Abstract
The nuclear envelope (NE) is continuous with the endoplasmic reticulum (ER), yet the NE carries out many functions distinct from those of bulk ER. This functional specialization depends on a unique protein composition that defines NE identity and must be both established and actively maintained. The NE undergoes extensive remodeling in interphase and mitosis, so mechanisms that seal NE holes and protect its unique composition are critical for maintaining its functions. New evidence shows that closure of NE holes relies on regulated de novo lipid synthesis, providing a link between lipid metabolism and generating and maintaining NE identity. Here, we review regulation of the lipid bilayers of the NE and suggest ways to generate lipid asymmetry across the NE despite its direct continuity with the ER. We also discuss the elusive mechanism of membrane fusion during nuclear pore complex (NPC) biogenesis. We propose a model in which NPC biogenesis is carefully controlled to ensure that a permeability barrier has been established before membrane fusion, thereby avoiding a major threat to compartmentalization.
INTRODUCTION
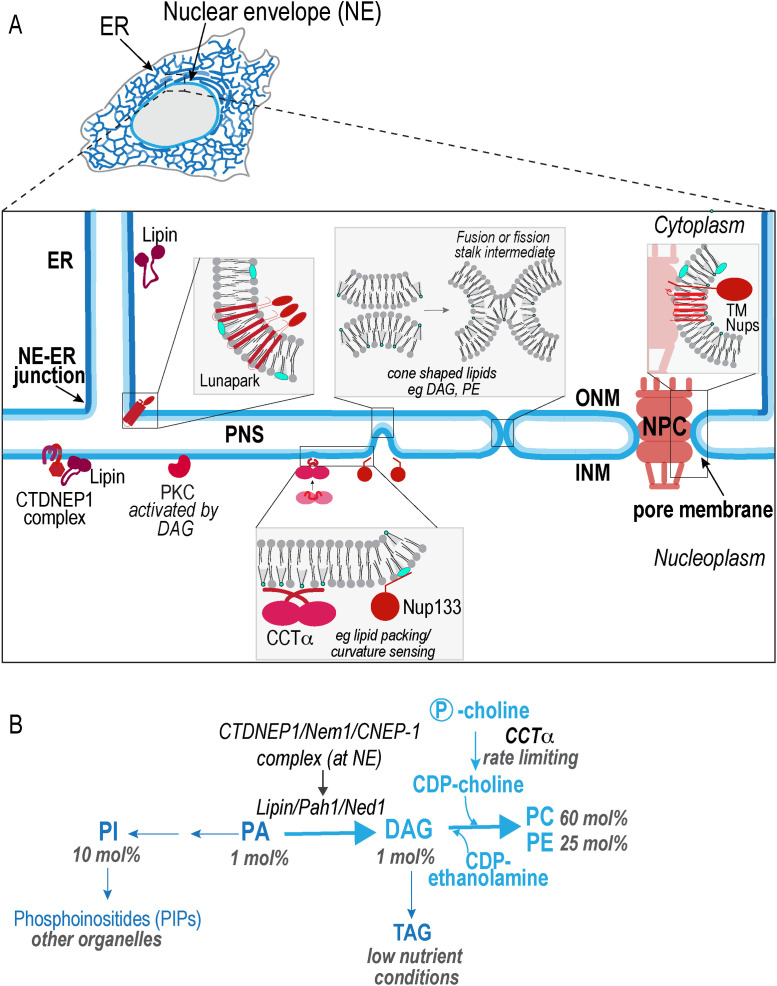
The nuclear envelope (NE) is a shared and essential feature of the endomembrane systems in all eukaryotes. It has long been appreciated that the NE is a defining structure for cellular organization as it is responsible for the separation of nucleoplasm from cytoplasm (Figure 1A) and the respective biological processes including transcription and translation that are operative therein. However, the NE is anything but a simple barrier. Instead, the inner nuclear membrane (INM) is endowed with a specialized proteome that is dedicated to a plethora of critical cellular functions (Korfali et al., 2012; Garapati and Mishra, 2018; Gerace and Tapia, 2018). These include the sensation and buffering of mechanical forces (Burke, 2018; Kirby and Lammerding, 2018), genome organization and regulation (Buchwalter et al., 2019a), and lipid metabolism (Bahmanyar et al., 2014; Bahmanyar, 2015; Barbosa et al., 2015, 2019; Haider et al., 2018; Romanauska and Kohler, 2018). From a toplogical perspective, the NE is a physical extension of the ER that encases chromatin via inner and outer nuclear membranes (INM/ONM) that have distinct protein compositions and are linked at sites of fusion where nuclear pore complexes (NPCs) reside (Ungricht and Kutay, 2017) (Figure 1A).
FIGURE 1:
Features of the nuclear envelope and ER and regulation of and functions for lipid asymmetry at the inner nuclear membrane (INM). (A) Schematic of the continuous NE and ER membranes. The inner nuclear membrane (INM) facing the nucleoplasm and outer nuclear membrane (ONM) physically linked to the ER at NE-ER junctions are separated by a lumen designated perinuclear space (PNS). A nuclear pore complex (NPC) is located at a fusion point between the INM/ONM to generate the pore membrane. Highlighted in different shades of red are proteins that may regulate lipid trafficking between the NE and ER as well as enzymes and proteins that are regulated by or sense bilayer lipid composition (PKC and Nup133), or that regulate de novo lipid synthesis (CTDNEP1/lipin and CCTα). The curvature of the membrane bilayers may also play a role in restricting diffusion of lipid species past NE-ER junctions (negative curvature) or the pore membrane (positive curvature). Schematic of a membrane fusion reaction (middle) highlights membrane bending at each intermediate step. (B) The de novo glycerolipid synthesis pathway. Mol% for lipid species specific to ER/NE membranes is shown (van Meer et al., 2008).
Given the diverse functionality of the NE, it is not surprising that a steadily growing and diverse list of human pathologies are caused by mutations in NE or INM-associated nuclear lamina proteins. These pathologies include movement disorders and myopathies (Dauer and Worman, 2009; Meinke and Schirmer, 2016), cases of severely reduced life span and progeria (Kubben and Misteli, 2017; Fichtman et al., 2019), embryonic lethality (Turner and Schlieker, 2016), and lipodystrophies (Shackleton et al., 2000). Disruption of NE stability is also common in cancer cells causing DNA damage, cancer-relevant chromosomal rearrangements, and the intiation of proinflammatory pathways (Lim et al., 2016; Umbreit and Pellman, 2017; Hatch, 2018).
Studies tackling NE pathologies, together with investigations centered on NE proteins in model organisms or tissue culture systems, revised the view of the NE to a dynamic membrane system that undergoes significant membrane remodeling even outside of open mitosis. This raises the question of what mechanisms are put to work to maintain or reestablish the NE permeabilty barrier, especially when NE integrity is perturbed. ESCRT (endosomal sorting complexes required for transport)-dependent processes are involved in the sealing of NE holes and have recently been discussed elsewhere (Campsteijn et al., 2016; Webster and Lusk, 2016; Gatta and Carlton, 2019; Vietri et al., 2020). We will discuss findings that connect lipid regulation and de novo lipid synthesis (see Figure 1B and more details below) to NE sealing (Kinugasa et al., 2019; Lee et al., 2020; Penfield et al., 2020). Another emerging principle underlying NE dynamics pertains to the sculpting of its proteome by quality control mechanisms. These serve to degrade unwanted or misfolded proteins to maintain INM identity and safeguard protein homeostasis (Smoyer and Jaspersen, 2019). Being situated close to the nuclear transcriptional machinery, proteolytic mechanisms at the INM would also be ideally positioned to relay a perceived physiological demand to a transcriptional output, for example in the context of lipid homeostasis.
Finally, the best established facet of NE dynamics is the phenomenon of nuclear transport. NPCs traverse the NE and create a selective passageway through the INM and ONM and the enclosed perinuclear space (PNS) (Figure 1A). Regulated transport relies on nuclear transport receptors that enable cargo to passage through a meshwork of phenylalanine–glycine repeat nucleoporins (FG-nups) that establish a permeablity barrier between cytosolic and nuclear compartments. However, it is not known how NPC assembly is coordinated. In the following, we argue that the selective permeability barrier function relies on the fidelity and timing of membrane fusion between the INM and ONM during nuclear pore biogenesis, because an uncoordinated process could create a NE breach (Ungricht and Kutay, 2017).
In this Perspective, we discuss mechanisms critical for maintaining the identity and genome barrier function of the NE, with a focus on emerging roles of lipid metabolism and regulation of NPC biogenesis. Connected to these processes is the discovery of proteolytic systems that survey the NE proteome and may additionally play roles in the sharpening of compartmental identity and regulation of activities involved in lipid metabolism.
SPATIAL PARTITIONING OF BILAYER LIPIDS WITHIN THE CONTINUOUS NE AND ER
It is well known that the INM contains a unique set of integral membrane proteins; however, whether it also contains specific lipid species that contribute to its distinct identity and functions is not known. While the targeting of specific lipid kinases and phosphatases spatially restricts unique lipids within membranes of noncontiguous organelles (Behnia and Munro, 2005), the free diffusion of lipids within the continuous membranes of the ER and NE has been presumed to prevent their spatial segregation (Berg et al., 2002). However, specific lipid species at the INM have been shown to support viral proliferation (Marschall et al., 2011), NE dynamics (Hatch and Hetzer, 2014), de novo lipid synthesis (Haider et al., 2018; Romanauska and Kohler, 2018) and NPC biogenesis (Drin et al., 2007). Thus, despite the direct continuity of the lipid bilayers of the NE and ER, mechanisms may exist to selectively enrich and regulate specific lipid species at the INM. Aspects of some of these processes as well as potential mechanisms that drive lipid asymmetry at the NE are discussed below.
Lipid metabolizing enzymes associated with the surface of ER membranes produce the bilayer lipids of the NE, yet whether the products of these reactions are differentially distributed within the NE and ER is not known (Baumann and Walz, 2001; Fagone and Jackowski, 2009). This is in part because of the difficulty inherent in purifying the NE to homogeneity. Mass spectrometry analysis shows that the ER and NE mainly contain unsaturated glycerophospholipids and very low levels of the precursors required for their de novo synthesis (Figure 1B) (van Meer et al., 2008). In metazoans, the de novo synthesis pathway for glycerophospholipids begins with formation of phosphatidic acid (PA) by the addition of two activated fatty acids to glycerol-3-phosphate, which can be dephosphorylated to form diacylglycerol (DAG) (Figure 1B). Both PA and DAG are precursors for the synthesis of membrane glycerophospholipids, phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylinositol (PI). Depending on nutrient conditions, DAG can also be converted to storage lipids (triglycerides) (Figure 1B) (Carman and Han, 2006; van Meer et al., 2008; Fagone and Jackowski, 2009). Cholesterol and the biosynthetic intermediates to sphingolipids, ceramides and sphingoid bases, are also synthesized on the surface of the ER but represent an extremely low percentage of steady state ER/NE lipids—these lipids physically interact to form microdomains that enrich in export vesicles that traffic to the Golgi apparatus for further modifications (van Meer et al., 2008). Because the length and degree of desaturation of fatty acids determines the flexibility and order of lipid bilayers, sphingolipids, which are composed of taller saturated fatty acids that pack tightly with cholesterol, are much more abundant in the thicker, stiffer plasma membrane than in internal membranes (Bigay and Antonny, 2012). Thus, the lipid composition of the ER/NE does not reflect its capacity for the synthesis of a diversity of lipids species, which is also under tight regulation by feedback mechanisms that safeguard its lipid content (Breslow, 2013; Goldstein and Brown, 2015).
The loose packing of unstaturated glycerolipids that populate the ER/NE provides a deformable environment that is suitable for insertion of newly synthesized proteins into ER membranes (Bigay and Antonny, 2012). Several lines of evidence suggest that regulation of fatty acid composition in the ER/NE may also support the function of the NE as an impermeable barrier to the genome. Overexpression of Elo2 to produce very long chain fatty acids suppresses NE ruptures in fission yeast (Kinugasa et al., 2019), and accumulation of long-chain sphingoid base precursors to ceramide suppress NE morphology defects induced by aneuploidy in budding yeast and human cells (Hwang et al., 2019). In addition, a sphingolipid hydrolase (Smpd4) that releases ceramide localizes to NPCs, suggesting a potential local role for sphingolipids and their precursors at the NE (Cheng et al., 2019). These taller saturated lipids interact with cholesterol and, although a link between cholesterol and NE stability has not been made, the INM protein LBR (lamin B receptor) catalyzes a reaction in the production of cholesterol, suggesting that sterol synthesis may be in part regulated at the INM (Tsai et al., 2016). A recent study showed that an increase in the production of lipids with unsaturated acyl chains by overexpression of Δ9 fatty acid desaturase Ole1 in Schizosaccharomyces japonicus rescues NE sealing defects, yet, unlike long-chain fatty acids, desaturated lipids are predicted to increase membrane fluidity (Lee et al., 2020). Clearly, fatty acid composition influences NE stability but because the degree of saturation of lipids in ER membranes could regulate lipid enzyme activity to control lipid content and abundance, more work is required to understand the direct role for lipid saturation in NE stability.
Several lines of evidence suggest that the signaling and structural lipid DAG has specific functions at the INM (Figure 1B). The DAG-activated protein kinase C (PKC) has been shown to phosphorylate lamins for local and global disassembly; many of these functions are reviewed elsewhere (Hatch and Hetzer, 2014). Experiments in budding yeast using PA and DAG sensors appended to nuclear localization signals also support a spatial enrichment of DAG at the INM (Romanauska and Kohler, 2018). Because budding yeast do not contain lamin-encoding genes (Peter and Stick, 2015), there are likely evolutionarily conserved functions for DAG at the INM that are independent of PKC activation for lamina disassembly. Consistent with this, Romanauska and Kohler (2018) showed that this pool of DAG can be utilized for de novo lipid synthesis and formation of lipid droplets at the INM, particularly under metabolic conditions that favor synthesis of storage lipids.
From yeast to metazoans, the reduction of lipin (Pah1 in Saccharomyces cerevisiae, Ned1 in fission yeasts) activity, the key enzyme that generates DAG for the production of glycerophospholipids or storage lipids (Figure 1B), causes abnormal NE expansion (Tange et al., 2002; Golden et al., 2009; Gorjanacz and Mattaj, 2009; Peterson et al., 2011; Bahmanyar et al., 2014) (Figure 1B). Lipin1 is regulated by multisite phosphorylation by the nutrient-sensing kinase mTORC1 and by the cyclin dependent kinase Cdk1 (Grimsey et al., 2008; Peterson et al., 2011). The highly conserved protein phosphatase complex CTDNEP1/NEP1R1 (Nem1/Spo7 in yeast and CNEP-1/NEPR-1 in Caenorhabditis elegans) dephosphorylates lipin (Santos-Rosa et al., 2005; Han et al., 2012; Bahmanyar et al. 2014) and in C. elegans CNEP-1 is enriched at the NE, suggesting that it locally regulates the NE-associated pool of lipin to produce DAG (Bahmanyar et al., 2014). In addition to its catalytic functions, lipin1 regulates the transcription of lipid synthesis genes in response to nutrient availability through an unknown mechanism (Santos-Rosa et al., 2005; Peterson et al., 2011).
Insight into a role for lipin in forming the NE independent of its function in transcription came from studies in transcriptionally quiescent C. elegans embryos (Golden et al., 2009; Gorjanacz and Mattaj, 2009; Bahmanyar et al., 2014; Penfield et al., 2020). CNEP-1 activation of lipin at the NE biases flux in the de novo glycerolipid synthesis pathway toward DAG and PC/PE synthesis at the expense of PI synthesis (Figure 1B) (Bahmanyar et al., 2014; Penfield et al., 2020). Ectopic ER sheets formed in cnep-1 mutant embryos invade NE holes and prevent NE sealing (Penfield et al., 2020). Loss of the NE adaptor protein CHMP7 for the ESCRT-III membrane scission machinery, which assembles on the negatively curved surface of NE holes to execute membrane fission (Vietri et al., 2020), did not cause NE sealing defects on its own, but exacerbated the invasion of ER membranes and the defects in sealing resulting from deletion of cnep-1. These results led to a model that regulation of lipid synthesis cooperates with the ESCRT machinery to mediate NE sealing by restricting and remodeling ER membranes that feed into NE holes (Penfield et al., 2020).
Depletion of PI producing enzymes rescued sealing defects in embryos deleted for cnep-1 and ESCRT components, emphasizing the importance of lipid regulation in membrane remodeling to form the NE (Penfield et al., 2020). It is not known whether the production of PI itself or PI derivatives or overall changes in the biophysical properties of the NE/ER affect ER structure and NE sealing under these conditions. The bulky head group of PI may prevent membrane fusion or impact signaling functions of PI or PI derivatives, which have been shown to play a role in ESCRT-dependent NE sealing (Ventimiglia et al., 2018). It also remains possible that depletion of PI enzymes restores flux toward DAG production to rescue sealing defects, suggesting a specific role for DAG or the ratio of PA to DAG in NE sealing (Figure 1B). In S. japonicus, overexpression of the essential enzyme Ole1 (SCD1 in humans), which introduces double bonds into fatty acids that are channeled into PA for glycerolipid synthesis (Figure 1B), rescues NE sealing defects caused by loss of CHMP7 (cmp7 in S. japonicus), further indicating a role for the de novo glycerolipid synthesis pathway in NE closure (Lee et al., 2020).
Another major enzyme in de novo lipid synthesis that is regulated at the INM is the rate-limiting enzyme in PC synthesis, CCTα (Figure 1A) (Cornell, 2016). Changes in the ratio of PC to PE at the INM causes lipid packing defects that relieve protein autoinhibiton of nuclear CCTα through absorption of its membrane-binding domain (Haider et al., 2018). The membrane-binding domain of CCTα locally senses changes in PC/PE ratios to restore PC homeostasis globally, suggesting that PE and PC rapidly diffuse and equilibrate between the ER and INM (Figure 1B). In support of this idea, Haider et al. (2018) found that CCTα does not have to localize to the INM to respond to lipid packing stress caused by an increase in the PE:PC ratio. Its localization to the nucleus confines its activity to INM lipids and keeps it from acting at other organelles. The flat surface of the INM relative to curved ER tubules may provide an environment more sensitive to changes in PC and PE levels (Cornell and Antonny, 2018).
Lipid synthesis and packing at the INM could facilitate the insertion of NPCs in interphase (Figure 1B). The nucleoporin Nup133 contains a membrane-binding domain ALPS motif that recognizes lipid packing defects (Drin et al., 2007). Membrane bending generated at the base of intermediate NPC structures (Figure 1B) (Otsuka et al., 2016; Otsuka and Ellenberg, 2018) is predicted to cause large packing defects (Bigay and Antonny, 2012) that may be sensed and stabilized by the Nup133 ALPS motif to promote pore assembly (Doucet et al., 2010). Whether a membrane bending protein or the local conversion of lipids induces lipid packing defects to initiate pore assembly is not known. Completion of pore assembly through accumulation of a fusogenic lipid such as DAG may induce negative curvature to assist in fusion between the INM and ONM (Figure 1B).
A unique lipid composition at the INM requires a way to spatially partition lipids within a continuous membrane system (Figure 1A). Two ideas discussed here are as follows: 1) a physical barrier that reduces the timescale of lateral diffusion for specific lipids from one area (peripheral ER/ONM) to the other (INM) and 2) spatial restriction of synthetic enzymes to generate a continuum of concentrations high in one area (e.g., peripheral ER) relative to the other (e.g., INM).
Our current knowledge of a gradient of phospholipid concentrations is based on the increased activation of the PA phosphatase lipin by the phosphatase CNEP-1/CTDNEP1 at the NE (Figure 1) (Kim et al., 2007; Bahmanyar et al., 2014). Because PA serves as a precursor to PI, the resultant lower PA phosphatase activity of lipin on ER tubules located toward the cell periphery may permit the local production of PI from PA. Continuously restricting PI production to peripheral ER tubules balanced with transport of PI to other organelles would support a gradient of high PI concentrations at the cell periphery and low PI concentrations at the INM. The spatial restriction of lipin activity prevents PI accumulation and formation of ectopic ER sheets that interfere with the formation of a sealed nuclear compartment (Penfield et al., 2020), a requirement for generating and maintaining NE identity.
In contrast to a gradient, a physical barrier would generate a distinct segregation of lipids between the peripheral ER and the INM. Two possible locations for a physical barrier are the fusion points between the ONM and INM (the pore membrane) or between the ONM and ER tubules (ER-NE junctions) (Figure 1B). The high negative curvature at ER-NE junctions might be sufficient to prevent the passage of inverted cone shaped lipids that prefer positive curvature, such as phosphoinositides, and more favorable to the diffusion of cone shaped lipids such as DAG and PE (Figure 1B) (McMahon and Boucrot, 2015). In contrast, the positive curvature of the nuclear pore membrane imposes the opposite constraints (Figure 1B). Because the curvature of the luminal leaflet is the opposite of that of the cytoplasmic leaflet, transbilayer movement or “flip-flop” could also impact the ability of lipids to bypass their curvature constraints (Tsuji et al., 2019).
In addition to membrane curvature, proteins could impose a physical barrier to the lateral diffusion of lipids (Trimble and Grinstein, 2015). The NPC is a stable structure associated with several transmembrane containing nucleoporins that might sterically hinder the diffusion of lipids with large or charged head groups (Figure 1B) (Rothballer and Kutay, 2013). Proteins that sit at ER-NE junctions may similarly serve as a critical intersection for the passage of lipids into the NE. The ER protein lunapark prefers the negatively curved membranes of three-way junctions and regulates their abundance in the ER, and so would be predicted to similarly control the number of ER-NE junctions that feed lipids into the NE (Figure 1B) (Chen et al., 2015; Wang et al., 2016). In fission yeast, lunapark regulates the bulk flow of lipids into the NE to support a constant nuclear-to-cytoplasmic-volume ratio (Kume et al., 2019). ER junctions that intersect the NE may be destabilized by an increase in surface tension at the NE—volume expansion may loosen the barrier to promote bulk membrane flow. Thus, the topology (tubule-sheet fusion) and position of these junctions at the interface of the NE and ER are poised to serve a key role in defining the identity of these compartments.
Future work will determine how barriers to the diffusion of specific lipid species might function in concert with spatial restriction of lipid biosynthetic enzymes to establish and maintain a unique lipid composition of the NE. How unique lipid species in turn control the local dynamics and specialized functions of the NE is a major and open area of research essential to our fundamental understanding of organelle biogenesis and function in normal and diseased cells.
PROTEIN TURNOVER AT THE INM: AN ENHANCER OF COMPARTMENTAL IDENTITY?
While membranes play a defining role for compartment identity, compartment-specific proteins carry out specific functions. This poses a key problem for the NE of eukaryotes with an open mitosis, as the repeated breakdown of the NE and nuclear lamina during every mitotic cycle leads to loss of compartment identity by mixing of integral proteins of the ER and INM (Ellenberg et al., 1997). The detailed events underlying NE reformation were reviewed recently (Ungricht and Kutay, 2017). Here we review mechanisms that maintain the NE proteome in interphase after the NE has formed, which is critical for cellular homeostasis since INM proteins that became effectively diluted due to cell duplication or are constantly removed via proteolysis (the average half-life of INM proteins is ∼3–4 d; Buchwalter et al., 2019b) need to be replaced. Equally important, particular physiological situations may require tailored compositions of INM proteins, for example in the context of cellular differentiation or stress responses, requiring dynamic adjustments through synthesis, nuclear import, and degradation.
Once the NE has reformed and the permeability barrier of the NPC has been established, INM components replenish the INM protein pool. Those INM proteins that bind lamin can diffuse from the ER to the INM and the reformed nuclear lamina, where they are locally retained by specific interaction with lamins and chromatin (Boni et al., 2015; Ungricht et al., 2015). Importantly, the NPC imposes a diffusion barrier effectively separating the ER and contiguous ONM from the INM by restricting the passage to membrane proteins with cytosolic domains of less than 40–60 kDa (Ohba et al., 2004; Ungricht et al., 2015). The molecular basis accounting for this property of the NPC is currently unknown. Structural information on the NPC-membrane interface at higher resolution is needed to resolve this conundrum. While this diffusion barrier can effectively exclude sizable membrane proteins, membrane proteins with smaller cytosolic domains may also need to be excluded from the INM as their presence there could potentially interfere with INM function.
One possible mechanism to enhance INM identity is to selectively remove proteins that do not belong to the INM and have “breached” the first selectivity filter. Indeed, a specialized branch of the ubiquitin (Ub)/proteasome system (UPS) is operative at the INM in both lower (Deng and Hochstrasser, 2006; Foresti et al., 2014; Khmelinskii et al., 2014) and higher (Tsai et al., 2016; Buchwalter et al., 2019b) eukaryotes, where it performs functions analogous to the ER-associated degradation (ERAD) machinery (Smoyer and Jaspersen, 2019). In brief, a typically polytopic Ub E3 ligase recognizes the substrate destined for degradation and mediates its ubiquitylation in conjunction with an Ub-conjugating enzyme. The ubiquitylated substrate is then extracted from the membrane and delivered to the 26S proteasome for degradation (Vembar and Brodsky, 2008). In yeast, the Asi1/2/3 complex and Doa10 are the E3 ligases responsible for turnover of INM proteins (Deng and Hochstrasser, 2006; Foresti et al., 2014; Khmelinskii et al., 2014) while the AAA+ ATPase Cdc48 and its human orthologue p97 mobilize the polyubiquitylated substrate from the INM in yeast and human cells, respectively (Foresti et al., 2014; Tsai et al., 2016) (Figure 2). The E3 ligases responsible for INM turnover in mammalian cells remain to be identified, though recently developed methodology relying on LBR-based model substrates for INM turnover may aid in their identification through proteomics-based or genome-wide screening approaches (Tsai et al., 2019). Intriguingly, proteasomes were directly observed in association with the NE and in juxtaposition to nuclear pores in Chlamydomonas reinhardtii via cryo-EM tomography (Albert et al., 2017), suggesting important roles for proteolytic systems at these sites. In yeast, the Asi2 subunit specifically recognizes transmembrane domains of orphan subunits of mislocalized ER proteins, leading to their removal from the INM through ubiquitylation, extraction by Cdc48, and subsequent turnover by the 26S proteasome (Natarajan et al., 2019). It will be interesting to test whether the concept of “proteolytic sharpening” of compartmental identity via removal of proteins that “leak” into INM territory extends to the NE of mammalian cells, or even to other compartments.
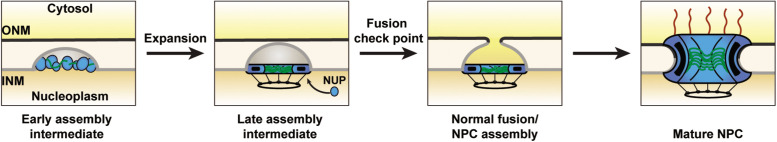
FIGURE 2:
A fusion checkpoint for nuclear pore biogenesis. Nuclear pore assembly after nuclear envelope reformation proceeds through an inside-out protrusion mechanism starting from the nuclear side. Early assembly intermediates contain NPC constituents that deform the inner nuclear membrane (INM). This structure needs to expand to bring the INM and outer nuclear membrane (ONM) in close proximity for fusion. Either the integrity of the NPC assembly intermediate is sensed or the transport competence of a late assembly intermediate is required to initiate fusion. This hypothetical checkpoint mechanism would prevent a transient perturbation of NE integrity and can potentially explain why multiple, distinct NPC assembly defects lead to NE blebs resembling late assembly intermediates that got arrested before fusion (see the text).
The turnover of polytopic membrane proteins from the INM of mammalian cells can be very rapid. The half-life of truncated LBR variants is about ∼10–15 min (Tsai et al., 2016), which is significantly faster than the turnover of unrelated polytopic ERAD substrates, typically in the range of hours (Cambridge et al., 2011). Thus, the INM-resident proteolytic system would be perfectly suited for rapid regulatory switches to initiate, for example, transcriptional responses following physiological stimuli. Given that regulatory proteolysis, such as the processing of transcription factors, plays key roles in mounting transcriptional responses to regulate lipid abundance (Goldstein and Brown, 2015) and phospholipid saturation (Rape et al., 2001), it is tempting to speculate that additional roles of the UPS in lipid sensing or regulation will be identified at the INM. The close proximity to the transcriptional machinery would make the NE an ideal relay station to sense and amplify changes in membrane composition, through either alterations in lipid composition or membrane fluidity caused by packing defects. Similar functions linking mechanosensation to a transcriptional output can be envisioned.
HOW TO INSTALL NPCS WHILE PREVENTING A BREACH: A FUSION CHECKPOINT?
The integrity of the NE and its permeability barrier are under constant threat from mechanical stress, due to defects in lipid packing and through manipulation by pathogens that breach the NE during their life cycle. Over the course of evolution, eukaryotes have therefore evolved multiple mechanisms to prevent or counteract these threats. Mechanical reinforcements are key to maintaining NE integrity. These rely in part on the nuclear lamina, a meshwork of lamin-class intermedate filaments that are connected to the INM by association with INM proteins, as well as LINC (linker of the nucleoskeleton and cytoskeleton) complexes that bridge the NE and connect the nuclear lamina to cytosolic components of the cytoskeleton (Burke and Stewart, 2014; Hatch and Hetzer, 2016; de Leeuw et al., 2018; Kirby and Lammerding, 2018; Stephens et al., 2019). In addition, surveillance mechanisms exist in both yeast and mammalian cells that detect defects in NE integrity and recruit ESCRT machinery to reseal the NE membrane (Olmos et al., 2015; Vietri et al., 2015; Thaller et al., 2019).
In higher eukaryotes, the occurrence of open mitosis by definition involves NE breakdown, representing a challenge for cellular compartmentalization. It is therefore not surprising that the reformation of the NE is an extremely rapid process that is completed within minutes after anaphase onset (Lu et al., 2011; Otsuka et al., 2018; Rampello et al., 2020). Cell division poses yet another problem for NE integrity: daughter cells need to replenish the number of NPCs that had been diluted in the process, and these new channels must perforate both membranes of the NE. At least two mechanistically distinct processes account for NPC assembly in dividing mammalian cells, referred to as postmitotic and interphase assembly, respectively (Doucet et al., 2010; Weberruss and Antonin, 2016; Otsuka et al., 2018). Postmitotic assembly occurs during NE reformation while interphase assembly starts immediately after the NE has reformed (Dultz and Ellenberg, 2010; Rampello et al., 2020). In mature NPCs resulting from either mechanism, a dense meshwork of FG repeats in the center of the NPC channel establishes a selective permeability barrier that allows the passage of only smaller proteins up to 30–60 KDa (Timney et al., 2016). Larger cargo requires appropriate sorting signals that are recognized by transport receptors (karyopherins) that mediate transport through the NPC (Wente and Rout, 2010; Schmidt and Gorlich, 2016). But what happens before the FG-nup network is completed? Stalled or slowly maturing NPC assembly intermediates could present a massive problem were they to traverse both NE membranes before the establishment of the FG-nup barrier, thus effectively creating a hole within the NE. Similarly, assembly defects might create long-lived, defective intermediates with properties similar to those of pore-forming toxins, a problem that can be counteracted by membrane sealing of defective assembly intermediates (Wente and Blobel, 1993; Webster et al., 2014). However, considering that a significant portion of newly formed NPCs are installed after the NE has reformed by interphase insertion in early G1 (Maul et al., 1972; Dultz and Ellenberg, 2010; Rampello et al., 2020), even unperturbed assembly could transiently create several hundreds of unscheduled, potentially toxic perforations of the NE in a mammalian cell. In what follows, we propose possible solutions to this problem that are informed by recent studies on NPC assembly.
Interphase NPC insertion relies on an inside-out budding mechanism in which NPC assembly is initiated at the INM and proceeds through a dome-shaped INM intermediate, with NPC precursors being situated at the base of the dome (Otsuka et al., 2016; Otsuka and Ellenberg, 2018) (Figure 2). The bulged INM then fuses with the ONM, and cytoplasmic fibrils are added only after this fusion has occurred (Figure 2). However, interphase NPC assembly is a rapid process that is difficult to resolve temporally by cryofixation of cells and EM tomography. The recent finding that the deletion of Torsin ATPases leads to an arrest in interphase NPC biogenesis offers a unique opportunity to characterize “frozen intermediates” of NPC assembly (Laudermilch et al., 2016; Rampello et al., 2020). Live cell imaging using lattice light sheet microscopy in Torsin-deficient cells suggests that the early stages of assembly at the INM happen synchronously and rapidly immediately after the NE has reformed (Rampello et al., 2020). This process is usually dynamic and would normally proceed to the step of INM/ONM fusion in an orderly manner (Otsuka et al., 2016). We propose that INM/ONM fusion represents a critical quality control checkpoint. In our model (Figure 2), fusion occurs only if NPC assembly has matured to a point where the permeability barrier requiring the presence of central channel FG nups is established, thereby avoiding perturbations of the barrier function of the NE. While a plethora of mutations in NPC components leads to NE blebs (for a review, see Thaller and Lusk, 2018), it is possible that many of these aberrant structures result from failure to pass a quality control step at or before the point of membrane fusion.
Going forward, it will be interesting to test whether such a quality control checkpoint does indeed occur and to explore how the completion of an NPC assembly intermediate that is ready for fusion can be perceived on the molecular level. Perhaps the presence of a component that is added at a late stage is perceived and relayed to the currently unknown fusogenic machinery, or one of the components added late is itself endowed with fusogenic properties. Alternatively, it may be that only when the assembly process proceeds properly that the ONM and INM come within a fusogenic distance (expansion phase, Figure 2).
As a variation of the latter model, one strategy would be to block the INM and ONM from achieving fusogenic proximity until the NPC precursor has acquired transport competence. (the two membranes are normally separated by up to 50 nm; Voeltz and Prinz, 2007). A specific transport event relying on the presence of FG-nups might expand the bleb or dome lumen and thereby bring the INM and ONM in close contact.The targeted delivery of a fusion activator or a fusogen from the nucleoplasm into the bleb lumen that depends on on the local presence of a dense FG-nup network might be another feasible strategy to build such a “fusion checkpoint.”
Indeed, the role of specific FG-nups is not restricted to the building a permeability barrier but also entails a “Velcro” function that is required for NPC assembly (Onischenko et al., 2017). Thus, the recruitment of factors implicated in fusion could similarly depend on the presence of specific FG-nups. Notably, fusogenic properties of RNAs have been described (Khvorova et al., 1999; Janas and Yarus, 2003), so the search for the fusogen—the “holy grail” of NPC biogenesis—does not necessarily need to be restricted to proteinaceous candidates.
While a role for Torsins as ER and NE-luminal ATPases in the context of fusogenic machinery was previously proposed (Laudermilch and Schlieker, 2016; Weberruss and Antonin, 2016; Chase et al., 2017), it may seem peculiar that unicellular eukaryotes do not have obvious Torsin orthologues and can still assemble NPCs. The question arises why interphase NPC biogenesis is stalled prior to membrane fusion in Torsin-deficient cells. Perhaps nature found alternative solutions for INM/ONM fusion, as for example in the case of endocytosis. In higher eukaryotes, dynamin GTPases mediate fission during endocytosis, whereas lower eukaryotes lack dynamins and employ actin-based mechanisms for fission (Ferguson and De Camilli, 2012; Lacy et al., 2018). In yeast, Brr6 and Brl1 locate to NPC assembly sites and their mutation elicits phenotypes consistent with a key role for INM/ONM fusion (Zhang et al., 2018); however, no direct homologues have so far been reported in mammalian organisms.
Another, not mutually exclusive, possibility is that Torsins are required to relieve an inhibition of fusion, for example, as part of the checkpoint mechanism outlined above. In a wider biological context, it may make sense to control the INM/ONM step beyond its possible checkpoint function in assembly as a means to regulate specialized transport. Notably, NE blebs with electron densities of dimensions at their base that are similar to NPCs and assembly intermediates of Torsin-deficient cells have been observed in zygotes and early embryos (see Szollosi and Szollosi, 1988; Laudermilch et al., 2016). Moreover, RNA-containing NE blebs were observed in NEs at the neuromuscular junction in Drosophila melanogaster (Speese et al., 2012) and were functionally linked to Torsins (Jokhi et al., 2013). Perhaps these structures do represent NPC assembly intermediates, but they may also serve to concentrate specialized nuclear export cargo that is stored in their lumen. These assemblies would therefore sit poised for fusion, which would occur rapidly and synchronously following appropriate stimuli. This type of mechanism could serve to sharpen temporal control of transcriptional programs in a manner reminiscent of synaptic signaling. As mentioned above, the lipid composition of the NE plays an important role in the process of INM/ONM fusion, and specialized lipids may therefore play additional roles in this fusion process. Notably, Torsins were recently linked to lipid metabolism (Grillet et al., 2016; Shin et al., 2019) and a TorsinA variant can deform unilamellar vesicles in vitro (Demircioglu et al., 2019). Lipid-specific probes suitable for in situ analysis would be particularly well-suited to shed light on these dynamic processes, as these could circumvent complications arising from biochemical fractionations of NE membranes and the inability to discern INM and ONM in NE isolates.
In conclusion, multiple mechanisms cooperate to shape the identity of the NE as a distinct subcompartment of the ER, either through limiting diffusion of proteins and lipids between those subcompartments or through localized remodeling and removal of undesired constituents. At the same time, the dynamic nature of the NE—for example during NPC biogenesis—requires fast-acting surveillance mechanisms and possibly checkpoints to preserve the separation of the cytoplasm and nucleoplasm that underpins eukaryotic organization. Among the major gaps in our knowledge of NE architecture is the absence of detailed information—both structural and compositional—of the ER/NE junctions and the NPC/pore membrane interface. An interdisciplinary effort is required to identify proteins that stabilize the membrane curvature at these sites and to identify mechanisms that restrict the diffusion of membrane proteins and lipids between the ER, ONM, and INM. Through these and other endeavors we will learn how separate identities are established to enable functional specification and diversification. Ultimately, a better understanding of these processes will allow us to rationalize how perturbations of this dynamic organization give rise to human pathologies, possibly informing strategies for therapeutic intervention.
Acknowledgments
We thank members of the Schlieker and Bahmanyar laboratories for comments on the manuscript and Anthony Rampello and Holly Merta for help with the figures. S.B. thanks J. Bewersdorf for helpful discussion. This work was supported by National Institutes of Health (NIH) grants R01GM114401 and R01GM126835 to C.S. and a National Science Foundation CAREER grant (1846010) and an NIH grant (R01GM131004) to S.B.
Abbreviations used:
- CCTα
choline phosphate cytidylyltransferase
- CHMP7
charged multivesicular body protein 7
- CNEP-1
C-terminal nuclear envelope phosphatase-1
- CTDNEP1
C-terminal domain nuclear envelope phosphatase 1
- Elo2
fatty acid elongase
- ESCRT
endosomal sorting complex required for transport
- FG nup
phenylalanine-glycine-rich nucleoporin
- INM
inner nuclear membrane
- NE
nuclear envelope
- NEP1R1
nuclear envelope phosphatase regulatory subunit 1
- NPC
nuclear pore complex
- ONM
outer nuclear membrane
- PNS
perinuclear space
- UPS
ubiquitin/proteasome system
Footnotes
REFERENCES
- Albert S, Schaffer M, Beck F, Mosalaganti S, Asano S, Thomas HF, Plitzko JM, Beck M, Baumeister W, Engel BD. (2017). Proteasomes tether to two distinct sites at the nuclear pore complex. Proc Natl Acad Sci USA , 13726–13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahmanyar S. (2015). Spatial regulation of phospholipid synthesis within the nuclear envelope domain of the endoplasmic reticulum. Nucleus , 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahmanyar S, Biggs R, Schuh AL, Desai A, Muller-Reichert T, Audhya A, Dixon JE, Oegema K. (2014). Spatial control of phospholipid flux restricts endoplasmic reticulum sheet formation to allow nuclear envelope breakdown. Genes Dev , 121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AD, Lim K, Mari M, Edgar JR, Gal L, Sterk P, Jenkins BJ, Koulman A, Savage DB, Schuldiner M, et al. (2019). Compartmentalized synthesis of triacylglycerol at the inner nuclear membrane regulates nuclear organization. Dev Cell , 755–766.e756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AD, Sembongi H, Su WM, Abreu S, Reggiori F, Carman GM, Siniossoglou S. (2015). Lipid partitioning at the nuclear envelope controls membrane biogenesis. Mol Biol Cell , 3641–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann O, Walz B. (2001). Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol , 149–214. [DOI] [PubMed] [Google Scholar]
- Behnia R, Munro S. (2005). Organelle identity and the signposts for membrane traffic. Nature , 597–604. [DOI] [PubMed] [Google Scholar]
- Berg JM, Tymoczko JL, Stryer L. (2002). Section 12.6, lipids and many membrane proteins diffuse rapidly in the plane of the membrane. In: Biochemistry, 5th ed, New York: WH Freeman. [Google Scholar]
- Bigay J, Antonny B. (2012). Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev Cell , 886–895. [DOI] [PubMed] [Google Scholar]
- Boni A, Politi AZ, Strnad P, Xiang W, Hossain MJ, Ellenberg J. (2015). Live imaging and modeling of inner nuclear membrane targeting reveals its molecular requirements in mammalian cells. J Cell Biol , 705–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK. (2013). Sphingolipid homeostasis in the endoplasmic reticulum and beyond. Cold Spring Harb Perspect Biol , a013326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwalter A, Kaneshiro JM, Hetzer MW. (2019a). Coaching from the sidelines: the nuclear periphery in genome regulation. Nat Rev Genet , 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwalter A, Schulte R, Tsai H, Capitanio J, Hetzer M. (2019b). Selective clearance of the inner nuclear membrane protein emerin by vesicular transport during ER stress. Elife , e49796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B. (2018). LINC complexes as regulators of meiosis. Curr Opin Cell Biol , 22–29. [DOI] [PubMed] [Google Scholar]
- Burke B, Stewart CL. (2014). Functional architecture of the cell’s nucleus in development, aging, and disease. Curr Top Dev Biol , 1–52. [DOI] [PubMed] [Google Scholar]
- Cambridge SB, Gnad F, Nguyen C, Bermejo JL, Kruger M, Mann M. (2011). Systems-wide proteomic analysis in mammalian cells reveals conserved, functional protein turnover. J Proteome Res , 5275–5284. [DOI] [PubMed] [Google Scholar]
- Campsteijn C, Vietri M, Stenmark H. (2016). Novel ESCRT functions in cell biology: spiraling out of control? Curr Opin Cell Biol , 1–8. [DOI] [PubMed] [Google Scholar]
- Carman GM, Han GS. (2006). Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem Sci , 694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase AR, Laudermilch E, Schlieker C. (2017). Torsin ATPases: harnessing dynamic instability for function. Front Mol Biosci , 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Desai T, McNew JA, Gerard P, Novick PJ, Ferro-Novick S. (2015). Lunapark stabilizes nascent three-way junctions in the endoplasmic reticulum. Proc Natl Acad Sci USA , 418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LC, Baboo S, Lindsay C, Brusman L, Martinez-Bartolome S, Tapia O, Zhang X, Yates JR, 3rd, Gerace L. (2019). Identification of new transmembrane proteins concentrated at the nuclear envelope using organellar proteomics of mesenchymal cells. Nucleus , 126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell R, Antonny B. (2018). CCTalpha commands phospholipid homeostasis from the nucleus. Dev Cell , 419–420. [DOI] [PubMed] [Google Scholar]
- Cornell RB. (2016). Membrane lipid compositional sensing by the inducible amphipathic helix of CCT. Biochim Biophys Acta , 847–861. [DOI] [PubMed] [Google Scholar]
- Dauer WT, Worman HJ. (2009). The nuclear envelope as a signaling node in development and disease. Dev Cell , 626–638. [DOI] [PubMed] [Google Scholar]
- de Leeuw R, Gruenbaum Y, Medalia O. (2018). Nuclear lamins: thin filaments with major functions. Trends Cell Biol , 34–45. [DOI] [PubMed] [Google Scholar]
- Demircioglu FE, Zheng W, McQuown AJ, Maier NK, Watson N, Cheeseman IM, Denic V, Egelman EH, Schwartz TU. (2019). The AAA + ATPase TorsinA polymerizes into hollow helical tubes with 8.5 subunits per turn. Nat Commun , 3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M, Hochstrasser M. (2006). Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature , 827–831. [DOI] [PubMed] [Google Scholar]
- Doucet CM, Talamas JA, Hetzer MW. (2010). Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell , 1030–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. (2007). A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol , 138–146. [DOI] [PubMed] [Google Scholar]
- Dultz E, Ellenberg J. (2010). Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J Cell Biol , 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, Lippincott-Schwartz J. (1997). Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol , 1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagone P, Jackowski S. (2009). Membrane phospholipid synthesis and endoplasmic reticulum function. J Lipid Res (Suppl), S311–S316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM, De Camilli P. (2012). Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol , 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichtman B, Zagairy F, Biran N, Barsheshet Y, Chervinsky E, Ben Neriah Z, Shaag A, Assa M, Elpeleg O, Harel A, Spiegel R. (2019). Combined loss of LAP1B and LAP1C results in an early onset multisystemic nuclear envelopathy. Nat Commun , 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresti O, Rodriguez-Vaello V, Funaya C, Carvalho P. (2014). Quality control of inner nuclear membrane proteins by the Asi complex. Science , 751–755. [DOI] [PubMed] [Google Scholar]
- Garapati HS, Mishra K. (2018). Comparative genomics of nuclear envelope proteins. BMC Genomics , 823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta AT, Carlton JG. (2019). The ESCRT-machinery: closing holes and expanding roles. Curr Opin Cell Biol , 121–132. [DOI] [PubMed] [Google Scholar]
- Gerace L, Tapia O. (2018). Messages from the voices within: regulation of signaling by proteins of the nuclear lamina. Curr Opin Cell Biol , 14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden A, Liu J, Cohen-Fix O. (2009). Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J Cell Sci , 1970–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS. (2015). A century of cholesterol and coronaries: from plaques to genes to statins. Cell , 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjanacz M, Mattaj IW. (2009). Lipin is required for efficient breakdown of the nuclear envelope in Caenorhabditis elegans. J Cell Sci , 1963–1969. [DOI] [PubMed] [Google Scholar]
- Grillet M, Dominguez Gonzalez B, Sicart A, Pottler M, Cascalho A, Billion K, Hernandez Diaz S, Swerts J, Naismith TV, Gounko NV, et al. (2016). Torsins are essential regulators of cellular lipid metabolism. Dev Cell , 235–247. [DOI] [PubMed] [Google Scholar]
- Grimsey N, Han GS, O’Hara L, Rochford JJ, Carman GM, Siniossoglou S. (2008). Temporal and spatial regulation of the phosphatidate phosphatases lipin 1 and 2. J Biol Chem , 29166–29174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider A, Wei YC, Lim K, Barbosa AD, Liu CH, Weber U, Mlodzik M, Oras K, Collier S, Hussain MM, et al. (2018). PCYT1A regulates phosphatidylcholine homeostasis from the inner nuclear membrane in response to membrane stored curvature elastic stress. Dev Cell , 481–495.e488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Bahmanyar S, Zhang P, Grishin N, Oegema K, Crooke R, Graham M, Reue K, Dixon JE, Goodman JM. (2012). Nuclear envelope phosphatase 1-regulatory subunit 1 (formerly TMEM188) is the metazoan Spo7p ortholog and functions in the lipin activation pathway. J Biol Chem , 3123–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch E, Hetzer M. (2014). Breaching the nuclear envelope in development and disease. J Cell Biol , 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM. (2018). Nuclear envelope rupture: little holes, big openings. Curr Opin Cell Biol , 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Hetzer MW. (2016). Nuclear envelope rupture is induced by actin-based nucleus confinement. J Cell Biol , 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Williams JF, Kneissig M, Lioudyno M, Rivera I, Helguera P, Busciglio J, Storchova Z, King MC, Torres EM. (2019). Suppressing aneuploidy-associated phenotypes improves the fitness of Trisomy 21 cells. Cell Rep , 2473–2488.e2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas T, Yarus M. (2003). Visualization of membrane RNAs. RNA , 1353–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokhi V, Ashley J, Nunnari J, Noma A, Ito N, Wakabayashi-Ito N, Moore MJ, Budnik V. (2013). Torsin mediates primary envelopment of large ribonucleoprotein granules at the nuclear envelope. Cell Rep , 988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A, Blaszczak E, Pantazopoulou M, Fischer B, Omnus DJ, Le Dez G, Brossard A, Gunnarsson A, Barry JD, Meurer M, et al. (2014). Protein quality control at the inner nuclear membrane. Nature , 410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Kwak YG, Tamkun M, Majerfeld I, Yarus M. (1999). RNAs that bind and change the permeability of phospholipid membranes. Proc Natl Acad Sci USA , 10649–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Gentry MS, Harris TE, Wiley SE, Lawrence JC, Jr, Dixon JE. (2007). A conserved phosphatase cascade that regulates nuclear membrane biogenesis. Proc Natl Acad Sci USA , 6596–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinugasa Y, Hirano Y, Sawai M, Ohno Y, Shindo T, Asakawa H, Chikashige Y, Shibata S, Kihara A, Haraguchi T, Hiraoka Y. (2019). The very-long-chain fatty acid elongase Elo2 rescues lethal defects associated with loss of the nuclear barrier function in fission yeast cells. J Cell Sci , jcs229021. [DOI] [PubMed] [Google Scholar]
- Kirby TJ, Lammerding J. (2018). Emerging views of the nucleus as a cellular mechanosensor. Nat Cell Biol , 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korfali N, Wilkie GS, Swanson SK, Srsen V, de Las Heras J, Batrakou DG, Malik P, Zuleger N, Kerr AR, Florens L, Schirmer EC. (2012). The nuclear envelope proteome differs notably between tissues. Nucleus , 552–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubben N, Misteli T. (2017). Shared molecular and cellular mechanisms of premature ageing and ageing-associated diseases. Nat Rev Mol Cell Biol , 595–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Cantwell H, Burrell A, Nurse P. (2019). Nuclear membrane protein Lem2 regulates nuclear size through membrane flow. Nat Commun , 1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy MM, Ma R, Ravindra NG, Berro J. (2018). Molecular mechanisms of force production in clathrin-mediated endocytosis. FEBS Lett , 3586–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudermilch E, Schlieker C. (2016). Torsin ATPases: structural insights and functional perspectives. Curr Opin Cell Biol , 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudermilch E, Tsai PL, Graham M, Turner E, Zhao C, Schlieker C. (2016). Dissecting Torsin/cofactor function at the nuclear envelope: a genetic study. Mol Biol Cell , 3964–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IJ, Stokasimov E, Dempsey N, Varberg JM, Jacob E, Jaspersen SL, Pellman D. (2020). Factors promoting nuclear envelope assembly independent of the canonical ESCRT pathway. J Cell Biol , e201908232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Quinton RJ, Ganem NJ. (2016). Nuclear envelope rupture drives genome instability in cancer. Mol Biol Cell , 3210–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Ladinsky MS, Kirchhausen T. (2011). Formation of the postmitotic nuclear envelope from extended ER cisternae precedes nuclear pore assembly. J Cell Biol , 425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall M, Feichtinger S, Milbradt J. (2011). Regulatory roles of protein kinases in cytomegalovirus replication. Adv Virus Res , 69–101. [DOI] [PubMed] [Google Scholar]
- Maul GG, Maul HM, Scogna JE, Lieberman MW, Stein GS, Hsu BY, Borun TW. (1972). Time sequence of nuclear pore formation in phytohemagglutinin-stimulated lymphocytes and in HeLa cells during the cell cycle. J Cell Biol , 433–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E. (2015). Membrane curvature at a glance. J Cell Sci , 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke P, Schirmer EC. (2016). The increasing relevance of nuclear envelope myopathies. Curr Opin Neurol , 651–661. [DOI] [PubMed] [Google Scholar]
- Natarajan N, Foresti O, Wendrich K, Stein A, Carvalho P. (2019). Quality control of protein complex assembly by a transmembrane recognition factor. Mol Cell , 108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T, Schirmer EC, Nishimoto T, Gerace L. (2004). Energy- and temperature-dependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J Cell Biol , 1051–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos Y, Hodgson L, Mantell J, Verkade P, Carlton JG. (2015). ESCRT-III controls nuclear envelope reformation. Nature , 236–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E, Tang JH, Andersen KR, Knockenhauer KE, Vallotton P, Derrer CP, Kralt A, Mugler CF, Chan LY, Schwartz TU, Weis K. (2017). Natively unfolded FG repeats stabilize the structure of the nuclear pore complex. Cell , 904–917.e919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka S, Bui KH, Schorb M, Hossain MJ, Politi AZ, Koch B, Eltsov M, Beck M, Ellenberg J. (2016). Nuclear pore assembly proceeds by an inside-out extrusion of the nuclear envelope. Elife . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka S, Ellenberg J. (2018). Mechanisms of nuclear pore complex assembly — two different ways of building one molecular machine. FEBS Lett , 475–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka S, Steyer AM, Schorb M, Heriche JK, Hossain MJ, Sethi S, Kueblbeck M, Schwab Y, Beck M, Ellenberg J. (2018). Postmitotic nuclear pore assembly proceeds by radial dilation of small membrane openings. Nat Struct Mol Biol , 21–28. [DOI] [PubMed] [Google Scholar]
- Penfield L, Shankar R, Szentgyorgyi E, Laffitte A, Mauro MS, Audhya A, Muller-Reichert T, Bahmanyar S. (2020). Regulated lipid synthesis and LEM2/CHMP7 jointly control nuclear envelope closure. J Cell Biol . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter A, Stick R. (2015). Evolutionary aspects in intermediate filament proteins. Curr Opin Cell Biol , 48–55. [DOI] [PubMed] [Google Scholar]
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, Sabatini DM. (2011). mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell , 408–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampello AJ, Laudermilch E, Vishnoi N, Prohet SM, Shao L, Zhao C, Lusk CP, Schlieker C. (2020). Torsin ATPase deficiency leads to defects in nuclear pore biogenesis and sequestration of MLF2. J Cell Biol, 10.1083/jcb.201910185. [DOI] [Google Scholar]
- Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S. (2001). Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell , 667–677. [DOI] [PubMed] [Google Scholar]
- Romanauska A, Kohler A. (2018). The inner nuclear membrane is a metabolically active territory that generates nuclear lipid droplets. Cell , 700–715.e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothballer A, Kutay U. (2013). Poring over pores: nuclear pore complex insertion into the nuclear envelope. Trends Biochem Sci , 292–301. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S. (2005). The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J , 1931–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HB, Gorlich D. (2016). Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem Sci , 46–61. [DOI] [PubMed] [Google Scholar]
- Shackleton S, Lloyd DJ, Jackson SN, Evans R, Niermeijer MF, Singh BM, Schmidt H, Brabant G, Kumar S, Durrington PN, et al. (2000). LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat Genet , 153–156. [DOI] [PubMed] [Google Scholar]
- Shin JY, Hernandez-Ono A, Fedotova T, Ostlund C, Lee MJ, Gibeley SB, Liang CC, Dauer WT, Ginsberg HN, Worman HJ. (2019). Nuclear envelope-localized torsinA-LAP1 complex regulates hepatic VLDL secretion and steatosis. J Clin Invest . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoyer CJ, Jaspersen SL. (2019). Patrolling the nucleus: inner nuclear membrane-associated degradation. Curr Genet , 1099–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, Li Y, Ataman B, Koon A, Chang YT, Li Q, et al. (2012). Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell , 832–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Banigan EJ, Marko JF. (2019). Chromatin’s physical properties shape the nucleus and its functions. Curr Opin Cell Biol , 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szollosi MS, Szollosi D. (1988). “Blebbing”’ of the nuclear envelope of mouse zygotes, early embryos and hybrid cells. J Cell Sci (Pt 2), 257–267. [DOI] [PubMed] [Google Scholar]
- Tange Y, Hirata A, Niwa O. (2002). An evolutionarily conserved fission yeast protein, Ned1, implicated in normal nuclear morphology and chromosome stability, interacts with Dis3, Pim1/RCC1 and an essential nucleoporin. J Cell Sci , 4375–4385. [DOI] [PubMed] [Google Scholar]
- Thaller DJ, Allegretti M, Borah S, Ronchi P, Beck M, Lusk CP. (2019). An ESCRT-LEM protein surveillance system is poised to directly monitor the nuclear envelope and nuclear transport system. Elife , e45284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaller DJ, Lusk CP. (2018). Fantastic nuclear envelope herniations and where to find them. Biochem Soc Trans , 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timney BL, Raveh B, Mironska R, Trivedi JM, Kim SJ, Russel D, Wente SR, Sali A, Rout MP. (2016). Simple rules for passive diffusion through the nuclear pore complex. J Cell Biol , 57–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimble WS, Grinstein S. (2015). Barriers to the free diffusion of proteins and lipids in the plasma membrane. J Cell Biol , 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PL, Zhao C, Schlieker C. (2019). Methodologies to monitor protein turnover at the inner nuclear membrane. Methods Enzymol , 47–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PL, Zhao C, Turner E, Schlieker C. (2016). The Lamin B receptor is essential for cholesterol synthesis and perturbed by disease-causing mutations. Elife , e16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji T, Cheng J, Tatematsu T, Ebata A, Kamikawa H, Fujita A, Gyobu S, Segawa K, Arai H, Taguchi T, et al. (2019). Predominant localization of phosphatidylserine at the cytoplasmic leaflet of the ER, and its TMEM16K-dependent redistribution. Proc Natl Acad Sci USA , 13368–13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner EM, Schlieker C. (2016). Pelger-Huët anomaly and Greenberg skeletal dysplasia: LBR-associated diseases of cholesterol metabolism. Rare Dis , e1241363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbreit NT, Pellman D. (2017). Cancer biology: genome jail-break triggers lockdown. Nature , 340–341. [DOI] [PubMed] [Google Scholar]
- Ungricht R, Klann M, Horvath P, Kutay U. (2015). Diffusion and retention are major determinants of protein targeting to the inner nuclear membrane. J Cell Biol , 687–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungricht R, Kutay U. (2017). Mechanisms and functions of nuclear envelope remodelling. Nat Rev Mol Cell Biol , 229–245. [DOI] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. (2008). Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol , 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. (2008). One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol , 944–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventimiglia LN, Cuesta-Geijo MA, Martinelli N, Caballe A, Macheboeuf P, Miguet N, Parnham IM, Olmos Y, Carlton JG, Weissenhorn W, Martin-Serrano J. (2018). CC2D1B coordinates ESCRT-III activity during the mitotic reformation of the nuclear envelope. Dev Cell , 547–563.e546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri M, Radulovic M, Stenmark H. (2020). The many functions of ESCRTs. Nat Rev Mol Cell Biol , 25–42. [DOI] [PubMed] [Google Scholar]
- Vietri M, Schink KO, Campsteijn C, Wegner CS, Schultz SW, Christ L, Thoresen SB, Brech A, Raiborg C, Stenmark H. (2015). Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature , 231–235. [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA. (2007). Sheets, ribbons and tubules — how organelles get their shape. Nat Rev Mol Cell Biol , 258–264. [DOI] [PubMed] [Google Scholar]
- Wang S, Tukachinsky H, Romano FB, Rapoport TA. (2016). Cooperation of the ER-shaping proteins atlastin, lunapark, and reticulons to generate a tubular membrane network. Elife , e18605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weberruss M, Antonin W. (2016). Perforating the nuclear boundary — how nuclear pore complexes assemble. J Cell Sci , 4439–4447. [DOI] [PubMed] [Google Scholar]
- Webster BM, Colombi P, Jager J, Lusk CP. (2014). Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell , 388–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster BM, Lusk CP. (2016). Border safety: quality control at the nuclear envelope. Trends Cell Biol , 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Blobel G. (1993). A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J Cell Biol , 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Rout MP. (2010). The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol , a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Neuner A, Ruthnick D, Sachsenheimer T, Luchtenborg C, Brugger B, Schiebel E. (2018). Brr6 and Brl1 locate to nuclear pore complex assembly sites to promote their biogenesis. J Cell Biol , 877–894. [DOI] [PMC free article] [PubMed] [Google Scholar]