Abstract
Chaenomeles (family Rosaceae) is a genus of five diploid species of deciduous spiny shrubs that are native to Central Asia and Japan. It is an important horticultural crop (commonly known as flowering quinces) in Europe and Asia for its high yield in fruits that are rich in juice, aroma, and dietary fiber. Therefore, the development of effective genetic markers of Chaenomeles species is advantageous for crop improvement through breeding and selection. In this study, we successfully assembled and analyzed the chloroplast genome of five Chaenomeles species. The chloroplast genomes of the five Chaenomeles species were very similar with no structural or content rearrangements among them. The chloroplast genomes ranged from 159,436 to 160,040 bp in length and contained a total of 112 unique genes, including 78 protein-coding genes, 30 tRNAs, and 4 rRNAs. Three highly variable regions, including trnR-atpA, trnL-F, and rpl32-ccsA, were identified. Phylogenetic analysis based on the complete chloroplast genome showed that Chaenomeles forms a monophyletic clade and had a close relationship with the genera Docynia and Malus. Analyses for phylogenetic relationships and the development of available genetic markers in future could provide valuable information regarding genetics and breeding mechanisms of the Chaenomeles species.
Subject terms: Phylogenetics, Plant evolution
Introduction
The genus Chaenomeles Lindley belongs to the tribe Maleae and is an ecologically and economically important part of the Rosaceae family1. Chaenomeles is closely related to the well-known fruit crop genera Cydonia (quince), Malus (apple), and Pyrus (pear). It comprises of five diploid (2n = 34) species: one species is endemic to Japan, and four originate from central Asia. Cultivation Chaenomeles plants as horticultural crops has been initiated in Europe and in Asia2,3.
Chaenomeles japonica (Thunb.) Lindl. ex Spach (Japanese quince) is a dwarf shrub that grows in central and south Japan, and is strongly self-incompatible that encourages outcrossing4. C. speciosa (Sweet) Nakai (flowering quince) is a large shrub (2–5 m) that grows at an altitude of 200–1,700 m in central and southern China, Tibet and Burma, and is traditionally used in medicines5. C. cathayensis (Hemsl.) Schneider (Chinese quince) is a large shrub or small tree (up to 6 m) that grows at an altitude of 900–2,500 m in southern China, Bhutan and Burma. C. speciosa and C. cathayensis are sympatric in the province of Yunnan, China6. C. thibetica Yü (Tibetan quince), is a large shrub that grows in Tibet and western Sichuan6. C. sinensis (Dum.Cours.) Koehne, also referred as Pseudocydonia sinensis (Chinese quince), is a shrub or small tree (5–10 m) that grows in central and southern China. Three of these species (C. cathayensis, C. japonica, and C. speciosa) have been used to create several interspecific hybrids for approximately 400 years, resulting in more than 500 cultivars3, with the aim of developing new ornamental cultivars.
There were less genetic information of Chaenomeles in public database. Understanding the genetic diversity among and within wild populations of Chaenomeles was effective for plant breeding and the development of ex situ conservation strategies for plant genetic resources. Isozymes, RAPDs, and several chloroplast genome markers have been used in population genetics studies2,7–9. However, these markers have low variation and reproducibility. Therefore, there is need to develop effective genetic markers to facilitate the identification, conservation, utilization and breeding of Chaenomeles species.
The chloroplast genome has a stable structure that conserves the size and gene content10. The chloroplast genome of most angiosperm plants is composed of two inverted repeats (IR), which separate the large (LSC) and the small (SSC) single copy regions. Furthermore, the size of a typical angiosperm chloroplast genome ranges from 115 to 165 kb and contain 110–130 genes, with about eighty protein-coding genes, four rRNA genes and thirty tRNA genes11,12. Complete chloroplast genome sequences have been widely used as a source of valuable data for understanding evolutionary biology13–16. For example, chloroplast genome data have been used extensively for plant phylogenetic analyses at family/genus/species levels and DNA chloroplast barcoding for accurate identification of plant species17–19. The development of DNA sequencing technology has resulted in the extensive use of chloroplast genomes for species identification and molecular phylogenetic studies.
In this study, we sequenced the chloroplast genome for the five species of Chaenomeles and a closely related species, Docynia delavayi. The specific aims of this study were to (1) understand the conservation and diversity of Chaenomeles chloroplast genome through comparative genomic approaches; (2) identify the most variable regions of these chloroplast genomes as DNA barcodes for future species identification and phylogeny studies for the species and genera of Rosaceae; and (3) determine their phylogenetic relationships using the chloroplast genome sequence data.
Materials and methods
Plant materials and DNA extraction
Fresh young leaves of C. cathayensis, C. japonica, and C. sinensis from Beijing Botanical Garden, Beijing (China), C. thibetica from Bomê County, Tibet (China), and D. delavayi from Kunming Institute of Botany, Yunnan (China) were obtained and subsequently dried with silica gel. Voucher specimens were deposited at the PE herbarium of the Institute of Botany, Chinese Academy of Sciences. The species’ DNA was extracted with a DNeasy Plant Mini Kit (Qiagen Co., Germany). The quality and quantity of the genomic DNA were measured on 1% agarose gel and by using a Thermo Scientific NanoDrop.
Illumina sequencing, assembly, and annotation
Purified DNA was used to generate short-insert (350 bp) paired-end sequencing libraries according to the Illumina standard protocol. The entire genome sequencing was carried out using a HiSeq X Ten system (Novogene, Beijing). Approximately 5 GB of raw data were generated from each genome with 150 bp paired-end read lengths.
Low-quality reads and adapters were filtered from the raw data by using Trimmomatic20. The clean paired-end reads were qualitatively assessed and assembled with SPAdes 3.6.121. The contigs were then checked using BLAST searches against the available complete chloroplast sequence of C. speciosa (KT932965). The relative position and direction of each contig were manually adjusted with Sequencher 5.4.5 according to the reference genome. Chloroplast genome annotation was performed with Plann22 using the C. speciosa reference sequence from Genbank. The annotated chloroplast genome sequences were submitted to GenBank under accession numbers MN506259–MN506262, and MN506264. A gene map of the annotated Chaenomeles chloroplast genome was drawn online using OGdraw23.
Genome comparison
To investigate the divergence in the chloroplast genome, the identity across the whole complete chloroplast (cp) genome was visualized using the mVISTA program for the five species, with the C. speciosa genome from GenBank as the reference. Default parameters were utilized to align the chloroplast genomes in Shuffle-LAGAN mode, and a sequence conservation profile was visualized using a mVISTA plot24. Any large structural events, such as gene order rearrangements and IR expansions/contractions, were recorded.
All five plant species’ chloroplast genomes were aligned using MAFFT v725, followed by an adjustment with Se-Al 2.026. To elucidate the level of sequence variation, SNP variation and k2p-distance among Chaenomeles chloroplast genomes were calculated using MEGA 6.0 software27.
To explore the diverging hotspot regions in Chaenomeles species and facilitate their utilization in identification, sliding window analysis was conducted to generate the nucleotide diversity of the chloroplast genome using the DnaSP v5.10 software28. The step size was set to 200 bp, with an 800 bp window length.
Analysis of tandem repeats and single sequence repeats
The REPuter program29 was used to identify repeats: forward, reverse, palindrome, and complement sequences. The following settings for repeat identification were used: (1) hamming distance equal to 3; (2) minimal repeat size set to 30 bp; and (3) maximum computed repeats set to 90 bp. Tandem repeats were identified using the web-based Tandem Repeats Finder (https://tandem.bu.edu/trf/trf.html), with 2, 7, and 7 set for the alignment parameters match, mismatch, and indel, respectively. Simple sequence repeats (SSRs) were detected using GMAT30 with thresholds of ten repeat units for mononucleotide SSRs, five repeat units for dinucleotide SSRs, four repeat units for trinucleotide SSRs, and three repeat units for tetra-, penta-, and hexa-nucleotide SSRs.
Phylogenetic reconstruction
We downloaded 28 published chloroplast genomes of Maleae from Genbank that were included in the analyses as the outgroup taxa to perform the phylogenetic reconstruction. A total of 34 chloroplast genomes were aligned using MAFFT v725. The gaps in the alignment were stripped. Phylogenetic trees were constructed using maximum likelihood (ML) and Bayesian analysis (BI) methods. The phylogenetic analyses used the best-fitting models of nucleotide substitution selected in ModelFinder31 under the Bayesian information criterion. The maximum likelihood (ML) analyses were performed in RAxML v.8.1.2432. The support branches (BS) were assessed with 1,000 rapid bootstrapping replicates. Bayesian inference was performed using MrBayes v3.2.233. The Markov chain Monte Carlo (MCMC) analysis was run for 2 × 5,000,000 generations. The fist 25% of the trees corresponding to the “burn-in” period were discarded, and the remaining tree parts were used to construct the majority-rule consensus tree. The stationarity series was considered to be reached when the average standard deviation of the split frequencies remained < 0.01.
Results and discussion
Chloroplast genomes features of Chaenomeles species
After Illumina paired-end sequencing, 25,235,314–28,277,676 reads were obtained for the five Chaenomeles species. Through de novo assembly, contig selection and second reference based assembly were then generated for the five complete chloroplast genomes. The assembled chloroplast genome of the five examined species had a high coverage depth of about 3,000×.
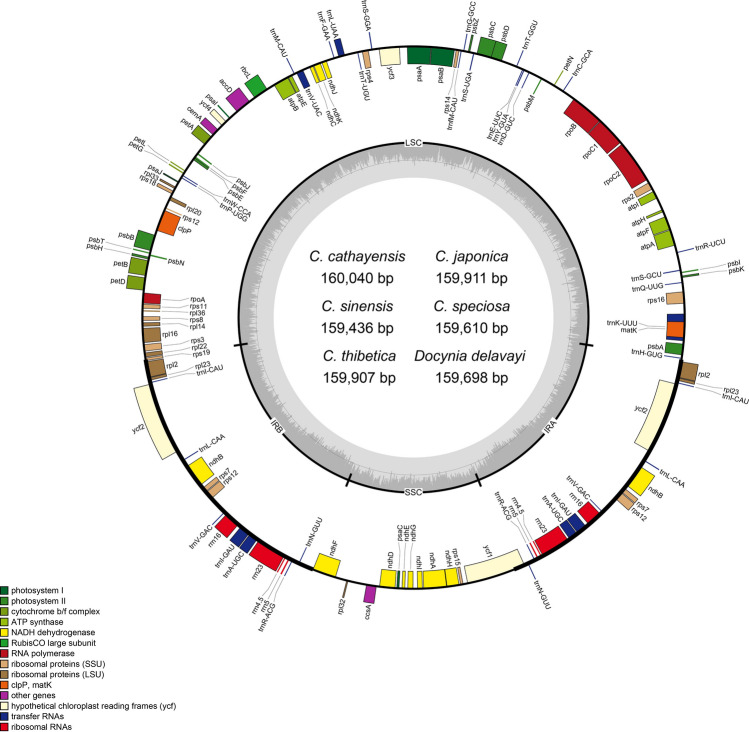
The complete chloroplast genomes of the five Chaenomeles species ranged from 159,436 bp (C. sinensis) to 160,040 bp (C. cathayensis) in length. All of the Chaenomeles chloroplast genomes displayed the typical quadripartite structure of angiosperm cpDNA (Fig. 1, Table 1), which consists of a pair of IR regions (26,300–26,393 bp) separated by a LSC region (87,476–87,937 bp) and a SSC region (19,229–19,345 bp). The overall guanine-cytosine (GC) content was 36.5–36.7%, indicating nearly identical levels among the Chaenomeles chloroplast genomes. GC content in the LSC, SSC and IR regions were 34.3–34.4%, 30.2–30.5% and 42.6–42.7%, respectively. The high GC content in the IR regions is due to the reduced presence of AT nucleotides in the four duplicate rRNA genes (rrn16, rrn23, rrn4.5, and rrn5). The GC content of the Chaenomeles chloroplast genome is close to that reported for other Rosaceae chloroplast genomes34,35. The Chaenomeles chloroplast genomes were compared to previously published data and showed highly similarity in genome structure34,36,37. With regard to the genome size, the length of complete chloroplast genome varies from 147 to 163 kb across Rosaceae34. The main reason for variation in genome length was expansions and contractions in IR regions and intergenic regions. In Chaenomeles, the junctions of IR and LSC or SSC have less variations, and exhibit the typical Rosaceae genome structure36.
Figure 1.
Chloroplast genomes of Chaenomeles. Genes on the inside are transcribed in a clockwise direction, while genes on the outside are transcribed in a counterclockwise direction.
Table 1.
Summary chloroplast genome features of five Chaenomeles species and Docynia delavayi.
| Gene features | C. cathayensis | C. japonica | C. sinensis | C. speciosa | C. thibetica | Docynia delavayi |
|---|---|---|---|---|---|---|
| Accession number in Genbank | MN506260 | MN506261 | MN506262 | KT932965 | MN506264 | MN506259 |
| Total cpDNA size (bp) | 160,040 | 159,911 | 159,436 | 159,610 | 159,907 | 159,698 |
| LSC length (bp) | 87,937 | 87,814 | 87,476 | 87,781 | 87,851 | 87,804 |
| SSC length (bp) | 19,345 | 19,311 | 19,246 | 19,229 | 19,298 | 19,156 |
| IR length (bp) | 26,379 | 26,393 | 26,357 | 26,300 | 26,379 | 26,369 |
| Total GC content (%) | 36.5 | 36.6 | 36.7 | 36.6 | 36.6 | 36.6 |
| LSC GC content (%) | 34.3 | 34.3 | 34.4 | 34.3 | 34.3 | 34.3 |
| SSC GC content (%) | 30.2 | 30.3 | 30.5 | 30.3 | 30.3 | 30.4 |
| IR GC content (%) | 42.6 | 42.6 | 42.7 | 42.6 | 42.6 | 42.7 |
| Total number of genes | 112 | 112 | 112 | 112 | 112 | 112 |
| Protein-coding genes | 78 | 78 | 78 | 78 | 78 | 78 |
| rRNA genes | 4 | 4 | 4 | 4 | 4 | 4 |
| tRNA genes | 30 | 30 | 30 | 30 | 30 | 30 |
The Chaenomeles chloroplast genome contained a total of 112 unique genes, including 78 protein-coding genes, 30 tRNAs, and four rRNAs. Nineteen genes were duplicated in the IR, including eight protein-coding genes, seven tRNA genes and four rRNA genes. Fifteen distinct genes had a single intron, and two genes (ycf3 and clpP) had two introns. The rps12 gene is trans-spliced with the 3′exon being duplicated in the IR, while the 5′ end is located at the LSC region. TrnK-UUU had the largest intron (2,561–2,570 bp) containing the matK gene. Compared with other Rosaceae chloroplast genomes, the number of unique genes were conserved35,36,38,39, and no significant changes of gene orders were found in the Chaenomeles.
Comparative analysis of the Chaenomeles plastomes
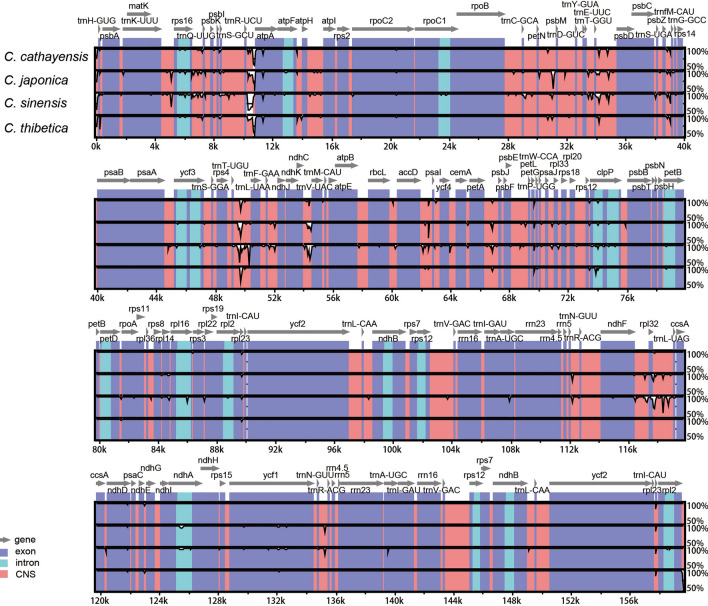
The mVISTA program was used to analyze the overall sequence identity of the chloroplast genome of the five Chaenomeles species, using the annotation for C. speciosa as a reference (Fig. 2). The Chaenomeles chloroplast genome displayed similar structure and gene order. The divergence level of the non-coding regions was higher than that of the coding regions. In addition, LSC and SSC regions had a larger divergence than the IR regions. A higher sequence divergence was found in the single copy regions than in the IRs and in the non-coding regions than in the coding regions, which is in accordance with the results found for other taxa40–42.
Figure 2.
Identity plot comparing the chloroplast genomes of the five Chaenomeles species, using C. speciosa as a reference sequence. Genome regions are color coded as protein-coding, rRNA, tRNA, intron, and conserved non-coding sequences (CNS).
To further understand the chloroplast genome sequence divergence among Chaenomeles species, the number of nucleotide substitutions and sequence k2p-distances were designated to represent the level of divergence. The five Chaenomeles chloroplast genomes were fully aligned, giving an alignment matrix of 161,903 bp; 843 variable sites and 147 information sites were found. The number of nucleotide substitutions in pairwise comparisons between the five species ranged from 63 to 695, and the k2p-distances ranged from 0.0004 to 0.0041 (Table 2). The lowest sequence divergence was between C. cathayensis and C. thibetica, while the highest sequence divergence occurred between C. sinensis and C. speciosa.
Table 2.
Numbers of nucleotide substitutions and sequence distances in five Chaenomeles complete chloroplast genomes.
| C. cathayensis | C. japonica | C. sinensis | C. speciosa | C. thibetica | |
|---|---|---|---|---|---|
| C. cathayensis | 282 | 633 | 695 | 63 | |
| C. japonica | 0.0018 | 588 | 304 | 284 | |
| C. sinensis | 0.0040 | 0.0037 | 651 | 631 | |
| C. speciosa | 0.0006 | 0.0019 | 0.0041 | 103 | |
| C. thibetica | 0.0004 | 0.0018 | 0.0040 | 0.0006 |
The upper triangle shows the number of nucleotide substitutions. The lower triangle indicates the number of sequence distances in complete chloroplast genomes.
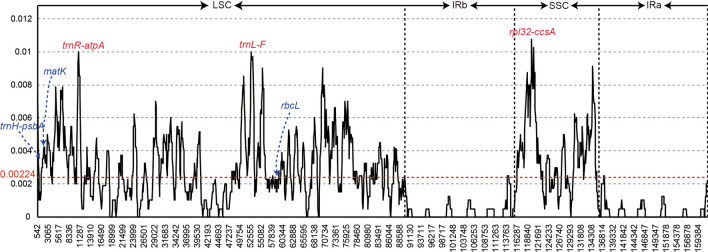
To identify the sequence divergence hotspots, the nucleotide diversity (pi) value within the slide window of 600 bp was calculated (Fig. 3). The pi value in the windows varied from 0 to 0.01075, with a mean of 0.00224. Three highly variable regions (pi > 0.01), including trnR-atpA, trnL-F, and rpl32-ccsA, were identified in the Chaenomeles chloroplast genomes. Among these regions, trnR-atpA and trnL-F were located in the LSC region, and rpl32-ccsA was in the SSC region. All nucleotide diversity values in the IR regions were less than 0.003 and no highly divergent sequences were found; therefore, these regions were considered to be conserved, whereas the universal DNA barcodes (matK, rbcL and trnH-psbA) had lower pi values. We compared these three highly variable markers in more detail (Table 3). The aligned length of the markers ranged from 816 bp for trnL-F to 1,603 bp for rpl32-ccsA. rpl32-ccsA showed the highest number of variable and informative sites. The average nucleotide diversity of the three rapidly evolving regions was 0.00986, which was 2.6 times higher than that of the universal DNA barcodes. The pi values of these regions showed 0.00373 (Table 3).
Figure 3.
Nucleotide diversity of the Chaenomeles chloroplast genomes.
Table 3.
Variability of nine variable markers and universal chloroplast DNA barcodes (rbcL and matK) in Chaenomeles.
| Markers | Length | Variable sites | Information sites | Nucleotide diversity | ||
|---|---|---|---|---|---|---|
| Numbers | % | Numbers | % | |||
| trnR-atpA | 1,142 | 26 | 2.28 | 3 | 0.26 | 0.01000 |
| trnL-F | 816 | 18 | 2.21 | 4 | 0.49 | 0.01000 |
| rpl32-ccsA | 1603 | 30 | 1.87 | 5 | 0.31 | 0.00969 |
| trnR-atpA+trnL-F+rpl32-ccsA | 3,561 | 74 | 2.08 | 12 | 0.34 | 0.00986 |
| rbcL | 1,427 | 9 | 0.63 | 0 | 0.00 | 0.00273 |
| matK | 1512 | 14 | 0.93 | 3 | 0.20 | 0.00410 |
| trnH-psbA | 354 | 4 | 1.13 | 2 | 0.56 | 0.00669 |
| rbcL+matK+trnH-psbA | 3,293 | 27 | 0.82 | 5 | 0.15 | 0.00373 |
Chloroplast genome markers are extensively used in plant phylogenetic studies to analyze relatedness and classify species. Some universal chloroplast regions, such as rbcL, matK, ndhF, trnH-psbA, psbK-psbI, and atpB-rbcL, have been used as markers in phylogenetic studies13,43–45. However, an increased number of studies have shown that it is inappropriate to use the universal markers to classify closely related species as they have lower variability. Comparative chloroplast genome analysis was a new strategy to identify the mutation hotspot markers13,41. The intergenic spacer trnL-F have a long time of use in plant phylogenetic and species identification studies46,47. In some groups this region often contains ploy A and T structures43 and affect sequence quality. The rpl32-ccsA marker includes two intergenic spacers (rpl32-trnL and trnL-ccsA) in the SSC region. More papers showed this region had higher variable sites44. trnR-atpA is less commonly used to reconstruct phylogenetic relationships or as DNA barcode. The highly variable makers discovered in this study could be regarded as potential molecular resources for species identification and applied in phylogenetic analyses of Rosaceae.
Analysis of repeat elements
Repetitive sequences in the chloroplast genome play an important key role in the genome rearrangement and stabilization, and they provide important information for understanding the evolutionary history of plant species and sequence divergence48–50. SSRs or microsatellites, and dispersed long repeats were the two main motifs in the chloroplast genomes.
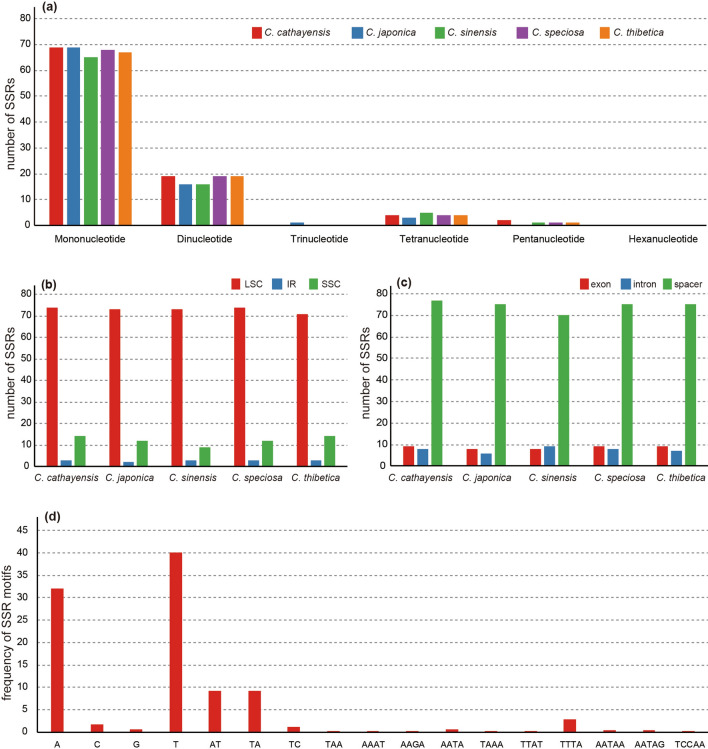
SSRs are important co-dominant molecular markers for evaluating germplasm, establishing phylogenetic and evolutionary relationships51, and they are widely present in the chloroplast genome41. Using GMAT analysis, mono-, di-, trin-, tetra-, penta-, and hexa-nucleotide SSRs were detected in every species, and each Chaenomeles chloroplast genome was found to contain 91 (C. thibetica) to 94 (C. cathayensis) SSRs. The number of SSRs are slightly lower than those reported in previous Rosaceae chloroplast genome studies with Hagenia (172)36 and Rubus (116)37.
All five Chaenomeles chloroplast genome had five types of SSRs, excluding the hexanucleotide SSR (Fig. 4a). In the five species examined, most of the SSRs were mononucleotide SSR (73.40%, 77.53%, 74.71%, 73.91%, and 73.63% in C. cathayensis, C. japonica, C. sinensis, C. speciosa, C. thibetica, respectively). SSRs were distributed more widely throughout the chloroplast genomes, and were usually located in the LSC regions (78.02–83.90%, Fig. 4b). Most of the SSRs were found in spacer regions (80.46–84.27%, Fig. 4c); only a few were located in the coding regions.
Figure 4.
SSR loci analysis of five Chaenomeles. (a) Number of different SSRs types; (b) number of SSRs in spacer, exon, and intron; (c) number of SSRs in LSC, SSC, and IR regions; (d) frequency of identified SSR motifs in the different repeat classes.
Almost all of the mononucleotide repeat sequences were comprised of A/T repeats (72.19%). Meanwhile, AT/TA repeats were the most common among dinucleotide SSRs (94.38%). In addition, two pentanucleotide repeats (AATAG and AATAG) were found in C. cathayensis and one in C. speciosa (AATAA), C. sinensis (TCCAA), and C. thibetica (AATAG) using our search criterion (Fig. 4d). In general, chloroplast genome sequences are highly conserved at the genus level, and in silico development of SSRs in chloroplast genomes has supported them as potentially transferable markers among species52,53. In addition SSRs are highly polymorphic and have been potential markers for establishing molecular evolutionary histories and demographic diversity54,55.
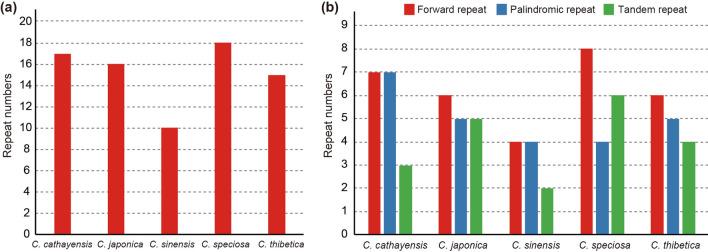
We classified sequence dispersed repeat motifs into five categories: forward, reverse, palindrome, complement and tandem repeats. In the Chaenomeles chloroplast genome, we identified three repeat motifs (Fig. 5). In general, the forward repeats were the most common, except for C. cathayensis and C. sinensis that had as many forward repeats as palindromic repeats. In total, 76 repeats with more than 30 bp were detected in the five Chaenomeles chloroplast genome. C. speciosa contained the most repeats (18) compared to the other four species (10, 15, 16, and 17 repeats, respectively, Fig. 5). The majority of repeats (63.16%) ranged in size from 31 to 35 bp. The longest repeat was a forward repeat of 82 bp in C. japonica. Repeat sequences are considered to play an important role in genome recombination and rearrangement and also have phylogenetic information in some groups50,56.
Figure 5.
Long repeat sequences in the chloroplast genomes of five Chaenomeles. (a) Number of repeats; (b) Number of different repeats types.
Phylogenomic analysis
Chloroplast genomes contain an abundance of phylogenetic information, which has been widely used for phylogeny reconstruction at different taxonomic levels, such as order, family, genus, and species in plants. Using chloroplast genome data, long-standing controversies related to various phylogenetically difficult groups have been resolved, supporting its importance in systematic studies.
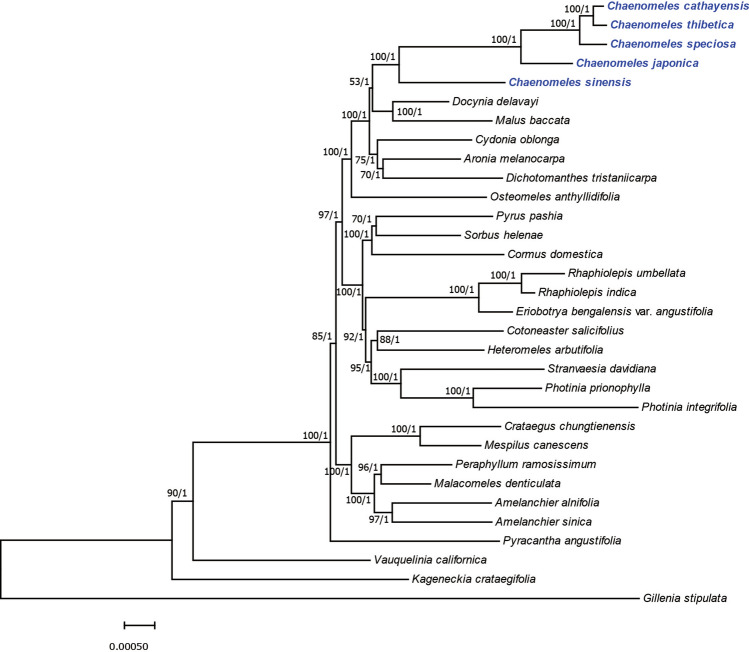
To better determine the phylogenetic position of Chaenomeles and further clarify the evolutionary relationships within the tribe Maleae, phylogenetic analyses was constructed based on the 32 Maleae complete chloroplast genomes, using Gillenia stipulata as an outgroup. The phylogenetic topologies of the ML and BI method were similar (Fig. 6), and most nodes were supported by high values (> 95%). However, some internal nodes tended to have poorer bootstrap support, indicating rapid radiation and/or incomplete lineage sorting. Chaenomeles was observed to be a sister lineage of Docynia and Malus based on low bootstrap support and posterior probability values (ML bootstrap support, BS = 53; posterior probability, PP = 1). The monophyly of Chaenomeles was strongly supported (BS = 100%, PP = 1). C. sinensis was the basal species in Chaenomeles. C. sinensis was once treated as a monotypic genera Pseudocydonia57. However, the morphological data and several chloroplast markers and ITS data57,58 did not support Pseudocydonia separated out of Chaenomeles. The result indicates the necessity of revising taxonomic boundaries of Chaenomeles and redefining taxonomic status of C. sinensis. The chloroplast genome phylogeny showed C. cathayensis was a sister species of C. thibetica. This results was congruent with Bartish et al.’s results, which recognized that C. thibetica appeared to be rather closely related to C. cathayensis using RAPDs and isozymes methods.
Figure 6.
Phylogenetic tree reconstruction of 32 taxa using maximum likelihood and Bayesian inference methods based on the complete chloroplast genome sequences. Number of the branches indicate ML bootstrap support value/Bayesian posterior probability.
Conclusions
In this study, we sequenced the total chloroplast genome of five Chaenomeles species by de novo sequencing, and showed that the chloroplast genome structure is well conserved throughout the genus. The comparative analyses revealed extremely low levels of sequence variability. However, repeat sequences, SSRs, and highly polymorphic regions were identified to be suitable for possible genetic markers. These markers could be considered for phylogenetic analysis and to resolve taxonomical discrepancies in Chaenomeles and potentially in other Rosaceae. Phylogenetic reconstruction based on the complete chloroplast genomes revealed the relationships among the five species of Chaenomeles. In summary, this study will be helpful for further research on the molecular evolution and speciation of this genus.
Supplementary information
Acknowledgements
This research was funded by Fundamental Research Funds for the Central Public Welfare Research Institutes, Grant number ZZXT201802.
Author contributions
Q.Y., L.G. and L.H. designed the experiment. Y.W. and Y.L collected samples and performed the experiment. J. S and Y.W. analyzed the data. J.S. and C.X. drafted the manuscript. All of the authors have read and approved the final manuscript.
Data availability
The complete chloroplast sequence generated and analyzed during the current study are available in GenBank (MN506259–MN506262, and MN506264).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jiahui Sun and Yiheng Wang.
Contributor Information
Qingjun Yuan, Email: yuanqingjun@icmm.ac.cn.
Lanping Guo, Email: glp01@126.com.
Luqi Huang, Email: huangluqi01@126.com.
Supplementary information
is available for this paper at 10.1038/s41598-020-67943-1.
References
- 1.Potter D, et al. Phylogeny and classification of Rosaceae. Plant Syst. Evol. 2007;266:5–43. [Google Scholar]
- 2.Garkava LP, Rumpunen K, Bartish IV. Genetic relationships in Chaenomeles (Rosaceae) revealed by isozyme analysis. Sci. Hortic. 2000;85:21–35. [Google Scholar]
- 3.Weber C. The genus Chaenomeles (Rosaceae) J. Arnold Arbor. 1964;45:161–205. [Google Scholar]
- 4.Rumpunen, K., Kviklys, D., Kaufmane, E. & Garkava, L. 484 edn 211–216 (International Society for Horticultural Science (ISHS), Leuven, Belgium).
- 5.Xianfei X, Xiaoqiang C, Shunying Z, Guolin Z. Chemical composition and antimicrobial activity of essential oils of Chaenomeles speciosa from China. Food Chem. 2007;100:1312–1315. [Google Scholar]
- 6.Gu C, Spongberg SA. In: Flora of China. Zheng-yi W, Peter HR, De-yuan H, editors. Beijing: Science Press; 2003. pp. 171–173. [Google Scholar]
- 7.Bartish IV, Garkava LP, Rumpunen K, Nybom H. Phylogenetic relationships and differentiation among and within populations of Chaenomeles Lindl. (Rosaceae) estimated with RAPDs and isozymes. Theoret. Appl. Genet. 2000;101:554–563. [Google Scholar]
- 8.Bartish IV, Rumpunen K, Nybom H. Combined analyses of RAPDs, cpDNA and morphology demonstrate spontaneous hybridization in the plant genus Chaenomeles. Heredity. 2000;85:383–392. doi: 10.1046/j.1365-2540.2000.00773.x. [DOI] [PubMed] [Google Scholar]
- 9.Kaneko Y, Nagaho I, Bang SW, Matsuzawa Y. Classification of flowering quince cultivars (genus Chaenomeles) using random amplified polymorphic DNA markers. Breed. Sci. 2000;50:139–142. [Google Scholar]
- 10.Dong W, Xu C, Cheng T, Lin K, Zhou S. Sequencing angiosperm plastid genomes made easy: A complete set of universal primers and a case study on the phylogeny of Saxifragales. Genome Biol. Evol. 2013;5:989–997. doi: 10.1093/gbe/evt063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniell H, Lin C-S, Yu M, Chang W-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:1–29. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olejniczak SA, Lojewska E, Kowalczyk T, Sakowicz T. Chloroplasts: State of research and practical applications of plastome sequencing. Planta. 2016;244:517–527. doi: 10.1007/s00425-016-2551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong W, Liu J, Yu J, Wang L, Zhou S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS One. 2012;7:e35071. doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong W, et al. Resolving the systematic positions of enigmatic taxa: Manipulating the chloroplast genome data of Saxifragales. Mol. Phylogenet. Evol. 2018;126:321–330. doi: 10.1016/j.ympev.2018.04.033. [DOI] [PubMed] [Google Scholar]
- 15.Ha Y-H, Kim C, Choi K, Kim J-H. Molecular phylogeny and dating of Forsythieae (Oleaceae) provide insight into the Miocene history of Eurasian temperate shrubs. Front. Plant Sci. 2018;9:20. doi: 10.3389/fpls.2018.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lloyd Evans D, Joshi SV, Wang J. Whole chloroplast genome and gene locus phylogenies reveal the taxonomic placement and relationship of Tripidium (Panicoideae: Andropogoneae) to sugarcane. BMC Evol. Biol. 2019;19:33. doi: 10.1186/s12862-019-1356-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong W, et al. Phylogenetic resolution in Juglans based on complete chloroplast genomes and nuclear DNA sequences. Front. Plant Sci. 2017;8:1148. doi: 10.3389/fpls.2017.01148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ross TG, et al. Plastid phylogenomics and molecular evolution of Alismatales. Cladistics. 2016;32:160–178. doi: 10.1111/cla.12133. [DOI] [PubMed] [Google Scholar]
- 19.Burke SV, Ungerer MC, Duvall MR. Investigation of mitochondrial-derived plastome sequences in the Paspalum lineage (Panicoideae; Poaceae) BMC Plant Biol. 2018;18:152. doi: 10.1186/s12870-018-1379-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bankevich A, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang DI, Cronk QCB. Plann: A command-line application for annotating plastome sequences. Appl. Plant Sci. 2015;3:1500026. doi: 10.3732/apps.1500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greiner S, Lehwark P, Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47:W59–W64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rambaut, A. Se-Al: Sequence alignment editor. version 2.0 (1996).
- 27.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 29.Kurtz S, et al. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Wang L. GMATA: An integrated software package for genome-scale SSR mining, marker development and viewing. Front. Plant Sci. 2016;7:1350. doi: 10.3389/fpls.2016.01350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang SD, et al. Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol. 2017;214:1355–1367. doi: 10.1111/nph.14461. [DOI] [PubMed] [Google Scholar]
- 35.Li W, et al. Development of chloroplast genomic resources for Pyrus hopeiensis (Rosaceae) Conserv. Genet. Resour. 2017;10:511–513. [Google Scholar]
- 36.Gichira AW, et al. The complete chloroplast genome sequence of an endemic monotypic genus Hagenia (Rosaceae): Structural comparative analysis, gene content and microsatellite detection. Peer J. 2017;5:e2846. doi: 10.7717/peerj.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang JY, Pak J-H, Kim S-C. The complete plastome sequence of Rubus takesimensis endemic to Ulleung Island, Korea: Insights into molecular evolution of anagenetically derived species in Rubus (Rosaceae) Gene. 2018;668:221–228. doi: 10.1016/j.gene.2018.05.071. [DOI] [PubMed] [Google Scholar]
- 38.Cheng H, et al. The complete chloroplast genome sequence of strawberry (Fragaria × ananassa Duch.) and comparison with related species of Rosaceae. Peer J. 2017;5:e3919. doi: 10.7717/peerj.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bao L, Li K, Liu Z, Han M, Zhang D. Characterization of the complete chloroplast genome of the Chinese crabapple Malus prunifolia (Rosales: Rosaceae: Maloideae) Conserv. Genet. Resour. 2016;20:1–3. [Google Scholar]
- 40.Dong W, et al. A chloroplast genomic strategy for designing taxon specific DNA mini-barcodes: A case study on ginsengs. BMC Genet. 2014;15:138. doi: 10.1186/s12863-014-0138-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W, et al. Interspecific chloroplast genome sequence diversity and genomic resources in Diospyros. BMC Plant Biol. 2018;18:210. doi: 10.1186/s12870-018-1421-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zong D, et al. Plastome sequences help to resolve deep-level relationships of populus in the family salicaceae. Front. Plant Sci. 2019;10:5. doi: 10.3389/fpls.2019.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shaw J, et al. The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005;92:142–166. doi: 10.3732/ajb.92.1.142. [DOI] [PubMed] [Google Scholar]
- 44.Shaw J, et al. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: The tortoise and the hare IV. Am. J. Bot. 2014;101:1987–2004. doi: 10.3732/ajb.1400398. [DOI] [PubMed] [Google Scholar]
- 45.Zhou S, et al. How many species of bracken (Pteridium) are there? Assessing the Chinese brackens using molecular evidence. Taxon. 2014;63:509–521. [Google Scholar]
- 46.Wallander E, Albert VA. Phylogeny and classification of Oleaceae based on rps16 and trnL-F sequence data. Am. J. Bot. 2000;87:1827–1841. [PubMed] [Google Scholar]
- 47.Zhang C, et al. Different maternal genome donor to Kengyilia species inferred from chloroplast trnL-F sequences. Biol. Plant. 2009;53:759–763. [Google Scholar]
- 48.Wang Z, Weber JL, Zhong G, Tanksley SD. Survey of plant short tandem DNA repeats. Theoret. Appl. Genet. 1994;88:1–6. doi: 10.1007/BF00222386. [DOI] [PubMed] [Google Scholar]
- 49.Lin WH, Kussell E. Evolutionary pressures on simple sequence repeats in prokaryotic coding regions. Nucleic Acids Res. 2012;40:2399–2413. doi: 10.1093/nar/gkr1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Milligan BG, Hampton JN, Palmer JD. Dispersed repeats and structural reorganization in subclover chloroplast DNA. Mol. Biol. Evol. 1989;6:355–368. doi: 10.1093/oxfordjournals.molbev.a040558. [DOI] [PubMed] [Google Scholar]
- 51.Sablok G, et al. ChloroMitoSSRDB: Open source repository of perfect and imperfect repeats in organelle genomes for evolutionary genomics. DNA Res. 2013;20:20. doi: 10.1093/dnares/dss038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diekmann K, Hodkinson TR, Barth S. New chloroplast microsatellite markers suitable for assessing genetic diversity of Lolium perenne and other related grass species. Ann. Bot. 2012;110:1327–1339. doi: 10.1093/aob/mcs044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoll A, et al. Development of microsatellite markers and assembly of the plastid genome in Cistanthe longiscapa (Montiaceae) based on low-coverage whole genome sequencing. PLoS One. 2017;12:e0178402. doi: 10.1371/journal.pone.0178402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu D, Abe J, Gai J, Shimamoto Y. Diversity of chloroplast DNA SSRs in wild and cultivated soybeans: Evidence for multiple origins of cultivated soybean. Theoret. Appl. Genet. 2002;105:645–653. doi: 10.1007/s00122-002-0972-7. [DOI] [PubMed] [Google Scholar]
- 55.Mariotti R, Cultrera NGM, Díez CM, Baldoni L, Rubini A. Identification of new polymorphic regions and differentiation of cultivated olives (Olea europaea L.) through plastome sequence comparison. BMC Plant Biol. 2010;10:211. doi: 10.1186/1471-2229-10-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greiner S, et al. The complete nucleotide sequences of the five genetically distinct plastid genomes of Oenothera, subsection Oenothera: I. Sequence evaluation and plastome evolution. Nucleic Acids Res. 2008;36:2366–2378. doi: 10.1093/nar/gkn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aldasoro JJ, Aedo C, Navarro C. Phylogenetic and phytogeographical relationships in Maloideae (Rosaceae) based on morphological and anatomical characters. Blumea. 2005;50:3–32. [Google Scholar]
- 58.Lo EYY, Donoghue MJ. Expanded phylogenetic and dating analyses of the apples and their relatives (Pyreae, Rosaceae) Mol. Phylogenet. Evol. 2012;63:230–243. doi: 10.1016/j.ympev.2011.10.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete chloroplast sequence generated and analyzed during the current study are available in GenBank (MN506259–MN506262, and MN506264).