Abstract
Background: Telomerase reverse transcriptase promoter (TERTp) mutations are related to a worse prognosis in various malignancies, including papillary thyroid carcinoma (PTC). Since mechanisms responsible for the poorer outcome of TERTp(+) patients are still unknown, searching for molecular consequences of TERTp mutations in PTC was the aim of our study. Methods: The studied cohort consisted of 54 PTCs, among them 24 cases with distant metastases. BRAF V600E, RAS, and TERTp mutational status was evaluated in all cases. Differences in gene expression profile between TERTp(+) and TERTp(−) PTCs were examined using microarrays. The evaluation of signaling pathways and gene ontology was based on the Gene Set Enrichment Analysis. Results: Fifty-nine percent (32/54) of analyzed PTCs were positive for at least one mutation: 27 were BRAF(+), among them eight were TERTp(+), and 1 NRAS(+), whereas five other samples harbored RAS mutations. Expression of four genes significantly differed in BRAF(+)TERTp(+) and BRAF(+)TERTp(−) PTCs. Deregulation of pathways involved in key cell processes was observed. Conclusions: TERTp mutations are related to higher PTC aggressiveness. CRABP2 gene was validated as associated with TERTp mutations. However, its potential use in diagnostics or risk stratification in PTC patients needs further studies.
Keywords: TERT, papillary thyroid carcinoma, gene expression profile
1. Introduction
Papillary thyroid carcinoma (PTC) is the most common type of thyroid cancer (TC), generally characterized by a good prognosis. Only a small percentage of PTCs demonstrates a higher aggressiveness and poor outcomes [1]. A proper stratification is crucial to select a group of patients at high risk of unfavorable PTC course and simultaneously to avoid overtreatment in low-risk cases. Moreover, the intra-tumor and interpatient heterogeneity observed in PTC, seem to have important implications for therapeutic approaches [2,3]. Despite significant progress in our understanding of the molecular background of PTCs, reliable prognostic and predictive molecular markers are still lacking. The Cancer Genome Atlas (TCGA) study, published in 2014, substantially reduced the ‘dark matter’ of PTC genome up to 3.5%, as genetic alterations were defined in about 96.5% PTC cases [4]. The most common ones, as reported previously by other authors, were related to genes of the RAS/RAF/MAPK pathway, with the BRAF V600E mutation being present in 61.7% of PTCs, RAS mutations in 12.9%, and RET/PTC rearrangements in 6.8% of cases. The mutation density in the analyzed cohort was low (0.41 nonsynonymous mutations per Mb, on average). It positively correlated with age, recurrence risk, and the distant Metastasis, patient Age, Completeness of resection, local Invasion, and tumor Size (MACIS) score. These results may reflect an indolent nature of the majority PTC cases on the one hand, and on the other hand, represent the molecular basis of aggressive PTC. Until recently, the BRAF V600E mutation was considered a marker of poor prognosis in PTC. Many studies were pointing to a possible relationship between its presence and the worse outcome [5,6]. Nevertheless, its role as a prognostic or predictive factor is still controversial [7]. The frequency of the BRAF V600E mutation ranges from 23 to 83% in different PTC cohorts. Its presence was demonstrated in both classic PTCs as well as in its aggressive subtypes [8]. According to the current ATA guidelines for diagnostics and treatment of differentiated thyroid carcinoma (DTC), the BRAF V600E mutation may be considered in risk stratification, however, in the context of clinical data only [9].
In 2013 two somatic mutations of telomerase reverse transcriptase (TERT) gene promoter were discovered in different solid cancers, including TCs [10,11,12]. Two alterations, c.-124C > T (C228T) and c.-146C > T (C250T) were reported in PTC with the prevalence of 11.3% [13]. Their frequency was higher in more aggressive histopathological PTC variants [10]. TERT encodes the catalytic subunit of telomerase, responsible for telomeres maintenance at the end of chromosomes. Telomerase, typically not expressed in somatic cells, is reactivated in cancer cells. It protects telomeres from further shortening and is also involved in cancer cell proliferation, resistance to apoptosis and antigrowth signals, increased angiogenesis, and metastatic potential [14]. It has been demonstrated that C228T and C250T TERT promoter (TERTp) mutations were much more prevalent in PTCs harboring BRAF V600E mutation. This co-occurrence was related to poor prognostic factors, including tumor size, older age at diagnosis, distant metastases, or shorter progression-free survival [13,15,16]. Also, the co-existence of RAS and TERTp mutations was associated with a worse prognosis. Shen et al. proposed a six-genotype genetic prognostic model for PTC, dividing patients into the following risk groups regarding the detected mutation: BRAF V600E/RAS mutation and TERTp mutation >>>> BRAF V600E = TERTp mutation alone > RAS mutation alone = wild-type genes [17].
Since the discovery of TERTp mutations in thyroid cancers, most studies have focused on their clinical implications. However, the data concerning their molecular consequences and potential differences/similarities to BRAF V600E-positive TCs are scarce. To our best knowledge, there was only one published study devoted to the transcriptomic analysis of TERTp-mutated PTCs [18]. Therefore, in the present study, we focused on the gene expression profile of TERTp-mutated PTC in BRAF(+) cases. We aimed to analyze the molecular consequences of TERTp alterations that could explain their impact on PTC outcomes. As our PTC cohort involved mainly patients showing unfavorable clinical course, we tried to find molecular mechanisms, responsible for PTC aggressiveness.
2. Results
2.1. Mutational Analysis for BRAFV600, TERTp, and RAS Somatic Mutations and Their Association with Histopathological and Clinical PTC Features
Thirty-two out of 54 PTC cases (59%) were positive for at least one of the analyzed genetic alterations. Initially, using the Sanger sequencing method, the BRAF V600E mutation was detected in 24 PTC samples. However, using the QPCR technique, enabled us to identify this genetic alteration in the other 3 PTC cases. Finally, 27/54 (50%) PTCs were BRAF(+). In nine out of 27 (33%) PTCs harboring BRAF V600E mutation, additional somatic alterations were detected: eight PTCs were positive for TERTp mutations (six cases with C228T and two with C250T mutation), and one PTC was positive for NRAS mutation. In a further five cases, mutations in RAS genes were found (two PTCs with HRAS mutations and three PTCs with NRAS mutations). However, we did not observe the co-existence of RAS and TERTp mutation. In the remaining 22 PTCs, no mutation was detected (NM-PTC). Considering distinct clinical and histopathological PTC features, the presence of BRAF, RAS, and TERTp mutations did not show any association with sex, histopathological PTC variant, multifocality, invasion of the tumor capsule, angioinvasion, or the presence of lymph node metastases (Table 1). In contrast, TERTp mutations (comparison between BRAF(+)TERTp(+) and BRAF(+)TERTP(−) PTCs) were significantly associated with tumor diameter, invasion of surrounding tissues, pN1b feature, the locally persistent disease after the 1st surgery, and distant metastases.
Table 1.
Characteristics of analyzed PTC cohort with BRAF, RAS and TERTp mutational status, and associations between clinico-histopathological and molecular features.
| Clinical/Histopathological Features | Total | NM-PTC | RAS(+) | BRAF(+)RAS(+) | BRAF(+)TERTp(−) | BRAF(+)TERTp(+) | Comparison Between the 5 Mutational Subgroups p-Value | BRAF(+)TERTp(+) vs. BRAF(+)TERTp(−) p-Value |
|---|---|---|---|---|---|---|---|---|
| No. 54 | No. 22 | No. 5 | No. 1 | No. 18 | No. 8 | |||
| Sex | 0.94 | 0.66 | ||||||
| Male | 21 (38.9%) | 9 (40.9%) | 2 (40.0%) | 0 (0.0%) | 6 (33.3%) | 4 (50.0%) | ||
| Female | 33 (61.1%) | 13 (59.1%) | 3 (60.0%) | 1 (100.0%) | 12 (66.7%) | 4 (50.0%) | ||
| Age at diagnosis | 0.005 | 0.14 | ||||||
| 48.5 (31.2–68.2) | 33.0 (21.0–46.8) | 49.0 (49.0–57.0) | 17.0 (17.0–17.0) | 55.0 (42.5–72.2) | 72.5 (56.5–77.2) | |||
| Classification according to the age at diagnosis | 0.0005 | 0.28 | ||||||
| <45 years | 19 (35.2%) | 14 (63.6%) | 0 (0.0%) | 1 (100.0%) | 4 (22.2%) | 0 (0.0%) | ||
| >45 years | 35 (64.8%) | 8 (36.4%) | 5 (100.0%) | 0 (0.0%) | 14 (77.8%) | 8 (100.0%) | ||
| Histopathological PTC classification | 0.15 | 1.00 | ||||||
| Classical variant | 46 (85.2%) | 17 (77.3%) | 3 (60.0%) | 1 (100.0%) | 17 (94.4%) | 8 (100.0%) | ||
| Follicular variant | 8 (14.8%) | 5 (22.7%) | 2 (40.0%) | 0 (0.0%) | 1 (5.6%) | 0 (0.0%) | ||
| Tumor diameter [mm] | 0.014 | 0.0008 | ||||||
| 24.0 (15.0–30.0) | 21.0 (13.5–30.0) | 30.0 (12.0–35.0) | 31.0 (31.0–31.0) | 18.0 (13.5–25.0) | 40.0 (29.5–61.2) | |||
| Multifocality | 0.59 | 0.40 | ||||||
| No | 26 (48.1%) | 9 (40.9%) | 2 (40.0%) | 1 (100.0%) | 11 (61.1%) | 3 (37.5%) | ||
| Yes | 28 (51.9%) | 13 (59.1%) | 3 (60.0%) | 0 (0.0%) | 7 (38.9%) | 5 (62.5%) | ||
| Invasion of the tumor capsule | 0.17 | 0.22 | ||||||
| No | 25 (46.3%) | 8 (36.4%) | 4 (80.0%) | 1 (100.0%) | 10 (55.6%) | 2 (25.0%) | ||
| Yes | 29 (53.7%) | 14 (63.6%) | 1 (20.0%) | 0 (0.0%) | 8 (44.4%) | 6 (75.0%) | ||
| Invasion of the surrounding tissues | 0.041 | 0.014 | ||||||
| No | 41 (75.9%) | 16 (72.7%) | 5 (100.0%) | 1 (100.0%) | 16 (88.9%) | 3 (37.5%) | ||
| Yes | 13 (24.1%) | 6 (27.3%) | 0 (0.0%) | 0 (0.0%) | 2 (11.1%) | 5 (62.5%) | ||
| Angioinvasion | 0.27 | 0.31 | ||||||
| No | 48 (88.9%) | 18 (81.8%) | 4 (80.0%) | 1 (100.0%) | 18 (100.0%) | 7 (87.5%) | ||
| Yes | 6 (11.1%) | 4 (18.2%) | 1 (20.0%) | 0 (0.0%) | 0 (0.0%) | 1 (12.5%) | ||
| pN1a | 0.25 | 0.20 | ||||||
| No | 22 (40.7%) | 7 (31.8%) | 2 (40.0%) | 0 (0.0%) | 11 (61.1%) | 2 (25.0%) | ||
| Yes | 32 (59.3%) | 15 (68.2%) | 3 (60.0%) | 1 (100.0%) | 7 (38.9%) | 6 (75.0%) | ||
| pN1b | 0.095 | 0.038 | ||||||
| No | 27 (50.0%) | 9 (40.9%) | 3 (60.0%) | 0 (0.0%) | 13 (72.2%) | 2 (25.0%) | ||
| Yes | 27 (50.0%) | 13 (59.1%) | 2 (40.0%) | 1 (100.0%) | 5 (27.8%) | 6 (75.0%) | ||
| Locally persistent disease after the 1st surgery | 0.012 | 0.005 | ||||||
| No | 47 (87.0%) | 19 (86.4%) | 5 (100.0%) | 1 (100.0%) | 18 (100.0%) | 4 (50.0%) | ||
| Yes | 7 (13.0%) | 3 (13.6%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 4 (50.0%) | ||
| Local recurrence | 0.01 | 0.086 | ||||||
| No | 49 (90.7%) | 21 (95.5%) | 4 (80.0%) | 0 (0.0%) | 18 (100.0%) | 6 (75.0%) | ||
| Yes | 5 (9.3%) | 1 (4.5%) | 1 (20.0%) | 1 (100.0%) | 0 (0.0%) | 2 (25.0%) | ||
| Distant metastases | 0.015 | 0.008 | ||||||
| Yes | 24 (44.4%) | 12 (54.5%) | 3 (60.0%) | 0 (0.0%) | 3 (16.7%) | 6 (75.0%) | ||
| No | 30 (55.6%) | 10 (45.5%) | 2 (40.0%) | 1 (100.0%) | 15 (83.3%) | 2 (25.0%) | ||
| Follow up | 95.9 (78.6–112.7) | 95.9 (75.5–110.2) | 104.6 (89.6–105.4) | 84.4 (84.4–84.4) | 101.7 (80.8–119.8) | 94.4 (87.1–100.7) | 0.86 | 0.50 |
| [months] | ||||||||
2.2. Impact of Studied Mutations on Gene Expression Profile of PTCs
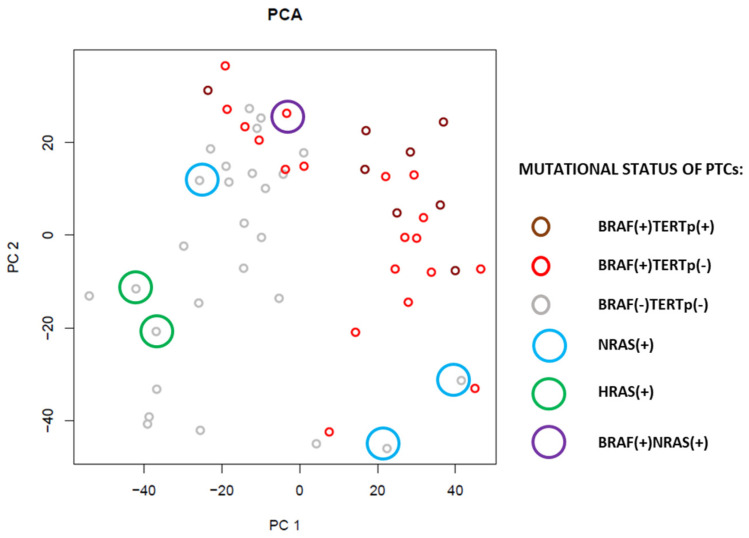
Principal Component Analysis (PCA) was performed in order to explore major sources of gene expression variation. In the first and second components, the main source of variation was the presence of the BRAF V600E mutation (Figure 1). The strongest similarity was observed between BRAF(+)TERTp(+) and BRAF(+)TERTp(−) PTCs. PTCs harboring HRAS and NRAS mutations, in turn, were grouped with NM-PTCs. However, they were not grouped as close as BRAF(+) cases.
Figure 1.
PCA analysis of PTCs with reference to BRAF, TERTp and RAS mutational status.
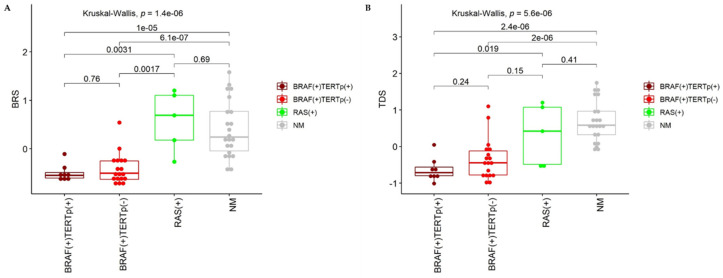
2.3. Mutational Status and TDS, and BRS Coefficients
We calculated the Thyroid Differentiation Score (TDS) and the BRAF-like RAS-like Score (BRS). BRS is a gene expression score that indicates whether a sample is more similar to samples harboring the BRAF V600E mutation (BRAF-like samples, low BRS score) or RAS mutation (RAS-like samples; high BRS score) (Figure 2). As expected, in our study, samples harboring the BRAF V600E mutation were more BRAF-like, whereas samples harboring RAS mutation were more RAS-like. The difference in BRS score between BRAF(+) and RAS(+) samples was statistically significant. NM-PTC samples were more RAS-like, and their BRS was significantly higher than in BRAF(+) samples (Figure 2A). However, the separation between NM-PTCs and BRAF(+) samples was not perfect. It may suggest that some of the samples without BRAF, TERTp, and RAS mutation, harbored other genetic alterations associated with gene expression profile similar to BRAF(+) PTCs.
Figure 2.
Analysis of BRS (A) and TDS (B). High BRS depicts RAS-like, whereas low BRS depicts BRAF-like samples. High TDS depicts high expression of thyroid specific genes and high thyroid differentiation. TDS: The Thyroid Differentiation Score, BRS: the BRAF-like RAS-like Score. p-values shown on plots were calculated with Kruskal-Wallis and two-tailed Mann-Whitney tests.
TDS analysis showed significant downregulation of thyroid-specific genes in PTCs harboring BRAF and TERTp mutation comparing to cases without these alterations (RAS(+) PTCs and NM-PTCs; Figure 2B). However, there was no statistically significant difference in TDS between BRAF(+)TERTp(+) and BRAF(+)TERTp(−) PTCs. Also, no statistically significant difference was observed between RAS(+) PTC cases and BRAF(+)TERTp(−) PTCs or NM-PTCs.
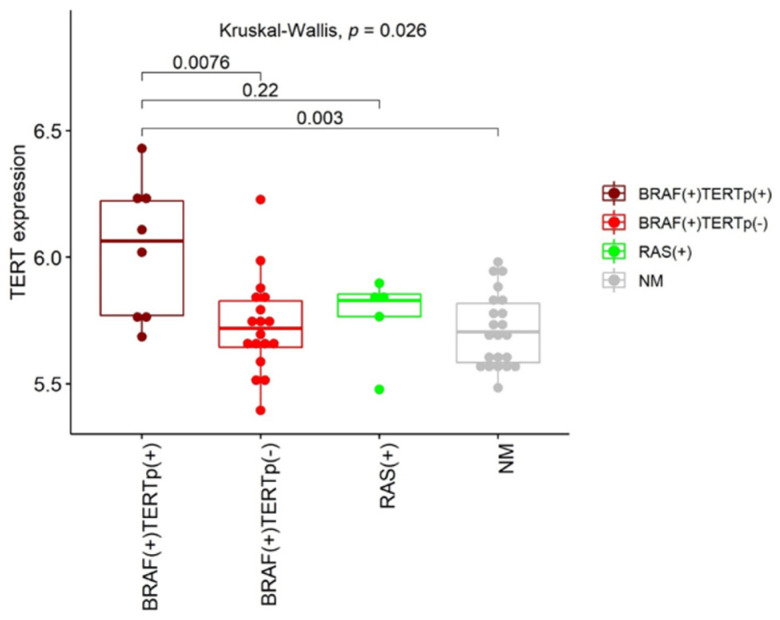
2.4. TERTp Mutations and Gene Expression Profile of PTCs
Our study mainly focused on the impact of TERTp mutations on gene expression profile. However, the presence of other somatic mutations was also considered. First, we analyzed TERT gene expression depending on the presence/absence of mutations within its promoter region. We observed a tendency of TERT up-regulation in PTCs harboring C228T or C250T mutation with significant differences limited to the following comparisons: BRAF(+)TERTp(+) vs. BRAF(+)TERTp(−) PTCs and BRAF(+)TERTp(+) vs. NM-PTCs (p = 0.0076 and p = 0.003, respectively; Figure 3). However, changes in the TERT gene expression between TERTp(+) and RAS(+) PTCs did not reach statistical significance (p = 0.22). Next, several comparative analyses were performed to find differences in gene expression that could result from the presence of a particular genetic alteration (BRAF, TERTp, or RAS mutations; Table 2). The highest number of significantly differentiating genes was obtained when PTCs harboring any of the studied mutations were compared to NM-PTCs (2543 genes; FDR < 0.05). Comparison of BRAF(+) PTCs with RAS(+) PTCs resulted in 709 differentially expressed genes (FDR < 0.05), whereas only four genes showed significant differences in their expression between BRAF(+)TERTp(+) and BRAF(+)TERTp(-) cases (FDR < 0.05) (Table 3). A small number of significantly changed genes was also found when RAS(+) PTCs and NM-PTCs were compared (3 genes; FDR < 0.05). The lists of all genes are given in Table S1.
Figure 3.
TERT gene expression in PTC sets with different mutations; RAS(+) cohort was BRAF(−) and TERTp(−). p-values shown on plot were calculated with Kruskal-Wallis and two-tailed Mann-Whitney tests.
Table 2.
Number of genes significantly deregulated depending on the molecular differentiating factor; M- the presence of BRAF or TERTp or RAS somatic mutation; NM- no somatic mutations within studied genes.
| Type of Comparison | Number of Differentiating Genes | |
|---|---|---|
| p < 0.001 | FDR < 0.05 | |
| M (32 PTCs) vs. NM (22 PTCs) | 1404 | 2543 |
| BRAF(+)TERTp(+) (8 PTCs) vs. BRAF(+)TERTp(−) (18 PTCs) | 126 | 4 |
| BRAF(+)TERTp(−) (18 PTCs) vs. RAS(+) (5 PTCs) | 523 | 709 |
| RAS(+) (5 PTCs) vs. NM (22 PTCs) | 97 | 3 |
Table 3.
Genes differentiating BRAF(+)TERTp(+) PTCs and BRAF(+)TERTp(−) PTCs.
| Gene Symbol | Entrez ID | Gene Name | BRAF(+)TERTp(+) Mean | BRAF(+)TERTp(−) Mean | Fold Change | p-Value | Adjusted p-Value |
|---|---|---|---|---|---|---|---|
| CRABP2 | 1382 | cellular retinoic acid binding protein 2 | 7.92 | 6.32 | 3.03 | 5.7 × 10−7 | 0.012 |
| ECM1 | 1893 | extracellular matrix protein 1 | 8.07 | 6.77 | 2.47 | 5.9 × 10−6 | 0.042 |
| KRT17 | 3872 | keratin 17 | 5.07 | 4.29 | 1.72 | 7.9 × 10−6 | 0.042 |
| MTMR3 | 8897 | myotubularin related protein 3 | 6.64 | 7.04 | 0.76 | 8.1 × 10−6 | 0.042 |
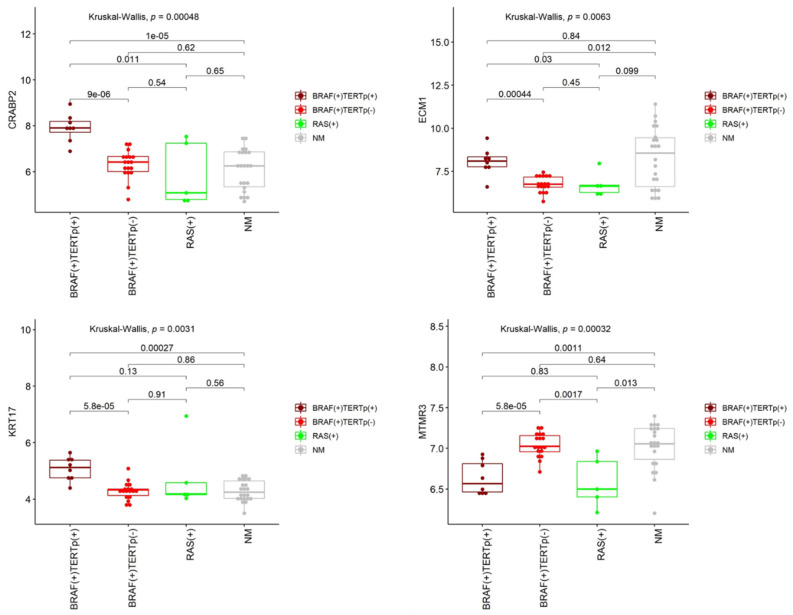
Of the four genes (CRABP2, ECM1, KRT17, and MTMR3) differentially expressed between TERTp(+) and TERTp(−) PTCs, all but one, MTMR3, were up-regulated in PTCs harboring TERTp mutation (Table 3; Figure 4). Besides, the MTMR3 gene showed significantly higher expression in BRAF(+)TERTp(−) PTCs comparing to RAS(+) ones. The ECM1 gene, in turn, was down-regulated in mutated PTCs in comparison to NM-PTCs.
Figure 4.
Expression of four genes significantly differentiating BRAF(+)TERTp(+) and BRAF(+)TERTp(−) PTCs in all analyzed PTC cases; RAS(+) cohort was BRAF(−) and TERTp(−). p-values shown on plots were calculated with Kruskal-Wallis and two-tailed Mann-Whitney tests.
2.5. Validation of TERTp-Dependent Genes
Validation of genes differentially expressed between TERTp(+) and TERTp(−) PTCs was performed on an independent PTC cohort, made available by the TCGA study [4]. In this analysis, two sets were considered: BRAF(+)TERTp(+) and BRAF(+)TERTp(−) PTCs (28 and 207 PTCs, respectively; with the exclusion of all PTCs harboring RAS mutations). Only one out of four genes studied, the CRABP2 gene was confirmed as significantly differentiating BRAF(+) PTCs with and without TERTp mutation (Table 4).
Table 4.
Validation of genes differentiating BRAF(+)TERTp(+) and BRAF(+)TERTp(−) PTCs performed on an independent set of PTC made available by the TCGA study.
| Gene Information | Our Study | TCGA Study | Compliance Between Our Study and TCGA Data | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene Symbol | Entrez ID | BRAF(+)TERTp(+) Mean Expression | BRAF(+)TERTp(−) Mean Expression | p-Value | Fdr-Adjusted p-Value | Fold Change | p-Value | FDR-Adjusted p-Value | Fold Change | |
| CRABP2 | 1382 | 7.92 | 6.32 | 5.7 × 10−7 | 0.012 | 3.03 | 5.8 × 10−8 | 1.0 × 10−5 | 2.21 | YES |
| ECM1 | 1893 | 8.07 | 6.77 | 5.9 × 10−6 | 0.042 | 2.47 | 0.33 | 0.69 | 1.27 | NO |
| KRT17 | 3872 | 5.07 | 4.29 | 7.9 × 10−6 | 0.042 | 1.72 | 0.10 | 0.41 | 0.53 | NO |
| MTMR3 | 8897 | 6.64 | 7.04 | 8.1 × 10−6 | 0.042 | 0.76 | 0.11 | 0.43 | 0.92 | NO |
2.6. Signaling Pathways and Gene Ontology Analysis
To evaluate potential differences in signaling pathways and changes in gene ontology (GO) terms resulting from the presence of TERTp mutations, the Gene Set Enrichment Analysis (GSEA) was used. Comparing BRAF(+)TERTp(+) and BRAF(+)TERTp(-) samples, we obtained 39 significantly enriched KEGG pathways, 644 significantly enriched GO Biological Process (BP) terms, 131 significantly enriched GO Cellular Component (CC) terms, and 209 significantly enriched GO Molecular Function (MF) terms (FDR < 0.05). Table 5 presents ten significantly enriched gene groups with the highest absolute normalized enrichment score (NES) for each analysis (Table 5).
Table 5.
Gene groups (pathways and gene ontology terms) significantly differentiating BRAF(+)TERTp(+) and BRAF(+)TERTp(−) PTCs obtained by the Gene Set Enrichment Analysis (FDR < 0.05). Ten gene groups with the highest NES were selected from each analysis.
| KEGG Pathways | |||||
| ID | Description | Set Size | NES | p-Value | Adjusted p-Value |
| hsa00562 | Inositol phosphate metabolism | 74 | −2.57 | 0.00366 | 0.04234 |
| hsa04070 | Phosphatidylinositol signaling system | 99 | −2.38 | 0.00408 | 0.04234 |
| hsa04120 | Ubiquitin mediated proteolysis | 134 | −2.16 | 0.00500 | 0.04234 |
| hsa04080 | Neuroactive ligand-receptor interaction | 333 | 2.11 | 0.00112 | 0.04234 |
| hsa04950 | Maturity onset diabetes of the young | 26 | 2.03 | 0.00156 | 0.04234 |
| hsa04740 | Olfactory transduction | 411 | 1.99 | 0.00110 | 0.04234 |
| hsa04141 | Protein processing in endoplasmic reticulum | 162 | −1.96 | 0.00510 | 0.04234 |
| hsa05217 | Basal cell carcinoma | 63 | 1.92 | 0.00141 | 0.04234 |
| hsa04666 | Fc gamma R-mediated phagocytosis | 87 | −1.83 | 0.00389 | 0.04234 |
| hsa04744 | Phototransduction | 28 | 1.83 | 0.00470 | 0.04234 |
| Gene Ontology Biological Process | |||||
| ID | Description | Set size | NES | p-Value | Adjusted p-Value |
| GO:0046856 | phosphatidylinositol dephosphorylation | 22 | −2.63 | 0.00029 | 0.01786 |
| GO:0031424 | Keratinization | 208 | 2.57 | 0.00012 | 0.01786 |
| GO:0032508 | DNA duplex unwinding | 73 | −2.56 | 0.00038 | 0.01786 |
| GO:0006261 | DNA-dependent DNA replication | 127 | −2.54 | 0.00047 | 0.01786 |
| GO:0046839 | phospholipid dephosphorylation | 27 | −2.53 | 0.00030 | 0.01786 |
| GO:0045005 | DNA-dependent DNA replication maintenance of fidelity | 32 | −2.49 | 0.00031 | 0.01786 |
| GO:0032878 | regulation of establishment or maintenance of cell polarity | 23 | −2.49 | 0.00029 | 0.01786 |
| GO:0050853 | B cell receptor signaling pathway | 50 | −2.47 | 0.00034 | 0.01786 |
| GO:0006895 | Golgi to endosome transport | 14 | −2.47 | 0.00027 | 0.01786 |
| GO:2000114 | regulation of establishment of cell polarity | 21 | −2.47 | 0.00029 | 0.01786 |
| Gene Ontology Cellular Components | |||||
| ID | Description | Set size | NES | p-Value | Adjusted p-Value |
| GO:0005882 | intermediate filament | 180 | 2.41 | 0.00012 | 0.00681 |
| GO:0045095 | keratin filament | 84 | 2.36 | 0.00013 | 0.00681 |
| GO:1990234 | transferase complex | 586 | −2.25 | 0.00225 | 0.01802 |
| GO:0042555 | MCM complex | 11 | −2.16 | 0.00026 | 0.00894 |
| GO:0010494 | cytoplasmic stress granule | 54 | −2.16 | 0.00035 | 0.00894 |
| GO:0005681 | spliceosomal complex | 155 | −2.15 | 0.00055 | 0.00894 |
| GO:0030990 | intraciliary transport particle | 22 | −2.15 | 0.00029 | 0.00894 |
| GO:0045111 | intermediate filament cytoskeleton | 218 | 2.14 | 0.00012 | 0.00681 |
| GO:0016607 | nuclear speck | 377 | −2.14 | 0.00116 | 0.01224 |
| GO:0000123 | histone acetyltransferase complex | 72 | −2.13 | 0.00038 | 0.00894 |
| Gene Ontology Molecular Function | |||||
| ID | Description | Set size | NES | p-Value | Adjusted p-Value |
| GO:0004386 | helicase activity | 125 | −2.86 | 0.00049 | 0.00730 |
| GO:0052866 | phosphatidylinositol phosphate phosphatase activity | 25 | −2.73 | 0.00029 | 0.00730 |
| GO:0061733 | peptide-lysine-N-acetyltransferase activity | 50 | −2.65 | 0.00035 | 0.00730 |
| GO:0034212 | peptide N-acetyltransferase activity | 55 | −2.62 | 0.00036 | 0.00730 |
| GO:0004402 | histone acetyltransferase activity | 48 | −2.61 | 0.00034 | 0.00730 |
| GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity | 20 | −2.60 | 0.00029 | 0.00730 |
| GO:0008094 | DNA-dependent ATPase activity | 70 | −2.52 | 0.00038 | 0.00730 |
| GO:0003678 | DNA helicase activity | 41 | −2.52 | 0.00033 | 0.00730 |
| GO:0005096 | GTPase activator activity | 194 | −2.50 | 0.00065 | 0.00778 |
| GO:0005085 | guanyl-nucleotide exchange factor activity | 193 | −2.47 | 0.00064 | 0.00778 |
Among significantly enriched (FDR < 0.05) gene groups, six groups were related to telomeres (Table 6).
Table 6.
Significantly enriched gene groups (FDR < 0.05) related to telomeres.
| Gene Ontology Biological Process | |||||
| ID | Description | Set Size | NES | p-Value | Adjusted p-Value |
| GO:0032204 | regulation of telomere maintenance | 75 | −1.81 | 0.00077 | 0.01884 |
| GO:0032205 | negative regulation of telomere maintenance | 32 | −1.79 | 0.00312 | 0.03647 |
| GO:0000723 | telomere maintenance | 147 | −1.72 | 0.00052 | 0.01786 |
| GO:0032200 | telomere organization | 159 | −1.57 | 0.00162 | 0.02471 |
| Gene Ontology Cellular Components | |||||
| ID | Description | Set Size | NES | p-Value | Adjusted p-Value |
| GO:0000781 | chromosome, telomeric region | 149 | −1.67 | 0.00053 | 0.00894 |
| GO:0000784 | nuclear chromosome, telomeric region | 115 | −1.55 | 0.00559 | 0.03593 |
3. Discussion
The presence of the TERTp mutation resulted in a changed expression of four genes only. It is the most important result of our analysis of the PTC gene expression profile. Moreover, only one of these genes was confirmed in an independent set of PTC samples made available by the TCGA [4] consortium. However, one should notice that the TCGA cohort consisted mainly of low-risk PTCs.
One of the hallmarks of malignant neoplasms is an evasion of replicative senescence and out-of-control proliferation, leading to immortalization of cancer cells [19]. The pivotal mechanism underlying this process is the reactivation of telomerase, an enzyme typically not expressed in somatic cells, but activated in about 80–90% of all malignant neoplasms [20]. Telomerase is a complex protein. However, its core is composed only of catalytic component TERT and internal telomerase RNA template (TERC) [21]. TERC is ubiquitously expressed in various human cells. On the contrary, TERT is repressed in somatic cells leading to telomerase silencing [22]. Its induction/telomerase activation not only provides telomeres stabilization but is also related to several oncogenic processes, independent of telomeres lengthening. For instance, it has been shown that TERT directly interacts with β-catenin and, as a consequence, stimulates epithelial-mesenchymal transition (EMT) and stemness of cancer cells, and, by interaction with NF-κB p65, up-regulates the expression of metalloproteinases (MMPs) in cancer cells [23,24]. Moreover, TERT contributes to survival signaling, growth signaling, invasion/metastasis, angiogenesis, DNA methylation, genetic aberrations, and even to radio/chemo-resistance, which make TERT an important factor related to a higher aggressiveness of cancer cells [22]. Also, in thyroid cancer, TERT induction has been linked to a poorer prognosis, higher risk of metastases, recurrence, and even death [13]. There are several mechanisms underlying TERT activation with TERTp somatic mutations being the most widely studied in many cancer types since their discovery in 2013 [11,12]. In TCs, their frequency is considered as intermediate. However, it increases dramatically from microcarcinomas (frequency reported at the level of about 5%) [25] to aggressive poorly differentiated and anaplastic TCs (48.8% and 41.8%, respectively) [20]. It seems to be more common in TCs harboring BRAF V600E mutation [26]. Although a lot of data concerning the clinical significance of TERTp mutations in TCs has been published, there is still much to discover in the field of their molecular consequences. To our best knowledge, this is the first study that analyses the impact of TERTp mutations on PTC transcriptome in BRAF(+) samples.
Our previous study showed that in Polish PTC patients, TERTp mutations are present in 8.5% of cases [16]. However, in the current analysis, in which almost half of the PTC cases presented distant metastases, the frequency of C228T and C250T mutations increased up to 14.8%. Of 30 non-metastatic cases, two PTCs were TERTp(+) (6.7%), whereas six out of 24 PTCs with distant metastases harbored a TERTp mutation (25%). Similar data obtained Gandolfi et al., who showed the presence of these alterations in 10% of non-metastatic PTCs and 33% of PTCs with distant metastases [27]. All PTCs harboring TERTp mutation in our study were BRAF V600E-positive. Moreover, in one PTC with BRAF mutation, NRAS point mutation was reported. However, none of the studied cases displayed the co-existence of TERTp and RAS somatic mutations.
In the current study, we focused mainly on genes and processes that are significantly altered in the presence of TERTp mutations. As indicated in previous studies, TERTp mutations were associated with elevated TERT expression [28] and, as presented by Fredriksson et al., this association was exceptional in its strength and was highest in copy number-stable cancers such as thyroid carcinoma [29]. Our results are in agreement with these data. We observed significant up-regulation of TERT transcript in TERTp(+) PTCs comparing to TERTp(−) ones harboring BRAF V600E mutation and NM-PTCs. Although there was no significant difference in TERT expression between TERTp(+) and RAS(+) PTCs, a tendency of TERTp mutations-dependent up-regulation of TERT mRNA was visible. However, a limited number of RAS(+) PTCs might impair the results and statistics.
The next step was a closer look at the whole gene expression profiles of our PTC set with a particular focus on cases with TERTp mutation. There was no surprise that all PTCs harboring BRAF mutation grouped close to each other in an unsupervised PCA analysis. It was the most potent differentiating factor within the first and second components. RAS(+) PTCs grouped with NM-PTC cases. PCA did not show differences between BRAF(+)TERTp(+) and BRAF(+)TERTp(−) samples. Moreover, according to the BRS, BRAF(+) PTCs, including all TERTp(+) cases, were BRAF-like and most RAS(+) and NM-PTCs were RAS-like. However, one RAS(+) and several NM-PTCs were more similar to BRAF-like PTCs. It might result from the presence of some additional alterations, and, in consequence, their gene expression profile was more similar to BRAF(+) PTCs. Some of the NM-PTCs may harbor RET fusions. In the TCGA study, RET rearrangements were present in 6.8% PTCs (33/484), and nearly all of them were weakly BRAF-like. However, we focused mainly on TERTp mutations and genetic alterations that most commonly co-exist with them. The presence of TERTp mutations, in turn, may impair the expression of thyroid-specific genes by down-regulating them. Although there were no differences in TDS among BRAF(+) PTCs with and without TERTp mutations, this association was significant in comparison of BRAF(+)TERTp(+) cases to RAS(+) and NM-PTCs. These data suggest that TERTp mutations may be crucial in PTC dedifferentiation and aggressiveness. It is supported by the association of TERTp mutations with poor prognostic factors, observed in our PTC cohort. Larger tumor size, invasion of the surrounding tissues, the locally persistent disease after the first surgery, pN1b, and distant metastases were significantly associated with mutated TERTp, which is in concordance with previous studies [13].
Taking into consideration significant diversity of BRAF-like PTCs, noticed in TCGA study [4] and a higher frequency of TERTp mutations in PTCs harboring BRAF V600E alteration [30,31], we focused on possible gene expression differences within BRAF(+) PTCs resulting from the presence of TERTp mutations. The observed difference was subtle. However, we did not expect any differences since all TERTp(+) cases were simultaneously BRAF(+). We are aware that not all BRAF(+) PTCs show poor outcomes, so there must be some additional factors responsible for disease aggressiveness. TERTp mutations certainly participate in this process. We obtained four genes differentiating BRAF(+)TERTp(+) PTCs and BRAF(+)TERTp(−) ones (CRABP2, ECM1, KRT17, and MTMR3). One out of these four genes, the CRABP2 gene, was positively validated on an independent PTC cohort, although with a lower number of high-risk cases. This result confirms that the TERTp(+)-dependent difference exists. The CRABP2 gene encodes the cellular retinoic acid-binding protein 2 that is responsible for retinoic acid (RA) transport to retinoic acid receptors (RARs) within the nucleus, inhibiting cell growth and proliferation [32]. That is why CRABP2 is considered as an antitumor agent. However, there are contradicting data suggesting the necessity of further studies on the CRABP2 role in tumorigenesis. For instance, CRABP2 protein was identified as a subtype-specific biomarker of ovarian cancer, since its expression positively correlated with tumor grade and cancer stage [33]. Similarly, higher CRABP2 expression corresponded to invasive retinoblastomas [34] and poorer prognosis in estrogen receptor-negative breast cancer [35]. Moreover, its overexpression was suggested to be a late event of pancreatic carcinogenesis that could be used as a marker to distinguish pancreatic ductal adenocarcinomas from other benign pancreatic conditions [36]. Kim et al. proposed plasma CRABP2 as a novel diagnostic and prognostic marker in non-small cell lung cancer [37]. In our study, the expression of CRABP2 was three times higher in the presence of TERTp mutation (in TCGA data FC was 2.2). Together with previous studies, it may support the hypothesis that an elevated CRABP2 expression is a late event, characteristic for more advanced and aggressive tumors. These data do not confirm the recognized main role of CRABP2 as an antitumor agent. An alternative mechanism of CRABP2 activity has been proposed. It has been proven that CRABP2 mediated proliferative activity not by interacting with RAR, but through PPARbeta/delta receptor in the presence of fatty acid binding protein 5 [32].
Two genes, KRT17 and ECM1, which expression was elevated in our TERTp(+) PTCs, demonstrated higher expression in a variety of cancer types. Keratins are components of the cytoskeleton and play a major role in cell protection and structural support. Keratin KRT17, belonging to type I keratin, was regenerated and highly expressed in many cancers [38], including gastric cancer [39], cervical cancer [40], oral squamous cell carcinoma [41], and breast cancer [42]. Moreover, KRT17 expression was proposed as a prognostic marker that can discriminate postoperative stage II patients with colorectal cancer with a high probability of disease recurrence, as support to available risk stratification factors [43]. Extracellular matrix protein 1 (ECM1), in turn, is a glycoprotein involved in cell proliferation, angiogenesis, migration, and metastases. Its elevated expression was observed in several malignancies, including thyroid, gastric, colorectal, lung carcinoma, invasive ductal breast carcinomas, hepatocellular cancer, and others [44,45]. A significantly high increase in ECM1 expression was observed in malignant epithelial tumors, especially in these tumors with distant metastases [44]. This observation would explain higher ECM1 expression in our TERTp(+) PTCs when compared to BRAF(+)TERTp(-) and RAS(+) PTCs. However, ECM1 occurred to be down-regulated in mutated PTCs in comparison to NM-PTCs. So, its role as a potential marker of TERTp(+) PTCs seems questionable.
Regarding the 4-gene signature of TERTp(+) PTCs in our study, the MTMR3 gene (encodes myotubularin-related protein 3) was the only down-regulated gene in TERTp(+) PTCs comparing to TERTp(−) ones. MTMR3 belongs to phosphoinositide 3-phosphatases with affinity to Ptdlns5P [46]. It may impair or enhance tumor growth in different types of cancer. For example, the lack of MTMR3 was shown to repress the proliferative and invasive potential of oral cancer cells [47]. Its exogenous expression inhibited the growth of transfected lung carcinoma cells [48]. On the contrary, its higher expression had negative effects on overall survival and relapse-free survival in patients with breast cancer [49]. Nevertheless, the role of MTMR3 in TERTp(+) PTCs requires further studies.
Our signaling pathways analysis revealed 39 enriched KEGG pathways differentially changed between BRAF(+)TERTp(+) and BRAF(+)TERTp(−) PTCs. The majority of pathways with the highest absolute NES (normalized enrichment score) value were enriched by down-regulated genes, among them Inositol phosphate metabolism, Phosphatidylinositol signaling system, and Ubiquitin mediated proteolysis pathways. The critical feature of cancer cells differentiating them from healthy ones is related to the reprogramming of fundamental pathways determining distinct processes, including proliferation, differentiation, and motility. The Ubiquitin pathway plays an essential role in the regulation of cell growth and cell proliferation through the control of key cell cycle proteins. Because of its involvement in crucial biochemical processes, it is a potential target for cancer-related deregulation. Impaired proteolysis of cell cycle regulators was reported in many human cancers as being contributed to tumorigenesis [50,51]. Phosphatidylinositol signaling system is also known as associated with cancer, mainly by its cooperation with the PI3K-Akt pathway that mediates cell proliferation, survival, and metabolism. Mainly, PI3K and PTEN play a key role in cancer, but also other members of this pathway seem to be implicated in the progression of tumors [52].
Among pathways with the highest NES value, enriched by up-regulated genes, we found the Neuroactive ligand-receptor interaction pathway, which includes receptors and ligands associated with intracellular and extracellular signaling pathways involved in the progression of the bladder, renal cell, and prostate cancer [53,54,55]. Up-regulated genes also enriched the Olfactory transduction pathway. Olfactory receptors (OR), playing a crucial role in healthy tissues, were also involved in multiple pathological processes, including hepatocarcinoma, non-small cell lung cancer, colorectal cancer, melanoma, and breast cancer [56]. Some of these ORs were proposed as markers in different cancer types to discriminate between cancer and healthy tissues. Moreover, comparing BRAF(+)TERTp(+) and BRAF(+)TERTp(−) PTCs, we found six significantly enriched gene groups related to telomeres. All of these gene groups had a negative NES value (i.e., they are enriched in genes downregulated in BRAF(+)TERTp(+) cases), which may suggest that the presence of TERTp mutations impairs processes involved in telomere maintenance.
Difference in gene expression profile found in our BRAF(+) PTCs as dependent on the mutated TERT promoter was subtle. However, positive verification of one out of four differentiating genes on an independent PTC cohort allows to hypothesize that obtained data will still be valid on a much larger tumor set. Especially since our and validation PTC sets differed in two important features- number of high-risk PTCs, that was incomparably larger in our cohort, and presence of TERTp mutations only as co-existing with BRAF V600E mutation in our PTC set. Despite these differences expression of CRABP2 gene was significantly up-regulated in TERTp(+) cases in both cohorts.
So far, there was only one published study that analyzed the impact of the presence of TERTp mutations on PTC transcriptome [18]. Chien et al., who used the data obtained from the TCGA study [4], found TERTp mutations to be associated with proliferative and metabolic alterations in PTC. Pathways related to DNA damage response and cell cycle regulation were enriched among up-regulated genes, whereas transporter and metabolic activities were overrepresented among down-regulated genes [18]. Although they analyzed a much larger group, they found no association between TERTp mutations and BRAF genetic alterations. No difference regarding thyroid differentiation genes was observed. Our PTC cohort was smaller. All TERTp(+) PTCs were BRAF(+). The main limitation of this study is related to the coexistence of the BRAF mutation in all TERTp(+) PTC samples. However, the presence of TERTp mutations, without co-existing BRAF or RAS genetic alterations is rare in PTC. In our previous study, 3 out of 189 PTCs (1.6%) were positive only for TERTp mutation (no BRAF and no RAS mutations detected). We found significantly lower TDS value in TERTp(+) PTCs comparing to RAS(+) and NM-PTCs. However, the limited number of samples and the presence of other than TERTp mutations in our analysis impaired the obtained results. In addition, because of the selection bias, almost half of our PTC patients had distant metastases. It increased the number of TERTp(+) PTCs in our cohort. It is also the crucial difference between our and TCGA PTC cohorts since the latter one included mainly low-risk PTCs (only 8 PTCs had distant metastases) [4]. Moreover, the use of a larger set of high-risk PTCs confined our cohort to cases with co-existing BRAF and TERTp mutations and obligated us to search for differences mainly within the BRAF(+) PTC set. On the one hand, it is a limitation of our analysis. On the other hand, it is important to find mechanisms responsible for PTC aggressiveness, since the BRAF V600E mutation as a prognostic marker in PTC is still controversial and is considered in treatment or risk stratification only regarding clinical features.
Despite mentioned limitations of the study, there are clear differences in gene expression profile between BRAF(+) PTC tumors carrying TERTp mutation and PTC tumors with only BRAF mutation. This suggests potential role of TERTp mutations in down-regulation of thyroid specific genes. Additionally, we confirmed that mutated TERT promoter is associated with poor prognosis of PTC and it might have a potential value as a prognostic marker.
4. Materials and Methods
4.1. Patients Characteristics
The study cohort consisted of 54 PTC cases, operated at Maria Sklodowska-Curie National Research Institute of Oncology Gliwice Branch, in Gliwice, Poland. The patients were selected based on their clinical course to obtain representative groups with and without distant metastases. The ‘metastatic cohort’ involved 24 PTC cases. The presence of distant metastases was confirmed by X-ray, computed tomography, scintigraphy, and/or histopathological examination. The ‘control group’, consisted of PTC patients without distant metastases. We intended to match the groups for gender, age, and primary advancement. However, the exact matching was not possible. The mean follow-up was 95.9 months (78.6–112.7 months). Clinical and histopathological characteristics are summarized in Table 1, together with the mutational status, given in the “Results” section.
The use of human tissue was approved by the Bioethics Committee at Maria Sklodowska-Curie National Research Institute of Oncology Gliwice Branch (Committee decision from 25 September 2001). Written informed consent to analyze the tissue was obtained from all patients. All clinical data were anonymized and de-identified before the analysis.
4.2. BRAF V600E and TERTp Mutation Analysis
DNA was isolated from the resected thyroids with the DNeasy Blood & Tissue Kit (Qiagen, GmbH, Hilden, Germany) according to the manufacturer protocol. Both BRAF and TERTp mutations were analyzed by PCR amplification and direct sequencing of the hot spots (V600E and C228T, C250T, respectively). A 160-bp region of the BRAF 15th exon, containing the codon 600, was amplified using following primers set: forward-5′-tgttttcctttacttactacacctca-3′; reverse-5′- gcctcaattcttaccatcca-3′, whereas the amplification of the 157-bp fragment of the TERTp containing C228T and C250T localizations was performed with the primers: forward-5′-ccccttcaccttccagctc-3′; reverse-5′-cagcgctgcctgaaactc-3′. The PCR steps were as follows: initial denaturation at 95 °C for 15 min, 34 cycles of denaturation at 95 °C for 30 s; primers annealing at 56.5 °C and 59.7 °C (for BRAF and TERTp, respectively) for 30 s; elongation at 72 °C for 30 s and the final elongation at 72 °C for 5 min. The PCR products were checked for the quality with gel electrophoresis, purified with enzymes, alkaline phosphatase (SAP) and exonuclease I (Life Technologies, Carlsbad, CA, USA), and then subjected to standard sequencing with ABI PRISMTM 1.1 BigDye Terminator Cycle Sequencing Ready Reaction Kit (Life Technologies) on the 3130xl Genetic Analyzer Applied Biosystems (Life Technologies). All samples were also analyzed for BRAF V600E mutations with the THDNA-RT64 kit (EntroGen, Woodland Hills, CA, USA). For the more detailed description of this method, follow the next subsection entitled “RAS mutations detection”.
4.3. RAS Mutations Detection
Quantitative real-time PCR (QPCR) was used in order to analyze RAS somatic mutations. The analysis was performed with the THDNA-RT64 kit (EntroGen) on the QuantStidio 12K Flex Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer protocols. This test is designed to analyze 15 most frequent somatic point mutations in PTCs within the RAS genes and V600E mutation in the BRAF gene. It provides information about whether there is a mutation in HRAS, NRAS, KRAS, or not, without pointing which mutation it is. The exact list of mutations analyzed with this test is given in Table S2.
4.4. Microarray Analysis
The GeneChip Human Gene 1.0 arrays (Affymetrix, Santa Clara, CA, USA) were used. The detailed methodology was described in our previous work [57].
Background correction, normalization and probe set summarization were done using the Robust Multichip Average (RMA) algorithm with library oligo v1.48.0 from R v3.6.0 environment, and custom CDF files from BrainArray (ENTREZG; v24) [58,59,60]. Principal component analysis (PCA) was performed with R/Bioconductor.
The selection of differentially expressed genes was performed using the limma 3.40.6 library [61]. Gene set analysis was performed using Gene-Set Enrichment Analysis (GSEA) implemented in the clusterProfiler 3.12.0 library [62,63]. The t-statistic was used as a gene ranking metric, and 10,000 gene set permutations were performed to calculate p-values and Normalized Enrichment Scores (NES). Pathways from the Kyoto Encyclopedia of Genes and Genomes (KEGG) and terms from the Gene Ontology (GO) with minimal gene size of 10 and maximal of 600 were used in GSEA [64,65].
All p-values were corrected for false discovery rate (FDR), according to Benjamini and Hochberg [66]. Corrected p-values < 0.05 were considered statistically significant.
4.5. TCGA Data Analysis
Gene expression data for TCGA PTC samples (based on RNA-seq data) were retrieved using the TCGAbiolinks 2.12.6 library [67]. The comparison was performed between 28 BRAF(+)TERTp(+) samples and 207 BRAF(+)TERTp(−) (only RAS-negative samples were analyzed) using edgeR 3.26.8 embedded in the TCGAbiolinks library [68]. The read counts were modeled using the negative binomial generalized log-linear model, and a likelihood ratio test was used to assess statistical significance [69]. p-values were corrected for false discovery rate (FDR), according to Benjamini and Hochberg [66]. Corrected p-values < 0.05 were considered statistically significant.
4.6. Calculation of Expression Scores
For each microarray sample, two scores based on gene expression were calculated: BRAF V600E-RAS Score (BRS) and Thyroid Differentiation Score (TDS) [4].
BRS is a 71-gene signature, developed in TCGA study [4], which quantifies the extent to which the gene expression profile of a given tumor resembles either the BRAF V600E or RAS mutant profiles. According to TCGA study, it comprises of 71 genes (13 genes up-regulated in RAS mutant samples and 58 genes up-regulated in BRAF V600E mutant samples). In our paper, BRS was calculated in three steps: (1) we calculated sum of expression of 13 genes up-regulated in RAS mutant samples minus the sum of expression of 58 genes up-regulated in BRAF V600E mutant samples; (2) we divided obtained measures by the number of genes in signature; (3) finally we centered obtained values at the mean across samples. Tumors with low BRS were defined BRAF V600E-like, while tumors with high BRS were defined as RAS-like.
TDS is a summarized expression level of 16 thyroid metabolism and function genes, developed in the TCGA study [4]. The expression values of each gene were first centered at the median across samples. Then, the mean across the 16 genes was calculated for each sample. Higher TDS depicts higher differentiation of the tumor.
4.7. Statistical Analysis of Clinico-Histopahological Data
Categorical data were summarized with numbers and percentages. Continuous data were summarized with medians and interquartile ranges. Comparisons of categorical variables were performed using Fisher’s exact test. Comparisons of continuous variables were performed using two-tailed Mann-Whitney U test (in case of two groups comparisons), and Kruskal-Wallis rank sum test (in case of comparisons of more than two groups). p values < 0.05 were considered statistically significant. Statistical analyses were performed using the R software version 3.6.0 and Gmisc package version 1.9.0 [70,71].
5. Conclusions
We found four genes differentiating TERTp(+) and TERTp(−) PTCs harboring BRAF mutation. It is a small but clear difference, since one of these genes, CRABP2, was validated on an independent set of PTCs. However, their potential use in diagnostics or risk stratification requires additional prospective studies on larger PTC cohorts. Our study is the first one in which the TERTp-dependent PTC gene expression profile was analyzed in BRAF V600E positive samples only. TERTp mutations may be associated with down-regulation of thyroid-specific genes, with their expression being significantly lower comparing to RAS(+) and NM-PTCs. TERTp mutations are related to higher PTC aggressiveness. These are not a frequent molecular event in PTCs, however, if present, always associated with poorer prognosis and a higher level of molecular aberrations.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/6/1597/s1, Table S1: The list of KRAS, NRAS, HRAS and BRAF mutations detectable by the Thyroid Cancer Mutation Analysis Kit (THDNA-RT64; EntroGen), Table S2: Lists of genes significantly deregulated depending on the molecular differentiating factor; M- presence of BRAF or TERTp or RAS somatic mutation; NM- lack of somatic mutations within studied genes.
Author Contributions
Conceptualization, D.R., A.P. (Aleksandra Pfeifer), and M.O.-W.; methodology, D.R.; software, A.P. (Aleksandra Pfeifer), M.C. (Marta Cieslicka); validation, A.P. (Aleksandra Pfeifer), M.C. (Marta Cieslicka); formal analysis, D.R.; investigation, D.R., M.K., A.P. (Agnieszka Pawlaczek), T.T., M.H., R.L.; resources, J.K., S.S.-U., A.C., E.Z.-N., M.C. (Mykola Chekan), D.H.-J., A.L., E.P.-C.; A.K.; data curation, S.S.-U.; writing—original draft preparation, D.R., A.P. (Aleksandra Pfeifer); writing—review and editing, D.R., A.P. (Aleksandra Pfeifer), J.K. and M.O.-W.; visualization, D.R., A.P. (Aleksandra Pfeifer); supervision, B.J. and M.O.-W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Centre for Research and Development project under the program “Prevention practices and treatment of civilization diseases” STRATEGMED (STRATEGMED2/267398/4/NCBR/2015).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Rusinek D., Chmielik E., Krajewska J., Jarzab M., Oczko-Wojciechowska M., Czarniecka A., Jarzab B. Current advances in thyroid cancer management. Are we ready for the epidemic rise of diagnoses? Int. J. Mol. Sci. 2017;18:1817. doi: 10.3390/ijms18081817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colombo C., Muzza M., Proverbio M.C., Tosi D., Soranna D., Pesenti C., Rossi S., Cirello V., De Leo S., Fusco N., et al. Impact of Mutation Density and Heterogeneity on Papillary Thyroid Cancer Clinical Features and Remission Probability. Thyroid. 2019;29:237–251. doi: 10.1089/thy.2018.0339. [DOI] [PubMed] [Google Scholar]
- 3.Chmielik E., Rusinek D., Oczko-Wojciechowska M., Jarzab M., Krajewska J., Czarniecka A., Jarzab B. Heterogeneity of Thyroid Cancer. Pathobiology. 2018 doi: 10.1159/000486422. [DOI] [PubMed] [Google Scholar]
- 4.Agrawal N., Akbani R., Aksoy B.A., Ally A., Arachchi H., Asa S.L., Auman J.T., Balasundaram M., Balu S., Baylin S.B., et al. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xing M. BRAF mutation in thyroid cancer. Endocr. Relat. Cancer. 2005;12:245–262. doi: 10.1677/erc.1.0978. [DOI] [PubMed] [Google Scholar]
- 6.Xing M., Westra W.H.H., Tufano R.P.P., Cohen Y., Rosenbaum E., Rhoden K.J.J., Carson K.A.A., Vasko V., Larin A., Tallini G., et al. BRAF mutation predicts a poorer clinical prognosis for papillary thyroid cancer. J. Clin. Endocrinol. Metab. 2005;90:6373–6379. doi: 10.1210/jc.2005-0987. [DOI] [PubMed] [Google Scholar]
- 7.Xing M., Alzahrani A.S., Carson K.A., Viola D., Elisei R., Bendlova B., Yip L., Mian C., Vianello F., Tuttle R.M., et al. Association between BRAF V600E mutation and mortality in patients with papillary thyroid cancer. JAMA. 2013;309:1493–1501. doi: 10.1001/jama.2013.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tavares C., Melo M., Cameselle-Teijeiro J.M., Soares P., Sobrinho-Simoes M. Genetic predictors of thyroid cancer outcome. Eur. J. Endocrinol. 2016;174:117–126. doi: 10.1530/EJE-15-0605. [DOI] [PubMed] [Google Scholar]
- 9.Haugen B.R., Alexander E.K., Bible K.C., Doherty G.M., Mandel S.J., Nikiforov Y.E., Pacini F., Randolph G.W., Sawka A.M., Schlumberger M., et al. 2015 American Thyroid Association Management Guidelines for Adult Patients with Thyroid Nodules and Differentiated Thyroid Cancer: The American Thyroid Association Guidelines Task Force on Thyroid Nodules and Differentiated Thyroid Cancer. Thyroid. 2016;26:1–133. doi: 10.1089/thy.2015.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X., Bishop J., Shan Y., Pai S., Liu D., Murugan A.K., Sun H., El-Naggar A.K., Xing M. Highly prevalent TERT promoter mutations in aggressive thyroid cancers. Endocr. Relat. Cancer. 2013;20:603–610. doi: 10.1530/ERC-13-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horn S., Figl A., Rachakonda P.S., Fischer C., Sucker A., Gast A., Kadel S., Moll I., Nagore E., Hemminki K., et al. TERT Promoter Mutations in Familial and Sporadic Melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 12.Huang F.W., Hodis E., Xu M.J., Kryukov G.V., Chin L., Garraway L.A. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu R., Xing M. TERT promoter mutations in thyroid cancer. Endocr. Relat. Cancer. 2016;23:143–155. doi: 10.1530/ERC-15-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Low K.C., Tergaonkar V. Telomerase: Central regulator of all of the hallmarks of cancer. Trends Biochem. Sci. 2013;38:426–434. doi: 10.1016/j.tibs.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Melo M., da Rocha A.G.G., Vinagre J.J., Batista R., Peixoto J., Tavares C., Celestino R., Almeida A., Salgado C., Eloy C., et al. TERT promoter mutations are a major indicator of poor outcome in differentiated thyroid carcinomas. J. Clin. Endocrinol. Metab. 2014;99:E754–E765. doi: 10.1210/jc.2013-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rusinek D., Pfeifer A., Krajewska J., Oczko-Wojciechowska M., Handkiewicz-Junak D., Pawlaczek A., Zebracka-Gala J., Kowalska M., Cyplinska R., Zembala-Nozynska E., et al. Coexistence of TERT Promoter Mutations and the BRAF V600E Alteration and Its Impact on Histopathological Features of Papillary Thyroid Carcinoma in a Selected Series of Polish Patients. Int. J. Mol. Sci. 2018;19:2647. doi: 10.3390/ijms19092647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen X., Liu R., Xing M. A six-genotype genetic prognostic model for papillary thyroid cancer. Endocr. Relat. Cancer. 2017;24:41–52. doi: 10.1530/ERC-16-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chien M.-N., Yang P.-S., Hsu Y.-C., Liu T.-P., Lee J.-J., Cheng S.-P. Transcriptome analysis of papillary thyroid cancer harboring telomerase reverse transcriptase promoter mutation. Head Neck. 2018;40:2528–2537. doi: 10.1002/hed.25385. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan D., Weinberg R.A.A. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Donati B., Ciarrocchi A. Telomerase and Telomeres Biology in Thyroid Cancer. Int. J. Mol. Sci. 2019;20:2887. doi: 10.3390/ijms20122887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shay J.W., Wright W.E. Telomeres and telomerase: Three decades of progress. Nat. Rev. Genet. 2019;20:299–309. doi: 10.1038/s41576-019-0099-1. [DOI] [PubMed] [Google Scholar]
- 22.Yuan X., Larsson C., Xu D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: Old actors and new players. Oncogene. 2019;38:6172–6183. doi: 10.1038/s41388-019-0872-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z., Li Q., Li K., Chen L., Li W., Hou M., Liu T., Yang J., Lindvall C., Bjorkholm M., et al. Telomerase reverse transcriptase promotes epithelial-mesenchymal transition and stem cell-like traits in cancer cells. Oncogene. 2013;32:4203–4213. doi: 10.1038/onc.2012.441. [DOI] [PubMed] [Google Scholar]
- 24.Ding D., Xi P., Zhou J., Wang M., Cong Y.-S. Human telomerase reverse transcriptase regulates MMP expression independently of telomerase activity via NF-kappaB-dependent transcription. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2013;27:4375–4383. doi: 10.1096/fj.13-230904. [DOI] [PubMed] [Google Scholar]
- 25.de Biase D., Gandolfi G., Ragazzi M., Eszlinger M., Sancisi V., Gugnoni M., Visani M., Pession A., Casadei G., Durante C., et al. TERT Promoter Mutations in Papillary Thyroid Microcarcinomas. Thyroid. 2015;25:1013–1019. doi: 10.1089/thy.2015.0101. [DOI] [PubMed] [Google Scholar]
- 26.Liu T., Yuan X., Xu D. Cancer-Specific Telomerase Reverse Transcriptase (TERT) Promoter Mutations: Biological and Clinical Implications. Genes. 2016;7:38. doi: 10.3390/genes7070038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gandolfi G., Ragazzi M., Frasoldati A., Piana S., Ciarrocchi A., Sancisi V. TERT promoter mutations are associated with distant metastases in papillary thyroid carcinoma. Eur. J. Endocrinol. 2015;172:403–413. doi: 10.1530/EJE-14-0837. [DOI] [PubMed] [Google Scholar]
- 28.Muzza M., Colombo C., Rossi S., Tosi D., Cirello V., Perrino M., De Leo S., Magnani E., Pignatti E., Vigo B., et al. Telomerase in differentiated thyroid cancer: Promoter mutations, expression and localization. Mol. Cell. Endocrinol. 2015;399:288–295. doi: 10.1016/j.mce.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 29.Fredriksson N.J., Ny L., Nilsson J.A., Larsson E. Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat. Genet. 2014;46:1258. doi: 10.1038/ng.3141. [DOI] [PubMed] [Google Scholar]
- 30.Landa I., Ganly I., Chan T.A., Mitsutake N., Matsuse M., Ibrahimpasic T., Ghossein R.A., Fagin J.A. Frequent somatic TERT promoter mutations in thyroid cancer: Higher prevalence in advanced forms of the disease. J. Clin. Endocrinol. Metab. 2013;98:E1562–E1566. doi: 10.1210/jc.2013-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X., Qu S., Liu R., Sheng C., Shi X., Zhu G., Murugan A.K., Guan H., Yu H., Wang Y., et al. TERT promoter mutations and their association with BRAF V600E mutation and aggressive clinicopathological characteristics of thyroid cancer. J. Clin. Endocrinol. Metab. 2014;99:E1130–E1136. doi: 10.1210/jc.2013-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schug T.T., Berry D.C., Shaw N.S., Travis S.N., Noy N. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell. 2007;129:723–733. doi: 10.1016/j.cell.2007.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toyama A., Suzuki A., Shimada T., Aoki C., Aoki Y., Umino Y., Nakamura Y., Aoki D., Sato T.-A. Proteomic characterization of ovarian cancers identifying annexin-A4, phosphoserine aminotransferase, cellular retinoic acid-binding protein 2, and serpin B5 as histology-specific biomarkers. Cancer Sci. 2012;103:747–755. doi: 10.1111/j.1349-7006.2012.02224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mallikarjuna K., Sundaram C.S., Sharma Y., Deepa P.R., Khetan V., Gopal L., Biswas J., Sharma T., Krishnakumar S. Comparative proteomic analysis of differentially expressed proteins in primary retinoblastoma tumors. Proteomics. Clin. Appl. 2010;4:449–463. doi: 10.1002/prca.200900069. [DOI] [PubMed] [Google Scholar]
- 35.Geiger T., Madden S.F., Gallagher W.M., Cox J., Mann M. Proteomic portrait of human breast cancer progression identifies novel prognostic markers. Cancer Res. 2012;72:2428–2439. doi: 10.1158/0008-5472.CAN-11-3711. [DOI] [PubMed] [Google Scholar]
- 36.Xiao W., Hong H., Awadallah A., Yu S., Zhou L., Xin W. CRABP-II is a highly sensitive and specific diagnostic molecular marker for pancreatic ductal adenocarcinoma in distinguishing from benign pancreatic conditions. Hum. Pathol. 2014;45:1177–1183. doi: 10.1016/j.humpath.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim D.J., Kim W.J., Lim M., Hong Y., Lee S.-J., Hong S.-H., Heo J., Lee H.-Y., Han S.-S. Plasma CRABP2 as a Novel Biomarker in Patients with Non-Small Cell Lung Cancer. J. Korean Med. Sci. 2018;33:e178. doi: 10.3346/jkms.2018.33.e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Troyanovsky S.M., Leube R.E., Franke W.W. Characterization of the human gene encoding cytokeratin 17 and its expression pattern. Eur. J. Cell Biol. 1992;59:127–137. [PubMed] [Google Scholar]
- 39.Hu H., Xu D.-H., Huang X.-X., Zhu C.-C., Xu J., Zhang Z.-Z., Zhao G. Keratin17 Promotes Tumor Growth and is Associated with Poor Prognosis in Gastric Cancer. J. Cancer. 2018;9:346–357. doi: 10.7150/jca.19838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Escobar-Hoyos L.F., Yang J., Zhu J., Cavallo J.-A., Zhai H., Burke S., Koller A., Chen E.I., Shroyer K.R. Keratin 17 in premalignant and malignant squamous lesions of the cervix: Proteomic discovery and immunohistochemical validation as a diagnostic and prognostic biomarker. Mod. Pathol. Off. J. United States Can. Acad. Pathol. Inc. 2014;27:621–630. doi: 10.1038/modpathol.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Regenbogen E., Mo M., Romeiser J., Shroyer A.L.W., Escobar-Hoyos L.F., Burke S., Shroyer K.R. Elevated expression of keratin 17 in oropharyngeal squamous cell carcinoma is associated with decreased survival. Head Neck. 2018;40:1788–1798. doi: 10.1002/hed.25164. [DOI] [PubMed] [Google Scholar]
- 42.Sorlie T., Perou C.M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M.B., van de Rijn M., Jeffrey S.S., et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ujiie D., Okayama H., Saito K., Ashizawa M., Thar Min A.K., Endo E., Kase K., Yamada L., Kikuchi T., Hanayama H., et al. KRT17 as a prognostic biomarker for stage II colorectal cancer. Carcinogenesis. 2019 doi: 10.1093/carcin/bgz192. [DOI] [PubMed] [Google Scholar]
- 44.Wang L., Yu J., Ni J., Xu X.-M., Wang J., Ning H., Pei X.-F., Chen J., Yang S., Underhill C.B., et al. Extracellular matrix protein 1 (ECM1) is over-expressed in malignant epithelial tumors. Cancer Lett. 2003;200:57–67. doi: 10.1016/S0304-3835(03)00350-1. [DOI] [PubMed] [Google Scholar]
- 45.Wang Z., Zhou Q., Li A., Huang W., Cai Z., Chen W. Extracellular matrix protein 1 (ECM1) is associated with carcinogenesis potential of human bladder cancer. Onco. Targets. Ther. 2019;12:1423–1432. doi: 10.2147/OTT.S191321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clague M.J., Lorenzo O. The myotubularin family of lipid phosphatases. Traffic. 2005;6:1063–1069. doi: 10.1111/j.1600-0854.2005.00338.x. [DOI] [PubMed] [Google Scholar]
- 47.Kuo Y.Z., Tai Y.H., Lo H.I., Chen Y.L., Cheng H.C., Fang W.Y., Lin S.H., Yang C.L., Tsai S.T., Wu L.W. MiR-99a exerts anti-metastasis through inhibiting myotubularin-related protein 3 expression in oral cancer. Oral Dis. 2014;20:e65–e75. doi: 10.1111/odi.12133. [DOI] [PubMed] [Google Scholar]
- 48.Yoo Y.D., Cho S.M., Kim J.S., Chang Y.S., Ahn C.M., Kim H.J. The human myotubularin-related protein suppresses the growth of lung carcinoma cells. Oncol. Rep. 2004;12:667–671. doi: 10.3892/or.12.3.667. [DOI] [PubMed] [Google Scholar]
- 49.Wang Z., Zhang M., Shan R., Wang Y.-J., Chen J., Huang J., Sun L.-Q., Zhou W.-B. MTMR3 is upregulated in patients with breast cancer and regulates proliferation, cell cycle progression and autophagy in breast cancer cells. Oncol. Rep. 2019;42:1915–1923. doi: 10.3892/or.2019.7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bashir T., Pagano M. Aberrant ubiquitin-mediated proteolysis of cell cycle regulatory proteins and oncogenesis. Adv. Cancer Res. 2003;88:101–144. doi: 10.1016/s0065-230x(03)88305-7. [DOI] [PubMed] [Google Scholar]
- 51.Spataro V., Norbury C., Harris A.L. The ubiquitin-proteasome pathway in cancer. Br. J. Cancer. 1998;77:448–455. doi: 10.1038/bjc.1998.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bunney T.D., Katan M. Phosphoinositide signalling in cancer: Beyond PI3K and PTEN. Nat. Rev. Cancer. 2010;10:342–352. doi: 10.1038/nrc2842. [DOI] [PubMed] [Google Scholar]
- 53.Fang Z.-Q., Zang W.-D., Chen R., Ye B.-W., Wang X.-W., Yi S.-H., Chen W., He F., Ye G. Gene expression profile and enrichment pathways in different stages of bladder cancer. Genet. Mol. Res. 2013;12:1479–1489. doi: 10.4238/2013.May.6.1. [DOI] [PubMed] [Google Scholar]
- 54.Liu X., Wang J., Sun G. Identification of key genes and pathways in renal cell carcinoma through expression profiling data. Kidney Blood Press Res. 2015;40:288–297. doi: 10.1159/000368504. [DOI] [PubMed] [Google Scholar]
- 55.He Z., Tang F., Lu Z., Huang Y., Lei H., Li Z., Zeng G. Analysis of differentially expressed genes, clinical value and biological pathways in prostate cancer. Am. J. Transl. Res. 2018;10:1444–1456. [PMC free article] [PubMed] [Google Scholar]
- 56.Weber L., Maßberg D., Becker C., Altmüller J., Ubrig B., Bonatz G., Wölk G., Philippou S., Tannapfel A., Hatt H., et al. Olfactory Receptors as Biomarkers in Human Breast Carcinoma Tissues. Front. Oncol. 2018;8:33. doi: 10.3389/fonc.2018.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rusinek D., Swierniak M., Chmielik E., Kowal M., Kowalska M., Cyplinska R., Czarniecka A., Piglowski W., Korfanty J., Chekan M., et al. BRAFV600E-Associated Gene Expression Profile: Early Changes in the Transcriptome, Based on a Transgenic Mouse Model of Papillary Thyroid Carcinoma. PLoS ONE. 2015;10:e0143688. doi: 10.1371/journal.pone.0143688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carvalho B.S., Irizarry R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dai M., Wang P., Boyd A.D., Kostov G., Athey B., Jones E.G., Bunney W.E., Myers R.M., Speed T.P., Akil H., et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.The Gene Ontology Consortium The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kanehisa M., Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 67.Colaprico A., Silva T.C., Olsen C., Garofano L., Cava C., Garolini D., Sabedot T.S., Malta T.M., Pagnotta S.M., Castiglioni I., et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Team R.C. A Language And Environment For Statistical Computing. R Foundation for Statistical Computing. [(accessed on 15 December 2017)];2013 Available online: https://www.r-project.org/
- 71.Gordon M. Gmisc: Descriptive Statistics, Transition Plots, and More. R package version 1.9.0. [(accessed on 16 June 2020)];2019 Available online: https://CRAN.R-project.org/package=Gmisc.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.