRTD categories could argue in favor of detection of latent virus. However, a large study of allogeneic HCT recipients with CMV LRTD demonstrated similar findings for CMV.6 In addition, the time periods with the highest proportion of HHV-6B+ BALF samples and highest viral loads correspond with the timing of blood and CNS HHV-6B reactivation and disease.28 Importantly, HHV-6B was detected in only one BALF sample (5%) at a low level from a historical cohort of 21 asymptomatic allogeneic HCT recipients who underwent a research BAL between days 35 and 45 after HCT at our center.5 This may reflect the incidence of pulmonary HHV-6B shedding or detection of latent virus in the absence of LRTD after HCT.
We also performed similar analyses using CMV+ BALF as the primary predictor of interest, given its close biologic relationship to HHV-6B and that it is an established pulmonary pathogen.29 First, we found that CMV was equally distributed among HHV-6B+ and HHV-6B− BALF samples and thus unlikely to be driving our primary findings. We demonstrated that CMV+ BALF was associated with an increased risk of overall mortality, similar to that of HHV-6B+ BALF—a finding that meets the Bradford Hill criteria of analogy.
Finally, we developed a RISH assay to detect markers of intraparenchymal HHV-6B replication in lung tissue. We identified HHV-6B RNA transcripts in lung tissue from all three tested subjects with HHV-6B+ BALF and well-preserved samples. Additional support for the significance of HHV-6B detection at the tissue level come from an animal model demonstrating that latent infection with murine roseolovirus, an HHV-6 homolog, can reactivate in murine lungs after HCT and induce an IPS-like phenotype.30
Limitations of this study include the retrospective, single-center design with the potential for residual confounding. Lack of broad availability of the new international standard for HHV-6B PCR31 limits generalizability of the viral load results. This cohort was a specifically defined group of allogeneic HCT recipients with LRTD undergoing BAL, which may explain clinical features such as the high rate of GVHD grades 3 and 4, and limits generalizability. Variability in the volume of saline injected and recovered from the BAL could affect quantification of HHV-6B viral load; we used the log10 scale for all viral load analyses, which may mitigate some of the differences in variable dilution. There was a possibility of misclassification of the cause of LRTD due to negative test results or lack of testing, particularly in the earlier period. However, we recapitulated our findings in analyses restricted to a more contemporary subgroup receiving a HCT on or after 2006, when comprehensive molecular testing was routinely performed. Analyses of the association of antivirals with outcomes were limited by small sample size and variable durations of antiviral administration. Given the recent approval of letermovir for CMV prophylaxis in CMV-seropositive adult allogeneic HCT recipients,32 which is limited in antiviral effect to CMV, the epidemiology of HHV-6B and other viral infections may change with reduced use of broader-spectrum antivirals.17
In conclusion, the molecular and clinical observations from this study demonstrate that HHV-6B can reactivate in the lungs after allogeneic HCT, and detection of HHV-6B in BALF from allogenic HCT recipients with LRTD is associated with increased mortality risk. The data presented here fulfill criteria to define pathogen-disease associations in the molecular era.18 Whether HHV-6B reactivation independently causes LRTD or is a copathogen reactivating in the context of pulmonary damage is unclear. Definitive evidence of causation will require a randomized prevention or treatment trial, although this has not been performed for most microorganisms accepted as pulmonary pathogens. Recognition of HHV-6B as a pulmonary pathogen has potential to improve outcomes after allogeneic HCT.
ACKNOWLEDGMENT
We thank the Infectious Disease Sciences Biorepository at Fred Hutchinson Cancer Research Center for providing samples for this study. We thank Louise Kimball, Sonia Goyal, Jennifer Schaeffer, Laurel Joncas-Schronce, Chris Davis, and Zach Stednick for help with data collection, in addition to Davide Myerson, Ratsamy (Savanh) Chanthaphavong, Sunni Farley, and David Woolston for tissue preparation, staining, and interpretation.
Footnotes
This study was presented in part at the 10th International Conference on Human Herpesviruses-6 and -7, Berlin, Germany, July 23-26, 2017.
Supported by the National Institutes of Health (Grants No. 5K23AI119133[JAH] and K24HL093294 [MB]), the American Society for Blood and Marrow Transplantation (New Investigator Award [JAH]), and a Dharam Ablashi Research Fund Grant from the HHV-6 Foundation. Additional resources were provided by the National Institutes of Health (Grants No. HL088021, CA78902, CA18029, CA015074, and HL122173).
AUTHOR CONTRIBUTIONS
Conception and design: Joshua A. Hill, Lisa K. Vande Vusse, Cecilia C.S. Yeung, Danielle M. Zerr, Lawrence Corey, Wendy M. Leisenring, Michael Boeckh
Administrative support: E. Lisa Chung, Cecilia C.S. Yeung, F. Mark Stewart, Keith R. Jerome, Lawrence Corey
Provision of study material or patients: E. Lisa Chung, Cecilia C.S. Yeung
Collection and assembly of data: Joshua A. Hill, Lisa K. Vande Vusse, E. Lisa Chung, Cecilia C.S. Yeung, Terry Stevens-Ayers, Meei-Li Huang, Keith R. Jerome, Danielle M. Zerr, Michael Boeckh
Data analysis and interpretation: Joshua A. Hill, Lisa K. Vande Vusse, Hu Xie, Cecilia C.S. Yeung, Sachiko Seo, Cynthia E. Fisher, F. Marc Stewart, Keith R. Jerome, Danielle M. Zerr, Wendy M. Leisenring, Michael Boeckh
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Human Herpesvirus 6B and Lower Respiratory Tract Disease After Hematopoietic Cell Transplantation
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Joshua A. Hill
Honoraria: Chimerix
Consulting or Advisory Role: Nohla Therapeutics, Amplyx Pharmaceuticals
Research Funding: Nohla Therapeutics, Karius
Travel, Accommodations, Expenses: Chimerix
Lisa K. Vande Vusse
Employment: University of Washington Medical Center
Cecilia C.S. Yeung
Consulting or Advisory Role: DiaCarta, OBI Pharma
Speakers' Bureau: Archer Biosciences
Research Funding: Pfizer
Cynthia E. Fisher
Research Funding: Gilead Sciences, Karius
F. Marc Stewart
Stock and Other Ownership Interests: Microbot Medical Device
Honoraria: The France Foundation (I), Physician Education Resource (I)
Consulting or Advisory Role: CVS Caremark (I), McKesson (I), Pfizer (I), Amgen (I)
Research Funding: Bristol-Myers Squibb (I), JW Pharmaceutical (I), Novartis (I), Pfizer (I), Glycomimetics (I), Trovagene (I), Invivoscribe (I), Aptose Biosciences (I), AbbVie (I), Trethera (I)
Patents, Royalties, Other Intellectual Property: Provisional patent filed: Application 62/694,874 filed July 6, 2018, entitled “High Throughput Drug Screening of Cancer Stem Cells” (I)
Danielle M. Zerr
Research Funding: Merck (Inst)
Lawrence Corey
Stock and Other Ownership Interests: Juno Therapeutics, Immune Design
Research Funding: Sanofi (Inst), Immune Design (Inst), Gilead Sciences (Inst)
Patents, Royalties, Other Intellectual Property: Coinventor listed on several patents involving potential herpes simplex virus vaccine development.
Michael Boeckh
Consulting or Advisory Role: Merck, Gilead Sciences, Shire, Chimerix, Janssen, VIR Biotechnology, Ansun, Ablynx
Research Funding: Merck (Inst), Chimerix (Inst), Vical (Inst), Astellas Pharma (Inst), Gilead Sciences (Inst), VIR Biotechnology (Inst), Ansun (Inst), Ablynx (Inst)
No other potential conflicts of interest were reported.
REFERENCES
- 1.Aguilar-Guisado M, Jiménez-Jambrina M, Espigado I, et al. Pneumonia in allogeneic stem cell transplantation recipients: A multicenter prospective study. Clin Transplant. 2011;25:E629–E638. doi: 10.1111/j.1399-0012.2011.01495.x. [DOI] [PubMed] [Google Scholar]
- 2.Panoskaltsis-Mortari A, Griese M, Madtes DK, et al. An official American Thoracic Society research statement: Noninfectious lung injury after hematopoietic stem cell transplantation: Idiopathic pneumonia syndrome. Am J Respir Crit Care Med. 2011;183:1262–1279. doi: 10.1164/rccm.2007-413ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gooley TA, Chien JW, Pergam SA, et al. Reduced mortality after allogeneic hematopoietic-cell transplantation. N Engl J Med. 2010;363:2091–2101. doi: 10.1056/NEJMoa1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukuda T, Hackman RC, Guthrie KA, et al. Risks and outcomes of idiopathic pneumonia syndrome after nonmyeloablative and conventional conditioning regimens for allogeneic hematopoietic stem cell transplantation. Blood. 2003;102:2777–2785. doi: 10.1182/blood-2003-05-1597. [DOI] [PubMed] [Google Scholar]
- 5.Seo S, Renaud C, Kuypers JM, et al. Idiopathic pneumonia syndrome after hematopoietic cell transplantation: Evidence of occult infectious etiologies. Blood. 2015;125:3789–3797. doi: 10.1182/blood-2014-12-617035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boeckh M, Stevens-Ayers T, Travi G, et al. Cytomegalovirus (CMV) DNA quantitation in bronchoalveolar lavage fluid from hematopoietic stem cell transplant recipients with CMV pneumonia. J Infect Dis. 2017;215:1514–1522. doi: 10.1093/infdis/jix048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Bolle L, Naesens L, De Clercq E. Update on human herpesvirus 6 biology, clinical features, and therapy. Clin Microbiol Rev. 2005;18:217–245. doi: 10.1128/CMR.18.1.217-245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krueger GR, Wassermann K, De Clerck LS, et al. Latent herpesvirus-6 in salivary and bronchial glands. Lancet. 1990;336:1255–1256. doi: 10.1016/0140-6736(90)92874-h. [DOI] [PubMed] [Google Scholar]
- 9.Ogata M, Oshima K, Ikebe T, et al. Clinical characteristics and outcome of human herpesvirus-6 encephalitis after allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant. 2017;52:1563–1570. doi: 10.1038/bmt.2017.175. [DOI] [PubMed] [Google Scholar]
- 10.Carrigan DR, Drobyski WR, Russler SK, et al. Interstitial pneumonitis associated with human herpesvirus-6 infection after marrow transplantation. Lancet. 1991;338:147–149. doi: 10.1016/0140-6736(91)90137-e. [DOI] [PubMed] [Google Scholar]
- 11.Mariotte E, Schnell D, Scieux C, et al. Significance of herpesvirus 6 in BAL fluid of hematology patients with acute respiratory failure. Infection. 2011;39:225–230. doi: 10.1007/s15010-011-0114-8. [DOI] [PubMed] [Google Scholar]
- 12.Cone RW, Hackman RC, Huang ML, et al. Human herpesvirus 6 in lung tissue from patients with pneumonitis after bone marrow transplantation. N Engl J Med. 1993;329:156–161. doi: 10.1056/NEJM199307153290302. [DOI] [PubMed] [Google Scholar]
- 13.Buchbinder S, Elmaagacli AH, Schaefer UW, et al. Human herpesvirus 6 is an important pathogen in infectious lung disease after allogeneic bone marrow transplantation. Bone Marrow Transplant. 2000;26:639–644. doi: 10.1038/sj.bmt.1702569. [DOI] [PubMed] [Google Scholar]
- 14.Nagate A, Ohyashiki JH, Kasuga I, et al. Detection and quantification of human herpesvirus 6 genomes using bronchoalveolar lavage fluid in immunocompromised patients with interstitial pneumonia. Int J Mol Med. 2001;8:379–383. doi: 10.3892/ijmm.8.4.379. [DOI] [PubMed] [Google Scholar]
- 15.Nishimaki K, Okada S, Miyamura K, et al. The possible involvement of human herpesvirus type 6 in obliterative bronchiolitis after bone marrow transplantation. Bone Marrow Transplant. 2003;32:1103–1105. doi: 10.1038/sj.bmt.1704269. [DOI] [PubMed] [Google Scholar]
- 16.Tomblyn M, Chiller T, Einsele H, et al. Guidelines for preventing infectious complications among hematopoietic cell transplantation recipients: A global perspective Biol Blood Marrow Transplant 151143–1238.2009[Erratum: Biol Blood Marrow Transplant 2010;16:294] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chemaly RF, Hill JA, Voigt S, et al. In vitro comparison of currently available and investigational antiviral agents against pathogenic human double-stranded DNA viruses: A systematic literature review. Antiviral Res. 2019;163:50–58. doi: 10.1016/j.antiviral.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Fredericks DN, Relman DA. Sequence-based identification of microbial pathogens: A reconsideration of Koch’s postulates. Clin Microbiol Rev. 1996;9:18–33. doi: 10.1128/cmr.9.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sedlak RH, Hill JA, Nguyen T, et al. Detection of human herpesvirus 6B (HHV-6B) reactivation in hematopoietic cell transplant recipients with inherited chromosomally integrated HHV-6A by droplet digital PCR. J Clin Microbiol. 2016;54:1223–1227. doi: 10.1128/JCM.03275-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris B, Lowy FD, Stover DE, et al. Diagnostic bronchoscopy in solid-organ and hematopoietic stem cell transplantation. Ann Am Thorac Soc. 2013;10:39–49. doi: 10.1513/AnnalsATS.201212-114FR. [DOI] [PubMed] [Google Scholar]
- 21.Zerr DM, Gooley TA, Yeung L, et al. Human herpesvirus 6 reactivation and encephalitis in allogeneic bone marrow transplant recipients. Clin Infect Dis. 2001;33:763–771. doi: 10.1086/322642. [DOI] [PubMed] [Google Scholar]
- 22.Baron EJ, Miller JM, Weinstein MP, et al. A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2013 Recommendations by the Infectious Diseases Society of America (IDSA) and the American Society for Microbiology (ASM)(a) Clin Infect Dis. 2013;57:e22–e121. doi: 10.1093/cid/cit278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Pauw B, Walsh TJ, Donnelly JP, et al. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis. 2008;46:1813–1821. doi: 10.1086/588660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang F, Flanagan J, Su N, et al. RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill JA, Ikoma M, Zerr DM, et al. RNA sequencing of the in vivo human herpesvirus 6B transcriptome to identify targets for clinical assays distinguishing between latent and active infections. J Virol. 2019;93:e01419-18. doi: 10.1128/JVI.01419-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill AB. The environment and disease: Association or causation? Proc R Soc Med. 1965;58:295–300. doi: 10.1177/003591576505800503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teira P, Battiwalla M, Ramanathan M, et al. Early cytomegalovirus reactivation remains associated with increased transplant-related mortality in the current era: A CIBMTR analysis. Blood. 2016;127:2427–2438. doi: 10.1182/blood-2015-11-679639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogata M, Satou T, Kadota J, et al. Human herpesvirus 6 (HHV-6) reactivation and HHV-6 encephalitis after allogeneic hematopoietic cell transplantation: A multicenter, prospective study. Clin Infect Dis. 2013;57:671–681. doi: 10.1093/cid/cit358. [DOI] [PubMed] [Google Scholar]
- 29.Erard V, Guthrie KA, Seo S, et al. Reduced mortality of cytomegalovirus pneumonia after hematopoietic cell transplantation due to antiviral therapy and changes in transplantation practices. Clin Infect Dis. 2015;61:31–39. doi: 10.1093/cid/civ215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. doi: 10.1164/rccm.201809-1635OC. Zhou X, O’Dwyer DN, Xia M, et al: First onset herpesviral infection and lung injury in allogeneic hematopoietic cell transplantation. Am J Respir Crit Care Med 200:63-74 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Govind S, Hockley J, Morris C, et al: Collaborative study to establish the 1st WHO international standard for human herpes virus 6B (HHV-6B) DNA for nucleic acid amplification technique (NAT)-based assays. WHO/BS/2017.2321. World Health Organization. https://apps.who.int/iris/handle/10665/260259.
- 32. doi: 10.1056/NEJMoa1706640. Marty FM, Ljungman P, Chemaly RF, et al: Letermovir prophylaxis for cytomegalovirus in hematopoietic-cell transplantation. N Engl J Med 370:1781-1789, 2017. [DOI] [PubMed] [Google Scholar]