Abstract
Irregular nuclear shapes characterized by blebs, lobules, micronuclei, or invaginations are hallmarks of many cancers and human pathologies. Despite the correlation between abnormal nuclear shape and human pathologies, the mechanism by which the cancer nucleus becomes misshapen is not fully understood. Motivated by recent evidence that modifying chromatin condensation can change nuclear morphology, we conducted a high-throughput RNAi screen to identify epigenetic regulators that are required to maintain normal nuclear shape in human breast epithelial MCF-10A cells. We silenced 608 genes in parallel using an epigenetics siRNA library and used an unbiased Fourier analysis approach to quantify nuclear contour irregularity from fluorescent images captured on a high-content microscope. Using this quantitative approach, which we validated with confocal microscopy, we significantly expand the list of epigenetic regulators that impact nuclear morphology.
INTRODUCTION
The nucleus of the mammalian cell organizes and protects the genome, and ensures normal gene expression and cell function. While the nuclei of most normal epithelial cell types have smooth circular or ellipsoidal shape, the nucleus of a cancer cell has irregularities in its contour including blebs, lobules, micronuclei, or invaginations. These nuclear irregularities are characteristic of many types of malignancies, including breast cancer (Abdalla et al., 2009; Williams et al., 2014), melanoma (Mijovic et al., 2013), squamous cell carcinoma (Speight, 2007), lung cancer (Petersen et al., 2009), and prostate cancer (Diamond et al., 1982; Epstein et al., 1984). Pathologists commonly evaluate nuclear shape irregularities to diagnose and prognose the severity of cancer in biopsies of human tissue (Gil et al., 2002; Kashyap et al., 2018). Additionally, irregular nuclear shapes are characteristic of several major human diseases, including laminopathies and progerias (Goldman et al., 2004; De Vos et al., 2010; Ho et al., 2011; Zwerger et al., 2011; Siu et al., 2012; Tariq et al., 2017; Steele-Stallard et al., 2018). Despite the well-established correlation between abnormal nuclear shape and human pathologies, the mechanisms by which nuclei of cells become misshapen are not fully understood.
Numerous cancers are characterized by a decrease in the expression of nuclear lamins (Capo-chichi et al., 2011a, b; Denais and Lammerding, 2014; Irianto et al., 2016). Nuclear lamins impart rigidity to the nucleus at nuclear extensions greater than 3 µm (Stephens et al., 2017) and down-regulation of lamins results in an abnormally shaped nucleus (Lammerding et al., 2004, 2006). Cytoskeletal forces that act on the nucleus can cause or amplify existing nuclear shape abnormalities (Hatch et al., 2013; Cain et al., 2014; Takaki et al., 2017; Tocco et al., 2018). There is also evidence that nuclear shape abnormalities can be caused by alterations to chromatin regulatory proteins. Knockdown of the chromatin remodeling enzyme BRG-1 (encoded by SMARCA4) increased the percentage of irregular nuclei in MCF-10A (human breast fibrocystic disease) cells, characterized by curves, bulges, folds, and invaginations in the nuclear contour (Imbalzano et al., 2013). Treatment with drugs that increase euchromatin or decrease heterochromatin resulted in nuclear blebbing in HT1080 cells and mouse embryonic fibroblasts, without altering lamins (Stephens et al., 2018a). Altering chromatin condensation through changes in histone modification states also mechanically softened nuclei, which may make them susceptible to developing abnormal morphologies (Stephens et al., 2018a). Conversely, condensing chromatin by increased levels of heterochromatin reduced the incidence of blebbed nuclei and partially rescued nuclear shape in Hutchinson-Gilford progeria syndrome (HGPS) cells, a disease caused by mutant lamin A (Stephens et al., 2018a, 2019b).
Motivated by these previous results, we conducted a high-throughput RNAi screen to identify individual epigenetic regulators that are required to maintain normal nuclear shape in noncancerous human breast epithelial MCF-10A cells. For our screen, we developed an automated elliptical Fourier analysis technique to quantify the elliptic Fourier coefficient (EFC) ratio which is a quantitative metric of nuclear shape abnormalities. Knockdown of a library of 608 epigenetic-related genes revealed novel gene products which caused statistically significant nuclear shape aberrations. Gene depletions with a mean EFC ratio below approximately 5.3 were visually abnormal. We used confocal microscopy to study nuclear abnormalities in a small set of 21 genes and found reductions in EFC ratio similar to results with the high-throughput screen. Our results offer new directions in understanding the mechanisms of aberrant nuclear shaping in malignancy.
RESULTS
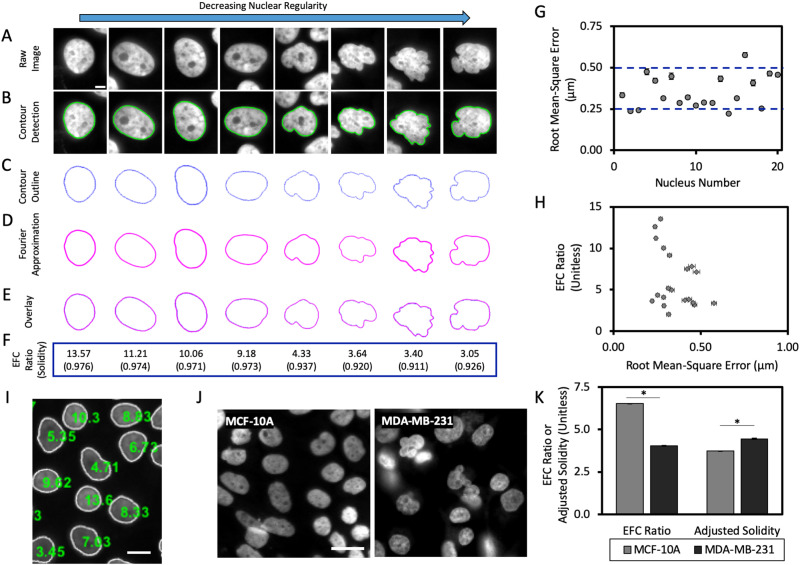
A Fourier analysis approach for quantification of nuclear contour irregularities
Commonly used methods to detect irregular nuclear shapes, such as manual classification, are subject to observer bias and poor reproducibility (Imbalzano et al., 2013). Here we used elliptical Fourier analysis to quantitatively measure nuclear morphology, which has previously been used to analyze irregular nuclear shapes (Diaz et al., 1989; Lammerding et al., 2006). This method approximates the nuclear contour with a series of diminishing harmonic ellipses, and an EFC ratio is calculated. The EFC ratio provides a relative estimate of the regularity of each nucleus, with larger EFC ratios corresponding to more circular or ellipsoidal (regular) nuclei, and smaller EFC ratios to nuclei with more complex, irregular shapes which require larger contributions of subsequent ellipses to accurately fit the nuclear contour. In our analysis of MCF-10A nuclei, a series of 15 harmonic ellipses were found to be sufficient for approximating nuclear contours (compare contour and reconstructed contour; Figure 1, A–E). The EFC ratio correlated well with visually obvious irregularities in the nuclear contour (Figure 1F), and there was a significant separation in EFC ratios between normal and visually irregular shapes (ranging from ∼10 for normal shapes to ∼2 for irregular shapes). In particular, nuclei with EFC ratios below approximately 5.3 appeared visually abnormal. Another objective shape descriptor commonly used to compare nuclear morphology in the literature is known as solidity, which is defined as the ratio of the measured area of the nucleus to that of a bounding convex shape. Therefore, nuclei with more irregular nuclear shapes would, in theory, have a significantly larger bounding convex area than regular nuclei with circular or ellipsoidal shapes. However, in contrast with the EFC ratio, the widely utilized metric of solidity (Choi et al., 2011; Schochlin et al., 2014; Hulsman et al., 2015; Jagielska et al., 2017; Verschuuren et al., 2017; Konstandinou et al., 2018; Stephens et al., 2018a, b) yielded relatively weak numerical separation for the same set of regular and visually irregular nuclei (Figure 1F).
FIGURE 1:
Quantification of nuclear contour irregularities with elliptical Fourier analysis. (A) Example images of single nuclei from nontransformed human MCF-10A mammary epithelial cells. Images of nuclei are arranged in descending order of regularity, from left to right. Scale bar is 5 µm. (B) Representative images of the detected nuclear contour in A overlaid onto the original raw image; nuclear contours were detected using MATLAB image processing algorithms. (C) Nuclear contours shown in B without the overlay of the original image (blue). (D) Elliptic Fourier approximations of the nuclear contours from C, calculated using 15 elliptic harmonics to fit each nuclear shape (Diaz et al., 1989; Lammerding et al., 2006). (E) Overlays of the detected nuclear contour and the elliptic Fourier approximation. (F) Representative EFC ratios (computed in MATLAB) and solidity measurements (where solidity = area/convex area; computed in ImageJ) for each nuclear shape depicted in A. (G) Quantification of the RMS error between the elliptic Fourier approximation and the nuclear boundary; each data point corresponds to a single nucleus (N = 3, n = 20 where N represents the number of biological replicates and n represents the total number of cells analyzed). (H) EFC ratio plotted against RMS error for each nucleus in (G; N = 3, n = 20). Error bars denote SEM. (I) Representative image after segmentation and quantification of EFC ratio. White outlines represent the detected nuclear boundary; green numbers overlaid onto nuclear centroids represent the EFC ratio for each respective nucleus. Scale bar is 20 µm. (J) Representative images of MCF-10A human breast epithelial cells and MDA-MB-231 human breast adenocarcinoma cells taken at 40× on a high-content microscope. Scale bar is 25 µm. (K) Mean EFC ratio and adjusted solidity (defined as [(1-solidity)*100]) for MCF-10A and MDA-MB-231 cells (N ≥ 15 and 3, n = 71,191 and 6952 for MCF-10A and MDA-MB-231, respectively). Error bars represent SEM. *p < 0.05 by the Bonferroni-corrected nonparametric Dunn’s test.
We quantitatively assessed the accuracy of the Fourier approximation of nuclear shapes by calculating the root mean-square (RMS) error; that is, the RMS difference between the Fourier reconstruction and the nuclear boundary. The RMS error was 500 nm or less (Figure 1G), which is comparable to the spatial resolution of the 40× objective (0.95 numerical aperture) used for imaging. The small RMS error in fitting the 2-D nuclear contour highlights the accuracy of the Fourier method and validates the visual agreement between estimated and measured contours in Figure 1, A–F.
If the error in estimation is dependent on the degree of nuclear irregularity, that would bias the Fourier approach and reduce its reliability. We therefore examined the correlation between the RMS error and the EFC ratio and found no correlation between these quantities (Figure 1H). Together with Figure 1G, these results validate the Fourier approximation method as an effective tool for detecting nuclear irregularities. We developed custom programs for automated quantification of the EFC ratio from fluorescent images of nuclei. The algorithm was able to detect the majority of isolated nuclei from fluorescent images (Figure 1I) and approximate nuclear contours through Fourier decomposition while excluding clumped nuclei.
Next, we performed imaging of MDA-MB-231 cancer nuclei, which are known to have abnormal shapes, using a high-throughput microscope and calculated EFC ratios. The mean EFC ratio of MDA-MB-231 cancer nuclei was 3.05 ± 0.03 and significantly lower than the mean EFC ratio of MCF-10A cells (EFC ratio of 6.5 ± 0.01), validating the use of the EFC ratio for quantifying nuclear abnormalities in cancer cells (Figure 1, J and K). Solidity, plotted using an adjusted solidity metric (defined as (1-solidity) multiplied by 100) which is inversely related to solidity in terms of nuclear regularity, was also statistically lower in MDA-MB-231 cells (mean adjusted solidity of 4.03 ± 0.03) than MCF-10A control nuclei (mean adjusted solidity of 3.74 ± 0.01), but the numerical separation between control and cancer nuclei was again much weaker (Figure 1K).
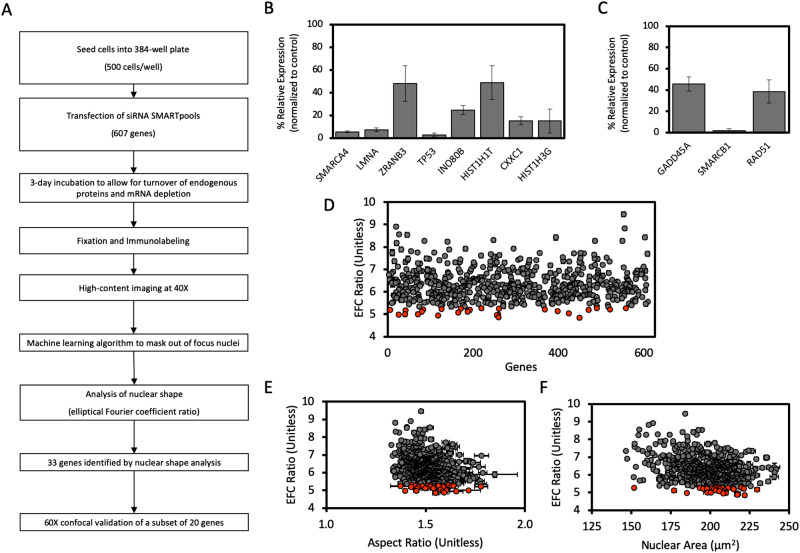
A high-throughput screen for nuclear shape abnormalities
We next developed an experimental and computational analysis pipeline for a high-throughput gene screen of the effects of depletion of chromatin proteins on nuclear shape (Figure 2A). MCF-10A cells were seeded into 384-well plates at an initial density of 500 cells/well. After seeding, cells were incubated with individual siRNAs at a concentration of 25 nM, which was found to maximize depletion of mRNA levels while minimizing loss of cell viability (Supplemental Figure S4), in 384-well plates for 72 h, which was determined to be sufficient time for knockdown via RT-qPCR, consistent with previous RNAi screens (Bolt et al., 2015) (Figure 2, B and C). Cells were then fixed and immunolabeled for target proteins (including lamin A), and fluorescent images of nuclei stained with Hoechst 33342 were acquired through high-throughput imaging at 40× resolution. Nuclear morphology was then quantified for in-focus nuclei using elliptical Fourier analysis. Gene depletions that resulted in abnormal nuclear shapes (decreased EFC ratios) were identified using nonparametric statistical analysis and a subsequent, refined criterion for mean EFC ratios below a cut-off value (Figure 2D).
FIGURE 2:
High-throughput nuclear imaging screen identifies epigenetic regulators required for maintenance of normal nuclear shape. (A) Schematic workflow of the siRNA screen for nuclear irregularities. (B) The efficacy of siRNA-mediated knockdown of expression in a subset of genes from the screen identified to significantly decrease EFC ratio, selected randomly, assayed with probe-based RT-qPCR. (C) The efficacy of siRNA-mediated knockdown of expression in a subset of genes from the screen which had no significant effect on EFC ratio, selected randomly, and assayed with probe-based RT-qPCR. Data for both B and C was first internally normalized to GAPDH expression, then externally normalized against a scrambled siRNA negative control, and subsequently analyzed using the 2-∆∆Ct method. Data represent mean of each condition from at least three biological replicates. Error bars represent SEM. (D) Results of the high-throughput screen for nuclear shape showing mean EFC ratio for each gene depletion condition plotted against arbitrary gene number (N = 3, n ≥ 216 for each condition). For D–F, gray data points represent gene depletions determined to have a statistically insignificant effect on the plotted parameter by the two-tailed Bonferroni-corrected nonparametric Dunn’s test; orange data points represent gene depletions which produce significant changes to nuclear shape (p < 0.05; all comparisons relative to the scrambled siRNA negative control). (E) Mean EFC ratio plotted against mean 2-D nuclear X-Y aspect ratio (length of major axis/length of minor axis) for each gene condition in the high-throughput screen (N = 3, n ≥ 216 for each condition). (F) Mean EFC ratio plotted against mean nuclear cross-sectional area (N = 3, n ≥ 216). Error bars represent SEM.
Using the analysis pipeline in Figure 2A, we screened 608 siRNAs targeting genes associated with epigenetic and chromatin regulatory roles. The full list of genes whose depletion caused statistically significant effects on EFC ratio is listed in Supplemental Table S2. The computational analysis was highly sensitive to nuclear shape abnormalities and therefore was able to detect subtle changes in nuclear shape. However, such subtle changes may not be physiologically impactful. Because nuclei below a mean value of approximately 5.3 looked visually abnormal (see Figure 1, A–F), we reasoned that imposing such a cut-off might be one way to narrow down the list of genes that significantly impact nuclear morphology. Genes that had visually detectable effects are listed in Supplemental Table S3. Using this criterion, our list of depletion conditions, which were found to statistically significantly induce irregular nuclear morphologies, was narrowed down to 33 genes, encoding a diverse array of proteins (Supplemental Table S3 and Supplemental Figure S5).
The degree of cellular spreading drives nuclear flattening and the assembly of actomyosin networks further generate mechanical force on the nuclear surface which can drive dysmorphia (Takaki et al., 2017). In addition, local motion of the cellular membrane can induce cancer nuclear deformations in 2-D cultures (Lele et al., 2018; Tocco et al., 2018; Kent et al., 2019). Consequently, potential confounding effects of gene depletions on cellular spreading may indirectly impact nuclear shape (Lele et al., 2018). We therefore quantified cell spreading area in a random subset of gene depletions. We found no significant correlation between cell spreading area and EFC ratio (Supplemental Figure S1A). Additionally, EFC ratio did not correlate significantly with nuclear aspect ratio (Figure 2E) or with nuclear cross-sectional area (Figure 2F). Nuclear contour irregularities, quantified by the EFC ratio, therefore appear largely independent of effects of siRNA transfection on cell spreading (Neelam et al., 2016).
Validation with confocal fluorescence microscopy
We next performed a limited confocal imaging analysis at 60× magnification (1.49 NA) on a subset of cells depleted for a random subset of 20 genes (Supplemental Table S4 and Supplemental Figure S6), 15 of which caused statistically significant effects on the EFC ratio in the high-throughput screen and 5 genes that had no effect on the EFC ratio (Table 1). As these experiments were done in 96-well plates (as opposed to the 384-well format in the high-throughput approach), we reconfirmed knockdown of gene expression using RT-qPCR in a subset of siRNA transfections (Supplemental Figure S1, B and C, for RT-qPCR results of genes which did or did not produce a statistically significant effect on EFC ratio, respectively). A series of confocal z-stack images were taken for each siRNA condition and used to create maximum intensity projections of nuclei, which were then used to calculate EFC ratios. Of siRNA conditions that were classified in the high-throughput screen to decrease EFC ratios (Supplemental Table S5), 60% were confirmed by confocal analysis to have the same effect (compare Table 1 with Table 2; Supplemental Table S6). On the other hand, 100% of siRNA conditions identified to have no effect on nuclear shape in the high-throughput screen (Supplemental Table S7) were confirmed to have no effect by confocal analysis (compare Table 1 with Table 2; Supplemental Table S8). These results validate the efficacy of the high-throughput screen in detecting effects on nuclear shape. Also, because siRNA knockdown efficiencies can be variable, we used three sets of siRNA smart pools that targeted unique sequences different from our original experiments (Supplemental Table S9) toward three top hits, TP53, DIDO1, and MTA2, and reperformed the knockdowns. We confirmed that the siRNA treatment caused a depletion of the mRNA levels for these genes (Supplemental Figure S2A) and also found as before that this depletion caused a statistically significant decrease in the EFC ratio (Supplemental Figure S2B).
TABLE 1:
Summary of results from the high-throughput screen for a subset of 21 randomly selected genes to be screen using confocal microscopy.
| High-throughput screen results | ||||
|---|---|---|---|---|
| Gene name | Number of nuclei | Mean EFC ratio | Standard error | Statistically significant? |
| CXXC1 | 805 | 4.85 | 0.07 | Yes |
| HIST1H3G | 1823 | 4.97 | 0.04 | Yes |
| HIST1H1T | 533 | 5.00 | 0.07 | Yes |
| SUZ12 | 807 | 5.03 | 0.06 | Yes |
| WSB2 | 1524 | 5.08 | 0.05 | Yes |
| MTA2 | 909 | 5.11 | 0.07 | Yes |
| ZRANB3 | 692 | 5.25 | 0.08 | Yes |
| INO80B | 1795 | 5.29 | 0.05 | Yes |
| FOXP1 | 505 | 5.34 | 0.10 | Yes |
| DIDO1 | 687 | 5.35 | 0.09 | Yes |
| ARID4A | 1015 | 5.37 | 0.06 | Yes |
| MAP3K7 | 2048 | 5.42 | 0.05 | Yes |
| TAF7 | 850 | 5.43 | 0.08 | Yes |
| RAD54L2 | 1946 | 5.48 | 0.04 | Yes |
| TP53 | 139 | 6.17 | 0.23 | Yes |
| RAD51 | 305 | 6.20 | 0.14 | No |
| LEO1 | 1283 | 6.30 | 0.07 | No |
| GADD45A | 902 | 6.83 | 0.08 | No |
| SCRAM | 15677 | 6.90 | 0.04 | N/A |
| FBL | 1036 | 6.92 | 0.08 | No |
| SMARCB1 | 819 | 7.04 | 0.09 | No |
Statistical significance of gene depletion EFC ratios, relative to the scrambled siRNA control, was determined by the two-tailed Bonferroni-corrected nonparametric Dunn’s test for p < 0.05.
TABLE 2:
Summary of results from the confocal microscopy screen for a subset of 21 randomly selected genes.
| Confocal screen results | |||||
|---|---|---|---|---|---|
| Gene name | Number of nuclei | Mean EFC ratio | Standard error | Statistically significant? | Hit in the high-throughput screen? |
| MTA2 | 176 | 4.62 | 0.15 | Yes | Yes |
| TAF7 | 255 | 5.07 | 0.10 | Yes | Yes |
| FOXP1 | 153 | 5.19 | 0.16 | Yes | Yes |
| DIDO1 | 146 | 5.14 | 0.15 | Yes | Yes |
| SUZ12 | 164 | 5.19 | 0.15 | Yes | Yes |
| TP53 | 235 | 5.35 | 0.13 | Yes | Yes |
| ARID4A | 176 | 6.01 | 0.20 | Yes | Yes |
| RAD54L2 | 165 | 5.49 | 0.13 | Yes | Yes |
| CXXC1 | 199 | 5.93 | 0.17 | Yes | Yes |
| MAP3K7 | 137 | 5.97 | 0.15 | No | Yes |
| FBL | 246 | 6.26 | 0.13 | No | No |
| RAD51 | 188 | 6.33 | 0.16 | No | No |
| WSB2 | 492 | 6.38 | 0.11 | No | Yes |
| HIST1H1T | 209 | 6.31 | 0.15 | No | Yes |
| HIST1H3G | 210 | 6.80 | 0.18 | No | Yes |
| ZRANB3 | 209 | 6.63 | 0.14 | No | Yes |
| LEO1 | 202 | 6.85 | 0.14 | No | No |
| SCRAM | 467 | 6.93 | 0.11 | N/A | N/A |
| GADD45A | 148 | 7.09 | 0.16 | No | No |
| SMARCB1 | 158 | 7.48 | 0.21 | No | No |
| INO80B | 124 | 7.79 | 0.23 | No | Yes |
Statistical significance of gene depletion EFC ratios, relative to the scrambled siRNA control, was determined by the two-tailed Bonferroni-corrected nonparametric Dunn’s test for p < 0.05.
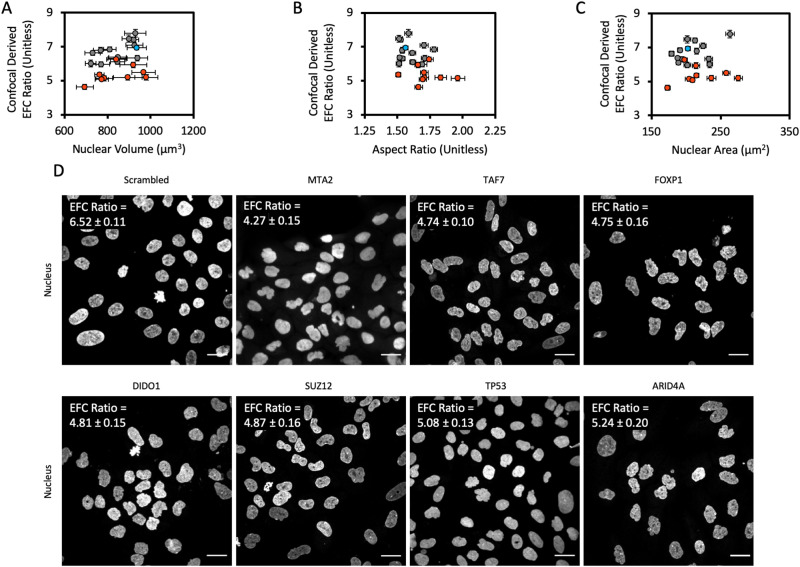
As nuclear volume is an important morphological parameter that can change during cancer progression, we quantified nuclear volume from confocal fluorescence images using our previously published methods (Tocco et al., 2018). We found no correlation between the EFC ratio and the nuclear volume (Figure 3A). Furthermore, in agreement with the results of the high-throughput screen, we found no correlation between the EFC ratio and the nuclear aspect ratio nor between the EFC ratio and the 2-D nuclear cross-sectional area (Figure 3, B and C). These results confirm the conclusions from the high-throughput screen and highlight the fact that the EFC ratio is a specific readout of nuclear contour irregularities.
FIGURE 3:
Confocal screen of selected genes from the high-throughput screen. (A) Mean EFC ratio plotted against mean 3-D nuclear volume. Nuclear volume was calculated from 3-D reconstructions created in ImageJ from x-y confocal scans at discrete z-focal planes, as has previously been described (Tocco et al., 2018). (B) Mean EFC ratio plotted against mean 2-D nuclear X-Y aspect ratio. (C) Mean EFC ratio plotted against mean nuclear cross-sectional area for a subset of genes screened by confocal microscopy. Gray data points represent gene depletions which were determined to have a statistically insignificant effect on EFC ratio by the two-tailed Bonferroni-corrected nonparametric Dunn’s test; orange data points represent gene depletions which had a statistically significant effect on EFC ratio (p < 0.05; all comparisons relative to the scrambled siRNA negative control). Blue data point denotes the scrambled siRNA negative control. Error bars are SEM. N = 3, n ≥ 127 for all data shown. (D) Representative images of the nucleus for chosen gene depletions. MTA2, TAF7, FOXP1, SUZ12, DIDO1, T53, and ARID4A were chosen based on statistically significant low EFC ratios caused by the corresponding siRNA transfection in the confocal screen. Scale bar is 25 µm. The EFC ratio in the upper left hand corner of each image panel represents the mean EFC ratio for that gene depletion condition, plus or minus the SEM (N = 3 for all gene depletion conditions; n = 478, 187, 268, 159, 178, 146, 241, and 176 for Scrambled, MTA2, TAF7, FOXP1, SUZ12, DIDO1, T53, and ARID4A, respectively).
To highlight the efficacy of the nuclear EFC ratio as a readout of nuclear contour, representative maximum projection images of nuclei for the scrambled control and the top seven gene depletion conditions from the confocal screen are shown in Figure 3D. Notably, depletion of each of these six genes with the lowest mean EFC ratios in the confocal screen produced visually pronounced nuclear aberrations compared with the scrambled siRNA negative control (compare MTA2, TP53, and Scrambled in Figure 3D and Table 2). Overall, these results significantly expand the list of epigenetic regulators that impact nuclear shape.
DISCUSSION
Nuclear pleomorphisms are characteristic of many types of cancers and have been used both diagnostically and prognostically in clinical settings (Papanicolaou and Traut, 1997; de Las Heras and Schirmer, 2014). Recent studies suggest that alterations to chromatin and epigenetic regulators may cause abnormal nuclear shapes (Imbalzano et al., 2013; Furusawa et al., 2015; Schreiner et al., 2015; Stephens et al., 2017, 2018a; Senigagliesi et al., 2019) associated with nuclear dysfunction (Stephens et al., 2019a). In this study, we used noncancerous MCF-10A breast epithelial cells in conjunction with high-throughput nuclear imaging and knockdown using an RNAi library to systematically screen epigenetic regulators of nuclear shape. The screen successfully revealed a number of genes encoding epigenetic regulatory proteins which caused nuclear shape abnormalities on depletion. Our findings that chromatin regulatory genes are implicated in abnormal nuclear shaping are in agreement with previous reports that knockdown of the SWI/SNF chromatin remodeling complex ATPase BRG1 (encoded by SMARCA4) increases the frequency of irregular nuclei (Imbalzano et al., 2013). A commonly used metric for quantifying nuclear shape abnormalities is solidity. We found that solidity does detect differences between nuclear shapes as expected. However, in comparison with solidity, the EFC ratio has a larger range of values that should allow more facile discrimination between normal and abnormal nuclear shapes. Another metric commonly used for nuclear morphology is roundness or circularity (defined as (4π*area)/(perimeter2)). The circularity metric for the genes in our confocal images similarly has a relatively narrow range of values (Supplemental Figure S3) and a weak correlation with the EFC ratio. These results suggest that the EFC ratio is a sensitive measure of nuclear shape abnormalities.
Our gene screen reveals one of the most well-known cancer-related genes, TP53, as a regulator of nuclear shape. TP53 is mutated in 92.98% of uterine carcinomas, 91.51% of ovarian serous cystadenocarcinomas, and 36.51% of all breast cancers with most mutations being loss-of-function (Grossman et al., 2016). A role for p53 in nuclear shape determination is consistent with an earlier study which reported increased nuclear shape variability in high-grade tumors in which p53 was frequently mutated (associated with loss-of-function) compared with low-grade tumors with infrequent p53 mutations (Friedrich et al., 1997). More recently, it was shown that p53 knockdown can cause nuclear rupture (Yang et al., 2017). Interestingly, another study has recently demonstrated that overexpression or activation of p53 induces nuclear deformation reminiscent of the irregular nuclear shapes found in HGPS cells and induces cellular senescence (Yoon et al., 2019). Our findings thus corroborate several studies which have linked dysregulation of p53 with aberrant nuclear shapes in a novel cell type.
Top hits that satisfied the EFC ratio less than ∼5.3 criterion in the high-throughput screen included TCF19, KMT5C, YAF2, RBBP5, POGZ, SIN3A, and CXXC1 (Supplemental Table S3). Our choice of a mean EFC ratio of below approximately 5.3 was based on visually noticeable contour irregularities, quantified by four independent observers, binned into binary “regular” and “irregular” groups based on the presence of concavities in the nuclear contour. If we were to not impose this cutoff, then the number of genes that induce statistically significant nuclear abnormalities is much larger. In the absence of a cutoff, of the 608 siRNAs screened (Supplemental Table S1), 71% (429/608 genes) significantly affected nuclear EFC ratios (mean EFC range of 4.85 ± 0.07 to 9.46 ± 0.09) compared with the scrambled control (mean EFC of 6.90 ± 0.04; Supplemental Table S2). A majority (58% of gene depletions or 354/608; Supplemental Table S5) decreased EFC ratio (indicating an increase in irregular nuclear shapes), whereas about 12% of gene depletions increased EFC ratio (Figure 2D and Supplemental Table S10). The reason this high-throughput screen is sensitive enough to detect subtle changes in nuclear abnormalities is that per gene depletion, hundreds of nuclei were analyzed on average, which vastly increases the statistical power. Genes which significantly decreased EFC ratio included histone variants, histone acetyltransferases, histone deacetylases, DNA binding proteins, transcription factors, and members of prominent chromatin remodeling complexes. However, it is likely that very subtle effects on nuclear shape, which are not immediately obvious to the eye, may not be physiologically relevant.
Interestingly, our high-throughput screen for epigenetic regulators of nuclear shape revealed several families of genes and protein complexes whose individual components have effects on nuclear shape. For instance, in the high mobility group nucleosomal binding domain (HMGN) family of genes, we found that depletion of HMGN2 significantly decreases EFC ratio (Supplemental Table S5), while HMGN3 significantly increased the EFC ratio (Supplemental Table S10). Members of this family of genes have previously been identified to regulate nuclear mechanics. For example, overexpression of HMGN5 was shown to decrease nuclear stiffness in mouse cardiomyocytes (Rochman et al., 2010; Furusawa et al., 2015), and expression of HMG-I/HMG-Y inversely correlates with nuclear stiffness in breast cancer (Senigagliesi et al., 2019). Interestingly, depletion of the HMGN5 was not found to significantly affect nuclear shape in our high-throughput screen of nuclear shape in MCF10A cells (Supplemental Table S7); this is in contrast to another study in which overexpression of the HMGN5 induced irregular nuclear shapes, increased nuclear size, and decreased nuclear rigidity in mouse cardiomyocytes (Rochman et al., 2010; Furusawa et al., 2015). The high-throughput screen also revealed that depletion of specific subunits of the ATP-dependent INO80 chromatin remodeling complex has differential effects on EFC ratio. Depletion of subunits encoded by INO80E and INO80D was found to significantly decrease EFC ratio (Supplemental Table S5), while depletion of INO80B was found to significantly increase EFC ratio (Supplemental Table S10).
Our results also implicate members of the histone family in regulation of nuclear shape. H1F0, which encodes the linker histone H1.0, has been shown to be down-regulated in cancer self-renewing cells in multiple cancer types. Loss of this particular histone was identified to destabilize AT-rich genomic regions thereby activating oncogenic pathways and has been correlated with worse patient outcomes across multiple cancers (Torres et al., 2016). While Torres et al. (2016) did not investigate nuclear shape, depletion of H1.0 significantly decreased the EFC ratio (mean EFC ratio of 5.40 ± 0.05) compared with the scrambled negative control (mean EFC ratio of 6.90 ± 0.04) in our high-throughput screen (Supplemental Table S7). It is possible that destabilization of AT-rich genomic regions by loss of H1.0 may induce these nuclear shape irregularities.
Validation of a subset of genes from the high-throughput screen using confocal microscopy revealed a number of genes which cause significant nuclear abnormalities on depletion. Top hits include MTA2, TAF7, FOXP1, SUZ12, DIDO1, TP53, RAD54L2, and CXXC1. Of these, MTA2 had the lowest mean EFC ratio of 4.62 ± 0.15 (mean EFC ratio of scrambled siRNA negative control was 6.93 ± 0.12) and resulted in visually apparent nuclear aberrations (Figure 3D; Supplemental Table S6). Similarly, SUZ12, DIDO1, and TP53 were also among top hits which resulted in visually aberrant nuclear morphologies (Figure 3D). SUZ12 tends to be overexpressed in cancer and is a subunit of the PRC2 complex that creates the repressive histone 3 lysine residue 27 trimethyl mark with the central enzymatic component EZH2; changes in nuclear shape might result from dysfunction of the PRC2 complex. DIDO1 located on chromosome 20q is a region that is frequently deleted in myeloproliferative neoplasms (Futterer et al., 2005). DIDO1 was recently shown to recognize the histone 3 lysine residue 4 trimethylation mark in a pH-dependent manner through a PHD domain (Tencer et al., 2017) and was identified as a possible germline variant in familial colorectal cancer (Thutkawkorapin et al., 2019). In a pan-cancer analysis in cBioPortal (Cerami et al., 2012; Gao et al., 2013), DIDO1 is mutated in ∼4% of cases, with frequent truncating mutations. Surprisingly, knockdown of lamin A/C, which is encoded by the LMNA gene, did not have comparably large effects on the mean EFC ratio in MCF-10A cells, even though the effects were statistically significant. The mean EFC ratio was 6.49 ± 0.04 for LMNA compared with 6.90 ± 0.04 for the scrambled siRNA in the high-throughput screen and 6.10 ± 0.13 for LMNA compared with 6.93 ± 0.12 for the scrambled siRNA in the confocal screen.
Since nuclear shape abnormalities can be induced purely by altering chromatin histone modification state (Stephens et al., 2018a), it is possible that a number of epigenetic regulators discovered in this paper impact the nucleus through modifications of chromatin structure. Given the significant number of genes revealed by our screen that can regulate nuclear shape, the mechanisms by which nuclear shape becomes abnormal in cancer are likely to be highly complex. Understanding the underlying mechanisms remains a central challenge.
MATERIALS AND METHODS
Cell culture
All cell types were maintained at 37°C in a humidified 5% (vol/vol) CO2 environment during culture. Human breast fibrocystic disease MCF-10A cells (ATCC CRL-10317) were cultured in DMEM/F-12 (Thermo Fisher Scientific) media supplemented with 5% donor horse serum (Fisher Scientific), 100 ng/ml cholera toxin (Sigma-Aldrich), 20 ng/ml epidermal growth factor (PeproTech Online), 0.5 mg/ml hydrocortisone (Sigma-Aldrich), 100 ng/ml insulin (Sigma-Aldrich), and 1% penicillin and streptomycin (Fisher Scientific). Human breast adenocarcinoma MDA-MB-231 cells were cultured in DMEM (Thermo Fisher Scientific) supplemented with 10% donor bovine serum (Invitrogen), 1% MEM nonessential amino acids (Corning), 1% 200 mM L-glutamine (Fisher Scientific), and 1% penicillin and streptomycin (Fisher Scientific). Cell cultures were maintained in T-75 flasks prior to experiments. To begin the high-throughput experiment, cells were trypsinized, counted, and diluted to 1 × 104 cells/ml in fresh cell media; 50 µl of this cell suspension were dispensed into each well of 384-well plates using a Matrix WellMate automatic cell dispenser (Thermo Fisher Scientific). Cells were allowed to spread overnight prior to transfection. For the confocal microscopy screen, cells were seeded manually at a density of 2000 cells per well into 96-well plates and allowed to adhere and spread overnight prior to transfection. MCF-10A cells were periodically validated with short tandem repeat analysis at the Interdisciplinary Center for Biotechnology at the University of Florida and checked for mycoplasma contamination.
Transfection of siRNAs
For high-throughput siRNA interference, MCF-10A cells were seeded into 384-well plates at a density of 500 cells/well in appropriate culture medium and incubated for 24 h. This seeding density was chosen to account for the mild cytotoxicity associated with transfection while ensuring that the cells were not confluent at the time of fixation. Prior to transfection, cells were washed once with 1× phosphate-buffered saline (PBS; Corning) and replenished with appropriate antibiotic-free culture medium. siRNA transfections were carried out using Dharmafect 1 (Dharmacon) in OptiMEM serum-free media (Thermo Fisher Scientific) per the manufacturer’s protocols, diluting to a final siRNA concentration of 25 nM. Posttransfection, cells were incubated for 72 h to allow siRNA-mediated interference and turnover of endogenous proteins to occur. The siGENOME SMARTpool siRNA library targeting 608 genes encoding epigenetic-associated proteins was obtained from Dharmacon. One additional condition was screened in quadruplicate wells per 384-well plate which served as a negative control: scrambled siRNA control (negative control; Dharmacon). For the confocal screen, cells were transfected for 72 h using the protocol described above, scaling initial cell seeding density and volumes proportionally to the increased culture dish surface area. For the siRNA optimization experiments, cells were seeded at an initial seeding density of 400 cells per well in 384-well plates and transfected using the protocol described above, varying final siRNA transfection concentration. Posttransfection, cells were incubated for 120 h to allow for siRNA-mediated interference and turnover of endogenous proteins to occur. A final siRNA transfection concentration of 25 nM was chosen to maximize development of phenotype while minimizing cell death associated with transfection (data not shown).
Fixing, immunofluorescence, and confocal imaging
MCF-10A cells in 384-well plates were fixed following the 72 h siRNA incubation period using an equal volume amount of 8% paraformaldehyde (Alfa Aesar) in PBS and added directly to the culture medium present in the well for 15 min. Cells in 96-well plates were washed once with 1× PBS (Corning) and then fixed using 4% paraformaldehyde (Alfa Aesar) diluted in 1× PBS. Cells in 384-well plates were then washed with 1× PBS using a Matrix WellMate high-throughput pipetting system (Thermo Fisher Scientific). Cells in 96-well plates were manually washed with 1× PBS. Cells were then simultaneously permeabilized and blocked using a permeabilization buffer consisting of 0.1% Triton X-100 (Sigma-Aldrich) supplemented with 1% bovine serum albumin (Fisher Scientific) for 45 min. Primary antibody toward E-cadherin (ab40772; Abcam) was diluted in the permeabilization buffer described above and used at 1:500 concentration. Following incubation at 4°C overnight in the dark, cells were washed once with 1× PBS and incubated with goat anti-rabbit Alexa Fluor 594 (Life Technologies) diluted in permeabilization buffer to a final concentration of 1:500 for 1 h at room temperature in the dark. Cells were then stained with Hoechst 33342 (Thermo Fisher Scientific) diluted to 1:200 and/or Alexa Fluor 594 Phalloidin (Thermo Fisher Scientific) diluted to 1:40 in 1× PBS for 1 h at room temperature in the dark. For confocal imaging, fixed and stained nuclei were imaged using a Nikon A1 laser scanning confocal microscope (Nikon), equipped with a 60× (1.49 NA) oil immersion objective and Nikon DU-4 CCD camera using Nikon NIS-Elements software. Z-stacks were acquired using an axial step size of 0.3 µm, and both maximum intensity projections and sum-of-slice projections were generated in ImageJ (National Institutes of Health [NIH]).
High-throughput imaging and image preprocessing
The 384-well plates were imaged using an Operetta CLS high-content imaging system (PerkinElmer) equipped with a 40× (NA of 0.95) air objective (PerkinElmer, #HH12000507). A total of 20 images, spanning the majority of the available well surface area excluding edges, were taken for each well; a minimum of three biological replicates were imaged for each siRNA condition. Images were corrected for illumination gradients using a custom CellProfiler (Carpenter Lab; Carpenter et al., 2006; Kamentsky et al., 2011) script, and both out-of-focus and mitotic nuclei were subsequently masked using a set of exclusion criteria generated from a machine learning algorithm in CellProfiler Analyst (Jones et al., 2009). For each masking process, a 15-parameter exclusion criterion was generated using a random forest classifier and training sets of approximately 800 nuclei, manually classified as either “in-focus” or “out-of-focus/mitotic.” The final images, corrected for uneven illumination and out-of-focus and mitotic nuclei, were used for all image analyses.
Nuclear morphometric analysis
To compute the EFC ratio in the high-throughput screen, raw images of Hoechst 33342-labeled nuclei imaged at the brightest equatorial plane were directly loaded into MATLAB (MATLAB 2018a and Image Processing Toolbox; MathWorks Inc) for processing. To compute the EFC ratio from confocal images, z-stack images were first loaded into ImageJ. Maximum-projection images of the Hoechst 33342 channel were then generated and saved. These maximum-projection images of the Hoechst channel were then loaded into MATLAB for processing.
Images were loaded into MATLAB (MATLAB2018a and Image Processing Toolbox; MathWorks Inc), and a built-in function was used to saturate the top and bottom 1% of all pixel values to improve image contrast. Nuclei touching the border of the image were then removed, and the image was segmented using an Otsu segmentation algorithm which automatically thresholds each image. A pixel area-size exclusion filter with empirically determined upper and lower bounds was then applied to segmented objects to exclude small imaging artifacts and large masses of touching nuclei which confound segmentation. Masks of the resulting nuclei were obtained and applied back onto the original image which was then fit with a Fourier decomposition using a custom MATLAB script. Elliptical Fourier analysis approximates nuclear shapes through decomposition of the shape into a series of harmonic ellipses (Diaz et al., 1989; Lammerding et al., 2006). The first four coefficients of each harmonic ellipse were used to calculate the major and minor axes of the harmonic ellipse fit. Nuclear contours were fit using a total of 15 harmonic ellipses, which were found to accurately approximate even the most complex, irregular nuclei. An EFC ratio was then calculated by the sum of the major and minor axes of the first harmonic ellipse, divided by the sum of the major and minor axes for the subsequent 14 harmonic ellipses. Physically, this measurement represents a relative ratio of the fit of the first harmonic ellipse to the following 14 harmonic ellipses; therefore, regular nuclear shapes (circular) will have larger EFC ratios than irregular nuclear shapes (nuclei containing blebs, invaginations, etc.).
Due to the sensitivity of elliptical Fourier analysis to subtle variations in nuclear contours, we opted to forego watershed algorithms which are traditionally employed to discern boundaries between closely bordering cells or nuclei in our method as inaccuracies in segmentation could propagate throughout the analysis and bias results. This analysis algorithm is ideal for high-throughput screening applications where image and cell numbers screened will be large to compensate for excluded nuclei.
To compute solidity (where solidity = area/convex area), raw images of Hoechst 33342-labeled nuclei imaged at the brightest equatorial plane were loaded into ImageJ. Nuclei were thresholded using the default ImageJ threshold algorithm, and holes in the resulting binary image were filled. ImageJ’s “Analyze Particles” feature was then used to quantify the shape descriptor solidity for each nucleus in Figure 1A. Adjusted solidity was calculated according to the following formula ([1-solidity]*100) to allow a measure of solidity to be plotted on the same axes as EFC ratio.
To compute circularity, defined as (4π*area)/(perimeter2), nuclear areas and nuclear perimeters were first measured from the final nuclear contour detected after the image processing algorithm described above for use in calculation of the EFC ratio in MATLAB. Circularity was then computed in MATLAB per the described definition and used for subsequent statistical analysis.
Cell spreading area and nuclear volume measurements
Cell spreading area was quantified for several gene depletions by manually tracing the boundary of individual cells in ImageJ and using ImageJ’s built-in measurement function. E-cadherin immunostained images were used to discern individual cell boundaries and quantify spreading area. The coordinates of the final traced cell boundary region of interest were saved and subsequently used to crop the Hoechst 33342-stained nucleus corresponding to the measured cell. These cropped images were then analyzed using the method described above to obtain corresponding EFC ratios. Automated algorithms were not used to detected cell boundaries due to difficulties in segmenting cells in monolayer cultures.
To measure nuclear volume, images of individual nuclei stained with Hoechst 33342 were cropped in ImageJ and nuclear volumes were measured using the “3D Objects Counter” tool after application of an intensity threshold calculated at the equatorial x-y focal plane of the nucleus, applied to all subsequent x-y planes in the z-stack image.
Validation of control siRNA knockdown with RT-qPCR
RT-qPCR experiments were run in parallel to the bulk RNAi screen to ensure that knockdown occurred in every plate screened with the following 11 randomly chosen genes: LMNA, SMARCA4, HIST1H1T, ZRNAB3, INO80B, RAD51, GADD45A, CXXC1, HIST1H3G, TP53, and SMARCB1. These genes were further tested for knockdown for the confocal screen as well, with one additional gene target, DIDO1. All SMARTpool siRNAs were obtained from Dharmacon (see Supplemental Table S1 for information on the siRNAs, including sequences targeted).
Whole cells were lysed using the SingleShot Probes One-Step Kit (Bio-Rad), designed for low-cell number inputs, 72 h posttransfection. Lysates were then combined with complete RT-qPCR reaction mix per the manufacturer’s protocol and predesigned fluorogenic probes targeting LMNA, SMARCA4, or GAPDH (Bio-Rad). Reactions were processed on a CFX-96 Real-time PCR detection system (Bio-Rad) using the following thermocycler conditions: 10 min at 50°C, 3 min at 95°C, followed by 40 cycles of 15 s at 95°C and 30 s at 60°C. All results were analyzed using the 2-∆∆Ct method and normalized against GAPDH expression (Livak and Schmittgen, 2001).
Statistics/replicates
Confidence in the EFC ratios was determined using a Kruskal–Wallis one-way analysis of variance nonparametric statistical test (Kruskal and Wallis, 1952) to determine if any of the measured gene conditions originated from a different distribution than other gene depletion conditions; specifically, comparisons to the scrambled siRNA negative control condition were used for interpretation of statistical signfiicance. Data derived from biological samples and cell culture experiments rarely follow normal distributions and, when maintaining consistent microscopy image acquisition conditions, rarely allow for equal sample numbers between conditions. Therefore, the Kruskal–Wallis one-way analysis of variance test was used for most analyses as this test does not assume equal sample sizes between conditions or that samples follow a specific distribution. Subsequently, a two-tailed Dunn’s test which allows for multiple comparisons was used to determine which, if any, of the gene depletion conditions originated from a different distribution than any other gene depletion condition; again, specifically, comparison to the scrambled siRNA negative control condition was used to determine statistical significance for our experiments. As the high-throughput screen has many unique gene depletions (genes screened), familywise error rates were corrected for using a two-tailed Bonferroni correction to a significance level of 5%. For our analysis, we were specifically interested if experimental conditions originated from a different distribution than our scrambled siRNA negative control condition. However, as all possible comparisons between conditions were still made and type I error was corrected for using Bonferroni corrections, our analysis represents a conservative listing of conditions identified to influence nuclear morphology. We note that there is high statistical power due to the large sample numbers in the high-throughput screen.
Supplementary Material
Acknowledgments
This work was supported by the NIH (R01 EB014869 to T.P.L.; K99GM123195 to A.D.S.), the National Cancer Institute (R01 CA172310 to H.L.; U54CA193419 to J.D.L), the National Cancer Institute Research Specialist Award (R50CA211487 to R.R.), and a supplement from the Northwestern PSOC to T.P.L. and J.D.L.
Abbreviations used:
- EFC
elliptic Fourier coefficient
- HGPS
Hutchinson-Gilford progeria syndrome
- PBS
phosphate-buffered saline
- RMS
root mean-square
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-09-0520) on April 22, 2020.
REFERENCES
- Abdalla F, Boder J, Markus R, Hashmi H, Buhmeida A, Collan Y. (2009). Correlation of nuclear morphometry of breast cancer in histological sections with clinicopathological features and prognosis. Anticancer Res , 1771–1776. [PubMed] [Google Scholar]
- Bolt MJ, Stossi F, Callison AM, Mancini MG, Dandekar R, Mancini MA. (2015). Systems level-based RNAi screening by high content analysis identifies UBR5 as a regulator of estrogen receptor-alpha protein levels and activity. Oncogene , 154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain NE, Tapley EC, McDonald KL, Cain BM, Starr DA. (2014). The SUN protein UNC-84 is required only in force-bearing cells to maintain nuclear envelope architecture. J Cell Biol , 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capo-chichi CD, Cai KQ, Simpkins F, Ganjei-Azar P, Godwin AK, Xu XX. (2011a). Nuclear envelope structural defects cause chromosomal numerical instability and aneuploidy in ovarian cancer. BMC Med , 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capo-chichi CD, Cai KQ, Smedberg J, Ganjei-Azar P, Godwin AK, Xu XX. (2011b). Loss of A-type lamin expression compromises nuclear envelope integrity in breast cancer. Chin J Cancer , 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol , R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov , 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Wang W, Ribeiro AJ, Kalinowski A, Gregg SQ, Opresko PL, Niedernhofer LJ, Rohde GK, Dahl KN. (2011). Computational image analysis of nuclear morphology associated with various nuclear-specific aging disorders. Nucleus , 570–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Las Heras JI, Schirmer EC. (2014). The nuclear envelope and cancer: a diagnostic perspective and historical overview. Adv Exp Med Biol , 5–26. [DOI] [PubMed] [Google Scholar]
- De Vos WH, Houben F, Hoebe RA, Hennekam R, van Engelen B, Manders EM, Ramaekers FC, Broers JL, Van Oostveldt P. (2010). Increased plasticity of the nuclear envelope and hypermobility of telomeres due to the loss of A-type lamins. Biochim Biophys Acta , 448–458. [DOI] [PubMed] [Google Scholar]
- Denais C, Lammerding J. (2014). Nuclear mechanics in cancer. Adv Exp Med Biol , 435–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond DA, Berry SJ, Umbricht C, Jewett HJ, Coffey DS. (1982). Computerized image-analysis of nuclear shape as a prognostic factor for prostatic-cancer. Prostate , 321–332. [DOI] [PubMed] [Google Scholar]
- Diaz G, Zuccarelli A, Pelligra I, Ghiani A. (1989). Elliptic fourier analysis of cell and nuclear shapes. Comput Biomed Res , 405–414. [DOI] [PubMed] [Google Scholar]
- Epstein JI, Berry SJ, Eggleston JC. (1984). Nuclear roundness factor - a predictor of progression in untreated stage A2 prostate-cancer. Laboratory Investigation , A18-A18. [DOI] [PubMed] [Google Scholar]
- Friedrich K, Dimmer V, Haroske G, Meyer W, Theissig F, Thieme B, Kunze KD. (1997). Morphological heterogeneity of p53 positive and p53 negative nuclei in breast cancers stratified by clinicopathological variables. Anal Cell Pathol , 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa T, Rochman M, Taher L, Dimitriadis EK, Nagashima K, Anderson S, Bustin M. (2015). Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat Commun , 6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futterer A, Campanero MR, Leonardo E, Criado LM, Flores JM, Hernandez JM, San Miguel JF, Martinez AC. (2005). Dido gene expression alterations are implicated in the induction of hematological myeloid neoplasms. J Clin Invest , 2351–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal , pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil J, Wu H, Wang BY. (2002). Image analysis and morphometry in the diagnosis of breast cancer. Microsc Res Tech , 109–118. [DOI] [PubMed] [Google Scholar]
- Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R, et al. (2004). Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA , 8963–8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, Staudt LM. (2016). Toward a shared vision for cancer genomic data. N Engl J Med , 1109–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. (2013). Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell , 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JC, Zhou T, Lai WH, Huang Y, Chan YC, Li X, Wong NL, Li Y, Au KW, Guo D, et al. (2011). Generation of induced pluripotent stem cell lines from 3 distinct laminopathies bearing heterogeneous mutations in lamin A/C. Aging (Albany NY) , 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsman M, Hulshof F, Unadkat H, Papenburg BJ, Stamatialis DF, Truckenmuller R, van Blitterswijk C, de Boer J, Reinders MJT. (2015). Analysis of high-throughput screening reveals the effect of surface topographies on cellular morphology. Acta Biomaterialia , 29–38. [DOI] [PubMed] [Google Scholar]
- Imbalzano KM, Cohet N, Wu Q, Underwood JM, Imbalzano AN, Nickerson JA. (2013). Nuclear shape changes are induced by knockdown of the SWI/SNF ATPase BRG1 and are independent of cytoskeletal connections. PLoS One , e55628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto J, Pfeifer CR, Ivanovska IL, Swift J, Discher DE. (2016). Nuclear lamins in cancer. Cell Mol Bioeng , 258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagielska A, Lowe AL, Makhija E, Wroblewska L, Guck J, Franklin RJM, Shivashankar GV, Van Vliet KJ. (2017). Mechanical strain promotes oligodendrocyte differentiation by global changes of gene expression. Front Cell Neurosci , 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TR, Carpenter AE, Lamprecht MR, Moffat J, Silver SJ, Grenier JK, Castoreno AB, Eggert US, Root DE, Golland P, Sabatini DM. (2009). Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning. Proc Natl Acad Sci USA , 1826–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE. (2011). Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics , 1179–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap A, Jain M, Shukla S, Andley M. (2018). Role of nuclear morphometry in breast cancer and its correlation with cytomorphological grading of breast cancer: a study of 64 cases. J Cytol , 41–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent IA, Zhang Q, Katiyar A, Li Y, Pathak S, Dickinson RB, Lele TP. (2019). Apical cell protrusions cause vertical deformation of the soft cancer nucleus. J Cell Physiol , 20675–20684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstandinou C, Glotsos D, Kostopoulos S, Kalatzis I, Ravazoula P, Michail G, Lavdas E, Cavouras D, Sakellaropoulos G. (2018). Multifeature quantification of nuclear properties from images of h&e-stained biopsy material for investigating changes in nuclear structure with advancing CIN grade. J Healthc Eng , 6358189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruskal WH, Wallis WA. (1952). Use of ranks in one-criterion variance analysis. J Am Stat Assoc , 583–621. [Google Scholar]
- Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, Lee RT. (2006). Lamins A and C but not lamin B1 regulate nuclear mechanics. J Biol Chem , 25768–25780. [DOI] [PubMed] [Google Scholar]
- Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. (2004). Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest , 370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lele TP, Dickinson RB, Gundersen GG. (2018). Mechanical principles of nuclear shaping and positioning. J Cell Biol , 3330–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods , 402–408. [DOI] [PubMed] [Google Scholar]
- Mijovic Z, Kostov M, Mihailovic D, Zivkovic N, Stojanovic M, Zdravkovic M. (2013). Correlation of nuclear morphometry of primary melanoma of the skin with clinicopathological parameters and expression of tumor suppressor proteins (p53 and p16(INK4a)) and bcl-2 oncoprotein. J BUON , 471–476. [PubMed] [Google Scholar]
- Neelam S, Hayes PR, Zhang Q, Dickinson RB, Lele TP. (2016). Vertical uniformity of cells and nuclei in epithelial monolayers. Sci Rep , 19689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanicolaou GN, Traut HF. (1997). The diagnostic value of vaginal smears in carcinoma of the uterus. 1941. Arch Pathol Lab Med , 211–224. [PubMed] [Google Scholar]
- Petersen I, Kotb WF, Friedrich KH, Schluns K, Bocking A, Dietel M. (2009). Core classification of lung cancer: correlating nuclear size and mitoses with ploidy and clinicopathological parameters. Lung Cancer , 312–318. [DOI] [PubMed] [Google Scholar]
- Rochman M, Malicet C, Bustin M. (2010). HMGN5/NSBP1: a new member of the HMGN protein family that affects chromatin structure and function. Biochim Biophys Acta , 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schochlin M, Weissinger SE, Brandes AR, Herrmann M, Moller P, Lennerz JK. (2014). A nuclear circularity-based classifier for diagnostic distinction of desmoplastic from spindle cell melanoma in digitized histological images. J Pathol Inform , 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner SM, Koo PK, Zhao Y, Mochrie SG, King MC. (2015). The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat Commun , 7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senigagliesi B, Penzo C, Severino LU, Maraspini R, Petrosino S, Morales-Navarrete H, Pobega E, Ambrosetti E, Parisse P, Pegoraro S, et al. (2019). The high mobility group A1 (HMGA1) chromatin architectural factor modulates nuclear stiffness in breast cancer cells. Int J Mol Sci . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu CW, Lee YK, Ho JC, Lai WH, Chan YC, Ng KM, Wong LY, Au KW, Lau YM, Zhang J, et al. (2012). Modeling of lamin A/C mutation premature cardiac aging using patient-specific induced pluripotent stem cells. Aging (Albany NY) , 803–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speight PM. (2007). Update on oral epithelial dysplasia and progression to cancer. Head Neck Pathol , 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele-Stallard HB, Pinton L, Sarcar S, Ozdemir T, Maffioletti SM, Zammit PS, Tedesco FS. (2018). Modeling skeletal muscle laminopathies using human induced pluripotent stem cells carrying pathogenic LMNA mutations. Front Physiol , 1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Banigan EJ, Adam SA, Goldman RD, Marko JF. (2017). Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol Biol Cell , 1984–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Banigan EJ, Marko JF. (2019a). Chromatin’s physical properties shape the nucleus and its functions. Curr Opin Cell Biol , 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Liu PZ, Banigan EJ, Almassalha LM, Backman V, Adam SA, Goldman RD, Marko JF. (2018a). Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol Biol Cell , 220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Liu PZ, Kandula V, Chen H, Almassalha LM, Backman V, O’Halloran T, Adam SA, Goldman RD, Banigan EJ, Marko JF. (2018b). Physicochemical mechanotransduction alters nuclear shape and mechanics via heterochromatin formation. bioRxiv 423442. [Google Scholar]
- Stephens AD, Liu PZ, Kandula V, Chen H, Almassalha LM, Herman C, Backman V, O’Halloran T, Adam SA, Goldman RD, et al. (2019b). Physicochemical mechanotransduction alters nuclear shape and mechanics via heterochromatin formation. Mol Biol Cell mbcE19050286T. [DOI] [PubMed] [Google Scholar]
- Takaki T, Montagner M, Serres MP, Le Berre M, Russell M, Collinson L, Szuhai K, Howell M, Boulton SJ, Sahai E, Petronczki M. (2017). Actomyosin drives cancer cell nuclear dysmorphia and threatens genome stability. Nat Commun , 16013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq Z, Zhang H, Chia-Liu A, Shen Y, Gete Y, Xiong ZM, Tocheny C, Campanello L, Wu D, Losert W, Cao K. (2017). Lamin A and microtubules collaborate to maintain nuclear morphology. Nucleus , 433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tencer AH, Gatchalian J, Klein BJ, Khan A, Zhang Y, Strahl BD, van Wely KHM, Kutateladze TG. (2017). A unique pH-dependent recognition of methylated histone H3K4 by PPS and DIDO. Structure , 1530–1539.e1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thutkawkorapin J, Lindblom A, Tham E. (2019). Exome sequencing in 51 early onset non-familial CRC cases. Mol Genet Genomic Med e605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocco VJ, Li Y, Christopher KG, Matthews JH, Aggarwal V, Paschall L, Luesch H, Licht JD, Dickinson RB, Lele TP. (2018). The nucleus is irreversibly shaped by motion of cell boundaries in cancer and non-cancer cells. J Cell Physiol , 1446–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres CM, Biran A, Burney MJ, Patel H, Henser-Brownhill T, Cohen AS, Li Y, Ben-Hamo R, Nye E, Spencer-Dene B, et al. (2016). The linker histone H1.0 generates epigenetic and functional intratumor heterogeneity. Science . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschuuren M, De Vylder J, Catrysse H, Robijns J, Philips W, De Vos WH. (2017). Accurate detection of dysmorphic nuclei using dynamic programming and supervised classification. PLoS One , PMID25803988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PA, Djordjevic B, Ayroud Y, Islam S, Gravel D, Robertson SJ, Parra-Herran C. (2014). Nuclear morphometry in flat epithelial atypia of the breast as a predictor of malignancy: a digital image-based histopathologic analysis. Anal Quant Cytopathol Histpathol , 305–313. [PubMed] [Google Scholar]
- Yang Z, Maciejowski J, de Lange T. (2017). Nuclear Envelope Rupture Is Enhanced by Loss of p53 or Rb. Mol Cancer Res , 1579–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon MH, Kang SM, Lee SJ, Woo TG, Oh AY, Park S, Ha NC, Park BJ. (2019). p53 induces senescence through Lamin A/C stabilization-mediated nuclear deformation. Cell Death Dis , PMID30728349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwerger M, Ho CY, Lammerding J. (2011). Nuclear mechanics in disease. Annu Rev Biomed Eng , 397–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.