Graphical abstract

Keywords: RNA-seq, Gene expression, Machine learning, Cancer
Abstract
Tumor biopsy is one of the most widely used materials in cancer diagnoses and molecular studies, where the purity of the biopsies (i.e., proportion of cells that are cancerous) is crucial for both applications. However, conventional approaches for tumor biopsy purity evaluation require experienced pathologists and/or various materials/experiments therefore were time-consuming and error prone. Rapid, easy-to-perform and cost-effective methods are thus still of demand. Recent studies had demonstrated that molecular signatures were informative to this task. Previously, we had developed GeneCT, a deep learning-based cancerous status and tissue-of-origin classifier for pan-tumor/tissue biopsies. In the current work, we applied GeneCT on datasets collected from various groups, where the experimental protocols and cancer types differed from each other. We found that GeneCT showed high accuracies on most datasets; for samples with unexpected results, in-depth investigations suggested that they might suffer from imperfect purity. In silico mixture experiments further showed that GeneCT classification was highly indicative in predicting the purity of the tumor biopsies. Considering that transcriptome profiling is a common and inexpensive experiment in molecular cancer studies, our deep learning-based GeneCT could thus serve as a valuable tool for rapid, preliminary tumor biopsy purity assessment.
1. Introduction
In clinical medicine, tissue biopsy is a widely used technique for disease (especially cancer) diagnoses and monitoring. Moreover, tumor biopsy is also one of the most frequently used materials in cancer-related studies, thus the purity of the biopsies (i.e., the proportion of cells that are cancerous) is critical for experimental designs and result interpretations [1]. In fact, tumor biopsies contain cancerous cells as well as various types of non-cancerous cells, such as immune cells, fibroblasts, blood vessels and adjacent non-cancerous cells. Conventional approaches for purity evaluations of tumor biopsies require experienced pathologists and/or various materials/experiments/instruments, therefore were time-consuming and error prone. In addition, cancer metastasis is very common and biopsies from such cases are also of interests in various molecular studies, while it is challenging to correctly determine the tissue origin and purity of biopsy samples obtained from the metastatic lesions. Rapid, easy-to-perform and cost-effective methods for purity assessment of tumor biopsies are thus still of demand.
Recently, various computational approaches had been developed to investigate the purities of tumor biopsies. These methods had successfully utilized the molecular signatures, such as gene expressions (e.g., the ESTIMATE algorithm [2]), copy number aberrations (e.g., the ABSOLUTE algorithm [3]) and DNA methylations (e.g., the LUMP algorithm [1]), to either estimate the purity [4], [5], [6], [7], or decode the cell compositions of biopsy samples [8], [9], [10], [11], [12], [13], [14]. Despite the high accuracy and consistency demonstrated in these studies, however, the majority of these methods only focused on biopsy samples from the TCGA (The Cancer Genome Atlas) project [15] and very few had been validated outside the TCGA datasets. In addition, most of the previous methods heavily relied on tissue- and/or tumor-specific biomarkers, therefore their performances in handling “novel” tissue/tumor types that had not been investigated in the original studies were also unexplored. Moreover, except for the infiltrating immune cells and stromal cells, few methods had modelled the existence of adjacent non-cancerous tissue cells (e.g., hepatocytes in liver tumors). Contaminations of these non-cancerous cells were also common and of particular interest in cancer metastasis cases. As a result, molecular-based computational approaches for tumor purity estimations are still under active investigations.
We and others had previously shown that the expression profile alone could indicate the cancerous status (i.e., cancerous or not) and tissue origin of the biopsy samples. For instance, we had built a deep learning based classifier, GeneCT (Generalizable Cancerous-status and Tissue-of-origin classifier), which showed high accuracy on the TCGA pan-cancer datasets [16]. More importantly, unlike other methods for this task, GeneCT does not use any cancer/tissue-type specific biomarkers to build the classification models. Instead, we utilized the common oncogenes and tumor suppressor genes to build the cancerous classification model, and transcription factors to build the tissue-of-origin classification model [16]. Such unique characteristic of GeneCT made us to explore the possibility of GeneCT as a generalizable tool in estimating the purities of tissue/tumor biopsies. We reasoned that cancerous and non-cancerous samples could be viewed as tumor biopsies of high and low purities. To this end, in this study, we applied GeneCT on a list of datasets generated from various non-TCGA sources. Our result showed that GeneCT is highly generalizable and held the potential to handle transcriptome datasets generated by various protocols and cancer types. More interestingly, for datasets with unexpected prediction results, further molecular investigations suggested that the poor accuracy was possibly related to impurity of the samples, thus demonstrating the potential of GeneCT as a rapid, preliminary purity evaluation tool for tumor biopsies.
2. Materials and methods
2.1. Transcriptome data processing
RNA-seq (whole transcriptome sequencing) data from various sources were collected from the literature. Sample information, RNA extraction and library preparation methods were summarized in Supplementary Table S1. Briefly, 10 paired clear cell renal carcinoma (ccRCC) tumor and adjacent normal kidney tissues, 17 breast tumors of 3 subtypes and 3 normal breast organoid samples, 29 primary liver tumor with adjacent normal tissue and 20 portal vein tumor thrombosis tissues, 10 basal cell carcinomas (BCCs) and 8 normal skin tissues, 10 pancreatic cancer tumors from the primary site or lung metastasis, 71 acute myeloid leukemia (AML) samples from bone marrow or peripheral blood, 23 primary colon tumors with their adjacent normal as well as liver metastasis and 5 normal adjacent liver tissues were collected. The RNA-seq data was processed following TCGA’s analysis pipeline. Briefly, raw sequencing reads were firstly pre-processed to remove sequencing adaptors and low-quality cycles using Ktrim [17] with default parameters; then the pre-processed reads were mapped to the human reference genome (NCBI build 37/UCSC hg19) using MapSplice (v12.07) [18] with default parameters; then gene expression was quantified and normalized using RSEM (v1.1.13) [19] against the UCSC gene annotation [20] with default parameters. Detailed information of the TCGA RNA-seq data processing pipeline could be found at https://webshare.bioinf.unc.edu/public/mRNAseq_TCGA/UNC_mRNAseq_summary.pdf.
2.2. In silico mixture experiments
To quantitatively evaluate the performance and behaviour of GeneCT on tumor samples with different purity levels, in silico mixture experiments were performed using RNA-seq data from tumor and adjacent normal tissue samples. For each mixture experiment, a total of 20 million reads were generated according to a pre-set tumor fraction (ranged from 0% to 100%). For instance, if the tumor fraction was 30%, then 6 million reads would be extracted from the RNA-seq data of the tumor sample while the rest 14 million reads would be extracted from the normal sample. In addition, two batch of mixture experiments were performed: the first batch used a primary colon tumor sample and its adjacent normal colon tissue, while the second batch used a colon tumor liver metastasis sample and its adjacent normal liver tissue. The in silico mixed sequencing reads were analysed following the TCGA’s RNA-seq analysis pipeline as described before, then the quantified and normalized gene expression values were analysed by GeneCT to predict the cancerous status and tissue origin. Note that besides the qualitative prediction result, GeneCT also provides confidence scores for the classifications, where a value close to 1 means it is likely to be cancerous (the closer the value to 1, the higher the confidence), while a value close to 0 mans it is likely to be non-cancerous (the closer the value to 0, the higher the confidence).
2.3. Building classification models
The detailed information on model building of GeneCT could be found in our previous work. Briefly, pan-cancer RNA-seq data from 11 common cancer types (~5300 samples in total) were collected from TCGA and separated into training and testing datasets. Due to the much higher number of tumor than adjacent normal samples, we randomly selected half number of normal samples and equal number of tumor samples to form the training dataset (~500 samples) and all the resting samples as testing dataset (~4800 samples). Notably, we did not use any cancer/tissue-type specific biomarkers. Instead, we utilized known oncogenes/tumor suppressor genes and transcription factors that showed high variability (i.e., not expressed constantly) in the RNA-seq data to build cancerous status and tissue origin classification models (the gene list could be found in Supplementary Table S2). Using artificial neural network (ANN) approach and the expression values of the variable oncogenes/tumor suppressor genes and transcription factors, we built two models for cancerous status and tissue origin determination of the biopsy samples, respectively. A 10-fold cross-validation was incorporated during training. Then the trained models were applied on the testing dataset to validate its performance, where our model demonstrated high accuracy (>98% in both cancerous status and tissue origin predictions), which was better than previous approaches. We also found that our models possessed high generalizability, i.e., its performance was not biased to any cancer types and it would work on “novel” cancer types that did not exist in the training dataset [16].
2.4. Statistical analysis
Statistical significance between two groups was determined by Mann-Whitney rank sum test. P < 0.05 was considered as statistically significant, and all probabilities were two-tailed.
3. Results
3.1. Application of GeneCT on cancer datasets collected from various sources
GeneCT classification models were built using TCGA’s pan-cancer transcriptome datasets, which data was generated under a unified protocol and platform. We thus wonder whether GeneCT possessed the ability to handle transcriptome data generated in different scenarios. To do this, a list of cancer transcriptome datasets from non-TCGA sources were collected from the literature. Notably, these datasets were generated using various protocols and library preparation kits. GeneCT prediction results were summarized in Table 1. Briefly, study from Yao et al. [21] contained 10 pairs of clear cell renal cell carcinoma (ccRCC) tumors and adjacent normal kidney tissues; GeneCT showed an accuracy of 100% in both cancerous status and tissue-of-origin classifications on this dataset. Similarly, study by Eswaran et al. [22] used 17 breast tumors (in 3 sub-types) and 3 adjacent normal breast tissues. GeneCT classified 16 out of 17 (94.1%) tumor samples as cancerous and 3 out of 3 (100.0%) normal tissues as non-cancerous; meanwhile all the samples (100.0%) were classified as breast origin. Yang et al. [23] used paired hepatocellular carcinoma (HCC) tumor, portal vein tumor thrombosis (PVTT) and adjacent normal liver tissues from 20 patients in their study. GeneCT successfully classified 16 (80%) tumors, 19 (95%) PVTT samples to be cancerous and 18 (90%) normal tissues as non-cancerous. Meanwhile, 59 out of the 60 biopsies were classified as liver origin, corresponding to an overall accuracy of 98.3% in tissue-of-origin classification on this dataset.
Table 1.
Prediction results of GeneCT on various non-TCGA datasets.
| Study | Sample type | Total no. of samples | Cancerous status prediction accuracy (%) | Tissue-of-origin prediction accuracy (%) |
|---|---|---|---|---|
| Yao et al. | Clear cell renal cell carcinoma | 10 | 100.0 | 100.0 |
| Normal kidney tissue | 10 | 100.0 | 100.0 | |
| Eswaran et al. | Breast cancer | 17 | 94.1 | 100.0 |
| Normal breast tissue | 3 | 100.0 | 100.0 | |
| Yang et al. | Hepatocellular carcinoma | 20 | 80.0 | 95.0 |
| Normal liver tissue | 20 | 95.0 | 100.0 | |
| portal vein tumor thrombosis | 20 | 90.0 | 100.0 | |
| Huang et al. | Hepatocellular carcinoma | 9 | 11.1 | 100.0 |
| Normal liver tissue | 9 | 55.5 | 100.0 | |
| McDonald et al. | Pancreatic cancer | 10 | 100.0 | NA |
| Garzon et al. | Acute Myeloid Leukemia | 71 | 83.1 | NA |
| Atwood et al. | Basal cell carcinoma | 13 | 100.0 | NA |
| Normal skin tissue | 8 | 62.5 | NA |
Another study by Huang et al. [24] included liver cancer samples from 9 pairs of tumors and adjacent normal tissues. Strikingly, GeneCT predicted only 1 out of 9 (11.1%) tumor samples as cancerous and 5 out of 9 (55.5%) adjacent normal tissues as non-cancerous, despite that all the samples (100.0%) were classified as liver origin. To dissect the reason behind the unexpected results on this dataset, we performed Principal Component Analysis (PCA) using the expression profile of all annotated genes to study the consistency among the samples [25]. As shown in Fig. 1A, the adjacent normal liver tissues which were predicted to be non-cancerous (grey dots) was closer to the tumor samples predicted as non-cancerous (blue dots), but not to those adjacent normal tissues predicted to be cancerous (red dots). In contrast, PCA analysis using Yang et al. dataset showed that adjacent normal tissues were clustered together and were not mixed with the tumor samples (Fig. 1B). Furthermore, we also investigated the expression of the ALB (Albumin) gene, the most commonly used marker gene in the liver tissue which is known to be frequently down-regulated in liver cancer [26]. As shown in Fig. 1C, the tumor samples predicted to be non-cancerous (blue dots) by GeneCT indeed showed a similar expression level to the adjacent normal tissues predicted to be non-cancerous (grey dots), while much higher than those adjacent normal tissues predicted to be cancerous (red dots; P = 0.016). Furthermore, we applied ESTIMATE software on the tumor samples in this dataset and found that those predicted as cancerous by GeneCT showed much lower ESTIMATE scores compared to others predicted as non-cancerous (Fig. 1D). The ESTIMATE score is a measurement of infiltrating stromal/immune cells in the tumors and higher scores indicate lower purity [2]; Fig. 1D thus suggested that GeneCT was consistent with ESTIMATE on these samples. Together, these results suggested that in Huang et al. dataset, the sample purity might not be perfect in the “mis”-classified samples (e.g., possibly due to cross-contamination during sample collection).
Fig. 1.
Troubleshooting of the liver cancer datasets. PCA result on (A) Huang et al. dataset and (B) Yang et al. dataset. The samples were colored according to cancerous status prediction results. (C) Expression of ALB gene among the samples. Expression was quantified as log2-scaled RPKM values. (D) ESTIMATE scores on the tumors grouped by GeneCT prediction result.
Furthermore, we also collected transcriptome datasets from cancer types that were not included in the training dataset when building GeneCT, in which scenario these tissue types were considered as “unknown” to further test GeneCT’s generalizability. For example, McDonald et al. [27] investigated 10 primary tumor and metastatic tumor samples from pancreatic cancer cases in their study, and GeneCT successfully classified all samples (100%) as cancerous. Similarly, GeneCT correctly classified 59 out of 71 (83.1%) acute myeloid leukemia (AML) cases as cancerous in a study by Garzon et al. [28]. Notably, the dataset was composed of 52 bone marrow and 19 peripheral blood biopsies, and GeneCT’s accuracies on biopsies from these two sources were not identical (88.5% and 68.4% on bone marrow and peripheral blood biopsies, respectively), which was in line with the fact that biopsies from bone marrow was usually preferred than peripheral blood in AML diagnoses and studies [29]. The last dataset from Atwood et al. [30] study contained 13 basal cell carcinoma (BCC) cases and 8 adjacent normal skin tissues. As a result, GeneCT successfully classified all the tumor cases (100%) as cancerous and 5 (62.5%) normal tissues as non-cancerous. These results thus demonstrated that GeneCT was highly generalizable and held the potential to be applied on any cancer types, even “unknown” ones.
3.2. Application of GeneCT on metastatic cancer samples
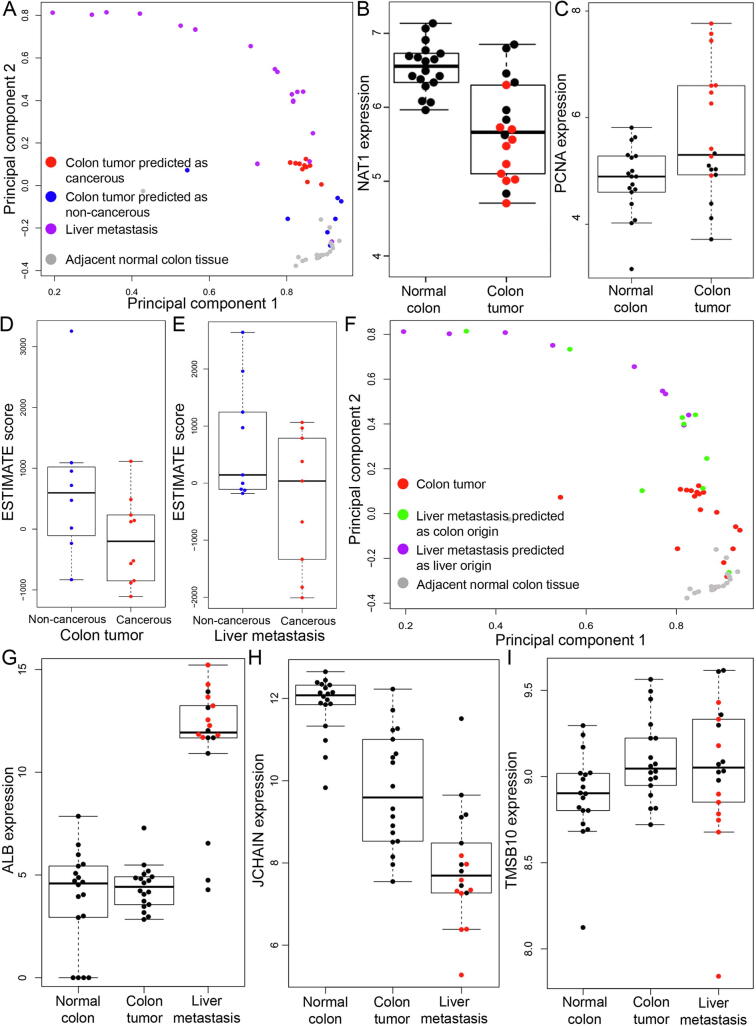
Metastatic cancer cases were one of the most challenging scenarios for quality control of the tissue biopsies. To evaluate the performance of GeneCT on such cases, two datasets with metastatic cancer samples were collected from the literature and the results were shown in Table 2. Both datasets were generated from colorectal cancer (CRC) with liver metastasis, which is one of the most common metastatic cancer types. Briefly, Lee et al. [31] employed 5 cases in their study and profiled the transcriptome of the primary colon tumor, metastatic tumor in liver, adjacent normal tissues of colon and liver for each case. GeneCT application led to 100% accuracy on this dataset in both cancerous-status and tissue-of-origin classifications (Table 2). In the other dataset, study from Kim et al. [32] recruited a larger cohort of colon-liver metastasis cases. GeneCT successfully classified all the adjacent normal colon tissues (100.0%) as non-cancerous and of colon origin. Meanwhile, 17 out of 18 (94.4%) of the colon tumors were predicted as colon origin; however, only 10 (55.6%) of them were predicted as cancerous. Moreover, only 50.0% (9 out of 18) of the liver metastatic samples were predicted as cancerous and 50.0% (9 out of 18) were predicted to be colon origin with the remaining 50.0% (9 out of 18) predicted as liver origin (Table 2). To confirm the prediction result, we performed PCA analysis using the expression profile of all annotated genes on this dataset, paying special attention to the colon tumors that were (mis-)classified as non-cancerous and the metastasis samples that were (mis-)classified as liver origin. The result (Fig. 2A) indicated that, indeed the colon tumor samples predicted as non-cancerous (blue dots) were closer to the adjacent normal colon tissues (grey dots) than those predicted as cancerous (red dots). Furthermore, NAT1 (N-Acetyltransferase 1; Fig. 2B) gene, known to be down-regulated in colon tumors [33] displayed significantly lower expression in the colon tumors classified as cancerous (red dots) compared to those classified as non-cancerous (P = 0.0014). Similarly, PCNA (Proliferating cell nuclear antigen; Fig. 2C) gene [33], known to be up-regulated [33], showed significantly higher expression in colon tumors classified as cancerous than those classified as non-cancerous (P = 0.021). These results led us to speculate that the purity of the colon tumors that were classified as non-cancerous might be not as high as those predicted as cancerous. Indeed, both colon tumors and liver metastasis samples predicted to be non-cancerous showed much higher ESTIMATE scores than those predicted as cancerous (Fig. 2D, E). In addition, we examined the expression profiles of top 10 up-regulated and 10 down-regulated genes in colon cancer mined from GEPIA database [34]. The results were shown in Supplementary Fig. S2. We found that for 15 (75%) out of 20 genes investigated, the tumors predicted as non-cancerous expressed in similar levels to the adjacent normal colon tissues which were significantly different from those predicted as cancerous. For the metastasis samples, we replotted the PCA result using the tissue-of-origin prediction result as the color scheme (Fig. 2F). The metastasis samples were not clustered together while those samples classified as colon origin (green dots) were closer to the colon tumors (red dots) and adjacent normal colon tissues (grey dots). The expression patterns of the liver marker gene, ALB, were shown in Fig. 2G. The metastasis samples classified as liver origin (red dots) showed a significantly higher expression (P = 0.024) than those classified as colon origin (grey dots). Furthermore, we mined Expression Atlas [35] and identified two highly expressed, colon-specific genes: TMSB10 (Thymosin Beta 10) and JCHAIN (Immunoglobulin J chain precursor). As shown in Fig. 2H and I, the metastasis samples that were classified as liver origin (red dots) showed lower expression than those classified as colon origin (grey dots; P = 0.050 and 0.011 for TMSB10 and JCHAIN, respectively). These results thus suggested that the metastatic tumor samples classified as liver origin might suffer from contamination of adjacent liver cells, and further demonstrated that GeneCT classifications was informative in evaluating the purity of the tissue biopsies in metastatic cancer samples.
Table 2.
Prediction results of GeneCT on cancer metastasis datasets.
| Study | Sample type | Total no. of samples | No. of samples predicted to be cancerous | Accuracy (%) | No. of samples predicted to be colon origin | Accuracy (%) |
|---|---|---|---|---|---|---|
| Lee et al. | Colon tumor | 5 | 5 | 100.0 | 5 | 100.0 |
| Liver metastasis | 5 | 5 | 100.0 | 5 | 100.0 | |
| Normal colon tissue | 5 | 0 | 100.0 | 5 | 100.0 | |
| Normal liver tissue | 5 | 0 | 100.0 | 0 | 100.0 | |
| Kim et al. | Colon tumor | 18 | 10 | 55.6 | 17 | 94.4 |
| Liver metastasis | 18 | 9 | 50.0 | 9 | 50.0 | |
| Normal colon tissue | 18 | 0 | 100.0 | 18 | 100.0 |
Fig. 2.
Troubleshooting of the colon cancer with liver metastasis datasets. (A) PCA result colored by the cancerous status prediction results. (B) Expression of NAT1 and (C) PCNA genes among the normal colon and colon tumor samples, respectively. The black and red dots represent the samples predicted as non-cancerous and cancerous, respectively. (D) ESTIMATE scores on colon tumors grouped by GeneCT prediction result. (E) ESTIMATE scores on liver metastasis samples grouped by GeneCT prediction result. (F) PCA result colored by the tissue-of-origin prediction results. (G) Expression of ALB, (H) JCHAIN and (I) TMSB10 genes among the normal colon, colon tumor and liver metastasis samples. The black and red dots represent the samples predicted to be colon and liver origin, respectively. Expression was quantified as log2-scaled RPKM values in B, C, G, H, and I. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Relationship between GeneCT classification and tumor purity
To further investigate the relationship between GeneCT prediction and tumor purity, two batches of in silico mixture experiments were performed using Lee et al. dataset. We first mixed the RNA-seq reads from a colon tumor and its adjacent normal colon tissue with various combinations. As a result, the proportion of tumor-derived reads in the in silico mixture data ranged from 0% to 100% with a gradient of 10% to simulate various levels of tumor purity. The mixture experiments were repeated 10 times and the GeneCT prediction results were shown in Fig. 3A. Note that the detailed cancerous status prediction score (a value between 0 and 1, where 1 means cancerous and 0 means non-cancerous) calculated by GeneCT were utilized. Fig. 3A showed that GeneCT prediction was qualitative in inferring the cancerous status of the samples that it would predict the sample as non-cancerous when the tumor fraction was below 80%. In the second batch, we performed mixture experiments using a liver metastasis and its adjacent normal liver tissue. The results were shown in Fig. 3B, which also showed a qualitative characteristic: GeneCT would predict the sample to be cancerous when the tumor fraction was higher than 30%. Moreover, when the tumor fraction increased, the tissue origin prediction turned from liver to colon. The results thus suggested that GeneCT classification was indeed indicative in predicting the purity of the tumor biopsies.
Fig. 3.
GeneCT prediction results on the in silico mixture data using (A) a colon tumor sample and its adjacent normal colon tissue, and (B) a colon tumor liver metastasis sample and its adjacent normal liver tissue. The y-axis was the scores in GeneCT cancerous status prediction, where 1 means cancerous and 0 means non-cancerous. Each dot represented one mixture experiment and the color of the dots indicated the tissue origin prediction result: black meant colon and red meant liver. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
In this study, we showed that our previously developed cancerous status and tissue-of-origin classifier, GeneCT, which utilized a deep learning approach to analyze transcriptome data, was able to work on various cancer types and serve as a rapid preliminary tool for tumor/tissue biopsy purity evaluations. It is notable that for the transcriptome datasets tested in this study, the RNA extraction protocols and library preparation kits are different from each other as well as from TCGA, which scenario may introduce adverse effects on the consistency of gene expression profiles [25], [36]. More importantly, considering that GeneCT was trained using TCGA datasets, these non-TCGA sources therefore allowed us to perform independent investigations and avoided the analysis to go around in circles. GeneCT showed high accuracy on most of these datasets. On the other hand, for those yielding a poor performance, further investigations indicated that the incorrect classifications might stem from the impurity of the samples. In fact, cross-contamination between tumors and adjacent normal tissues frequently occurs during biopsy collection; even TCGA could only guarantee that 80% cells in their tumor samples are cancerous. Impurity is especially detrimental for cancer metastasis studies because it may lead to incorrect interpretation of the results [1]. We think that the purity issue may not affect the results and conclusions in Huang et al. and Kim et al. datasets tested here, while may limit the sensitivity of their assays in discovering informative genes/pathways for downstream functional studies.
One valuable characteristic of GeneCT is that it is only based on few common oncogenes, tumor suppressor genes and transcription factors to do the analysis. Such genes are known to be frequently altered in various cancer types, therefore purity estimation using these genes should introduce minor bias in downstream cancer type specific differentially expressed genes mining, which is the most widely performed investigations in molecular cancer studies. In addition, we believe that generalizability is another valuable characteristic for any purity evaluation tools, especially the ability to handle “unknown” tissue types. We think that the high generalizability of GeneCT originates from the feature genes that we used to build it. Unlike most other methods [37], [38], [39], [40], [41], GeneCT does not require any cancer/tissue-type specific biomarkers for its classification models. We reasoned that the numbers of cancer/tissue-type specific biomarkers usually vary significantly among different cancer/tissue types [34], [35] thus might introduce biases in the classification models. In addition, currently it is infeasible to include all cancer types to build one universal classifier due to the large number of existing and ever-growing newly discovered cancer types, therefore for classifiers trained with cancer-type specific biomarkers, it could be risky to apply them on biopsies with unclassified or unknown cancer types considering that their underline biomarkers are only informative to specific cancer types during training. In contrast, oncogenes and tumor suppressor genes used by GeneCT are usually not specific to one cancer type instead frequently altered in multiple cancer types [42], [43], [44]. Similarly, even though most of the transcription factors do not show strong specificity toward certain tissue type [45], [46], however, their expression pattern is highly related to the tissue identity [46], [47], thus promises the generalizability. The high performance on datasets of various (including “unknown”) cancer types from non-TCGA sources indeed demonstrated the wide applicability of our approach as well as the generalizability of our method.
The in silico mixture experiments showed that GeneCT prediction results were indeed indicative to the purity of the tumor biopsies. The data also suggested that deep learning technology could play roles in biopsy purity evaluation fields. However, the results on the two batches of mixture experiments also showed that GeneCT prediction might only serve as a preliminary, qualitative assessment of tumor purity.
5. Conclusion
In conclusion, considering the high generalizability and requirement of transcriptome data only, we believe that GeneCT could serve as a valuable tool for rapid, preliminary purity evaluation of pan-cancer tumor biopsies with minor request on materials and cost. Further works towards a quantitative method to accurately deduce the purity level of the tumor biopsies using deep learning approaches would be valuable in the future (e.g., using various purity levels of tumor biopsies to training the models).
Acknowledgments
Acknowledgements
This study has been supported by Shenzhen Bay Laboratory and Guangdong Basic and Applied Basic Research Foundation (2019A1515110173).
Author contributions
Conceived of the study: K.S., H.W. and H.S.; Performed study: F.F., D.C., Y.Z., H.W., H.S. and K.S.; Result interpretation: F.F., H.S. and K.S.; Wrote the paper: H.W. and K.S.
Conflict of interest
None declared.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.06.007.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Aran D., Sirota M., Butte A.J. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. doi: 10.1038/ncomms9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshihara K., Shahmoradgoli M., Martinez E., Vegesna R., Kim H., Torres-Garcia W. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter S.L., Cibulskis K., Helman E., McKenna A., Shen H., Zack T. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30(5):413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benelli M., Romagnoli D., Demichelis F. Tumor purity quantification by clonal DNA methylation signatures. Bioinformatics. 2018;34(10):1642–1649. doi: 10.1093/bioinformatics/bty011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng X., Zhao Q., Wu H.J., Li W., Wang H., Meyer C.A. MethylPurify: tumor purity deconvolution and differential methylation detection from single tumor DNA methylomes. Genome Biol. 2014;15(8):419. doi: 10.1186/s13059-014-0419-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johann P.D., Jager N., Pfister S.M., Sill M. RF_Purify: a novel tool for comprehensive analysis of tumor-purity in methylation array data based on random forest regression. BMC Bioinf. 2019;20(1):428. doi: 10.1186/s12859-019-3014-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng X.L., Moffitt R.A., Torphy R.J., Volmar K.E., Yeh J.J. De novo compartment deconvolution and weight estimation of tumor samples using DECODER. Nat Commun. 2019;10(1):4729. doi: 10.1038/s41467-019-12517-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z., Wu H. TOAST: improving reference-free cell composition estimation by cross-cell type differential analysis. Genome Biol. 2019;20(1):190. doi: 10.1186/s13059-019-1778-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moss J., Magenheim J., Neiman D., Zemmour H., Loyfer N., Korach A. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun. 2018;9(1):5068. doi: 10.1038/s41467-018-07466-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun K., Jiang P., Chan K.C.A., Wong J., Cheng Y.K., Liang R.H. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci U S A. 2015;112(40):E5503–E5512. doi: 10.1073/pnas.1508736112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahmani E., Schweiger R., Shenhav L., Wingert T., Hofer I., Gabel E. BayesCCE: a Bayesian framework for estimating cell-type composition from DNA methylation without the need for methylation reference. Genome Biol. 2018;19(1):141. doi: 10.1186/s13059-018-1513-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun K., Jiang P., Cheng S.H., Cheng T.H.T., Wong J., Wong V.W.S. Orientation-aware plasma cell-free DNA fragmentation analysis in open chromatin regions informs tissue of origin. Genome Res. 2019;29(3):418–427. doi: 10.1101/gr.242719.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gai W., Sun K. Epigenetic biomarkers in cell-free DNA and applications in liquid biopsy. Genes (Basel) 2019;10(1):32. doi: 10.3390/genes10010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun K., Wang J., Wang H., Sun H. GeneCT: a generalizable cancerous status and tissue origin classifier for pan-cancer biopsies. Bioinformatics. 2018;34(23):4129–4130. doi: 10.1093/bioinformatics/bty524. [DOI] [PubMed] [Google Scholar]
- 17.Sun K. Ktrim: an extra-fast and accurate adapter- and quality-trimmer for sequencing data. Bioinformatics. 2020;36(11):3561–3562. doi: 10.1093/bioinformatics/btaa171. [DOI] [PubMed] [Google Scholar]
- 18.Wang K., Singh D., Zeng Z., Coleman S.J., Huang Y., Savich G.L. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38(18) doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu F., Kent W.J., Clawson H., Kuhn R.M., Diekhans M., Haussler D. The UCSC known genes. Bioinformatics. 2006;22(9):1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 21.Yao X., Tan J., Lim K.J., Koh J., Ooi W.F., Li Z. VHL deficiency drives enhancer activation of oncogenes in clear cell renal cell carcinoma. Cancer Discov. 2017;7(11):1284–1305. doi: 10.1158/2159-8290.CD-17-0375. [DOI] [PubMed] [Google Scholar]
- 22.Eswaran J., Cyanam D., Mudvari P., Reddy S.D., Pakala S.B., Nair S.S. Transcriptomic landscape of breast cancers through mRNA sequencing. Sci Rep. 2012;2:264. doi: 10.1038/srep00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y., Chen L., Gu J., Zhang H., Yuan J., Lian Q. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat Commun. 2017;8:14421. doi: 10.1038/ncomms14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y., Zheng J., Chen D., Li F., Wu W., Huang X. Transcriptome profiling identifies a recurrent CRYL1-IFT88 chimeric transcript in hepatocellular carcinoma. Oncotarget. 2017;8(25):40693–40704. doi: 10.18632/oncotarget.17244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danielsson F., James T., Gomez-Cabrero D., Huss M. Assessing the consistency of public human tissue RNA-seq data sets. Brief Bioinform. 2015;16(6):941–949. doi: 10.1093/bib/bbv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong I.H., Lau W.Y., Leung T., Johnson P.J. Quantitative comparison of alpha-fetoprotein and albumin mRNA levels in hepatocellular carcinoma/adenoma, non-tumor liver and blood: implications in cancer detection and monitoring. Cancer Lett. 2000;156(2):141–149. doi: 10.1016/s0304-3835(00)00473-0. [DOI] [PubMed] [Google Scholar]
- 27.McDonald O.G., Li X., Saunders T., Tryggvadottir R., Mentch S.J., Warmoes M.O. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet. 2017;49(3):367–376. doi: 10.1038/ng.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garzon R., Volinia S., Papaioannou D., Nicolet D., Kohlschmidt J., Yan P.S. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci U S A. 2014;111(52):18679–18684. doi: 10.1073/pnas.1422050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Percival M.E., Lai C., Estey E., Hourigan C.S. Bone marrow evaluation for diagnosis and monitoring of acute myeloid leukemia. Blood Rev. 2017;31(4):185–192. doi: 10.1016/j.blre.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atwood S.X., Sarin K.Y., Whitson R.J., Li J.R., Kim G., Rezaee M. Smoothened variants explain the majority of drug resistance in basal cell carcinoma. Cancer Cell. 2015;27(3):342–353. doi: 10.1016/j.ccell.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J.R., Kwon C.H., Choi Y., Park H.J., Kim H.S., Jo H.J. Transcriptome analysis of paired primary colorectal carcinoma and liver metastases reveals fusion transcripts and similar gene expression profiles in primary carcinoma and liver metastases. BMC Cancer. 2016;16:539. doi: 10.1186/s12885-016-2596-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim S.K., Kim S.Y., Kim J.H., Roh S.A., Cho D.H., Kim Y.S. A nineteen gene-based risk score classifier predicts prognosis of colorectal cancer patients. Mol Oncol. 2014;8(8):1653–1666. doi: 10.1016/j.molonc.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu F., Ji F., Ji Y., Jiang Y., Sun X., Lu Y. In-depth analysis of the critical genes and pathways in colorectal cancer. Int J Mol Med. 2015;36(4):923–930. doi: 10.3892/ijmm.2015.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang Z., Li C., Kang B., Gao G., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kapushesky M, Emam I, Holloway E, Kurnosov P, Zorin A, Malone J, et al. Gene expression atlas at the European bioinformatics institute. Nucleic Acids Res 2010;38(Database issue):D690–8. [DOI] [PMC free article] [PubMed]
- 36.Sun Z., Asmann Y.W., Nair A., Zhang Y., Wang L., Kalari K.R. Impact of library preparation on downstream analysis and interpretation of RNA-Seq data: comparison between Illumina PolyA and NuGEN Ovation protocol. PLoS ONE. 2013;8(8) doi: 10.1371/journal.pone.0071745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang W., Wan S., Yang Z., Teschendorff A.E., Zou Q. Tumor origin detection with tissue-specific miRNA and DNA methylation markers. Bioinformatics. 2018;34(3):398–406. doi: 10.1093/bioinformatics/btx622. [DOI] [PubMed] [Google Scholar]
- 38.Li Y., Kang K., Krahn J.M., Croutwater N., Lee K., Umbach D.M. A comprehensive genomic pan-cancer classification using The Cancer Genome Atlas gene expression data. BMC Genomics. 2017;18(1):508. doi: 10.1186/s12864-017-3906-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Q., Chen J., Ni S., Tan C., Xu M., Dong L. Pan-cancer transcriptome analysis reveals a gene expression signature for the identification of tumor tissue origin. Mod Pathol. 2016;29(6):546–556. doi: 10.1038/modpathol.2016.60. [DOI] [PubMed] [Google Scholar]
- 40.Peng L., Bian X.W., Li D.K., Xu C., Wang G.M., Xia Q.Y. Large-scale RNA-Seq transcriptome analysis of 4043 cancers and 548 normal tissue controls across 12 TCGA cancer types. Sci Rep. 2015;5:13413. doi: 10.1038/srep13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei I.H., Shi Y., Jiang H., Kumar-Sinha C., Chinnaiyan A.M. RNA-Seq accurately identifies cancer biomarker signatures to distinguish tissue of origin. Neoplasia. 2014;16(11):918–927. doi: 10.1016/j.neo.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee E.Y., Muller W.J. Oncogenes and tumor suppressor genes. Cold Spring Harb Perspect Biol. 2010;2(10) doi: 10.1101/cshperspect.a003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.An O., Pendino V., D'Antonio M., Ratti E., Gentilini M., Ciccarelli F.D. NCG 4.0: the network of cancer genes in the era of massive mutational screenings of cancer genomes. Database (Oxford) 2014;2014:bau015. doi: 10.1093/database/bau015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao M., Kim P., Mitra R., Zhao J., Zhao Z. TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res. 2016;44(D1):D1023–D1031. doi: 10.1093/nar/gkv1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D'Alessio A.C., Fan Z.P., Wert K.J., Baranov P., Cohen M.A., Saini J.S. A systematic approach to identify candidate transcription factors that control cell identity. Stem Cell Rep. 2015;5(5):763–775. doi: 10.1016/j.stemcr.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun K., Wang H., Sun H. mTFkb: a knowledgebase for fundamental annotation of mouse transcription factors. Sci Rep. 2017;7(1):3022. doi: 10.1038/s41598-017-02404-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.