Abstract
Circular RNAs (circRNAs) are covalently closed, endogenous non-coding RNAs and certain circRNAs are linked to human tumors. Owing to their circular form, circRNAs are protected from degradation by exonucleases, and therefore, they are more stable than linear RNAs. Many circRNAs have been shown to sponge microRNAs, interact with RNA-binding proteins, regulate gene transcription, and be translated into proteins. Mounting evidence suggests that circRNAs are dysregulated in cancer tissues and can mediate various signaling pathways, thus affecting tumorigenesis, metastasis, and remodeling of the tumor microenvironment. First, we review the characteristics, biogenesis, and biological functions of circRNAs, and describe various mechanistic models of circRNAs. Then, we provide a systematic overview of the functional roles of circRNAs in gynecological cancers. Finally, we describe the potential future applications of circRNAs as biomarkers for prognostic stratification and as therapeutic targets in gynecological cancers. Although the function of most circRNAs remains elusive, some individual circRNAs have biologically relevant functions in cervical cancer, ovarian cancer, and endometrial cancer. Certain circRNAs have the potential to serve as biomarkers and therapeutic targets in gynecological cancers.
Keywords: circular RNA, gynecological cancer, biomarker, non-coding RNA, cancer diagnosis, cancer treatment
1. Introduction
The Encyclopedia of DNA Elements (ENCODE) project consortium reported that only 2% of the transcribed RNAs are translated into proteins, whereas the majority of other transcripts have no apparent coding potential [1]. Increasing evidence suggests that a significant part of such non-protein coding RNAs (ncRNAs) participate in complex regulatory networks composed of other nucleic acids or proteins [2,3]. The complexity of these networks allows ncRNAs to regulate the expression of a broad spectrum of genes [2,3].
Generally, ncRNAs can be separated into small non-coding RNAs (small ncRNAs), which are shorter than 50 nt, medium non-coding RNAs, which are between 50–200 nt, and long non-coding RNAs (lncRNAs), which are longer than 200 nt [4]. MicroRNAs (miRNAs) are the most studied small ncRNAs and they typically bind to the 3′-untranslated regions (3′-UTR) of protein-coding mRNA by imperfect sequence-specific recognition to either degrade it or repress its translation [5]. LncRNAs can be further subdivided into various categories, including linear RNAs and circular RNAs (circRNAs) [4]. Linear lncRNAs are involved in a broad range of biological processes and are key regulators of cancer metastasis [6,7].
CircRNAs were first discovered in the 1970s [8,9] and were initially thought to be a non-functional by-product of aberrant splicing in cells [10]. However, recent advances in sequencing technologies have revealed that large numbers of circRNAs are broadly expressed in a wide range of mammalian tissue [11]. Over 30,000 circRNAs have already been found in human tissues [12]. Due to their circular form and lack of free ends, circRNAs are resistant to RNA degradation by an exonuclease and have greater stability than linear RNAs [13]. Although the functions of most circRNAs are still unknown, at least some individual circRNAs are novel players in normal physiological and pathological conditions [14,15].
In this review, we discuss the characteristics, biogenesis, biological functions, and working mechanisms of circRNAs, with a particular emphasis on important findings concerning gynecological cancers.
2. General Characteristics of circRNAs
CircRNAs are covalently-closed RNA molecules, and they are produced by a non-canonical splicing process called “back-splicing”, which takes place in the nucleus [16]. Unlike typical linear RNAs, the 5′ cap and 3′ polyadenylation tail (poly-(A) tail) are absent in circRNAs. Therefore, circRNAs are not easily degraded by an exonuclease and are considerably more stable than linear RNAs. The average half-life of circRNAs in mammary cells is at least 2.5–4.8 times longer than the median half-life of their linear counterparts [13,17]. Following biogenesis, the majority of circRNAs, with the exception of intron-containing circRNAs that mainly exist in the nucleus, are exported to the cytoplasm in a size-dependent manner [18].
On average, circRNAs contain 1–5 exons and are 500 nt long [16]. The expression of a circular RNA does not often correlate with the expression of its cognate linear mRNA [16]. Of note, some circRNAs are expressed at a much lower or higher level than their linear counterparts [16]. Many circRNAs are highly conserved across mammals [17]. circRNAs exhibit dynamic expression levels during development and are expressed in a tissue-specific manner [19,20,21].
3. Mechanisms of circRNA Formation and Degradation
Although the mechanism of circRNA formation is still unclear, the majority of circRNAs are considered to originate from exons in the coding region of a gene, and the rest originate from the 5′- or 3′-UTRs, introns and intergenic regions, as well as from antisense RNAs [15]. According to their structural domains, circRNAs can be divided into four categories: exonic circRNA (ecircRNA), circular intronic RNA (ciRNA), exonic-intronic circRNA (EIciRNA) and intergenic circRNA [22]. EcircRNAs appear to be the most abundant circRNA type, accounting for over 80% of known circRNAs [23]. There are currently three hypothetical models to explain the formation of ecircRNAs: lariat-driven circularization, intron-pairing-driven circularization, and RNA-binding protein-mediated circularization [24,25]. The generation of ecircRNAs is a result of pre-mRNA splicing when the 3′ splice donor attaches to the 5′ splice acceptor [24,25]. Researchers have also demonstrated that RNA-binding proteins can serve as activators or inhibitors in the formation of circRNAs [24,25]. For instance, Quaking preferentially binds to a particular sequence motif on a linear RNA and bridges the two bracketing introns together, thus facilitating the formation of circRNAs in human mammary epithelial cells [26].
Compared to the mechanisms of circRNA formation, our understanding of circRNA degradation is limited. Some miRNAs can direct Ago2 to degrade circRNAs in human lung cancer cells and HEK-293 cells [27,28]. A previous study reported that an endonuclease, RNase P/MRP, cuts N6-methyladenosine (m6A)-containing circRNA in a sequence-dependent manner [29]. Upon viral infection, the activation of the endonuclease RNase L is responsible for circRNAs degradation [30]. A good number of circRNAs in human tumor cells are predicted to be highly structured, and their decay is regulated by UPF1 and G3BP1, two RNA-binding proteins [31]. In addition, human tumor cells can remove cytoplasmic circRNAs by releasing cargo-bearing extracellular vesicles, such as microvesicles and exosomes [32,33].
4. Biological Functions and Mechanistic Models of circRNAs in Tumors
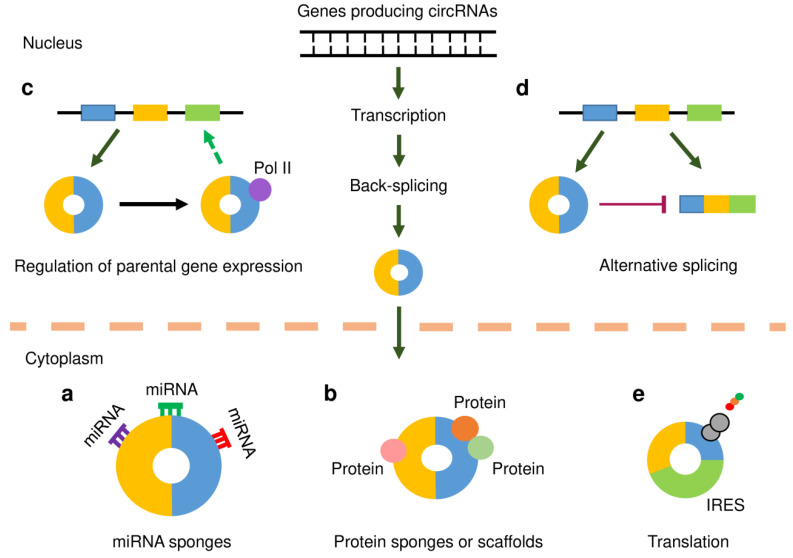
In human tumor cells, circRNAs are believed to serve as miRNA (or protein) sponges, scaffolds for proteins, regulators of transcription or splicing, and a few circRNAs can be translated into peptides or proteins under certain conditions (Figure 1). Some circRNAs can regulate cancer metastasis and affect the tumor microenvironment.
Figure 1.
Functional mechanisms of circular RNAs (circRNAs). CircRNAs, like mRNAs, are transcribed from protein-coding genes by RNA polymerase II (Pol II). However, they are spliced in a non-canonical splicing process known as back-splicing. Following biogenesis, the majority of circRNAs are exported to the cytoplasm in a size-dependent manner. (a) CircRNAs may act as microRNA (miRNA) sponges by binding to miRNAs and inhibiting their functions, thereby releasing downstream target genes from miRNA-mediated repression. (b) CircRNAs that harbor binding sites for RNA-binding proteins (RBPs) may function as protein sponges and thus regulate gene expression. In addition, circRNAs may work as scaffolds in the assembly of protein complexes. (c) CircRNAs may associate with Pol II to enhance the transcription of their parental genes. (d) CircRNAs may compete with their linear counterparts against canonical pre-mRNA splicing, thus suppressing the expression of their parent genes. (e) CircRNAs that contain internal ribosomal entry site (IRES) elements and open reading frame may be translated into protein or polypeptide.
4.1. MiRNA Sponge
One of the most frequently studied functions of circRNA is its ability to act as a miRNA sponge. Recent observations have found that some lncRNAs act as competing endogenous RNAs (ceRNAs), which regulate other RNA transcripts by competing for shared miRNAs [25] (Figure 1a). CircRNAs that harbor multiple binding sites for miRNAs can function as efficient miRNA sponges to indirectly regulate the expression of protein-coding genes [34]. For example, circHIPK3 modulates tumor cell growth by silencing nine miRNAs [23]. Despite the consistency of these findings, there are a few examples that challenge this ceRNA hypothesis for circRNAs [35,36]. For example, bioinformatics analysis has indicated that miRNA-binding sites are generally not enriched in circRNAs any more than could be expected by chance, with the exception of the reported circRNA, ciRS-7 [35]. In another study, the circRNAs downloaded from the circBase database were screened for Ago2-bound regions [36]. As a result, 58,063 circRNAs were found to own Ago2-bound regions. Six circRNAs were selected, and the potential ability of these circRNAs to interact with miRNAs was explored using RNA-binding protein immunoprecipitation-PCR experiments. The results revealed that these six circRNAs did not exhibit strong potential to serve as miRNA sponges [36].
4.2. Protein Sponge and Scaffold
RNA-binding proteins are a class of proteins that bind RNAs to regulate the metabolic processing of RNAs by mediating their maturation, transport, localization, translation and turnover [37] (Figure 1b). CircRNAs that contain multiple complementary binding sites to one or multiple proteins might function as protein sponges. Several circRNAs were suggested to sequester HuR, an extensively studied RNA-binding protein, which consequently affects its action on target gene regulation [38,39,40]. The interactions between circRNAs and RNA-binding proteins have been shown to influence cancer progression [41]. In addition, circRNAs were observed to act as dynamic scaffolds that bring different proteins together to form a complex [41]. In mouse fibroblast cells, circ-Foxo3 can interact with both p21 and CDK2 to form a ternary complex that results in the inhibition of cell cycle progression [42].
4.3. Transcription and Splicing
Unlike ecircRNAs, which are located in the cytoplasm, ciRNAs and EIciRNAs are primarily enriched in the nucleus. Previous studies have shown that a class of EIciRNAs interact with RNA polymerase II (Pol II) and accumulate at transcription sites, thereby promoting the transcription of their parental genes [43,44] (Figure 1c). Moreover, certain circRNA can compete with their linear counterparts against canonical pre-mRNA splicing, thus suppressing the expression of their parent genes [45,46] (Figure 1d).
4.4. Translation of circRNAs
CircRNAs were initially regarded as ncRNAs that lack protein-coding abilities, owing to the absence of a 5′ cap structure and poly-(A) tail. However, subsequent studies have revealed that some circRNAs carry open reading frames, and can be translated into peptides or proteins [47,48] (Figure 1e). The presence of internal ribosomal entry sites (IRES) was shown to drive circRNA translation through a cap-independent mechanism [49]. N6-methyladenosine (m6A) refers to the methylation that occurs in the mRNAs, and the presence of m6A on circRNA has the potential to initiate cap-independent translation [50]. The peptides or proteins encoded by circRNAs have critical functions in mediating cancer development [51,52]. Therefore, it seems necessary to determine whether the proteins originating from circRNA translation are functional in gynecological cancers.
5. Methods for Detecting and Characterizing circRNAs
The detailed description of bioinformatic tools, laboratory techniques and general workflow used for circRNA research has been reviewed elsewhere [53,54,55,56,57].
Previous studies have demonstrated that tumor-adjacent tissues have similar molecular changes to those found in the tumor tissues or present a unique intermediate state between healthy and tumor tissues [58,59,60]. One example is miR-29c, which is significantly upregulated in tumor-adjacent tissues when compared with gastric cancer tissues as well as non-cancerous tissues [60]. Most of the previous studies that analyze circRNA global expression in human cancer tissues have only used matched tumor-adjacent samples as normal control [61,62]. The results indicate that the expression of circRNAs in cancer tissues is frequently downregulated when compared with the matched tumor-adjacent tissues [61,62]. However, some circRNAs are actually overexpressed in tumor-adjacent tissues in comparison to cancer samples and normal samples without cancer [63], suggesting that the use of tumor-adjacent tissues for comparison purposes may lead to experimental biases regarding the true circRNA expression profile associated with tumors [63].
The functional characterization of circRNAs is commonly conducted by following two types of strategies: RNA interference (RNAi)-induced circRNA silencing and gain-of-function studies using circRNA expression plasmids. One of the challenges for circRNA overexpression is that the vectors used for overexpressing circRNA might generate many undesired transcripts, such as linear RNAs or concatenated and/or intertwined circRNAs [64,65]. Therefore, it might be difficult to determine if a specific circRNA has an impact on a particular phenotype in tumor cells. Although recently developed expression vectors were believed to efficiently drive circRNA formation, but not linear RNAs [66], the combination of RNAi-based gene silencing with vector-mediated overexpression approaches may present more opportunities to fully understand the exact functions of circRNAs in tumor cells.
Recently, the novel CRISPR/Cas13-based approaches using guide RNAs, which target the circRNA-specific back-splicing junction sites, have been used to achieve precise and robust circRNA silencing without disturbing their linear cognate mRNAs [67,68]. These studies suggest that the application of CRISPR/Cas-based circRNA engineering systems may be a promising method for performing functional studies of circRNA in human cancers.
6. Evidence Acquisition
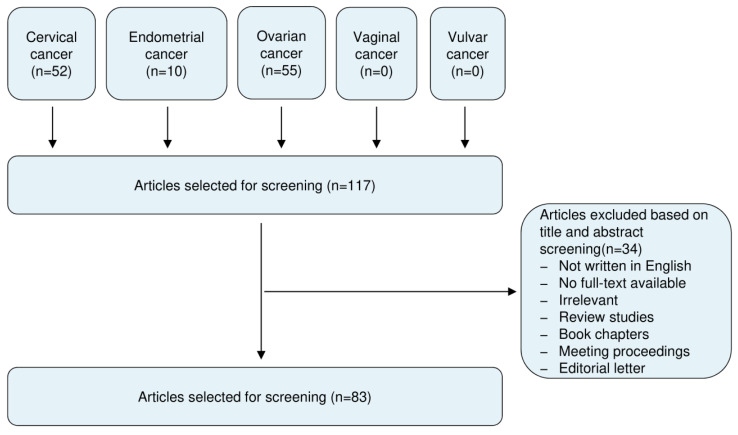
This review included literature from the PubMed and Google Scholar databases published up to 20 February 2020, using the following keywords: circular RNA, circRNA, cervical cancer, endometrial cancer, ovarian cancer, vaginal cancer and vulvar cancer. All studies recognized were assessed for relevance by two authors by checking the title and abstract. After excluding the irrelevant articles, studies without access to the full-text of the publication, case reports, letters, expert opinions, meeting proceedings, review articles and non-English articles, our screening identified 83 articles that addressed the expression, roles and mechanisms of circRNAs in cervical cancer (CC), ovarian cancer (OC) and endometrial cancer (EC). These selected articles were evaluated independently by two authors. A flow diagram of the study selection process was shown in Figure 2.
Figure 2.
Summary of literature search, screening and selection.
7. The Expression, Functions and Mechanisms of circRNAs in Gynecological Cancers
7.1. CircRNAs in CC
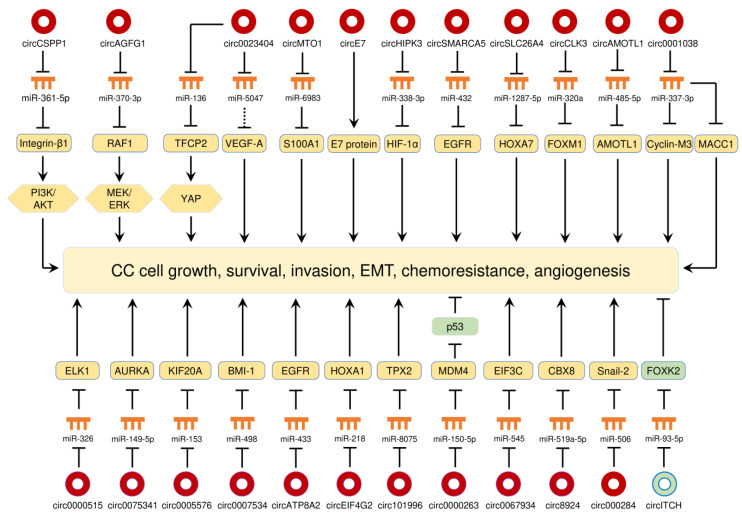
CC is the fourth most common female malignancy in the world and kills more than 300,000 deaths worldwide each year [69]. High-risk subtypes (16 and 18) of human papillomavirus (HPV) are well established as the causative agents responsible for CC [69]. Massive studies have shown that a large number of circRNAs were differentially expressed in CC tissues compared with adjacent normal cervical tissues, and play oncogenic or tumor-suppressive roles in the initiation and progression of CC [70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105]. All of the circRNAs that are abnormally expressed in CC are summarized in Table 1, and their functions in CC are shown in Figure 3. As demonstrated by many studies, circRNAs function as miRNA sponges to regulate different aspects of CC progression through regulating multiple signaling pathways (Figure 3). These studies demonstrate that the circRNA-miRNA-mRNA regulatory networks and an HPV E7 oncoprotein-encoding circRNA play crucial roles in controlling CC tumorigenesis and progression.
Table 1.
The expression and mechanisms of circRNAs in cervical cancer (CC).
| CircRNA | Samples or Cell Lines | Expression | Function | Mechanism | Ref. |
|---|---|---|---|---|---|
| circ0018289 | CC and adjacent normal tissues; HeLa and SiHa cells | Up | Oncogene | Sponges miR-497 | [76] |
| circ000284 | HeLa and SiHa cells | Up | Oncogene | Increases Snail-2 expression via sponging miR-506 | [77] |
| circ0023404 | CC and adjacent normal tissues; HeLa and SiHa cells | Up | Oncogene | Activates YAP pathway via promoting TFCP2 expression by sponging miR-136 | [78] |
| circ8924 | CC and adjacent normal tissues; HeLa and SiHa cells | Up | Oncogene | Increases CBX8 expression via sponging miR-519a-5p | [79] |
| circ0067934 | CC and adjacent normal tissues; HeLa and SiHa cells | Up | Oncogene | Increases EIF3C expression via sponging miR-545 | [80] |
| circ0000263 | HeLa and C33A cells | Up | Oncogene | Inhibits p53 levels by upregulating MDM4 expression via sponging miR-150-5p | [81] |
| circSMARCA5 | CC and adjacent normal tissues; HeLa and C33A cells | Down | TS | Sponges miR-620 | [82] |
| circ101996 | CC and adjacent normal tissues; SiHa and CaSki cells | Up | Oncogene | Increases TPX2 expression by sponging miR-8075 | [83] |
| circEIF4G2 | CC and adjacent normal tissues; HeLa and C33A cells | Up | Oncogene | Increases HOXA1 expression by sponging miR-218 | [84] |
| circATP8A2 | CC and adjacent normal tissues; HeLa and SW756 cells | Up | Oncogene | Increases EGFR expression by sponging miR-433 | [85] |
| circ0023404 | CC and adjacent normal tissues; HeLa and SiHa cells | Up | Oncogene | Increases VEGF-A expression by sponging miR-5047 | [86] |
| circE7 | CaSki cells | - | Oncogene | Codes HPV E7 oncoprotein | [87] |
| circMTO1 | HeLa and SiHa cells | Up | Oncogene | Increase S100A1 expression by sponging miR-6983 | [88] |
| circ0000745 | CC and adjacent normal tissues; CaSki and SiHa cells | Up | Oncogene | Inhibits E-cadherin expression | [89] |
| circ0007534 | CC and adjacent normal tissues; HeLa and SiHa cells | Up | Oncogene | Increases BMI-1 expression by sponging miR-498 | [90] |
| circ0005576 | CC and normal cervical tissues; HeLa and SiHa cells | Up | Oncogene | Increases KIF20A expression by sponging miR-153 | [91] |
| circ0000285 | CC and adjacent normal tissues; C33A cells | Up | Oncogene | Increases FUS expression | [92] |
| circAGFG1 | HeLa and SiHa cells | Up | Oncogene | Activates MEK/ERK pathway via promoting RAF1 expression by sponging miR-370-3p | [93] |
| circ0075341 | CC and normal cervical tissues; CaSki and SiHa cells | Up | Oncogene | Increases AURKA expression by sponging miR-149-5p | [94] |
| circ0000515 | CC and adjacent normal tissues; HeLa and SiHa cells | Up | Oncogene | Increases ELK1 expression by sponging miR-326 | [95] |
| circ0001038 | CC and adjacent normal tissues; HeLa and SW756 cells | Up | Oncogene | Increases Cyclin-M3/MACC1 expression by sponging miR-337-3p | [96] |
| circAMOTL1 | CC, adjacent normal tissues and normal cervical tissues; CaSki and HeLa cells | Up | Oncogene | Increases AMOTL1 expression by sponging miR-485-5p | [97] |
| circMYBL2 | CC, and adjacent normal tissues; CaSki and HeLa cells | Up | Oncogene | Sponges miR-361-3p | [98] |
| circCLK3 | CC and adjacent normal tissues; HeLa and SiHa cells | Up | Oncogene | Induces EMT via promoting FoxM1 expression by sponging miR-320a | [99] |
| ciRS-7 | CC, cervical intraepithelial neoplasia and normal cervical tissues, HeLa, CaSki, C33A and SiHa cells | Up | Oncogene | Increases N-cadherin/Vimentin expression and decreases E-cadherin expression | [100] |
| circSLC26A4 | CC and adjacent normal tissues; CaSki and HeLa cells | Up | Oncogene | Increases HOXA7 expression by sponging miR-1287-5p | [101] |
| circSMARCA5 | CC and adjacent normal tissues; CaSki and HeLa cells | Up | Oncogene | Increases EGFR expression by sponging miR-432 | [102] |
| circITCH | CC and adjacent normal tissues; HeLa cells | Down | TS | Increases FOXK2 expression by sponging miR-93-5p | [103] |
| circHIPK3 | CC and adjacent normal tissues; HeLa and C4I cells | Up | Oncogene | Increases HIF-1α expression by sponging miR-338-3p | [104] |
| circCSPP1 | CC and adjacent normal tissues; HeLa cells | Up | Oncogene | Increases Integrin β1 expression and activate the PI3K/AKT pathway via sponging miR-361-5p | [105] |
TS: tumor suppressor; up: upregulation; down: downregulation.
Figure 3.
The circRNA-miRNA-mRNA regulatory networks and an HPV E7 oncoprotein-encoding circRNA play crucial roles in controlling CC progression, angiogenesis and chemoresistance. Red circles: oncogenic circRNAs; green circles: tumor-suppressive circRNAs.
7.1.1. CircRNAs as Oncogenes in CC
CircRNAs Promote CC Growth, Survival, Invasion, EMT and Chemoresistance
Diverse circRNAs (such as circEIF4G2, circ0007534, circ0005576, circ0075341, circ0000515, circAMOTL1, and circSLC26A4) were previously described to promote the tumorigenesis of CC by increasing the respective downstream targets mRNAs, including HOXA1, BMI-1, KIF20A, AURKA, ELK1, AMOTL1 and HOXA7 via sponging miRNAs [84,90,91,94,95,97,101]. Additionally, circRNAs (such as circ0018289, circ0000285 and circMYBL2,) were found to be overexpressed in CC tissues, and their upregulation enhanced CC cell invasion and metastasis [76,92,98].
The PI3K/AKT signaling pathway controls multiple cellular processes including proliferation, growth, invasion and chemoresistance, and it is one of the most frequently dysregulated pathways in human cancers, including CC [106,107]. Integrin β1 can promote cancer initiation and progression through the activation of multiple downstream pathways, including the PI3K/AKT signaling [108]. By sponging miR-361-5p, circCSPP1 stimulates the expression of integrin β1, allowing downstream activation of the PI3K/AKT pathway, which contributes to CC cell proliferation and migration [105]. The RAF/MEK/ERK cascade plays a critical role in the development and progression of cancer [109]. A recent study suggested that circAGFG1 was upregulated in CC cell lines compared with normal cervical cells, and it could induce CC cell proliferation and migration by elevating RAF1 expression and activating the MEK/ERK pathway, via sponging miR-370-3p (a suppressor of RAF1) [93]. Epithelial-mesenchymal transition (EMT) is a biological process in which epithelial cells acquire mesenchymal phenotypes, thus conferring the malignant properties to cancer cells including invasive behaviors, cancer stem cell properties, and greater resistance to cancer treatment [110]. Experimental studies have revealed that circRNAs affect the EMT program in human cancers (including CC) through different mechanisms [77,80,89,99,100,104,111]. For instance, circ000284 indirectly increases Snail-2 expression to promote EMT and invasion of CC cells through sponging miR-506 [77]. CircCLK3 induces the levels of FOXM1 via sponging miR-320a, and overexpression of FOXM1 could trigger EMT-like changes, including the acquisition of Vimentin and N-cadherin expression and reduction of E-cadherin expression [99]. Also, the upregulation of circ0067934, circ0000745, ciRS-7 and circHIPK3 in CC cells was shown to increase the expression of mesenchymal markers and decrease the expression of epithelial markers [80,89,100,104]. Inactivation of p53 in human tumors occurs through missense mutations in the DNA-binding domain of the p53 protein, and can also result from the amplification/overexpression of its specific inhibitors (such as MDM2 and MDM4) [112]. Circ0000263 can increase the expression of MDM4 by sponging miR-150-5p, thus causing degradation of p53 in CC cells [81].
Circ0023404 enhanced chemoresistance of CC cells to cisplatin by regulating autophagy signaling [86]. Moreover, another circRNA, circMTO1, was shown to upregulate S100A1 expression through sponging miR-6893, thereby promoting tumorigenesis and chemoresistance in CC [88].
CircRNA Encodes HPV E7 Oncoprotein
The oncoproteins E6 and E7 encoded by HPV16 and 18 contribute to the neoplastic transformation of squamous cervical epithelium [113]. The E6 protein from high-risk HPVs binds the p53 tumor suppressor protein for degradation [114], whereas the E7 protein interacts with the retinoblastoma protein and inactivates this cellular protein [115]. A recent report showed that oncogenic HPV16 generates a 472nt circular RNA called circE7, which contains the entire E7 open reading frame [87]. In this study, circE7 was shown to be translated to the E7 oncoprotein, and knockdown of circE7 in CC-derived CaSki cells reduced E7 protein levels and suppresses tumor growth and malignancy both in vitro and in vivo [87]. These results suggest that circE7 appears to be essential for E7 protein expression and the transformation of CaSki cells.
CircRNAs as Tumor Suppressors in CC
Recent data suggest that circSMARCA5 was downregulated in CC tissues compared with adjacent normal tissues, and overexpression of circSMARCA5 suppresses the proliferation and invasion of HeLa cells by sponging miR-620 [82]. However, controversial findings have been reported in another study showing that circSMARCA5 was expressed at higher levels in CC tissues, and knockdown of circSMARCA5 suppressed cellular growth and invasion [102]. The oncogenic functions of circSMARCA5 were mediated through a miR-432/EGFR axis [102]. Besides, the expression of circITCH was significantly decreased in CC tissues compared with the adjacent normal tissues [103]. CircITCH acts as an endogenous miR-93-5p sponge to induce the levels of FOXK2 in CC cells, leading to the suppression of the malignant behaviors [103].
7.2. CircRNAs in OC
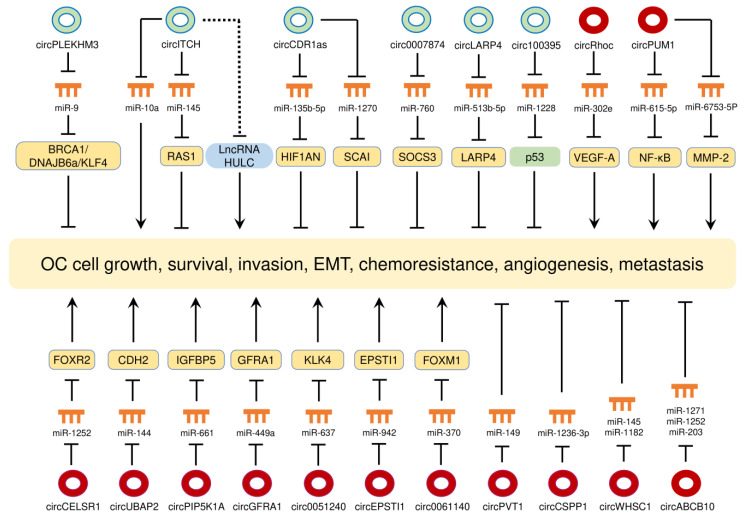
Expression profiling studies and bioinformatics analysis have revealed significant changes in circRNA expression between OC tissues and normal ovarian tissues [116,117], between miliary-type OC and non-miliary OC [118], between serum samples from OC patients and those samples from healthy controls [119], between primary and metastatic sites of OC [120], and between OC cells and non-malignant human ovarian surface epithelial cells [62]. The expression and mechanisms of circRNAs in OC are presented in Table 2 and their biological roles are shown in Figure 4.
Table 2.
The expression and mechanisms of circRNAs in ovarian cancer (OC).
| CircRNA | Samples or Cell Lines | Expression | Function | Mechanism | Ref. |
|---|---|---|---|---|---|
| circEPSTI1 | OC and adjacent normal tissues; OV119 and A2780 cells | Up | Oncogene | Increases EPSTI1 expression by sponging miR-942 | [121] |
| circGFRA1 | OC and adjacent normal tissues; OV119 and A2780 cells | Up | Oncogene | Increases GFRA1 expression by sponging miR-449a | [122] |
| circ0051240 | OC and adjacent normal tissues; OVCAR-3 and H8910 cells | Up | Oncogene | Increases KLK4 expression by sponging miR-637 | [123] |
| circPIP5K1A | OC and adjacent normal tissues; SKOV-3 and A2780 cells | Up | Oncogene | Increases IGFBP5 expression by sponging miR-661 | [124] |
| circPVT1 | SKOV-3 and CAOV3 cells | Up | Oncogene | Sponges miR-149 | [125] |
| circABCB10 | OC and adjacent normal tissues; SKOV-3 cells | Up | Oncogene | Inhibits miR-1271, miR-1252 and miR-203 expression | [126] |
| circSMAD7 | OC and adjacent normal tissues; TOV112D cells | Up | Oncogene | Inhibits KLF6 expression | [127] |
| circANKRD12 | SKOV-3 cells | Up | Oncogene | Increases Cyclin-D1 expression | [128] |
| circCSPP1 | OC, borderline tumor and normal ovarian tissues; OVCAR-3 and A2780 cells | Up | Oncogene | Sponges miR-1236-3p | [129] |
| circ0061140 | SKOV-3 and A2780 cells | Up | Oncogene | Promotes EMT by increasing FOXM1 expression via sponging miR-370 | [130] |
| circUBAP2 | OC and adjacent normal tissues; OVCAR-3 and H8910 cells | Up | Oncogene | Increases CDH2 expression by sponging miR-144 | [131] |
| circWHSC1 | OC and normal ovarian tissues; OVCAR-3 and CAOV3 cells | Up | Oncogene | Increases MUC1/hTERT expression by sponging miR-145/1182 | [132,133,134] |
| circVPS13C- has-circ-001567 | OC and adjacent normal ovarian tissues; SKOV-3 and OV-1063 cells | Up | Oncogene | Increases N-cadherin expression and decreases E-cadherin expression | [135] |
| circCELSR1 | Drug-resistant/sensitive OC tissues; SKOV3 and HeyA-8 cells | Up | Oncogene | Increases FOXR2 expression by sponging miR-1252 | [136] |
| circPUM1 | OC and normal ovarian tissues; OVCAR-3 and A2780 cells | Up | Oncogene | Increases NF-κB/MMP2 expression by sponging miR-615-5p/miR-6753-5p | [137] |
| circRhoc | OC and normal ovarian tissues; CAOV3 and A2780 cells | Up | Oncogene | Increases VEGF-A expression by sponging miR-302e | [138,139,140] |
| circITCH | OC and adjacent normal ovarian tissues; CAOV3 and SKOV-3 cells | Down | TS | Increases RASA1 expression via sponging miR-145 | [141] |
| circITCH | SKOV-3 cells | Down | TS | Sponges miR-10a-α | [142] |
| circITCH | OC and normal ovarian tissues; UWB1.289 and UWB1.289 + BRCA1 cells | Down | TS | Inhibits lncRNA HULC expression | [143] |
| circITCH | OC and adjacent normal ovarian tissues; SKOV-3 cells | Down | TS | - | [144] |
| circCDR1as | OC and normal ovarian tissues; H8910 and A2780 cells | Down | TS | Increases HIF1AN expression by sponging miR-135b-5p | [145] |
| circCDR1as | Drug-resistant/sensitive OC tissues; SKOV3 and A2780 cells | Down | TS | Increases SCAI expression by sponging miR-1270 | [146] |
| circ0007874 | SKOV3 and A2780 cells | Down | TS | Increases SOCS3 expression by sponging miR-760 | [147] |
| circLARP4 | SKOV3 and A2780 cells | Down | TS | Increases LARP4 expression by sponging miR-513b-5p | [148] |
| circPLEKHM3 | Primary OC, metastatic OC and normal ovarian tissues; A2780 and MDAH2274 cells | Down | TS | Inactivates the PI3K/AKT and Wnt/β-catenin pathways via promoting BRCA1, DNAJB6a and KLF4 expression by sponging miR-9 | [149] |
| circ100395 | OC and adjacent normal ovarian tissues; SKOV-3 and ES-2 cells | Down | TS | Increases E-cadherin expression and reduces N-cadherin and Snail expression via promoting p53 expression by sponging miR-1228 | [150] |
TS: tumor suppressor; up: upregulation; down: downregulation.
Figure 4.
The circRNA-miRNA-mRNA regulatory networks play essential roles in mediating ovarian cancer progression, angiogenesis and chemoresistance. Red circles: oncogenic circRNAs; green circles: tumor-suppressive circRNAs.
7.2.1. CircRNAs as Oncogenes in OC
CircRNAs Promote CC Growth, Survival, Invasion, EMT, Chemoresistance and Metastasis
Several circRNAs (such as circEPSTI1, circGFRA1, circ0051240, circPIP5K1A, circPVT1 and circABCB10) have been found to increase the proliferative and invasive capacities of OC cells by acting as miRNA sponges [121,122,123,124,125,126]. Knockdown of circSMAD7 by short-hairpin RNA (shRNA) was associated with decreased cell proliferation, migration and invasion and with increased expression of KLF6 in CC cells [127]. Silencing of circANKRD12 reduced OC cell proliferation, probably through downregulating the expression of cyclin D1 [128].
CircCSPP1 was shown to be highly expressed in OC tissues and borderline tumor tissues when compared with normal ovarian tissues [129]. Further investigations suggest that circCSPP1 sponged miR-1236-p to increase the expression of ZEB1, leading to increased cell invasion and EMT phenotypes [129]. Circ0061140, which was upregulated in OC cell lines relative to normal ovarian epithelial cells, was reported to stimulate OC cell proliferation, invasion and EMT properties, probably via binding to miR-370 [130]. Overexpression of FOXM1, a target of miR-370, rescued the EMT properties that were reduced by either circ0061140 silencing or miR-370 overexpression [130]. By sponging miR-144, circUBAP2 promoted the expression of N-cadherin in OC cells [131]. CircWHSC1 was increasingly expressed in OC tissues compared with normal ovarian tissues, and there was a tendency towards increased levels of circWHSC1 expression in poorly- and moderately-differentiated OCs compared with well-differentiated OCs [132]. By interacting with miR-145/miR-1182, circWHSC1 was shown to elevate the levels of two EMT-inducer genes (MUC1/hTERT) in OC cells [132,133,134]. Inhibition of a circRNA VPS13C-has-circ-001567 by shRNA attenuated cellular invasion, resulting in the downregulation of N-cadherin and upregulation of E-cadherin [135].
CircCELSR1 acts as a sponge for the tumor suppressor miR-1252, and suppression of miR-1252, in turn, contributes to paclitaxel resistance in OC cells, possibly by inducing the expression of its target gene FOXR2 [136].
CircPUM1 was found to be highly expressed in OC tissues compared with normal ovarian tissues, and higher circPUM1 expression was positively correlated with advanced FIGO stage [137]. Studies in both OC cell lines and mouse OC models suggest that circPUM1 promoted cell invasiveness and growth by sponging miR-615-5p and miR-6753-5p to upregulate NF-κB and MMP2 expression [137]. Interestingly, circPUM1 was present in OC cell-derived extracellular vesicles, and exosomal circPUM1 facilitated tumor peritoneal dissemination in vivo [137].
CircRNAs Promotes Angiogenesis in OC
Angiogenesis is influenced by the microenvironment and tightly regulated by a balance between pro- and anti-angiogenic factors [138]. Vascular endothelial growth factor (VEGF-A) has a prominent role in tumor angiogenesis [139]. CircRhoC, a circRNA derived from the RhoC mRNA, was overexpressed in OC tissues relative to normal ovarian tissues, and higher circRhoC expression was detected in advanced-stage tumors compared with early-stage tumors [140]. Suppression of circRhoC with shRNA significantly attenuated cell migration and invasion in vitro, and inhibited the intraperitoneal dissemination of OC cells in vivo [140]. By performing mechanistic analysis, the same study indicated that circRhoC can serve as a sponge for miR-302e to upregulate the expression of VEGF-A, which potentially promotes OC angiogenesis and metastasis [140].
7.2.2. CircRNAs as Tumor Suppressors in OC
Some circRNAs exert their tumor-suppressive roles in OC cells [141,142,143,144,145,146,147,148,149,150] (Table 2 and Figure 4). According to these reports, the “miRNA” sponge seems to be the most common mechanism by which circRNAs regulate the malignant properties of OC cells. Interestingly, one study proposed a different potential mechanism whereby circITCH might inhibit the proliferation of OC cells by downregulating the levels of lncRNA HULC, although the interaction between circITCH and HULC has not yet been defined [143]. Aberrant activation of the Wnt/β-catenin signaling plays a key role in promoting the cancer stem cell self-renewal, metastasis and chemoresistance in OC [151]. By sponging miR-9, circPLEKHM3 functions as a tumor suppressor to enhance the expression of BRCA1, DNAJB6 and KLF4, which consequently inactivate the PI3K/AKT and Wnt/β-catenin signaling pathways in OC cells [149].
7.3. CircRNAs in EC
To date, very few reports on circRNAs in EC have been published [152,153,154,155,156]. The expression profile of circRNAs was found to be altered in EC tissues compared with the adjacent normal tissues [152]. Similarly, another study identified 62,167 circRNAs that were differentially expressed in grade 3 EC tissues when compared with the adjacent normal tissues [153]. A recent study indicated a positive correlation between RNA-binding protein QKI and circRNAs, and a negative correlation of QKI with the activity of specific miRNAs, suggesting a potential model where QKI, circRNAs, and miRNAs form a regulatory feedback loop in EC [154]. Another study identified differentially expressed circRNAs (including 209 upregulated circRNAs and 66 downregulated circRNAs) in extracellular vesicles isolated from the serum of EC patients compared with healthy controls [155]. CircPUM1 was significantly overexpressed in EC tissues and EC cell lines, and it promoted the development of EC by regulating the miR-136/NOTCH3 axis [156].
8. CircRNAs as Potential Biomarkers in Gynecological Cancers
Early studies have provided evidence that circRNAs could be used as biomarkers in distinguishing cancer tissues from normal tissues and predicting the progression and prognosis, and assessment of response to therapy in many cancer types [15]. Dysregulation of circRNAs has been reported to be closely related to the clinicopathological characteristics of patients with CC, OC and EC [79,80,94,117,149,153]. In addition, levels of some circRNAs have been associated with survival times in patients with CC and OC [78,80,141,142,157,158,159,160]. In an in silico analysis of an OC dataset obtained from the GEO database, the expression of six-circRNAs was found to be an independent predictor of overall survival in patients with OC [160].
Moreover, circRNAs can be released into the extracellular space, and subsequently detected in body fluids (such as blood, plasma, serum and exosomes) from patients with gynecological cancers [117,119,137,161,162,163]. Higher serum levels of circSETDB1 have been found in patients with OC compared to healthy controls [161]. Serum levels of circSETDB1 were positively correlated with chemoresistance and shorter survival in patients with OC [161]. CircBNC2 was downregulated in the plasma of patients with OC compared to healthy individuals, and plasma levels of circBNC2 provided a potential tool to distinguish patients with early-stage OC from healthy individuals [164].
The circRNAs that might serve as potential prognostic biomarkers in gynecological cancers are summarized in Table 3.
Table 3.
CircRNAs as potential biomarkers in gynecological cancers.
| circRNA | Cancer | Expression | Sample | Clinicopathologic Features | Prognosis | Ref. |
|---|---|---|---|---|---|---|
| circ0023404 | CC | Up | Tissue | - | OS | [78] |
| circ8924 | CC | Up | Tissue | Size, stage, tumor invasion | - | [79] |
| circ0067934 | CC | Up | Tissue | Stage, LNM | OS | [80] |
| circ101996 | CC | Up | Tissue | Stage, LNM, size | OS | [83] |
| circEIF4G2 | CC | Up | Tissue | Size, LNM | OS | [84] |
| circATP8A2 | CC | Up | Tissue | Stage, LNM, tumor invasion | OS | [85] |
| circ0000745 | CC | Up | Tissue | Grade, vascular/lymphatic invasion | - | [89] |
| circ0005576 | CC | Up | Tissue | Stage, LNM | OS | [91] |
| circ0075341 | CC | Up | Tissue | Size, stage, LNM | - | [94] |
| circ0000515 | CC | Up | Tissue | - | OS | [95] |
| circ0001038 | CC | Up | Tissue | LNM, tumor invasion | OS | [96] |
| circAMOTL1 | CC | Up | Tissue | Tumor metastasis, stage | OS | [97] |
| circMYBL2 | CC | Up | Tissue | Stage, size, LNM | - | [98] |
| circCLK3 | CC | Up | Tissue | Grade, stage, tumor invasion | OS, DFS | [99] |
| ciRS-7 | CC | Up | Tissue | Size, stage, tumor invasion, LNM, HPV infection | - | [100] |
| circSLC26A4 | CC | Up | Tissue | - | OS | [101] |
| circ1656 | OC | Down | Tissue | Stage | - | [117] |
| circ0051240 | OC | Up | Tissue | Tumor metastasis, LNM, stage | OS | [123] |
| circPIP5K1A | OC | Up | Tissue | - | OS | [124] |
| circABCB10 | OC | Up | Tissue | Grade, size, stage | OS | [126] |
| CircCSPP1 | OC | Up | Tissue | Stage, grade | - | [129] |
| circUBAP2 | OC | Up | Tissue | Stage | OS | [131] |
| circWHSC1 | OC | Up | Tissue | Grade | - | [132] |
| circVPS13C-has-circ-001567 | OC | Up | Tissue | Stage, LNM | - | [135] |
| circCELSR1 | OC | Up | Tissue | Chemoresistance | - | [136] |
| circPUM1 | OC | Up | Tissue | Stage | - | [137] |
| circRhoC | OC | Up | Tissue | Stage, grade | - | [140] |
| circITCH | OC | Down | Tissue | - | OS | [141] |
| circITCH | OC | Down | Tissue | Size, stage, grade | OS | [142] |
| circCDR1as | OC | Down | Tissue | chemoresistance | - | [145] |
| circPLEKHM3 | OC | Down | Tissue | Tumor metastasis | OS, DFS | [149] |
| circ100395 | OC | Down | Tissue | LNM, tumor metastasis, stage | OS | [150] |
| circ0039569 | EC | Up | Tissue | Grade | - | [153] |
| circLARP4 | OC | Down | Tissue | Stage, LNM | OS, DFS | [157] |
| circHIPK3 | OC | Up | Tissue | LNM, stage | OS, DFS | [158] |
| circEXOC6B | OC | Down | Tissue | Age, LNM | OS | [159] |
| circN4BP2L2 | OC | Down | Tissue | Age, stage, LNM | PFS | [159] |
| circSETDB1 | OC | Up | Serum | Stage, LNM, chemoresistance | PFS | [161] |
| circBNC2 | OC | Down | Plasma | Grade, histological subtype, LNM, tumor metastasis | - | [164] |
Up: upregulation; down: downregulation; CC: cervical cancer; EC: endometrial cancer; OC: ovarian cancer; LNM: lymph node metastasis; OS: overall survival; DFS: disease-free survival; PFS: progress-free survival.
9. CircRNAs as Potential Therapeutic Targets for Gynecological Cancers
Recent data have proved that several circRNAs can influence the growth, survival, invasion, EMT, angiogenesis, and drug resistance in gynecological cancers. Manipulation of circRNA abundance has been shown to possess anti-tumor activities in experimental xenograft models of CC [88,91,95], OC [121,123,124] and EC [156].
The introduction of circRNAs with binding sites for oncogenic miRNAs (or proteins) might sequester miRNAs (or proteins) from native targets, and consequently inhibit the biological activities of tumor-promoting miRNAs (or oncoproteins) in tumor cells [15]. Generally, numerous oncogenic miRNAs are overexpressed in a given tumor, thus artificial circRNAs that contain multiple binding sites for multiple oncogenic miRNAs would be expected to simultaneously bind and inhibit the functions of different tumor-causing miRNAs. Alternatively, circRNAs might be used as stable carriers that deliver tumor-suppressive miRNAs (or proteins) to tumor cells.
In addition, exosomal circRNAs could modulate the progression of cancer, the remodeling of tumor microenvironment, anti-tumor immune response, and the occurrence of metastasis [165]. The discovery that mRNAs from tumor cells can be transfected into dendritic cells and further translated into proteins to function as antigens are intriguing [166,167]. Similar to mRNA, circRNA can be detected in exosomes. Thus, the circRNAs with protein-coding potential might function as potential antigens and be used as potent targets for cancer immunotherapy [168]. Collectively, the research indicates the potential use of circRNAs as therapeutic targets for the treatment of gynecological cancers.
10. Conclusions
Despite many exciting advances in the circRNA research area, there are still arguments regarding the functionality of circRNAs [169]. At least, some individual circRNAs have biologically relevant functions in ovarian, cervical and endometrial cancer cells [87,137,156]. Their biogenesis, functions, and action mechanisms in gynecological cancer cells need to be fully investigated. Recent studies suggest that circRNAs may fulfill a regulatory role in gynecological cancers by sponging miRNAs, although the possibility of other mechanisms (such as scaffolds for protein complexes) also exists. CircRNAs have recently been recognized as emerging prognostic biomarkers for gynecological cancers. The unique ability of circRNAs to sponge RNAs (or proteins) and code for proteins make them valuable as potential therapeutic tools to treat gynecological cancers.
Acknowledgments
We thank Zhujie Xu for her contribution in preparing the figures.
Author Contributions
Writing—review and editing, P.D., D.X., Y.X., J.Y., K.I., Y.K., and H.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a grant from JSPS Grant-in-Aid for Scientific Research (C) (18K09278 and 19K09769) and an NIH/NCI grant 1R21CA216585-01A1 to J. Yue.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cech T.R., Steitz J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Mongelli A., Martelli F., Farsetti A., Gaetano C. The Dark That Matters: Long Non-coding RNAs as Master Regulators of Cellular Metabolism in Non-communicable Diseases. Front. Physiol. 2019;10:369. doi: 10.3389/fphys.2019.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dahariya S., Paddibhatla I., Kumar S., Raghuwanshi S., Pallepati A., Gutti R.K. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol. Immunol. 2019;112:82–92. doi: 10.1016/j.molimm.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 5.Dong P., Konno Y., Watari H., Hosaka M., Noguchi M., Sakuragi N. The impact of microRNA-mediated PI3K/AKT signaling on epithelial-mesenchymal transition and cancer stemness in endometrial cancer. J. Transl. Med. 2014;12:231. doi: 10.1186/s12967-014-0231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fatima F., Nawaz M. Vesiculated Long Non-Coding RNAs: Offshore Packages Deciphering Trans-Regulation between Cells, Cancer Progression and Resistance to Therapies. Noncoding RNA. 2017;3:10. doi: 10.3390/ncrna3010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong P., Xiong Y., Yue J., JB Hanley S., Kobayashi N., Todo Y., Watari H. Exploring lncRNA-Mediated Regulatory Networks in Endometrial Cancer Cells and the Tumor Microenvironment: Advances and Challenges. Cancers. 2019;11:234. doi: 10.3390/cancers11020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu M.T., Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 9.Sanger H.L., Klotz G., Riesner D., Gross H.J., Kleinschmidt A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 11.Chen L., Wang C., Sun H., Wang J., Liang Y., Wang Y., Wong G. The bioinformatics toolbox for circRNA discovery and analysis. Brief. Bioinform. 2020 doi: 10.1093/bib/bbaa001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X., Han P., Zhou T., Guo X., Song X., Li Y. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci. Rep. 2016;6:34985. doi: 10.1038/srep34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enuka Y., Lauriola M., Feldman M.E., Sas-Chen A., Ulitsky I., Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370–1383. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aufiero S., Reckman Y.J., Pinto Y.M., Creemers E.E. Circular RNAs open a new chapter in cardiovascular biology. Nat. Rev. Cardiol. 2019;16:503–514. doi: 10.1038/s41569-019-0185-2. [DOI] [PubMed] [Google Scholar]
- 15.Kristensen L.S., Hansen T.B., Venø M.T., Kjems J. Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang C., Shan G. What happens at or after transcription: Insights into circRNA biogenesis and function. Transcription. 2015;6:61–64. doi: 10.1080/21541264.2015.1071301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang C., Liang D., Tatomer D.C., Wilusz J.E. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32:639–644. doi: 10.1101/gad.314856.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/annotation/f782282b-eefa-4c8d-985c-b1484e845855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maass P.G., Glažar P., Memczak S., Dittmar G., Hollfinger I., Schreyer L., Sauer A.V., Toka O., Aiuti A., Luft F.C., et al. A map of human circular RNAs in clinically relevant tissues. J. Mol. Med. 2017;95:1179–1189. doi: 10.1007/s00109-017-1582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou T., Xie X., Li M., Shi J., Zhou J.J., Knox K.S., Wang T., Chen Q., Gu W. Rat BodyMap transcriptomes reveal unique circular RNA features across tissue types and developmental stages. RNA. 2018;24:1443–1456. doi: 10.1261/rna.067132.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lei B., Tian Z., Fan W., Ni B. Circular RNA: A novel biomarker and therapeutic target for human cancers. Int J. Med. Sci. 2019;16:292–301. doi: 10.7150/ijms.28047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G., et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong Y., He D., Peng Z., Peng W., Shi W., Wang J., Li B., Zhang C., Duan C. Circular RNAs in cancer: An emerging key player. J. Hematol. Oncol. 2017;10:2. doi: 10.1186/s13045-016-0370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shang Q., Yang Z., Jia R., Ge S. The novel roles of circRNAs in human cancer. Mol. Cancer. 2019;18:6. doi: 10.1186/s12943-018-0934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 27.Wan L., Zhang L., Fan K., Cheng Z.X., Sun Q.C., Wang J.J. Circular RNA-ITCH Suppresses Lung Cancer Proliferation via Inhibiting the Wnt/β-Catenin Pathway. Biomed. Res. Int. 2016;2016:1579490. doi: 10.1155/2016/1579490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen T.B., Wiklund E.D., Bramsen J.B., Villadsen S.B., Statham A.L., Clark S.J., Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park O.H., Ha H., Lee Y., Boo S.H., Kwon D.H., Song H.K., Kim Y.K. Endoribonucleolytic Cleavage of m6A-Containing RNAs by RNase P/MRP Complex. Mol. Cell. 2019;74:494–507. doi: 10.1016/j.molcel.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 30.Liu C.X., Li X., Nan F., Jiang S., Gao X., Guo S.K., Xue W., Cui Y., Dong K., Ding H., et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell. 2019;177:865–880. doi: 10.1016/j.cell.2019.03.046. [DOI] [PubMed] [Google Scholar]
- 31.Fischer J.W., Busa V.F., Shao Y., Leung A.K.L. Structure-Mediated RNA Decay by UPF1 and G3BP1. Mol. Cell. 2020;78:70–84. doi: 10.1016/j.molcel.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lasda E., Parker R. Circular RNAs Co-Precipitate with Extracellular Vesicles: A Possible Mechanism for circRNA Clearance. PLoS ONE. 2016;11:e0148407. doi: 10.1371/journal.pone.0148407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao M., Xu J., Zhong S., Liu Y., Xiao H., Geng L., Liu H. Expression profiles and potential functions of circular RNAs in extracellular vesicles isolated from radioresistant glioma cells. Oncol Rep. 2019;41:1893–1900. doi: 10.3892/or.2019.6972. [DOI] [PubMed] [Google Scholar]
- 34.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 35.Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Militello G., Weirick T., John D., Döring C., Dimmeler S., Uchida S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief. Bioinform. 2017;18:780–788. doi: 10.1093/bib/bbw053. [DOI] [PubMed] [Google Scholar]
- 37.Hentze M.W., Castello A., Schwarzl T., Preiss T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018;19:327–341. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 38.Abdelmohsen K., Panda A.C., Munk R., Grammatikakis I., Dudekula D.B., De S., Kim J., Noh J.H., Kim K.M., Martindale J.L., et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X.X., Xiao L., Chung H.K., Ma X.X., Liu X., Song J.L., Jin C.Z., Rao J.N., Gorospe M., Wang J.Y. Interaction between HuR and circPABPN1 Modulates Autophagy in the Intestinal Epithelium by Altering ATG16L1 Translation. Mol. Cell Biol. 2020;40:e00492-19. doi: 10.1128/MCB.00492-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y., Yang F., Fang E., Xiao W., Mei H., Li H., Li D., Song H., Wang J., Hong M., et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26:1346–1364. doi: 10.1038/s41418-018-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang A., Zheng H., Wu Z., Chen M., Huang Y. Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics. 2020;10:3503–3517. doi: 10.7150/thno.42174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic. Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang G., Li S., Yang N., Zou Y., Zheng D., Xiao T. Recent progress in circular RNAs in human cancers. Cancer Lett. 2017;404:8–18. doi: 10.1016/j.canlet.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 44.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 45.Wang M., Yu F., Wu W., Zhang Y., Chang W., Ponnusamy M., Wang K., Li P. Circular RNAs: A novel type of non-coding RNA and their potential implications in antiviral immunity. Int J. Biol Sci. 2017;13:1497–1506. doi: 10.7150/ijbs.22531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 47.Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E., et al. Translation of CircRNAs. Mol. Cell. 2017;66:9–21. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M., et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Granados-Riveron J.T., Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim. Biophys. Acta. 2016;1859:1245–1251. doi: 10.1016/j.bbagrm.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 50.Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y., et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang M., Zhao K., Xu X., Yang Y., Yan S., Wei P., Liu H., Xu J., Xiao F., Zhou H., et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018;9:4475. doi: 10.1038/s41467-018-06862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye F., Gao G., Zou Y., Zheng S., Zhang L., Ou X., Xie X., Tang H. circFBXW7 Inhibits Malignant Progression by Sponging miR-197-3p and Encoding a 185-aa Protein in Triple-Negative Breast Cancer. Mol. Ther. Nucleic Acids. 2019;18:88–98. doi: 10.1016/j.omtn.2019.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szabo L., Salzman J. Detecting circular RNAs: Bioinformatic and experimental challenges. Nat. Rev. Genet. 2016;17:679–692. doi: 10.1038/nrg.2016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jakobi T., Dieterich C. Computational approaches for circular RNA analysis. Wiley Interdiscip Rev. RNA. 2019;10:e1528. doi: 10.1002/wrna.1528. [DOI] [PubMed] [Google Scholar]
- 55.Barrett S.P., Salzman J. Circular RNAs: Analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carrara M., Fuschi P., Ivan C., Martelli F. Circular RNAs: Methodological challenges and perspectives in cardiovascular diseases. J. Cell Mol. Med. 2018;22:5176–5187. doi: 10.1111/jcmm.13789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bonizzato A., Gaffo E., Te Kronnie G., Bortoluzzi S. CircRNAs in hematopoiesis and hematological malignancies. Blood Cancer J. 2016;6:e483. doi: 10.1038/bcj.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aran D., Camarda R., Odegaard J., Paik H., Oskotsky B., Krings G., Goga A., Sirota M., Butte A.J. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat. Commun. 2017;8:1077. doi: 10.1038/s41467-017-01027-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Assumpção M.B., Moreira F.C., Hamoy I.G., Magalhães L., Vidal A., Pereira A., Burbano R., Khayat A., Silva A., Santos S., et al. High-Throughput miRNA Sequencing Reveals a Field Effect in Gastric Cancer and Suggests an Epigenetic Network Mechanism. Bioinform. Biol. Insights. 2015;9:111–117. doi: 10.4137/BBI.S24066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pereira A.L., Magalhães L., Moreira F.C., Reis-das-Mercês L., Vidal A.F., Ribeiro-Dos-Santos A.M., Demachki S., Anaissi A.K.M., Burbano R.M.R., Albuquerque P., et al. Epigenetic Field Cancerization in Gastric Cancer: MicroRNAs as Promising Biomarkers. J. Cancer. 2019;10:1560–1569. doi: 10.7150/jca.27457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shang X., Li G., Liu H., Li T., Liu J., Zhao Q., Wang C. Comprehensive Circular RNA Profiling Reveals That hsa_circ_0005075, a New Circular RNA Biomarker, Is Involved in Hepatocellular Crcinoma Development. Medicine. 2016;95:e3811. doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bachmayr-Heyda A., Reiner A.T., Auer K., Sukhbaatar N., Aust S., Bachleitner-Hofmann T., Mesteri I., Grunt T.W., Zeillinger R., Pils D. Correlation of circular RNA abundance with proliferation-exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci. Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vidal A.F., Ribeiro-Dos-Santos A.M., Vinasco-Sandoval T., Magalhães L., Pinto P., Anaissi A.K.M., Demachki S., de Assumpção P.P., Dos Santos S.E.B., Ribeiro-Dos-Santos Â. The comprehensive expression analysis of circular RNAs in gastric cancer and its association with field cancerization. Sci. Rep. 2017;7:14551. doi: 10.1038/s41598-017-15061-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilusz J.E. Circular RNAs: Unexpected outputs of many protein-coding genes. RNA Biol. 2017;14:1007–1017. doi: 10.1080/15476286.2016.1227905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cortés-López M., Miura P. Emerging Functions of Circular RNAs. Yale J. Biol. Med. 2016;89:527–537. [PMC free article] [PubMed] [Google Scholar]
- 66.Kramer M.C., Liang D., Tatomer D.C., Gold B., March Z.M., Cherry S., Wilusz J.E. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168–2182. doi: 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y., Nguyen T.M., Zhang X.O., Phan T., Clohessy J.G., Pandolfi P.P. Optimized RNA-targeting CRISPR/Cas13d technology outperforms shRNA in identifying essential circRNAs. bioRxiv. 2020 doi: 10.1101/2020.03.23.002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li S.Q., Li X., Xue W., Zhang L., Cao S.M., Lei Y.N., Yang L.Z., Guo S.K., Zhang J.L., Gao X., et al. Screening for functional circular RNAs using the CRISPR-Cas13 system. bioRxiv. 2020 doi: 10.1101/2020.03.23.002865. [DOI] [PubMed] [Google Scholar]
- 69.Cohen P.A., Jhingran A., Oaknin A., Denny L. Cervical cancer. Lancet. 2019;393:169–182. doi: 10.1016/S0140-6736(18)32470-X. [DOI] [PubMed] [Google Scholar]
- 70.Li S., Teng S., Xu J., Su G., Zhang Y., Zhao J., Zhang S., Wang H., Qin W., Lu Z.J., et al. Microarray is an efficient tool for circRNA profiling. Brief. Bioinform. 2019;20:1420–1433. doi: 10.1093/bib/bby006. [DOI] [PubMed] [Google Scholar]
- 71.Zheng S.R., Zhang H.R., Zhang Z.F., Lai S.Y., Huang L.J., Liu J., Bai X., Ding K., Zhou J.Y. Human papillomavirus 16 E7 oncoprotein alters the expression profiles of circular RNAs in Caski cells. J. Cancer. 2018;9:3755–3764. doi: 10.7150/jca.24253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu D., Li Y., Ming Z., Wang H., Dong Z., Qiu L., Wang T. Comprehensive circular RNA expression profile in radiation-treated HeLa cells and analysis of radioresistance-related circRNAs. Peer J. 2018;6:e5011. doi: 10.7717/peerj.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang H., Zhao Y., Chen M., Cui J. Identification of Novel Long Non-coding and Circular RNAs in Human Papillomavirus-Mediated Cervical Cancer. Front. Microbiol. 2017;8:1720. doi: 10.3389/fmicb.2017.01720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yi Y., Liu Y., Wu W., Wu K., Zhang W. Reconstruction and analysis of circRNA-miRNA-mRNA network in the pathology of cervical cancer. Oncol. Rep. 2019;41:2209–2225. doi: 10.3892/or.2019.7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Di Liddo A., de Oliveira Freitas Machado C., Fischer S., Ebersberger S., Heumüller A.W., Weigand J.E., Müller-McNicoll M., Zarnack K. A combined computational pipeline to detect circular RNAs in human cancer cells under hypoxic stress. J. Mol. Cell Biol. 2019;11:829–844. doi: 10.1093/jmcb/mjz094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gao Y.L., Zhang M.Y., Xu B., Han L.J., Lan S.F., Chen J., Dong Y.J., Cao L.L. Circular RNA expression profiles reveal that hsa_circ_0018289 is up-regulated in cervical cancer and promotes the tumorigenesis. Oncotarget. 2017;8:86625–86633. doi: 10.18632/oncotarget.21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ma H.B., Yao Y.N., Yu J.J., Chen X.X., Li H.F. Extensive profiling of circular RNAs and the potential regulatory role of circRNA-000284 in cell proliferation and invasion of cervical cancer via sponging miR-506. Am. J. Transl Res. 2018;10:592–604. [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang J., Zhao X., Zhang J., Zheng X., Li F. Circular RNA hsa_circ_0023404 exerts an oncogenic role in cervical cancer through regulating miR-136/TFCP2/YAP pathway. Biochem. Biophys. Res. Commun. 2018;501:428–433. doi: 10.1016/j.bbrc.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 79.Liu J., Wang D., Long Z., Liu J., Li W. CircRNA8924 Promotes Cervical Cancer Cell Proliferation, Migration and Invasion by Competitively Binding to MiR-518d-5p/519-5p Family and Modulating the Expression of CBX8. Cell Physiol. Biochem. 2018;48:173–184. doi: 10.1159/000491716. [DOI] [PubMed] [Google Scholar]
- 80.Hu C., Wang Y., Li A., Zhang J., Xue F., Zhu L. Overexpressed circ_0067934 acts as an oncogene to facilitate cervical cancer progression via the miR-545/EIF3C axis. J. Cell Physiol. 2019;234:9225–9232. doi: 10.1002/jcp.27601. [DOI] [PubMed] [Google Scholar]
- 81.Cai H., Zhang P., Xu M., Yan L., Liu N., Wu X. Circular RNA hsa_circ_0000263 participates in cervical cancer development by regulating target gene of miR-150-5p. J. Cell Physiol. 2019;234:11391–11400. doi: 10.1002/jcp.27796. [DOI] [PubMed] [Google Scholar]
- 82.Tian J.D.C., Liang L. Involvement of circular RNA SMARCA5/microRNA-620 axis in the regulation of cervical cancer cell proliferation, invasion and migration. Eur Rev. Med. Pharmacol. Sci. 2018;22:8589–8598. doi: 10.26355/eurrev_201812_16622. [DOI] [PubMed] [Google Scholar]
- 83.Song T., Xu A., Zhang Z., Gao F., Zhao L., Chen X., Gao J., Kong X. CircRNA hsa_circRNA_101996 increases cervical cancer proliferation and invasion through activating TPX2 expression by restraining miR-8075. J. Cell Physiol. 2019;234:14296–14305. doi: 10.1002/jcp.28128. [DOI] [PubMed] [Google Scholar]
- 84.Mao Y., Zhang L., Li Y. circEIF4G2 modulates the malignant features of cervical cancer via the miR-218/HOXA1 pathway. Mol. Med. Rep. 2019;19:3714–3722. doi: 10.3892/mmr.2019.10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ding L., Zhang H. Circ-ATP8A2 promotes cell proliferation and invasion as a ceRNA to target EGFR by sponging miR-433 in cervical cancer. Genes. 2019;705:103–108. doi: 10.1016/j.gene.2019.04.068. [DOI] [PubMed] [Google Scholar]
- 86.Guo J., Chen M., Ai G., Mao W., Li H., Zhou J. Hsa_circ_0023404 enhances cervical cancer metastasis and chemoresistance through VEGFA and autophagy signaling by sponging miR-5047. Biomed. Pharmacother. 2019;115:108957. doi: 10.1016/j.biopha.2019.108957. [DOI] [PubMed] [Google Scholar]
- 87.Zhao J., Lee E.E., Kim J., Yang R., Chamseddin B., Ni C., Gusho E., Xie Y., Chiang C.M., Buszczak M., et al. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat. Commun. 2019;10:2300. doi: 10.1038/s41467-019-10246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen M., Ai G., Zhou J., Mao W., Li H., Guo J. circMTO1 promotes tumorigenesis and chemoresistance of cervical cancer via regulating miR-6893. Biomed. Pharm. 2019;117:109064. doi: 10.1016/j.biopha.2019.109064. [DOI] [PubMed] [Google Scholar]
- 89.Jiao J., Zhang T., Jiao X., Huang T., Zhao L., Ma D., Cui B. hsa_circ_0000745 promotes cervical cancer by increasing cell proliferation, migration, and invasion. J. Cell Physiol. 2020;235:1287–1295. doi: 10.1002/jcp.29045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rong X., Gao W., Yang X., Guo J. Downregulation of hsa_circ_0007534 restricts the proliferation and invasion of cervical cancer through regulating miR-498/BMI-1 signaling. Life Sci. 2019;235:116785. doi: 10.1016/j.lfs.2019.116785. [DOI] [PubMed] [Google Scholar]
- 91.Ma H., Tian T., Liu X., Xia M., Chen C., Mai L., Xie S., Yu L. Upregulated circ_0005576 facilitates cervical cancer progression via the miR-153/KIF20A axis. Biomed. Pharmacother. 2019;118:109311. doi: 10.1016/j.biopha.2019.109311. [DOI] [PubMed] [Google Scholar]
- 92.Chen R.X., Liu H.L., Yang L.L., Kang F.H., Xin L.P., Huang L.R., Guo Q.F., Wang Y.L. Circular RNA circRNA_0000285 promotes cervical cancer development by regulating FUS. Eur Rev. Med. Pharmacol. Sci. 2019;23:8771–8778. doi: 10.26355/eurrev_201910_19271. [DOI] [PubMed] [Google Scholar]
- 93.Wu F., Zhou J. CircAGFG1 promotes cervical cancer progression via miR-370-3p/RAF1 signaling. BMC Cancer. 2019;19:1067. doi: 10.1186/s12885-019-6269-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shao S., Wang C., Wang S., Zhang H., Zhang Y. Hsa_circ_0075341 is up-regulated and exerts oncogenic properties by sponging miR-149-5p in cervical cancer. Biomed. Pharm. 2020;121:109582. doi: 10.1016/j.biopha.2019.109582. [DOI] [PubMed] [Google Scholar]
- 95.Tang Q., Chen Z., Zhao L., Xu H. Circular RNA hsa_circ_0000515 acts as a miR-326 sponge to promote cervical cancer progression through up-regulation of ELK1. Aging. 2019;11:9982–9999. doi: 10.18632/aging.102356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Y., Wang L., Wang W., Guo X. Overexpression of circular RNA hsa_circ_0001038 promotes cervical cancer cell progression by acting as a ceRNA for miR-337-3p to regulate cyclin-M3 and metastasis-associated in colon cancer 1 expression. Gene. 2020;733:144273. doi: 10.1016/j.gene.2019.144273. [DOI] [PubMed] [Google Scholar]
- 97.Ou R., Lv J., Zhang Q., Lin F., Zhu L., Huang F., Li X., Li T., Zhao L., Ren Y., et al. circAMOTL1 Motivates AMOTL1 Expression to Facilitate Cervical Cancer Growth. Mol. Ther. Nucleic Acids. 2020;19:50–60. doi: 10.1016/j.omtn.2019.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang J., Li H., Liang Z. circ-MYBL2 Serves as a Sponge For miR-361-3p Promoting Cervical Cancer Cells Proliferation and Invasion. Onco Targets Ther. 2019;12:9957–9964. doi: 10.2147/OTT.S218976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hong H., Zhu H., Zhao S., Wang K., Zhang N., Tian Y., Li Y., Wang Y., Lv X., Wei T., et al. The novel circCLK3/miR-320a/FoxM1 axis promotes cervical cancer progression. Cell Death Dis. 2019;10:950. doi: 10.1038/s41419-019-2183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhou Y., Shen L., Wang Y.Z., Zhou C.C. The potential of ciRS-7 for predicting onset and prognosis of cervical cancer. Neoplasma. 2020;67:312–322. doi: 10.4149/neo_2019_190415N334. [DOI] [PubMed] [Google Scholar]
- 101.Ji F., Du R., Chen T., Zhang M., Zhu Y., Luo X., Ding Y. Circular RNA circSLC26A4 Accelerates Cervical Cancer Progression via miR-1287-5p/HOXA7 Axis. Mol. Ther. Nucleic Acids. 2020;19:413–420. doi: 10.1016/j.omtn.2019.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang P., Qi B., Yao H., Zhang L., Li Y., Li Q. Circular RNA cSMARCA5 regulates the progression of cervical cancer by acting as a microRNA-432 sponge. Mol. Med. Rep. 2020;21:1217–1223. doi: 10.3892/mmr.2020.10910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li J., Guo R., Liu Q., Sun J., Wang H. Circular RNA Circ-ITCH Inhibits the Malignant Behaviors of Cervical Cancer by microRNA-93-5p/FOXK2 Axis. Reprod. Sci. 2020;27:860–868. doi: 10.1007/s43032-020-00140-7. [DOI] [PubMed] [Google Scholar]
- 104.Qian W., Huang T., Feng W. Circular RNA HIPK3 Promotes EMT of Cervical Cancer through Sponging miR-338-3p to Up-Regulate HIF-1α. Cancer Manag. Res. 2020;12:177–187. doi: 10.2147/CMAR.S232235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang W., Xie T. Hsa_circ_CSPP1/MiR-361-5p/ITGB1 Regulates Proliferation and Migration of Cervical Cancer (CC) by Modulating the PI3K-Akt Signaling Pathway. Reprod. Sci. 2020;27:132–144. doi: 10.1007/s43032-019-00008-5. [DOI] [PubMed] [Google Scholar]
- 106.Shi X., Wang J., Lei Y., Cong C., Tan D., Zhou X. Research progress on the PI3K/AKT signaling pathway in gynecological cancer (Review) Mol. Med. Rep. 2019;19:4529–4535. doi: 10.3892/mmr.2019.10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bahrami A., Hasanzadeh M., Hassanian S.M., ShahidSales S., Ghayour-Mobarhan M., Ferns G.A., Avan A. The Potential Value of the PI3K/Akt/mTOR Signaling Pathway for Assessing Prognosis in Cervical Cancer and as a Target for Therapy. J. Cell Biochem. 2017;118:4163–4169. doi: 10.1002/jcb.26118. [DOI] [PubMed] [Google Scholar]
- 108.Cruz da Silva E., Dontenwill M., Choulier L., Lehmann M. Role of Integrins in Resistance to Therapies Targeting Growth Factor Receptors in Cancer. Cancers (Basel) 2019;11:692. doi: 10.3390/cancers11050692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dhillon A.S., Hagan S., Rath O., Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 110.Lu W., Kang Y. Epithelial-Mesenchymal Plasticity in Cancer Progression and Metastasis. Dev. Cell. 2019;49:361–374. doi: 10.1016/j.devcel.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shang B.Q., Li M.L., Quan H.Y., Hou P.F., Li Z.W., Chu S.F., Zheng J.N., Bai J. Functional roles of circular RNAs during epithelial-to-mesenchymal transition. Mol. Cancer. 2019;18:138. doi: 10.1186/s12943-019-1071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bieging K.T., Mello S.S., Attardi L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pal A., Kundu R. Human Papillomavirus E6 and E7: The Cervical Cancer Hallmarks and Targets for Therapy. Front. Microbiol. 2020;10:3116. doi: 10.3389/fmicb.2019.03116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Scheffner M., Werness B.A., Huibregtse J.M., Levine A.J., Howley P.M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 115.Dyson N., Howley P.M., Münger K., Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 116.Teng F., Xu J., Zhang M., Liu S., Gu Y., Zhang M., Wang X., Ni J., Qian B., Shen R., et al. Comprehensive circular RNA expression profiles and the tumor-suppressive function of circHIPK3 in ovarian cancer. Int. J. Biochem. Cell Biol. 2019;112:8–17. doi: 10.1016/j.biocel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 117.Gao Y., Zhang C., Liu Y., Wang M. Circular RNA profiling reveals circRNA1656 as a novel biomarker in high grade serous ovarian cancer. Biosci. Trends. 2019;13:204–211. doi: 10.5582/bst.2019.01021. [DOI] [PubMed] [Google Scholar]
- 118.Bachmayr-Heyda A., Auer K., Sukhbaatar N., Aust S., Deycmar S., Reiner A.T., Polterauer S., Dekan S., Pils D. Small RNAs and the competing endogenous RNA network in high grade serous ovarian cancer tumor spread. Oncotarget. 2016;7:39640–39653. doi: 10.18632/oncotarget.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang J., Wu A., Yang B., Zhu X., Teng Y., Ai Z. Profiling and bioinformatics analyses reveal differential circular RNA expression in ovarian cancer. Gene. 2020;724:144150. doi: 10.1016/j.gene.2019.144150. [DOI] [PubMed] [Google Scholar]
- 120.Ahmed I., Karedath T., Andrews S.S., Al-Azwani I.K., Mohamoud Y.A., Querleu D., Rafii A., Malek J.A. Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma. Oncotarget. 2016;7:36366–36381. doi: 10.18632/oncotarget.8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xie J., Wang S., Li G., Zhao X., Jiang F., Liu J., Tan W. circEPSTI1 regulates ovarian cancer progression via decoying miR-942. J. Cell Mol. Med. 2019;23:3597–3602. doi: 10.1111/jcmm.14260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu J., Yu F., Wang S., Zhao X., Jiang F., Xie J., Deng M. circGFRA1 Promotes Ovarian Cancer Progression by Sponging miR-449a. J. Cancer. 2019;10:3908–3913. doi: 10.7150/jca.31615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang M., Xia B., Xu Y., Zhang Y., Xu J., Lou G. Circular RNA (hsa_circ_0051240) promotes cell proliferation, migration and invasion in ovarian cancer through miR-637/KLK4 axis. Artif. Cells Nanomed. Biotechnol. 2019;47:1224–1233. doi: 10.1080/21691401.2019.1593999. [DOI] [PubMed] [Google Scholar]
- 124.Sun Y., Li X., Chen A., Shi W., Wang L., Yi R., Qiu J. circPIP5K1A serves as a competitive endogenous RNA contributing to ovarian cancer progression via regulation of miR-661/IGFBP5 signaling. J. Cell Biochem. 2019;120:19406–19414. doi: 10.1002/jcb.29055. [DOI] [PubMed] [Google Scholar]
- 125.Sun X., Luo L., Gao Y. Circular RNA PVT1 enhances cell proliferation but inhibits apoptosis through sponging microRNA-149 in epithelial ovarian cancer. J. Obstet. Gynaecol. Res. 2020;46:625–635. doi: 10.1111/jog.14190. [DOI] [PubMed] [Google Scholar]
- 126.Chen Y., Ye X., Xia X., Lin X. Circular RNA ABCB10 correlates with advanced clinicopathological features and unfavorable survival, and promotes cell proliferation while reduces cell apoptosis in epithelial ovarian cancer. Cancer Biomark. 2019;26:151–161. doi: 10.3233/CBM-190064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhao Y., Qin X.P., Lang Y.P., Kou D., Shao Z.W. Circular RNA circ-SMAD7 promoted ovarian cancer cell proliferation and metastasis by suppressing KLF6. Eur. Rev. Med. Pharm. Sci. 2019;23:5603–5610. doi: 10.26355/eurrev_201907_18294. [DOI] [PubMed] [Google Scholar]
- 128.Karedath T., Ahmed I., Al Ameri W., Al-Dasim F.M., Andrews S.S., Samuel S., Al-Azwani I.K., Mohamoud Y.A., Rafii A., Malek J.A. Silencing of ANKRD12 circRNA induces molecular and functional changes associated with invasive phenotypes. BMC Cancer. 2019;19:565. doi: 10.1186/s12885-019-5723-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li Q.H., Liu Y., Chen S., Zong Z.H., Du Y.P., Sheng X.J., Zhao Y. circ-CSPP1 promotes proliferation, invasion and migration of ovarian cancer cells by acting as a miR-1236-3p sponge. Biomed. Pharmacother. 2019;114:108832. doi: 10.1016/j.biopha.2019.108832. [DOI] [PubMed] [Google Scholar]
- 130.Chen Q., Zhang J., He Y., Wang Y. hsa_circ_0061140 Knockdown Reverses FOXM1-Mediated Cell Growth and Metastasis in Ovarian Cancer through miR-370 Sponge Activity. Mol. Ther. Nucleic Acids. 2018;13:55–63. doi: 10.1016/j.omtn.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 131.Sheng M., Wei N., Yang H.Y., Yan M., Zhao Q.X., Jing L.J. CircRNA UBAP2 promotes the progression of ovarian cancer by sponging microRNA-144. Eur. Rev. Med. Pharmacol. Sci. 2019;23:7283–7294. doi: 10.26355/eurrev_201909_18833. [DOI] [PubMed] [Google Scholar]
- 132.Zong Z.H., Du Y.P., Guan X., Chen S., Zhao Y. CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182. J. Exp. Clin. Cancer Res. 2019;38:437. doi: 10.1186/s13046-019-1437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ponnusamy M.P., Seshacharyulu P., Lakshmanan I., Vaz A.P., Chugh S., Batra S.K. Emerging role of mucins in epithelial to mesenchymal transition. Curr. Cancer Drug Targets. 2013;13:945–956. doi: 10.2174/15680096113136660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu Z., Li Q., Li K., Chen L., Li W., Hou M., Liu T., Yang J., Lindvall C., Björkholm M., et al. Telomerase reverse transcriptase promotes epithelial-mesenchymal transition and stem cell-like traits in cancer cells. Oncogene. 2013;32:4203–4213. doi: 10.1038/onc.2012.441. [DOI] [PubMed] [Google Scholar]
- 135.Bao L., Zhong J., Pang L. Upregulation of Circular RNA VPS13C-has-circ-001567 Promotes Ovarian Cancer Cell Proliferation and Invasion. Cancer Biother. Radiopharm. 2019;34:110–118. doi: 10.1089/cbr.2018.2641. [DOI] [PubMed] [Google Scholar]
- 136.Zhang S., Cheng J., Quan C., Wen H., Feng Z., Hu Q., Zhu J., Huang Y., Wu X. circCELSR1 (hsa_circ_0063809) Contributes to Paclitaxel Resistance of Ovarian Cancer Cells by Regulating FOXR2 Expression via miR-1252. Mol. Ther. Nucleic Acids. 2020;19:718–730. doi: 10.1016/j.omtn.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Guan X., Zong Z.H., Liu Y., Chen S., Wang L.L., Zhao Y. circPUM1 Promotes Tumorigenesis and Progression of Ovarian Cancer by Sponging miR-615-5p and miR-6753-5p. Mol. Nucleic Acids. 2019;18:882–892. doi: 10.1016/j.omtn.2019.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Krill L.S., Tewari K.S. Exploring the therapeutic rationale for angiogenesis blockade in cervical cancer. Clin. Ther. 2015;37:9–19. doi: 10.1016/j.clinthera.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chung A.S., Lee J., Ferrara N. Targeting the tumour vasculature: Insights from physiological angiogenesis. Nat. Rev. Cancer. 2010;10:505–514. doi: 10.1038/nrc2868. [DOI] [PubMed] [Google Scholar]
- 140.Wang L.L., Zong Z.H., Liu Y., Guan X., Chen S., Zhao Y. CircRhoC promotes tumorigenicity and progression in ovarian cancer by functioning as a miR-302e sponge to positively regulate VEGFA. J. Cell Mol. Med. 2019;23:8472–8481. doi: 10.1111/jcmm.14736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hu J., Wang L., Chen J., Gao H., Zhao W., Huang Y., Jiang T., Zhou J., Chen Y. The circular RNA circ-ITCH suppresses ovarian carcinoma progression through targeting miR-145/RASA1 signaling. Biochem. Biophys. Res. Commun. 2018;505:222–228. doi: 10.1016/j.bbrc.2018.09.060. [DOI] [PubMed] [Google Scholar]
- 142.Luo L., Gao Y.Q., Sun X.F. Circular RNA ITCH suppresses proliferation and promotes apoptosis in human epithelial ovarian cancer cells by sponging miR-10a-α. Eur. Rev. Med. Pharmacol. Sci. 2018;22:8119–8126. doi: 10.26355/eurrev_201812_16503. [DOI] [PubMed] [Google Scholar]
- 143.Yan H., Xiang H., Sun B., Feng F., Chen P. Circular RNA-ITCH Inhibits the Proliferation of Ovarian Carcinoma by Downregulating lncRNA HULC. Reprod. Sci. 2020;27:375–379. doi: 10.1007/s43032-019-00049-w. [DOI] [PubMed] [Google Scholar]
- 144.Luo L., Gao Y., Sun X. Circ-ITCH correlates with small tumor size, decreased FIGO stage and prolonged overall survival, and it inhibits cells proliferation while promotes cells apoptosis in epithelial ovarian cancer. Cancer Biomark. 2018;23:505–513. doi: 10.3233/CBM-181609. [DOI] [PubMed] [Google Scholar]
- 145.Chen H., Mao M., Jiang J., Zhu D., Li P. Circular RNA CDR1as acts as a sponge of miR-135b-5p to suppress ovarian cancer progression. Oncol. Targets Ther. 2019;12:3869–3879. doi: 10.2147/OTT.S207938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhao Z., Ji M., Wang Q., He N., Li Y. Circular RNA Cdr1as Upregulates SCAI to Suppress Cisplatin Resistance in Ovarian Cancer via miR-1270 Suppression. Mol. Ther. Nucleic Acids. 2019;18:24–33. doi: 10.1016/j.omtn.2019.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 147.Li L., Yu P., Zhang P., Wu H., Chen Q., Li S., Wang Y. Upregulation of hsa_circ_0007874 suppresses the progression of ovarian cancer by regulating the miR-760/SOCS3 pathway. Cancer Med. 2020;9:2491–2499. doi: 10.1002/cam4.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lin W., Ye H., You K., Chen L. Up-regulation of circ_LARP4 suppresses cell proliferation and migration in ovarian cancer by regulating miR-513b-5p/LARP4 axis. Cancer Cell Int. 2020;20:5. doi: 10.1186/s12935-019-1071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang L., Zhou Q., Qiu Q., Hou L., Wu M., Li J., Li X., Lu B., Cheng X., Liu P., et al. CircPLEKHM3 acts as a tumor suppressor through regulation of the miR-9/BRCA1/DNAJB6/KLF4/AKT1 axis in ovarian cancer. Mol. Cancer. 2019;18:144. doi: 10.1186/s12943-019-1080-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Li X., Lin S., Mo Z., Jiang J., Tang H., Wu C., Song J. CircRNA_100395 inhibits cell proliferation and metastasis in ovarian cancer via regulating miR-1228/p53/epithelial-mesenchymal transition (EMT) axis. J. Cancer. 2020;11:599–609. doi: 10.7150/jca.35041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nguyen V.H.L., Hough R., Bernaudo S., Peng C. Wnt/β-catenin signalling in ovarian cancer: Insights into its hyperactivation and function in tumorigenesis. J. Ovarian Res. 2019;12:122. doi: 10.1186/s13048-019-0596-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen B.J., Byrne F.L., Takenaka K., Modesitt S.C., Olzomer E.M., Mills J.D., Farrell R., Hoehn K.L., Janitz M. Analysis of the circular RNA transcriptome in endometrial cancer. Oncotarget. 2017;9:5786–5796. doi: 10.18632/oncotarget.23534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ye F., Tang Q.L., Ma F., Cai L., Chen M., Ran X.X., Wang X.Y., Jiang X.F. Analysis of the circular RNA transcriptome in the grade 3 endometrial cancer. Cancer Manag. Res. 2019;11:6215–6227. doi: 10.2147/CMAR.S197343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dou Y., Kawaler E.A., Zhou D.C., Gritsenko M.A., Huang C., Blumenberg L., Karpova A., Petyuk V.A., Savage S.R., Satpathy S., et al. Proteogenomic Characterization of Endometrial Carcinoma. Cell. 2020;180:729–748.e26. doi: 10.1016/j.cell.2020.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Xu H., Gong Z., Shen Y., Fang Y., Zhong S. Circular RNA expression in extracellular vesicles isolated from serum of patients with endometrial cancer. Epigenomics. 2018;10:187–197. doi: 10.2217/epi-2017-0109. [DOI] [PubMed] [Google Scholar]
- 156.Zong Z.H., Liu Y., Chen S., Zhao Y. Circ_PUM1 promotes the development of endometrial cancer by targeting the miR-136/NOTCH3 pathway. J. Cell Mol. Med. 2020;24:4127–4135. doi: 10.1111/jcmm.15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zou T., Wang P.L., Gao Y., Liang W.T. Circular RNA_LARP4 is lower expressed and serves as a potential biomarker of ovarian cancer prognosis. Eur. Rev. Med. Pharm. Sci. 2018;22:7178–7182. doi: 10.26355/eurrev_201811_16250. [DOI] [PubMed] [Google Scholar]
- 158.Liu N., Zhang J., Zhang L.Y., Wang L. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur. Rev. Med. Pharm. Sci. 2018;22:3713–3718. doi: 10.26355/eurrev_201806_15250. [DOI] [PubMed] [Google Scholar]
- 159.Ning L., Long B., Zhang W., Yu M., Wang S., Cao D., Yang J., Shen K., Huang Y., Lang J. Circular RNA profiling reveals circEXOC6B and circN4BP2L2 as novel prognostic biomarkers in epithelial ovarian cancer. Int. J. Oncol. 2018;53:2637–2646. doi: 10.3892/ijo.2018.4566. [DOI] [PubMed] [Google Scholar]
- 160.Guo Q., He Y., Sun L., Kong C., Cheng Y., Zhang G. In silico detection of potential prognostic circRNAs through a re-annotation strategy in ovarian cancer. Oncol Lett. 2019;17:3677–3686. doi: 10.3892/ol.2019.10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang W., Wang J., Zhang X., Liu G. Serum circSETDB1 is a promising biomarker for predicting response to platinum-taxane-combined chemotherapy and relapse in high-grade serous ovarian cancer. Onco Targets Ther. 2019;12:7451–7457. doi: 10.2147/OTT.S220700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang Y.M., Huang L.M., Li D.R., Shao J.H., Xiong S.L., Wang C.M., Lu S.M. Hsa_circ_0101996 combined with hsa_circ_0101119 in peripheral whole blood can serve as the potential biomarkers for human cervical squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2017;10:11924–11931. [PMC free article] [PubMed] [Google Scholar]
- 163.Fan C.M., Wang J.P., Tang Y.Y., Zhao J., He S.Y., Xiong F., Guo C., Xiang B., Zhou M., Li X.L., et al. circMAN1A2 could serve as a novel serum biomarker for malignant tumors. Cancer Sci. 2019;110:2180–2188. doi: 10.1111/cas.14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hu Y., Zhu Y., Zhang W., Lang J., Ning L. Utility of Plasma circBNC2 As A Diagnostic Biomarker In Epithelial Ovarian Cancer. Onco Targets Ther. 2019;12:9715–9723. doi: 10.2147/OTT.S211413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang Y., Liu J., Ma J., Sun T., Zhou Q., Wang W., Wang G., Wu P., Wang H., Jiang L., et al. Exosomal circRNAs: Biogenesis, effect and application in human diseases. Mol. Cancer. 2019;18:116. doi: 10.1186/s12943-019-1041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lee J., Boczkowski D., Nair S. Programming human dendritic cells with mRNA. Methods Mol. Biol. 2013;969:111–125. doi: 10.1007/978-1-62703-260-5_8. [DOI] [PubMed] [Google Scholar]
- 167.Vik-Mo E.O., Nyakas M., Mikkelsen B.V., Moe M.C., Due-Tønnesen P., Suso E.M., Sæbøe-Larssen S., Sandberg C., Brinchmann J.E., Helseth E., et al. Therapeutic vaccination against autologous cancer stem cells with mRNA-transfected dendritic cells in patients with glioblastoma. Cancer Immunol. Immunother. 2013;62:1499–1509. doi: 10.1007/s00262-013-1453-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Feng Z., Meng S., Zhou H., Xu Z., Tang Y., Li P., Liu C., Huang Y., Wu M. Functions and Potential Applications of Circular RNAs in Cancer Stem Cells. Front. Oncol. 2019;9:500. doi: 10.3389/fonc.2019.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Dragomir M., Calin G.A. Circular RNAs in Cancer—Lessons Learned From microRNAs. Front. Oncol. 2018;8:179. doi: 10.3389/fonc.2018.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]