Abstract
The dynamic N6-methyladenosine (m6A) modification of mRNA plays a role in regulating gene expression and determining cell fate. However, the functions of m6A mRNA modification in bladder cancer stem cells (BCSCs) have not been described. Here, we show that global RNA m6A abundance and the expression of m6A-forming enzyme METTL3 are higher in BCSCs than those in non-CSCs of bladder cancer (BCa) cells. The depletion of the METTL3 inhibited the self-renewal of BCSCs, as evidenced by decreased ALDH activity and sphere-forming ability. Mechanistically, METTL3 regulates the m6A modification and thereby the expression of AF4/FMR2 family member 4 (AFF4), knockdown of which phenocopies the METTL3 ablation and diminishes the tumor-initiating capability of BCSCs in vivo. AFF4 binds to the promoter regions and sustains the transcription of SOX2 and MYC which have critical biological functions in BCSCs. Collectively, our results demonstrate the critical roles of m6A modification in self-renewal and tumorigenicity of BCSCs through a novel signaling axis of METTL3-AFF4-SOX2/MYC.
1. Introduction
Cancer stem cells (CSCs, also known as tumor-initiating cells), a relatively rare population of cancer cells, have characteristics of self-renewal capability, tumorigenic capacity, and pluripotency, which contribute to the driving force of tumorigenesis and metastasis. These stemness properties make CSCs resistant to conventional chemotherapies and cause subsequent recurrence, leading to clinical treatment failure [1]. Effective therapeutics and strategies targeting CSCs are desperately needed, whereas our knowledge of the CSCs is still incomplete so far.
Bladder carcinoma (BCa) is one of the most common malignancies and is characterized by rapid progression and high risk of recurrence [2, 3]. To better understand and eventually eliminate the bladder cancer stem cells (BCSCs), we and other groups have successfully identified several different BCSCs and determined their roles in BCa progression in vivo [4–7]. Moreover, we have found low-dose decitabine (a DNA methyltransferase inhibitor) could diminish the stemness of BCSCs without causing severe cytotoxicity [8], suggesting an important role of epigenetic regulation in BCSCs.
Besides the DNA methylation, recently we and others have found that aberrant N6-methyladenosine (m6A) methylation was also implicated in BCa progression [9–11]. RNA m6A is the most prevalent chemical mark observed in approximately 25% of eukaryotic mRNAs [12–14]. In mammalian cells, this dynamic modification is catalyzed by a methyltransferase complex consisting of several “writers,” which include methyltransferase-like 3 (METTL3), METTL14, Wilms tumor 1-associated protein (WTAP), VIRMA (KIAA1429), and RBM15 [15–19], and removed by two “erasers”: fat mass and obesity-associated protein (FTO) [20] and alkylation repair homolog protein 5 (ALKBH5) [21]. Aberrant m6A modification plays crucial roles in the progression of different types of cancer [22], especially as the modulator of CSCs of breast cancer [23], glioblastoma [24, 25], and leukemia [26, 27]. However, its function and mechanism in regulating CSCs seem to be context-dependent and have not been described in BCSCs so far.
In our previous study, we found that m6A abundance of both MYC and AFF4 mRNAs was regulated by aberrantly expressed METTL3 in BCa cells [11]. As a core component of the super elongation complex (SEC), AFF4 is involved in the regulation of transcription elongation of many genes encoding the pluripotency factors [28, 29]. For instance, AFF4 could upregulate SOX2 transcription to promote the tumor-initiation capacity of head and neck squamous cell carcinoma (HNSCC) [30], and MYC is another known target of AFF4 [11, 31]. Inspired by these results, we hypothesized that m6A plays a role in promoting the stemness of BCa cells by regulating AFF expression.
Here, we provide unequivocal evidences that the expression of METTL3 and RNA m6A level is significantly higher in the CSCs relative to the non-CSCs of BCa; METTL3 promote the self-renewal capability of BCSCs by regulating the mRNA m6A level and therefore the expression of AFF4, which in turn bind to the promoter regions of SOX2 and MYC to activate their transcription. Our findings reveal the role and mechanism of RNA m6A in regulating the stemness of BCSCs and will inspire future studies regarding their applications in clinical treatment.
2. Materials and Methods
2.1. Cell Culture, Flow Cytometry, and Sphere Formation Assays
BCa cell lines 5637 (ATCC NO. HTB-9) and UM-UC-3 (ATCC NO. CRL-1749) were purchased from the Chinese Academy of Cell Resource Center (Shanghai, China) and maintained as previously described [32]. Cell lines were routinely tested for mycoplasma and not cultured for longer than 20 passages. Specific siRNAs were transfected into cells by Lipofectamine™ RNAiMAX Transfection Reagent (13778-075) according to the manufacturer's instructions.
For flow cytometry analysis, BCa cells were stained using the ALDEFLUOR assay kit (StemCell Technologies) according to the manufacturer's instructions. Acquisition and sorting were then performed using the BECKMAN Moflo XDP (Beckton Dickson, Mountain View, CA). Gates for fluorescence fractionations were established using unstained and isotype controls.
For sphere formation assays, FACS-sorted cells were cultured in 24-well ultralow attachment plates (Corning Inc., Corning, NY, USA) at a density of 1,000 cells per well. Cells were cultured in serum-free DMEM/F12 supplemented with growth factors EGF, β-FGF, and IGF-1 at a concentration of 20 ng/ml (PeproTech, Rocky Hill, NJ, USA). Spheres with a diameter of over 20 μm were counted 7 days after plating.
2.2. Detect Gene Expression
For mRNA level examination, total RNA of BCa cells was extracted using Trizol reagent (Invitrogen). Complementary DNA (cDNA) synthesis was performed with the PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, RR047A) using 1 μg RNA per sample. qPCR reactions were performed using TB Green® Premix Ex Taq™ (Takara RR820A) to determine mRNA transcript levels. Primers for qRT-PCR are listed in Supplementary Table S1, siRNAs are used to knockdown METTL3, and AFF4 expression is listed in Supplementary Table S2.
For Western blotting, BCa cells were lysed with RIPA buffer as a standard protocol. The cell lysate was then mixed with loading buffer and incubated at 100°C for 5 min and subjected to conventional Western analysis. Antibodies are listed in Supplementary Table S3. The relative levels of proteins were quantified using densitometry with the Gel-Pro Analyzer (Media Cybernetics, Rockville, MD, USA). The target bands were densitometrically quantified and indicated under each band.
2.3. m6A Quantification
RNA m6A levels were evaluated by the m6A RNA quantification kit (Epigentek, P-9005) according to the manufacturer's protocol. Briefly, 200 ng total RNA of each sample was bound to the strip well of a 96-well plate, followed by m6A antibody capture and washing. After incubated with the substrate for 5 min before the reaction was stopped, the absorbance of each well was read on a microplate reader (Multiskan FC, Thermo Scientific) at 450 nm.
2.4. CHIP Assay
Chromatin immunoprecipitation (ChIP) assay was performed using a Simple ChIP Assay Kit (Cell Signaling Technology, Danvers, MA) according to the manufacturer's instruction. The precipitated DNA samples were purified and measured by qPCR. Results were shown as the percentage of input controls. Primers and antibodies used for CHIP assay are listed in the supplementary Table S1 and Table S2, respectively.
2.5. Limiting Dilution Transplantation Assay
Stable AFF4 knockdown 5637 cells and control cells were serially diluted (1 × 105-2.7 × 106), resuspended in 50 μl of Matrigel (Corning, 354230), and injected subcutaneously into BALB/cASlac-nu nude mice (Shanghai Laboratory Animals Center, SLAC). Subsequent tumors were monitored weekly until mice presented signs of distress, and the mice were sacrificed. All animal procedures were performed under a protocol approved by the Laboratory Animal Center of Anhui Medical University.
Paraffin sections of samples from xenografts were antigen retrieved, blocked, and processed as described before [33]. The intensity of immunostaining was measured by Image-Pro Plus 6.0 image analysis software (Media Cybernetics). The intensity of each image was calculated by normalizing the average integrated optical density (IOD) with the total selected area of interest (AOI).
2.6. Statistics
All experiments were performed at least three times, unless otherwise noted. Data are presented as the means ± standard deviation (S.D.) or standard error (S.E.). All of the statistical analyses were performed using Excel (Microsoft, Redmond, WA) or Prism (GraphPad Software Inc., La Jolla, CA). The two-tailed Student's t-test was used, and a p value of <0.05 was considered significant. For limiting dilution assay, a statistical test was performed as described previously [34].
3. Results
3.1. RNA m6A Levels Are Elevated in BCSCs
To estimate the potential role of RNA m6A modification in regulating the stemness of BCa, we examined the global RNA m6A levels of CSCs and non-CSCs of BCa. Aldehyde dehydrogenase 1 family, member A1 (ALDH1A1) was used as a marker to isolate BCSCs [35] from two established cancer cell lines, 5637 and UM-UC-3, by flow cytometry (Figure 1(a)), and RNA m6A methylation abundance was evaluated by the m6A RNA quantification kit. The results showed that ratios of m6A RNA/total RNA in ALDH1-positive (ALDH1+) cells isolated from both 5637 and UM-UC-3 were significantly higher than those in ALDH-negative (ALDH1−) proportion (Figure 1(b)). We then checked the expression patterns of known m6A writers (i.e., METTL3, METTL14, and WTAP) and erasers (i.e., FTO and ALKBH5) by quantitative RT-PCR to determine which subunit may account for m6A dysregulation of CSCs and found that the expression of METTL3 rather than other regulators was significantly elevated in ALDH1+ BCa cells (Figure 1(c)). The protein level of METTL3 was further validated to be higher in ALDH1+ BCa cells by Western blot (WB) (Figure 1(d)). All these data indicate that METTL3 is upregulated in BCSCs and may be implicated in self-renewal.
Figure 1.
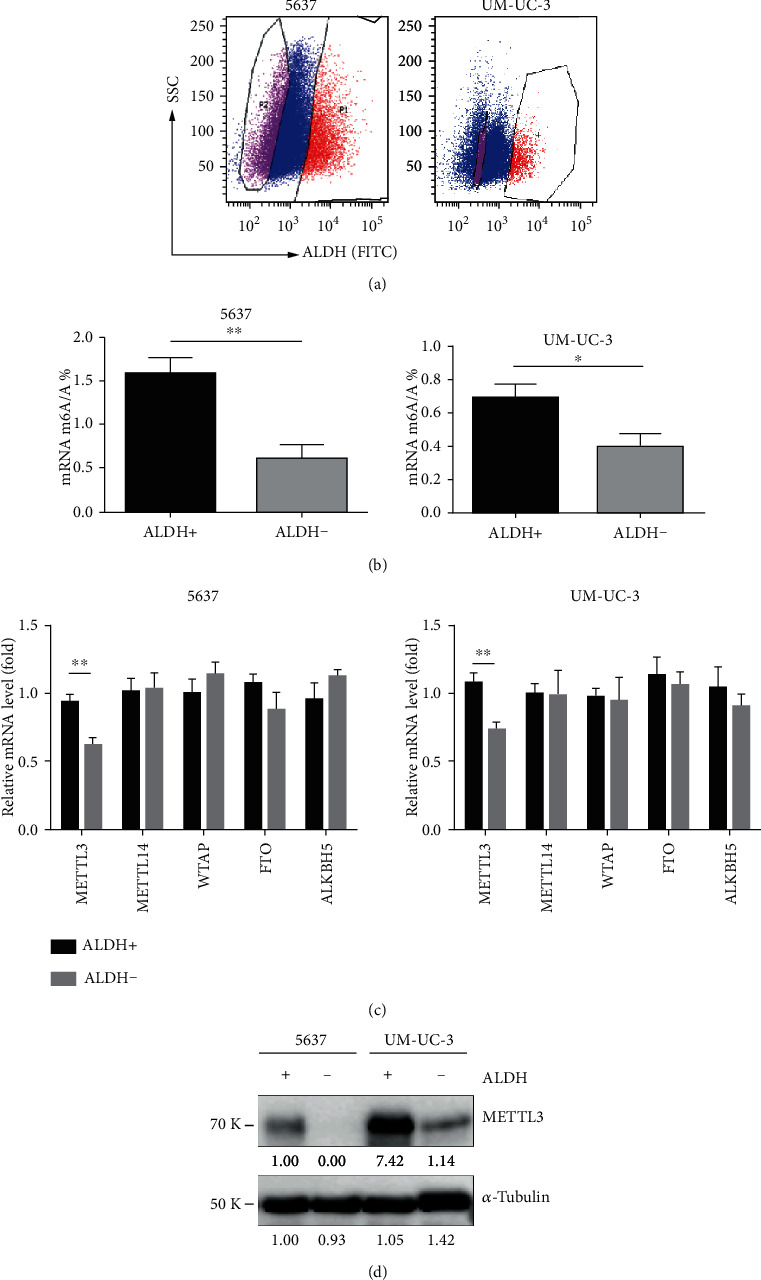
Differential m6A levels between CSCs and non-CSCs of BCa. (a) Representative gating scheme for FACS sorting of ALDH1-stained 5637 and UM-UC-3 cells. (b) Quantification of m6A levels in ALDH1-positive and ALDH1-negative BCa cells (∗p < 0.05, ∗∗p < 0.01, Student t-test). (c) mRNA levels of RNA m6A writers and erasers in ALDH1-positive and ALDH1-negative BCa cells (∗∗p < 0.01, Student t-test). (d) Protein levels of RNA m6A methyltransferase METTL3 in ALDH1-positive and ALDH1-negative BCa cells.
3.2. Targeting METTL3 Expression Impairs BCSC Self-Renewal
To determine whether METTL3 is important to BCSC self-renewal, we used two distinct siRNAs (si-METTL3-1 and si-METTL3-2) to ablate METTL3 expression in 5637 and UM-UC-3 cells. Compared with a nontargeting control siRNA (si-GFP), both specific siRNAs significantly reduced METTL3 mRNA and protein levels (Figures 2(a) and 2(b)). Two characteristics to identify populations of BCSCs are the ability to generate clusters of daughter cells when they are cultured on ultralow adherence plates (sphere assay) and high ALDH activity which can be quantified by flow cytometry using a fluorogenic substrate [7].
Figure 2.
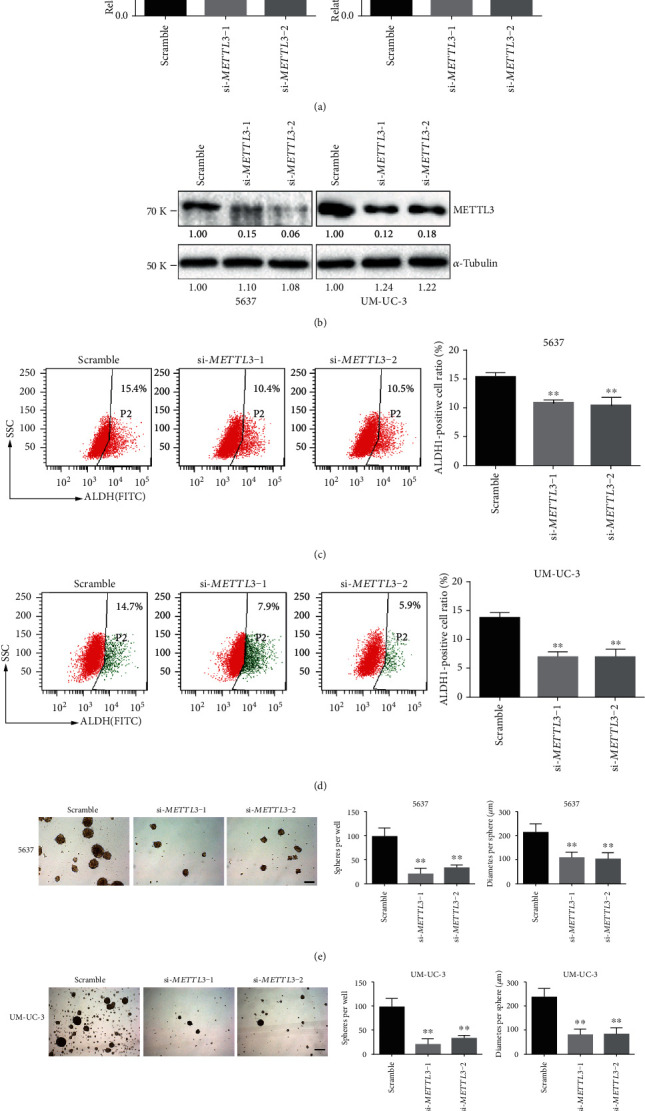
METTL3 is required to sustain self-renewal of BCa cells. The knockdown effect of specific siRNAs (si-METTL3-1 and si-METTL3-2) in 5637 and UM-UC-3 cells was verified at both the mRNA ((a) by qRT-PCR) and protein levels ((b) by Western blot). Ratio of cells with high ALDH activity (c, d); number and size of spheres formed in stem cell medium (e, f) of the BCa cells transfected with indicated siRNAs are plotted, and representative images are presented. ∗∗p < 0.01 compared to the scramble group, by Student t-test. Scale bar, 250 μm.
36 hours after siRNA transfection, cells with high ALDH activity were examined and sorted by flow cytometry, and the same amount of cells with high ALDH activity from different transfection groups was further transferred to ultralow adherence plates with stem cell medium. One week later, the number of formed spheres was counted. Both the percentages of cells with high ALDH activity (Figures 2(c) and 2(d)) and sphere formation frequency (Figures 2(e) and 2(f)) were significantly decreased upon METTL3 knockdown. With the above evidences, we concluded that METTL3 is required for the BCSC self-renewal in vitro.
3.3. AFF4 Is Regulated by METTL3 in BCSCs
In the previous study, we performed transcriptome sequencing and m6A sequencing followed by a series validation in 5637 cells, which proved the m6A modification and expression of AFF4 mRNA were directly regulated by METTL3 [11]. To identify if AFF4 was also the target of METLL3 in BCSCs, we then checked both mRNA and protein levels of AFF4 in ALDH1+ and ALDH1− cells from 5637 and UM-UC-3, respectively. Not surprisingly, a significantly higher level of AFF4 expression was observed in the ALDH1+ proportion compared to the corresponding ALDH1− counterpart in BCa cells (Figures 3(a) and 3(b)). Moreover, gene-specific m6A-qPCR using primers to amplify either the m6A peak region (indicated by our m6A-sequencing results) or a control (non-peak) region showed a markedly increased m6A abundance of AFF4 mRNA in ALDH1+ BCa cells (Figures 3(c) and 3(d)), which suggest the difference of AFF4 expression between CSCs and non-CSCs is regulated by METTL3-mediated m6A modification primarily. To validate if AFF4 acted downstream of METTL3 in BCSCs, we further analyzed the effect of AFF4 deficiency on the BCa self-renewal using a similar strategy to METTL3 knockdown. With effective ablation of AFF4 expression by siRNAs in both 5637 and UM-UC-3 cells (Figures 4(a) and 4(b)), both ALDH activity (Figures 4(c) and 4(d)) and sphere formation frequency (Figures 4(e) and 4(f)) showed a significant decrease upon AFF4 knockdown, which mimic the phenotype resulting from METTL3 knockdown and indicate the regulatory relationship between AFF4 and METTL3 in BCSC self-renewal.
Figure 3.
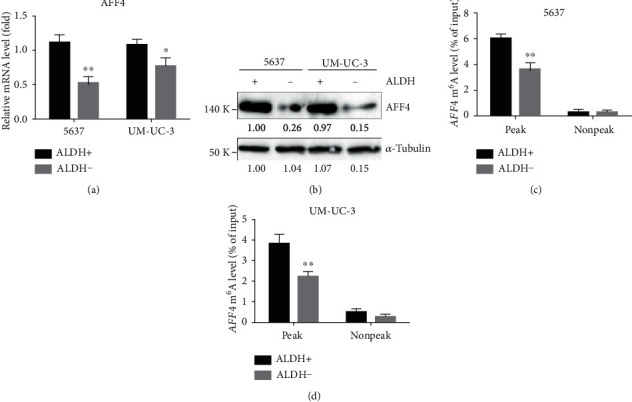
Differential expression level and m6A levels of AFF4 between CSCs and non-CSCs of BCa. mRNA levels (a) and protein levels (b) of AFF4 in ALDH1-positive and ALDH1-negative BCa cells. m6A modification in specific regions of AFF4 transcripts in ALDH1-positive and ALDH1-negative 5637 (c) and UM-UC-3 (d) cells was tested by gene-specific m6A-qPCR assay. ∗p < 0.05, ∗∗p < 0.01 compared to the scramble group, by Student t-test.
Figure 4.
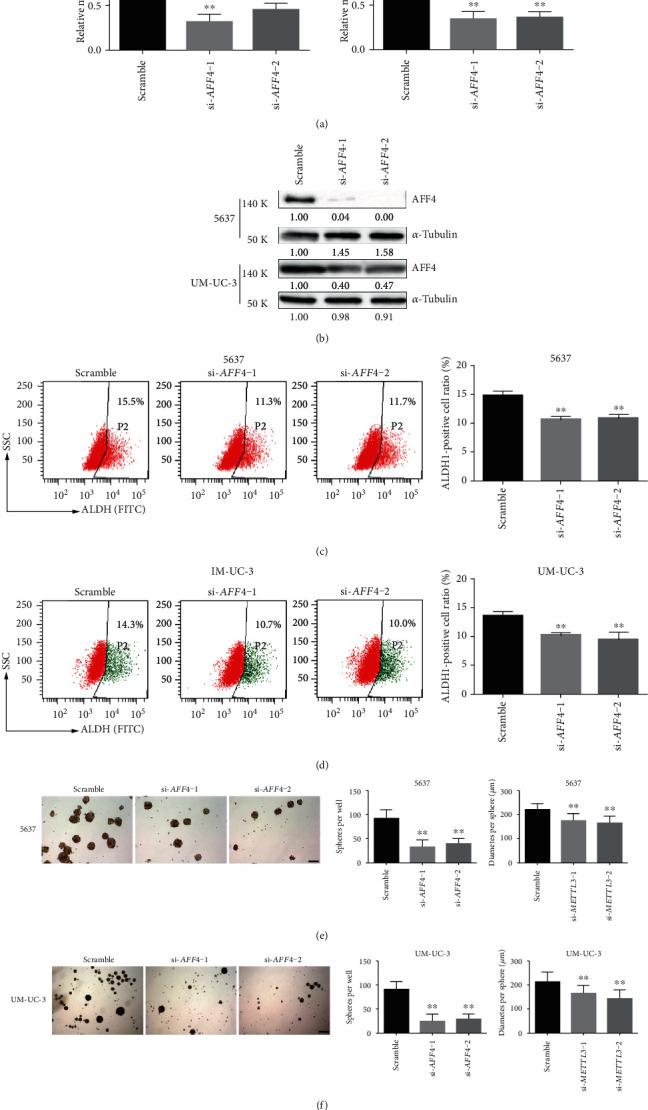
AFF4 mimic the phenotype of METTL3 in regulating the stemness of BCSCs. The knockdown effect of specific siRNAs (si-AFF4-1 and si-AFF4-2) in 5637 and UM-UC-3 cells was verified at both the mRNA ((a) by qRT-PCR) and protein levels ((b) by Western blot). Ratio of cells with high ALDH activity (c, d); number and size of spheres formed in stem cell medium (e, f) of the BCa cells transfected with indicated siRNAs are plotted, and representative images are presented. ∗p < 0.05, ∗∗p < 0.01 compared to the scramble group, by Student t-test. Scale bar, 250 μm.
3.4. AFF4 Directly Regulates MYC and SOX2 Gene Expression in BCa Cells
As an essential component of SEC, AFF4 can bind to DNA directly and regulate the transcription elongation of many genes. MYC and SOX2, well-known pluripotency factors of CSCs, have been reported to be regulated by AFF4 in BCa [11] and HNSCC [30], respectively. To investigate whether MYC and SOX2 are effectors of AFF4 in regulating the self-renewal capability of BCSCs, we performed CHIP assay in 5637 and UM-UC-3 cells and found AFF4 directly bound to MYC and SOX2 promoter regions, which were barely detectable after AFF4 knockdown (Figure 5(a)). Besides, we also confirmed the expression of MYC and SOX2 in response to AFF4 knockdown by qRT-PCR and Western blot. The results showed knockdown of AFF4 drastically reduced the expression of these two genes at both mRNA level and protein level (Figures 5(b) and 5(c)).
Figure 5.
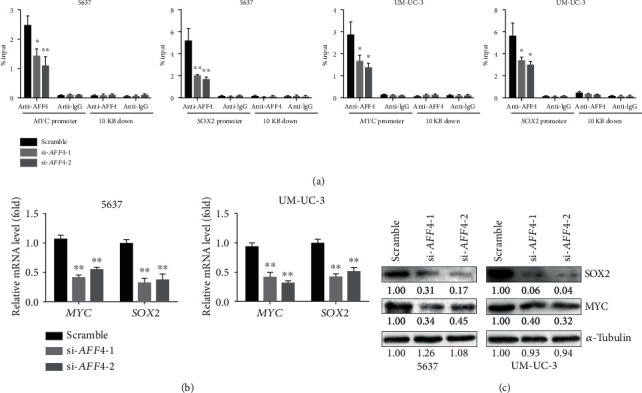
AFF4 regulates SOX2 and MYC expression in BCa cells. (a) ChIP assay showed the recruitment of AFF4 at MYC and SOX2 promoter regions in 5637 and UM-UC-3 cells transfected with indicated siRNAs at 48 h posttransfection. Expression of MYC and SOX2 in 5637 and UM-UC-3 cells transfected with indicated siRNAs (si-AFF4-1 and si-AFF4-2) was verified at both the mRNA ((b) by qRT-PCR) and protein levels ((c) by Western blot). ∗p < 0.05, ∗∗p < 0.01 relative to the scramble group by Student t-test.
3.5. AFF4 Promotes BCSC Self-Renewal In Vivo and Is a Negative Prognostic Factor for BCa Patients
To further evaluate the effect of AFF4 depletion on the self-renewal capacity of BCSCs in vivo, we conducted limiting dilution transplantation assay, a method widely used to assess cancer stem cell content. AFF4 expression was stably ablated by short hairpin RNA (sh-AFF4) in 5637 cells, which were then injected subcutaneously into immune-deficient mice, and tumor growth was measured over time. Consistent with the in vitro results, AFF4-deficient cells exhibited a significantly lower tumor-propagating potential than the control cells (sh-GFP) comprising the tumor bulk (Figures 6(a) and 6(b)). Following ALDH1 staining and FACS analysis showed a clear reduction of ALDH-positive ratio (Figure 6(c)), along with AFF4, SOX2, and MYC expression in xenografts generated from sh-AFF4 5637 cells relative to the control tumors (Figure 6(d)).
Figure 6.
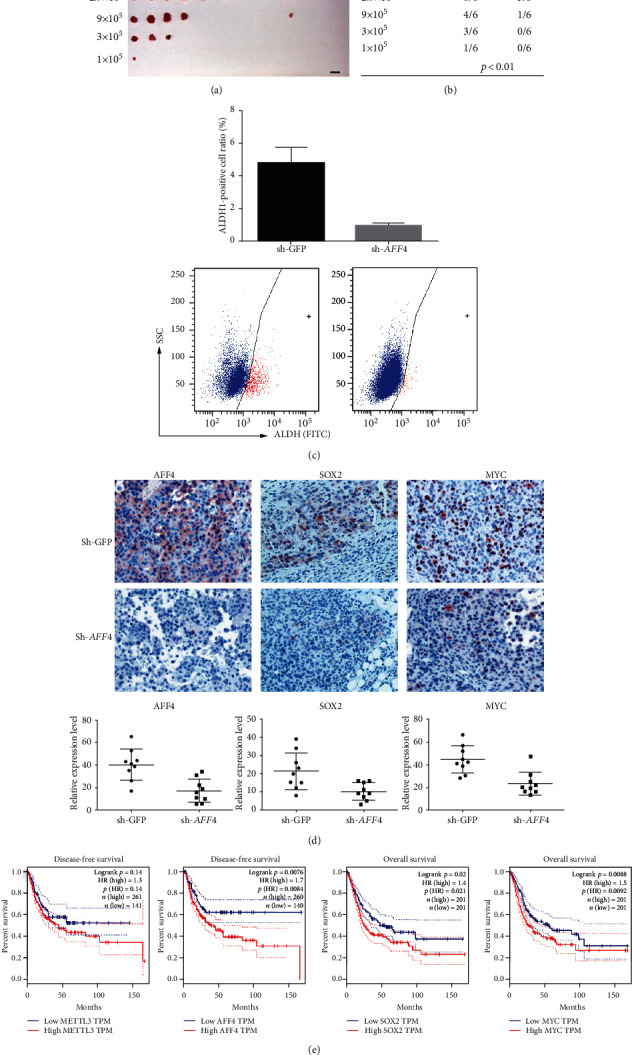
AFF4 is essential for BCa tumor propagating in vivo. Graph (a) and quantification (b) of the percentage of tumor-free mice 30 days after subcutaneous injection of different dilutions of AFF4 knockdown 5637 cells or control cells into immunodeficient mice (n = 6 for each dilution). (c) Ratio of ALDH-positive cells from the xenografts. ∗∗p < 0.01 by Student t-test. (d) Quantitative measurement and representative images of AFF4, SOX2, and MYC expression in xenografts generated by AFF4 stable knockdown BCa cells and control cells. (∗∗p < 0.01 by Student t-test). Scale bar, 50 μm. (e) Correlation between METTL3, AFF4, SOX2, and MYC mRNA expression and survival of BCa patients in TCGA dataset. Disease-free or overall patient survival in groups of high and low expression was analyzed by the Kaplan-Meier survival curve and compared by the log-rank test.
The cancer stemness properties of BCSCs contribute to the chemoresistance, metastasis, and recurrence, which are often related to poor clinical outcome. We queried The Cancer Genome Atlas (TCGA) database and analyzed the survival curve of BCa with the help of the GEPIA online tool [36]. A worse disease-free survival was found in the METTL3 high expression group than that in the METTL3 low expression group, and the p value shows a certain trend toward significance (Figure 6(e), p = 0.11), while higher expression of AFF4 was clearly a significant indicator of poor prognosis of BCa (Figure 6(e), p = 0.008). Besides, the higher expression of SOX2 (Figure 6(e), p = 0.02) and MYC (Figure 6(e), p = 0.009) was also significantly associated with worse overall survival. Taken together, aberrant expression of AFF4 is associated with BCSCs within the tumor bulk which may lead to poor prognosis.
4. Discussion
We have shown in our previous study that METTL3 plays a critical role in the pathogenesis of BCa, by positively regulating the expression of IKBKB, RELA, AFF4, and MYC through m6A-based posttranscriptional regulation [11]. Here, we demonstrate that mRNA m6A modification is critical for maintaining BCSC self-renewal and tumor development. The knockdown of METTL3 expression reduced the self-renewal of BCSCs. Emerging data have suggested that the global abundance of m6A and expression levels of its regulators, including writers, erasers, and readers, are often dysregulated in various types of cancers and are critical for cancer initiation, progression, metastasis, and drug resistance and cancer relapse [22]. Intriguingly, reasons of m6A dysregulation in CSCs are different among various types of cancer, considering the roles of FTO, ALKBH5, and METTL3 in glioblastoma stem cells [24, 25] and of METTL14 and FTO in leukemia stem cells [26, 27]. In BCa, our data shows METTL3 is the only regulator that is aberrantly expressed and critical for BCa pathogenesis and BCSC maintenance. This study uncovered a critical role of mRNA m6A modification in regulating BCSCs self-renewal and tumorigenesis. Nevertheless, the reason for aberrant METTL3 expression in BCa is still unknown and awaits further investigation.
AFF4 is a core component and required for SEC stability and activity, by acting as a scaffold to assemble the SEC [37, 38]. Evidences showing that AFF4 might play a role in regulating pluripotency include its involvement in the osteogenic differentiation of human mesenchymal stem cells [28] and odontogenic differentiation of human dental pulp cells [29]. AFF4 is also required for the tumor-initiating capacity of stem-like cells in HNSCC [30]. In our previous study, AFF4 was indicated by our transcriptome and m6A sequencing data to be a direct target of METTL3 in BCa cells; we then demonstrated that AFF4 mRNA is regulated by METTL3 in a m6A-dependent manner [11]. In the current study, we reveal that both the m6A abundance and the expression level of AFF4 mRNA are elevated in BCSCs, which is consistent with the expression pattern of METTL3. Moreover, ALDH activity and sphere-forming ability in vitro as well as tumor-initiating capacity in vivo were all abrogated upon AFF4 knockdown. Besides, there was a clear correlation between AFF4 expression and BCa invasion potential [11], which is another commonly used indicator of tumorigenicity. Taken together, our data suggest AFF4 is a bona fide target of METTL3 in regulating the self-renewal capacity of BCSCs.
Our previous work proved Sox2 as a marker for stem-like tumor cells of BCa in vivo [7]. Besides, there are evidences indicated that downregulation of c-Myc suppressed CSC differentiation in BCa, and overexpression of c-Myc increased the levels of stem cell markers including SOX2 [39]. Therefore, SOX2 and MYC both are master regulators of self-renewal and differentiation of CSCs and are essential for BCa initiation and progression. SOX2 mRNA was reported to contain m6A modification in embryonic stem cells [40] and glioblastoma stem cells [24], and m6A modification of MYC mRNA was found in the CSCs of acute myeloid leukemia. Indeed, we have also confirmed MELLT3 could regulate MYC expression by promoting the m6A modification of its mRNA in BCa cells [11]. It is likely that METTL3 promote the expression of SOX2 and MYC through m6A-based posttranscriptional regulation as well as AFF4-mediated regulation at the transcriptional level, which reinforces the signal activating the tumor-initiating and self-renewal capabilities of BCa cells. Meanwhile, methyltransferase METTL3 has a global effect on many RNAs; just like AFF4/SEC, MYC and SOX2 exert a broad effect on the expression of various pluripotency-related genes by binding to multiple sites of DNA. Therefore, the role of METTL3 regulating BCSCs might not merely rely on AFF4. Other potential target genes involved in BCa initiation and self-renewal need to be investigated.
In summary, we found m6A modification of AFF4 RNA was upregulated by METTL3 and their expression was elevated in BCSCs, which in turn promotes the expression of SOX2 and MYC to enhance tumorigenesis and tumor-initiating capacity of BCa. Our findings indicate AFF4 may serve as a biomarker and a potential target of therapies for patients with BCa.
Acknowledgments
The authors thank the Center for Scientific Research of Anhui Medical University and Facilities of School of Life Science for valuable help in our experiment. This work was supported by the National Natural Science Foundation of China (81872313 and 81672776 to YL, 81802391 to QG, and 31501838 to XH Z), the Anhui University Natural Science Research Project (KJ2019A0236 to YY Z), and the Anhui Provincial Natural Science Foundation (1808085QH266 to QG).
Contributor Information
Yingyin Zhang, Email: zhangyingyin@ahmu.edu.cn.
Yang Li, Email: liyang@ahmu.edu.cn.
Data Availability
All data is available upon request by contacting the corresponding authors: Yang Li, Ph.D. Department of Genetics, School of Life Science, Anhui Medical University, Hefei, Anhui 230031, China; Tel.: 86 551-65160327; E-mail: liyang@ahmu.edu.cn; and Yingyin Zhang, Department of Genetics, School of Life Science, Anhui Medical University, Hefei, Anhui 230031, China; Tel.: 86 551-65169646; E-mail: liyang@ahmu.edu.cn.
Conflicts of Interest
All the authors have declared no conflict of interest.
Authors' Contributions
Qian Gao and Jin Zheng contributed equally to this work.
Supplementary Materials
Supplementary Table 1: information of primers. Supplementary Table 2: information of siRNAs. Supplementary Table 3: information of antibodies. Supplementary Figure 1: backbone of the plasmids used for shRNA construct.
References
- 1.Batlle E., Clevers H. Cancer stem cells revisited. Nature medicine. 2017;23(10):1124–1134. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2020. CA: a cancer journal for clinicians. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 3.Zigeuner R. News in diagnosis, treatment, and risk group assessment. Nature reviews Urology. 2017;14(2):74–76. doi: 10.1038/nrurol.2016.264. [DOI] [PubMed] [Google Scholar]
- 4.Chan K. S., Espinosa I., Chao M., et al. Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor-initiating cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(33):14016–14021. doi: 10.1073/pnas.0906549106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Batavia J., Yamany T., Molotkov A., et al. Bladder cancers arise from distinct urothelial sub-populations. Nature Cell Biology. 2014;16(10):982–991. doi: 10.1038/ncb3038. 1-5. [DOI] [PubMed] [Google Scholar]
- 6.Papafotiou G., Paraskevopoulou V., Vasilaki E., Kanaki Z., Paschalidis N., Klinakis A. KRT14 marks a subpopulation of bladder basal cells with pivotal role in regeneration and tumorigenesis. Nature Communications. 2016;7(1) doi: 10.1038/ncomms11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu F., Qian W., Zhang H., et al. SOX2 is a marker for stem-like tumor cells in bladder cancer. Stem cell reports. 2017;9(2):429–437. doi: 10.1016/j.stemcr.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu M., Sheng L., Cheng M., et al. Low doses of decitabine improve the chemotherapy efficacy against basal-like bladder cancer by targeting cancer stem cells. Oncogene. 2019;38(27):5425–5439. doi: 10.1038/s41388-019-0799-1. [DOI] [PubMed] [Google Scholar]
- 9.Xie H., Li J., Ying Y., et al. METTL3/YTHDF2 m6A axis promotes tumorigenesis by degrading SETD7 and KLF4 mRNAs in bladder cancer. Journal of cellular and molecular medicine. 2020;24(7):4092–4104. doi: 10.1111/jcmm.15063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han J., Wang J. Z., Yang X., et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Molecular cancer. 2019;18(1):p. 110. doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng M., Sheng L., Gao Q., et al. The m6A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-κB/MYC signaling network. Oncogene. 2019;38(19):3667–3680. doi: 10.1038/s41388-019-0683-z. [DOI] [PubMed] [Google Scholar]
- 12.Desrosiers R., Friderici K., Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proceedings of the National Academy of Sciences of the United States of America. 1974;71(10):3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 14.Meyer K. D., Patil D. P., Zhou J., et al. 5′ UTR m6A Promotes Cap-Independent Translation. Cell. 2015;163(4):999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bokar J. A., Rath-Shambaugh M. E., Ludwiczak R., Narayan P., Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. The Journal of biological chemistry. 1994;269(26):17697–17704. [PubMed] [Google Scholar]
- 16.Liu J., Yue Y., Han D., et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA _N_ 6-adenosine methylation. Nature chemical biology. 2014;10(2):93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ping X. L., Sun B. F., Wang L., et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell research. 2014;24(2):177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y., Li Y., Toth J. I., Petroski M. D., Zhang Z., Zhao J. C. _N_ 6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nature cell biology. 2014;16(2):191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Y., Xie L., Wang M., et al. Mettl3-mediated m6A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nature Communications. 2018;9(1):p. 4772. doi: 10.1038/s41467-018-06898-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia G., Fu Y., Zhao X., et al. _N_ 6-Methyladenosine in nuclear RNA is a major substrate of the obesity- associated FTO. Nature chemical biology. 2011;7(12):885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng G., Dahl J. A., Niu Y., et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular Cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H., Weng H., Chen J. m6A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell. 2020;37(3):270–288. doi: 10.1016/j.ccell.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang C., Samanta D., Lu H., et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(14):E2047–E2056. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang S., Zhao B. S., Zhou A., et al. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell. 2017;31(4):591–606.e6. doi: 10.1016/j.ccell.2017.02.013. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui Q., Shi H., Ye P., et al. m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Reports. 2017;18(11):2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vu L. P., Pickering B. F., Cheng Y., et al. The _N_ 6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nature medicine. 2017;23(11):1369–1376. doi: 10.1038/nm.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weng H., Huang H., Wu H., et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m6A Modification. Cell Stem Cell. 2018;22(2):191–205.e9. doi: 10.1016/j.stem.2017.11.016. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou C.-c., Xiong Q.-c., Zhu X.-x., et al. AFF1 and AFF4 differentially regulate the osteogenic differentiation of human MSCs. Bone research. 2017;5(1) doi: 10.1038/boneres.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y., Xiao Q., Wu Z., Xu R., Zou S., Zhou C. AFF4 enhances odontogenic differentiation of human dental pulp cells. Biochemical and biophysical research communications. 2020;525(3):687–692. doi: 10.1016/j.bbrc.2020.02.122. [DOI] [PubMed] [Google Scholar]
- 30.Deng P., Wang J., Zhang X., et al. AFF4 promotes tumorigenesis and tumor-initiation capacity of head and neck squamous cell carcinoma cells by regulating SOX2. Carcinogenesis. 2018;39(7):937–947. doi: 10.1093/carcin/bgy046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo Z., Lin C., Guest E., et al. The super elongation complex family of RNA polymerase II elongation factors: gene target specificity and transcriptional output. Molecular and cellular biology. 2012;32(13):2608–2617. doi: 10.1128/MCB.00182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., Deng H., Lv L., et al. The miR-193a-3p-regulated ING5 gene activates the DNA damage response pathway and inhibits multi-chemoresistance in bladder cancer. Oncotarget. 2015;6(12):10195–10206. doi: 10.18632/oncotarget.3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang Y., Zhu F., Zhang H., et al. Conditional ablation of TGF-β signaling inhibits tumor progression and invasion in an induced mouse bladder cancer model. Scientific Reports. 2016;6(1) doi: 10.1038/srep29479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Y., Smyth G. K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. Journal of immunological methods. 2009;347(1-2):70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Su Y., Qiu Q., Zhang X., et al. Aldehyde dehydrogenase 1 A1-positive cell population is enriched in tumor-initiating cells and associated with progression of bladder cancer. Cancer Epidemiology Biomarkers & Prevention. 2010;19(2):327–337. doi: 10.1158/1055-9965.EPI-09-0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Research. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin C., Smith E. R., Takahashi H., et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Molecular Cell. 2010;37(3):429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mueller D., Garcia-Cuellar M. P., Bach C., Buhl S., Maethner E., Slany R. K. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biology. 2009;7(11):p. e1000249. doi: 10.1371/journal.pbio.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Q., Yin Q., Mao Y., et al. Hsa_circ_0068307 mediates bladder cancer stem cell-like properties via miR-147/c-Myc axis regulation. Cancer Cell International. 2020;20(1):p. 151. doi: 10.1186/s12935-020-01235-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Batista P. J., Molinie B., Wang J., et al. m6A RNA Modification Controls Cell Fate Transition in Mammalian Embryonic Stem Cells. Cell Stem Cell. 2014;15(6):707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: information of primers. Supplementary Table 2: information of siRNAs. Supplementary Table 3: information of antibodies. Supplementary Figure 1: backbone of the plasmids used for shRNA construct.
Data Availability Statement
All data is available upon request by contacting the corresponding authors: Yang Li, Ph.D. Department of Genetics, School of Life Science, Anhui Medical University, Hefei, Anhui 230031, China; Tel.: 86 551-65160327; E-mail: liyang@ahmu.edu.cn; and Yingyin Zhang, Department of Genetics, School of Life Science, Anhui Medical University, Hefei, Anhui 230031, China; Tel.: 86 551-65169646; E-mail: liyang@ahmu.edu.cn.


