Abstract
In order to investigate enterobacteria presence involved in the secondary infections in Porcine Reproductive and Respiratory Syndrome (PRRS) pigs with different viral co-infections, we identified enterobacteria for guiding clinical treatment. Twenty-one diseased pigs were diagnosed with the PRRS virus (PRRSV) and other 7 virus primers by PCR/RT-PCR in the lung and spleen samples. Enterobacteria were isolated using MacConkey agar from 5 visceral samples of PRRS pigs, and identified by 16S rDNA sequencing. PRRSV was positive in 100% of the lung samples and 81.0% of the spleen samples. Seven diseased pigs were diagnosed with only PRRSV infection (33.3%), 7 pigs with PRRSV and 1 or 2 other viruses (33.3%) and 7 pigs with PRRSV and more than 2 types of other viruses (33.3%). PRRSV was more inclined to co-infect pigs with porcine group A rotavirus (PARV) with the co-infection rate of 52.4% (11/21). Approximately 13 types of bacteria were successfully isolated from lung, spleen, liver, kidney and lymph node samples of different PRRS pigs. Enterobacteria were isolated in 100% of lung, liver and lymph samples from pigs infected with PRRSV alone. However, the isolation rates were significantly decreased in the more than 3 viruses co-infection group. Escherichia coli was the most prevalent bacterium, followed by Morganella, Proteus, Shigella, Salmonella, Klebsiella and Aeromonas. Most of the isolated enterobacteria were opportunistic pathogens. Therefore, timely combination with antimicrobial agents is necessary for effective treatment of PRRS-infected pigs.
Keywords: Porcine reproductive and respiratory syndrome, Enterobacteria, Viral co-infection, PCR, 16S rDNA sequencing
Highlights
-
•
PRRSV was more inclined to co-infect with PARV.
-
•
Pigs co-infected with more virus, less enterobacteria were isolated from their viscera.
-
•
Enterobacteria isolated from viscera of PRRS pigs were opportunistic pathogens.
-
•
coli, Morganella and Proteus were the most prevalent enterobacteria.
-
•
Secondary infection of enterobacteria should be considered in PRRS treatment.
1. Introduction
Porcine reproductive and respiratory syndrome (PRRS) is a swine disease caused by a virus, which poses a significant economic threat to the swine industry worldwide [1]. The PRRS virus (PRRSV) can infect pigs of various ages and cause different clinical symptoms. When the virus infects suckling pigs, it usually causes death, while there may be no significant features except for reproductive failure in some sows when the virus infects adult pigs. When the virus infects nursery pigs, it causes respiratory disorder, but does not directly result in death if no secondary infection occurs, unless pigs infected with some highly pathogenic PRRSV isolates [2]. Therefore, secondary infection is usually one of the main reasons for death in PRRS infected pigs [3,4].
In practical production, farmers focus more attention on ways to prevent and treat infected pigs. Farmers employ vaccine inoculation and isolated rearing to prevent infection [5]. However, there seem to be no effective treatment options. The infection of PRRSV itself could not cause death in nursery pigs. Secondary infection is the direct cause of death, but the pathogens of secondary infection have not been understood to date; thus, there is no effective treatment.
In the present study, to investigate bacteria involved in the secondary infection during the PRRS development, we isolated and identified enterobacteria in different viscera of PRRS pigs, and analysed the enterobacterial proliferation from the gut, a natural strain reservoir, to other viscera of PRRS pigs with different viral co-infections, to provide a reference therapy for secondary infection in pigs with PRRS.
2. Materials and methods
2.1. Samples collection
All the procedures involving animals in this study were carried out in accordance with The Care and Use Guidelines of Experimental Animals established by the Ministry of Agriculture of China, and the animal protocol for this study approved by the Ethics Committee of Jiangxi Agricultural University (protocol number JXAULL-2018004). The clinical manifestations of involved diseased pigs at 40–50 days old included fever (40–41 °C), coughing, purple ears, breathing difficulties, loss of appetite, diarrhoea and other surface symptoms. The autopsy manifestations of the above pigs were lung oedema with fibrinous exudate, groin and mesenteric lymph node enlargement and hemorrhage, mild spleen swelling, renal enlargement and paleness, and petechial hemorrhage. The samples of porcine lung, liver, spleen, kidney and lymph nodes were collected and transported to a lab under refrigerated conditions on the day of collection. Samples were collected from 21 diseased pigs and 2 healthy pigs from 5 different breeding farms in the Jiangxi Province in China.
2.2. Virus detection
The lung and spleen samples from pigs with PRRS were cut into pieces and homogenised to be used for virus detection, respectively. Viral RNA and DNA were extracted by using TaKaRa MiniBEST Viral RNA/DNA Extraction Kit (TaKaRa Inc, Dalian, China) according to the manufacturer's instructions. Portion of each extraction was subjected to RNA reverse transcription by using PrimeScript™ 1st strand cDNA synthesis kit (Takara Inc). The extractions were then used for DNA virus detection, and the cDNAs were used for RNA virus detection. The specific primers along with the PCR fragment sizes for each virus are listed in Table 1 [[5], [6], [7], [8], [9], [10], [11], [12]]. The PCR reaction was performed as follows: 1 μL of DNA/cDNA as template, 2 μL of Primer-F and Primer-R (10 μM), 12.5 μL of Premix Taq™ DNA Polymerase and addition of H2O to total 25 μL. A 3-step cycling protocol was used for polymerase chain reaction (PCR) as follows: 94 °C for 5min, 30 cycles of 94 °C for 30s, corresponding annealing temperature for 30 s, and 72 °C for 30 s, then 72 °C for 10min. The PCR productions were examined by agarose gel electrophoresis.
Table 1.
Primer sequences and PCR fragment sizes used for virus and bacterial identification in this study.
| Pathogen target | Primer sequence(5′-3′) | Fragment size | Reference |
|---|---|---|---|
| PRRSV | F: TGAYGGGCGACAATGTCC | 319 bp | [6] |
| R: CGCAGACAAATCCAGAVG | |||
| PEDV | F:TTCTGAGTCACGAACAGCCA | 651 bp | [7] |
| R: CATATGCAGCCTGCTCTGAA | |||
| TGEV | F: GTGGTTTTGGTYRTAAATGC | 859 bp | [7] |
| R: CACTAACCAACGTGGARCTA | |||
| PARV | F: AAAGATGCTAGGGACAAAATTG | 308 bp | [8] |
| R: TTCAGATTGTGGAGCTATTCCA | |||
| CSFV | F: AGACGGCCTGTACCATAATA | 610 bp | [9] |
| R: GTATAAGATGTCCACGG | |||
| PCV2 | F: GAAGAATGGAAGAAGCGG | 360 bp | [10] |
| R: CTCACAGCAGTAGACAGGT | |||
| PRV | F: GGTGGACCGGCTGCTGAACGA | 455 bp | [11] |
| R: GCTGCTGGTAGAACGGCGTCA | |||
| PPV | F: AAATGAATCTGGGGGTGGGG | 316 bp | [12] |
| R: CCAGTCCGCTGGATTGAACC | |||
| Bacterial 16s rDNA | 27F: AGAGTTTGATCCTGGCTCAG | 1466 bp | [13] |
| 1492R:TACGGTTACCTTGTTACGACTT |
2.3. Enterobacteria isolation and identification
The viscera, including lung, spleen, liver, kidney and lymph nodes from the pigs with PRRS were used for enterobacteria isolation. Firstly, the visceral specimen was removed from the plastic bag to a sterile plate under the biosafety cabinets, and 6 sections were cut by using a sterile blade at different positions of the specimen after the specimen surface was sterilized using 75% alcohol. An inoculating loop was used to scrape the different newly cut surfaces of sections and inoculate the samples onto MacConkey agar (Haibo Inc, Qingdao, China) individually. The enterobacteria from each visceral specimen were cultured on the Maconkey agars at 37 °C for 16 h. The colonies with different morphologies were chosen to inoculate another MacConkey agar for bacterial purification. After the colonies were purified, they were used for DNA extraction. The bacteria were identified using 16S rDNA sequencing (Tsingke Inc, Qingdao, China) with primers 27F and 1492R. The 16S rDNA primer sequences and PCR fragment sizes are also listed in Table 1.
2.4. Statistical analysis
The statistical software SPSS (version 19.0) (International Business Machines Corporation, New York, USA) was used for data analysis. The viral positive detection rates between lung and spleen samples, and the bacterial positive isolation rates among different viscera were analysed using the chi-square test. The bacterial positive isolation rates among different viral infection groups were analysed using one-way analysis of variance (ANOVA) test. Differences were considered statistically significant at p ≤ 0.05.
3. Results
3.1. Virus detection in lung and spleen samples of suspected PRRS-positive pigs
Forty-six lung and spleen samples from 21 suspected PRRS pigs and 2 healthy pigs were individually examined by PCR or RT-PCR for the presence of PRRSV and other possible viruses. The viral detection results are shown in Table 2 . All suspected PRRS pigs showed positive detection for PRRSV in the lung samples, which confirmed that the diseased pigs were indeed infected with PRRSV. No virus was detected in healthy samples. PRRSV was positive in 100% of lung samples, and in 81.0% (17/21) of spleen samples, indicating that PRRSV was significantly easier to be detected in porcine lungs than in spleens (p < 0.05, Fig. 1 ).
Table 2.
Virus detection results in the lung and spleen samples of diseased pigs.
| Code | PRRSV(L/S) | PEDV(L/S) | TGEV(L/S) | PARV(L/S) | CSFV(L/S) | PCV2(L/S) | PRV(L/S) | PPV(L/S) |
|---|---|---|---|---|---|---|---|---|
| 1 | +/+ | −/− | −/− | −/− | −/− | −/− | −/− | −/− |
| 2 | +/+ | −/− | −/− | −/− | −/− | −/− | −/− | −/− |
| 3 | +/+ | −/− | −/− | −/− | −/− | −/− | −/− | −/− |
| 4 | +/+ | −/− | −/− | −/− | −/− | −/− | −/− | −/− |
| 5 | +/+ | −/− | −/− | −/− | −/− | −/− | −/− | −/− |
| 6 | +/+ | −/− | −/− | −/− | −/− | −/− | −/− | −/− |
| 7 | +/+ | −/− | −/− | −/− | −/− | −/− | −/− | −/− |
| 8 | +/+ | −/− | −/− | +/+ | −/− | −/− | −/− | −/− |
| 9 | +/+ | −/+ | −/− | −/− | −/− | −/− | −/− | −/− |
| 10 | +/+ | −/− | −/− | +/+ | −/− | +/− | −/− | −/− |
| 11 | +/+ | −/− | −/− | −/+ | −/− | −/− | −/− | +/+ |
| 12 | +/+ | −/− | −/− | −/+ | −/− | +/+ | −/− | −/− |
| 13 | +/− | −/+ | −/− | −/− | −/− | +/+ | −/− | −/− |
| 14 | +/+ | +/+ | −/− | −/− | −/− | +/+ | −/− | −/− |
| 15 | +/+ | −/− | −/− | −/+ | −/− | +/+ | −/− | +/+ |
| 16 | +/+ | −/− | −/− | −/+ | −/− | +/+ | −/− | +/+ |
| 17 | +/+ | −/+ | −/− | +/+ | −/− | −/− | −/− | +/+ |
| 18 | +/+ | +/+ | −/− | +/+ | −/− | +/− | −/− | −/− |
| 19 | +/− | −/− | −/− | +/+ | −/− | +/+ | −/− | +/+ |
| 20 | +/− | −/+ | −/− | −/+ | −/− | +/+ | −/− | +/+ |
| 21 | +/− | +/+ | −/− | +/+ | −/− | +/+ | −/− | +/+ |
| 22 | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− |
| 23 | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− |
S: Spleen; L: Lung; +: Positive; -: Negative.
Fig. 1.
Detection of different types of viruses in lung and spleen samples of pigs afflicted with PRRS. PRRSV and the other 7 viruses may cause symptoms like breathing difficulties or diarrhoea syndrome, such as PEDV, TGEV, PARV, CSFV, PCV2, PRV and PPV being detected in lung and spleen samples using RT-PCR. *p < 0.05.
In addition to PRRSV, some other viruses that could cause respiratory or digestive symptoms were also detected. These viruses included porcine epidemic diarrhoea virus (PEDV), transmissible gastroenteritis coronavirus (TGEV), porcine group A rotavirus (PARV), classical swine fever virus (CSFV), porcine circovirus type 2 (PCV2), pseudorabies virus (PRV) and porcine parvovirus (PPV). From the results of Table 2, TEGV, CSFV and PRV could not be detected from any of the lung or spleen samples, but PEDV, PARV, PCV2 and PPV had varying extents of positive detection. The positive rates of PEDV and PARV were higher in the spleen (33.3% and 52.4%) than in the lung samples (14.3% and 28.6%), while the positive rates of PCV2 were higher in the lung (47.6%) than in the spleen samples (38.1%). The positive rates of PPV were the same (33.3%) in both types of samples.
We further investigated the co-infection of PRRSV with other related viruses. As long as any of the lung or spleen samples were detected with the virus, the corresponding pig was thought to be infected with PRRSV. Seven diseased pigs were detected with only PRRSV (33.3%), 7 pigs detected with 2 or 3 types of viruses, and 7 pigs detected with 4 or 5 types of viruses. PRRS pigs were more inclined to be co-infected with PARV and PCV2, with co-infection rates of 52.4% (11/21) and 47.6% (10/21), respectively.
3.2. Enterobacteria from the visceral samples of pigs with PRRS
Through the above examination, we confirmed that the diseased pigs were indeed infected with PRRSV, simultaneously. We further determined that some other viruses may infect simultaneously with PRRSV. Nevertheless, the real causes of death among the nursery pigs with PRRS may be the pathogens causing secondary infections [3,4]. Therefore, we further investigated the enterobacterial proliferation from the gut to the surrounding viscera, such as lung, spleen, liver, kidney and lymph nodes. From the enterobacterial isolation and identification results (Table 3 ), approximately 13 types of bacteria were successfully isolated from several viscera of different PRRS-positive pigs, and no enterobacterium was isolated in healthy samples.
Table 3.
Enterobacterial isolation from viscera of the diseased pigs.
| Code/Organs | Lung | Spleen | Liver | Kidney | Lymph |
|---|---|---|---|---|---|
| 1 | Ec, Sasp | Ec, Pm, Spsp, Sasp | Ec, Pm, Sasp | Ec, Sasp | Ec, Pm, Mm, Sasp, Pr |
| 2 | Ec | Ko | Ec, Pm, Mm | – | Ec, Pm, Mm |
| 3 | Ec, Pm | – | Ec, Pm, Mm | – | Ec, Pm |
| 4 | Ec, Pm, Pa | Ec, Mm | Ec, Asp | Kp, Mm | Ec, Mm, Pc |
| 5 | Ec, Pm, Shsp, Asp | Ec, Mm, Kp | Ec | Ec, Kp | Ec, Kp, Eh |
| 6 | Ec, Pm, Sasp | Ec, Sasp | Ec, Pm, Sasp | Ec, Mm, Sasp | Ec, Pm, Mm, Sasp |
| 7 | Ec, Shsp, Ac | Ec | Ef, Ah, Cf | – | Ec |
| 8 | Ec, Pm, Mm, Eh, Cf | Ec, Mm, Ac, Sasp | Shsp | Ec, Pm, Mm, Psp | Ec, Ac, Sasp |
| 9 | Ec, Mm, Kp, Shsp | Ec, Mm, Shsp | Mm, Kp | Ec, Mm, Eh | Ec, Mm, Enc |
| 10 | – | – | Ec, Pm | Ec, Pv | Ec |
| 11 | Ec, Pm, Mm, Pv | Ec, Pm, Pv | Ec, Pm | – | Ec, Pm, Ko |
| 12 | Ec, Shsp | Ec, Pm, Ef, Ac | Ec | – | Ec, Mm, Sasp |
| 13 | Shsp | – | Shsp | – | Ec, Mm, Shsp, Ef, Stsp |
| 14 | Ec, Mm, Kp, Ac, Asp, | – | Ec | – | Ec, Pm |
| 15 | – | Ec, Kp, Cs | Ec, Mm, Kp, Asp, Cf | Mm, Kp | Ec, Mm, Kp |
| 16 | – | Ec, Enc | – | – | – |
| 17 | Enc, Ec | Shsp | – | – | – |
| 18 | Pa | Kp | – | Mm | Ec, Sasp |
| 19 | Ah | – | Ec | – | – |
| 20 | – | Pm | – | – | Ec, Pm, Mm, Eh, Sasp |
| 21 | Ec, Mm, Eb | Pm, Mm | – | – | – |
| 22 | – | – | – | – | – |
| 23 | – | – | – | – | – |
Ac: Aeromonas caviae; Ah: Aeromonas hydrophila; Asp: Acinetobacter sp.; Cf: Citrobacter freundii; Cs: Cronobacter sakazakii; Eb: Enterobacteriaceae bacterium; Ec: Escherichia coli; Ef: Escherichia fergusonii;Eh: Enterobacter hormaechei; Enc: Enterobacter cloacae; Ko: Klebsiella oxytoca; Kp: Klebsiella pneumonia; Mm: Morganella morganii; Pa: Pasteurella aerogenes; Pc: Pecto-bacterium carotovorum; Pm: Proteus mirabilis; Pr: Providencia rettgeri; Psp: Providencia sp.; Pv: Proteus vulgaris; Sasp: Salmonella sp.; Shsp: Shigella sp.; Spsp: Sphingomonas sp. Stsp: Stenotrophomonas sp.
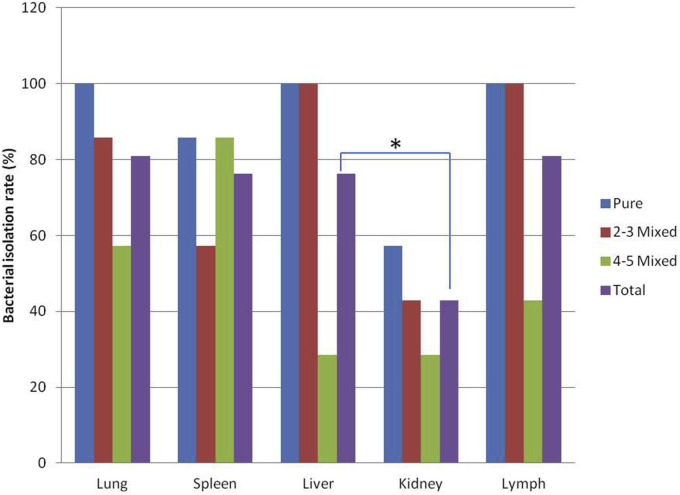
If only one type of enterobacterium was isolated, the corresponding organ was supposed to be infiltrated by the gut bacteria. Based on this analysis (Fig. 2 ), we found that the total enterobacterial isolation rate from kidneys (42.9%, 9/21) were significantly lower (p < 0.05), and the total rates from the other 4 viscera were comparative in levels. We also found that when pigs were co-infected with more types of viruses, the lower bacterial isolation rate was obtained (p < 0.05). In the PRRSV alone infection group, enterobacteria were detected with a 100% positive rate in lung, liver and lymph samples, whereas in the co-infection group with more than 3 types of virus species, the isolation rates in lung, liver, kidney and lymph were significantly decreased. Only the rates from spleens were comparative.
Fig. 2.
Detection of total enterobacteria in 5 different viscera of pigs afflicted with PRRS. Enterobacteria were isolated using MacConkey agar from lung, spleen, liver, kidney and lymphnode samples of PRRS pigs with different viral co-infections. Pure: pigs infected with only PRRSV; 2–3 Mixed: pigs infected with 2–3 types of viruses; 4–5 Mixed: pigs infected with 4–5 types of viruses. *p < 0.05.
Among the isolated enterobacteria, Escherichia coli was the most prevalent bacterium (Fig. 3 ). The total isolation rate of E. coli from lymph nodes was highest (81.0%, 17/21), while the rate from kidneys was lowest (28.6%, 6/21). Lower E. coli isolation rates were obtained in conjunction with infection with more types of viruses. Besides E. coli, Morganella morganii was found to have a higher positive isolation rate in almost all viscera than other 11 types of bacteria, especially in lymph nodes. Proteus sp., including Proteus mirabilis and Proteus vulgaris, were found to have a sub-higher positive isolation rate in the samples. In addition, some common pathogens, such as Salmonella, Shigella and Klebsiella were detected in the visceral samples (Fig. 4 ), and the positive detection rate of Shigella in lung samples was up to 23.8%, and the rate of Salmonella in lymph nodes was up to 28.6%. Aeromonas sp. was another bacterium with a positive isolation rate above 10%, although only in lung samples.
Fig. 3.
E. coli isolation in 5 different viscera of pigs afflicted with PRRS. The identification of E. coli in lung, spleen, liver, kidney and lymph node samples of PRRS pigs with different viral co-infections. Pure: pigs infected with only PRRSV; 2–3 Mixed: pigs infected with 2–3 types of viruses; 4–5 Mixed: pigs infected with 4–5 types of viruses. *p < 0.05.
Fig. 4.
Other major enterobacteria identified in 5 different viscera of pigs afflicted with PRRS: lungs, spleens, livers, kidneys and lymph nodes.
4. Discussion
PRRS is a panzootic and economically important disease in pigs. In nursery pigs, PRRS can cause severe respiratory tract symptoms, and may even lead to death. However, pure PRRSV infection is not the main reason accounting for porcine death. Multiple organ failure caused by secondary infection may be the etiologic reason [3,4]. As is well known, the gut is a natural microbiota reservoir, and the opportunistic pathogens in the gut of PRRS pigs may be very easy to proliferate to the surrounding viscera, resulting in the secondary infection. Thus, in the present study, we isolated and identified 13 strains of enterobacteria in different visceral samples from PRRS pigs. Most of them were opportunistic pathogens. We further investigated the association of the bacterial detection with the viral detection results, to try to find a new treatment strategy for PRRS.
In the current study, we first detected 7 other porcine viruses in addition to PRRSV in the lung and spleen samples to investigate the most common viruses that tend to co-infect with PRRSV. The result showed that PRRSV could be detected in 100% of lung samples, indicating the PRRSV infection was really occurring. However, the PRRSV detection rate in lung was higher than in the spleen, which confirmed that it is easier for PRRSV to invade the respiratory system and then affect other organs, such as the spleen [14]. For the 7 detected viruses, PARV (52.4%) was the most common co-infecting virus, followed by PCV2 (47.6%), and then PEDV and PPV (33.3%). PCV2 and PPV were reported to tend to co-infect with PRRSV previously [15]. PRV was also another common co-infecting virus with PRRSV [16]. However in our study, PRV was not detected from any lung or spleen samples. The reason for this may be associated with sampling location or time, resulting in different viral detection. Nevertheless, PARV was firstly reported to co-infect with PRRSV in the current study. PARV is a virus causing vomiting and diarrhoea. Therefore, PARV should also be considered as a potential cause responsible for the diarrhoea symptoms of PRRS pigs.
In addition to PRRSV, we isolated and identified the enterobacteria in different viscera from PRRS pigs. We found that at least 1 strain of enterobacteria could be isolated from one of the viscera of the PRRS-positive pigs. The isolation rates of enterobacteria in lung and lymph nodes samples were tied highest (81.0%), followed by those in liver and spleen samples (both 76.2%). Only the isolation rate from kidneys was lower, indicating that the kidney may not be easily colonized for enterobacteria from the gut infiltrate. Moreover, we found that more viral co-infections resulted in less enterobacterial isolation, especially when there were 4 or 5 types of co-infecting viruses, and the enterobacterial isolation rates from livers and lymph nodes were significantly decreased. The reason accounted for this may be possibility that more infecting viruses cause more severe symptoms in diseased pigs, so that the enterobacteria may not have enough time to proliferate from the gut to other viscera prior to death. However, the isolation rates in spleens from different viral co-infected pigs were paralleled possibly because the spleen as the largest immune organ in the body was easier for pathogens to infiltrate.
Generally, more than one enterobacterium could be isolated in the visceral samples of PRRSV pigs, but the most prevalent bacterium was E. coli. E. coli was found in 66.7% (14/21) lung samples and in 81.0% (17/21) lymph node samples respectively. The positive detection rate of E. coli was similar to those of total enterobacteria. Besides E. coli, other pathogenic or opportunistic pathogenic enterobacteria were also detected in 5 different viscera. Morganella morganii was the second type of enterobacteria isolated from the diseased samples. M. morganii is a commensal bacterium in animal intestinal tracts, however, it is also considered an uncommon cause of community-acquired infection, such as urinary tract infections [17]. Proteus spp., especially Proteus mirabilis, were the third type of isolated enterobacteria. P. mirabilis is widely distributed in soil and water, and can lead to urinary tract infection, wound infections, septicaemia, and pneumonia [18,19]. The next bacterium isolated at a higher level was Salmonella. Salmonella is an enterobacterium affecting animals and the environment worldwide, some strains of which can cause illnesses, such as typhoid fever, paratyphoid fever, and food poisoning [20]. From the specimens of healthy pigs, no enterobacteria were isolated, but which does not mean that there are no bacteria in the visteria, only because enterobacteria were examined in this study.
From our results, we presumed that when the pigs were infected with PRRSV, the internal viscera were damaged, and the immunity was decreased. Then, bacteria such as M. morganii, P. mirabilis or Salmonella in the gut have the opportunity to spread out and infiltrate the lung, liver, spleen and kidney. Lymph nodes, as whole body distributed immunity organ, have the ability to respond to the bacteria initially. Thus, the positive detection rates of the above 3 bacteria in lymph node samples were higher, but this is not always the case. For opportunistic pathogens, such as Klebsiella [21], Shigella and Aeromonas [22], the positive detection rates were higher in lung samples. Anti-microbial drug sensitivity analysis can be conducted on these pathogenic bacteria or conditional pathogenic bacteria, in order to clarify the effective drugs against these bacteria, so as to timely prevent them from spreading to other organs to cause secondary infection when the PRRSV infection is happened [23]. Of course, for opportune and effective preventing the secondary infection, it is also suggested to try to directly use some antibiotic with a quite broader antibacterial spectrum to treat the diseased pigs or to prevent other pigs in the same pigpen to develop infection, if there is indeed no enough time to conduct the anti-microbial drug sensitivity analysis.
In the current study, some other opportunistic enterobacteria could also be detected in different viscera. These enterobacteria included Providencia rettgeri, causing diarrhoea [24] or urinary tract infections [25]; Sphingomonas, causing mostly nosocomial, non-life-threatening infections [26]; Acinetobacter, causing various infections in unhealthy individuals [27]; Escherichia fergusonii, causing wound infections and even bacteraemia [28]; Providencia, causing infections associated with gastroenteritis and bacteraemia [29]; Stenotrophomonas, an opportunistic pathogen in highly debilitated patients [30]; and Cronobacter sakazakii, causing bacteraemia, meningitis and necrotising enterocolitis in infants [31]. At the same time, some commensal bacteria were isolated, such as Pasteurella aerogenes, Citrobacter freundii, Enterobacter cloacae and Enterobacter hormaechei. Among the isolated enterobacteria, Sphingomonas and Escherichia fergusonii were firstly reported to be isolated from swine samples. The bacterial identification results demonstrated that timely intervention with antimicrobial agents is necessary for effective treatment of PRRS pigs.
5. Conclusions
Summarily, in pigs with typical PRRS symptoms, PRRSV not only co-infected with other types of viruses, but was also found to result in enterobacterial proliferation from the gut to other viscera, and thus cause the secondary infections that threaten the lives of pigs. In our study, we identified the common enterobacterial pathogens causing secondary infections in PRRS pigs co-infected with other different viruses. Our results may provide some guidance for the clinical treatment of PRRS.
Author statement
Ge Zhao, Liheng Liu, and Junwei Wang were responsible for the conception, and design of the study. Charles Li participated in the experimental design and provided the guidance. Lujie Zhang and Liheng Liu was responsible for samples collection. Na Liu, Jianmei Zhao and Yuehua Li actively identified and analyzed the samples. Ge Zhao, Liheng Liu, and Junwei Wang contributed to drafting this manuscript. All authors have read and approved the final manuscript.
Funding
This work was funded by the National Key Research and 281 Development Program of China [grant No. 2018YFD0500505], the Technology Foundation for Selected Overseas Chinese Scholar, Ministry of Personnel of China [grant No. 2016176], as well as the Doctoral Scientific Research Fundation of Jiangxi Agriculture University [grant No. 3173].
Declaration of competing interest
The authors declared that they have no conflict of interest to this work.
References
- 1.Albina E. Epidemiology of porcine reproductive and respiratory syndrome (PRRS): An overview. Vet. Microbiol. 1997;55:309–316. doi: 10.1016/s0378-1135(96)01322-3. [DOI] [PubMed] [Google Scholar]
- 2.Tian K., Yu X., Zhao T., Feng Y., Cao Z., Wang C., Hu Y., Chen X., Hu D., Tian X., Liu D., Zhang S., Deng X., Ding Y., Yang L., Zhang Y., Xiao H., Qiao M., Wang B., Hou L., Wang X., Yang X., Kang L., Sun M., Jin P., Wang S., Kitamura Y., Yan J., Gao G.F. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One. 2007;6:e526. doi: 10.1371/journal.pone.0000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solano G.I., Bautista E., Molitor T.W., Segales J., Pijoan C. Effect of porcine reproductive and respiratory syndrome virus infection on the clearance of Haemophilus parasuis by porcine alveolar macrophages. Can. J. Vet. Res. 1998;62:251–256. [PMC free article] [PubMed] [Google Scholar]
- 4.Xu M., Wang S., Li L., Lei L., Liu Y., Shi W., Wu J., Li L., Rong F., Xu M., Sun G., Xiang H., Cai X. Secondary infection with Streptococcus suis serotype 7 increases the virulence of highly pathogenic porcine reproductive and respiratory syndrome virus in pigs. Virol. J. 2010;7:184. doi: 10.1186/1743-422X-7-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma J., Chen Y., Li S., Sun S. Diagnosis and treatment of porcine reproductive and respiratory syndrome. J. Anim. Sci. Vet. Med. 2017;36:140–142. [Google Scholar]
- 6.Yang K., Li Y., Duan Z., Guo R., Liu Z., Zhou D., Yuan F., Tian Y. A one-step RT-PCR assay to detect and discriminate porcine reproductive and respiratory syndrome viruses in clinical specimens. Gene. 2013;531:199–204. doi: 10.1016/j.gene.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 7.Kim S.Y., Song D.S., Park B.K. Differential detection of transmissible gastroenteritis virus and porcine epidemic diarrhea virus by duplex RT-PCR. J. Vet. Diagn. Invest. 2001;13:516–520. doi: 10.1177/104063870101300611. [DOI] [PubMed] [Google Scholar]
- 8.Elschner M., Prudlo J., Hotzel H., Otto P., Sachse K. Nested reverse transcriptase-polymerase chain reaction for the detection of group A rotaviruses. J. Vet. Med. B. 2002;49:77–81. doi: 10.1046/j.1439-0450.2002.00510.x. [DOI] [PubMed] [Google Scholar]
- 9.Choi C., Chae C. Detection of classical swine fever virus in boar semen by reverse transcription-polymerase chain reaction. J. Vet. Diagn. Invest. 2003;15:35–41. doi: 10.1177/104063870301500108. [DOI] [PubMed] [Google Scholar]
- 10.Yang J.S., Song D.S., Kim S.Y., Lyoo K.S., Park B.K. Detection of porcine circovirus type 2 in feces of pigs with or without enteric disease by polymerase chain reaction. J. Vet. Diagn. Invest. 2003;15:369–373. doi: 10.1177/104063870301500412. [DOI] [PubMed] [Google Scholar]
- 11.Perez L.J., Perera C.L., Frias M.T., Nunez J.I., Ganges L., de Arce H.D. A multiple SYBR Green I-based real-time PCR system for the simultaneous detection of porcine circovirus type 2, porcine parvovirus, pseudorabies virus and Torque teno sus virus 1 and 2 in pigs. J. Virol. Methods. 2012;179:233–241. doi: 10.1016/j.jviromet.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 12.Kim J., Chae C. A comparison of virus isolation, polymerase chain reaction, immunohistochemistry, and in situ hybridization for the detection of porcine circovirus 2 and porcine parvovirus in experimentally and naturally coinfected pigs. J. Vet. Diagn. Invest. 2004;16:45–50. doi: 10.1177/104063870401600108. [DOI] [PubMed] [Google Scholar]
- 13.Lane D.J. 16S/23S rRNA sequencing. In: Stackebrandt E., Goodfellow M., editors. Vol. 89. John Wiley and Sons; New York: 1991. pp. 115–175. (Nucleic Acid Techniques in Bacterial Systematic). [Google Scholar]
- 14.Rossow K.D., Collins J.E., Goyal S.M., Nelson E.A., Christopher-Hennings J., Benfield D.A. Pathogenesis of porcine reproductive and respiratory syndrome virus infection in gnotobiotic pigs. Vet. Pathol. 1995;32:361–373. doi: 10.1177/030098589503200404. [DOI] [PubMed] [Google Scholar]
- 15.Jia Y., Lu H., Zhang S., Zhou B., Chen P. Investigation on epidemiology of Co-infection of porcine circovirus type 2, porcine reproductive and respiratory disease syndrome virus and porcine parvovirus. Virol. Sin. 2004;19:467–470. [Google Scholar]
- 16.Wang H., Song H., Shi L., Wang W., Zhao M., Ma B., Cui Y., Zhang C. Detection and analysis of porcine circovirus, pseudorabies virus mixed infection with porcine reproductive and respiratory syndrome virus. China Anim. Husbandry Vet. Med. 2012;124:185–191. [Google Scholar]
- 17.Falagas M.E., Kavvadia P.K., Mantadakis E., Kofteridis D.P., Bliziotis I.A., Saloustros E., Maraki S., Samonis G. Morganella morganii infections in a general tertiary hospital. Infection. 2006;34:315–321. doi: 10.1007/s15010-006-6682-3. [DOI] [PubMed] [Google Scholar]
- 18.Coker C., Poore C.A., Li X., Mobley H.L. Pathogenesis of Proteus mirabilis urinary tract infection. Microb. Infect. 2000;2:1497–1505. doi: 10.1016/s1286-4579(00)01304-6. [DOI] [PubMed] [Google Scholar]
- 19.Gonzales G. eMedicine from WebMD; 2008. Proteus Infections (Last Ed) [Google Scholar]
- 20.Ryan K.J., Ray C.G. fourth ed. McGraw Hill; 2004. Sherris Medical Microbiology; pp. 362–368. [Google Scholar]
- 21.Beckingsale A.B., Williams D., Gibson J.M., Rosenthal A.R. Klebsiella and acute anterior uveitis. Br. J. Ophthalmol. 1984;68:866–868. doi: 10.1136/bjo.68.12.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker J.L., Shaw J.G. Aeromonas spp. clinical microbiology and disease. J. Infect. 2011;62:109–118. doi: 10.1016/j.jinf.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Li Y., Li X., Gong S., Ao Y., Zhu L., Xu Z. Etiology analysis of the bacterial secondary infection of PRRSV in Sichuan Province from 2013 to 2016. Acta Agricul. Zhejiang. 2017;9:1437–1444. [Google Scholar]
- 24.Yoh M. Importance of Providencia species as a major cause of travellers' diarrhea. J. Med. Microbiol. 2005;54:1077–1082. doi: 10.1099/jmm.0.45846-0. [DOI] [PubMed] [Google Scholar]
- 25.Jones B.D., Mobley H.L. Genetic and biochemical diversity of ureases of Proteus, Providencia, and Morganella species isolated from urinary tract infection. Infect. Immun. 1987;55:2198–2203. doi: 10.1128/iai.55.9.2198-2203.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maragakis L.L., Chaiwarith R., Srinivasan A., Torriani F.J., Avdic E., Lee A., Ross T.R., Carroll K.C., Perl T.M. Sphingomonas paucimobilis bloodstream infections associated with contaminated intravenous fentanyl. Emerg. Infect. Dis. 2009;15:12–18. doi: 10.3201/eid1501.081054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dent L.L., Marshall D.R., Pratap S., Hulette R.B. Multidrug resistant Acinetobacter baumannii: a descriptive study in a city hospital. BMC Infect. Dis. 2010;10:196. doi: 10.1186/1471-2334-10-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahapatra A., Mahapatra S., Mahapatra A. Escherichia fergusonii: an emerging pathogen in South Orissa. Indian J. Med. Microbiol. 2005;23:204. doi: 10.4103/0255-0857.16598. [DOI] [PubMed] [Google Scholar]
- 29.Armbruster C.E., Smith S.N., Yep A., Mobley H.L. Increased incidence of urolithiasis and bacteremia during Proteus mirabilis and Providencia stuartii coinfection due to synergistic induction of urease activity. J. Infect. Dis. 2014;209:1524–1532. doi: 10.1093/infdis/jit663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calza L., Manfredi R., Chiodo F. Stenotrophomonas (Xanthomonas) maltophilia as an emerging opportunistic pathogen in association with HIV infection: a 10-year surveillance study. Infection. 2003;31:155–161. doi: 10.1007/s15010-003-3113-6. [DOI] [PubMed] [Google Scholar]
- 31.Hunter C.J., Petrosyan M., Ford H.R., Prasadarao N.V. Enterobacter sakazakii: an emerging pathogen in infants and neonates. Surg. Infect. 2008;9:533–539. doi: 10.1089/sur.2008.006. [DOI] [PMC free article] [PubMed] [Google Scholar]