Abstract
Bovine spongiform encephalopathy (BSE) is a prion disease characterized by spongiform degeneration and astrocytosis in the brain. Unlike classical BSE, which is caused by prion-disease-contaminated meat and bone meal, the cause of atypical BSE has not been determined. Since previous studies have reported that the somatic mutation in the human prion protein gene (PRNP) has been linked to human prion disease, the somatic mutation of the PRNP gene was presumed to be one cause of prion disease. However, to the best of our knowledge, the somatic mutation of this gene in cattle has not been investigated to date. We investigated somatic mutations in a total of 58 samples, including peripheral blood; brain tissue including the medulla oblongata, cerebellum, cortex, and thalamus; and skin tissue in 20 individuals from each breed using pyrosequencing. In addition, we estimated the deleterious effect of the K211 somatic mutation on bovine prion protein by in silico evaluation tools, including PolyPhen-2 and PANTHER. We found a high rate of K211 somatic mutations of the bovine PRNP gene in the medulla oblongata of three Holsteins (10% ± 4.4%, 28% ± 2%, and 19.55% ± 3.1%). In addition, in silico programs showed that the K211 somatic mutation was damaging. To the best of our knowledge, this study is the first to investigate K211 somatic mutations of the bovine PRNP gene that are associated with potential BSE progression.
Keywords: E211K, E200K, bovine spongiform encephalopathy, prion, somatic mutation, prion protein gene (PRNP)
1. Introduction
Bovine spongiform encephalopathy (BSE) is a well-known prion disease characterized by the accumulation of abnormal prion protein (PrPSc), which shows resistance to proteinase K [1,2,3,4,5,6,7,8,9,10,11,12,13]. Classical BSE was first reported in the United Kingdom (UK) in 1986 and has been propagated by PrPSc-contaminated meat and bone meal. Since being controlled by more thorough surveillance of contaminated feed, the number of classical BSE cases has dramatically decreased [14,15]. However, novel forms of BSE and atypical BSEs have been reported in many countries. Unlike classical BSE, the cause of atypical BSE has not been determined [16,17,18].
In humans, prion diseases are classified as sporadic, familial, and infectious forms. Approximately 85% of all human prion diseases are sporadic Creutzfeldt–Jakob disease (CJD). Approximately 15% of all human prion diseases, including familial CJD, Gerstmann–Straussler–Scheinker syndrome (GSS), and fatal familial insomnia (FFI), are known to be caused by a germline mutation of the prion protein gene (PRNP) [19,20]. Additionally, <1% of all human prion diseases, including iatrogenic CJD, variant CJD, and kuru, are acquired by transmission. However, accurate causes of sporadic CJD have not been investigated to date [21,22]. Somatic mutations of the PRNP gene have been reported in the blood and brain tissue of sporadic cases of prion diseases. D178N and P102L somatic mutations have been reported in sporadic CJD patients and sporadic GSS symptom patients, respectively [23,24]. The commonality of these patients is that they were caused by somatic mutations, and not germline mutations inherited from their parents. Thus, somatic mutation of the PRNP gene was assumed to be one cause of sporadic prion diseases, in which no cause has been identified to date. In cattle, the E211K mutation of the bovine PRNP gene, which is homologous to the E200K mutation of the human PRNP gene found in familial CJD, was first reported in atypical BSE in the United States in 2006 [25,26,27]. At present, since atypical BSEs, which may refer to sporadically occurring BSE, have been occurring steadily in many countries, it is important to investigate the somatic mutation on E211K of the PRNP gene in cattle.
In this study, we investigated the E211K somatic mutation of the PRNP gene in Korean cattle using pyrosequencing. We collected peripheral whole blood and several types of tissue in cattle, including the medulla oblongata, cerebellum, cortex, thalamus, and skin tissue. We purified the genomic DNA from each tissue and blood and performed pyrosequencing to identify the E211K somatic mutation of the bovine PRNP gene. Furthermore, we estimated a deleterious effect of the E211K somatic mutation on bovine prion protein by in silico evaluation tools such as PolyPhen-2 and PANTHER [28,29].
2. Results
2.1. Sample Preparation
We collected samples from peripheral blood and five tissues, including the medulla oblongata, cerebellum, cortex, thalamus, and skin. Detailed information on the collected samples is described in Table 1. DNA extraction was performed with peripheral blood and brain tissues, and all DNA samples were subjected to quality checks using the A260/A280 ratio with 1.8–2.0 values.
Table 1.
Detailed information of the bovine samples used.
| Inspection Site | Sex | Hanwoo | Holstein | |
|---|---|---|---|---|
| Brain | Medulla oblongata | Male | 5 | 5 |
| Female | 5 | 5 | ||
| Cerebellum | Male | 3 | ||
| Female | 3 | |||
| Cortex | Male | 3 | ||
| Female | 3 | |||
| Thalamus | Male | 3 | ||
| Female | 3 | |||
| Peripheral blood | NA | 5 | 5 | |
| Skin tissue | NA | 5 | 5 | |
NA: not available.
2.2. Validation of the Inspection Tool
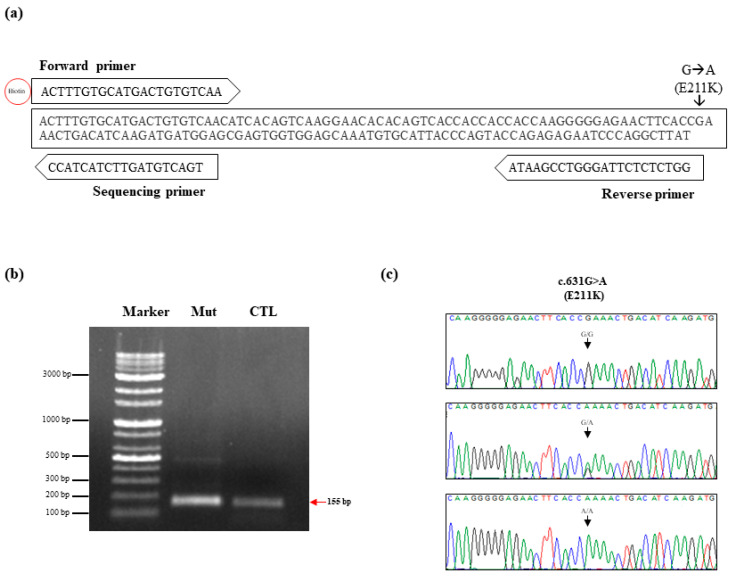
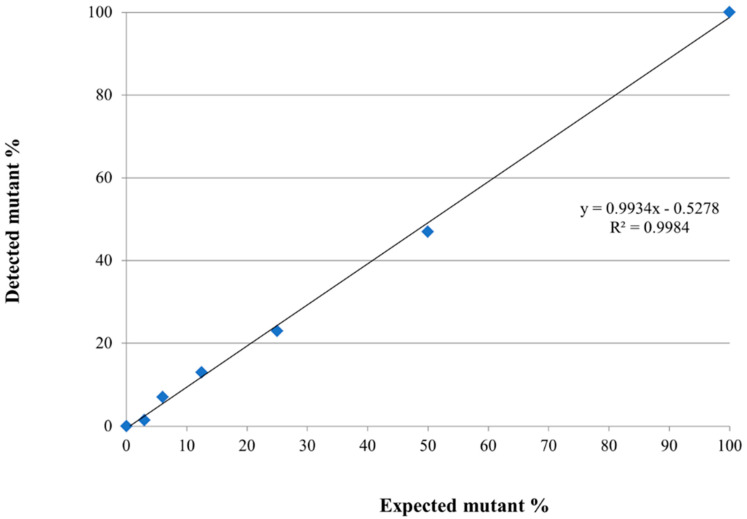
To investigate the somatic mutation of K211 in the bovine PRNP gene, we performed pyrosequencing analysis, which is known as a convenient tool to measure somatic mutations with low frequencies. Detailed information is described in Figure 1a. The polymerase chain reaction (PCR) products containing the mutation site at codon 211 of the bovine PRNP gene were amplified using biotinylated forward and non-biotinylated reverse primers (Table 2). The pyrosequencing primer set (forward, reverse, and sequencing primer) was generated using PyroMark Assay Design (QIAGEN, Germany). To verify the accuracy of the detection of pyrosequencing, we synthesized the K211 mutant gene using PCR-based site-directed mutagenesis. Biotinylated PCR products were confirmed on a gel with a length of 155 bp (Figure 1b). Before the pyrosequencing analysis, we confirmed the K211 mutation of the standard material by Sanger sequencing (Figure 1c). Then, we mixed standard material of the E211 normal gene with that of K211 mutant genes at specific ratios. According to the manufacturer’s recommendation, reliable detection and quantification of sequence variation using pyrosequencing is above 5% mutation. Thus, we excluded the dilutions of extremely low levels (0.1–3%). Each experiment was performed in duplicate. The mutation rate, which was observed using pyrosequencing, showed a high correlation with the expected mutation rate (R2 = 0.9984, Figure 2).
Figure 1.
(a) Schematic design of pyrosequencing to detect the E211K somatic mutation of the bovine prion protein gene (PRNP). (b) Gene-specific primers were used to obtain 155 bp amplicons of the bovine PRNP gene containing K211 mutation and E211 wild type. Marker: 100 bp DNA ladder marker; Mut: K211 mutation sample; CTL: E211 normal sample. (c) Electropherograms of Sanger sequencing results. The upper panel indicates Sanger sequencing results of the DNA amplicon with 100% E211 normal sample. The middle panel indicates Sanger sequencing results of mixed DNA amplicons with 50% E211 normal sample and 50% K211 mutation sample. The lower panel indicates Sanger sequencing results of DNA amplicon with 100% K211 mutation sample.
Table 2.
Detailed information on the primer set used.
| Purpose | Name | Sequence | Size | Annealing Temperature | |
|---|---|---|---|---|---|
| Site-directed mutagenesis | Round 1 | F1_F | ATGGTGAAAAGCCACATAGGCAG | 652 bp | 55 °C |
| F1_R | CCATCATCTTGATGTCAGTTTtGGTGAAGTTCTC | ||||
| Round 1 | F2_F | GAGAACTTCACCaAAACTGACATCAAGATGATGG | 204 bp | 55 °C | |
| F2_R | GATAATGAAAACAGGAAGGTTGCCCC | ||||
| Round 2 | F1_F | ATGGTGAAAAGCCACATAGGCAG | 820 bp | 55 °C | |
| F2_R | GATAATGAAAACAGGAAGGTTGCCCC | ||||
| Pyrosequencing | Amplification | Biotin_PF | ACTTTGTGCATGACTGTGTCAA | 155 bp | 58 °C |
| PR | ATAAGCCTGGGATTCTCTCTGG | ||||
| Sequencing | Seq_R | CCATCATCTTGATGTCAGT | |||
Figure 2.
Validation of the detection accuracies of pyrosequencing. The scatter plot indicates the average of the observed somatic mutation rates of the standard materials with mutation rates of 100%, 50%, 25%, 12.5%, 6%, 3%, and 0%. Regression analysis was performed to determine the correlation between the observed somatic mutation rates and the expected somatic mutation rates.
2.3. Investigation of K211 Somatic Mutation of Bovine PRNP Gene in Korean Cattle
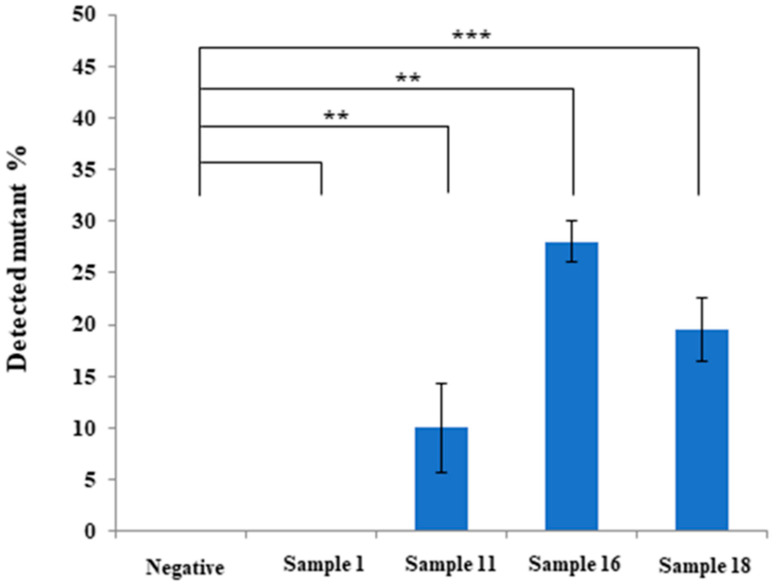
We performed pyrosequencing to examine the K211 somatic mutation of the PRNP gene in 58 brain and blood samples of Hanwoo and Holstein cattle. Detailed information on the pyrosequencing results is shown in Table 3. Among the 58 samples, three samples of the medulla oblongata of Holstein cattle, including samples 11, 16, and 18, showed somatic mutation rates of >10% (Table 3). The comparison of detection results between 0% negative standard and four bovine samples was analyzed using the Student’s t-test. Notably, samples 11 (10% ± 4.4%), 16 (28% ± 2%), and 18 (19.55% ± 3.1%) showed significantly high K211 somatic mutation rates (Table 3 and Figure 3).
Table 3.
K211 somatic mutation rates of the prion protein gene (PRNP) observed in Hanwoo and Holstein cattle.
| Sample No. | Breeds | Sex | Inspection Site | * AVG of Detected Mut. (%) | ** STD |
|---|---|---|---|---|---|
| Sample 1 | Hanwoo | Female | Medulla oblongata | 0 | 0 |
| Sample 2 | Hanwoo | Female | Medulla oblongata | 0 | 0 |
| Sample 3 | Hanwoo | Female | Medulla oblongata | 0 | 0 |
| Sample 4 | Hanwoo | Female | Medulla oblongata | 0 | 0 |
| Sample 5 | Hanwoo | Female | Medulla oblongata | 0 | 0 |
| Sample 6 | Hanwoo | Male | Medulla oblongata | 0 | 0 |
| Sample 7 | Hanwoo | Male | Medulla oblongata | 0 | 0 |
| Sample 8 | Hanwoo | Male | Medulla oblongata | 0 | 0 |
| Sample 9 | Hanwoo | Male | Medulla oblongata | 0 | 0 |
| Sample 10 | Hanwoo | Male | Medulla oblongata | 0 | 0 |
| Sample 11 | Holstein | Female | Medulla oblongata | 10 | 4.4 |
| Sample 12 | Holstein | Female | Medulla oblongata | 0 | 0 |
| Sample 13 | Holstein | Female | Medulla oblongata | 0 | 0 |
| Sample 14 | Holstein | Female | Medulla oblongata | 0 | 0 |
| Sample 15 | Holstein | Female | Medulla oblongata | 0 | 0 |
| Sample 16 | Holstein | Male | Medulla oblongata | 28 | 2 |
| Sample 17 | Holstein | Male | Medulla oblongata | 0 | 0 |
| Sample 18 | Holstein | Male | Medulla oblongata | 19.55 | 3.1 |
| Sample 19 | Holstein | Male | Medulla oblongata | 0 | 0 |
| Sample 20 | Holstein | Male | Medulla oblongata | 0 | 0 |
| Sample 21 | Hanwoo | Female | Cerebellum | 0 | 0 |
| Sample 22 | Hanwoo | Female | Cerebellum | 0 | 0 |
| Sample 23 | Hanwoo | Female | Cerebellum | 0 | 0 |
| Sample 24 | Holstein | Male | Cerebellum | 0 | 0 |
| Sample 25 | Holstein | Male | Cerebellum | 0 | 0 |
| Sample 26 | Holstein | Male | Cerebellum | 0 | 0 |
| Sample 27 | Hanwoo | Female | Cortex | 0 | 0 |
| Sample 28 | Hanwoo | Female | Cortex | 0 | 0 |
| Sample 29 | Hanwoo | Female | Cortex | 0 | 0 |
| Sample 30 | Holstein | Male | Cortex | 0 | 0 |
| Sample 31 | Holstein | Male | Cortex | 0 | 0 |
| Sample 32 | Holstein | Male | Cortex | 0 | 0 |
| Sample 33 | Hanwoo | Female | Thalamus | 0 | 0 |
| Sample 34 | Hanwoo | Female | Thalamus | 0 | 0 |
| Sample 35 | Hanwoo | Female | Thalamus | 0 | 0 |
| Sample 36 | Holstein | Male | Thalamus | 0 | 0 |
| Sample 37 | Holstein | Male | Thalamus | 0 | 0 |
| Sample 38 | Holstein | Male | Thalamus | 0 | 0 |
| Sample 39 | Hanwoo | NA | Peripheral blood | 0 | 0 |
| Sample 40 | Hanwoo | NA | Peripheral blood | 0 | 0 |
| Sample 41 | Hanwoo | NA | Peripheral blood | 0 | 0 |
| Sample 42 | Hanwoo | NA | Peripheral blood | 0 | 0 |
| Sample 43 | Hanwoo | NA | Peripheral blood | 0 | 0 |
| Sample 44 | Holstein | NA | Peripheral blood | 0 | 0 |
| Sample 45 | Holstein | NA | Peripheral blood | 0 | 0 |
| Sample 46 | Holstein | NA | Peripheral blood | 0 | 0 |
| Sample 47 | Holstein | NA | Peripheral blood | 0 | 0 |
| Sample 48 | Holstein | NA | Peripheral blood | 0 | 0 |
| Sample 49 | Hanwoo | NA | Skin tissue | 0 | 0 |
| Sample 50 | Hanwoo | NA | Skin tissue | 0 | 0 |
| Sample 51 | Hanwoo | NA | Skin tissue | 0 | 0 |
| Sample 52 | Hanwoo | NA | Skin tissue | 0 | 0 |
| Sample 53 | Hanwoo | NA | Skin tissue | 0 | 0 |
| Sample 54 | Holstein | NA | Skin tissue | 0 | 0 |
| Sample 55 | Holstein | NA | Skin tissue | 0 | 0 |
| Sample 56 | Holstein | NA | Skin tissue | 0 | 0 |
| Sample 57 | Holstein | NA | Skin tissue | 0 | 0 |
| Sample 58 | Holstein | NA | Skin tissue | 0 | 0 |
* AVG of detected mut. (%): average of detected percentage of K211 mutations; ** STD: standard deviation; NA: not available.
Figure 3.
Representative results on the K211 somatic mutation rates of the bovine prion protein gene (PRNP) using pyrosequencing. Negative: standard material with 0% somatic mutation rates; Sample 1: Hanwoo, medulla oblongata; Sample 11: Holstein, medulla oblongata; Sample 16: Holstein, medulla oblongata; Sample 18: Holstein, medulla oblongata. All experiments were repeated three times. ** indicates p < 0.01 and *** indicates p < 0.001.
2.4. In Silico Estimation of the Deleterious Effect of K211 Somatic Mutation of Bovine Prion Protein
To validate the prediction effectiveness of the annotation software, prior to the analysis of the K211 somatic mutation of the bovine PRNP gene, we analyzed the previously reported pathogenic PRNP mutations related to human genetic prion diseases. Although six mutations of human PRNP (D167N, V180I, V203I, I215V, M232R, and M232T) were predicted to be deleterious by only one program, the remaining 34 pathogenic mutations were predicted to be deleterious by PolyPhen-2 and PANTHER (Supplementary Table S1). We estimated the biological impact of the somatic mutation c.631G>A (E211K) of the bovine PRNP gene using in silico programs, including PolyPhen-2 and PANTHER. PolyPhen-2 and PANTHER predicted that E211K was “probably damaging” with a score of 0.993 and “possibly damaging” with a score of 361, respectively (Table 4).
Table 4.
In silico evaluation of the impact of K211 somatic mutation on the bovine prion protein.
| Somatic Mutation | Methods | Score | Prediction |
|---|---|---|---|
| c.631G > A (E211K) |
PolyPhen-2 | 0.993 | Probably damaging |
| PANTHER | 361 | Possibly damaging |
3. Discussion
Although the number of classical BSE cases caused by contaminated meat and bone meal has decreased dramatically due to global efforts, the number of atypical BSE cases increased [30,31]. In humans, genetic prion disease makes up approximately 10% to 15% of cases with germline mutations of the PRNP gene [19]. However, although sporadic prion disease accounts for 85% of human prion diseases, the exact cause of sporadic prion disease has not been revealed to date. In recent studies, somatic mutations in the PRNP gene have been identified in human sporadic prion diseases and have been suggested as one cause of sporadic prion disease. In cattle, the E211K germline mutation of the bovine PRNP gene was first reported in the United States in 2006 [25,26,27]. The bovine E211K mutation showed homology in the region with the E200K mutation of the human PRNP gene, which is most frequently observed in human familial prion diseases [20,22]. Thus, we investigated the E211K somatic mutation of the bovine PRNP gene, which may be considered a novel risk factor for BSE in Korean cattle.
In our previous study, the germline mutation at codon 211 of the bovine PRNP gene was not observed in 384 Hanwoo cattle and 152 Holstein cattle [5]. Remarkably, we found high rates of K211 somatic mutation of the bovine PRNP gene in three Holstein cattle (Table 3). In Korea, since classical and atypical BSEs have never been reported in Hanwoo, the absence of K211 somatic mutations in Hanwoo is notable. In addition, although we tested K211 somatic mutations, including whole blood and four brain regions, somatic mutations of this gene were detected only in the medulla oblongata. The expected level of K211 mutations of the bovine PRNP gene, which can initiate atypical BSE, is elusive. A previous study in early-onset Alzheimer’s disease reported that 14% of mutations of the presenilin 1 (PSEN1) gene in brain cells are responsible for the initiation of this disease [32]. In another study, the somatic mutations of amyloid precursor protein (APP), nicastrin (NCSTN), sortilin-related receptor (SORL1), and microtubule affinity-regulating kinase 4 (MARK4) were observed in sporadic Alzheimer’s disease patients in 0.2–10.5% [33]. We observed three samples containing over 10% K211 mutations (Figure 3). Thus, BSE inspection of these samples seems essential to elucidate the expected level of K211 mutations of the PRNP gene, which can initiate atypical BSE in the future.
According to previous studies, since the medulla oblongata showed a prominent accumulation of PrPSc, the diagnosis of BSE was performed in the medulla oblongata region [34]. However, since the three Holstein cattle carrying the K211 somatic mutation in the medulla oblongata were not investigated as to whether K211 somatic mutation could be observed in other brain regions, it is difficult to conclude that this somatic mutation was only found in the medulla oblongata. In addition, since atypical BSE has shown to be a prominent brain pathology in the frontal cortex [17,18], if the atypical BSE was caused by a somatic mutation of the PRNP gene, it is expected that the somatic mutation was also found in the cerebral cortex. Thus, further investigation of the K211 somatic mutation is highly desirable in other brain regions, including the cerebral cortex of the three Holstein cattle carrying K211 somatic mutation in the medulla oblongata. We also performed in silico analysis using PolyPhen-2 and PANTHER to evaluate whether the mutation of K211 of the bovine PRNP gene affects the bovine PrP protein. Notably, the K211 somatic mutation of the bovine PRNP gene was evaluated as “damaging”. Since PolyPhen-2 can estimate structural variation, the prediction can be interpreted that K211 somatic mutation can contribute to the conformational change to the susceptible structure of the PrPSc. In addition, the wild-type PrP with E211 allele was predicted to be of high preservation time (361, Table 4) and K211 somatic mutation was estimated to be deleterious by using the PANTHER program. The prediction of PANTHER indicated that K211 mutation has been very rare and has not been observed in ancestral proteins for a long time. To confirm the effect of K211 mutation on prion disease, investigation of symptoms of prion disease in bovine PrP transgenic mice carrying the K211 allele is highly desirable in the future. It seems possible that a high somatic mutation rate contributes to sporadic prion disease in cattle.
Previous studies have reported that prion disease is accelerated in transgenic (Tg) mice expressing wild-type bank vole prion protein (BVPrP) containing E200K, which is associated with human familial CJD [35]. In addition, recent studies have demonstrated that a human PrP Tg mouse model with the E200K mutation can lead to spontaneous prion disease [36]. In cattle, the E211K mutation, which is a homologous region of human E200K, is expected to have a deleterious effect on BSE, and several in silico programs in the present study are expected to cause structural changes in the bovine prion protein (Table 4). To determine whether the E211K mutation affects the susceptibility to prion disease, brain samples with high mutation rates of E211K should be investigated by protein misfolding cyclic amplification (PMCA) or Western blotting (WB) in the future.
4. Materials and Methods
4.1. Ethics Statement
Peripheral whole blood, brain, and skin tissues were collected from slaughterhouses in the Republic of Korea. All experimental procedures were approved by the Institute of Animal Care and Use Committee of Jeonbuk National University (JBNU 2018-079) and approved on 1 October 2018. All experiments using cattle were performed following the Korean Experimental Animal Protection Act.
4.2. Genomic DNA Extraction
Genomic DNA was extracted from 20 mg of tissue using a Tissue Genomic DNA Isolation kit (QIAGEN, Germantown, MD, USA) and 200 µL of blood using a Blood Genomic DNA Isolation kit (QIAGEN, USA) following the manufacturer’s instructions.
4.3. PCR
PCR was performed using Taq DNA Polymerase (QIAGEN, Hilden, Germany) following the manufacturer’s instructions. The PCR mixture contained 20 pmol of each primer, 5 µL of 10× Taq DNA polymerase buffer, 1 μL of 10 mM dNTP mixture, and 2.5 units of Taq DNA polymerase. The PCR conditions for each primer are described in Table 1. PCR was performed using an S-1000 Thermal Cycler (Bio-Rad, Hercules, CA, USA).
4.4. Site-Directed Mutagenesis
PCR-based site-directed mutagenesis was performed to obtain a DNA fragment of mutant type K211 of the bovine PRNP gene, which was used as a positive control. Briefly, 4 oligonucleotide primers were synthesized to carry out site-directed mutagenesis. Detailed information on the primers and experimental conditions is described in Table 1. In the first round of PCR, each pair (F1 pair and F2 pair) was amplified from a targeted region of the bovine PRNP gene. The second round of PCR was performed by F1_F and F2_R primer pairs, and amplicons amplified by the F1 pair and F2 pair were used as templates to obtain a DNA fragment of mutant K211 of the bovine PRNP gene.
4.5. Sanger Sequencing
The PCR products were purified using a PCR Purification Kit (Thermo Fisher Scientific, Bridgewater, NJ, USA) and directly analyzed with an ABI 3730 automatic sequencer (ABI, Foster City, CA, USA). Sequencing results were read by Finch TV software (Geospiza Inc., Seattle, WA, USA).
4.6. Pyrosequencing
Pyrosequencing was performed using biotinylated amplicons of the bovine PRNP gene by PCR. Briefly, the genomic DNA (10 ng) isolated from cattle was amplified by PCR using a biotinylated forward primer and a non-biotinylated reverse primer. Detailed information on the primers and PCR conditions is described in Table 1. The PCR products (25 µL) were mixed with 2 µL Streptavidin Sepharose High-Performance medium (GE Healthcare, Madison, WI, USA), 40 µL PyroMark Binding Buffer (QIAGEN, USA), and 8.5 µL high-purity autoclaved water, and centrifuged for 15 min at 14,000 rpm. The resulting products were washed with 70% ethanol for 10 s, PyroMark denaturation buffer (QIAGEN, USA) for 10 s, and PyroMark wash buffer (QIAGEN, USA) for 10 s, and then mixed with 24.2 µL PyroMark annealing buffer (QIAGEN, USA) and 0.8 µL sequencing primer (2 µM). Finally, the prepared samples were loaded into the PyroMark Q24 (QIAGEN, USA) and operated in the allele quantification (AQ) mode according to the manufacturer’s protocol.
4.7. Statistical Analysis
Three independent experiments were carried out, and the data were reported as the mean values ± standard deviation (SD). Statistical significance using the p-value was calculated with SAS version 9.4 (SAS Institute Inc., Cary, NC, USA) using a two-tailed Student’s t-test for single comparisons. The symbols **, and *** indicate p < 0.01, and p < 0.001, respectively.
4.8. In Silico Estimation of the Impact of E211K Somatic Mutation of the Bovine Prion Protein
A deleterious effect of somatic mutation E211K in bovine prion protein was evaluated by the PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/index.shtml) and PANTHER (http://www.pantherdb.org/) programs. PolyPhen-2 analyzed the impact of amino acid substitution on the protein of interest based on various features, including the functional or binding properties of sequence, phylogenetic property, and 3D structural information characterizing the substitution. PANTHER predicted the likelihood of a specific amino acid substitution in causing a functional impact on the protein by calculating the length of time that a given amino acid has been evolutionarily preserved to the protein of interest. The length of the preservation time is proportional to the magnitude of the likelihood of the functional impact.
5. Conclusions
In this study, we first identified a total of three high somatic mutation rates of E211K in three Holsteins using pyrosequencing. Next, we performed in silico estimation of the E211K mutation of the bovine PRNP gene using PolyPhen-2 and PANTHER, and it was predicted to be damaging. To the best of our knowledge, this study was the first to investigate potential BSE-associated K211 somatic mutations of the bovine PRNP gene.
Acknowledgments
This work was supported by an NRF (National Research Foundation of Korea) Grant funded by the Korean Government (NRF-2019-Fostering Core Leaders of the Future Basic Science Program/Global Ph.D. Fellowship Program).
Abbreviations
| APP | amyloid precursor protein |
| AQ | allele quantification |
| BSE | bovine spongiform encephalopathy |
| BVPrP | bank vole prion protein |
| PrPSc | abnormal prion protein |
| UK | United Kingdom |
| CJD | Creutzfeldt–Jakob disease |
| GSS | Gerstmann–Straussler–Scheinker syndrome |
| FFI | fatal familial insomnia |
| MARK4 | microtubule affinity-regulating kinase 4 |
| NCSTN | nicastrin |
| PRNP | prion protein gene |
| PCR | polymerase chain reaction |
| PMCA | protein misfolding cyclic amplification |
| SD | standard deviation |
| SORL1 | sortilin-related receptor |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/12/4246/s1.
Author Contributions
S.-Y.W., Y.-C.K., and B.-H.J. conceived and designed the experiments. S.-Y.W. and Y.-C.K. performed the experiments. S.-Y.W., Y.-C.K. and B.-H.J. analyzed the data. S.-Y.W., Y.-C.K. and B.-H.J. wrote the paper. All authors read and approved the final manuscript.
Funding
This research was supported by the Basic Science Program through the National Research Foundation (NRF) of Korea, funded by the Ministry of Education, Science, and Technology (2018R1D1A1B07048711). This research was supported by the Basic Science Research Program through the National Research Foundation (NRF) of Korea, funded by the Ministry of Education (2017R1A6A1A03015876). This work was supported by an NRF (National Research Foundation of Korea) Grant funded by the Korean Government (NRF-2019-Fostering Core Leaders of the Future Basic Science Program/Global Ph.D. Fellowship Program).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Prusiner S.B. The prion diseases. Brain Pathol. 1998;8:499–513. doi: 10.1111/j.1750-3639.1998.tb00171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prusiner S.B. Prions. Proc. Natl. Acad. Sci. USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeong B.H., Jin H.T., Carp R.I., Kim Y.S. Bovine spongiform encephalopathy (BSE)-associated polymorphisms of the prion protein (PRNP) gene in Korean native cattle. Anim. Genet. 2013;44:356–357. doi: 10.1111/age.12004. [DOI] [PubMed] [Google Scholar]
- 4.Kim Y.C., Jeong B.H. Bovine spongiform encephalopathy (BSE) associated polymorphisms of the prion-like protein gene (PRND) in Korean dairy cattle and Hanwoo. J. Dairy Res. 2018;85:7–11. doi: 10.1017/S0022029917000814. [DOI] [PubMed] [Google Scholar]
- 5.Kim Y.C., Jeong B.H. Lack of germline mutation at codon 211 of the prion protein gene (PRNP) in Korean native cattle—Short communication. Acta Vet. Hung. 2017;65:147–152. doi: 10.1556/004.2017.015. [DOI] [PubMed] [Google Scholar]
- 6.Kim Y.C., Jeong B.H. First report of prion-related protein gene (PRNT) polymorphisms in cattle. Vet. Rec. 2018;182:717. doi: 10.1136/vr.104123. [DOI] [PubMed] [Google Scholar]
- 7.Kim Y.C., Won S.Y., Jeong B.H. Absence of single nucleotide polymorphisms (SNPs) in the open reading frame (ORF) of the prion protein gene (PRNP) in a large sampling of various chicken breeds. BMC Genomics. 2019;20:922. doi: 10.1186/s12864-019-6315-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Won S.Y., Kim Y.C., Kim S.K., Jeong B.H. The First Report of Genetic and Structural Diversities in the SPRN Gene in the Horse, an Animal Resistant to Prion Disease. Genes. 2019;11:39. doi: 10.3390/genes11010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Won S.Y., Kim Y.C., Kim K., Kim A.D., Jeong B.H. The First Report of Polymorphisms and Genetic Features of the prion-like Protein Gene (PRND) in a Prion Disease-Resistant Animal, Dog. Int. J. Mol. Sci. 2019;20:1404. doi: 10.3390/ijms20061404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim S.K., Kim Y.C., Won S.Y., Jeong B.H. Potential scrapie-associated polymorphisms of the prion protein gene (PRNP) in Korean native black goats. Sci. Rep. 2019;9:15293. doi: 10.1038/s41598-019-51621-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y.C., Kim S.K., Jeong B.H. Scrapie susceptibility-associated indel polymorphism of shadow of prion protein gene (SPRN) in Korean native black goats. Sci. Rep. 2019;9:15261. doi: 10.1038/s41598-019-51625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim Y.C., Jeong B.H. The first report of polymorphisms and genetic characteristics of the prion protein gene (PRNP) in horses. Prion. 2018;12:245–252. doi: 10.1080/19336896.2018.1513316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim Y.C., Jeong M.J., Jeong B.H. The first report of genetic variations in the chicken prion protein gene. Prion. 2018;12:197–203. doi: 10.1080/19336896.2018.1471922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collee J.G., Bradley R. BSE: A decade on—Part 2. Lancet. 1997;349:715–721. doi: 10.1016/S0140-6736(96)08496-6. [DOI] [PubMed] [Google Scholar]
- 15.Collee J.G., Bradley R. BSE: A decade on—Part I. Lancet. 1997;349:636–641. doi: 10.1016/S0140-6736(96)01310-4. [DOI] [PubMed] [Google Scholar]
- 16.Gurgul A., Polak M.P., Larska M., Slota E. PRNP and SPRN genes polymorphism in atypical bovine spongiform encephalopathy cases diagnosed in Polish cattle. J. Appl. Genet. 2012;53:337–342. doi: 10.1007/s13353-012-0102-4. [DOI] [PubMed] [Google Scholar]
- 17.Costassa E.V., Iulini B., Mazza M., Acutis P., Maurella C., Meloni D., Pautasso A., Capucci L., Bozzetta E., Simmons M.M., et al. Pathogenesis and Transmission of Classical and Atypical BSE in Cattle. Food Saf. 2016;4:130–134. doi: 10.14252/foodsafetyfscj.2016018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dudas S., Czub S. Atypical BSE: Current Knowledge and Knowledge Gaps. Food Saf. 2017;5:10–13. doi: 10.14252/foodsafetyfscj.2016028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovacs G.G., Puopolo M., Ladogana A., Pocchiari M., Budka H., van Duijn C., Collins S.J., Boyd A., Giulivi A., Coulthart M., et al. Genetic prion disease: The EUROCJD experience. Hum. Genet. 2005;118:166–174. doi: 10.1007/s00439-005-0020-1. [DOI] [PubMed] [Google Scholar]
- 20.Lloyd S., Mead S., Collinge J. Genetics of prion disease. Top. Curr. Chem. 2011;305:1–22. doi: 10.1007/128_2011_157. [DOI] [PubMed] [Google Scholar]
- 21.Uttley L., Carroll C., Wong R., Hilton D.A., Stevenson M. Creutzfeldt-Jakob disease: A systematic review of global incidence, prevalence, infectivity, and incubation. Lancet Infect. Dis. 2020;20:e2–e10. doi: 10.1016/S1473-3099(19)30615-2. [DOI] [PubMed] [Google Scholar]
- 22.Jeong B.H., Kim Y.S. Genetic studies in human prion diseases. J. Korean Med. Sci. 2014;29:623–632. doi: 10.3346/jkms.2014.29.5.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alzualde A., Moreno F., Martínez-Lage P., Ferrer I., Gorostidi A., Otaegui D., Blázquez L., Atares B., Cardoso S., López de Munain A. Somatic mosaicism in a case of apparently sporadic Creutzfeldt-Jakob disease carrying a de novo D178N mutation in the PRNP gene. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2010;153:1283–1291. doi: 10.1002/ajmg.b.31099. [DOI] [PubMed] [Google Scholar]
- 24.Kojović M., Glavač D., Ožek B., Zupan A., Popović M. De novo P102L mutation in a patient with Gerstmann-Sträussler-Scheinker disease. Eur. J. Neurol. 2011;18:e152–e153. doi: 10.1111/j.1468-1331.2011.03531.x. [DOI] [PubMed] [Google Scholar]
- 25.Nicholson E.M., Brunelle B.W., Richt J.A., Kehrli M.E., Jr., Greenlee J.J. Identification of a heritable polymorphism in bovine PRNP associated with genetic transmissible spongiform encephalopathy: Evidence of heritable BSE. PLoS ONE. 2008;3:e2912. doi: 10.1371/journal.pone.0002912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greenlee J.J., Smith J.D., West Greenlee M.H., Nicholson E.M. Clinical and pathologic features of H-type bovine spongiform encephalopathy associated with E211K prion protein polymorphism. PLoS ONE. 2012;7:e38678. doi: 10.1371/journal.pone.0038678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore S.J., West Greenlee M.H., Smith J.D., Vrentas C.E., Nicholson E.M., Greenlee J.J. A Comparison of Classical and H-Type Bovine Spongiform Encephalopathy Associated with E211K Prion Protein Polymorphism in Wild-Type and EK211 Cattle Following Intracranial Inoculation. Front. Vet. Sci. 2016;3:78. doi: 10.3389/fvets.2016.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013;76:7–20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang H., Thomas P.D. PANTHER-PSEP: Predicting disease-causing genetic variants using position-specific evolutionary preservation. Bioinformatics. 2016;32:2230–2232. doi: 10.1093/bioinformatics/btw222. [DOI] [PubMed] [Google Scholar]
- 30.Kumagai S., Daikai T., Onodera T. Bovine Spongiform Encephalopathy—A Review from the Perspective of Food Safety. Food Saf. 2019;7:21–47. doi: 10.14252/foodsafetyfscj.2018009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hagiwara K., Sato Y., Yamakawa Y., Hara H., Tobiume M., Okemoto-Nakamura Y., Sata T., Horiuchi M., Shibata H., Ono F. Tracking and clarifying differential traits of classical- and atypical L-type bovine spongiform encephalopathy prions after transmission from cattle to cynomolgus monkeys. PLoS ONE. 2019;14:e0216807. doi: 10.1371/journal.pone.0216807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beck J.A., Poulter M., Campbell T.A., Uphill J.B., Adamson G., Geddes J.F., Revesz T., Davis M.B., Wood N.W., Collinge J., et al. Somatic and germline mosaicism in sporadic early-onset Alzheimer’s disease. Hum. Mol. Genet. 2004;13:1219–1224. doi: 10.1093/hmg/ddh134. [DOI] [PubMed] [Google Scholar]
- 33.Nicolas G., Acuna-Hidalgo R., Keogh M.J., Quenez O., Steehouwer M., Lelieveld S., Rousseau S., Richard A.C., Oud M.S., Marguet F., et al. Somatic variants in autosomal dominant genes are a rare cause of sporadic Alzheimer’s disease. Alzheimers Dement. 2018;14:1632–1639. doi: 10.1016/j.jalz.2018.06.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Keulen L., Schreuder B., Meloen R., Poelen-Van Den Berg M., Mooij-Harkes G., Vromans M., Langeveld J.P.M. Immumohistochemical detection and localization of prion protein in brain tissue of sheep with natural scrapie. Vet. Pathol. 1995;32:299–308. doi: 10.1177/030098589503200312. [DOI] [PubMed] [Google Scholar]
- 35.Watts J.C., Giles K., Bourkas M.E., Patel S., Oehler A., Gavidia M., Bhardwaj S., Lee J., Prusiner S.B. Towards authentic transgenic mouse models of heritable PrP prion diseases. Acta Neuropathol. 2016;132:593–610. doi: 10.1007/s00401-016-1585-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedman-Levi Y., Meiner Z., Canello T., Frid K., Kovacs G.G., Budka H., Avrahami D., Gabizon R. Fatal prion disease in a mouse model of genetic E200K Creutzfeldt-Jakob disease. PLoS Pathog. 2011;7:e1006294. doi: 10.1371/journal.ppat.1002350. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.