Abstract
Weight regulation and the magnitude of weight loss after a Roux-en-Y gastric bypass (RYGB) can be genetically determined. DNA methylation patterns and the expression of some genes can be altered after weight loss interventions, including RYGB. The present study aimed to evaluate how the gene expression and DNA methylation of PIK3R1, an obesity and insulin-related gene, change after RYGB. Blood samples were obtained from 13 women (35.9 ± 9.2 years) with severe obesity before and six months after surgical procedure. Whole blood transcriptome and epigenomic patterns were assessed by microarray-based, genome-wide technologies. A total of 1966 differentially expressed genes were identified in the pre- and postoperative periods of RYGB. From these, we observed that genes involved in obesity and insulin pathways were upregulated after surgery. Then, the PIK3R1 gene was selected for further RT-qPCR analysis and cytosine-guanine nucleotide (CpG) sites methylation evaluation. We observed that the PI3KR1 gene was upregulated, and six DNA methylation CpG sites were differently methylated after bariatric surgery. In conclusion, we found that RYGB upregulates genes involved in obesity and insulin pathways.
Keywords: DNA methylation, insulin, obesity, microarray, gene expression, bariatric surgery, weight loss
1. Introduction
The worldwide increase in the obesity epidemic is a consequence of an obesogenic environment characterized by an imbalance between energy intake and expenditure [1]. Such environmental factors have been linked to a rapid weight gain; however, genetic alterations may also contribute substantially to obesity [2,3]. Consequently, obesity can result from genetic and epigenetic factors that influence body weight maintenance [4], in addition to an unbalanced glucose and insulin homeostasis. Different metabolic pathways affect the development of this complex disease, namely energy expenditure regulation, adipocyte differentiation, adipose tissue accumulation, and pathways related to eating behavior [5].
The Roux-en-Y gastric bypass (RYGB)—the most common surgical procedure to treat obesity after the failure of conventional treatments [6]—can also improve comorbidities related to obesity [7]. Furthermore, given the role of genetics in weight regulation, the magnitude of weight loss after surgery can also be genetically determined [8]. As a consequence, numerous biological pathways may be altered in obese patients, and bariatric surgery could restore the normal functions of regulatory networks [9].
Epigenetic modifications may also indicate critical relationships between genetic, environmental, and lifestyle factors. DNA methylation patterns are currently well characterized for chronic diseases; methylation is characterized by the presence of methyl (−CH3) groups at cytosine-guanine nucleotide (CpG) sites. However, it remains unclear how DNA methylation influences gene transcription, obesity and its related comorbidities [10].
The phosphatidylinositol 3-kinase (PIK3) signaling pathway plays a vital role in insulin metabolism. Several reports show that the canonical pathway PIK3/AKT is disrupted in obese subjects with and without diabetes compared with non-obese subjects [11]. PIK3 has both catalytic (PIK3CA and PIK3CD) and regulatory subunits (PIK3R1), being the last one critical for obesity studies since its expression is linked to insulin metabolism and initiation of inflammation processes [12].
Currently, the patterns of individual CpG sites of PIK3R1 before and after bariatric surgery remain uncharacterized. Thus, in the present study, we aimed to determine the impacts of RYGB on the expression and DNA methylation patterns of the PIK3R1 gene. We chose to use whole blood as the target tissue as it reflects the systemic response to metabolic alterations and is an essential biological tissue for biomarker discovery [13]. As far as we know this was the first study to analyze the pattern of individual CpG sites of PIK3R1 before and after bariatric surgery.
2. Results
2.1. Anthropometrics Data and Biochemical Profile
The mean preoperative weight of the 13 investigated female patients was 115.3 ± 19.4 kg (BMI: 42.5 ± 7 kg/m²), and a 6-month weight loss corresponded to 24.7% of initial weight and 54.4% of excess body weight (EBW). Four patients (12.5%) had type 2 diabetes in the preoperative period and were taking metformin as the primary treatment. After surgery, we observed that blood glucose concentration decreased from 93.3 ± 13.5 to 83.6 ± 28.4 mg/dL. Table 1 shows the anthropometric data and biochemical profile of obese patients before and six months after RYGB. Data for comorbidities, life habits, and family history of obesity were previously reported by our group [14].
Table 1.
Anthropometric data and biochemical profile of obese patients in preoperative and postoperative (six months after Roux-en-Y gastric bypass (RYGB)).
| Variables | Preoperative | Postoperative |
|---|---|---|
| N | 13 | 13 |
| Age (years) | 35.9 ± 9.2 | 36.2 ± 9.3 |
| Weight (kg) | 115.3 ± 19.4 | 85.3 ± 13.8 a |
| BMI (kg/m²) | 42.5 ± 7.0 | 31.8 ± 5.4 a |
| TC (mg/dL) | 177.4 ± 32.7 | 152.5 ± 28.4 a |
| HDL-C (mg/dL) | 41.3 ± 6.0 | 44.3 ± 8.1 |
| LDL-C (mg/dL) | 112.4 ± 27.6 | 88.6 ± 23.0 a |
| TG (mg/dL) | 129.7 ± 42.7 | 75.0 ± 27.0 a |
| Glucose (mg/dL) | 93.3 ± 13.5 | 83.6 ± 28.4 a |
Mean ± standard deviation; BMI: body mass index; %EWL: excess weight loss; TC: total cholesterol; HDL-C: high-density lipoprotein cholesterol; LDL-C: low-density lipoprotein cholesterol; TG: triglycerides; a: p-value < 0.05 compared to preoperative.
2.2. Gene Expression
The comparison of gene expression profiles in obese patients before and after RYGB revealed that 1188 genes were upregulated (Supplementary File 1). Gene Ontology (GO) enrichment analysis showed that 13 genes were significantly involved with the onset of obesity and insulin-related pathways (p-value < 0.05). Then we selected genes with p-values ≤0.05 and fold change cutoff of ≥2 are presented in Table 2. The description of the pipeline for GO calculations can be accessed at http://geneontology.org/docs/go-enrichment-analysis/.
Table 2.
Upregulated genes after six months of RYGB that are involved in the onset of obesity and insulin pathway (p-value < 0.05). The fold change results were derived from the microarray assay.
| Symbol | Gene Name | Fold Change * |
|---|---|---|
| Obesity | ||
| EPS8 | Epidermal growth factor receptor pathway substrate 8 | 5.57 × 106 |
| PTPN2 | Protein tyrosine phosphatase, non-receptor type 2 | 2.79 × 106 |
| ACTR3 | ARP3 actin-related protein 3 homolog | 2.32 × 106 |
| RPS6KA3 | Ribosomal protein S6 kinase | 2.45 × 106 |
| MAPK9 | Mitogen-activated protein kinase 9 | 2.26 × 106 |
| RAP1B | RAP1B member of RAS oncogene family | 5.51 × 106 |
| PLA2G4A | Phospholipase A2 | 9.65 × 106 |
| PIK3R1 | Phosphoinositide-3-kinase, regulatory subunit 1 | 6.55 × 106 |
| Insulin Pathway | ||
| EIF4E | Eukaryotic translation initiation factor 4E | 3.76 × 106 |
| IKBKB | Inhibitor of kappa light polypeptide gene enhancer in B-cells kinase beta | 3.90 × 106 |
| PHKB | Phosphorylase kinase beta | 4.13 × 106 |
| MAPK9 | Mitogen-activated protein kinase 9 | 2.26 × 106 |
| PPP1CB | Protein phosphatase 1, catalytic subunit, beta isozyme | 2.19 × 106 |
| SOCS4 | Suppressor of cytokine signaling 4 | 4.13 × 106 |
| PIK3R1 | Phosphoinositide-3-kinase, regulatory subunit 1 alpha | 6.55 × 106 |
* p-value < 0.05.
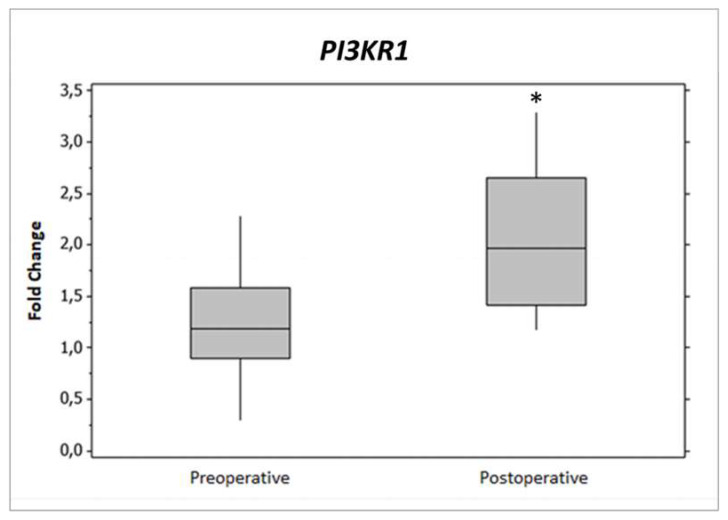
PIK3R1 gene expression was significantly enriched in both lists (p < 0.05) from Table 2, because it was chosen to be validated and analyzed more deeply by DNA methylation. PIK3R1 gene expression was verified by quantitative RT-qPCR (Figure 1).
Figure 1.
Relative gene expression of phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1) in pre- and postoperative periods. Both periods were composed of 13 patients. The fold change was calculated with the equation 2−∆∆Ct. * p < 0.05.
We compared results for PIK3R1 gene expression, including and excluding the data from two patients that had blood glucose values above the recommended. Since we did not find any difference in such comparison, the patients were kept for further analyses.
2.3. CpG Methylation of PIK3R1
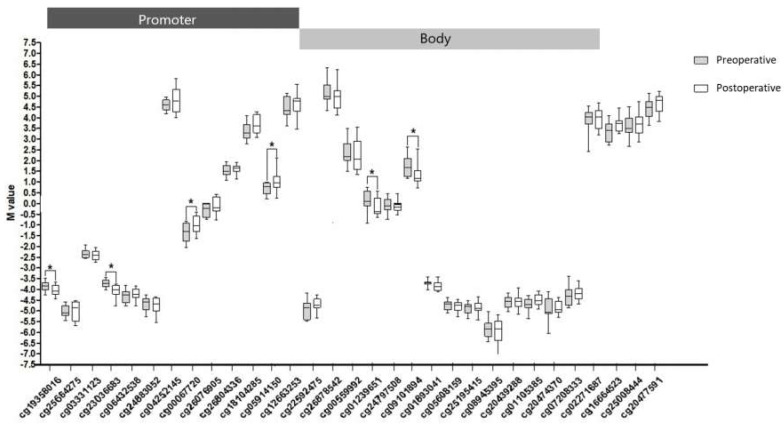
After quality control checks, we obtained 410.586 valid probes. Using iMETHYL [15], we found that 31 probes were mapped to the PIK3R1 gene. M values for all probes are shown in Figure 2. When comparing the data before and after RYGB, we found six differentially methylated CpG sites (cg19358016, cg23036683, cg00067720, cg05914150, cg01239651, and cg09101894). These probes were primarily hypomethylated in patients after surgery, and the majority of them were located in CpGs of the promoter region (TSS 200 and 1500) and 5’ UTR of PIK3R1 gene.
Figure 2.
M values of evaluated CpG sites in promoter and body regions of PI3KR1 gene in pre- and postoperative periods of RYGB. * p < 0.05 shows CGs statistically significant in patients.
2.4. Relationship between CpG Methylation and PI3KR1 Gene Expression
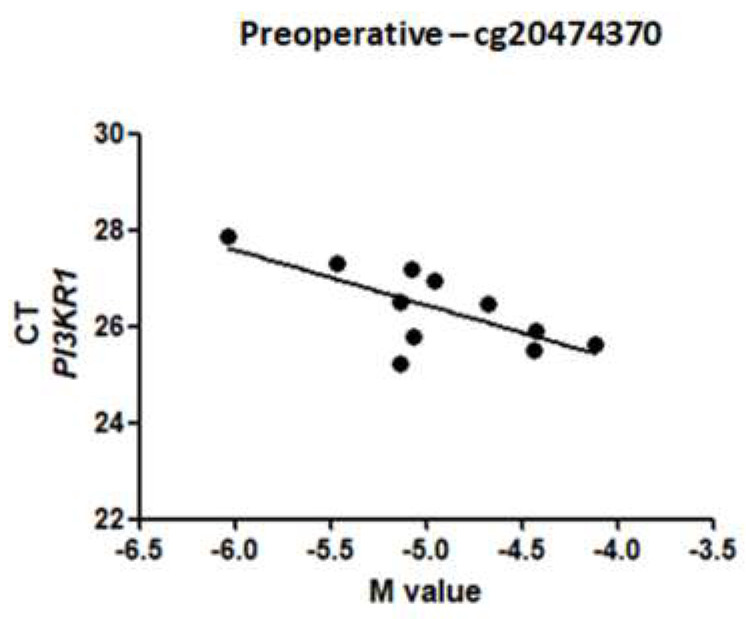
After stepwise linear regressions, three CpG sites were associated with RT-qPCR values (p < 0.005): cg22592475, cg26878542, and cg20474370. However, only cg20474370 strongly correlated with gene expression in the preoperative group (Pearson correlation, r = −0.709, p = 0.001) (Figure 3). To build a multivariable linear regression model, we included the cycle threshold (CT) gene expression data values and the M values for DNA methylation. Moreover, the model was adjusted by the endogenous control expression data. The β values, multiple r², and p-values are shown in Table 3. The β values represent the direction of the association and r² the percentage of explanation of the model.
Figure 3.
Correlation between gene expression data (RT-qPCR) and the M value in the site cg20474370 of PI3KR1 in before RYGB (r = −0.709, p < 0.01). CT: cycle threshold.
Table 3.
Multiple linear regression showing the contribution of cg20474370 in gene expression.
| Gene Symbol (Gene ID) | Site | Gene Region | β | Adjusted Multiple r² | p-Value |
|---|---|---|---|---|---|
| PI3KR1 (ENSG00000145675) | cg20474370 | Body | −0.59 | 0.70 | 0.003 |
β: regression coefficient, r²: coefficient of determination. Regression was adjusted by endogenous control (RT-qPCR data).
3. Discussion
We have longitudinally determined whether surgical weight loss alters the transcriptome of leucocytes in obese women. In this report, we show that genes involved in obesity and insulin pathways are upregulated after RYGB. Our results are in keeping with the findings from a pilot study showing that the expression levels of 200 transcripts linked to obesity and type 2 diabetes change significantly after bariatric surgery [16]. Genetic and environmental factors alter signaling and enzymatic networks that control glucose and lipid metabolism, resulting in obesity and diabetes development and maintenance [17]. Here, we show that PIK3R1 expression levels have increased significantly after RYGB.
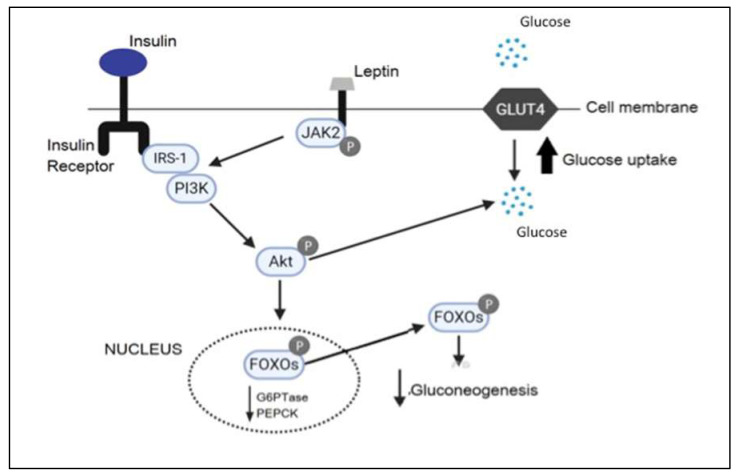
PIK3 signaling is one of the main pathways by which leptin and insulin alter neuronal excitability in the hypothalamus [18]. The deletion of PIK3 in POMC neurons suppresses food intake [19]. Moreover, PIK3 signaling partially modulates the action of leptin and insulin on food intake [20]. On the other hand, peripheral inflammation due to obesity is a key component for developing type 2 diabetes in these patients. The excess of adipose tissue increases secretion of inflammatory cytokines such as TNF-alpha and IL-6 that inhibit phosphorylation of insulin receptors (IRS-1), promoting insulin resistance [21]. Therefore, JAK2 autophosphorylation leads to the recruitment and phosphorylation of IRS-1, which then recruits PIK3 and activates downstream signals [22]. For this reason, we hypothesized that reduced adipose tissue after RYGB improves insulin resistance and activates PIK3. This might decrease weight and increase hyperglycemia through activation of FOXO1 and GLUT4 (Figure 4).
Figure 4.
RYGB improves insulin and leptin resistance with consequent IRS-1 phosphorylation to initiate a signaling cascade that involves PIK3. PIK3 activation leads to Akt downstream signaling. The phosphorylation of IRS-1 triggered by PIK3 allows glucose to enter the cell improving blood glucose levels.
We selected the PIK3R1 gene for an in-depth analysis since it is critical for obesity, insulin resistance, and may also be affected by DNA methylation [23]. Some authors suggest that activation of PIK3 can affect the response of cells to energy balance modulation; however, this mechanism is still not fully understood [24]. The downregulation of several genes has already been reported to initiate an inflammatory response in the adipose tissue, decreasing insulin sensitivity [12] (p. 2). We hypothesized that DNA methylation would be a critical factor in explaining the increased gene expression in the postoperative period.
After bariatric surgery, several DNA methylation sites were differentially methylated, and the majority were hypomethylated. M-values close to 0 indicate that the site is half methylated, positive M-values indicate that the site is hypermethylated, and negative values indicate the opposite [25]. A strong negative correlation was observed between cg20474370 methylation and PIK3R1 expression data in the preoperative period. Some other weak and medium (<0.6) correlations were found, but those correlations changed after surgery. This result suggests that epigenetic triggers are different in obese patients after bariatric surgery.
The presence of significant metabolic pathway changes in the postoperative period may per se be an epigenetic stimulus to change the methylome. Zuo et al. (2012) suggested that the activation of PIK3 might influence downstream epigenetic control in cultured cancer cells [26]. This statement suggests that PIK3 may both be affected and promote epigenetic alterations. Using a multivariate linear regression model, we found that the methylation levels of cg20474370 could explain around 70% of PIK3R1 gene expression when corrected using endogenous controls in the preoperative period. Nevertheless, in the postoperative period, correlations were weaker, suggesting that epigenetic mechanisms that control PIK3R1 gene expression are altered after surgery. The punctual effects of DNA methylation are still unclear and should be further examined.
We found a weak correlation between PIK3R1 gene expression and methylation. However, one cannot disregard that DNA methylation has effects on PIK3R1 expression since we observed differentially methylated probes located at the promoter and 5′ UTR regions. These regions control gene expression and are part of the traditional DNA methylation paradox [27]. Indeed, studies have verified that PIK3 expression is influenced by DNA methylation [28]. It is known that methylation changes in CpGs present in the gene body can promote biological effects [27] (p. 6). Thus, experiments with higher resolution of CpG sites may increase the accuracy of our findings. Similarly, next-generation sequencing experiments may provide a better overview of the transcriptomic changes after RYGB.
4. Materials and Methods
We investigated 13 women with severe obesity (Body Mass Index, BMI > 40 kg/m2) before and six months after RYGB. Patients who were not submitted to the standard surgical technique, patients who lost follow-up with the multidisciplinary team, pregnant women, and patients with thyroid disease, cancer, and psychiatric disorders were excluded. Anthropometric measurements (weight, height, and BMI) and blood samples were obtained before and after surgery. All subjects signed an informed consent term before participating in the study. The study was conducted following the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of the Clinical Hospital of Ribeirão Preto Medical School (Certificate of Presentation for Ethical Consideration—CAAE: 18973913.0.0000.5440, approval date: 14 June 2016).
4.1. RNA Isolation
Total RNA was extracted and purified from blood cells using the phenol-chloroform extraction method. The quality and concentration of RNA were determined with the Agilent 2100 Bioanalyzer and the Agilent RNA 6000 Nano Kit (Agilent, Amstelveen, Netherlands). To evaluate gene expression, samples with RNA integrity number (RIN) higher than 7.0 were used in the analysis.
4.2. Gene Expression Analysis
RNA amplification and labeling were performed with an Epicenter Target Amp Nano-g Biotin a-RNA kit (Labeling kit for Illumina®) that produced amplified biotinylated cRNAs with an input of 500 ng of total RNA. The hybridization protocol used 750 ng cRNA on HumanHT-12_v4 slides in specific buffer at 58 °C for 16 h, which was followed by washings and streptavidin-Cy3 staining. The slides were scanned with the Illumina Bead Array Reader (iSCAN®) on green channel only. The global gene expression analysis of these patients was previously reported by our group [14].
The first-line check and background correction were performed with the GenomeStudio Software (Illumina®). Samples with less than 6000 significantly detected probes (detection p-value < 0.01) were excluded from further analyses. Differentially expressed genes obtained between pre- and post-RYGB were identified after a cutoff for fold change >2 in gene expression and of a t-test p-value of less than 0.05. These p-values were corrected for multiple comparisons by using a false discovery rate (FDR) of 0.05 [11] (p. 2). The microarray data were deposited in the Gene Expression Omnibus database under access number GSE83223 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE83223).
4.3. Validation Analysis
Microarray gene expression was confirmed through RT-qPCR using TaqMan®Gene Expression Assays (Thermo Fisher, Foster City, CA) for three genes (PIK3R1, GAPDH and ACTINB). First, total RNA was reverse transcribed into cDNA using the High Capacity cDNA Reverse Transcription Kit (ThermoFisher). The TaqMan technology suitably quantifies relative gene expression [13] (p. 2). GAPDH and ACTINB were used as reference genes since their expression does not vary, as corroborated by the data obtained from the microarray analysis. Triplicates and positive and negative controls were included in all reactions. Table 4 presents the details of expression assays used. The final reaction was 20 uL, and each reaction was used as an input of 50 ng of total RNA.
Table 4.
RT-qPCR TaqMan®Gene Expression Assays.
| Gene | ID (Gene Expression Assay) | Fragment (pb) |
|---|---|---|
| PIK3R1 | Hs00933163 | 82 |
| GAPDH | Hs99999905 | 122 |
| ACTINB | Hs99999903 | 171 |
4.4. DNA Extraction
The blood for DNA methylation analysis was collected after 12 hours of fasting. DNA was extracted from peripheral blood leukocytes and used to hybridize 450k BeadChip according to the manufacturer’s instructions.
4.5. DNA Methylation Analysis
The methylation data were analyzed using the Infinium Human Methylation 450k BeadChip array following Illumina Methylation HD BeadChip protocol, as previously described by our group in the same set of patients [29]. The Beadchips were scanned with the Illumina IScanSQ system, and the image intensities were extracted with the Genome Studio 2.0. The methylation level was expressed as a beta (β) value—calculated as the intensity of the methylated channel divided by the total intensity β values range from 0 (unmethylated) to 1 (fully methylated). β values can be broadly interpreted as the percentage of CpG methylation.
Quality control and normalization of β values (BMIQ method) were performed using the Champ package for R. We filtered out probes with p-values above 0.01 and beadcount <3 in at least 5%, confounding starts (NoCG, SNPS, Multhit, and XY). For statistical analyses, β values were transformed in M values using the equation M value = log2 (β/1 − β), as described elsewhere [30]. Comparison of β values and M values methods for quantifying methylation levels by microarray analysis were performed. Further, we mapped the CpGs of interest located in genomic regions of the three targets validated by RT-qPCR using the integrative multi-omics database iMETHYL [12] (p. 2).
4.6. Statistical Analysis
Statistical analyses were performed using Statistical Package for the Social Sciences- SPSS v.20. The Shapiro-Wilk test was used to verify if clinical data were normally distributed, and values were compared using parametric or equivalent non-parametric tests. p-values <0.05 were considered as statistically significant. For RT-qPCR data, 2−ΔΔCt was calculated based on Livak recommendations, geometric average for GAPDH, and ACTINB were used as endogenous control [14]. Eutrophic women were used as reference groups for the analyses. To investigate the relationship between gene expression and DNA methylation data, we used M values. Individual linear regressions were conducted to check associations before building multivariable linear regression models [31]. All CpG sites significantly associated with gene expression data (p-value < 0.05) were included in the final model. Glycemia was tested but did not change r2 considerably; thus, endogenous control genes were used to adjust the final model. The linear regression models were conducted using the lm( ) function in R v.3.5.2 (RStudio, Inc., Boston, MA, USA).
5. Conclusions
The weight loss induced by bariatric surgery can alter the expression of genes involved in insulin metabolism and obesity pathways. Overexpression of genes related to these pathways may be associated with the improvement of glucose metabolism, and DNA methylation impacts it partially.
Acknowledgments
São Paulo Research Foundation and National Council for Scientific and Technological Development.
Abbreviations
| RYGB | Roux-en-Y Gastric Bypass |
| CAAE | Certificate of Presentation for Ethical Consideration |
| BMI | Body Mass Index |
| RIN | RNA Integrity Number |
| FDR | False Discovery Rate |
| GO | Gene Ontology |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/12/4476/s1.
Author Contributions
Conceptualization: M.A.S.P., C.F.N., N.Y.N., J.S.M., D.R.S.S., and C.B.N.; methodology: C.B.N., M.A.S.P., C.F.N., N.Y.N., W.S.J., C.C.-O., and B.A.P.d.O.; software: N.Y.N. and V.A.B.P.; validation: M.A.S.P. and N.Y.N.; formal analysis: M.A.S.P., C.F.N., N.Y.N., V.A.B.P., B.A.P.d.O.; resources: W.S.J., C.B.N., J.S.M., D.R.S.S., and F.B.J.; data curation: M.A.S.P., C.F.N., C.C.-O., N.Y.N., B.A.P.d.O.; writing—original draft preparation: M.A.S.P., N.Y.N., C.F.N., V.A.B.P.; writing—review and editing: M.A.S.P., N.Y.N., C.F.N., C.B.N.; supervision: J.S.M., C.B.N.; project administration: C.B.N.; funding acquisition, F.B.J., C.B.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by São Paulo Research Foundation (FAPESP) (#grants numbers: 2018/24069-3, 2018/08784-4 and 2013/12819-4] and National Council for Scientific and Technological Development (CNPq) [#grant No. 480763/2013-5].
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- 1.Swinburn B., Egger G., Raza F. Dissecting obesogenic environments: The development and application of a framework for identifying and prioritizing environmental interventions for obesity. Prev. Med. 1999;29:563–570. doi: 10.1006/pmed.1999.0585. [DOI] [PubMed] [Google Scholar]
- 2.Corbin L.J., Timpson N.J. Body mass index: Has epidemiology started to break down causal contributions to health and disease? Obesity. 2016;24:1630–1638. doi: 10.1002/oby.21554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Llewellyn C., Wardle J. Behavioral susceptibility to obesity: Gene environment interplay in the development of weight. Physiol. Behav. 2015;152:494–501. doi: 10.1016/j.physbeh.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Rao K.R., Lal N., Giridharan N.V. Genetic & epigenetic approach to human obesity. Indian J. Med. Res. 2014;140:589–603. [PMC free article] [PubMed] [Google Scholar]
- 5.Xue J., Ideraabdullah F.Y. An assessment of molecular pathways of obesity susceptible to nutrient, toxicant and genetically induced epigenetic perturbation. J. Nutr. Biochem. 2016;30:1–13. doi: 10.1016/j.jnutbio.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Lorenzo A., Soldati L., Sarlo F., Calvani M., Di Lorenzo N., Di Renzo L. New obesity classification criteria as a tool for bariatric surgery indication. World J. Gastroenterol. 2016;14:681–703. doi: 10.3748/wjg.v22.i2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerron D.A., Portenier D.D. Patient Selection and Surgical Management of High-Risk Patients with Morbid Obesity. Surg. Clin. N. Am. 2016;96:743–762. doi: 10.1016/j.suc.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 8.Still C.D., Wood G.C., Chu X., Erdman R., Manney C.H., Benotti P.N., Petrick A.T., Strodel W.E., Mirshahi U.L., Mirshahi T., et al. High allelic burden of four obesity SNPs is associated with poorer weight loss outcomes following gastric bypass surgery. Obesity. 2011;19:1676–1683. doi: 10.1038/oby.2011.3. [DOI] [PubMed] [Google Scholar]
- 9.González-Plaza J.J., Gutiérrez-Repiso C., García-Serrano S., Rodriguez-Pacheco F., Garrido-Sánchez L., Santiago-Fernández C., García-Arnés J., Moreno-Ruiz F.J., Rodríguez-Cañete A., García-Fuentes E. Effect of Roux-en-Y gastric bypass-induced weight loss on the transcriptomic profiling of subcutaneous adipose tissue. Surg. Obes. Relat. Dis. 2016;12:257–263. doi: 10.1016/j.soard.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Nicoletti C.F., Nonino C.B., de Oliveira B.A., de Souza Pinhel M.A., Mansego M.L., Milagro F.I., Zulet M.A., Martinez J.A. DNA Methylation and Hydroxymethylation Levels in Relation to Two Weight Loss Strategies: Energy-Restricted Diet or Bariatric Surgery. Obes. Surg. 2016;26:603–611. doi: 10.1007/s11695-015-1802-8. [DOI] [PubMed] [Google Scholar]
- 11.Hochberg Y., Benjamini Y. More powerful procedures for multiple significance testing. Stat. Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 12.Curdy C.E.M., Schenk S., Holliday M.J., Philp A., Julie AHouck Patsouris D., Majka S.M. Attenuated Pik3r1 Expression Prevents Insulin Resistance and Adipose Tissue Macrophage Accumulation in Diet-Induced Obese Mice. Diabetes. 2012;61:2495–2505. doi: 10.2337/db11-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubaux R., Santos DD B., Enfield KS S., Rowbotham D., Lam S., Lam W.L., Martinez V.D. Molecular features in arsenic-induced lung tumors. Rev. Mol. Cancer. 2013;12:3–11. doi: 10.1186/1476-4598-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Souza Pinhel M.A., Noronha N.Y., Nicoletti C.F., de Oliveira B.A., Cortes-Oliveira C., Pinhanelli V.C., Junior W.S., Machry A.J., da Silva Junior W.A., Souza D.R., et al. Changes in Global Transcriptional Profiling of Women Following Obesity Surgery Bypass. Obes. Surg. 2018;28:176–186. doi: 10.1007/s11695-017-2828-x. [DOI] [PubMed] [Google Scholar]
- 15.Komaki S., Shiwa Y., Furukawa R., Hachiya T., Ohmomo H., Otomo R., Satoh M., Hitomi J., Sobue K., Sasaki M., et al. iMETHYL: An integrative database of human DNA methylation, gene expression, and genomic variation. Off. J. Jpn. Soc. Hum. Genet. 2018;5:1–4. doi: 10.1038/hgv.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berisha S.Z., Serre D., Schauer P., Kashyap S.R., Smith J.D. Changes in whole blood gene expression in obese subjects with type 2 diabetes following bariatric surgery: A pilot study. PLoS ONE. 2011;6:e16729. doi: 10.1371/journal.pone.0016729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doria A., Patti M.E., Kahn C.R. The emerging genetic architecture of type 2 diabetes. Cell Metab. 2008;8:186–200. doi: 10.1016/j.cmet.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon O., Kim K.W., Kim M.S. Leptin signalling pathways in hypothalamic neurons. Cell Mol. Life Sci. 2016;73:1457–1477. doi: 10.1007/s00018-016-2133-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill J.W., Xu Y., Preitner F., Fukuda M., Cho Y.R., Luo J., Balthasar N., Coppari R., Cantley L.C., Kahn B.B., et al. Phosphatidyl inositol 3-kinase signaling in hypothalamic proopiomelanocortin neurons contributes to the regulation of glucose homeostasis. Endocrinology. 2009;150:4874–4882. doi: 10.1210/en.2009-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams K.W., Margatho L.O., Lee C.E., Choi M., Lee S., Scott M.M., Elias C.F., Elmquist J.K. Segregation of acute leptin and insulin effects in distinct populations of arcuate proopiomelanocortin neurons. J. Neurosci. 2010;30:2472–2479. doi: 10.1523/JNEUROSCI.3118-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wellen K.E., Hotamisligil G.S. Inflammation, stress, and diabetes. J. Clin. Investig. 2005;115:1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donato J., Jr., Frazão R., Elias C.F. The PI3K signaling pathway mediates the biological effects of leptin. Arq. Bras. Endocrinol. Metabol. 2010;54:591–602. doi: 10.1590/S0004-27302010000700002. [DOI] [PubMed] [Google Scholar]
- 23.Vidal E., Sayols S., Moran S., Guillaumet A.A., Schroeder S.A., Royo R., Orozco M. A DNA methylation map of human cancer at single base-pair resolution. Oncogenomics. 2017;36:5648–5657. doi: 10.1038/onc.2017.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikki A., FordKaylyn L., Devlin L.M. Deconvoluting the Obesity and Breast Cancer Link: Secretome, Soil and Seed Interactions. J. Mammary Gland Biol. Neoplasia. 2013;18:267–275. doi: 10.1007/s10911-013-9301-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du P., Zhang X., Huang C.C., Jafari N., Kibbe W.A., Hou L., Lin S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuo T. Epigenetic Silencing Mediated Through Activated PI3K/AKT Signaling in Breast Cancer. Cancer Res. 2012;5:1752–1762. doi: 10.1158/0008-5472.CAN-10-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones P.A. The DNA methylation paradox. Trends Genet. 1999;15:35–49. doi: 10.1016/S0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- 28.Guénard F. Methylation and Expression of Immune and Inflammatory Genes in the Offspring of Bariatric Bypass Surgery Patients. J. Obes. 2013;2013:2–9. doi: 10.1155/2013/492170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicoletti C.F., Pinhel M.A., Diaz-Lagares A., Casanueva F.F., Jácome A., Pinhanelli V.C., de Oliveira B.A., Crujeiras A.B., Nonino C.B. DNA methylation screening after roux-en Y gastric bypass reveals the epigenetic signature stems from genes related to the surgery per se. BMC Med. Genom. 2019;12:72. doi: 10.1186/s12920-019-0522-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian Y. ChAMP: Updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics. 2017;15:3982–3984. doi: 10.1093/bioinformatics/btx513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davé V., Yousefi P., Huen K., Volberg V., Holland N. Relationship between expression and methylation of obesity-related genes in children. Mutagenesis. 2015;30:411–420. doi: 10.1093/mutage/geu089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.