SUMMARY
For almost 50 years, sleep laboratories around the world have been collecting massive amounts of polysomnographic (PSG) physiological data to diagnose sleep disorders, the majority of which are not utilized in the clinical setting. Only a small fraction of the information available within these signals is utilized to generate indices. For example, the apnea hypopnea index (AHI) remains the primary tool for diagnostic and therapeutic decision-making for obstructive sleep apnea (OSA) despite repeated studies showing it to be inadequate in predicting clinical consequences. Today, there are many novel approaches to PSG signals, making it possible to extract more complex metrics and analyses that are potentially more clinically relevant for individual patients. However, the pathway to implement novel PSG metrics/analyses into routine clinical practice is unclear. Our goal with this review is to highlight some of the novel PSG metrics/analyses that are becoming available. We suggest that stronger academic-industry relationships would facilitate the development of state-of-the-art clinical research to establish the value of novel PSG metrics/analyses in clinical sleep medicine. Collectively, as a sleep community, it is time to reinvent how we utilize the polysomnography to move us towards Precision Sleep Medicine.
Keywords: precision medicine, precision sleep medicine, polysomnography, physiological signals
INTRODUCTION
One of the goals of Precision Medicine is to predict an individual’s risk of morbidity and mortality using different sources of data, from clinical history and physical examination, to physiological, imaging and molecular biomarkers, all to maximize health. Implementing “Precision Sleep Medicine” would utilize these principles and include the use of physiological data from the polysomnogram (PSG), both conventional and novel metrics, and combine them with imaging, questionnaires, genetic and -OMIC data to fully understand sleep and sleep disorders in the individual. One sleep disorder area where precision medicine is already being developed is obstructive sleep apnea (OSA) [1–7].
Statement of Problem.
When sleep medicine emerged as a new field in the 1960s, scoring criteria for sleep stages for analog PSG was established by Rechtschaffen and Kales [8], and was based on human visual inspection of physiological signals recorded during sleep. Since then, consensus on sleep stages and scoring rules by sleep experts culminated in the establishment of the American Academy of Sleep Medicine (AASM) Scoring Manual. Originally published in 2007 [9], there has been one major update, Version 2.0 in 2012 [10], with six subsequent updates (Versions 2.1 to 2.6, last in January 2020) to standardize calibrations, nomenclature and other particular aspects of the test. While these scoring criteria allowed PSG data to be evaluated uniformly between labs, it has potentially oversimplified assessment of sleep disorders. One example of this is seen in OSA, where we have reduced the entire PSG to one main index, the apnea-hypopnea index (AHI), i.e. the average number of apneas and hypopneas per hour of sleep [10, 11]. The AHI allows hypopneas to have the same weight as apneas, and it does not differentiate whether the respiratory events are obstructive or central in nature, long or short, or whether they are associated with an arousal, an increase in heart rate or a drop in oxygenation. Furthermore, despite OSA being a heterogeneous disorder where patients have different symptoms [12, 13], pathophysiology [3–6] and clinical outcomes, patients are simply grouped into ‘OSA, adult’ or ‘OSA, pediatric.’ Lastly, irrespective of origin or severity, we tend to treat adult OSA patients with continuous positive airway pressure (CPAP) treatment as the first line of therapy, despite the availability of other treatments.
Potential Solution.
As technology advances, it is inevitable that Sleep Medicine will undergo an evolution that changes current practices. Today, PSG signals are digitized and contain an enormous amount of information such that visual inspection is no longer enough to realize or analyze all the signals. Mathematical and computational methods that can more comprehensively and consistently analyze digitized PSG signals have been available for decades, yet we continue to use the same scoring criteria designed for analog based recordings. Thus, it is time to ask the question, how do we enhance clinical care by harnessing all the PSG information? In the example of OSA, one solution is to combine conventional and novel PSG metrics/analyses to identify more OSA subtypes that would better predict the best first line of therapy for an individual with the goal of preventing clinical consequences. With so many novel automated PSG tools and analysis methods available, now is the opportune time to consider how to conduct the necessary research to prove their validity as important physiological biomarkers.
Our goal with this narrative review is to outline how the PSG can be “reinvented” in the age of Precision Medicine using signals already being collected. We propose that there are two important advances to consider. First, is the identification of novel PSG metrics that can be obtained from raw PSG signals, such as sleep depth, sleep drive, arousal intensity and hypoxic burden. Our literature search strategy was to identify manuscripts that described new metrics that might contribute to assessment of the physiological consequences of OSA. Second, is the development of novel PSG analytical approaches to analyze multi-dimensional data, for example, autoscore sleep stages or diagnose narcolepsy using the PSG (without the multiple sleep latency test) or score movement disorders on the polysomnogram. Here we searched for manuscripts that reported machine learning approaches to multi-dimensional data. It may be the role of sleep researchers that have expertise in signal processing and computational analyses of big data to further develop PSG metrics/analyses, but equally important is the academic-industry partnerships between software companies to make the novel PSG metrics/analyses readily available so clinician investigators can assess their value for the practice of sleep medicine. Only by working together as a sleep community, can we advance and realize “Precision Sleep Medicine” of the future.
Based on these concepts, this narrative review will: 1) Describe novel PSG metrics that have been proposed to assess physiological disturbances during sleep with a focus on sleep-disordered breathing; 2) Explain different machine learning strategies; 3) Describe examples of application of machine learning approaches to the analysis of signals from the polysomnogram because both novel PSG metrics and new machine learning approaches are required to advance our understanding of sleep and sleep disorders; and 4) Outline future directions and steps needed to take advantage of the opportunities that these developments provide.
“PRECISION SLEEP MEDICINE”
Precision Medicine is an approach to healthcare, endorsed by the United States government [14], that aims to predict an individual’s risk using different sources of data – from genetic predisposition to environment and lifestyle choices, as well as symptoms/questionnaires, physiological, molecular and imaging biomarkers – then tailoring an intervention to maximize benefit for individual subjects [15]. This is based on the concept that there are major individual differences between patients with the same disorder. Management of patients should be cognizant of these differences and treatment directed at the individual. Other fields, such as “Precision Oncology,” “Precision Psychiatry” and “Precision Pulmonary” have already made significant advances toward precision medicine with proven benefits in patient outcomes [16–19]. Understanding disease subtypes is one of the first steps towards precision medicine that subsequently allows clinicians to tailor management for each subtype.
An exciting shift towards “Precision Sleep Medicine” is already underway with most of the work so far being done within OSA [20]. For example, OSA is a heterogeneous disorder [21] for which the use of conventional PSG metrics to diagnose OSA has not changed much in the last 40 years. Consequently, OSA heterogeneity or OSA subtypes cannot be fully captured. While the AHI is the current measure clinicians use to diagnose OSA, it is of limited value to inform Precision Medicine [22–24]. The AHI cannot capture the heterogeneity of underlying pathophysiological mechanisms including arousal threshold, ventilatory instability, upper airway collapsibility, upper airway muscle responsiveness, heart rate variability, and changes in sleep depth [25]. Just as supine-dependent OSA is a recognized subtype that can be treated with positional therapy, there are likely many other OSA subtypes that exist based on the pathways to disease [21, 26], clinical symptom presentation [12, 27–29], physiological expression [30], susceptibility to comorbidities [31, 32] and treatment response [20, 33, 34], but have not yet been clinically recognized. While we use conventional PSG parameters such as total sleep time, percentages of sleep stages, sleep latency, sleep efficiency and the arousal index to give context to a patient’s sleep history, these measures are rarely utilized to inform clinical decisions. Researchers have shown, PSG parameters such as oxygen desaturation indices (ODI) and total time spent with oxygen saturation less than 88%, are better than the AHI at informing clinical consequences and have been associated with excessive sleepiness and hypertension [35, 36] and cardiovascular/cerebrovascular regulation [37]. However, these associations are typically quite modest, suggesting that a more nuanced approach to assessment of physiological abnormalities could provide more robust identification. Furthermore, despite the known limitations of Positive Airway Pressure (PAP) as a first line therapy, irrespective of the OSA subtype, severity or risk of specific comorbidities, and the availability of alternative therapies, as a field, we still have not agreed on a validated clinical algorithm that tailors therapy to the individual’s endotype. By systematically understanding which OSA pathophysiological endotypes responds to specific therapeutic options [26], clinicians would be able to offer more individualized OSA therapy. In short, by condensing the rich physiological data of the PSG to the AHI, we have been unable to fully characterize OSA heterogeneity and identify meaningful subtypes. Thus, the first step to implementing precision sleep medicine for OSA is to recognize that the ‘one-size-fits-all’ approach is not effective, and a more comprehensive characterization of the disease is necessary. One important domain of a more comprehensive characterization of diseases like OSA are the physiological changes that occur during sleep.
NOVEL PSG METRICS
Recently, there have been major advances in the application of computational methods to derive novel physiological metrics that can be extracted automatically from the PSG. However, while the clinical value of these novel metrics is currently unknown, this review aims to provide reasons why and how more in-depth studies are needed to assess these novel PSG metrics. For us to understand the clinical value of these novel PSG metrics, we need more sleep clinicians to accept that novel PSG metrics could provide more clinical value and we need software companies to incorporate novel PSG metrics into existing software to make them more readily available for clinical research. Then, as a sleep community, we can perform the much-needed longitudinal studies to assess whether these novel PSG metrics provide enhanced prognostic information by identifying who is at increased risk of specific clinical outcomes. Given that novel PSG metrics provide information from different domains, it seems reasonable to argue that they will help identify subtypes of different sleep disorders and their underlying pathophysiological mechanisms. Examples of novel PSG metrics include sleep depth (e.g. Odds Ratio Products [ORP] [38] and cardiopulmonary coupling [39]), sleep drive (e.g. ORP-9) [40], arousal intensity and heart rate response to arousals [41], arousal threshold [42], and a more detailed characterization of respiratory events including hypoxic burden [43–46]. Collectively, novel PSG metrics will likely provide a more in-depth understanding of normal and abnormal sleep physiology. In this section we briefly review some novel PSG metrics and describe their potential role in assessment of sleep and characterization of sleep disorders, with a focus on OSA. Some of these are specific to OSA (e.g. hypoxic burden [44–47]), while others that assess sleep depth [39, 48] are relevant not only to OSA but other sleep disorders. While some studies have been performed to assess the clinical utility of these novel PSG metrics, larger validation studies are still necessary before applying them to clinical practice.
Sleep Depth (ORP and Cardio-pulmonary coupling) and sleep drive (ORP-9)
While sleep quantity is important for sleep health, sleep quality is equally important [49]. Critical to determining sleep quantity and sleep quality is scoring sleep stages. Conventionally, sleep stages are scored using EEG, EOG and EMG criteria for 30-second epochs that are a carryover from when scoring was performed on analog-based PSGs. Even though PSGs have been digitized for decades, we continue to use the 50% rule [9]. This means if wake EEG is present in less than half of a 30-second epoch, it is not accounted for in sleep fragmentation, sleep depth or sleep quality when in fact, sleep state changes continuously rather than an arbitrary 30-second epoch. Furthermore, the EEG delta, theta, alpha/sigma, and beta waves used to score a stage of sleep most likely does not capture the variability of sleep stages both within one patient and between patients. For example, a patient’s EEG signal patterns in stage 2 sleep may vary from the beginning to the end of the night and may vary when compared to another patient’s stage 2 sleep, yet currently, we equally classify it all as stage 2. In sleep disorders such as insomnia, OSA, and narcolepsy, where sleep becomes increasingly fragmented, the current scoring criteria fails to account for more rapidly changing sleep stages and sleep depth. This leads to the loss of clinically relevant data regarding sleep architecture, as well as masking the potential impact of arousal dynamics and the physiologic impact of arousals [25, 38, 40]. While traditional scoring rules and 30-second epochs cannot capture this variability in staging sleep, if we used shorter epochs and automated scoring, we could capture this variability. Automated EEG-analysis using spectral analysis makes it possible to quantify EEG power density and develop novel sleep quality indices. This argues that a continuous measure of sleep depth and sleep drive is likely to be more informative.
Odds Ratio Product (ORP):
Younes et al [38] initially set out to determine continuous measures of sleep depth specifically in OSA patients. Using Fourier transform of EEG signals, they developed a continuous index of sleep depth - the odds ratio product (ORP). ORP is based on spectral analysis from the Fourier transform of the EEG evaluating the relative power in different frequency ranges (delta, theta, alpha/sigma, and beta). Thus, the ORP moves beyond measuring delta power during sleep which has not become part of routine clinical assessment. With ORP, relative power is calculated for 3-second consecutive epochs, then each frequency range is ranked between 0 (less intense) and 9 (most intense). Each epoch is then assigned a 4digit number (Bin #), which represents, in order, the ranks of delta, theta, alpha/sigma, and beta powers. For example, an epoch with a Bin# 2189 indicates a power spectrum with low delta and theta powers and high alpha and beta powers. For each Bin #, the probability of being “awake” in the next 30-second epochs staged awake is determined and scaled by 40 (wake epochs in the development files represented 40% of all epochs), resulting in a continuous measure of sleep depth – the ORP, ranging from 0 (always asleep) to 2.5 (always awake). ORP has recently been used by Goldschmied et al [50] to characterize sleep depth during recovery sleep after 36 hours of sleep deprivation in 200 healthy subjects. They found that the ORP values remained lower throughout the first night of recovery sleep compared to baseline sleep, indicating that as expected, sleep is deeper during recovery sleep, supporting the concept that ORP is an adequate measure of sleep depth. Meza-Vargas et al. [51] measured ORP in 30 patients with overnight PSG and MSLT. They found that higher levels of ORP during non-REM sleep i.e., lighter sleep, correlated with excessive daytime sleepiness, as determined by the MSLT [51]. In addition, using a large sample of monozygotic (N=59 pairs) and dizygotic (N=41 pairs) twins, our group has evaluated ORP based on repeated overnight PSG before and after sleep deprivation (manuscript in preparation). We observed that ORP is at least moderately stable from night to night within an individual patient, with intraclass correlation coefficients (ICCs) of 0.55 during non-REM and 0.70 during REM sleep. Thus, ORP behaves as an individual trait-like characteristic.
Cardio-pulmonary coupling:
Cardio-pulmonary coupling is another measure of sleep depth, but it does not require EEG signals; thus, it will be easier to deploy in the ambulatory setting. Thomas et al [39] derived a sleep spectrogram from a single-lead electrocardiogram, which could then be used to track cardiopulmonary interactions. This cardio-pulmonary coupling during sleep was extracted from the normal-to-normal sinus inter-beat interval series and a corresponding electrocardiogram-derived respiration signal. Then using Fourier-based techniques, coherence (or cross-power) of these 2 simultaneous signals was used to generate a spectrographic representation of cardio-pulmonary coupling dynamics during sleep. This technique showed that non-REM sleep in adults demonstrates spontaneous abrupt transitions between high- and low-frequency cardio-pulmonary coupling regimes, which have characteristic EEG, respiratory, and heart-rate variability signatures in both health and disease. Since cardiopulmonary coupling demonstrates a closer relationship to cyclic and non-cyclic alternating pattern states (measure of sleep instability, further discussed below) than standard sleep staging, it represents a novel measure of sleep depth during non-rapid eye sleep. Cardio-pulmonary coupling has been used in the setting of mandibular advancement devices in OSA patients [52], and wearable devices [53]. Lee et al [52] compared baseline measures and after 3 months use of mandibular advancement devices. Not only did they find that all respiratory indices improved, but also, when using cardio-pulmonary coupling, sleep depth also improved. Thomas et al [53] evaluated a wearable device, the M1, and used novel computational analyses of cardiopulmonary coupling to estimate sleep quality over multiple nights. Focusing on night-to-night variability, both healthy participants and sleep apnea patients treated with PAP demonstrated stable breathing and stable sleep, while participants with insomnia demonstrated less stable sleep. This study demonstrated how the combination of wearable devices and novel analyses could provide new insights into the management of sleep disorders.
Odds Ratio Product (ORP-9):
Using the ORP, Younes et al [40] also developed an index, called ORP-9 that averages the ORP, 9 seconds after an arousal/awakening. This reflects the ability to return to sleep after the arousal, i.e., sleep drive. Lower ORP-9 indicates an ability to rapidly return to deeper sleep. Patients with higher ORP-9 are more susceptible to repeat arousals/awakenings within the next 30 seconds than those with lower values. This study suggests that determining an OSA patient’s ability to return to deep sleep following an arousal/awakening may characterize one’s susceptibility to sleep fragmentation and overall sleep depth. Collectively, ORP and ORP-9 can be used to evaluate patients’ depth of sleep and sleep drive, which may explain different symptom presentation and underlying pathophysiology in OSA, insomnia and narcolepsy patients.
Arousal intensity and heart rate response to arousals
Cortical arousals that occur at the end of respiratory events are a significant cause of sleep disruption and sleep fragmentation [54]. Conventionally, AASM scoring criteria for an arousal is based on an abrupt shift in the EEG to higher frequencies for at least 3 seconds preceded by at least 10 seconds of stable sleep [9, 55]. The issue with human scoring of an arousal is that its appearance may vary considerably between individuals and within an individual across the night resulting in poor inter-scorer reliability [56].
Arousal intensity:
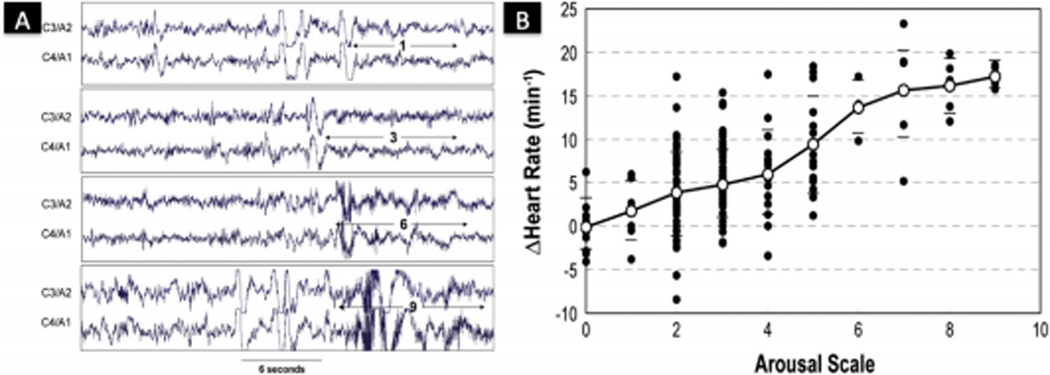
Azarbarzin et al [41] set out to automate arousal detection and devised an intensity scale to catergorize arousals. They applied a two-level discrete wavelet transform to all arousal episodes using the two central electrodes (C3/A2 and C4/A1) (Figure 1A). Then, every arousal was given an intensity scale between 0 (no relative change in EEG) and 9 (most intense arousal). The arousal intensity scale was strongly associated with arousal-related tachycardia (Figure 1B) and respiratory and pharyngeal muscle responses [57]. This strong association between arousal intensity and physiological consequences can not be captured using non-automated (e.g. visual inspection) arousal detection. This suggests that the intensity of cortical arousals is an independent trait that could reflect different underlying pathophysiologies of OSA (i.e., instability of respiratory control) or different susceptibility to developing cardiovascular complications, or explain unrefreshing sleep in patients with insomnia. Furthermore, within healthy young adults, the arousal intensity was reproducible within an individual [58], and this trait has been shown to be heritable [59].
Fig. 1.
Arousal Intensity. (A) Examples of arousals with different intensities within one patient. C3/A2 and C4/A1 are central electroencephalograms. The numbers 1, 3, 6, 9 reflect the different identified arousals intensities [39]. (B) There is an increased change in heart rate as arousals increase in intensity within one patient. Each dot represents one or more arousals. Horizontal bars are ± standard deviation. The solid line represents the average response. Figure reproduced with permission from Ref. [39].
Heart rate response to arousals:
Heart rate response to arousals is the difference between the maximum heart rate in the 8 seconds following arousal onset and the baseline pre-arousal heart rate. The magnitude of the heart rate response to arousal varies with intensity of arousal with more intense arousals resulting in larger increases in heart rate (Figure 1B). Thus, comparing heart rate response to arousal between subjects requires evaluating either the slope of the line describing heart rate response to intensity of arousal (on a scale of 1 to 9) or the heart rate response to arousal at a fixed intensity, e.g., arousal intensity 5, 8, etc. The heart rate response to arousal is quite stable within subjects when assessed on two separate nights of study but quite different between subjects, i.e., it is a biological trait [58]. Studies in twins for arousal intensity threshold of the change in heart rate controlled for age, sex, and race indicate that it is a heritable trait [59] as the intraclass correlations between monozygotic and dizygotic pairs is 0.449 and 0 respectively. Thus it seems likely, although currently unproven, that the heart rate response to arousals will be a measure of sympathetic response [60] and would be a clinically useful surrogate.
It is also relevant to note that arousals may not be the only determinant of heart rate increase after obstructive events in OSA patients. In a study conducted by Azarbarzin et al. [61], the authors placed 20 patients with severe OSA on CPAP and performed dial-down experiments that reduced CPAP pressure to produce different degrees of obstructive events. The authors evaluated arousals and heart rate changes following airway obstruction events terminated by an increase in CPAP pressure versus events terminated by the spontaneous opening of the airway. Dial-down obstructive events that demonstrated spontaneous opening were associated with arousals and the highest change in heart rate. Events that were terminated by increases in CPAP pressure also demonstrated increases in heart rate but without an arousal and had a dose-response behavior according to the severity of obstruction. This study suggests that arousals may not be the only contributor to increased heart rate after severe obstructive events.
Arousal threshold
An arousal threshold is interconnected with respiratory drive which may explain an arousal from sleep in response to obstructed breathing [21, 62] (apnea or hypopnea). The variability of arousal threshold between patients is most likely determined by one’s genetics or by the medications one takes for various diseases [63, 64]. Thus, individuals with low arousal thresholds may wake from very small respiratory perturbations contributing to the instability of sleep and could benefit from sedatives to increase arousal threshold thereby enhancing sleep stability and improving sleep quality [65]. Edwards et al [65] developed a novel noninvasive approach to estimate arousal threshold using data from PSG signals. They obtained the gold standard arousal threshold by instrumenting subjects with epiglottic pressure catheters in the supine position, while obtaining standard PSG measures. A low arousal threshold was correctly predicted in 84.1% of subjects using 3 standard PSG variables (AHI < 30, oxygen saturation nadir > 82.5%, and fraction of hypopneas >58.3%) using a multiple logistic regression model. Utilizing leave-one-out cross-validation, the study confirmed the predictive ability of this simple algorithm with a sensitivity of 82.2% and a specificity of 84.0%. While of interest, this tool requires further validation in independent datasets to be implemented into clinical practice (see further below).
Cyclic Alternating Pattern
Cyclic alternating pattern (CAP) [66] are sequences of long-lasting periodic activity that alternates between two EEG patterns that has been associated with sleep instability. An increase in CAP rate (more frequent changes in EEG pattern) correlates with increased sleep instability and has been associated with fatigue and sleepiness in adults with upper airway resistance syndrome [67]. It has been demonstrated within OSA patients that an increase in CAP rate and duration [68] is better at detecting respiratory events compared to arousal related respiratory events [69]. Interestingly, CAP rate has also been found to correlate with symptoms of fibromyalgia [70] and periodic limb movement in sleep patients [68]. While combining CAP parameters with other features of EEG may yield higher performing metrics, the clinical utility of CAP to diagnosis and treat OSA or other sleep disorders is still uncertain. Larger studies are needed to determine whether CAP based measures during non-REM sleep are beneficial before applying these algorithms clinically.
Respiratory events and hypoxic burden
Conventionally, all OSA respiratory events are treated equally when calculating the AHI, whether there is a complete cessation of breathing (apnea) or a partial flow limitation (hypopnea) that terminates with either an arousal or an oxygenation drop. Hypopneas that terminate with an arousal are counted based on early work by Douglas and Martin that demonstrated induced “arousals” in normal subjects increased blood pressure and sleepiness [71]. It is recognized that apneas and hypopneas differ in event duration and level of oxygen desaturation [43] that is heritable [72], as well as the total area under the oxygen desaturation curve [44]. At this point what degree of hypoxic burden is pathological or what is normal is still unclear but may depend on an individual’s genetic ability to respond to hypoxemia (e.g. antioxidant capacity, arousal threshold) and comorbidities (e.g. heart and lung disease).
Desaturation duration and event duration:
A Finnish group set out to provide better characterization of respiratory events (apneas and hypopneas). First [45] they used linear modeling strategies to propose an adjusted AHI (AHIAdjusted) as a function of the severity of obstructive events, defined as
The severity of the obstructive events (ObsSev) was defined as a function of the duration of the hypopnea and apnea events (HypDur, ApDur), the corresponding desaturation areas (DesArea), and the the total analysed time (T)
where the number of hypopneas is M and the number of apneas is N. The authors demonstrated that the adjusted AHI was associated with all-cause mortality and non-fatal cardiovascular events. Most recently [73] they demonstrated that conventional AHI did not perform as well as the adjusted AHI in predicting all-cause mortality. Then, they calculated respiratory event duration and identified 2 novel features-desaturation duration (respiratory event was defined as having at least ≥3% drop in SaO2 and duration was measured between the onset of the SaO2 drop to the lowest SaO2 before the signal returned back to baseline), and event duration (defined as apneas/hypopneas divided into 8 different categories based on duration) [46]. They found that increased event duration correlated to increased drops in SaO2 desaturation and made a case for giving longer event duration more weight than shorter events when estimating OSA severity and associated long term cardiovascular risk.
Respiratory event duration:
Koch et al [43] evaluated whether the presence of hypoxia during respiratory events were associated with different clinical outcomes, specifically, hypertension and sleepiness. They analyzed data from the Wisconsin Sleep Cohort and used an automated algorithm to mark respiratory events with desaturation (hypoxia-breathing disturbance index-H-BDI) and without desaturation (nonhypoxia-breathing disturbance index-NH-BDI) regardless of arousal. Increased H-BDI was associated with hypertension prevalence, defined as blood pressure ≥ 140/90 mmHg or requiring antihypertensives. In contrast, a two-fold increase of NH-BDI was associated with more objective sleepiness (β = −0.52 minutes on MSLT, p < 0.001), despite no difference in subjective sleepiness (β = 0.12 on Epworth Sleepiness Scale score, p = 0.10). Thus, in sleep disordered breathing, NH-BDI could be an objective measure of daytime sleepiness, independent of hypoxia. Lastly, longitudinal analyses over a 4-year followup period found that baseline NH-BDI was associated with increasing H-BDI, suggesting that hypoxia severity may evolve over time.
Hypoxic burden:
Azarbarzin et al [44] evaluated the hypoxic burden of sleep apnea as a way to quantify severity and associate it with clinical consequences. The investigators used data from 2 cohort studies, The Outcomes of Sleep Disorders in Older Men (MrOS) and the Sleep Heart Health Study (SHHS) and used all-cause and cardiovascular disease (CVD)-related mortality as outcome measures. Hypoxic burden was determined by measuring the area under the oximetry desaturation curve associated with respiratory events. They found that in both cohorts, a higher hypoxic burden was associated with increased risk of CVD deaths. However, when using traditional measures of OSA severity, severe OSA (defined by AHI >30) was associated with mortality in 40–70-year-old men within the SHHS, however, AHI >30 within the MrOS cohort did not demonstrate an association with mortality. Collectively this suggests that hypoxic burden which incorporates event duration and severity of oxygen desaturation during respiratory events is better at predicting CVD-related mortality than the AHI.
NOVEL PSG ANALYTIC APPROACHES
In addition to novel PSG metrics, application of established mathematical and computational analysis from other fields to physiological data from the PSG will also help us characterize the heterogeneity of sleep disorders. This novel implementation of mathematical/computational analyses has been well established in fields such as physics, astronomy and economics, but it was not until recently that engineers and mathematicians wanted to solve problems within the biological sciences and medicine [74, 75]. We now see an increasing number of applications that support the development of a “new age” where systems biology and big data drive Precision Medicine. PSG data with its multiple signals across the sleep period is ideal to applying signal-processing techniques and predictive modeling that will advance Precision Sleep Medicine. Signal processing techniques and machine learning are established analysis methods that can be used to potentially identify features to predict, for example, OSA subtypes. In general terms, developers use machine learning to train a model to predict a desired outcome using labeled examples (i.e. sleep stages N1, N2, N3, REM) and a set of features (i.e. variables, or predictors such as spindles, eye movements or EEG spectral power), in what is defined as “training data.” To ensure that the trained model is robust to generalization, it is then deployed in what is defined as “testing” or “validation” data with known outcome labels that were not used to train the model. If the predictive performance is acceptable and sustained across the training and testing data, the model is considered successful.
Examples of successful implementation of machine learning models include Google’s DeepMind [76] to play chess, shoji and go [77], as well as in medicine to process medical scans [78, 79], detect diabetic retinopathy [80], detect metastases in lymph nodes [81], and determine whether a tumor is malignant or benign [82]. Today there are several examples of successful implementation of machine learning within sleep. First, as highlighted by recent studies, the problem of poor inter-rater reliability of scoring sleep stages using current guidelines [83, 84] is solved with automated scoring. Second, as previously discussed, the problem of conventional PSG metrics unable to identify OSA subtypes has started to be solved with preliminary use of novel analytic methods of raw PSG signals to identify OSA subtypes. Lastly, today, to make the diagnose and treat narcolepsy, we require a multiple sleep latency test (MSLT) [85] that is both labor intensive and time consuming. Early research using novel analytic methods have found a way to identify narcolepsy using only the PSG. In this section we briefly review machine learning approaches and then give examples of how the application of mathematical/computational analysis has been applied to the autoscoring of sleep stages, identification of OSA subtypes, the identification of narcolepsy from the PSG without the MSLT and diagnosing movement disorders during sleep.
Machine learning: unsupervised vs supervised
Machine learning is the umbrella term that encompasses computational approaches that deal with large, complex, multi-dimensional data to perform specific tasks without the use of explicit instructions. While machine learning has been utilized for decades in other fields, it has only recently been gaining traction and popularity in the biological sciences and medicine. PSGs generate a large amount of data, on the order of 0.1–1TB and it is complex and multidimensional, ranging from 1 Hz on the SaO2 monitor to 512 Hz in one EEG channel, depending on the configuration. Signal processing techniques using machine learning has been used to derive relevant features from the PSG data to autoscore sleep stages and better characterize respiratory events [86]. While it is appropriate to be cautious of a computer analyzing PSGs, automated tools and analyses can be useful to identify subtle patterns within the PSG signals that cannot be detected by human visual inspection. Machine learning algorithms can be divided into two broad groups: unsupervised and supervised learning.
Unsupervised learning is an algorithm that formulates inferences from data that are not labeled. For example, a researcher is interested in staging epochs of sleep. In unsupervised learning, the researcher will not explicitly label each epoch, but let the algorithm label each epoch according to input features e.g. power spectrum density of different bands of the EEG, EMG and EOG. The algorithm would then label the epochs into two or more groups, and then the researcher would name the groups according to the differences in the features across the groups. Consistent with the current understanding of sleep stages, we might expect from this example that 4 groups would be identified by the algorithm and the researcher would name the 4 groups N1, N2, N3 and REM. However, it is possible that more (or less) groups would be identified by the algorithm, suggesting that there may be different sub-classification of sleep stages in this hypothetical example. The strength of unsupervised learning is that there is no bias by labeling data, with potential to find patterns of commonalities not previously discovered. In unsupervised learning, experts are still needed to interpret the results, which may not always be obvious. Examples of unsupervised methods include cluster analyses, anomaly detection, and some neural networks. As early as the mid-90s, investigators such as Pardey et al [87] set out to demonstrate the limitations of rule-based sleep staging by using artificial neural networks to develop a 10-dimensional feature vector to characterize sleep on a continuous scale. More recently, many groups have utilized cluster analyses to identify OSA subtypes. We have used cluster analysis to identify 3 different OSA subtypes based on symptom data of unlabeled patients with OSA (AHI>15) [28] and validated these OSA subtypes in 2 additional studies [12, 27]. Another group used cluster analysis with conventional PSG metrics of OSA and demographic data and identified 7 OSA subtypes (will be discussed below) [1].
Supervised learning is different in that the researcher uses labeled data (e.g., epochs labeled as N1, N2, N3 and REM), and the machine learning algorithm establishes a rule that separates these groups as best as possible. Herein “garbage in, garbage out” is apparent as supervised learning is critically dependent on a very robust set of expertly labeled examples to enable the algorithm to find the rule that separates the groups as accurately as possible. Logically, the supervised algorithm using labeled examples cannot identify the presence of other, novel groups. While many machine learning classification methods are dependent on the features that are included in the algorithm, it is important to note that identifying features requires prior expert knowledge (e.g. clinical experience). Features included in an algorithm can be extracted from, for example, the PSG, using signal processing techniques on time series signals (e.g. EEG). An advantage of feature extraction is that it reduces the dimensionality of the data and can help reduce the number of training examples needed to satisfactorily calculate the rule for separating the groups. Automatic feature selection techniques can be used to select discriminating features from a pool of candidate features. An emerging area in supervised machine learning in the era of Big Data is the use of methods that improve feature selection that is not limited by hand-engineered features [88]. One such method that learns its own features is the convolutional neural network (CNN), but has the disadvantage of requiring larger data sets to train than algorithms using hand-engineered features [89]. The top performing systems in the 2018 Computing in Cardiology Physionet competition of identifying non-apnea arousals from the PSG all used CNNs which outperformed systems using hand engineered features [90, 91]. The competition organizers provided approximately 1000 overnight PSGs for training algorithms, suggesting this may be the minimum number of PSGs to automate feature selection. Examples of supervised methods include logistic regression, support vector machine, classification and regression trees, random forests, artificial neural networks and deep learning. We now describe recent specific examples of application of machine learning using data from PSGs.
Example: Machine learning and Autoscoring Sleep Stages
Olesen et al [92] set out to produce autoscoring of sleep stages using machine learning techniques, specifically, the use of deep residual neural networks of unprocessed PSG signals. In brief, unprocessed data were passed through 50 convolutional layers, which performed their own feature extraction before classification into one of five sleep stages. Three model configurations were trained on 1850 polysomnogram recordings and subsequently tested on 230 independent recordings. The best performing model yielded an accuracy of 84.1% with most of the errors made on non-REM stage 1 and 3 decisions, which may reflect how we define sleep stages today. This group went on to fully automate sleep staging using random-forest algorithms [93] to better detect REM sleep using only EEG and EOG signals. Using the publicly available Institute of Systems and Robotics from Coimbra (ISRUC)-Sleep database [94] a model was trained and tested on 100 subjects with different sleep disorders which was also manually scored by two individual experts. EOG and EEG signals were divided into overlapping moving 33-s epochs with steps of 3s, then several time- and frequency-domain features were computed. The features were used to train a random forest classifier that labelled each 33-s epoch with the probabilities of being wake, REM and non-REM. The performance of the model was tested using a 20-fold cross validation scheme. For the epochs that the scorers agreed on, the classification achieved an overall accuracy of 92.6%. Most recently, Zhang et al [95] used deep learning to autoscore sleep stages and found that it outperformed human agreement on sleep staging. Although accuracy for autoscoring is repeatedly high, the question is: are we ready as a field, to accept automated sleep stage scoring? The advantage of using automated sleep scoring is three-fold: (1) we will have consistent staging across labs both clinically and in research, for which we can develop treatment algorithms and build upon each other’s findings without questioning the reliability of sleep stages (2) automated scoring will reduce the time needed by technologists and physicians to process EEG recordings and (3) we may move away from current sleep staging and discover altogether novel sleep stages. The disadvantages of using current generation automated sleep scoring are a distrust of automatic staging by clinicians and potentially an inability of automated algorithms to adapt to an individual patient. To address the latter, more tests of validity will be required. It will also require continued efforts to ensure that quality data are collected and further standardization of signal collection.
Example: Cluster analysis identifies OSA subtypes based on conventional PSG variables
Zinchuk et al. used cluster analysis to identify OSA subtypes based on physiological characteristics using 29 PSG measures, demographics and comorbidities [1]. They identified seven OSA subtypes (Table 1) within conventional categories of OSA severity; two subtypes of none/mild conventional OSA severity (Cluster A: mild; Cluster B: periodic limb-movement syndrome [PLMS]), two in moderate conventional OSA severity (Cluster C: non-rapid eye movement [NREM]-related apneas with poor sleep; Cluster D: REM-related apneas with increased hypoxic burden), and three in severe conventional OSA severity (Cluster E: Hypopneas with significant desaturations; Cluster F: Arousals and poor sleep; Cluster G: Combined severe subtype). The most interesting finding was that certain clusters exhibited greater risk for cardiovascular (CV) outcomes. Specifically, Cluster B (PLMS) had a twofold higher risk than Cluster A (Mild) for Acute Coronary Syndrome, Transient Ischemic Attack, stroke or death from any cause. Interestingly, Cluster D (REM and hypoxia) did not have an increase in cardiovascular outcomes compared to Cluster A (Mild). While conventional OSA severity (based on AHI) was not associated with CV outcomes, analysis within each OSA subtype did reveal some association with CV outcomes. In regular CPAP users (vs. non-regular CPAP users) there was an attenuation in CV risk within the PLMS (cluster B) and hypopnea/hypoxia cluster (cluster E) (marked with a * red asterisk in Table 1). Therefore, PSG clusters may also inform which OSA patients would benefit from CPAP therapy with respect to CV outcomes. The authors concluded that there is evidence of PSG heterogeneity within traditional OSA severity categories that can be extracted using unsupervised cluster analyses of which only some subtypes are associated with CV morbidity/mortality.
Table 1.
Description of and labels for the potysomnographic clusters based on distinguishing features.
| Cluster (n) | Cluster label | Median AHIa (events/h) | Conventional OSA severitya |
|---|---|---|---|
| A (533) | Mild | 4 | None\mild |
| B (119)* | PLMS | 10 | |
| C (186) | NREM and poor sleep | 19 | Moderate |
| D (168) | REM and hypoxia | 19 | |
| E (75)* | Hypopnea and hypoxia | 44 | Severe |
| F (42) | Arousal and poor sleep | 68 | |
| G (124) | Combined severe | 84 |
AHI, apnea-hypopnea index: NREM, non-rapid eye movement: OSA, obstructive sleep apnea: PLMS, periodic limb movements of sleep: REM, rapid eye movement.
In regular CPAP users (vs. non-regular CPAP users) there was an attenuation in CV risk within the PLMS (cluster B) and hypopnea/hypoxia clusters (cluster E). Table reproduced with permission from Ref. [ 1 ].
OSA severity definitions: none/mild (AHI < 15). moderate (15 ≤ AHI < 30) and severe (AH ≥ 30). AHI was not used in generating patient clusters. Median AH Is and severity categories based on median AHI for each cluster are shown for descriptive purposes only (mean AHIs were 7.5, 13.6, 24.0, 25.0, 47.6, 72.6 and 82.4 for clusters A,B,C,D,E,F and G, respectively).
Example: Algorithm to determine OSA pathophysiological subtypes based on physiology extracted from clinical PSG
A novel mathematical approach to determine OSA pathology is based on a model of ventilatory control [21]. The goal of this effort was to extract assessments of key physiological risk factors for OSA from the PSG, i.e., upper airway collapsibility, arousal threshold, overall loop gain (a measure of stability of ventilatory control system), and upper airway muscle responsiveness to negative intraluminal pressure [5, 42, 96]. These physiological mechanisms have been identified using complex experimental protocols that require insertion of fine wires to measure genioglossus activity, catheters to assess epiglottic pressure, as well as use of multiple step-down tests where CPAP pressure is acutely reduced to precipitate upper airway collapse [97–99]. Given the complexity of the protocols, these assessments were done in a small sample of patients and cannot be widely used in clinical practice. However, the goal is to obtain the same information from standard assessments done during the overnight sleep study using some assumptions extrapolated from these small studies to other larger samples. For example, one assumption is that there is a single value for these 4 pathophysiological mechanisms for each subject when most likely these mechanisms will change across the night in response to sleep stages, body position, etc., and across time with trajectory of disease. Thus, while this is an interesting concept to personalize pathophysiological mechanisms and therapy, it would seem essential to assess both within and between night variability in individual subjects to determine the stability of these pathophysiological traits before this approach is applied clinically in large numbers of subjects.
Example: Machine learning algorithm of PSG to diagnose Narcolepsy
To make the diagnosis to treat Type-1 Narcolepsy (T1N) currently requires an overnight PSG and next day Multiple Sleep Latency Test (MSLT) [85]. The question is whether there are features in the overnight PSG that make the MSLT unnecessary. To address this, Stephansen et al [100] used a supervised machine learning algorithm, convolutional neural network (CNN) with a Long Short-Term Memory capability (LSTM), on raw PSG signals to identify T1N patients using sleep/wake stage mixing/dissociation, a characterization that has been found to be higher in narcoleptic subjects compared to non-narcoleptic subjects [101–105]. Sleep/wake stage mixing/dissociation uses 38 features from EOG, EMG and EEG to differentiate wake, stage N1, N2 N3 and REM sleep then provides a composite measure of 8 prominent features that differentiate narcolepsy and controls (Figure 2) [102]. The investigators used the overnight PSG to identify a T1N marker based on higher than normal overlap between probabilities of different sleep stages, a highly predictive feature of sleep stage mixing/dissociation based on descriptive statistics and the persistence of a set of new time series data generated from the geometric mean of every permutation of the set of sleep stages. A T1N PSG physiological biomarker based on increased sleep/wake stage mixing/dissociation overlaps achieved a specificity of 96% and a sensitivity of 91% to diagnose narcolepsy. This biomarker was trained on 645 PSGs (7 datasets), then tested on 444 different PSGs (5 datasets) then replicated on 321 different PSGs (2 datasets not involved in training or testing). The addition of HLA-DQB1*06:02 genotyping further increased specificity to 99%. If adopted, this algorithm would not only reduce time spent in sleep labs (no longer require MSLT) but enables the possibility of diagnosing T1N using Type 2 home sleep studies.
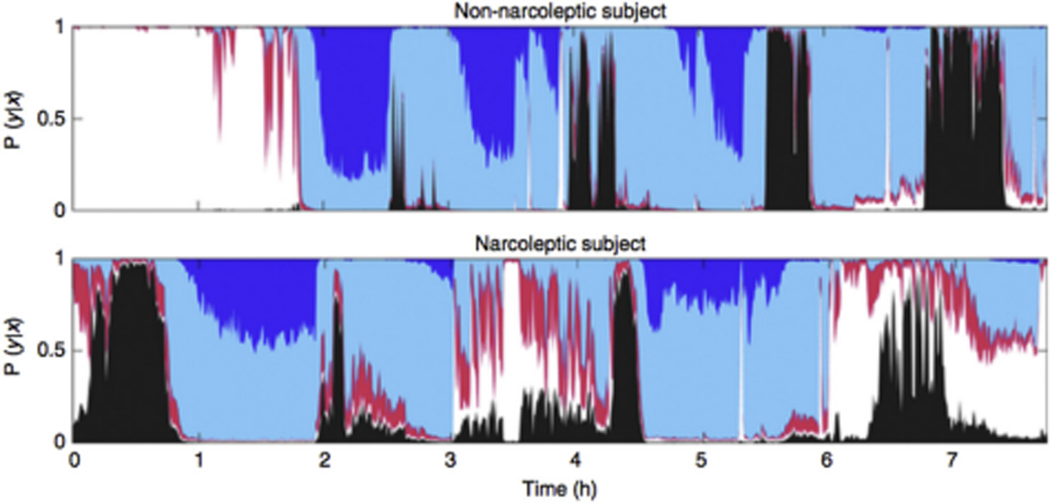
Fig. 2.
Example of hypnodensity graph in a patient without narcolepsy (top panel) and a patient with narcolepsy (bottom panel) [95]. A hypnodensity graph is a hypnogram generated by an algorithm that uses raw sleep study data to identify features that provides more information about sleep trends (probability of occurrence of each sleep state for each epoch because each epoch of sleep within the same stage is not identical) in addition to single sleep stage. Narcolepsy, a condition characterized by sleep/wake stage mixing/dissociation [96–100], has a greater than normal overlap between stages and this may be a biomarker for narcolepsy on the PSG, without the need for an MSLT. Color codes: white: wake; red: N1; light blue: N2; dark blue: N3, black: REM. Figure reproduced with permission from Ref. [95].
These investigators also used neural networks in 2,784 PSGs of normal and abnormal (e.g. insomnia, OSA, restless legs syndrome, periodic leg movement and narcolepsy) sleep subjects to automate sleep stage scoring to generate the hypnodensity graph. The accuracy of sleep stage scoring was then validated in 70 subjects as assessed by six scorers to independently validate their best automated scoring algorithm. They found that their best algorithm for autoscoring achieved 87% accuracy when compared to 5 scorers and performed better than any individual scorer. Lastly, they were able to score at higher resolution i.e., 5 second epochs, making the outdated 30 second epoch no longer necessary.
Example: Machine learning algorithm of PSG to diagnose movement disorders during sleep
Since there is a high prevalence of movement disorders during sleep [106], could limb EMG processing be a part of reinventing the PSG? For example, patients are unable to consistently report periodic limb movement disorder, a leg movement disorder during sleep. And, while leg movements are a common finding in patients with sleep disordered breathing, leg movements are not always causally linked to respiratory events [107]. To address this, the validated automatic scoring program MATPLM1 [108] was used to identify candidate leg movements according to standards put forth by the World Association of Sleep Medicine [109, 110]. If the recommended rules for algorithms are implemented, a high sensitivity and specificity for diagnosing periodic limb movements during sleep can be reached.
FUTURE DIRECTIONS
In this review we have discussed how novel PSG metrics and analyses can improve our understanding of sleep and sleep disorders. We are at an exciting crossroad of combining these novel approaches to modernize physiological biomarkers from the PSG which could then be integrated with other data such as -OMIC data, questionnaires, genetic data and imaging data to fully understand mechanisms and its relationship to clinical outcomes. One step is to make centralized databases publicly available to perform the research to determine clinical value in diagnosing subtypes and tailor treatment. Another step is to start considering how the sleep community would translate the science into clinical practice with the goal of developing Precision Sleep Medicine.
Integrate physiologic biomarkers from the PSG with -OMICs and other data towards Precision Sleep Medicine:
Although advanced PSG analysis will likely achieve benefits in its own right, the ultimate goal for Precision Sleep Medicine is to integrate multiple sources of data to develop a more complete picture of the individual and their health trajectory. Such sources of data extend beyond the clinical and biological (specimens) domains and should include continuous or serial measures of lifestyle, mood, stress, relationships, and health behaviors. The advent of wearable technologies and a variety of health apps has revolutionized our ability to capture such rich data on a large scale. The challenge, however, is developing methods for aligning and analyzing such datasets that is clinically applicable and ethically acceptable. One of the first steps toward this challenge, is to centralize large databases and make them publicly available so it will be clearer as to which PSG metrics/analyses has stronger associations between a patient’s data and clinical outcomes. Lastly, discussions will be needed as to how to implement them into existing software packages.
Centralized large databases need to be publicly available:
Large databases that connect multiple hospital systems within a region, state, country and across multiple countries, are required to leverage advances in Precision Sleep Medicine. Data integration is a method of bringing information from different sources (e.g. Electronic Health Records from hospitals and clinics, -OMIC data and raw PSG signals) in chronological order. Obviously there are many patient privacy/security concerns in creating such a database that links individuals across private and public sources from storing to distributing the data [34]. Some progress has been made to provide researchers access to large resources to identify what questions should be asked and in what order. In the United States, the National Institute of Health has funded 2 large sleep databases. The National Heart, Lung, Blood/Sleep Institute has funded The National Sleep Research Resource [34,35], to improve access to sleep data, including overnight physiological signals. The National Institute of General Medical Sciences and the National Institute of Biomedical Imaging and Bioengineering has funded PhysioNet, which offers free web access to large collections of recorded physiologic signals including sleep data. In Canada, the Canadian Sleep and Circadian Network has been collecting PSGs and banking samples. The SIESTA group is a comprehensive service provider that supports the measurement of sleep, wake and brain activity in clinical trials and research. They have compiled a database of triple scored sleep recordings from 200 healthy subjects, two nights each and 100 patients with various sleep disorders [111]. The European Sleep Apnea Database (ESADA) group collects sleep recordings of in-lab and home sleep studies, all on sleep apnea patients in order to identify risk factors and phenotype sleep apnea patients [112]. International Consortiums like the Sleep Apnea Global Interdisciplinary Consortium (SAGIC) has been collecting well-curated sleep data and questionnaire data for over 5 years [12] and is currently collecting genotype data on a subset. The challenge remains not only how to integrate all these databases and make them publicly available, but also how to translate findings into clinical practice.
Translating the science to clinical practice:
When sleep medicine first emerged as a field, understandably, there was little to no science to guide clinical practice. Today, almost 50 years later, we have a wealth of science based on modern technologies but translating this to clinical practice has been slow. One action to improve translation is to take a note from the pulmonary and cardiology communities, where governing societies have built an infrastructure to include both a research arm and a clinical arm to more quickly translate information between the two arms. Another action to improve translation is to encourage more software companies to partner with researchers to embed a “research only” tab of novel PSG metrics and analyses into existing software. Only by having it commercially available can more clinical investigators be involved to do the needed studies to correlate these novel PSG metrics and analyses to clinically meaningful outcome measures and accelerate progress.
CONCLUSION
It is exciting to be a part of a new age of Precision Medicine where there is a shift from a “one-size-fits-all” mentality to personalized medicine for the individual. Precision Sleep Medicine would involve unraveling the heterogeneity of sleep disorders using a “systems” medicine approach that includes novel PSG metrics/analyses (capturing physiological data), imaging data (capturing anatomy data), questionnaire data (capturing symptoms and demographics), longitudinal clinical information (capturing clinical co-morbidities and consequences), genetic data, and biological biomarkers (e.g. -OMICs of blood, saliva, urine). The use of automated scoring of PSGs that uses machine-learning algorithms to standardize large curated datasets will greatly enhance our ability to identify subtypes of different sleep disorders, understand disease progression, prognosis, and treatment response. It is time to reinvent how we utilize the polysomnogram towards the advancement of Precision Sleep Medicine.
Practice Points.
An exciting shift towards Precision Sleep Medicine is underway which involves the application of Precision Medicine concepts to sleep disorders.
A first step towards Precision Sleep Medicine is identifying subtypes. Some Obstructive Sleep Apnea subtypes using symptoms and physiological data have already been identified.
Research Agenda.
Understanding differences and similarities between sleep disease subtypes is a first step towards precision sleep medicine that will lead to tailored management and prevention of clinical consequences. To identify disease subtypes, big sleep-centric databases that align clinical consequences to raw signals of polysomnogram, imaging, genetic data and – OMIC biomarkers are needed.
While novel polysomnogram metrics and analyses are already available, the sleep community would benefit from having access to them within commercially available software that will facilitate clinical research in these areas thereby translating the science into clinical practice.
ACKNOWLEDGEMENTS
The authors represent the Sleep Apnea Global Interdisciplinary Consortium (SAGIC) (https://www.med.upenn.edu/sleepctr/sagic.html) which is a collaboration of 10 sites, conducting research projects worldwide on a variety of topics related to the common disorder, obstructive sleep apnea. Major foci of activity include advancement of personalized sleep medicine using craniofacial risk factors and genetics of OSA. The 10 sites and members at each site include: University of Pennsylvania (Allan I. Pack, Richard Schwab, Diane C. Lim, Greg Maislin, Brendan T. Keenan, Olivia J. Veatch, Diego Mazzotti, Mary Boland, Francis Pack, Jinyoung Kim, now at the University of Nevada, Las Vegas), The Ohio State University (Ulysses J. Magalang, Jesse Mindel, M. Melanie Lyons, Steven Holfinger, Samantha Rojas), the University of Iceland (Thorarinn Gislason, BryndSs BenediktsdLttir), Charite Universitatsmedizin Berlin (Thomas Penzel, Bernd Sanner, Ingo Fietze, Maria Franczyk, Naima Laharnar, Hua Qin), Peking University (Fang Han, Adele Liyue Xu, Jing Jing Guo), Shanghai University (Qing Yun Li, Yingni Lin), Chang Gung Memorial Hospital (Ning-hung Chen, Li-Pang Chuang, Yu-Sheng Lin, Shih-Wei Lin, Hung-Yu Huang), Korea University (Chol Shin, Seung Ku Lee), University of Sydney (Peter A. Cistulli, Philip deChazal, Kate Sutherland), University of Western Australia (Bhajan Singh, Nigel McArdle, Peter Eastwood). Funding support: American Academy of Sleep Medicine Foundation (# 194-SR-18, PI: DRM) and NIH grants P01 HL094307 and R01 HL134015 (PI: AIP).
Glossary of terms:
- Machine learning
is the umbrella term that encompasses computational approaches that deal with large, complex, multi-dimensional data to perform specific tasks without the use of explicit instructions
- Supervised learning
is when the researcher uses labeled data (e.g., epochs labeled as N1, N2, N3 and REM), and the machine learning algorithm establishes a rule that separates these groups as best as possible
- Unsupervised learning
is an algorithm that formulates inferences from data that are not labeled
Abbreviations:
- AASM
American Academy of Sleep Medicine
- AHI
apnea-hypopnea index
- CAP
cyclic alternating pattern
- CNN
convolutional neural network
- CVD
cardiovascular disease
- EEG
electroencephalogram
- EOG
electrooculography
- EMG
electromyogram
- H-BDI
hypoxia-breathing disturbance index
- Hz
hertz
- MSLT
multiple sleep latency test
- ODI
oxygen desaturation index
- ORP
odds ratio product
- OSA
obstructive sleep apnea
- PAP
positive airway pressure
- PLMS
period limb movement syndrome
- PSG
polysomnogram
- REM
rapid eye movement
- SAGIC
Sleep Apnea Global Interdisciplinary Consortium
- SaO2
oxygen saturation
- T1N
type 1 narcolepsy
- TB
terabyte
Footnotes
CONFLICT OF INTERESTS
The following authors declare financial interests/personal relationships: Jinyoung Kim has received research support (equipment) from Cerebra. Peter A. Cistulli holds an endowed academic chair at the University of Sydney, established through funding from ResMed and is a consultant/adviser/received research support from ResMed, SomnoMed, Zephyr Sleep Technologies. He has a pecuniary interest in SomnoMed related to a previous role in R&D (2004). He has received speaker fees from Nox Medical and ResMed. Philip de Chazal holds an endowed academic chair at the University of Sydney, established through funding from ResMed and has received research support from ResMed and SpaceLabs. Kate Sutherland has received research support (equipment) from SomnoMed. The following SAGIC members (not authors) declare the following financial interests/personal relationships: Richard Schwab receives grant funding/research support from ResMed, Inspire and CryOSA. Þórarinn Gislason and Bryndís Benediktsdóttir have a pecuniary interest in Nox Holding ehf and in Nox Health Inc. Nigel McArdle received research funding support from Oventus Pty Ltd, Nyxoah Pty Ltd and Zelda Therapeutics Pty Ltd.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES (* recommended reading)
- *[1].Zinchuk AV, Jeon S, Koo BB, Yan X, Bravata DM, Qin L, Selim BJ, Strohl KP, Redeker NS, Concato J and Yaggi HK 2018. Polysomnographic phenotypes and their cardiovascular implications in obstructive sleep apnoea Thorax 73 472–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zinchuk A and Yaggi HK 2019. Phenotypic Subtypes of OSA: A Challenge and Opportunity for Precision Medicine Chest [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bamagoos AA, Cistulli PA, Sutherland K, Madronio M, Eckert DJ, Hess L, Edwards BA, Wellman A and Sands SA 2019. Polysomnographic Endotyping to Select Patients with Obstructive Sleep Apnea for Oral Appliances Ann Am Thorac Soc 16 1422–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sands SA, Edwards BA, Terrill PI, Butler JP, Owens RL, Taranto-Montemurro L, Azarbarzin A, Marques M, Hess LB, Smales ET, de Melo CM, White DP, Malhotra A and Wellman A 2018. Identifying obstructive sleep apnoea patients responsive to supplemental oxygen therapy Eur Respir J 52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sands SA, Edwards BA, Terrill PI, Taranto-Montemurro L, Azarbarzin A, Marques M, Hess LB, White DP and Wellman A 2018. Phenotyping Pharyngeal Pathophysiology using Polysomnography in Patients with Obstructive Sleep Apnea Am J Respir Crit Care Med 197 1187–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sands SA, Terrill PI, Edwards BA, Taranto Montemurro L, Azarbarzin A, Marques M, de Melo CM, Loring SH, Butler JP, White DP and Wellman A 2018. Quantifying the Arousal Threshold Using Polysomnography in Obstructive Sleep Apnea Sleep 41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mazzotti DR, Keenan BT, Lim DC, Gottlieb DJ, Kim J and Pack AI 2019. Symptom Subtypes of Obstructive Sleep Apnea Predict Incidence of Cardiovascular Outcomes Am J Respir Crit Care Med 200 493–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rechstaffen A and Kales A 1968. A Manual of Standardized Terminology: Techniques of Scoring System for Sleep states of Human Subjects: Brain Information Services/Brain Research Institute, University of California, Los Angeles: ) [Google Scholar]
- [9].Iber C, Ancoli-Israel S, Chesson A and Quan SF 2007. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology, and Technical Specification (Westchester, IL: American Academy of Sleep Medicine; ) [Google Scholar]
- [10].Berry RB, Budhiraja R, Gottlieb DJ, Gozal D, Iber C, Kapur VK, Marcus CL, Mehra R, Parthasarathy S, Quan SF, Redline S, Strohl KP, Davidson Ward S L, Tangredi MM and American Academy of Sleep M 2012. Rules for scoring respiratory events in sleep: update of the 2007 AASM Manual for the Scoring of Sleep and Associated Events. Deliberations of the Sleep Apnea Definitions Task Force of the American Academy of Sleep Medicine J Clin Sleep Med 8 597–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Guilleminault C, Tilkian A and Dement WC 1976. The sleep apnea syndromes Annu. Rev. Med. 27 465–84 [DOI] [PubMed] [Google Scholar]
- [12].Keenan BT, Kim J, Singh B, Bittencourt L, Chen NH, Cistulli PA, Magalang UJ, McArdle N, Mindel JW, Benediktsdottir B, Arnardottir ES, Prochnow LK, Penzel T, Sanner B, Schwab RJ, Shin C, Sutherland K, Tufik S, Maislin G, Gislason T and Pack AI 2018. Recognizable clinical subtypes of obstructive sleep apnea across international sleep centers: a cluster analysis Sleep 41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kim J, Keenan BT, Lim DC, Lee SK, Pack AI and Shin C 2018. Symptom-Based Subgroups of Koreans With Obstructive Sleep Apnea J Clin Sleep Med 14 437–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Collins FS and Varmus H 2015. A new initiative on precision medicine N Engl J Med 372 793–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Haendel MA, Chute CG and Robinson PN 2018. Classification, ontology, and precision medicine N. Engl. J. Med. 379 1452–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yates LR, Seoane J, Le Tourneau C, Siu LL, Marais R, Michiels S, Soria JC, Campbell P, Normanno N, Scarpa A, Reis-Filho JS, Rodon J, Swanton C and Andre F 2018. The European Society for Medical Oncology (ESMO) precision medicine glossary Ann. Oncol. 29 30–5 [DOI] [PubMed] [Google Scholar]
- [17].Walko C, Kiel PJ and Kolesar J 2016. Precision medicine in oncology: New practice models and roles for oncology pharmacists Am. J. Health. Syst. Pharm. 73 1935–42 [DOI] [PubMed] [Google Scholar]
- [18].Kraft M 2011. Asthma phenotypes and interleukin-13--moving closer to personalized medicine N. Engl. J. Med. 365 1141–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fajt ML and Wenzel SE 2015. Asthma phenotypes and the use of biologic medications in asthma and allergic disease: the next steps toward personalized care J. Allergy Clin. Immunol. 135 299–310; quiz 1 [DOI] [PubMed] [Google Scholar]
- [20].Lim DC, Sutherland K, Cistulli PA and Pack AI 2017. P4 medicine approach to obstructive sleep apnoea Respirology 22 849–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *[21].Eckert DJ, White DP, Jordan AS, Malhotra A and Wellman A 2013. Defining phenotypic causes of obstructive sleep apnea. Identification of novel therapeutic targets Am. J. Respir. Crit. Care Med. 188 996–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Butler MP, Emch JT, Rueschman M, Sands SA, Shea SA, Wellman A and Redline S 2019. Apnea-hypopnea event duration predicts mortality in men and women in the Sleep Heart Health Study Am. J. Respir. Crit. Care Med. 199 90312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Borsini E, Nogueira F and Nigro C 2018. Apnea-hypopnea index in sleep studies and the risk of over-simplification Sleep Sci 11 45–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kendzerska T and Leung RS 2016. Going beyond the apnea-hypopnea index Chest 149 1349–50 [DOI] [PubMed] [Google Scholar]
- [25].Mazzotti DR, Lim DC, Sutherland K, Bittencourt L, Mindel JW, Magalang U, Pack AI, de Chazal P and Penzel T 2018. Opportunities for utilizing polysomnography signals to characterize obstructive sleep apnea subtypes and severity Physiol. Meas. 39 09TR1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zinchuk AV, Gentry MJ, Concato J and Yaggi HK 2017. Phenotypes in obstructive sleep apnea: A definition, examples and evolution of approaches Sleep Med Rev 35 113–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kim J, Keenan BT, Lim DC, Lee SK, Pack AI and Shin C 2018. Symptom-based subgroups of Koreans with obstructive sleep apnea Journal of Clinical Sleep Medicine 14 437–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *[28].Ye L, Pien GW, Ratcliffe SJ, Bjornsdottir E, Arnardottir ES, Pack AI, Benediktsdottir B and Gislason T 2014. The different clinical faces of obstructive sleep apnoea: a cluster analysis Eur Respir J 44 1600–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mazzotti DR, Keenan BT, Lim DC, Gottlieb DJ, Kim J and Pack AI 2019. Symptom subtypes of obstructive sleep apnea predict incidence of cardiovascular outcomes Am. J. Respir. Crit. Care Med. doi: 10.1164/rccm.201808-1509OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bosi M, Milioli G, Riccardi S, Melpignano A, Vaudano AE, Cortelli P, Poletti V and Parrino L 2018. Arousal responses to respiratory events during sleep: the role of pulse wave amplitude J. Sleep Res. 27 259–67 [DOI] [PubMed] [Google Scholar]
- [31].Lacedonia D, Carpagnano GE, Patricelli G, Carone M, Gallo C, Caccavo I, Sabato R, Depalo A, Aliani M, Capozzolo A and Foschino Barbaro MP 2018. Prevalence of comorbidities in patients with obstructive sleep apnea syndrome, overlap syndrome and obesity hypoventilation syndrome Clin Respir J 12 1905–11 [DOI] [PubMed] [Google Scholar]
- [32].Bonsignore MR, Baiamonte P, Mazzuca E, Castrogiovanni A and Marrone O 2019. Obstructive sleep apnea and comorbidities: a dangerous liaison Multidiscip Respir Med 14 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pien GW, Ye L, Keenan BT, Maislin G, Bjornsdottir E, Arnardottir ES, Benediktsdottir B, Gislason T and Pack AI 2018. Changing faces of obstructive sleep apnea: treatment effects by cluster designation in the Icelandic Sleep Apnea Cohort Sleep 41 zsx201, 10.1093/sleep/zsx201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bonsignore MR, Suarez Giron MC, Marrone O, Castrogiovanni A and Montserrat JM 2017. Personalised medicine in sleep respiratory disorders: focus on obstructive sleep apnoea diagnosis and treatment European Respiratory Review 26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ren R, Li Y, Zhang J, Zhou J, Sun Y, Tan L, Li T, Wing YK and Tang X 2016. Obstructive sleep apnea with objective daytime sleepiness is associated with hypertension Hypertension 68 1264–70 [DOI] [PubMed] [Google Scholar]
- [36].Martynowicz H, Skomro R, Gac P, Mazur G, Porebska I, Brylka A, Nowak W, Zielinski M, Wojakowska A and Poreba R 2017. The influence of hypertension on daytime sleepiness in obstructive sleep apnea Journal of the American Society of Hypertension : JASH 11 295–302 [DOI] [PubMed] [Google Scholar]
- [37].Beaudin AE, Waltz X, Hanly PJ and Poulin MJ 2017. Impact of obstructive sleep apnoea and intermittent hypoxia on cardiovascular and cerebrovascular regulation Exp. Physiol. 102 743–63 [DOI] [PubMed] [Google Scholar]
- *[38].Younes M, Ostrowski M, Soiferman M, Younes H, Raneri J and Hanly P 2015. Odds ratio product of sleep EEG as a continuous measure of sleep state Sleep 38 641–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *[39].Thomas RJ, Mietus JE, Peng CK and Goldberger AL 2005. An electrocardiogram-based technique to assess cardiopulmonary coupling during sleep Sleep 28 1151–61 [DOI] [PubMed] [Google Scholar]
- [40].Younes M and Hanly PJ 2016. Immediate postarousal sleep dynamics: an important determinant of sleep stability in obstructive sleep apnea J Appl Physiol (1985) 120 801–8 [DOI] [PubMed] [Google Scholar]
- *[41].Azarbarzin A, Ostrowski M, Hanly P and Younes M 2014. Relationship between arousal intensity and heart rate response to arousal Sleep 37 645–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sands SA, Terrill PI, Edwards BA, Taranto Montemurro L, Azarbarzin A, Marques M, de Melo CM, Loring SH, Butler JP, White DP and Wellman A 2018. Quantifying the arousal threshold using polysomnography in obstructive sleep apnea Sleep 41 doi: 10.1093/sleep/zsx183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Koch H, Schneider LD, Finn LA, Leary EB, Peppard PE, Hagen E, Sorensen HBD, Jennum P and Mignot E 2017. Breathing disturbances without hypoxia are associated with objective sleepiness in sleep apnea Sleep 40 doi: 10.1093/sleep/zsx152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *[44].Azarbarzin A, Sands SA, Stone KL, Taranto-Montemurro L, Messineo L, Terrill PI, Ancoli-Israel S, Ensrud K, Purcell S, White DP, Redline S and Wellman A 2019. The hypoxic burden of sleep apnoea predicts cardiovascular disease-related mortality: the Osteoporotic Fractures in Men Study and the Sleep Heart Health Study Eur Heart J 40 1149–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Muraja-Murro A, Kulkas A, Hiltunen M, Kupari S, Hukkanen T, Tiihonen P, Mervaala E and Toyras J 2014. Adjustment of apnea-hypopnea index with severity of obstruction events enhances detection of sleep apnea patients with the highest risk of severe health consequences Sleep Breath 18 641–7 [DOI] [PubMed] [Google Scholar]
- *[46].Kulkas A, Duce B, Leppanen T, Hukins C and Toyras J 2017. Severity of desaturation events differs between hypopnea and obstructive apnea events and is modulated by their duration in obstructive sleep apnea Sleep Breath 21 829–35 [DOI] [PubMed] [Google Scholar]
- [47].Koch H, Schneider LD, Finn LA, Leary EB, Peppard PE, Hagen E, Sorensen HBD, Jennum P and Mignot E 2017. Breathing Disturbances Without Hypoxia Are Associated With Objective Sleepiness in Sleep Apnea Sleep 40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Younes M, Ostrowski M, Soiferman M, Younes H, Younes M, Raneri J and Hanly P 2015. Odds ratio product of sleep EEG as a continuous measure of sleep state Sleep 38 641–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Buysse DJ 2014. Sleep health: can we define it? Does it matter? Sleep 37 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Goldschmied J, Kuna ST, Maislin G, Pack AI and Younes M 2019. Changes in sleep depth following sleep deprivation assessed by three methods Sleep & Breathing 42 A128 [Google Scholar]
- [51].Meza-Vargas S, Giannouli E and Younes M 2016. Enhancements to the multiple sleep latency test Nature and science of sleep 8 145–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lee WH, Ahn JC, We J, Rhee CS, Lee CH, Yun PY, Yoon IY and Kim JW 2014. Cardiopulmonary coupling analysis: changes before and after treatment with a mandibular advancement device Sleep Breath 18 891–6 [DOI] [PubMed] [Google Scholar]
- [53].Thomas RJ, Wood C and Bianchi MT 2017. Cardiopulmonary coupling spectrogram as an ambulatory clinical biomarker of sleep stability and quality in health, sleep apnea and insomnia Sleep pii: 4718136. doi: 10.1093/sleep/zsx196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kimoff RJ 1996. Sleep fragmentation in obstructive sleep apnea Sleep 19 S61–6 [DOI] [PubMed] [Google Scholar]
- [55].1992 EEG arousals: scoring rules and examples: a preliminary report from the Sleep Disorders Atlas Task Force of the American Sleep Disorders Association Sleep 15 173–84 [PubMed] [Google Scholar]
- [56].Loredo JS, Clausen JL, Ancoli-Israel S and Dimsdale JE 1999. Night-to-night arousal variability and interscorer reliability of arousal measurements Sleep 22 916–20 [DOI] [PubMed] [Google Scholar]
- [57].Amatoury J, Azarbarzin A, Younes M, Jordan AS, Wellman A and Eckert DJ 2016. Arousal intensity is a distinct pathophysiological trait in obstructive sleep apnea Sleep 39 2091–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Azarbarzin A, Ostrowski M, Younes M, Keenan BT, Pack AI, Staley B and Kuna ST 2015. Arousal responses during overnight polysomnography and their reproducibility in healthy young adults Sleep 38 1313–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Gao X, Azarbarzin A, Keenan BT, Ostrowski M, Pack FM, Staley B, Maislin G, Pack AI, Younes M and Kuna ST 2017. Heritability of heart rate response to arousals in twins Sleep 40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Horner RL 1996. Autonomic consequences of arousal from sleep: mechanisms and implications Sleep 19 S193–5 [DOI] [PubMed] [Google Scholar]
- [61].Azarbarzin A, Ostrowski M, Moussavi Z, Hanly P and Younes M 2013. Contribution of arousal from sleep to postevent tachycardia in patients with obstructive sleep apnea Sleep 36 881–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Younes M 2004. Role of arousals in the pathogenesis of obstructive sleep apnea Am. J. Respir. Crit. Care Med. 169 623–33 [DOI] [PubMed] [Google Scholar]
- [63].Younes M, Park E and Horner RL 2007. Pentobarbital sedation increases genioglossus respiratory activity in sleeping rats Sleep 30 478–88 [DOI] [PubMed] [Google Scholar]
- [64].Berry RB and Gleeson K 1997. Respiratory arousal from sleep: mechanisms and significance Sleep 20 654–75 [DOI] [PubMed] [Google Scholar]
- [65].Edwards BA, Eckert DJ, McSharry DG, Sands SA, Desai A, Kehlmann G, Bakker JP, Genta PR, Owens RL, White DP, Wellman A and Malhotra A 2014. Clinical predictors of the respiratory arousal threshold in patients with obstructive sleep apnea Am. J. Respir. Crit. Care Med. 190 1293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Terzano MG, Mancia D, Salati MR, Costani G, Decembrino A and Parrino L 1985. The cyclic alternating pattern as a physiologic component of normal NREM sleep Sleep 8 137–45 [DOI] [PubMed] [Google Scholar]
- [67].Guilleminault C, Lopes MC, Hagen CC and da Rosa A 2007. The cyclic alternating pattern demonstrates increased sleep instability and correlates with fatigue and sleepiness in adults with upper airway resistance syndrome Sleep 30 641–7 [DOI] [PubMed] [Google Scholar]
- [68].Parrino L, Boselli M, Buccino GP, Spaggiari MC, Di Giovanni G and Terzano MG 1996. The cyclic alternating pattern plays a gate-control on periodic limb movements during non-rapid eye movement sleep J. Clin. Neurophysiol. 13 314–23 [DOI] [PubMed] [Google Scholar]
- [69].Milioli G, Bosi M, Grassi A, Riccardi S, Terzano MG, Cortelli P, Poletti V and Parrino L 2015. Can sleep microstructure improve diagnosis of OSAS? Integrative information from CAP parameters Arch. Ital. Biol. 153 194–203 [DOI] [PubMed] [Google Scholar]
- [70].Rizzi M, Sarzi-Puttini P, Atzeni F, Capsoni F, Andreoli A, Pecis M, Colombo S, Carrabba M and Sergi M 2004. Cyclic alternating pattern: a new marker of sleep alteration in patients with fibromyalgia? J. Rheumatol. 31 1193–9 [PubMed] [Google Scholar]
- [71].Douglas NJ and Martin SE 1996. Arousals and the sleep apnea/hypopnea syndrome Sleep 19 S196–7 [PubMed] [Google Scholar]
- [72].Liang J, Cade BE, Wang H, Chen H, Gleason KJ, Larkin EK, Saxena R, Lin X, Redline S and Zhu X 2016. Comparison of heritability estimation and linkage analysis for multiple traits using principal component analyses Genet. Epidemiol. 40 222–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Korkalainen H, Toyras J, Nikkonen S and Leppanen T 2019. Mortality-risk-based apnea-hypopnea index thresholds for diagnostics of obstructive sleep apnea J. Sleep Res. e12855 [DOI] [PubMed] [Google Scholar]
- [74].Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, Cui C, Corrado G, Thrun S and Dean J 2019. A guide to deep learning in healthcare Nat Med 25 24–9 [DOI] [PubMed] [Google Scholar]
- [75].Topol EJ 2019. High-performance medicine: the convergence of human and artificial intelligence Nat Med 25 44–56 [DOI] [PubMed] [Google Scholar]
- [76].Mnih V, Kavukcuoglu K, Silver D, Rusu AA, Veness J, Bellemare MG, Graves A, Riedmiller M, Fidjeland AK, Ostrovski G, Petersen S, Beattie C, Sadik A, Antonoglou I, King H, Kumaran D, Wierstra D, Legg S and Hassabis D 2015. Human-level control through deep reinforcement learning Nature 518 529–33 [DOI] [PubMed] [Google Scholar]
- [77].Silver D, Huang A, Maddison CJ, Guez A, Sifre L, van den Driessche G, Schrittwieser J, Antonoglou I, Panneershelvam V, Lanctot M, Dieleman S, Grewe D, Nham J, Kalchbrenner N, Sutskever I, Lillicrap T, Leach M, Kavukcuoglu K, Graepel T and Hassabis D 2016. Mastering the game of Go with deep neural networks and tree search Nature 529 484–9 [DOI] [PubMed] [Google Scholar]
- [78].Erickson BJ, Korfiatis P, Akkus Z and Kline TL 2017. Machine learning for medical imaging Radiographics 37 505–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Klang E 2018. Deep learning and medical imaging Journal of thoracic disease 10 1325–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, Kim R, Raman R, Nelson PC, Mega JL and Webster DR 2016. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs JAMA 316 2402–10 [DOI] [PubMed] [Google Scholar]
- [81].Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, van der Laak J, the CC, Hermsen M, Manson QF, Balkenhol M, Geessink O, Stathonikos N, van Dijk MC, Bult P, Beca F, Beck AH, Wang D, Khosla A, Gargeya R, Irshad H, Zhong A, Dou Q, Li Q, Chen H, Lin HJ, Heng PA, Hass C, Bruni E, Wong Q, Halici U, Oner MU, Cetin-Atalay R, Berseth M, Khvatkov V, Vylegzhanin A, Kraus O, Shaban M, Rajpoot N, Awan R, Sirinukunwattana K, Qaiser T, Tsang Y W, Tellez D, Annuscheit J, Hufnagl P, Valkonen M, Kartasalo K, Latonen L, Ruusuvuori P, Liimatainen K, Albarqouni S, Mungal B, George A, Demirci S, Navab N, Watanabe S, Seno S, Takenaka Y, Matsuda H, Ahmady Phoulady H, Kovalev V, Kalinovsky A, Liauchuk V, Bueno G, Fernandez-Carrobles MM, Serrano I, Deniz O, Racoceanu D and Venancio R 2017. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer JAMA 318 2199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Cheng JZ, Ni D, Chou YH, Qin J, Tiu CM, Chang YC, Huang CS, Shen D and Chen CM 2016. Computer-aided diagnosis with deep learning architecture: applications to breast lesions in US images and pulmonary nodules in CT scans Scientific reports 6 24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Younes M, Kuna ST, Pack AI, Walsh JK, Kushida CA, Staley B and Pien GW 2018. Reliability of the American Academy of Sleep Medicine Rules for Assessing Sleep Depth in Clinical Practice J Clin Sleep Med 14 205–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Younes M, Raneri J and Hanly P 2016. Staging Sleep in Polysomnograms: Analysis of Inter-Scorer Variability J Clin Sleep Med 12 885–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Krahn LE, Hershner S, Loeding LD, Maski KP, Rifkin DI, Selim B and Watson NF 2015. Quality measures for the care of patients with narcolepsy J Clin Sleep Med 11 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Mikkelsen KB, Ebajemito JK, Bonmati-Carrion MA, Santhi N, Revell VL, Atzori G, Della Monica C, Debener S, Dijk DJ, Sterr A and de Vos M 2019. Machine-learning-derived sleep-wake staging from around-the-ear electroencephalogram outperforms manual scoring and actigraphy J. Sleep Res. 28 e12786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Pardey J, Roberts S, Tarassenko L and Stradling J 1996. A new approach to the analysis of the human sleep/wakefulness continuum J. Sleep Res. 5 201–10 [DOI] [PubMed] [Google Scholar]
- [88].Urbanowicz RJ, Meeker M, La Cava W, Olson RS and Moore JH 2018. Relief-based feature selection: Introduction and review J Biomed Inform 85 189–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].de Chazal P, Sutherland K and Cistulli PA 2019. Advanced polysomnographic analysis for OSA: A pathway to personalized management? Respirology [DOI] [PubMed] [Google Scholar]
- [90].Ghassemi MM, Moody BE, Lehman LH, Song C, Li Q, Sun H, Mark RG, Westover MB, Clifford GD. 2018. You snooze, you win: the PhysioNet/Computing in Cardiology Challenge 2018 Comput. Cardiol 45 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].de Chazal P, Sadr N. 2019. Automatic Scoring of Non-Apnoea Arousals Using Hand-Crafted Features From the Polysomnogram Physiol Meas 40 [DOI] [PubMed] [Google Scholar]
- *[92].Olesen AN, Jennum P, Peppard P, Mignot E and Sorensen HBD 2018. Deep residual networks for automatic sleep stage classification of raw polysomnographic waveforms Conference proceedings: … Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Annual Conference 2018 1–4 [DOI] [PubMed] [Google Scholar]
- [93].Klok AB, Edin J, Cesari M, Olesen AN, Jennum P and Sorensen HBD 2018. A new fully automated Random-Forest algorithm for sleep staging Conference proceedings: … Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Annual Conference 2018 4920–3 [DOI] [PubMed] [Google Scholar]
- [94].Khalighi S, Sousa T, Santos JM and Nunes U 2016. ISRUC-Sleep: A comprehensive public dataset for sleep researchers Comput. Methods Programs Biomed. 124 180–92 [DOI] [PubMed] [Google Scholar]
- [95].Zhang L, Fabbri D, Upender R and Kent D 2019. Automated sleep stage scoring of the Sleep Heart Health Study using deep neural networks Sleep [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Terrill PI, Edwards BA, Nemati S, Butler JP, Owens RL, Eckert DJ, White DP, Malhotra A, Wellman A and Sands SA 2015. Quantifying the ventilatory control contribution to sleep apnoea using polysomnography Eur. Respir. J. 45 408–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Smith PL, Wise RA, Gold AR, Schwartz AR and Permutt S 1988. Upper airway pressure-flow relationships in obstructive sleep apnea J Appl Physiol (1985) 64 789–95 [DOI] [PubMed] [Google Scholar]
- [98].Gleadhill IC, Schwartz AR, Schubert N, Wise RA, Permutt S and Smith PL 1991. Upper airway collapsibility in snorers and in patients with obstructive hypopnea and apnea Am. Rev. Respir. Dis. 143 1300–3 [DOI] [PubMed] [Google Scholar]
- [99].Schwartz AR, Smith PL, Wise RA, Gold AR and Permutt S 1988. Induction of upper airway occlusion in sleeping individuals with subatmospheric nasal pressure J. Appl. Physiol. 64 535–42 [DOI] [PubMed] [Google Scholar]
- *[100].Stephansen JB, Olesen AN, Olsen M, Ambati A, Leary EB, Moore HE, Carrillo O, Lin L, Han F, Yan H, Sun YL, Dauvilliers Y, Scholz S, Barateau L, Hogl B, Stefani A, Hong S C, Kim TW, Pizza F, Plazzi G, Vandi S, Antelmi E, Perrin D, Kuna ST, Schweitzer PK, Kushida C, Peppard PE, Sorensen HBD, Jennum P and Mignot E 2018. Neural network analysis of sleep stages enables efficient diagnosis of narcolepsy Nature communications 9 5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Christensen JA, Carrillo O, Leary EB, Peppard PE, Young T, Sorensen HB, Jennum P and Mignot E 2015. Sleep-stage transitions during polysomnographic recordings as diagnostic features of type 1 narcolepsy Sleep Med 16 1558–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Olsen AV, Stephansen J, Leary E, Peppard PE, Sheungshul H, Jennum PJ, Sorensen H and Mignot E 2017. Diagnostic value of sleep stage dissociation as visualized on a 2-dimensional sleep state space in human narcolepsy J. Neurosci. Methods 282 9–19 [DOI] [PubMed] [Google Scholar]
- [103].Jensen JB, Sorensen HB, Kempfner J, Sorensen GL, Knudsen S and Jennum P 2014. Sleep-wake transition in narcolepsy and healthy controls using a support vector machine J. Clin. Neurophysiol. 31 397–401 [DOI] [PubMed] [Google Scholar]
- [104].Vassalli A, Dellepiane JM, Emmenegger Y, Jimenez S, Vandi S, Plazzi G, Franken P and Tafti M 2013. Electroencephalogram paroxysmal theta characterizes cataplexy in mice and children Brain 136 1592–608 [DOI] [PubMed] [Google Scholar]
- [105].Pizza F, Vandi S, Iloti M, Franceschini C, Liguori R, Mignot E and Plazzi G 2015. Nocturnal sleep dynamics identify narcolepsy type 1 Sleep 38 1277–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Leary EB, Moore HEt, Schneider LD, Finn LA, Peppard PE, and Mignot E 2018. Periodic limb movements in sleep: Prevalence and associated sleepiness in the Wisconsin Sleep Cohort Clin Neurophysiol 129 2306–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Ferri R 2017. Leg movements during sleep and respiratory events are not causally linked. Sleep Med 34 254–5 [DOI] [PubMed] [Google Scholar]
- [108].Huang AS, Skeba P, Yang MS, Sgambati FP, Earley CJ and Allen RP 2015. MATPLM1, A MATLAB script for scoring of periodic limb movements: preliminary validation with visual scoring Sleep Med 16 1541–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Ferri R, Fulda S, Allen RP, Zucconi M, Bruni O, Chokroverty S, Ferini-Strambi L, Frauscher B, Garcia-Borreguero D, Hirshkowitz M, Hogl B, Inoue Y, Jahangir A, Manconi M, Marcus CL, Picchietti DL, Plazzi G, Winkelman JW, Zak RS, International and European Restless Legs Syndrome Study G 2016. World Association of Sleep Medicine (WASM) 2016 standards for recording and scoring leg movements in polysomnograms developed by a joint task force from the International and the European Restless Legs Syndrome Study Groups (IRLSSG and EURLSSG) Sleep Med 26 86–95 [DOI] [PubMed] [Google Scholar]
- [110].Skeba P, Fulda S, Hiranniramol K, Earley CJ and Allen RP 2016. Defining morphology of periodic leg movements in sleep: an evidence-based definition of a minimum window of sustained activity Sleep Breath 20 1293–9 [DOI] [PubMed] [Google Scholar]
- [111].Klosch G, Kemp B, Penzel T, Schlogl A, Rappelsberger P, Trenker E, Gruber G, Zeitlhofer J, Saletu B, Herrmann WM, Himanen SL, Kunz D, Barbanoj MJ, Roschke J, Varri A and Dorffner G 2001. The SIESTA project polygraphic and clinical database IEEE Eng. Med. Biol. Mag. 20 51–7 [DOI] [PubMed] [Google Scholar]
- [112].Hedner J, Grote L, Bonsignore M, McNicholas W, Lavie P, Parati G, Sliwinski P, Barbe F, De Backer W, Escourrou P, Fietze I, Kvamme JA, Lombardi C, Marrone O, Masa JF, Montserrat JM, Penzel T, Pretl M, Riha R, Rodenstein D, Saaresranta T, Schulz R, Tkacova R, Varoneckas G, Vitols A, Vrints H and Zielinski J 2011. The European Sleep Apnoea Database (ESADA): report from 22 European sleep laboratories Eur. Respir. J. 38 635–42 [DOI] [PubMed] [Google Scholar]