Abstract
The world is facing health and economic havoc due to the Corona Virus Disease-2019 (COVID-19) pandemic. Given the number of affected people and the mortality rate, the virus is undoubtedly a serious threat to humanity. By analogy with earlier reports about Severe Acute Respiratory Syndrome (SARS-CoV) and Middle East Respiratory Syndrome (MERS-CoV) - viruses, the novel Coronavirus’ replication mechanism is likely well understood. The structure of an endoribonuclease (NSP15) of SARS-CoV-2 was reported recently. This enzyme is expected to play a crucial role in replication. In this work, attempts were made to identify inhibitors of this enzyme. To achieve the goal, high throughput in silico screening and molecular docking procedures were performed. From an Enamine database of a billion compounds, 3978 compounds with potential antiviral activity were selected for screening and induced fit docking that funneled down to eight compounds with good docking score and docking energy. Detailed analysis of non-covalent interactions at the active site and the apparent match of the molecule with the shape of the binding pocket were assessed. All the compounds show significant interactions for tight binding. Since all the compounds are synthetic with favorable drug-like properties, these may be considered for immediate optimization and downstream applications.
Keywords: Endoribonuclease, Coronavirus, COVID-19, NSP15 inhibitors
1. Introduction
In recent times viral infections have become a deadly threat to human society globally, because of the major improvements in scale and speed of transportation. Novel viruses emerge and they are implicated for all severe acute respiratory syndromes. Among them, SARS-CoV and MERS-CoV have caused high fatality rates. At present, SARS-CoV-2 is causing a rapidly spreading pandemic. The mortality rate caused by COVID-19 is very high among the infected population. Besides, the pandemic demands a major mitigation effort and this and direct costs to health care systems have caused a recession. Animal to human transmissions were witnessed in the past, in the last few years, human to human transmissions are also reported [1,2] and form the basis for the current pandemic.
Though vaccines are under development, only the supporting health care system can currently play a role in the treatment of infections. In considering ways to treat infection one has to understand in detail virus replication, potential reservoirs, mechanisms of transmission and approaches to treat general symptoms. The symptoms of COVID-19 infection are often mild and very similar to that of influenza rather than the symptoms resulting from SARS and MERS viral infections [3]. These mild cases of the disease are thought to play a role in its spreading.
To design drugs for COVID-19, several strategies can be employed as reported in recent times [4]. The first strategy is to design compounds based on the existing broad spectrum of anti-virals. This approach has the advantage of having compounds with established pharmaceutical properties that have a history of use in people and so can be readily used. However, this strategy is strictly limited to a finite library of compounds and the likelihood that a powerful therapy will result directly from a member of such a family is very uncertain. Inhibitors such as ribavirin and cyclophilin used to treat coronavirus pneumonia fall in this category. The second is to use available databases that may have compounds with therapeutic effects against the corona virus for screening and the third strategy is based on the genomic information and pathological properties. Among these, the first one is a faster approach as we have a huge number of compounds in anti-viral databases for initial screening. Also, there are reports regarding small molecules against crucial protein targets that are essential for viral replication with proven potency. Recently, there are several reports cited on designing drugs for SARS-CoV-2 employing in silico virtual screening methods [[5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15]].
In the case of SARS and MERS, other than papain-like proteases, Non-structural proteins (NSPs) have been studied and reported as effective targets for small molecule drugs [16]. Paralogs in coronaviruses having several conserved sequence motifs and have been convincingly presented as a drug target [2]. Now, several details are emerging about the SARS-CoV-2 entry into host cells and replication. Functions of different NSPs and open reading frames (ORFs) of the virus are elucidated such as DNA replication, epigenetic and gene expression regulation, vesicle trafficking, lipid modification, RNA processing and regulation, ubiquitin ligation, signaling, nuclear transport, construction of cytoskeleton, mitochondria, and extracellular matrix [17]. Recently, NSP15 of COVID-19, uridylate-specific endoribonuclease (NendoU) structure was reported and suggested as a drug target based on the high sequence similarity with NSP15 of SARS and MERS. NSP15 is reported to be involved in RNA replication and processing of subgenomic RNAs but the function is still not clearly understood.
2. Materials and methods
2.1. Sequence analysis
Primary amino acid sequences corresponding to NSP15 of SARS-CoV-2, SARS and MERS were retrieved from sequence databases. Multiple sequence alignment was done using ClustalW and figures were prepared using ESPript 3.0 [18].
2.2. Protein preparation and grid generation
The three-dimensional crystal structure of NSP15 was retrieved from the Protein Data Bank (PDB ID: 6VWW) [19]. To perform the docking studies, the protein structure was optimized using the protein preparation wizard available in the Schrödinger software suite [20,21]. The protein model preparation involved two steps. In the first step, hydrogen atoms were added and side-chain atoms were neutralized. The second step is the refinement, in which the water molecules were removed and the model was minimized. Finally, energy minimization was carried out through the conjugate gradient method using the OPLS-2005 force field to have a unique low-energy minimum structure. A grid box was generated comprising important active site residues responsible for NSP15 endonuclease activity. Active site residues are His235, His250, Lys290, Ser294, Tyr343, and Thr341. Glide grid module embedded in Grid Generation panel of Schrödinger software suite was employed.
2.3. Anti-viral library preparation
A set of 3978 molecules with anti-viral activity was prepared by retrieving from the Enamine database (https://enamine.net/). All these compounds were prepared using the Ligprep module of Schrodinger [22] for geometry optimization and energy minimization.
2.4. Structure-based virtual screening (SBVS)
Initially, a library of anti-viral compounds was docked into NSP15 of COVID-19; a new potential drug target using the HTVS algorithm (Glide, Schrodinger). The grid of about 20 × 20 × 20 Å3 was selected to cover the entire catalytic site. Molecular docking was carried out in two steps; standard precision (SP) and extra precision (XP) [23] (Schrodinger LLC, 2014) to analyze interactions between the protein and ligands and thereby to rank them based on docking score and glide energy. Based on the docking score and glide energy, the top 20% of docked compounds were further re-docked using standard precision (SP) algorithm. Extra precision (XP) docking procedure was carried out with 20% of SP docking results. During docking, the active site residues were kept rigid (which include H235, H250, K290, S294, T341 and Y343) and ligands were allowed to be flexible. Since there were large numbers of molecules in the library, only one docked pose per ligand was considered in several stages of virtual screening. To analyze binding characteristics, induced-fit docking was performed for selected molecules.
2.5. Induced fit docking studies of selected compounds against NSP15 endoribonuclease (NSP15)
To have dynamic binding characteristics of the proposed ligands (from earlier steps) with NSP15, flexible docking was carried out through induced fit docking (IFD) procedures. For each ligand, twenty binding poses were considered and the best pose was selected based on the interactions, docking score and glide energy. All molecular modeling studies were carried out using the OPLS-AA (Optimized Potential for Liquid Simulation - All-Atom) force field. All docked complexes were analyzed for ligand interactions using 2D maestro view and superposition analysis was carried out using chimera visualization software.
3. Results and discussion
3.1. Sequence alignment
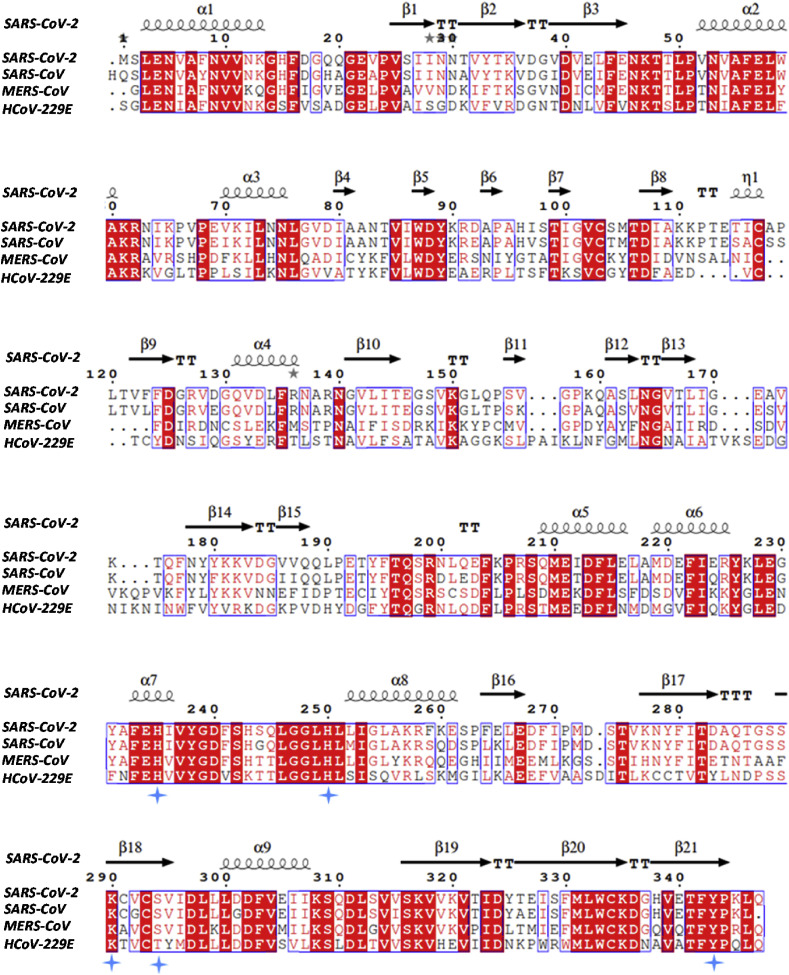
The sequence comparison of NSP15 (COVID-19) with SARS-CoV-2, MERS-CoV and H–CoV-229E (Human Corona Virus) reveals a high similarity (Fig. 1 ). The full-length sequence alignment shows NSP15 to have 87%, 52% and 44% identity with the above-mentioned viruses respectively. When only the catalytic domains (235–343 of NSP15 of COVID-19) are aligned, a high degree of conservation exists with 89%, 61% and 50% of three virus proteins. The conserved regions are found to be distributed throughout the length of the protein, in particular, the conservation is notably high among the catalytic domains (Fig. 1) which can be attributed to functional similarity. Such high sequence similarity among catalytic domain regions strongly supports functional inferences that can be drawn among all studies of this protein family. The catalytic triad residues, His235, His250 and Lys290 are conserved in all viruses belonging to the coronavirus family. These residues are implicated in the ribonuclease activity as well as functional hexamer formation [24,25] and of course, this area of the protein is a potential drug target for that reason.
Fig. 1.
Multiple sequence alignment of NSP15. Blue stars represent catalytic residues. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Screening anti-viral compounds against NSP15 using SBVS approach
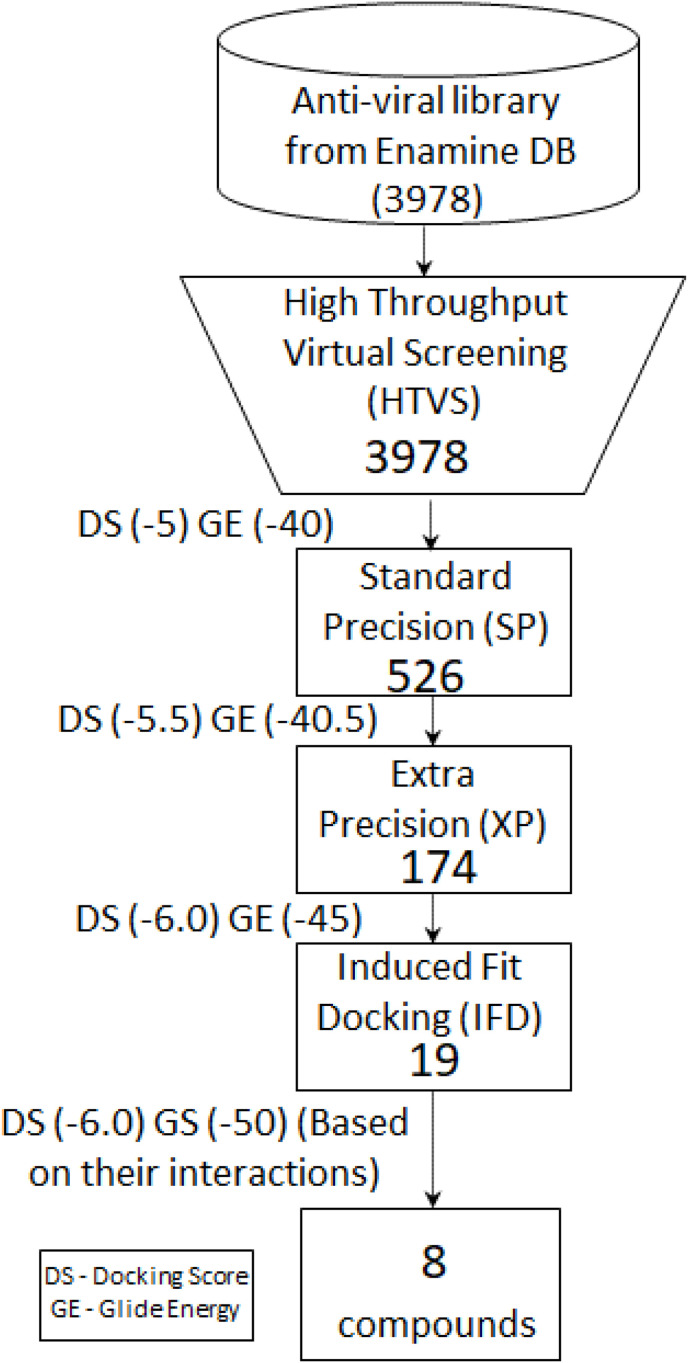
In total, 3978 compounds with anti-viral activity from the enamine database were downloaded for specific docking. Computer-aided screening protocol of the binding region of NSP-15 of SARS-CoV-2 was employed using a virtual screening workflow with the docking program Glide (Schrodinger suite). A flexible docking approach was run which automatically generates different conformers for each ligand molecule. A total of 3978 ligands were used for screening using multiple docking filters of HTVS. Based on the docking score, 526 compounds were shortlisted with values better than a docking score (DS) of −5.00 and −40.00 kcal/mol of glide energy (GE), followed by SP docking resulted in 174 compounds (DS -5.5 and GE -40.5 kcal/mol). Finally, 174 compounds were subjected to XP (extra precision) mode of screening that resulted in 19 compounds (DS -6.0, GE -46.0). The detailed workflow for SBVS is shown in Fig. 2 .
Fig. 2.
Structure Based Virtual Screeing - Flow chart. Three dimensional structure of the target protein NSP15 was downloaded from wwPDB with ID: 6VWW.
3.3. Interaction of ligands with NSP15
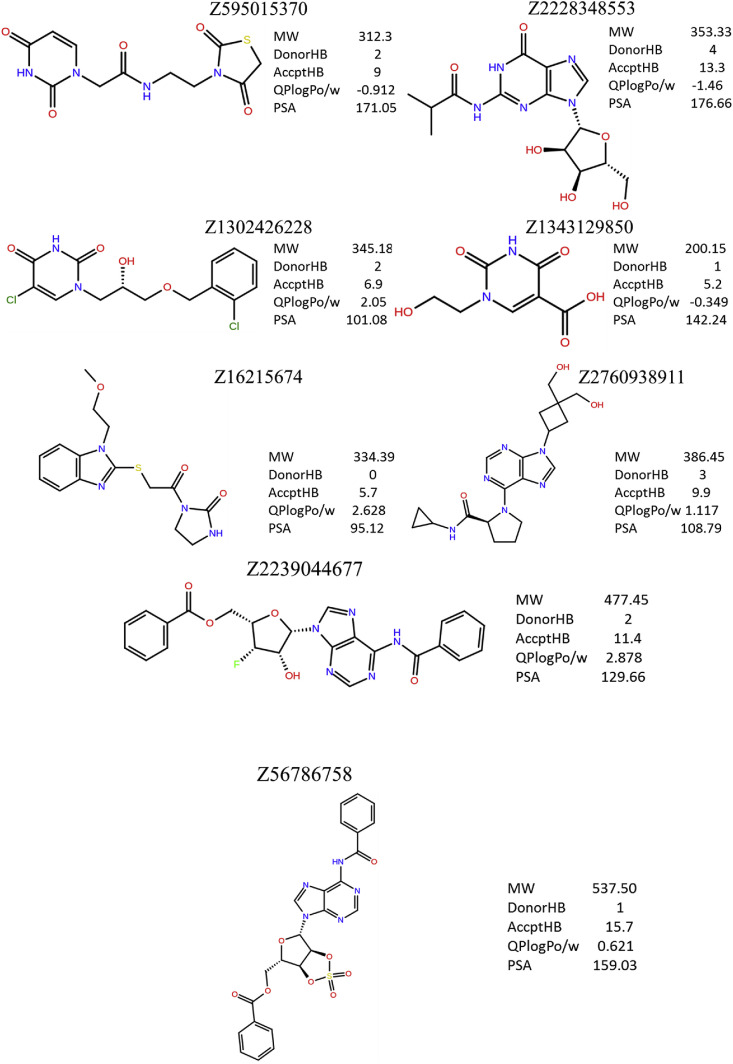
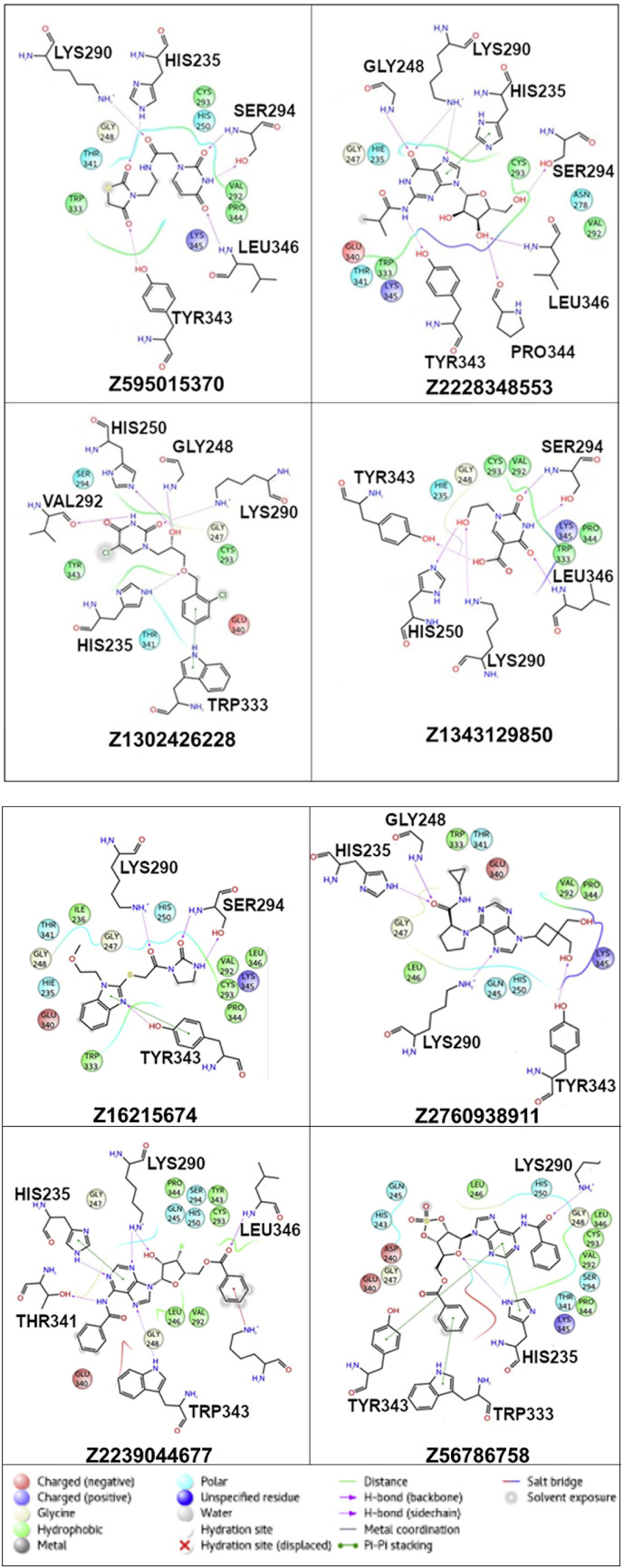
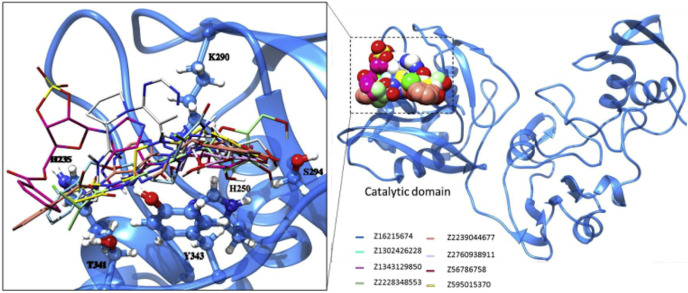
Among nineteen compounds, from flexible docking through IFD, eight compounds that exhibit significant binding at the active site of NSP15 were selected. The two-dimensional scheme and their physicochemical properties are shown in Fig. 3 . Predicted binding interactions were also compared in terms of glide score and energy (Table 1 and Table 2 ). From these tables, it is interesting to note that all the compounds do interact with the important catalytic residue Lys290 which is involved predominantly in the hydrolysis of a nucleoside as understood from SARS and MERS NSP15 [24,25]. The residue His235 which is essential for hydrolysis is found to have interaction with 5 out of 8 compounds. Hydroxyl groups of Ser294 and Tyr343 residues which are close to the catalytic residues and may be responsible for the binding of the substrate also make hydrogen bonds with ligands in the models. It is also noted that, in the case of MERS (NSP15), the importance of the residue Tyr343 (Y343A) for ribonuclease activity has been established by mutational studies [26]. Compounds, Z595015370, Z1343129850 and Z2760938911 exhibit four interactions at the active site for favorable binding. Compounds except Z56786758 are found to form many interactions with catalytic residues and their vicinity that show their potential binding characteristics. From figure (Fig. 4 ), it can be seen that the modeling has resulted in proposed binding situations which are qualitatively what would be expected for inhibitors of such an enzyme. Also, it is noted that all the compounds are located within the active site suggesting that the docking performed without unusual steric problems (Fig. 5 ).
Fig. 3.
Schematic representation of identified new lead molecules. Physico-chemical properties also provided.
Table 1.
Docking Score and Glide energy (in kcal/mol) of identified lead compounds against NSP15 (PDBID: 6VWW).
| Compounds | Docking score (kcal/mol) | Glide energy (kcal/mol) | Interactions (D-H … A) |
|---|---|---|---|
| Z595015370 | −10.50 | −51.37 |
Ser294 (N–H⋯O) (N–H⋯O)Ser294 Lys290 (N–H⋯O) His235(N–H⋯O) Tyr343 (O–H⋯O) Leu346(N–H⋯O) |
| Z2228348553 | −9.87 | −50.03 | (N–H⋯O)Tyr343 Gly248 (N–H⋯O) Lys290(N–H⋯O) Lys290(N–H⋯N) Leu346 (N–H⋯O) (O–H⋯O)Ser294 (O–H⋯O) Pro 344 |
| Z1302426228 | −9.68 | −44.33 |
His235 (N–H⋯O) Gly248 (N–H⋯O) (O–H⋯N)His250 Lys290 (N–H⋯O) (N–H⋯O)Val 292 |
| Z1343129850 | −9.42 | −40.01 | (O–H⋯O)Tyr343 Leu346 (N–H⋯O) (N–H⋯O)Ser294 Ser294 (N–H⋯O) (O–H⋯N)His250 (O–H⋯O)Tyr343 Lys290 (N–H⋯O) |
| Z16215674 | −9.07 | −49.35 |
(N–H⋯O)Ser294 Ser294(N–H⋯O) Lys290 (N–H⋯O) Tyr343 (O–H⋯N) |
| Z2760938911 | −8.65 | −55.95 |
Ser294 (N–H⋯O) (O–H⋯O)Leu346 Tyr343 (O–H⋯O) (N–H⋯N)Lys290 His235 (N–H⋯O) Gly248 (N–H⋯O) |
| Z2239044677 | −6.74 | −60.19 | Trp 333 (N–H⋯N) (N–H⋯O)Thr341 His235 (N–H⋯N) Lys290 (N–H⋯N) Lys290 (N–H⋯O) Leu346 (N–H⋯O) |
| Z56786758 | −5.44 | −56.21 |
Lys290 (N–H⋯O) His235 (N–H⋯O) |
Table 2.
Catalytic site interactions of identified lead compounds.
| Compounds | H235 | H250 | K290 | S294 | T341 | Y343 |
|---|---|---|---|---|---|---|
| Z595015370 | Y | X | Y | Y | X | Y |
| Z2228348553 | X | X | Y | Y | X | Y |
| Z1302426228 | Y | X | Y | X | X | X |
| Z1343129850 | X | Y | Y | Y | X | Y |
| Z16215674 | X | X | Y | Y | X | Y |
| Z2760938911 | Y | X | Y | Y | X | Y |
| Z2239044677 | Y | X | Y | X | Y | X |
| Z56786758 | Y | X | Y | X | X | X |
*Interactions rendered by the ligands with catalytic residues (Y indicates “yes”and X indicates “no”).
Fig. 4.
Docked complexes (2D maestro view): All the ligands are found to interact with active site with many variety of favourable interactions.
Fig. 5.
Superposition of all identified compounds at the active site of NSP15. All the compounds found to fit well with the catalytic site.
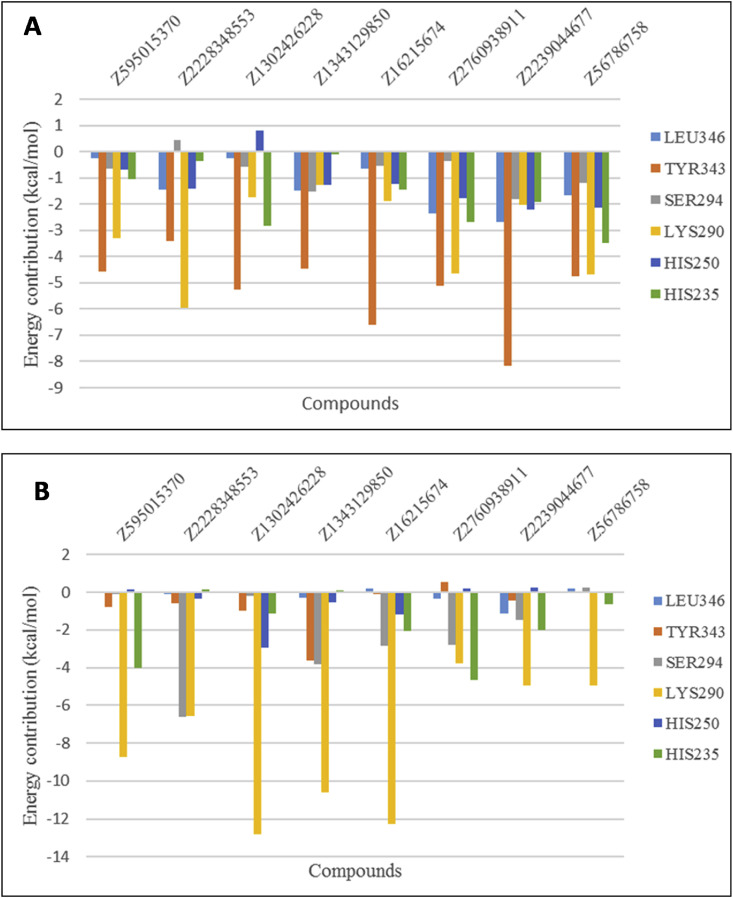
The compound Z595015370 which has a docking score of −10.50 kcal/mol and glide energy of −51.37 kcal/mol establishes many distinct types of interactions with NSP15 and hence would be expected to exhibit tight binding. To understand the binding free energy of docked complexes and contributions, per residual decomposition energy analysis was performed and it is found that many active site residues significantly contribute through van der Waals and electrostatic interactions in addition to hydrogen bonding interactions for favorable binding (Fig. 6 ).
Fig. 6.
Per residual decomposed energy analysis of identified lead compounds. (A) van der Waals (B) Electrostatics.
Endoribonuclease (NSP15) of SARS, is studied in detail and a triad of His-His-Lys (corresponding to His235, His250 and Lys290 of COVID-19) is implicated in catalytic activity and many residues near the active site environment are reported to perform the recycling of hexamers from monomers or trimers. Mutational analyses on these residues make NSP15 unable to associate to exhibit hydrolysis activity. Also, H235A, H250A and K290A mutants are reported to severely reduce the endoribonuclease activity. In the case of MERS-CoV NSP15 also, Tyr339 (Tyr343 in COVID-19) mutant (Y339A) are shown to have decreased the activity [24,25].
4. Conclusion
In this work, with the urgency to address the recent global COVID-19 pandemic (as WHO's declaration) and the high demand for new drug candidates for treating the viral infection, an important endoribonuclease which is reported very recently is considered for structure-based drug design efforts. To make screening effective and quick, compounds with putative antiviral activities were retrieved from the enamine database and hierarchical filtering was used to shortlist compounds that show tight binding at the active site of NSP15. Identified compounds show favorable drug-like properties which include solubility. From docking procedures, the apparent interactions seem to compare well with expectations of good inhibitor active site interactions. Hence, these compounds may be useful to inhibit NSP15 endoribonuclease activity and in turn arrest virus replication. In addition, since these compounds are already synthesized, they can be considered readily for further studies.
Disclosure statement
The authors declare that they have no conflict of interests.
Data availability statement
“Data available on request from the authors”.
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Author A.D. thanks Indian Council of Medical Research (ICMR), India for supporting his research by awarding Senior Research Fellowship. The authors also extend sincere thanks to Dr. Eric Anderson, Ph.D., Head, Peptide Centric Mass Spectrometry, The National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD 20892, USA for his help with language correction and for providing helpful suggestions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2020.100392.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Song H.-D., Tu C.-C., Zhang G.-W., Wang S.-Y., Zheng K., Lei L.-C. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci Unit States Am. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rahaman J., Siltberg-Liberles J. Avoiding regions symptomatic of conformational and functional flexibility to identify antiviral targets in current and future coronaviruses. Genome Biol Evol. 2016;8:3471–3484. doi: 10.1093/gbe/evw246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao S., Cao P., Chong M.K., Gao D., Lou Y., Ran J. The time-varying serial interval of the coronavirus disease (COVID-19) and its gender-specific difference: a data-driven analysis using public surveillance data in Hong Kong and Shenzhen, China from January 10 to February 15, 2020. Infect Control Hosp Epidemiol. 2020:1–8. doi: 10.1017/ice.2020.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benítez-Cardoza C.G., Vique-Sánchez J.L. Potential inhibitors of the interaction between ACE2 and SARS-CoV-2 (RBD), to develop a drug. Life Sci. 2020;256 doi: 10.1016/j.lfs.2020.117970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandra A., Gurjar V., Qamar I., Singh N. Identification of potential inhibitors of SARS-COV-2 endoribonuclease (EndoU) from FDA approved drugs: a drug repurposing approach to find therapeutics for COVID-19. J Biomol Struct Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1775127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choudhury A., Mukherjee S. In silico studies on the comparative characterization of the interactions of SARSCoV2 spike glycoprotein with ACE2 receptor homologs and human TLRs. J Med Virol. 2020:1–9. doi: 10.1002/jmv.25987. jmv.25987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enmozhi S.K., Raja K., Sebastine I., Joseph J. Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: an in silico approach. J Biomol Struct Dyn. 2020:1–7. doi: 10.1080/07391102.2020.1760136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng S., Luan X., Wang Y., Wang H., Zhang Z., Wang Y. Eltrombopag is a potential target for drug intervention in SARS-CoV-2 spike protein. Infect Genet Evol. 2020 doi: 10.1016/j.meegid.2020.104419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta M.K., Vemula S., Donde R., Gouda G., Behera L., Vadde R. In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. J Biomol Struct Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1751300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hage-Melim Li da S., Federico L.B., de Oliveira N.K.S., Francisco V.C.C., Correia L.C., de Lima H.B. Virtual screening, ADME/Tox predictions and the drug repurposing concept for future use of old drugs against the COVID-19. Life Sci. 2020;256:117963. doi: 10.1016/j.lfs.2020.117963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J., Song W., Huang H., Sun Q. Pharmacological therapeutics targeting RNA-dependent RNA polymerase, proteinase and spike protein: from mechanistic studies to clinical trials for COVID-19. J Clin Med. 2020;9:1131. doi: 10.3390/jcm9041131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iftikhar H., Ali H.N., Farooq S., Naveed H., Shahzad-ul-Hussan S. Identification of potential inhibitors of three key enzymes of SARS-CoV2 using computational approach. Comput Biol Med. 2020;122 doi: 10.1016/j.compbiomed.2020.103848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah B., Modi P., Sagar S.R. In silico studies on therapeutic agents for COVID-19: drug repurposing approach. Life Sci. 2020;252 doi: 10.1016/j.lfs.2020.117652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islam R., Parves M.R., Paul A.S., Uddin N., Rahman M.S., Mamun A Al. A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. J Biomol Struct Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1761883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Báez-Santos Y.M., St John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon David E., Jang Gwendolyn M., Bouhaddou Mehdi, Xu Jiewei, Obernier Kirsten, O’Meara Matthew J., Guo Jeffrey Z., Swaney Danielle L., Tummino Tia A., Hüttenhain Ruth, Kaake Robyn M., Richards Alicia L., Tutuncuoglu Beril, Foussard Helene, Batra Jyoti, Krogan N.J. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gouet P. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Youngchang Kim, Robert Jedrzejczak, Natalia I. Maltseva ME, Adam Godzik, K M and A J. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2 2020:1–18. [DOI] [PMC free article] [PubMed]
- 20.Madhavi Sastry G., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 21.Schrödinger Release 2014-1 . Epik, Schrödinger, LLC; New York, NY: 2014. Protein preparation Wizard. ; Impact, Schrödinger, LLC, New York, NY, 2014; Prime, Schrödinger, LLC, New York, NY. 2014. [Google Scholar]
- 22.Schrödinger Release . LigPrep, Schrödinger, LLC; New York, NY: 2020. [n.d] [Google Scholar]
- 23.Glide Schrödinger. Schrödinger, LLC; New York, NY: 2014. Version 6.0. [n.d] [Google Scholar]
- 24.Deng X., Hackbart M., Mettelman R.C., O'Brien A., Mielech A.M., Yi G. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci U S A. 2017;114:E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L., Li L., Yan L., Ming Z., Jia Z., Lou Z. Structural and biochemical characterization of endoribonuclease Nsp15 encoded by Middle East respiratory syndrome coronavirus. J Virol. 2018;92 doi: 10.1128/jvi.00893-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guarino L.A., Bhardwaj K., Dong W., Sun J., Holzenburg A., Kao C. Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. J Mol Biol. 2005;353:1106–1117. doi: 10.1016/j.jmb.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
“Data available on request from the authors”.
The data that support the findings of this study are available from the corresponding author upon reasonable request.