Highlights
-
•
Laboratory changes reflect systemic inflammation and impairment in patients.
-
•
Lymphopenia can be used to discriminate against patients at risk for SARS.
-
•
Changes in hemostatic patterns are associated with complications of COVID-19.
Keywords: SARS-CoV-2, COVID-19, Clinical features, Laboratory findings
Abstract
In the last decades, coronaviruses have been a major threat to public health worldwide. SARS-CoV-2 is the third known coronavirus that causes fatal respiratory diseases in humans. The initial clinical features of SARS-CoV-2 infection are quite nonspecific and not all suspected patients can be tested to exclude or confirm the diagnosis. Increasing scientific evidence has shown that abnormalities in routine laboratory tests, particularly hematological tests, have the potential to indicate, in a quick, practical and economical way, the need for specific laboratory tests for the diagnosis of SARS-CoV-2 infection, besides assisting in the prognosis of the disease and in the optimization of its clinical monitoring. In order to address in a simple and practical way the various aspects related to SARS-CoV-2 infection, this review reports the history of the virus, the epidemiology and pathophysiology of COVID-19, with emphasis on its laboratory diagnosis, particularly in hematological changes found during the course of the disease.
1. General aspects of SARS-CoV-2 infection
Coronaviruses (CoVs) are a large viruses group belonging to the Coronaviridae family [1], [2], presenting a single-stranded RNA genome [3]. The genome is surrounded by a helical capsid and a lipoprotein envelope containing several spicules of glycoprotein that together give the virus a crown appearance. Hence comes the word “corona” which, in Latin, means crown [4]. When infecting humans, CoVs can cause diseases of varying severity, from upper respiratory tract infections similar to a common cold, to liver, enteric, neurological diseases and lower respiratory tract infections such as pneumonia, bronchitis and severe acute respiratory syndrome (SARS) [1], [3], [5]. SARS can be caused by the severe acute respiratory syndrome coronavirus (SARS-CoV) [6], by the coronavirus of the Middle East respiratory syndrome (MERS-CoV) [7], and recently by the coronavirus of severe acute respiratory syndrome 2 (SARS-CoV-2) [8].
On December 31, 2019, the Wuhan Municipal Health Commission, Hubei Province, China, reported the existence of 27 cases of patients with pneumonia of unknown etiology, epidemiologically related to a local wholesale market for wildlife and seafood [8]. After laboratory investigations, on January 7, 2020, the causative agent of these infections was identified, considered a new CoV in 2019 and officially designated by the World Health Organization (WHO) as 2019-nCoV [9]. Subsequently, the International Virus Taxonomy Committee renamed 2019-nCoV as SARS-CoV-2 [10], [11].
SARS-CoV-2 was quickly transmitted among humans, spreading to different countries around the world, threatening human life and generating many financial losses [4]. On January 30, 2020, WHO issued a worldwide public health alert regarding the emergence of a new epidemic viral disease [12]. On February 11, 2020, WHO announced the name for the epidemic disease caused by SARS-CoV-2: coronavirus disease 2019 (COVID‐19) and declared, on March 11, 2020, a pandemic state [13].
SARS-CoV-2 spread occurs by inhalation or ingestion of viral droplets. Thus, the main sources of human infection are contact with any contaminated surfaces (viral droplets can spread from one to two meters and settle on surfaces) [14] or with the respiratory droplets of infected people (e.g. through sneezing, coughing or physical contact). SARS-CoV-2 infection can also occur by touching the nose, eyes or mouth with hands contaminated with the virus [15]. A recent study identified high SARS-CoV-2 RNA concentration in aerosols present in bathroom areas of patients at two hospitals in Wuhan, dedicated to COVID-19 cases, and in public areas prone to agglomeration, raising the concern to evaluate the potential of transmission of this virus by aerosols [16]. Therefore, the correct hand hygiene, use of personal protective equipments and social isolation are very important strategies in combating the transmission of SARS-CoV-2 [15]. Quarantine measures should be established to restrict the movement of uninfected people in regions where there is an epidemic outbreak and infected people, who can act as spreading the virus agents as long as the symptoms last until clinical recovery [14].
Currently, there is no proven antiviral treatment for COVID-19 [15] and knowledge about SARS-CoV-2 is still scarce. Daily, reported cases and deaths number increase considerably in numerous regions of the planet. In this context, early diagnosis and infections prevention has become one of the priorities for the control of this coronaviruses [17].
SARS-CoV-2 incubation period is up to two weeks, usually ranging from three to seven days after infection. In most cases, SARS-CoV-2 infection is asymptomatic and, in that case, the individual will not need medical assistance (up to 80%). However, asymptomatic patients are an important source of spread of CoV, who, as they continue with their normal daily activities, can spread the virus to those in contact [10], [18].
In symptomatic cases, symptoms of COVID-19 are nonspecific [15] and the clinical presentation is similar to SARS-CoV infection. The most commonly reported symptoms are fever [19], dry cough, dyspnoea and fatigue [15], [17]. Non-respiratory symptoms (e.g. diarrhea, nausea, vomiting, headache and muscle pain) are usually uncommon [17].
It is known that in some people the infection can get worse, requiring hospitalization and even referral to the intensive care unit (ICU) [10], [20]. In these cases, COVID-19 may progress to acute respiratory distress syndrome, followed by septic shock, refractory metabolic acidosis, coagulation dysfunction, multiple organ failure and, consequently, death [15].
SARS-CoV-2 can cause serious clinical complications, especially in elderly patients and in those with previous comorbidities, especially diabetes [20]; cardio and cerebrovascular diseases [21], [22]; obesity; cancer and digestive, endocrine, nervous and respiratory systems pathologies [23], constituting 50% to 75% of deaths [14]. Porcheddu et al. (2020), when comparing mortality rates of SARS-CoV-2 patients in China and Italy, they realized that in China most deaths occurred in the age group above 50 years, while in Italy over 60 and in both countries, comorbidities have been shown to be important contributors to death [24].
2. Laboratory diagnosis
For COVID-19 laboratory diagnosis, to date (July/2020), tests used are based on the following methods: real-time reverse transcription-polymerase chain reaction (RT-PCR); serologic tests for SARS-CoV-2 (Anti-SARS-CoV-2 IgA, IgM and/or IgG), utilizing ELISA, chemiluminescence or immunochromatography assays; in addition to SARS-CoV-2 antigen immunochromatographic test in upper respiratory tract specimens [25], [26], [27].
The RT-PCR is the gold standard for SARS-CoV-2 detection and it is the laboratory test of choice for the diagnosis of symptomatic patients in the acute phase [28].
The diagnosis made by real time RT-PCR uses the RNA extracted from samples of the respiratory tract, such as nasopharyngeal swab/oropharyngeal, tracheal aspirate, sputum and bronchoalveolar lavage [14], [29], [30]. It is important to note that the reliability of the results of laboratory tests depends, among other factors, that the collection of biological material be carried out at an infection stage allowing identification of the pathogen (as the viral load is usually higher in the first week of the disease, RT-PCR should be performed, preferably, between the 3rd and 5th day of symptoms onset); the biological sample is of good quality, which must be maintained with proper handling and transportation [31]. Wang et al. demonstrated, in a study with 1070 samples of 205 patients with COVID-19, that the sensitivity of the test is 32%, 63%, 72% and 93% in oropharyngeal swab, nasopharyngeal swab, sputum and bronchoalveolar lavage [32].
The serological method is mainly adopted for retrospective diagnosis and its sensitivity is generally lower than that of the molecular method. Serological tests are based on the detection of the specific antibody (IgA, IgM and IgG) produced against SARS-CoV-2 or the antigen of the virus [31]. Serology can be performed when the availability of molecular tests is limited or if there are at least 14 days after the onset of symptoms [30]. Less than 50% of patients with COVID-19 have detectable antibodies in the serum before 7 to 10 days after the onset of symptoms [33].
As the identification of SARS-CoV-2 infection has become a major challenge in clinical practice, for social, financial, logistical and analytical reasons, the search for abnormalities in routine, low-cost and suggestive (or pathognomonic) laboratory parameters is extremely important to assist in confirming COVID-19 diagnosis. In this context, we performed the present literature review on hematological and biochemical changes in SARS-CoV-2 infection, whose knowledge by health professionals could be useful in the management of the disease.
3. Hematological and biochemical abnormalities during SARS-CoV-2 infection
It is known that in SARS-CoV-2 virus infections, as well as in other infectious diseases such as influenza [34], varicella [35], [36], dengue [37], [38], [39], acquired human immunodeficiency virus (HIV) [40], [41], SARS-CoV [42], [43], [44], [45] and MERS-CoV [46], hematological changes can occur and often present, the potential to optimize the monitoring of infectious process or to indicate the suspicion of their severity.
Laboratory abnormalities, particularly hematological changes, allow checking the status of SARS-CoV-2 infection, since the hematopoietic system and hemostasis suffer significant impacts during the evolution of COVID-19 [47].
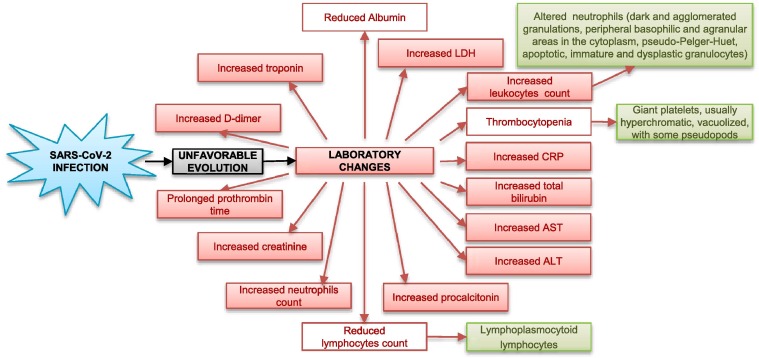
The most common hematological findings include lymphocytopenia [19], [48], [49], [50], neutrophilia [23], [51], [52], eosinopenia [50], [53], [54], mild thrombocytopenia [50] and, less frequently, thrombocytosis [48], [55]. The presence of reactive lymphocytes has been reported only occasionally [56]. The leukocyte count may be normal, reduced [19], [50], [57] or increased [22], [23], [58]. According to a meta-analysis [59], leukocytosis, lymphopenia and thrombocytopenia are associated with greater severity and even fatality in COVID-19 cases. Main laboratory changes in patients with an unfavorable evolution of SARS-CoV-2 infection are shown in Fig. 1 .
Fig. 1.
Main laboratory changes in patients with an unfavorable evolution of SARS-CoV-2 infection.
As already reviewed by Terpos et al. [60], such abnormalities have been reported by several authors and are associated, in different parts of the world, the need for ICU admission and SARS development. Still according to these authors, during the first days of the disease, when patients manifest non-specific symptoms, the leukocyte count and the absolute value of lymphocytes are normal or slightly reduced. Posteriorly, around the 7th to the 14th day of infection, the disease begins to affect organs with greater SARS-CoV-2 cell receptor expression, the angiotensin-converting enzyme 2 (ACE2) [61], such as the lungs, heart and gastrointestinal tract, with characteristic clinical symptoms and expressive increase in the levels of inflammatory mediators and cytokines [9]. At this disease stage, more expressive hematological changes are highlighted, particularly a significant reduction in the lymphocytes number. This finding was more evident in those who suffered death compared to those who survived. The latter showed their nadir for lymphocytopenia around the 7th day of symptoms, with subsequent recovery [22].
Thus, on the basis of Terpos et al. [60], it is possible to admit that the dynamics of the absolute lymphocyte count, that is, its serial count may be predictive of the clinical outcome of patient. An analysis of the literature revealed that among all the hematological abnormalities, lymphocytopenia has been highlighted as the most frequent since admission to death [19], [62], [63], [64], [65], [66], [67]. From data based on the complete blood count, it is possible to calculate ratios between its parameters, whose interpretation has considerable clinical value. Thus, a decreased lymphocyte/leukocyte count ratio has already been reported indicating severe disease and/or fatal outcomes [65], [68]. Similarly, increased neutrophil/lymphocyte and neutrophil/platelets ratio may be indicative of myocardial injury and increased mortality [69]. Therefore, it is important to monitor the hematological parameters in order to try to assess the progression and prognosis of COVID-19.
According Terpos et al. [60], possible explanations for significant reduction in lymphocytes count include: (a) direct infection in these cells, causing their lysis by SARS-CoV-2, since lymphocytes have ACE2 receptors on the surface; (b) possible lymphocyte apoptosis caused by the systemic inflammatory process with consequent large cytokines production; (c) atrophy of lymphoid organs, such as the spleen, impairing lymphocyte turnover and (d) lactic acidosis inhibiting lymphocyte proliferation, which is more evident in cancer patients, a risk group for COVID-19 complications.
Regarding the lymphocytes morphological aspect, as reported by Fan et al. [56] and Chng et al. [70], in most patients with low lymphocyte count, these were shown to be reactive with lymphoplasmocytoid characteristics. In addition to the lymphocytes morphological abnormalities, Zini et al. [71], after blood film microscopic observation of 40 patients with COVID-19 on admission, before administering antiviral and anti-inflammatory treatment, observed the presence of marked morphological abnormalities of the neutrophil lineage. These changes have included presence of numerous dark and agglomerated granulations (similar to toxic granulations) and peripheral basophilic and agranular areas in the cytoplasm, and grossly grouped chromatin in the nucleus; pseudo-Pelger-Huet neutrophils segmented or not; apoptotic neutrophils; immature and dysplastic granulocytes, especially small myelocytes and metamielocytes, as well as promyelocytes. Platelet morphology also showed peculiar and frequent anomalies, consisting mainly in the presence of giant platelets, usually hyperchromatic, vacuolized, with some showing pseudopods, not only in patients with thrombocytosis but also in those with thrombocytopenia. The authors have suggested that these abnormalities may indicate a severe, transient and reversible myelopoiesis disorder, especially in accelerated and disordered granulopoiesis, in patients with severe symptomatic COVID-19. This disorder may be related to the cytokine “storm” and hyperinflammation, that appear in the progression o COVID-19 pneumonia, possibly as secondary hemophagocytic lymphohistiocytosis, leading to organ failure [68], [71], [72], [73].
During the COVID-19 course, changes in hemostasis tests have also been reported, such as prolonged prothrombin and activated partial thromboplastin times and increased D-dimer levels. In cases of worsening COVID-19, D-dimer levels become rised, with formation of microthrombi in peripheral blood vessels and recurrent coagulation disorders [57], [74], [75].
In a retrospective study of 183 patients with COVID-19 [76], prothrombin time, D-dimer and the degradation products of fibrin/fibrinogen, measured at hospital admission, were higher in non-surviving patients than in survivors, respectively. In those hospitalized late, antithrombin activity (AT) and fibrinogen levels were significantly lower in non-survivors, suggesting that conventional blood coagulation parameters during the course of the disease may be associated with prognosis.
Most of the patients who died had diagnostic criteria for disseminated intravascular coagulation (DIC). Lippi e Plebani [77] stressed that the evaluation of the hemostatic system by specific tests should integrate the routine clinical monitoring of the patient with COVID-19, in view of fulfillment of the laboratory criteria for the diagnosis of DIC in nearly three quarters of patients who died.
Other laboratory abnormalities included increased erythrocyte sedimentation rate (ESR), increased levels of lactic dehydrogenase (LDH), C-reactive protein (CRP) and muscle enzymes, in addition to changes in cardiac, renal and liver functions, among others [75].
The following is a summary of some main studies carried out with emphasis on the description of laboratory data obtained.
Huang et al. [57] , in a study in Wuhan, China, reported the epidemiological, clinical, laboratory and radiological characteristics, treatment and clinical results of 41 patients infected with SARS-CoV-2. In hospital admission, blood count showed leukopenia with leukocyte count below 4 × 109/L [10 (25%) of 40 patients]; and lymphopenia with a lymphocyte count lower than 1 × 109/L [26 (65%) of 40 patients]. Mean D-dimer values [2.4 mg/L (0.6–14.4)]; and prothrombin time [12.2 s (11.2–13.4)] were higher in patients admitted to ICU compared to those not admitted to ICU [0.5 mg/L (0.3–0.8)], p = 0.0042 para D-dimer; 10.7 s (9.8–12.1), p = 0.012 for prothrombin time). According to Salamanna et al. (2020), these laboratory changes show an existing hypercoagulable state in critically ill patients infected with SARS-CoV-2 infection, which could favor the formation of microthrombi in different organs, mainly in the lungs. In pulmonary microthrombosis, damage to endothelial cells leads to overactivation, aggregation and retention of platelets in the injured regions, which can reduce the number of megakaryocytes and, consequently, platelets, in addition to increasing their consumption [78]. As reviewed by Frater et al., 2020 [67], several critically-ill patients have been reported to develop coagulopathy, antiphospholipid antibodies and increased arterial and venous thrombotic events such as cerebral infarction. According the same authors, abnormal coagulation tests can be useful to predict the severity of the disease, support therapy guidance and improve patients' clinical outcome.
In addition, hypersensitive troponin I levels increased significantly in five patients, in whom the diagnosis of virus-related cardiac injury was made. Aspartate aminotransferase levels (AST) increased in 15 (37%) of 41 patients, including eight (62%) of 13 patients in ICU and seven (25%) of 28 patients not admitted to ICU. Compared to patients not admitted to ICU, those admitted to ICU had higher plasma levels of interleukin (IL) 2, IL-7, IL-10, tumor necrosis factor alpha (TNF-α), granulocyte colony stimulating factor (G-CSF), gamma interferon-induced protein 10 (IP-10), type 1 monocyte chemoattractant protein (MCP-1) and macrophage inflammatory protein-1 alpha (MIP-1α). Similar results were found by Liu et al. [79], who reported that the severity of COVID-19 can be predicted by lymphopenia, neutrophilia, hypoalbuminemia, high levels of LDH and CRP. In addition, when compared to healthy controls, plasma levels of angiotensin II in patients infected with SARS-CoV-2 were significantly higher and strongly associated with lung injury and viral load. According Jin et al. [75], it is necessary to pay close attention to patient with an absolute lymphocyte value less than 0.8 × 109/L or a significant reduction in the number of CD4 and CD8 T cells, being recommend to evaluate the hematological alterations again after three days. Main laboratory changes, especially hematological ones, related to the diagnosis and/or prognosis of SARS-CoV infection are shown in Table 1 .
Table 1.
Main laboratory abnormalities related to diagnosis and/or prognosis of SARS-CoV-2 infection.
| Laboratory parameters | Abnormalities | References |
|---|---|---|
| Lymphocytes | Reduction | Chan et al., 2020; Chen et al., 2020; Guan et al., 2020; Huang et al., 2020; Liu et al., 2020; Wang et al. 2020; Zhou et al., 2020; Young et al., 2020; Sun et al., 2020 |
| Platelets | Reduction | Chan et al., 2020; Chen et al., 2020; Guan et al., 2020 |
| Increase | Ruan et al., 2020; Lippi et al., 2020 | |
| Neutrophils | Increase | Chen et al., 2020; Wang et al. 2020 |
| Reduction | Liu et al., 2020 | |
| Eosinophils | Reduction | Sun et al, 2020; Zhang et al, 2020; Liu et al., 2020. |
| Leukocytes | Reduction | Guan et al., 2020; Huang et al., 2020 |
| Increase | Chen et al., 2020; Wang et al., 2020; Zhou et al., 2020 | |
| C-reactive protein(CRP) | Increase | Chan et al., 2020; Chen et al., 2020; Guan et al., 2020; Liu et al., 2020; Ruan et al., 2020; Young et al., 2020 |
| Ferritin | Increase | Chen et al., 2020; Zhou et al., 2020 |
| Procalcitonin | Increase | Chen et al., 2020; Wang et al. 2020; Zhou et al., 2020 |
| Lactate dehydrogenase (LDH) | Increase | Chan et al., 2020; Chen et al., 2020; Liu et al., 2020; Wang et al. 2020; Zhou et al., 2020 |
| D-dimer | Increase | Chen et al., 2020; Huang et al., 2020; Tang et al., 2020; Wang et al. 2020; Zhou et al., 2020 |
| Hemoglobin (Hb) | Reduction | CHEN et al., 2020 |
| Troponin | Increase | Huang et al., 2020; Ruan et al., 2020; Wang et al. 2020; Zhou et al., 2020 |
| Myoglobin | Increase | Ruan et al., 2020 |
| Angiotensin II | Increase | Liu et al., 2020 |
| Cytokines | Increase | Fu et al., 2020; Huang et al., 2020; Ruan et al., 2020; Zhou et al., 2020 |
| Prothrombin time | Increase | Huang et al., 2020; Tang et al., 2020; Zhou et al., 2020 |
| Aspartate aminotransferase (AST) | Increase | Chen et al., 2020; Huang et al., 2020; Wang et al. 2020 |
| Alanine aminotransferase (ALT) | Increase | Chen et al., 2020; Wang et al. 2020; Zhou et al., 2020 |
| Total bilirubin | Increase | Wang et al., 2020 |
| Creatinine | Increase | Wang et al., 2020; Zhou et al., 2020 |
| Albuin | Reduction | Liu et al., 2020 |
| Creatine kinase (CK) | Increase | Wang et al., 2020; Zhou et al., 2020 |
| Urea | Increase | Wang et al. 2020 |
Guan et al. [19] extracted data from COVID-19 patients (N = 1099) in 552 hospitals, in 30 provinces, autonomous regions and municipalities in China. Upon admission, the disease was classified as severe in 173 patients and non severe in 926 patients. The first group had major laboratory abnormalities, such as lymphopenia and leukopenia than those with non severe disease. Of these patients, 83.2% had lymphopenia; 36.2% thrombocytopenia; 33.7% leukopenia and most of them had high levels of CRP. In a retrospective cohort study [22], with 191 patients at Jinyintan Hospital and Wuhan Pulmonary Hospital, China, lymphopenia occurred in 40% of these patients, in addition to leukocytosis, increased LDH, ALT, highly sensitive cardiac troponin I, serum ferritin, creatine kinase (CK), D-dimer, prothrombin time, creatinine, IL-6 and procalcitonin, whose parameters were shown to be associated with hospital death. D-dimer levels above 1 µg/mL at admission were associated with a greater chance of death. Lymphocyte count was significantly higher in surviving patients than in non survivors. It is noteworthy that in survivors, the lymphocyte count was lower on the 7th day after onset of the disease and improved during hospitalization, while severe lymphopenia was observed until death in non survivors. D-dimer levels, high sensitivity cardiac troponin I, LDH, IL-6 and ferritin were higher in non survivors compared to survivors and increased with disease progression. Thus, the levels of D-Dimer and highly sensitive cardiac troponin may indicate a greater or lesser risk of death for patients with SARS-CoV-2 infection [80]. Furthermore, lymphopenia seems to be one of the most relevant hematological abnormalities in COVID-19, its use being suggested as a severity biomarker of this infection [81].
For both groups, survivors and non survivors, LDH increased at the beginning of COVID-19, but decreased from day 13 in those who survived. Lippi and Plebani [77] stated that on hospital admission of patients with SARS-CoV-2 infection, procalcitonin does not appear to be significantly altered, however, its progressive increase may suggest a worse prognosis. They have also suggested that the measurement of other biomarkers, such as presepsin, would probably help to increase the accuracy in the identification of serious cases of COVID-19 and would improve the current approach to predict the risk of mortality. Wang et al. [58] conducted a retrospective study with hospitalized patients with SARS-CoV-2 infection (N = 138) at Zhongnan Hospital, Wuhan University, China. When comparing the laboratory characteristics of patients admitted to ICU with those not admitted, several significant differences were identified, such as a higher leukocyte and neutrophil count, and elevated levels of D-Dimer, CK, urea, creatinine, hypersensitive troponin I, procalcitonin, LDH, AST, ALT and total bilirubin in the first group. In addition, it was found that during hospitalization, most patients had marked lymphopenia and in non survivors lymphopenia worsened over time. Leukocyte and neutrophil counts, and D-Dimer levels were higher in non survivors than in survivors, findings similar to those identified by Zhou el al. [22]. With COVID-19 evolution and the worsening of clinical condition, the urea and creatinine levels increased progressively until death. These changes suggest, according to the researchers, that SARS-CoV-2 infection may be related to coagulation activation, cellular immune deficiency, and myocardium, liver and kidneys lesions.
Regarding the platelet changes observed during the course of COVID-19, a possible mechanism for abnormal thrombopoiesis related to SARS-CoV-2 infection has been proposed by Yang et al. [45]. According to these researchers, CoV can directly infect bone marrow hematopoietic and stromal cells, inducing cell growth inhibition and apoptosis; in addition to stimulating a low-grade DIC state, as evidenced by prolonged clotting times and increased D-Dimer levels. These findings explain thrombocytopenia possibly aggravated by consumption, and may cause immune damage to blood cells, inducing the synthesis of immune complexes and auto antibodies. Thrombopoiesis can occur in the lungs, where platelets are released from mature megakaryocytes. Thus, it is speculated that in the damaged lung tissue there is an increase in the consumption of platelets (e.g. due to platelet aggregation or thrombus formation) [82] and thrombopoiesis reduction. Finally, these authors reinforce that the elucidation of the mechanisms of how CoV enters blood cells is important to better understand SARS pathogenesis and for pharmacological therapies development.
It is worth mentioning that future researches allowing to know other hematological and biochemical abnormalities during the infection by SARS-CoV-2 are of great importance for a better understanding of the pathogenesis of COVID-19, its clinical evolution and monitoring. Based on the clinical events described so far in patients with the severe form of the disease, studies focusing on the effects of SARS-CoV-2 infection on endothelial cells and their interactions with platelets may bring new insights. Likewise, investigating the degree of hyperactivation of both platelets and coagulation, at different stages of the disease, could add value to the existing knowledge about hemostatic changes that often result in thrombotic complications and worsening of the patient’s clinical condition. Platelet indices such as MPV, PDW and immature platelet fraction, are some examples of markers that deserve to be further explored, given the ease of obtaining in modern blood cell analyzers, at no additional cost. Studies using complementary methodologies, such as flow cytometry to evaluate platelet microparticles (in addition to endothelial, monocytic and lymphocyte microparticles) and fluorimetry to measure the endogen thrombin potential (ETP) may also be promising. In view of the coagulopathy evidenced in the most severe cases of COVID-19, thrombin generation assays (TGA) could be an useful tool, as they are able to estimate both phenotypes, that is, hyper and hypocoagulability [83], [84]. Evaluation of the potential of thrombin generation (CAT method) deserve to be explored in new experimental studies, given the state of hypercoagulability described in patients with COVID-19. We are not aware of studies on COVID-19 using TGA to assess patients' hemostatic status. Although TGA is not yet an adequately standardized and validated technique for clinical use, it is possible that its use in research can bring important information about hemostatic abnormalities at different stages of the disease.
As mentioned before, it is necessary to monitor the COVID-19 course through laboratory tests as a prognostic tool and guidance to the medical team. Using a combination of clinical and laboratory data and sophisticated statistical methodologies and machine learning, it is suggested that substantial efforts be made to develop an index or algorithm with good prognostic accuracy for patients with this disease. Statistical and machine learning models with high predictive power and consistency with the reality known and accepted by medical specialists in relation to SARS-CoV-2 infection would be extremely welcome in the current pandemic scenario that may continue for a time not yet predictable. We believe that a tool with such characteristics would have a potential value in clinical practice, considering its efficiency, the ease and speed in obtaining laboratory data, in addition to its low cost.
Finally, in the light of reports in the literature about the importance of the blood group in the clinical manifestations of COVID-19 [85], we also suggest that the ABO blood group be incorporated into the variety of tests requested. This recommendation is based on studies by Ellinghaus et al., 2020 [85], whose authors have reported the involvement of ABO blood groups in susceptibility to COVID-19. According to the same authors, blood group “O” is associated with a lower risk of infection by SARS-CoV-2 compared to non O blood groups, while blood group A was associated with a greater risk than those non A.
4. Concluding remarks
Since December 2019, the SARS-CoV-2 was rapidly spreading among humans and it became a pandemic. Currently, scientific knowledge about the SARS-CoV-2 origin, virulence and spread is still not well established, which corroborates the existence of several gaps, ranging from the understanding of COVID-19 pathophysiology to the discovery of an effective antiviral drug therapy and a vaccine.
SARS-CoV-2 infection identification has become a major challenge in clinical practice, as it presents clinical manifestations that are confused with a simple cold or flu, and the difficulty of performing RT-PCR (gold standard), as well as serological tests. Thus, a large number of suspected individual samples are awaiting diagnosis, which is fundamental for adoption of adequate measures of isolation, treatment and others. Therefore, the search for hematological, biochemical or other suggestive laboratory parameters is extremely welcome in this scenario in which SARS-CoV-2 infection is spreading with enormous speed around the world and the availability of specific tests is still quite limited.
According to the literature available at the time on SARS-CoV-2 infection, there is scientific evidence of major laboratory changes reflecting systemic inflammation and impairment that spans, to a lesser or greater extent the renal, hepatic, cardiac, immune, hemostatic, bone marrow, and peripheral blood systems, among others. For example, among the hematological parameters, lymphopenia can be used to discriminate patients at risk for SARS.
Available data suggest that several hematological parameters may change in the course of SARS-CoV-2 infection and that some of them can be considered significant predictors of unfavorable clinical outcomes, such as admission to ICU or even death. Given the above, the present literature review may be relevant, not only to disseminate what is already known, but it may also serve as a valuable base for future investigations. The search for routine hematological and biochemical variables and others that may assist in the clinical diagnosis of patients suspected of being infected with SARS-CoV-2, or that can predict the severity of the disease or even serve for its monitoring is highly desirable at a time of great challenge imposed by the pandemic.
However, this literature review article has some limitations. Because COVID-19 is a very recent pandemic, several studies of patients infected with SARS-CoV-2 are still being carried out or are awaiting to be published. Furthermore, this review focused on studies practically from the same region, due to the fact that the first patients diagnosed with SARS-CoV-2 infection were in China.
Finally, still in the laboratory context, it is essential that good clinical laboratory and biosafety practices prevail and that the patient be the center of attention. Additional training of laboratory personnel responsible for collecting, transporting and handling biological samples and carrying out the various laboratory tests for patients with COVID-19 is recommended. Training for new demands and development of skills in data interpretation are also required, as well as cooperation and collaboration among laboratory professionals. In today's challenging times, more than ever, the clinical laboratory needs to be patient-centered and with qualified leadership to be safe, efficient, effective and timely.
Acknowledgments
Acknowledgments
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brazil (CAPES) - Finance Code 001; Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil (CNPq) (Research Fellowship MGC 311185/2019-3)
References
- 1.P.C. Woo, S.K. Lau, Y. Huang, K.Y. Yuen, Coronavirus diversity, phylogeny and interspecies jumping, Exp. Biol. Med. (Maywood) 234(10) (2009) 1117–1127. [DOI] [PubMed]
- 2.Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16(1):69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hui D.S. An overview on severe acute respiratory syndrome (SARS) Monaldi Arch. Chest Dis. 2005;63(3):149–157. doi: 10.4081/monaldi.2005.632. [DOI] [PubMed] [Google Scholar]
- 6.Cheng V.C., Lau S.K., Woo P.C., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007;20(4):660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28(2):465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li G., Fan Y., Lai Y., Han T., Li Z., Zhou P., Pan P., Wang W., Hu D., Liu X., Zhang Q., Wu J. Coronavirus infections and immune responses. J. Med. Virol. 2020;92(4):424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cossarizza A., De Biasi S., Guaraldi G., Girardis M., Mussini C. SARS-CoV-2, the Virus that Causes COVID-19: Cytometry and the New Challenge for Global Health. Cytometry A. 2020;97(4):340–343. doi: 10.1002/cyto.a.24002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorbalenya A., Baker S., Baric R., de Groot R., Drosten C., Gulyaeva A., Haagmans B., Lauber C., Leontovich A., Neuman B., Penzar D., Perlman S., Poon L., Samborskiy D., Sidorov I., Sola I., Ziebuhr J. Severe acute respiratory syndrome-related coronavirus: The species and its viruses -a statement of the Coronavirus. Study Group, bioRxiv. 2020:1–15. [Google Scholar]
- 12.Bouadma L., Lescure F.X., Lucet J.C., Yazdanpanah Y., Timsit J.F. Severe SARS-CoV-2 infections: practical considerations and management strategy for intensivists. Intensive Care Med. 2020;46(4):579–582. doi: 10.1007/s00134-020-05967-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.WHO, WHO Director-General's opening remarks at the media briefing on COVID-19 - 11 March 2020, (2020).
- 14.Singhal T. A Review of Coronavirus Disease-2019 (COVID-19) Indian J. Pediatr. 2020;87(4):281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.She J., Jiang J., Ye L., Hu L., Bai C., Song Y. 2019 novel coronavirus of pneumonia in Wuhan, China: emerging attack and management strategies. Clin. Transl. Med. 2020;9(1):19. doi: 10.1186/s40169-020-00271-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y., Ning Z., Chen Y., Guo M., Gali N.K., Sun L., Duan Y., Cai J., Westerdahl D., Liu X., Xu K., Ho K.F., Kan H., Fu Q., Lan K. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020 doi: 10.1038/s41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- 17.Shanmugaraj B., Malla A., Phoolcharoen W. Emergence of Novel Coronavirus 2019-nCoV: Need for Rapid Vaccine and Biologics Development. Pathogens. 2020;9(2) doi: 10.3390/pathogens9020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munster V.J., Koopmans M., van Doremalen N., van Riel D., de Wit E. A Novel Coronavirus Emerging in China - Key Questions for Impact Assessment. N. Engl. J. Med. 2020;382(8):692–694. doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- 19.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.L., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z.J., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Zhong N.S. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bloomgarden Z.T. Diabetes and COVID-19. J. Diab. 2020;12(4):347–348. doi: 10.1111/1753-0407.13027. [DOI] [PubMed] [Google Scholar]
- 21.Zheng Y.Y., Ma Y.T., Zhang J.Y., Xie X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 2020;17(5):259–260. doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porcheddu R., Serra C., Kelvin D., Kelvin N., Rubino S. Similarity in Case Fatality Rates (CFR) of COVID-19/SARS-COV-2 in Italy and China. J. Infect. Dev. Ctries. 2020;14(2):125–128. doi: 10.3855/jidc.12600. [DOI] [PubMed] [Google Scholar]
- 25.WHO Use of laboratory methods for SARS diagnosis. World Health Organization, Geneva. 2020 [Google Scholar]
- 26.Vashist S.K. In, Vitro Diagnostic Assays for COVID- 19: Recent Advances and Emerging Trends. Diagnostics (Basel) 2020;10(4) doi: 10.3390/diagnostics10040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.CDC, Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens from Persons for Coronavirus Disease 2019 (COVID-19), Centers of Disease and Control and Prevention, Georgia, 2020.
- 28.Brasil, Acurácia dos testes diagnósticos registrados para a COVID-19 Ministério da Saúde, Brasília, 2020.
- 29.Zhang N., Wang L., Deng X., Liang R., Su M., He C., Hu L., Su Y., Ren J., Yu F., Du L., Jiang S. Recent advances in the detection of respiratory virus infection in humans. J. Med. Virol. 2020;92(4):408–417. doi: 10.1002/jmv.25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin Y., Wunderink R.G. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23(2):130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brasil Guia. Brasília; Ministério da Saúde: 2020. de Vigilância Epidemiológica - Emergência de Saúde Pública de Importância Nacional pela Doença pelo Coronavírus 2019. [Google Scholar]
- 32.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020 doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.W. Tan, Y. Lu, J. Zhang, J. Wang, Y. Dan, Z. Zhaoxia Tan, X. He, C. Qian, Q. Sun, Q. Hu, H. Liu, S. Ye, X. Xiang, Y. Zhou, W. Zhang, Y. Guo, X. Wang, W. He, X. Wan, F. Sun, Q. Wei, C. Chen, G. Pan, J. Xia, Q. Mao, Y. Chen, G. Deng, Viral Kinetics and Antibody Responses in Patients with COVID-19, medRxiv, 2020.
- 34.Dengler L., Kuhn N., Shin D.L., Hatesuer B., Schughart K., Wilk E. Cellular changes in blood indicate severe respiratory disease during influenza infections in mice. PLoS One. 2014;9(7) doi: 10.1371/journal.pone.0103149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ali N., Anwar M., Majeed I., Tariq W.U. Chicken pox associated thrombocytopenia in adults. J. Coll. Phys. Surg. Pak. 2006;16(4):270–272. [PubMed] [Google Scholar]
- 36.Shahid I., Ul Ain Q., Rashid T., Azam A. An observational study about abnormal hematological changes occurs in chicken pox in adult patients, Indo American. J. Pharmaceut. Sci. 2018;5(8):7319–7322. [Google Scholar]
- 37.Hamed M.A. Hematological changes among children with dengue fever in Saudi Arabia. Egypt. J. Haematol. 2017;42:129–133. [Google Scholar]
- 38.Kularatnam G.A.M., Jasinge E., Gunasena S., Samaranayake D., Senanayake M.P., Wickramasinghe V.P. Evaluation of biochemical and haematological changes in dengue fever and dengue hemorrhagic fever in Sri Lankan children: a prospective follow up study. BMC Pediatr. 2019;19(1):87. doi: 10.1186/s12887-019-1451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rashmi M.V. Hamsaveena, Haematological and biochemical markers as predictors of dengue infection. Malays. J. Pathol. 2015;37(3):247–251. [PubMed] [Google Scholar]
- 40.Jackson B.S., Pretorius E. Pathological Clotting and Deep Vein Thrombosis in Patients with HIV. Semin. Thromb. Hemost. 2019;45(2):132–140. doi: 10.1055/s-0038-1676374. [DOI] [PubMed] [Google Scholar]
- 41.Aziz N., Quint J.J., Breen E.C., Oishi J., Jamieson B.D., Martinez-Maza O., Detels R. 30-Year Longitudinal Study of Hematological Parameters of HIV-1 Negative Men Participating in Los Angeles Multicenter AIDS Cohort Study (MACS) Lab. Med. 2019;50(1):64–72. doi: 10.1093/labmed/lmy044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He Z., Zhao C., Dong Q., Zhuang H., Song S., Peng G., Dwyer D.E. Effects of severe acute respiratory syndrome (SARS) coronavirus infection on peripheral blood lymphocytes and their subsets. Int. J. Infect Dis. 2005;9(6):323–330. doi: 10.1016/j.ijid.2004.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang M., Hon K.L., Li K., Fok T.F., Li C.K. The effect of SARS coronavirus on blood system: its clinical findings and the pathophysiologic hypothesis. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2003;11(3):217–221. [PubMed] [Google Scholar]
- 44.Yang M., Ng M.H., Li C.K., Chan P.K., Liu C., Ye J.Y., Chong B.H. Thrombopoietin levels increased in patients with severe acute respiratory syndrome. Thromb. Res. 2008;122(4):473–477. doi: 10.1016/j.thromres.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang M., Ng M.H., Li C.K. Thrombocytopenia in patients with severe acute respiratory syndrome (review) Hematology. 2005;10(2):101–105. doi: 10.1080/10245330400026170. [DOI] [PubMed] [Google Scholar]
- 46.Al-Tawfiq J.A., Hinedi K., Abbasi S., Babiker M., Sunji A., Eltigani M. Hematologic, hepatic, and renal function changes in hospitalized patients with Middle East respiratory syndrome coronavirus. Int. J. Lab. Hematol. 2017;39(3):272–278. doi: 10.1111/ijlh.12620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Debuc B., Smadja D.M. Is COVID-19 a New Hematologic Disease? Stem Cell. Rev. Rep. 2020 doi: 10.1007/s12015-020-09987-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020 doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang F., Nie J., Wang H., Zhao Q., Xiong Y., Deng L., Song S., Ma Z., Mo P., Zhang Y. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J. Infect. Dis. 2020 doi: 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun S., Cai X., Wang H., He G., Lin Y., Lu B., Chen C., Pan Y., Hu X. Abnormalities of peripheral blood system in patients with COVID-19 in Wenzhou, China. Clin. Chim. Acta. 2020;507:174–180. doi: 10.1016/j.cca.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qian G.Q., Yang N.B., Ding F., Ma A.H.Y., Wang Z.Y., Shen Y.F., Shi C.W., Lian X., Chu J.G., Chen L., Ren D.W., Li G.X., Chen X.Q., Shen H.J., Chen X.M. A retrospective, multi-centre case series, QJM; China: 2020. Epidemiologic and Clinical Characteristics of 91 Hospitalized Patients with COVID-19 in Zhejiang. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mo P., Xing Y., Xiao Y., Deng L., Zhao Q., Wang H., Xiong Y., Cheng Z., Gao S., Liang K., Luo M., Chen T., Song S., Ma Z., Chen X., Zheng R., Cao Q., Wang F., Zhang Y. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.J.J. Zhang, X. Dong, Y.Y. Cao, Y.D. Yuan, Y.B. Yang, Y.Q. Yan, C.A. Akdis, Y.D. Gao, Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China, Allergy, 2020. [DOI] [PubMed]
- 54.Liu F., Xu A., Zhang Y., Xuan W., Yan T., Pan K., Yu W., Zhang J. Patients of COVID-19 may benefit from sustained lopinavir-combined regimen and the increase of eosinophil may predict the outcome of COVID-19 progression. Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lippi G., Plebani M., Henry B.M. Thrombocytopenia is associated with severe coronavirus disease, (COVID-19) infections: A meta-analysis. Clin. Chim. Acta. 2019;506(2020):145–148. doi: 10.1016/j.cca.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fan B.E., Chong V.C.L., Chan S.S.W., Lim G.H., Lim K.G.E., Tan G.B., Mucheli S.S., Kuperan P., Ong K.H. Hematologic parameters in patients with COVID-19 infection. Am J Hematol. 2020 doi: 10.1002/ajh.25774. [DOI] [PubMed] [Google Scholar]
- 57.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Dawei, Hu Bo, Hu Chang, Zhu Fangfang, Liu Xing, Zhang Jing, Wang Binbin, Xiang Hui, Cheng Zhenshun, Xiong Yong, Zhao Yan, Li Yirong, Wang Xinghuan, Peng Zhiyong. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA. 2020;323(11):1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Henry B.M., de Oliveira M.H.S., Benoit S., Plebani M., Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin. Chem. Lab. Med. 2020 doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 60.Terpos E., Ntanasis-Stathopoulos I., Elalamy I., Kastritis E., Sergentanis T.N., Politou M., Psaltopoulou T., Gerotziafas G., Dimopoulos M.A. Hematological findings and complications of COVID-19. Am. J. Hematol. 2020 doi: 10.1002/ajh.25829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020 doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan L., Wang Q., Zhang D., Ding J., Huang Q., Tang Y.Q., Miao H. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct. Target Ther. 2020;5:33. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bhatraju P.K., Ghassemieh B.J., Nichols M., Kim R., Jerome K.R., Nalla A.K., Greninger A.L., Pipavath S., Wurfel M.M., Evans L., Kritek P.A., West T.E., Luks A., Gerbino A., Dale C.R., Goldman J.D., O'Mahony S., Mikacenic C. Covid-19 in Critically Ill Patients in the Seattle Region - Case Series. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deng Y., Liu W., Liu K., Fang Y.Y., Shang J., Zhou L., Wang K., Leng F., Wei S., Chen L., Liu H.G. Clinical characteristics of fatal and recovered cases of coronavirus disease 2019 (COVID-19) in Wuhan, China: a retrospective study. Chin. Med. J. (Engl.) 2020 doi: 10.1097/CM9.0000000000000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arentz M., Yim E., Klaff L., Lokhandwala S., Riedo F.X., Chong M., Lee M. Characteristics and Outcomes of 21 Critically Ill Patients With COVID-19 in Washington State. JAMA. 2020 doi: 10.1001/jama.2020.4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frater J.L., Zini G., d'Onofrio G., Rogers H.J. COVID-19 and the clinical hematology laboratory. Int. J. Lab. Hematol. 2020;42(Suppl 1):11–18. doi: 10.1111/ijlh.13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.T. Guo, Y. Fan, M. Chen, X. Wu, L. Zhang, T. He, H. Wang, J. Wan, X. Wang, Z. Lu, Cardiovascular Implications of Fatal Outcomes of Patients With Coronavirus Disease 2019 (COVID-19), JAMA Cardiol., 2020. [DOI] [PMC free article] [PubMed]
- 70.Chng W.J., Lai H.C., Earnest A., Kuperan P. Haematological parameters in severe acute respiratory syndrome. Clin. Lab. Haematol. 2005;27(1):15–20. doi: 10.1111/j.1365-2257.2004.00652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zini G., Bellesi S., Ramundo F., d'Onofrio G. Morphological anomalies of circulating blood cells in COVID-19. Am. J. Hematol. 2020 doi: 10.1002/ajh.25824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Siddiqi H., Mehra M. COVID-19 Illness in Native and Immunosuppressed States: A Clinical-Therapeutic Staging Proposal [published online ahead of print. J. Heart Lung Transplant. 2020 doi: 10.1016/j.healun.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., Tsoi H.W., Lo S.K., Chan K.H., Poon V.K., Chan W.M., Ip J.D., Cai J.P., Cheng V.C., Chen H., Hui C.K., Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jin Y.H., Cai L., Cheng Z.S., Cheng H., Deng T., Fan Y.P., Fang C., Huang D., Huang L.Q., Huang Q., Han Y., Hu B., Hu F., Li B.H., Li Y.R., Liang K., Lin L.K., Luo L.S., Ma J., Ma L.L., Peng Z.Y., Pan Y.B., Pan Z.Y., Ren X.Q., Sun H.M., Wang Y., Wang Y.Y., Weng H., Wei C.J., Wu D.F., Xia J., Xiong Y., Xu H.B., Yao X.M., Yuan Y.F., Ye T.S., Zhang X.C., Zhang Y.W., Zhang Y.G., Zhang H.M., Zhao Y., Zhao M.J., Zi H., Zeng X.T., Wang X.H. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version) Mil. Med. Res. 2020;7(1):4. doi: 10.1186/s40779-020-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tang N., Li D., Wang X., Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020;18(4):844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lippi G., Plebani M. Laboratory abnormalities in patients with COVID-2019 infection. Clin. Chem. Lab. Med. 2020 doi: 10.1515/cclm-2020-0198. [DOI] [PubMed] [Google Scholar]
- 78.F. Salamanna, M. Maglio, M.P. Landini, M. Fini, Platelet functions and activities as potential hematologic parameters related to Coronavirus Disease 2019 (Covid-19), Platelets, 2020, pp. 1–6. [DOI] [PubMed]
- 79.Liu Y., Yang Y., Zhang C., Huang F., Wang F., Yuan J., Wang Z., Li J., Feng C., Zhang Z., Wang L., Peng L., Chen L., Qin Y., Zhao D., Tan S., Yin L., Xu J., Zhou C., Jiang C., Liu L. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci. China Life Sci. 2020;63(3):364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kavsak P.A., de Wit K., Worster A. Emerging key laboratory tests for patients with COVID-19. Clin. Biochem. 2020;81:13–14. doi: 10.1016/j.clinbiochem.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.F.B. Duarte, R.P.G. Lemes, I.A. Duarte, B.A. Duarte, J.V.A. Duarte, Hematological changes in Covid-19 infections, Rev. Assoc. Med. Bras. (1992) 66(2) (2020) 99. [DOI] [PubMed]
- 82.Yang J., Yang M., Xu F., Li K., Lee S.K., Ng P.C., Tam J.S., Yuen P.M., Fok T.F. Effects of oxygen-induced lung damage on megakaryocytopoiesis and platelet homeostasis in a rat model. Pediatr. Res. 2003;54(3):344–352. doi: 10.1203/01.PDR.0000079186.86219.29. [DOI] [PubMed] [Google Scholar]
- 83.Hemker H.C., Giesen P., Al Dieri R., Regnault V., de Smedt E., Wagenvoord R., Lecompte T., Beguin S. Calibrated automated thrombin generation measurement in clotting plasma. Pathophysiol. Haemost. Thromb. 2003;33(1):4–15. doi: 10.1159/000071636. [DOI] [PubMed] [Google Scholar]
- 84.Lecut C., Peters P., Massion P.B., Gothot A. Is there a place for thrombin generation assay in routine clinical laboratory? Ann. Biol. Clin. (Paris) 2015;73(2):137–149. doi: 10.1684/abc.2014.1018. [DOI] [PubMed] [Google Scholar]
- 85.D. Ellinghaus, F. Degenhardt, L. Bujanda, M. Buti, A. Albillos, P. Invernizzi, J. Fernandez, D. Prati, G. Baselli, R. Asselta, M.M. Grimsrud, C. Milani, F. Aziz, J. Kassens, S. May, M. Wendorff, L. Wienbrandt, F. Uellendahl-Werth, T. Zheng, X. Yi, R. de Pablo, A.G. Chercoles, A. Palom, A.E. Garcia-Fernandez, F. Rodriguez-Frias, A. Zanella, A. Bandera, A. Protti, A. Aghemo, A. Lleo, A. Biondi, A. Caballero-Garralda, A. Gori, A. Tanck, A. Carreras Nolla, A. Latiano, A.L. Fracanzani, A. Peschuck, A. Julia, A. Pesenti, A. Voza, D. Jimenez, B. Mateos, B. Nafria Jimenez, C. Quereda, C. Paccapelo, C. Gassner, C. Angelini, C. Cea, A. Solier, D. Pestana, E. Muniz-Diaz, E. Sandoval, E.M. Paraboschi, E. Navas, F. Garcia Sanchez, F. Ceriotti, F. Martinelli-Boneschi, F. Peyvandi, F. Blasi, L. Tellez, A. Blanco-Grau, G. Hemmrich-Stanisak, G. Grasselli, G. Costantino, G. Cardamone, G. Foti, S. Aneli, H. Kurihara, H. ElAbd, I. My, I. Galvan-Femenia, J. Martin, J. Erdmann, J. Ferrusquia-Acosta, K. Garcia-Etxebarria, L. Izquierdo-Sanchez, L.R. Bettini, L. Sumoy, L. Terranova, L. Moreira, L. Santoro, L. Scudeller, F. Mesonero, L. Roade, M.C. Ruhlemann, M. Schaefer, M. Carrabba, M. Riveiro-Barciela, M.E. Figuera Basso, M.G. Valsecchi, M. Hernandez-Tejero, M. Acosta-Herrera, M. D'Angio, M. Baldini, M. Cazzaniga, M. Schulzky, M. Cecconi, M. Wittig, M. Ciccarelli, M. Rodriguez-Gandia, M. Bocciolone, M. Miozzo, N. Montano, N. Braun, N. Sacchi, N. Martinez, O. Ozer, O. Palmieri, P. Faverio, P. Preatoni, P. Bonfanti, P. Omodei, P. Tentorio, P. Castro, P.M. Rodrigues, A. Blandino Ortiz, R. de Cid, R. Ferrer, R. Gualtierotti, R. Nieto, S. Goerg, S. Badalamenti, S. Marsal, G. Matullo, S. Pelusi, S. Juzenas, S. Aliberti, V. Monzani, V. Moreno, T. Wesse, T.L. Lenz, T. Pumarola, V. Rimoldi, S. Bosari, W. Albrecht, W. Peter, M. Romero-Gomez, M. D'Amato, S. Duga, J.M. Banales, J.R. Hov, T. Folseraas, L. Valenti, A. Franke, T.H. Karlsen, Genomewide Association Study of Severe Covid-19 with Respiratory Failure, N. Engl. J. Med., 2020. [DOI] [PMC free article] [PubMed]