Abstract
Protein aggregation, once believed to be a harbinger and/or consequence of stress, age, and pathological conditions, is emerging as a novel concept in cellular regulation. Normal versus pathological aggregation may be distinguished by the capacity of cells to regulate the formation, modification, and dissolution of aggregates. We find that Caenorhabditis elegans aggregates are observed in large cells/blastomeres (oocytes, embryos) and in smaller, further differentiated cells (primordial germ cells), and their analysis using cell biological and genetic tools is straightforward. These observations are consistent with the hypothesis that aggregates are involved in normal development. Using cross-platform analysis in Saccharomyces cerevisiae, C. elegans, and Xenopus laevis, we present studies identifying a novel disaggregase family encoded by animal genomes and expressed embryonically. Our initial analysis of yeast Arb1/Abcf2 in disaggregation and animal ABCF proteins in embryogenesis is consistent with the possibility that members of the ABCF gene family may encode disaggregases needed for aggregate processing during the earliest stages of animal development.
INTRODUCTION
Amyloid research has historically emphasized severe age-related neuropathologic conditions such as Alzheimer’s and Huntington’s diseases (Knowles et al., 2014); however, a growing number of examples in yeast, snails, fruit flies, and bacterial biofilms shows that proteins in amyloid conformations can provide important benefits (Newby and Lindquist, 2013). Conformational switching between native and amyloid states of certain proteins influences cellular phenotype (Chernoff et al., 1995; Saupe, 2000; Balguerie et al., 2003; Fowler et al., 2006; Rambaran and Serpell, 2008; Halfmann et al., 2012), and we now know that many proteins form (Maji et al., 2009; Si et al., 2010; Newby and Lindquist, 2013), or can form amyloids (Kato et al., 2012). Amyloid particles are resistant to denaturants and proteolytic cleavage and are thus useful in protein assembly and storage. Importantly, amyloid assembly is reversible; released proteins can refold into their native state (Tompa, 2012), as exemplified by amyloid-based storage of mammalian peptide hormones in presecretory vesicles, which revert to a soluble, functional monomeric form on secretion and exposure to the extracellular environment (Maji et al., 2009).
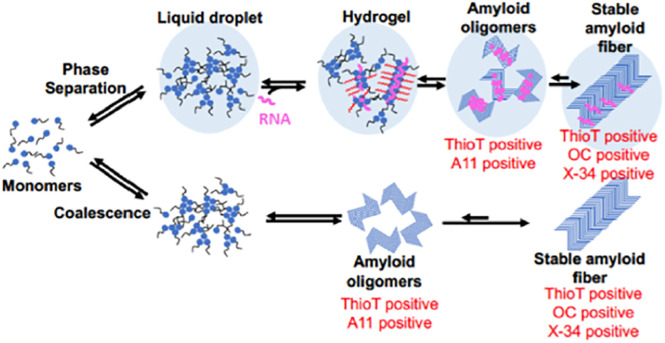
However, not all protein aggregates are amyloid. Protein aggregates are a heterogenous collection of protein and protein–RNA coalescences that can differ in material state. Phase-separated condensates known as liquid droplets are quite dynamic, while gel-like semisolids formed by the addition of RNA are more stable. Many reserve the term amyloid for the most stable fibrillar aggregates, though it is evident that the cross-beta structure often used as the defining structure of amyloids shows up in oligomers and gels that are part of a structural continuum (Figure 1) (Kato and McKnight, 2018). Defining the material state of aggregates in vivo remains challenging, at least in part because aggregates may be heterogeneous and capable of transitions between states. Regardless of the material state in which they occur, structurally distinctive properties in aggregates can be identified using specific reagents including dyes like Congo Red and Thioflavin T (ThioT) and antibodies that recognize epitopes in amyloid oligomers or fibers (Molliex et al., 2015).
FIGURE 1:
Simplified overview of aggregation pathways. Intrinsically disordered monomeric proteins self-associate to create an assortment of transient higher-order structures including liquid droplets, hydrogels, and the highly stable solid amyloids. These structures are differentially detected by stains such as ThioT and X-34 and conformer specific antibodies including the oligomer-specific A11 antibody and the fiber-specific OC antibody. Modified from Hayes et al. (2018) and Wu and Fuxreiter (2016).
Disaggregases participate in the recovery of proteins aggregated as a result of stress or chemical exposure and in the recovery of active monomers from highly ordered amyloid fibers. For example, the fungal amyloid-remodeling chaperone, Hsp104, is a dynamic ring-translocase and hexameric AAA+ protein that couples ATP hydrolysis with disassembly and reactivation of proteins trapped in soluble preamyloid oligomers, disordered protein aggregates, and stable amyloid or prion conformers (Sweeny and Shorter, 2016). In addition, Hsp104 converts long fibers into small oligomeric “propagons” that support amyloid maintenance and transmission to daughter cells (Shorter and Lindquist, 2004; Romanova and Chernoff, 2009). Although protein aggregation is found in all eukaryotes, there is no HSP104 homologue in animal genomes. The absence of a metazoan counterpart to the fungal HSP104 gene (Erives and Fassler, 2015) has prompted investigators to look for alternative animal proteins that fulfill its functions. These efforts led to the identification of the Hsp110, Hsp70, and Hsp40 chaperones that work together to extract and refold some proteins from denatured aggregates (Shorter, 2011; Rampelt et al., 2012) although more slowly than Hsp104 (Duennwald et al., 2012). In addition, the small Hsps, Hsp26 and Hsp42 in yeast, and HspB5 in human (Nillegoda et al., 2018), were found to both enhance Hsp104 catalyzed disaggregation and potentiate disaggregation by the Hsp110-70-40 complex (Duennwald et al., 2012; Gao et al., 2015). Finally, as summarized in recent reviews (Lackie et al., 2017; Stroo et al., 2017), a small number of additional genes have likewise been found to contribute to solubilization of amyloid proteins in animal models of human amyloid diseases in an Hsp104/Hsp110-independent manner.
In addition to HSP104, the yeast genome includes a fungal-specific disaggregase encoded by the NEW1 gene (Inoue et al., 2011). In in vitro studies, the New1 protein exhibits Hsp104-independent disaggregase activity toward the well-characterized Sup35 [PSI+] amyloid (prion) (Inoue et al., 2011). Based on their close phylogenetic relationship to the New1 disaggregase, we hypothesized that ABCF genes, present in animals as well as fungi, might encode an animal disaggregase. New1 and the ABCF proteins are members of the ATP binding cassette (ABC) superfamily. Interestingly, the ABCF proteins, the related New1, and the phylogenetically unrelated Hsp104 are all members of the P-loop domain superfamily (Snider and Houry, 2008) characterized by a P loop NTPase domain consisting of a conserved nucleotide phosphate-binding motif referred to as the Walker A motif and a second more variable region known as the Walker B motif (Walker et al., 1982; Saraste et al., 1990; Koonin et al., 2000). The AAA+ subfamily proteins (which include Hsp104) contain a 200–250 amino acid (aa) 4-helix bundle AAA+ module which, together with the P loop domain, sandwiches the nucleotide while the nucleotide binding folds of prototypical ABC ATPase subfamily proteins consists of characteristic Walker A and B boxes separated by ∼120 aa residues including the ABC signature motif (LSGGQ) (Snider and Houry, 2008). Most ABC domain proteins are membrane associated; however, the “F” subfamily, encoded in all eukaryotic genomes (Kerr, 2004), is soluble, lacking the transmembrane domains, but retaining ATP binding and hydrolysis activities. Several ABCF proteins have been implicated in ribosome biogenesis and translation (Marton et al., 1997; Kerr, 2004; Dong et al., 2005; Boel et al., 2014).
Here we show that the yeast Abcf2 protein encoded by the ARB1 gene also promotes aggregate processing. This novel activity for ABCF proteins is supported by mass spectrometry analysis of the Arb1 interactome (Dong et al., 2005) showing association of Arb1 with Zuo1, Ydj1, and Ssa2 chaperones, each involved in different aspects of recovery from or prevention of protein misfolding. The dnaJ domain protein, Zuo1, is particularly interesting because it works together with Ssz1 and Ssb1/2 Hsp70s in the folding of nascent polypeptide chains at the ribosome and also (like Arb1) plays a role in ribosome biogenesis (Gautschi et al., 2001; Albanese et al., 2010; Koplin et al., 2010; Preissler and Deuerling, 2012).
Mammalian Abcf2 proteins have been studied in a number of contexts. Abcf2 was found to interact with the injected enteropathogenic Escherichia coli effector protein EspF in the mitochondria. These studies find that EspF facilitates host cell death by interfering with a protective function provided by Abcf2. Mammalian Abcf2 also interacts with nonmuscle alpha-actinin (Ando-Akatsuka et al., 2012) to control the regulatory volume decrease process in human epithelial cells.
The relationship between the Abcf proteins and the yeast New1 disaggregase, coupled with the ability of New1 to solubilize amyloid proteins (Inoue et al., 2011), prompted us to consider the possibility that ABCF proteins may play a role in aggregate processing. We find that yeast Arb1 makes a contribution to the disaggregation of disordered (heat-denatured) aggregates and to the cleavage or maintenance and propagation of ordered amyloid fibers, that animal ABCF proteins have related functions; and finally, that reduced ABCF levels in Caenorhabditis elegans and Xenopus laevis lead to defects in early development.
RESULTS
Aggregates are present in the C. elegans germline and early embryo
Amyloids have primarily been studied in fully developed or in aging tissue. In this study, we ask whether the presence of amyloids in oocytes as seen in Xenopus (Hayes and Weeks, 2016) is a ubiquitous feature of early animal development, and if so, whether they are crucial to a successful developmental program, and finally, what function(s) they may carry out. To examine this, we first looked for amyloid-like particles in C. elegans germlines and embryos using well-accepted methods for amyloid detection. These include: 1) ThioT, a nontoxic chemical dye that fluoresces when bound to amyloid (Vassar and Culling, 1959; Biancalana and Koide, 2010; Groenning, 2010); 2) X-34, a fluorescent derivative of Congo red which binds to beta-sheet structures (Styren et al., 2000; Link et al., 2001) and two amyloid-specific antibodies; 3) A11, an antibody that specifically detects the conformation of amyloid oligomers regardless of their amino acid sequence (Kayed et al., 2007); and 4) OC, which recognizes amyloid fibrils (Kayed et al., 2007) (Figure 1). We observed distinct puncta in the germline or early embryo with ThioT and X-34 (Supplemental Figure S1), as well as with A11 and OC antibodies. Here we focus on patterns we observe in A11-stained samples as the A11 antibody provides excellent signal to background resolution. We find that A11 antibody stains both centrosomes of dividing blastomeres (Figure 2, B and C white arrows, B’ and C’ show centrosomes in the same embryos marked by anti-γ-tubulin), a pattern also seen with the OC antibody (Figure 2D), the nuclear membrane in germline oocytes and embryos (Figure 2, E and inset, and F, white arrowheads), and perinuclear puncta that localize to the P cell lineage (Figure 2, G black arrows, and H–H’’’), consistent with the pattern of RNA processing bodies known as P granules.
FIGURE 2:
Endogenous C. elegans amyloids. (A) Hermaphrodite gonad cartoon showing germline and early embryos. (B, C) A11 antibody positive puncta (green, arrows) in a 1-cell zygote (B) and an eight-cell embryo (C). (B’, C’) γ-Tubulin staining (red) shows these puncta colocalize with centrosomes. (D) A similar pattern is also seen with the OC antibody (green). (E, F) A11 (green) stains nuclear membranes in germline oocytes (arrowheads in E), all cells in early embryos (E, inset arrowheads), and select cells in late embryos (arrowheads in F). Cells in F identified as PGCs by the PES-1 (red) expression in the adjacent somatic gonadal precursors (arrows). (G) Noncentrosomal A11 puncta in the P2 cell that become visible with longer exposure (black arrows). (A–F) Blue, DAPI-stained DNA. (H–H’’’) A subdomain of C. elegans P granules displays amyloid character. Embryonic P cell stained for A11 (green in H), P granule marker, PGL-1 (red in H’), and DAPI-stained DNA (blue in H’’). (H–H’’’) Arrows denote P granules that are A11 negative, while arrowheads denote P granules that are A11-positive; c, centrosome. Frequencies of depicted phenotypes are as follows: (B,C, E, G) 100% n > 100; (D) 100% n = 63; (F) 100% n = 17; (H and H’’) 100% n > 100; (H’’’) 77% of P granules are positive for A11 n > 100. Scale bar: 12 um in B–D and G; 20 um in E; 44 um in E, inset; 21 µM in F; and 6 µM in H–K.
Thus, C. elegans embryos begin life with an appreciable amount of protein in A11-positive protein deposits, a characteristic shared with Xenopus (Hayes and Weeks, 2016). Interestingly, newly fertilized worm oocytes and early embryos exhibit ubiquitous A11-positive centrosomes and nuclear membranes, while only two cells exhibit A11 staining (Figure 2F, arrowheads) at the late embryonic stage, suggesting cell type-specific solubilization of most aggregates has occurred during embryonic development. We found PES-1 expression in the adjacent somatic gonad cells (Figure 2F, arrows), indicating that cells with persistent A11 staining are the two primordial germ cells (PGCs), and that the embryonic germline may already be converting (or continuing to retain) germline proteins to A11-positive status via aggregation at this early stage.
The staining we observe is consistent with reported amyloid-like aggregates at various subcellular locales (Ader et al., 2010; Kato et al., 2012; Zwicker et al., 2014; Hayes and Weeks, 2016), but the P granule pattern was somewhat surprising since P granules are reported to display liquidlike behaviors and have been described as homogeneous highly dynamic liquid droplets that lack solid components (Brangwynne et al., 2009) and would therefore not be expected to stain with A11. However, recent reports using lattice sheet microscopy suggest that P granules are nonhomogeneous with subdomains consisting of a more solid, gel-like and nondynamic condensate (Putnam et al., 2018). We tested the idea that P granules have distinct A11-positive containing compartments by costaining P granules with A11 and a P granule marker, anti-PGL-1. We find P granules can be positive (Figure 2, H and H’’’ arrowheads) or negative (Figure 2H-H’’’ arrows) for A11 and that when present, A11 stains the interior of the P granule (Figure 2H’’’). We also observe heterogeneity at centrosomes,which exhibit a large A11 domain with an internal OC reactive compartment, suggesting that centrosomes may consist of a fibrillar amyloid core embedded in an oligomeric domain (Figure 2, B, D, and H; Supplemental Figure S1, D–G).
A role for embryonic amyloid in development
Given the absence of HSP104 from animal genomes, we speculated that stable aggregates might be required during development. We hypothesized that the regulated assembly and disassembly of embryonic aggregates are controlled by endogenous disaggregases with specific clients and that the introduction of broad spectrum disaggregase like Hsp104 might perturb development. To test this, we introduced a variant of fungal HSP104 with increased activity in worms (“potentiated” Hsp104 A503V; generous gift of J. Shorter) (Jackrel et al., 2014) via mRNA injection into the worm germline (Table 1 and Figure 3). As a control, we conducted injections of GFP mRNA, which resulted in minimal defects in oocytes and embryos and normal A11 embryonic patterning (for examples, see Figure 3, A and B). In contrast, potentiated Hsp104 mRNA injections resulted in oocytes and embryos with perturbed A11 patterns (Figure 3, K–N). We also see altered germline morphology and disrupted germline A11 staining specifically after potentiated Hsp104 misexpression (see Table 1). In embryos injected with potentiated Hsp104, we note extra A11-positive nuclear puncta (Figure 3K), decreased/aberrant A11 staining of the centrosome (Figure 3, L–N), and/or absent/faint nuclear membrane staining (Figure 3, K–N) in 40% (n = 110) of oocytes or embryos after potentiated Hsp104 injection.
TABLE 1:
Effect of Hsp104 misexpression in C. elegans.
| Phenotype | GFP mRNA | Mutant HSP104 mRNA | Potentiated HSP104 mRNA | HSP104 protein |
|---|---|---|---|---|
| Germline organization defect | 0% (n = 13) | 0% (n = 23) | 22% (n = 77) | 6% (n = 17) |
| Altered A11 germline distribution | 0% (n = 13) | 0% (n = 23) | 48% (n = 77) | 65% (n = 17) |
| Embryonic cell division defect | 3% (n = 30) | 26% (n = 82) | 30% (n = 110) | 23% (n = 22) |
| Altered A11 embryonic distribution | 3% (n = 30) | 1% (n = 82) | 40% (n = 110) | 41% (n = 22) |
FIGURE 3:
Effects of HSP104 misexpression in C. elegans. (A–J) Typical control-injected embryos after GFP mRNA injection (A, B) or buffer injection (C–E) showing normal distribution of A11 on oocyte nuclear membrane (A) and embryonic nuclear membranes and centrosomes at the 1- (B), 4- (C), 16- (D), and 32-cell stage (E). (F–J) Corresponding DAPI-stained DNA images of A–E. (K–N) A11-stained oocyte (K) and embryos (L–N) after potentiated HSP104 mRNA injections. (K) Oocyte exhibiting dim nuclear membrane A11 but additional A11 nuclear puncta (arrowheads). (L) Dividing one-cell embryo with dim centrosomal (arrows) and nuclear membrane A11. (M) Four- to eight-cell embryo with dim and misshapen centrosomes (arrows). (N) Thirty-two-cell embryo with variably dim centrosomes (arrows). (O–R) Corresponding DAPI-stained DNA images of K–N. (S–U) A11-stained embryos after mutant HSP104 mRNA injections. (S, T) Aberrant three-cell embryo with two cells in telophase and one in anaphase (S) and six-cell embryo (T) but exhibiting normal A11 patterns. (U) Thirty-two-cell embryo showing normal A11 and cell division patterns after mutant HSP104 mRNA injection. (V–X) Corresponding DAPI-stained DNA images of (S–U). (Y–Z) Sixteen-cell embryo with dim centrosomal A11 puncta (arrows) after injection of wild-type Hsp104 protein. (Y’) Same A11 sample as Y, but with 70% brighter exposure. (Z) Corresponding DAPI-stained DNA image of Y and Y’. Frequencies of depicted phenotypes are as follows: (A–E) 97% n = 30; (K–N) 40% abnormal A11 patterning and 30% defective cell division defects, n = 110; (S–U) 99% normal A11 pattern n = 82; (S, T) 26% abnormal cell division defects n = 82; (U) 74% normal cell division pattering n = 82; (Y–Z) 41% abnormal A11 n = 22. Scale bar, 10 µm
Since ATPase activity is not required for all aspects of Hsp104 function (Zhang et al., 2019), we also injected a mutant form that has reduced (8%) ATPase activity (Torrente et al., 2016). Interestingly, although germline morphology and all A11 patterns were unaffected, 26% of embryos (n = 82) exhibited cell cycle timing defects and spindle alignment defects including three-celled embryos (Figure 3S) as well as elongated embryos (Figure 3T). The remaining 74% of mutant Hsp104 mRNA injections produced normal embryos (Figure 3U). Similar levels (30%, n = 110 embryos) of cell cycle defects were observed among embryos injected with potentiated Hsp104 (Table 1). These cell cycle defects were observable because C. elegans embryonic blastomeres divide nearly simultaneously with invariant spindle orientation, such that odd numbers of cells or embryos with variable cell arrangements are never ordinarily observed. Importantly, though expression of either form of Hsp104 led to developmental defects, only active Hsp104 showed defective A11 patterns. Together these data indicate that the A11-positive aggregates we observe in early development are sensitive to disaggregase activity and that unregulated expression of Hsp104 causes developmental defects, though some aspects of this phenotype may not require ATP hydrolysis.
While these results suggest that potentiated Hsp104 disrupts early development, we were concerned that the severe germline and embryonic cell division defects in many of the mRNA-injected worms may have had pleiotropic effects. Therefore, we turned to commercially available Hsp104 protein injections to circumvent the delay required for mRNA translation and nascent protein folding and to avoid possible dysregulation of RNA metabolism in the germline. Although commercial Hsp104 is not a potentiated protein, Hsp104 has been reported to stimulate disaggregation in vitro by interacting with Hsp110-Hsp70-Hsp40 complexes (Shorter, 2011). Like the mRNA injections, Hsp104 protein injections into worm germlines gave rise to aberrant embryonic cell division patterns and eventual embryonic lethality (23% of embryos had defective cell division patterns; n = 22) but protein injections had milder effects on P0 germlines (Hsp104 protein injections caused morphological defects in 6% of P0 germlines compared with 22% for mRNA injections, Table 1) and embryonic defects occurred later in development. Thus, effects of Hsp104 activity on A11-positive aggregate accumulation could be observed in germlines and embryos lacking obvious morphological defects. Importantly, even mildly affected germlines and embryos exhibited reduced A11 staining (increased exposure times are required to visualize the A11 expression pattern in 41% [9/22] embryos quantified) indicating a specific effect on amyloid structures in the early embryo (Figure 3, Y and Z). These phenotypes are not seen in buffer-injected embryos (e.g., Figure 3, C–E). The above data are consistent with the idea that the HSP104 gene is incompatible with animal development and requisite protein aggregation patterns.
Phylogenic and genetic analyses of yeast ABCF genes encoding a novel candidate disaggregase protein family
In addition to HSP104, the yeast genome encodes a fungal-specific disaggregase encoded by the NEW1 gene (Inoue et al., 2011). In in vitro studies, the New1 protein exhibits Hsp104-independent disaggregase activity toward the well-characterized Sup35 [PSI+] amyloid (prion) (Inoue et al., 2011). Phylogenetically, the taxonomically ubiquitous ABCF proteins are sister to the New1/EF3 clade (Boel et al., 2014) (Supplemental Figure S2). Based on their phylogenetic relationship, we hypothesized that ABCF proteins might encode one or more animal disaggregases. Animal genomes include three ABCF family members, ABCF1, ABCF2, and ABCF3. ABCF3 is orthologous to yeast GCN20 and ABCF2 is orthologous to the yeast ARB1 gene. While strains lacking ARB1 are inviable, strains depleted for Arb1 protein are viable, but slow growing. Such strains exhibit defects in 35S pre-rRNA, 20S rRNA processing, and other rRNA processing events in the 60S biogenesis pathway (Dong et al., 2005). Consistent with these defects, Arb1 is physically associated with various ribosomal proteins and ribosomal biogenesis factors among others (Dong et al., 2005).
To study the effects of reduced levels of the essential Arb1 protein, we used strains expressing an unstable Arb1 protein (Arb1-d) under the control of the carbon source-regulated UASG element (Dong et al., 2005). We used α-FLAG Western analysis to investigate the impact of carbon source (2 h) on Arb1 protein expression from the UASG-ARB1-d construct relative to expression from a similarly FLAG-tagged construct in which ARB1 is under the control of its own promoter (Dong et al., 2005). We found that protein expression from the UASG-ARB1-d construct, even grown in permissive galactose conditions, is fivefold lower than expression from PARB1 ARB1 expression plasmid and that a shift to glucose reduces expression levels at minimum another 10-fold (Figure 4A). Hence the three constructs represent something of an allelic series with respect to Arb1 protein levels. As expected, viability and growth rate were reduced on glucose media but not on galactose media (Figure 4B) (Dong et al., 2005). Hence the reduction in steady state Arb1 protein levels in cells expressing UASG-ARB1-d as their only source of Arb1 is tolerated. Depletion of Arb1 caused temperature sensitivity; the poor growth phenotype of glucose grown cells was exacerbated at 26 and 37°C (Figure 4C). In addition, we found that the Arb1-depleted strain is sensitive to the aminoglycoside antibiotic, Hygromycin B (HB) which stabilizes the tRNA-ribosomal acceptor site, thus inhibiting translocation and polypeptide synthesis (Figure 4C).
FIGURE 4:
Phenotypic analysis of Arb1-depleted strains. (A) Western analysis using α-FLAG antibody to gauge levels of Arb1 protein expressed from an arb1 deletion strain carrying the UASG-ARB1-d depletion construct [pDH25.1] (Dong et al., 2005) cultivated in galactose or glucose, relative to Arb1 protein levels from pDH29 (Dong et al., 2005), in which the ARB1 gene is expressed under the control of its own promoter. (B) Growth of the arb1 deletion strain carrying pDH25.1 (ARB1-d) or pDH22 (ARB1) (Dong et al., 2005) after serial dilutions on YPD (glucose) or YP Gal plates (galactose). (C) Arb1 depletion causes temperature and HB sensitivity. YPD plates containing the indicated concentrations of HB were incubated at 26, 30, or 37°C for 2–5 d as appropriate. (D) Arb1 depletion strain phenotypes are exacerbated by the absence of the New1 disaggregase. Strains were grown to early log phase in YP Gal Raff media at 30°C and serial dilutions spotted on YP Gal Raff (Gal Raff) or YPD (glucose) plates with 0 or 30 μg/ml HB which were incubated at 26°C (4 d) or 18°C (7 d). arb1Δ [pDH25.1] (YDH226) (Dong et al., 2005); hsp104Δ (JF2498); new1Δ arb1Δ [pDH25.1] (JF2510); hsp104Δ arb1Δ [pDH25.1] (JF2511).
Because of the close phylogenetic relationship between Arb1 and New1 (Supplemental Figure S2), we tested for potential functional relationships with disaggregase type chaperones including Hsp104, the mitochondrially localized Hsp104 paralogue, Hsp78, and New1. Deletion mutations in each gene were crossed into the arb1 deletion strain carrying the UASG-ARB1-d plasmid. Double mutants were identified by colony PCR and their growth was compared with single mutants on glucose or galactose-raffinose media at 26°C with or without HB. The new1Δ mutation increased HB and low temperature sensitivity of the arb1 (UASG-ARB1-d) strain, whereas the hsp104Δ (Figure 4D) and hsp78Δ (unpublished data) mutations had no effect. These results are consistent with the possibility that New1 and Arb1 represent distinct disaggregation pathways although other more complex genetic interactions are more likely since disaggregation phenotypes (below) seen in the Arb1-depleted strains were not exacerbated in the double mutants.
ABCF2 proteins exhibit disaggregase activity
Protein disaggregases are among the least understood components of the proteostasis network; however, the yeast Hsp104 prototype is known to couple ATP hydrolysis to the rapid dissolution and reactivation of diverse proteins trapped in disordered aggregates as well as ordered stress-induced assemblies, preamyloid oligomers, amyloids, and prions (Shorter, 2017). To investigate disaggregase type chaperone activities for Arb1, we first tested whether Arb1 plays a role in reversing disordered aggregates using heat-denatured firefly luciferase as a test case. We examined the effect of Arb1 depletion on resolubilization of GFP puncta caused by heat denaturation of an FFL-GFP fusion protein (Figure 5). Firefly luciferase in this construct has been sensitized to heat denaturation, while GFP is heat stable (Abrams and Morano, 2013) and continues to fluoresce throughout and following heat treatment. Greater than 95% of cells of all strains regardless of genotype contained at least one, and typically many aggregates immediately following heat treatment (time 0) and 80–90% of cells of all genotypes contained aggregates 30 min later. At 60 min postheat treatment, differences among the strains start becoming evident, with the Arb1-depleted strain (ARB1-d glucose) lagging in its recovery (dissolution of puncta caused by heat denaturation) relative to wild-type (wt, 1x Arb1), Arb1 from galactose-grown cultures (ARB1-d galactose, ∼20% Arb1), and new1Δ strains. The Arb1-depleted strain (∼2% Arb1) continued to dissolve aggregates from 60 to 90 min, but significantly slower than other strains. Strains lacking HSP104 (hsp104Δ), used as a control, show no reduction in cells with aggregates throughout the entire time course. As expected from the absence of any defect in luciferase resolubilization in the new1 mutant, the new1 mutation had no impact on the Arb1 depletion phenotype when tested in double mutants (unpublished data). Hence normal Arb1 levels appear to facilitate or increase the rate of disaggration of disordered aggregates like luciferase-GFP (Figure 5).
FIGURE 5:
Arb1 depletion delays the dissolution of heat-denatured FFL-GFP aggregates. (A) Quantitation of the percentage of cells with aggregates remaining after 30, 60, or 90 min of recovery time following heat treatment. Strains of different genotypes, all carrying the p426Met25-FFL-GFP aggregation reporter plasmid, were grown to early log phase at 30°C in selective media with galactose and raffinose as carbon sources. Cultures were then switched to selective media with glucose as carbon source for 2.5 h to deplete Arb1 and then to media lacking methionine for 1.5 h to express the FFL-GFP gene. Finally, cultures were heat treated (25 min, 42°C) and allowed to recover at 30°C for the indicated times before imaging. A minimum of 100 cells of each genotype were tabulated for each time point. Data for each strain and time point are the averages of three to six trials involving at least three transformants. Error bars are the SD of the means. ANOVA analysis with Tukey post hoc tests were used to determine the significance of observed difference in phenotype of the wild-type strain relative to strains of all other genotypes (excluding hsp104Δ) at each time point (30′, 60′, and 90′). The ARB1-d (glucose) sample was significantly different (**P < 0.01) from the wild-type strain at all three times. Other comparisons (not indicated in the figure for clarity) were nonsignificant with the exception of new1 and ARB1-d (glucose and galactose) (p < 0.01, 90′) and the arb1(ARB1) control and ARB1-d (glucose and galactose (p < 0.01, 90′). (B) Representative images for strain YDH226 under Arb1 depletion (glucose) and Arb1 expressing conditions (galactose) at indicated times following heat shock. Strains were wild type (BY4730), UASG-ARB1 (YDH226), new1Δ (JF2475), and hsp104Δ (JF2473) transformed with the pRS426 UASMET25FFL-GFP plasmid.
Htt-Q 97 processing is affected by Arb1 depletion
Given the involvement of Arb1 in disaggregation of disordered luciferase aggregates, we were interested in evaluating Arb1 for a potential role in amyloid processing. To investigate this, we took advantage of a set of constructs consisting of exon 1 of the human Huntingtin protein (Htt) fused to GFP (Krobitsch and Lindquist, 2000). Constructs differ in the length of the Htt exon 1 glutamine tract, having either the normal short Q tract length (∼20) or various longer lengths including 46 glutamines, and longer pathological lengths such as 97 (Q97). Expression of each Htt-Qx genes is driven by the upstream activating sequence from the TDH3 gene (formerly known as GPD) which is active in glucose grown cultures at normal temperatures. In preliminary comparisons of wild-type strains carrying Htt-Q25, Htt-Q46 and Htt-Q97, only Htt-Q97 containing strains had appreciable levels of GFP puncta and the low level (∼10% of cells) of puncta observed in wild-type strains carrying Htt-Q46 was not significantly influenced by other genotypes (Supplemental Figure S3), so the analysis of the shorter reporters was not pursued.
Consistent with previous studies, we found that Htt-Q97 is almost completely soluble in strains lacking Hsp104 (Krobitsch and Lindquist, 2000). This is because in addition to its disaggregation activity, Hsp104 is involved both directly and indirectly in the maintenance of amyloids (Chernoff et al., 1995; Shorter and Lindquist, 2004, 2006; Aron et al., 2007; Bardill et al., 2009; Bardill and True, 2009). We therefore anticipated that a reduction in the number of cells with aggregates might also be seen in the Arb1-depleted cultures.
The aggregation behavior of the Htt-Q97-GFP fusion protein was compared in haploid strains of varying genotypes including arb1Δ carrying an ARB1 expression plasmid, new1Δ, hsp104Δ, Arb1-d depleted (glucose, down arrow), and Arb1-d expressing (galactose, up arrow) strains. As expected, most cells (80%) in the wild-type and wild-type-like (arb1Δ carrying an ARB1 expression plasmid) cultures contained visible aggregates compared with only 1–5% of cells in the hsp104Δ culture. By contrast, in strains lacking New1 or depleted for Arb1 (ARB1-d, glucose), 60–65% of cells had visible aggregates, a significant (p < 0.01) reduction compared with wt controls (Figure 6A). A new1 arb1 (glucose) double mutant was not significantly different from the single mutants. Our analysis suggests that ARB1 is required for efficient maintenance of the Htt-Q97-GFP reporter. These results are consistent with an Hsp104-like role for Arb1 in aggregate processing.
FIGURE 6:
Arb1 depletion causes a defect in maintenance (propagation) of Htt-Q97 amyloids. (A) Percentage of cells maintaining Htt-Q97 –GFP aggregates in the presence or absence of candidate chaperones. Cells were grown to early log phase in selective media containing galactose (up arrow) at 30°C at which point the culture was split and galactose was washed out and replaced with selective media containing glucose (down arrow) prior to growth for another 4 h. Data for each strain are the averages of three transformants and two to three trials. Error bars are the SD of the means. Strains were arb1Δ with pDH22 (ARB1), new1Δ (JF2475), UASG-ARB1-d (arb1Δ with pDH25, YDH226) (Dong et al., 2005), and hsp104Δ (JF2473). All strains carried the p416-Htt-Q97-GFP plasmid. The significance of differences in phenotype between the wt strain and strains of other genotypes was assessed by ANOVA-Tukey HSD analysis (**p < 0.01). (B, C) Representative SDD-AGE (B) and SDS–PAGE (C) anti-GFP Western analysis of extracts prepared from haploid strains wt (BY4730), hsp104Δ (JF2473), new1 Δ (JF2475), and the arb1Δ strain YDH226 carrying the pDH25.1 Arb1 depletion plasmid. In later experiments, an arb1Δ (pARB1) strain was also included (not shown) as it proved to be a better control than the galactose grown pDH25.1, which expresses reduced levels of Arb1 due to protein destabilization (Dong et al., 2005) (Figure 4 and Supplemental Figure S4B). Three transformants were analyzed in each experiment. Approximately equal loading of extracts across lanes was evaluated by quantitation of the SDS–PAGE gel following REVERT (LI-COR) staining. In the SDD-AGE experiment, wild-type extracts were loaded at 0.5× so that the signal was more even across the blot. The difference in loading was accounted for in the analysis. (D) Aggregate load was calculated as the percentage of aggregated GFP signal over the total signal (SDS–PAGE + SDD-AGE). Significance was evaluated by ANOVA-Tukey HSD analysis (Tukey, 1949) per experiment (three transformants) to reduce the impact of technical (extract to extract) variability and keep the focus on biological variability. The hsp104Δ was significantly different (P < 0.01) from all other strains. The wt and the arb1Δ(pARB1) control strains were significantly different (*p < 0.05) from all other strains. Differences between the new1Δ and ARB1-d strains were not significant.
Use of microscopy to count puncta-positive cells does not permit distinctions between cells that have a single visible puncta and those that may contain many dozens. To better investigate total aggregate load in the Arb1-depleted and new1 mutants, we used semidenaturing detergent agarose gel electrophoresis (SDD-AGE) (Kryndushkin et al., 2003), which is suitable for characterizing large protein polymers including amyloids that are stable in 2% SDS at room temperature. Extracts were analyzed in parallel on SDD-AGE and SDS–PAGE gels (for quantitative detection of monomeric reporter protein) and then subjected to Western blot analysis with anti-GFP antibody. The relative abundance of aggregated GFP (SDD-AGE, Figure 6B) was normalized to the total GFP (SDD-AGE + SDS–PAGE, Figure 6C) signal. Results of these analyses were comparable to microscopy results. Strains with reduced levels of Arb1 (ARB1-d, galactose, and glucose) had significantly reduced aggregate load (approximately 60%; p < 0.05) compared with wild-type strains. The aggregated fraction of Htt-Q97-GFP was also significantly reduced in strains lacking NEW1 although neither Arb1 depleted nor new1Δ phenotypes were as severe as hsp104Δ strains (Figure 6D).
Animal ABCF proteins complement ARB1 depletion and GCN20 deletion phenotypes
To examine the potential for animal ABCF genes to confer disaggregation activity, we evaluated the impact of animal ABCF expression vectors on luciferase-GFP and Htt-Q97-GFP phenotypes described earlier (Figures 5 and 6). Expression plasmids were constructed in which X. laevis ABCF2 (XlABCF2), C. elegans abcf-2 (CeABCF2), and human ABCF2 (HsABCF2) cDNAs were placed under the control of the yeast ADH1 promoter. Expression of each animal gene was confirmed by Western analysis (Supplemental Figure S4) and the plasmids were introduced into strains carrying either the FFL-GFP or the Htt-Q97-GFP reporters.
To circumvent the need for changes in media (galactose to glucose), we used haploinsufficient diploids in which Arb1 levels are constitutively reduced. Diploids were constructed from strains carrying the so-called DAmP (Decreased Abundance by mRNA Perturbation) (Breslow et al., 2008) allele (ARB1-DAmP) and an arb1Δ strain kept alive with an ARB1 (URA3) expression plasmid. Cells lacking the ARB1 plasmid were then isolated by counterselection for the URA3 plasmid marker. As we observed in haploids, the rate of luciferase-GFP resolubilization was significantly slower (P < 0.01) in the DAmP/ arb1Δ strain relative to a DAmP/ARB1 diploid (not shown) or ARB1/ARB1 diploid ( Figure 7A).
FIGURE 7:
Aggregate processing phenotypes in yeast strains with low levels of Arb1 or lacking Gcn20/Abcf3 are partially complemented by animal ABCF genes. (A) Plasmids carrying each of three different animal ABCF2 cDNAs under yeast PADH1 control (Xl, X. laevis [high copy[, Ce, Caenorhabditis elegans [high copy], and Hs, Homo sapiens [low copy]) were introduced into the ARB1-DAmP/arb1Δ diploid strain (JF2639) carrying the pRS426 UASMET25FFL-GFP aggregation reporter plasmid and resolubilization following heat treatment monitored over time (0, 60, 90, and 120 min). (B) As for A, resolubilization of the FFL-GFP reporter was examined in the ARB1-DAmP/arb1Δ diploid strain (JF2639) expressing Xenopus ABCF2 (pSS2182) or two versions of the the corresponding Walker Domain (Walker A-2, K431W) mutant (pSS2198 and pSS2199). (C) Similar to A, resolubilization of the FFL-GFP reporter was examined in the ARB1-DAmP/arb1Δ diploid strain (JF2639) expressing human ABCF1, ABCF2, and ABCF3 cDNAs. For data in A–C, a minimum of 100 cells of each genotype were tabulated for each time point, and data for each strain and time point are the averages of three to six trials involving at least three transformants. (D) Amyloid disaggregation was evaluated as described in the legend to Figure 6. Strains were wt (BY4730), hsp104 (JF2473), and gcn20 (from the deletion collection). High and low copy human ABCF1, ABCF2, and ABCF3 expression plasmids as well as the yeast ARB1 plasmid, pDH129, were introduced into the gcn20Δ strain carrying the amyloid reporter plasmid, p416-Htt-Q97-GFP, and evaluated for their effect on amyloid propagation. For all experiments, significance was determined using ANOVA-Tukey HSD analysis (Tukey, 1949) (*p < 0.05, **p < 0.01). In A, the resolubilization phenotype of DAmP/arb1Δ strains (carrying an empty vector) was complemented by expression of animal ABCF2 genes at 90 and 120 min. In B, complementation by expression of the Xenopus Walker domain mutant (abcf2-K431W) was significantly reduced relative to expression of the wild-type Xenopus ABCF2. In C, complementation by the human ABCF1 and ABCF2 plasmids (but not ABCF3) was significantly better than the same DAmP/arb1Δ strain carrying an empty vector. In D, wild-type aggregation values were significantly different from hsp104Δ, gcn20Δ, and gcn20Δ + pARB1. In addition, aggregation values for the gcn20 strain carrying HsABCF1 (low and high copy), HsABCF2 (high copy), and HsABCF3 (low and high copy) were significantly different from gcn20Δ + vector and gcn20Δ = pARB1. (**p < 0.01; *p < 0.05).
All three animal ABCF2 expression plasmids complemented the disaggregase phenotype in the DAMP/arb1Δ strain (Figure 7A, significant at 90′ and 120′). We further investigated the basis for the complementation by mutating the Walker A motif (pattern G-x(4)-GK-[TS]) (Supplemental Figure S2) in the XlABCF2 expression plasmid. The lysine (K) residue in the Walker A motif is crucial for nucleotide binding (Hanson and Whiteheart, 2005) and the mutation is expected to cause loss of any ATP-dependent functions. The mutated (K431W; G-x(4)-GW-S) expression plasmid failed to complement the luciferase-GFP reporter resolubilization phenotype (Figure 7B). The extent of luciferase-GFP resolubilization (t = 90′, 120′) was not significantly different in comparisons of the DAmP/arb1Δ strain carrying an empty vector (unpublished data) or the XlABCF2-K431W plasmid.
The activity of other ABCF gene family members was tested by introducing human ABCF1, ABCF2, and ABCF3 genes into the same DAmP/arb1Δ strain background, again carrying the luciferase-GFP aggregation reporter (Figure 7C). Like HsABCF2, HsABCF1 complemented the resolubilization phenotype while differences between the DAMP/arb1Δ phenotype and HsABCF3 transformants were not significant. We conclude that two of the three human ABCF genes have disaggregation activity with respect to disordered aggregates that is comparable to that of yeast Arb1.
Given the lack of complementation of ARB1/ABCF2 mutant phenotypes by HsABCF3, we investigated a yeast gcn20Δ mutant, which lacks yeast ABCF3. Luciferase resolubilization was not significantly compromised in the gcn20Δ strain (Supplemental Figure S6); however, Htt-Q97-GFP aggregation was significantly compromised relative to wt (Figure 7D). Introduction of (additional) ARB1 did not complement the aggregation phenotype (Figure 7D); however, both HsABCF1 and HsABCF3 (high and low copy) expression plasmids exhibited statistically significant levels of complementation. Interestingly, high levels of HsABCF2 did as well. Hence, we find that animal ABCF genes are capable of disaggregating both disordered (luciferase) and ordered (Htt-Q97) aggregates.
A role for ABCF proteins in Xenopus development
The yeast data suggested that animal ABCF proteins have disaggregase activity. We next asked whether these proteins play a role in animal development. During Xenopus oogenesis abcf2 mRNA is several times more abundant than either abcf1 or abcf3 (Collart et al., 2014) and is also the most abundant Abcf protein found in oocytes (Peshkin et al., 2015). Several lines of evidence suggest that Abcf2 may have a role in early development. First, abcf2 is an animal hemisphere localized message in Xenopus oocytes and early embryos (Supplemental Figure S5) and the maternal mRNA is first activated via polyadenylation at the midblastula transition (Collart et al., 2014). The embryonic cells that inherit the most maternal abcf2 mRNA arise from the animal hemisphere of the embryo and are the cells that will eventually give rise to ectoderm and mesoderm. They include the cells that are mobilized during gastrulation. Second, oligonucleotide-mediated reduction of abcf2 mRNA (Figure 8A) resulted in the failure of the embryo to gastrulate, which is shown clearly by the absence or diminished formation of a blastopore lip (Figure 8B, lower panels). Third, although we noted little difference in cell cycle timing in the first five rounds of embryonic cleavage, embryos with lower maternal abcf2 mRNA began to divide more slowly when compared with uninjected or control-injected embryos.
FIGURE 8:
De novo synthesis of the localized mRNA ABCF2 is required for gastrulation and embryonic development in Xenopus. (A) Oligonucleotide-mediated reduction in ABCF2 mRNA relative to the Histone H4 control as assayed by rtPCR (left). RNA was isolated from embryos 2 h postinjection with the indicated levels of DMED-modified anti-ABCF2 oligonucleotide. (B) A comparison of uninjected and injected embryos assayed 12 h after injection shows the dose-dependent inhibition of formation of the blastopore lip and sequential progression toward gastrulation. (C) Spatially directed reduction of ABCF2 shows the importance of localized maternal inheritance of ABCF2 mRNA and the ability of human ABCF2 to replace lost maternal ABCF2 mRNA. Injection of DMED-modified anti-ABCF2 oligonucleotide was delayed until embryos had completed three cleavage cycles, forming an eight-cell embryo with visually distinct animal and vegetal tiers of cells; 0.1 ng of oligonucleotide, human ABCF2 mRNA, or both were injected into either the four animal hemisphere cells or four vegetal hemisphere cells as indicated. Clusters of embryos were photographed at the indicated time. By 24 h, the delay in cell cleavage and failure to gastrulate can be seen in embryos injected with anti-ABCF2 alone when injections were directed to the animal hemisphere tier of cells. Scale bars in B and C: 1 mm.
We further examined if directing antisense oligonucleotides to the animal hemisphere alone would replicate the effect of injecting at the one cell stage. We delayed injection until the eight cell stage and targeted the 4 animal hemisphere blastomeres with antisense alone, a rescue mRNA encoding human abcf2 alone or both antisense and rescue mRNA. Antisense treatment of the animal hemisphere blastomeres is sufficient to both slow early embryo development and inhibit gastrulation (0/30 embryos gastrulated). The loss of Xenopus abcf2 mRNA can be rescued by co-injecting with human abcf2 mRNA restoring nearly normal completion of gastrulation (Figure 8C) in 22/30 embryos. In contrast, injection of antisense-abcf2 oligonucleotide into the vegetal hemisphere had a much less severe effect. Most of these embryos initiated gastrulation with only slight delay compared with controls (20/30 embryos). We note that vegetally injected embryos were less likely to form normal neurula (only 8/30 vs. 25/30 for controls, 18/30 for embryos injected with human abcf2 mRNA, and 15/30 for embryos injected with both antisense-abcf2 and human abcf2 mRNA). It is possible that by injecting at the eight-cell stage we may have altered maternal levels of abcf2 found in the marginal zone, something that future studies will address. These experiments indicate that new translation of the maternally inherited abcf2 mRNA is required for normal development of Xenopus embryos by providing an essential function required for establishing normal gastrulation. Importantly, when Xenopus maternal mRNA is reduced the developmental consequences are rescued by providing mRNA encoding human abcf2.
A role for ABCF genes in C. elegans development
We also tested the role of abcf genes in C. elegans development. We found that ABCF-2 knockdown in C. elegans had no discernible effect on development so we turned to a deletion allele (ok2388), which was reported to have deleterious phenotypes requiring balancing. However, after backcrossing, ok2388 homozygotes showed no major linked phenotypes, though a low percentage of developmental defects, including vulval bursting and sterility, was observed. Therefore we analyzed a second ABCF family member, ABCF-1, which, like ABCF-2, is reported to be expressed during early development (Hashimshony et al., 2012). RNAi knockdown of C. elegans ABCF-1 (fed at L1 larval stage) caused loss of oocyte production (Figure 9, A and D) and subsequent 100% germline sterility (n > 200). We also uncovered a developmental role for ABCF-1 by performing feeding RNAi at the L4 stage; using this technique germline sterility is circumvented but the F1 progeny show defects in larval development. Specifically, we fed L4 N2 hermaphrodites ABCF-1 or empty vector RNAi on individual plates. After 24 h, we removed the adults and followed each cohort of progeny. Using this “L4 fed ABCF-1 RNAi” strategy we find that embryonic lethality is rare (1/210; Figure 9A, left graph). The resulting larvae showed developmental delays leading to larval arrest (86% of F1 progeny) and larval lethality (9%) such that after 3–4 d 100% of the control plates contained only adult worms, but only 4% of the L4 fed ABCF-1 RNAi F1 progeny became adults, and the total number of worms was half that of the control plates. As with L1 feeding, any F1 larvae from these L4 fed ABCF-1 RNAi experiments reaching adulthood were sterile. In contrast the empty vector plates contained F1 progeny that lacked embryonic lethality and resulting larvae developed with normal timing and were 100% fertile. While these experiments show a role for ABCF-1 in larval development, we suspected that, by selecting embryos that were laid in just the first 24 h after L4 feeding commenced, a role for ABCF-1 in embryonic development might be masked by delayed RNAi efficacy. To address this, we followed the ABCF-1 RNAi parental generation by moving them to fresh RNAi plates every 24 h and examined the resulting cohorts for embryonic defects (Figure 9A, middle and right graphs). After 2 d of ABCF-1 RNAi, embryonic lethality did become evident, 6.6% (17/259) of embryos died before hatching, and the remaining larvae died or arrested at the L2/L3 stages (1 adult out of 258 worms after 1 wk). After 3–4 d of ABCF-1 RNAi, embryonic lethality is very severe at 92% (134/146) and the larvae that did hatch arrested earlier at the L1/L2 stage. Average brood size for the ABCF-1 RNAi hermaphrodites was 100 with total embryonic lethality at 15.3%. The larvae that did develop also showed a marked developmental delay (Supplemental Table S1). In contrast, N2 worms fed control RNAi gave rise to an average brood size of 248 with just 0.6% embryonic lethality. The abcf-2(ok2388) homozygotes, with and without ABCF-1 RNAi, showed no reproducible differences compared with controls (Figure 9A), though ABCF-2 loss did worsen the germline defect of abcf-1(L1 fed RNAi) animals (see below).These data indicate that ABCF-1 plays a critical role in germline/oocyte development as well as larval development. Longer knockdown periods reveal a role for ABCF-1 in embryonic development.
FIGURE 9:
C. elegans ABCF loss of function causes germline arrest and increased amyloid production. (A) Top, experimental scheme to assess ABCF loss of function effects on development. Bottom, phenotypic quantitation. (B–E) Wild-type (B, D) or abcf-2(ok2388) (C, E) adult germlines exposed to control (B, C) or abcf-1 (D, E) RNAi and stained for amyloid oligomers (A11, green) and DNA (DAPI, blue). Frequencies of depicted phenotypes are as follows: (B) 100% n > 100; (C) 100% n = 20; (D) 100% n = 30; (E) 71% (n = 17) of germlines showed expansion of A11-positive structures into the distal germline and 76% (n = 17) showed enlarged or fused oocytes. Arrowheads denote A11 puncta. Arrows in E denote expanded A11 puncta compared with the localization in abcf-1 single loss of function animals (arrowheads in D and E). EV, empty RNAi vector. Scale bar, 50 µm.
Developmental defects after ABCF-1 knockdown are associated with changes in aggregation patterns
Since ABCF-1 RNAi produced germline and early embryonic phenotypes, we examined A11 patterns in germlines of adult animals deficient for ABCF function. While abcf-2(ok2388) germlines showed a mild increase in diffuse A11 staining (Figure 9C), germlines of abcf-1(L1 RNAi) animals exhibited extra A11 puncta in the few oocytes present in the proximal region (Figure 9D, inset). The double mutant germlines, abcf-2(ok2388);abcf-1(RNAi), displayed both the single mutant phenotypes and an expansion of A11-positive structures into the distal germline (Figure 9E arrows, 71% of germlines examined). Additionally, abcf-1;abcf-2 double loss of function animals had grossly expanded proximal germlines, with enlarged or fused oocytes (Figure 9E asterisk, 76% of germlines), suggesting that ABCF-1 and ABCF-2 may have redundant roles in germline patterning. Together, these data suggest that one or more ABCF proteins are required during germline and early embryonic development and have a role in A11-positive aggregate processing.
DISCUSSION
Endogenous aggregates are abundant during C. elegans oogenesis and early embryonic development
We find amyloid positive aggregates in young, healthy C. elegans individuals at multiple subcellular locales including the nuclear membrane and nonmembrane bound organelles such as P granules and centrosomes. These observations extend our previous report of aggregates in Xenopus oocytes (Hayes and Weeks, 2016). While Xenopus germinal vesicles represent excellent opportunities to visualize aggregation in nuclei, the presence of yolk in postvitellogenic oocytes precludes analysis of cytoplasmic structures. In contrast, the transparency of C. elegans tissues enables the use of amyloid probes in the cytoplasm. The analysis of both animal systems demonstrates that amyloid formation during development may be a general phenomenon across the animal kingdom.
While the specificity of A11 antibody for the oligomeric form of amyloid aggregates has recently been challenged with some studies suggesting that amyloid oligomer conformation is present in a group of natively folded proteins (Yoshiike et al., 2008), our use of A11 staining as a first indicator of amyloid-like aggregates is supported by additional observations including: detection of centrosomes and P granules by the OC antibody, which recognizes a more fibrillar form of amyloid (Figure 2; Supplemental Figure S1); alterations in A11 staining in vivo on heterologous expression of the yeast disaggregase, Hsp104 (Figure 3); and expansion of A11-positive bodies after loss of candidate endogenous disaggregases of the ABCF family.
Interestingly, by midembryogenesis, only the PGCs remained positive for A11 (Figure 2). Whether this is due to tissue-specific retention of the original inherited aggregates, ubiquitous loss followed by de novo aggregate formation in the newly formed germline, or reduction of particle size below the resolution of our imaging methodology remains unclear. However, the idea of tissue-specific solubilization to promote somatic development is an intriguing one. We hypothesize that one or more specific proteins form amyloids in the maternal germline to carry out important maternal and/or embryonic functions. The severe phenotypes caused by forced expression of HSP104 suggests disaggregase-sensitive substrates are present in early C. elegans embryos and that regulation of their disaggregation is required for normal development.
HSP104 misexpression in C. elegans disrupted the pattern of amyloid aggregates marked by A11 and caused embryonic cell cycle defects and embryonic lethality. The division defects were also observed in catalytically mutant HSP104 indicating that both amyloid regulation as well as a potential “holdase” function (Zhang et al., 2019) or the sequestering of endogenous chaperones may be incompatible with animal development.
What disaggregase functions are compatible with development?
Evidence of an ancient physiological role for protein aggregation has strong support from studies in both prokaryotes and eukaryotes. However, strategies for regulating aggregate formation and resolution may have evolved independently in animals. For example, the Hsp100 type disaggregase (Hsp104 in Saccharomyces cerevisiae), a robust disaggregase found in most branches of life, subcellular compartments including the plastid and mitochondria as well as the cytoplasm, is not present in animal genomes (Erives and Fassler, 2015).
The Hsp110-Hsp70-Hsp40 chaperone network has emerged together with small HSPs as one major animal disaggregase activity (Duennwald et al., 2012; Torrente and Shorter, 2013; Nillegoda et al., 2015; Nillegoda et al., 2018). However, RNAi knockdown of Hsp110 in C. elegans revealed no developmental phenotypes in the absence of proteotoxic stresses (heat, aging) (Nillegoda and Bukau, 2015; Kirstein et al., 2017). In addition, Hsp110 has not yet been shown to exhibit any activity with respect to endogenous aggregates. Hence, while this network constitutes an important poststress activity, Hsp110-Hsp70-Hsp40 network does not satisfy the criteria for a developmental disaggregase.
We have focused on the ABCF proteins as disaggregase candidates that might regulate the developmentally important amyloids present in C. elegans and Xenopus. The ABCF proteins emerge as candidates due to their phylogenetic relationship to New1, an Hsp104-independent amyloid disaggregase in yeast (Inoue et al., 2011) (Supplemental Figure S2) and conservation across species.
Since Hsp104 has aggregate processing activities with respect to both ordered (amyloid) and disordered aggregates (Murray and Kelly, 2012), we evaluated the yeast New1, Arb1/Abcf2, and Gcn20 (Abcf3) proteins for both activities. The FFL-GFP fusion combines FFL which forms disordered aggregates on chemical or heat (39–45°C) treatment (Nathan et al., 1997) and GFP, which is highly resistant to protein unfolding stresses (Penna et al., 2004) and thus allows intracellular deposits to be visualized with fluorescence microscopy. We found that strains with depleted levels of Arb1 are compromised in the refolding of heat-denatured FFL, although less so than strains lacking Hsp104 (Abrams and Morano, 2013). The New1 protein is dispensable for resolubilization of disordered aggregates showing that Arb1 and New1 have different activity profiles. The specificity of this phenotype was further examined in strains deleted for the HEF3 and GCN20 genes. Like New1, the closely related Hef3 (Supplemental Figure S2) exhibited no resolubilization defects (Supplemental Figure S6). Likewise, GCN20, which encodes the yeast Abcf3 counterpart (Supplemental Figure S2), exhibited only weak activity, at best, in this assay (Supplemental Figure S6). Introduction of C. elegans, human and Xenopus ABCF proteins into yeast strains with reduced Arb1 levels confirmed that both animal ABCF2 and ABCF1 are capable of disaggregating disordered FFL-GFP aggregates.
To ascertain whether Arb1 shares an ability to resolubilize and/or propagate amyloid aggregates with Hsp104 and New1, we turned to the human Huntingtin protein, a well-studied amyloid model in yeast as well as in transgenic animals. Only the N-terminal fragment of the 350-kDa Htt protein is needed to produce amyloid positive aggregates that depend on the length of the Q tract (Krobitsch and Lindquist, 2000), and plasmids encoding fusions between the Htt N- terminus and GFP (e.g., Htt-Q(25-97)-GFP constructs; Krobitsch and Lindquist, 2000) are common tools in studies of amyloidogenesis. The severe aggregation phenotype seen in wild-type strains expressing Htt-Q97-GFP is dramatically affected by deletion of HSP104, which virtually eliminates aggregation (Krobitsch and Lindquist, 2000). We found that new1Δ mutants exhibited an increase in the percentage of cells with soluble reporter relative to wild type. New1 is a useful positive control since in vitro assays previously established that New1 has Hsp104-independent activity in disaggregation of Sup35 amyloid fibers (Inoue et al., 2011). Our assays also revealed an increase in soluble Htt-Q97-GFP in the Arb1(Abcf2)-depleted strain and in strains lacking Gcn20 (Abcf3), suggesting that the ABCF proteins themselves have, or facilitate, amyloid-directed activity. Each of the human ABCF genes was capable of complementing the Htt-Q97 aggregation phenotype of the gcn20 mutant confirming that all three members of the ABCF gene family encode proteins with disaggregation activity directed at ordered aggregates, and that this activity is robust even in a heterologous context.
The role of ABCF disaggregases in development
The essential nature of Arb1/Abcf2 in S. cerevisiae was reproduced in our studies on ABCF proteins in animal development. We uncovered developmental roles for ABCFs in two different metazoans. In C. elegans, temporally controlled ABCF-1 RNAi caused embryonic lethality and sterility, indicating that the stable production of the ABCF-1 protein is essential for normal development. Although ABCF-2 effects were less pronounced, the extreme phenotype caused by the combined loss of ABCF-1 and ABCF-2 suggests some functional redundancy. Complementary analyses in Xenopus embryos showed that de novo synthesis of ABCF2 was essential during early development. In Xenopus the onset of embryonic gene transcription begins only after the initial 12 cell divisions. Hence, any spatial deposition of maternal mRNAs is reinforced by cell division. We found that reduction in abcf2 mRNA in the animal but not the vegetal hemisphere by targeted antisense oligonucleotide injections prevented gastrulation. We have yet to determine which protein aggregates may be the natural target of ABCF proteins. However, of the many gene products that affect gastrulation (Chen et al., 2004), the phenotype we see most resembles that reported in FUS gene analysis (Dichmann and Harland, 2012). Intriguingly, the FUS protein, which is a regulator of RNA processing and splicing, is known to form protein aggregates and is encoded during early development by a maternal mRNA with a similar distribution to ABCF2. It will be interesting in future studies to examine if FUS protein might be a client of the ABCF2 disaggregase. The expansion of A11-reactive puncta after ABCF loss could reflect expansion of the RNA processing bodies (P granules) in the C. elegans germline. This would be consistent with the idea that control of protein aggregation is important for proper management of RNA processing and subsequent germline function or specification in adult and embryonic germline tissues. Therefore, the management of RNA processing proteins may be common to animal disaggregase activity.
Several observations suggest that specific aggregates and ABCF disaggregases are required for assembly and function of animal centrosomes or other cell division machinery. We not only detect aggregates at C. elegans centrosomes, which are required to form and properly align the spindle during development, but also find that forced overexpression of Hsp104 disaggregase disrupts cell division timing and cell division orientation and that ABCF loss causes germline arrest in C. elegans and a slowed cell cycle in late blastula stage Xenopus embryos.
Why do ribosomal biogenesis proteins participate in disaggregation?
Prior characterization of Arb1 revealed striking ribosome biogenesis phenotypes (Dong et al., 2005). We therefore considered whether the disaggregation phenotypes of Arb1 and New1 might be a consequence of a global slowdown in translation or rRNA synthesis. Several observations suggest this is not the case. First, Western analysis showed no reduction in FFL-GFP expression in strains with reduced Arb1 or New1 levels despite the impact on aggregation (Supplemental Figure S7). Second, loss of ABCF2 in Xenopus embryos had very little effect on the timing of the midblastula transition, (which is dependent on translation) as judged by the onset of transcription of the embryonic gene GS17 (Krieg and Melton, 1985) (unpublished data) and finally, the effects we observe in Xenopus occur prior to the onset of new ribosomal RNA synthesis, which recommences in late stage gastrula (Newport and Kirschner, 1982). Although we are not able to completely rule out an effect of translation on protein folding in the arb1 and new1 mutants, the close phylogenetic relationship between the disaggregase protein New1 and the soluble ABC protein Arb1 may hint at the reverse, that the ribosome biogenesis defects are secondary to deficiencies in protein folding.
Ribosome-associated chaperones including RAC (Hsp70-Ssz1 and Hsp40-Zuo1), which interacts with and stimulates ATP hydrolysis by ribosome-associated Hsp70s to promote substrate binding, and NAC (a heterodimer consisting of alpha [Btt1 and Egd1] and beta [Egd2] subunits) bind to the large ribosomal subunit and interact with nascent polypeptides to support early folding events and prevent misfolding and aggregation (Preissler and Deuerling, 2012). Loss of these functions leads to aggregation of ribosomal proteins and ribosomal biogenesis factors (Koplin et al., 2010) as well as ribosome biogenesis defects. Arb1 cosediments with polysomes and is required for biogenesis of the small ribosomal subunit (Inoue et al., 2011). Hence ribosome-associated ABC proteins may play a role in the control of protein folding and aggregation activities in the earliest stages of protein synthesis, perhaps by negatively regulating RAC or NAC activity, or by interfering with the ability of the ribosome to carry out its own protein folding program, and the ribosome biogenesis phenotypes for the which the Arb1 protein was named (ATPase involved in ribosome biogenesis) may be a secondary consequence of reduced unfoldase activity in Arb1-depleted strains.
What is the role of aggregates in development?
Finally, it is interesting to speculate on what role(s) aggregates might play in early development. One possibility is that aggregates are storage structures preventing the premature availability of temporally restricted proteins. Amyloid type aggregates have been proposed to be a storage mechanism in pituitary secretory granules where peptide hormones amyloidize until their eventual release into the extracellular space, whereupon they disaggregate (Maji et al., 2009; Otzen and Riek, 2019). C. elegans aggregates may eventually be solubilized in the embryonic soma to promote proper somatic development and soma/germline differentiation. This idea is supported by our observations of widespread aggregates in the nuclear envelope and centrosomes of all early embryonic blastomeres in C. elegans, a pattern that becomes restricted to the PGCs by midembryogenesis (Figure 2). In this “amyloid storage” scenario, the C. elegans ABCF mutant phenotype would be due to a decrease in soluble protein(s) required for somatic development. Alternatively, the aggregate itself may function in the germline, a situation reminiscent of Aplysia memory formation (Si et al., 2010), the strengthening of arthropod silk fibers (Iconomidou et al., 2011), and during stress response (Halfmann and Lindquist, 2010; Halfmann et al., 2012; Newby and Lindquist, 2013). Here, amyloid expansion after ABCF loss leads to defects caused by inappropriate gain of this function in the soma that is incompatible with further development and cell fate specification.
Another attractive model is that aggregate deposition or the ability to modulate aggregate structure is regulated spatially. For example, the early restriction of mRNA encoding ABCF2 in Xenopus could ensure that aggregates in the presumptive ectoderm and mesoderm are disaggregated at the appropriate time, while lower levels of ABCF2 in the presumptive endoderm (which is also the site of future germ cells) may ensure specific aggregates and their co-aggregates are retained. Regardless of which mechanisms are operating, our results strongly suggest that proper assembly/disassembly of aggregates in the oocytes and early embryos of animals is necessary for normal progression through the earliest stages of animal development.
MATERIALS AND METHODS
Strains
Yeast strains.
Strains used in this study include YDH209 (OY305) and YDH226 (OY306), which were a generous gift from the Hinnebusch lab (Dong et al., 2005). All strains are derivatives of S288C (Mortimer and Johnston, 1986). Strains hsp104Δ (JF2473), new1Δ (JF2475), and arb1Δ carrying pDH25.1, which expresses an unstable Arb1 protein (YDH226); new1Δ arb1Δ [pDH25.1] (pJF2510), and hsp104Δ arb1Δ [pDH22] (JF2511) were constructed by crosses followed by tetrad dissection and genotypes verification by colony PCR. The DAmP diploid strains, JF2614 and JF2639, were constructed starting with the heterozygous ARB1-DAmP diploid (Dharmacon) (Yan et al., 2008). This strain was dissected and tested for genotype. Haploid JF2612 was mated with BY4730 to generate JF2614 and with YDH209 to generate JF2639. JF2639 was plated on media containing 5-FOA to allow for the loss of the ARB1 plasmid, pDH22. See Table 2 for additional information on strains used in this study.
TABLE 2:
Yeast strains.
| Strain name (storage name) | Relevant genotype | Source |
|---|---|---|
| YDH209 (OY305) | MATA his3Δ1 leu2Δ0 ura3Δ0 arb1Δ::KanMX4; pDH22 [URA3] | Dong et al. (2005) |
| YDH226(OY306) | MATA his3Δ1 leu2Δ0 ura3Δ0 arb1Δ::KanMX4; pDH25.1 [LEU2] | Dong et al. (2005) |
| JF2473 | MATA met15Δ0 leu2Δ0 ura3Δ0 hsp104Δ::KanMX4 | This study |
| JF2475 | MATA met15Δ0 his3Δ1 leu2Δ0 ura3Δ0 new1Δ::KanMX4 | This study |
| JF2498 | MATα leu2Δ0 ura3Δ0 hsp104Δ::kanMX4 | This study |
| JF2501 | MATα new1Δ::KanMX4 met15Δ0 leu2Δ0 ura3Δ0 | This study |
| JF2510 | MATA ura3Δ0 met15Δ0 leu2Δ0 new1Δ::KanMX4 arb1Δ::KanMX4; pDH25.1 | This study |
| JF2511 | MAT(A or α) his3Δ1 ura3Δ0 leu2Δ0 hsp104Δ::KanMX4 arb1Δ::kanMX4; pDH25.1 | This study |
| JF2612 | MATA his3Δ1 met15Δ0 ura3Δ0 leu2Δ0 arb1-DAmP | This study |
| JF2614 | MATa/α leu2Δ0/+ ura3Δ0/ ura3Δ0 met15Δ0/met15Δ0 arb1-DAmP/ARB1 | BY4730 x JF2612 |
| JF2639 | MATA/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ ura3Δ0 met15Δ0/+ arb1-DAmP/arb1Δ | This study YDH209 x JF2612 after 5-FOA |
| BY4730(OY208) | MATA met15Δ0 leu2Δ0 ura3Δ0 | Brachmann et al. (1998) |
| (OY337) | MATA his3Δ1 ura3Δ0 leu2Δ0 met15Δ0 pURA3-2μ-LEU2d UASG PCYC1 GST-6xHis-ARB1 | Dharmacon Zhu et al. (2001); Zhu and Snyder (2001) |
| (OY336) | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ ura3Δ0 met15Δ0/met15Δ0 CYH2+/cyh2 arb1-DAmP/ARB1 | Breslow et al. (2008) |
| JF2693 | MATa/α leu2Δ0/leu2Δ0 ura3Δ0/ ura3Δ0 met15Δ0/MET15 LYS2/lys2Δ0 | BY4730 x BY4739 |
| (OY350) | MATα his3Δ 1 leu2Δ 0 ura3Δ 0 gcn20Δ::kanMX4 | Deletion collection |
C. elegans strains.
C. elegans strains used in this study includes: N2 [wildtype]; JK3013 [qIs61(Ppes-1::GFP); SA264 [unc-119(ed3)III; tjIs71(Ppie-1::mCherry::H2B, Ppie-1:2xmCherry::tbg-1, unc-119(+)]; BTP233 [tag-164 abcf-2(ok2388)III].
Media.
YP media were prepared with or without 2% agar and included 2% glucose (YPD) or 2% galactose (YPGal) or 2% galactose plus 1% raffinose (YPGR). HB stocks were filter sterilized and added after autoclaving to various final concentrations as indicated. Synthetic complete media lacked uracil, leucine, or both and were prepared with glucose (SC) or galactose and raffinose (SCGR) as carbon sources. SC media lacking methionine were used for induction of the MET25-driven FFL-GFP reporter.
Culture conditions and phenotype testing.
Overnight cultures were prepared in synthetic complete or rich media containing galactose and raffinose. Strains were then subcultured into the same media and grown to log phase for competent cells and all reporter assays. Analysis of temperature, HB, and cycloheximide sensitivity was conducted using fresh stationary phase cultures, which were serially diluted in water and spotted on media as indicated in each figure legend. Arb1 depletion in the OY306 strain following 2 h of incubation in glucose media was verified by Western blot analysis.
Phylogenetic analysis.
Maximum likelihood phylogenetic analysis was conducted using MEGA 7.0 (Tamura et al., 2007; Kumar et al., 2016). The evolutionary history was inferred by using the Maximum Likelihood method based on the General Reverse Transcriptase + Freq. model (Dimmic et al., 2002). The tree with the highest log likelihood (–27503.3961) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 29 aa sequences. All positions with less than 50% site coverage were eliminated. There were a total of 710 positions in the final dataset. Accession numbers for the sequences used are shown in the legend to Figure S2.
Aggregation assays
Luciferase-GFP disaggregation assay.
FFL-luciferase-GFP renaturation assays were performed essentially as described (Abrams and Morano, 2013). Samples were fixed after each time point to prevent additional refolding during microscopy. Resolubilization was calculated at various time points after heat treatment as the percentage of cells lacking detectable aggregates in the form of GFP puncta.
Htt-Q 97 amyloid disaggregation assay.
Strains of various genotypes were transformed with a plasmid carrying the constitutively expressed PTDH3-Htt-Q97-GFP fusion gene. All strains carrying the ARB1 depletion plasmid were grown in duplicate to log phase in dropout media containing galactose. At log phase, one of the duplicates was imaged or spun down for protein extraction (see below). The second duplicate was washed in water, resuspended in the same volume of glucose media, and incubated for a further 4 h, after which it was imaged or spun down for protein extraction.
Blind counts.
Aggregation assay results were spot checked for accuracy and reproducibility by blind counting. Here, a set of slides corresponding to multiple genotypes and times (where pertinent) were assigned a random number by individuals not involved in this research. Each slide was then inspected by an author of this paper and assigned aggregation percentages based on hundreds of cells. Finally, a second author decoded the slides and calculated averages, standard deviations, and significance.
Microscopy.
Yeast cultures expressing GFP reporters were fixed at the appropriate time points as described (Bloom et al., 2018). Microscopy of yeast cells was conducted using ZEISS Axio Imager D2 compound fluorescent scope and Zeiss Zen software. Images were captured using GFP and DIC filters. Exposure time for FFL-Luciferase-GFP detection and Htt-Q97-GFP was 42 and 100 ms, respectively; 100 ms was used for both reporters in diploid strains. A minimum of 100 cells over three fields was counted and tabulated using ImageJ software with a “minimum displayed value” setting of 15 in order to reduce the contribution of cells whose fluorescence level was insufficient to evaluate. As a result of this setting, 10–20% of sampled cells for any given genotype were not counted (Schindelin et al., 2012; Schneider et al., 2012; Rueden et al., 2017). Blind counting tests were conducted to confirm the reliability of the measurements. C. elegans microscopy was conducted on a Zeiss Axio Imager D2 motorized compound fluorescence upright microscope with DIC and a Zeiss Axiocam MRm with ZEN image capture software. Images of Xenopus embryos were acquired with a Zeiss Discovery V8 dissecting microscope using an AxioCam MrC 5 camera and AxioVision software (Zeiss).
Protein extracts.
Strains carrying either the Htt-Q97-GFP or the FFL-Luciferase-GFP reporter were cultured as described above, washed in 10 mM, Tris 1 mM EDTA, pH 8 (TE), and pelleted before freezing at –80°C. The best quality extracts were prepared from samples that had been stored at –80°C for less than 1 wk. After thawing on ice, ∼100 µl of glass beads and 100 µl of extraction buffer (30 mM Tris, pH 8.5, 5 mM EDTA, 3 mM DTT, 5% glycerol plus 2.5 µl of 40 mM PMSF, and 2.5 µl of PIC; Sigma, P8215) was added to pellets. Cells were disrupted with 7 × 30 s Qiagen Tissue Lyser LT cycles alternating with 1 min on ice. The mixture was centrifuged at 2000 rpm for 1 min and the beads and pellet were discarded. The supernatant protein concentration was determined using the Bradford Protein Assay (Bio-Rad) using a bovine serum albumin (BSA) standard curve.
Gel electrophoresis
SDD–AGE.
SDD-AGE gels were 1.8% agarose prepared in 1× TAE, 0.1% SDS, and 0.4 cm thickness as described in Campbell (2014). Htt-Q97-GFP extracts were incubated at 37°C for 10 min in 4× sample buffer: 2× TAE, 20% glycerol, 8%SDS, and 1% Bromophenol Blue. Electrophoresis was at 3 V/cm at 4°C in 1× TAE and 0.1% SDS buffer for 6 h. Protein was transferred to nitrocellulose overnight using the horizontal capillary method as described (Campbell, 2014) and processed for Western blotting as described below (GFP). The aggregated fraction per genotype was calculated by 1) summing the GFP signal (per microgram of protein) in the corresponding lanes of SDS–PAGE and SDD-AGE gels, 2) normalizing the GFP numbers to account for lane to lane differences in loading using several bands after total protein detection using REVERT staining of the SDS–PAGE gel, and finally 3) dividing the aggregated GFP signal detected on the SDD-AGE gel over the normalized total GFP signal. The reported values are averages over three biological and two technical replicates.
SDS–AGE.
SDS–PAGE running and stacking gels were made with 8 and 4% 29:1 acrylamide, bisacrylamide, and 0.1% SDS. Protein extracts were incubated in 5× LSB: 15% SDS, 0.575 M sucrose, 0.325 M Tris-HCl, pH 6.8, 5% beta Mercaptoethanol, and 0.002% Bromophenol Blue at 37°C for 10 min prior to loading. Gels were run at 5 V/cm for 2 h, then 15 V/cm for 1.5 h at 4°C in running buffer (25 mM Tris, 192 mM glycine, and 0.1% SDS). Protein was transferred to nitrocellulose in Tris-glycine buffer (25 mM Tris, 192 mM glycine, 20% methanol) by electroblotting at 25 V at 4°C OVN. Posttransfer membranes were incubated with Ponceau S (0.5% [wt/vol] in 10% glacial acetic acid) for 5 min at room temperature followed by a water rinse to examine lane to lane loading differences. REVERT protein strain (LI-COR) was used when normalization between lane loading for Western analysis was needed. Membranes were washed once in phosphate-buffered saline (PBS)-T (1× PBS with 0.1% Tween-20) to remove the stain prior to blocking. Membranes were incubated in REVERT protein strain for 5 min, then washed 2× with wash buffer: 6.7% glacial acetic acid and 30% methanol. Membranes were rinsed with water before signal detection using the LI-COR Odyssey imaging system (700 nm).
Western blot analysis
Detection of GFP.
Membranes were incubated with PBS-T plus 5% nonfat milk followed by anti-GFP (anti-mouse monoclonal antibody; SC-9996 lot# K2102; Santa Cruz Biotechnology) at a 1:1000 dilution in PBS-T plus 3% BSA at 4°C OVN. Secondary antibody (IRDye 800LT Goat anti-Mouse IgG [H + L]) was diluted 1:20,000 in TBS-T plus 5% milk at RT for 1 h in the dark. Signal was detected with a LI-COR Odyssey imaging system (800 nm).
Detection of ABCF2.
Membranes were incubated in PBS-T plus 5% nonfat milk followed by anti-ABCF2 (polyclonal antibody ab102618, PA3-16796; Thermo-Fisher Scientific) at a 1:500 dilution in PBS-T plus 3% BSA at 4°C OVN. Secondary antibody (Alexa Fluor Plus 800 Goat anti-Rabbit IgG; Invitrogen) was diluted 1:20,000 dilution in TBS-T plus 5% milk at RT for 1 h in the dark. Signal was detected with a LI-COR Odyssey imaging system (800 nm).
Detection of the FLAG epitope.
FLAG Westerns were conducted as described for ABCF2, but with a mouse anti-FLAG primary antibody (# SLBT7654; Sigma-Aldrich).
Plasmids and plasmid construction
Yeast plasmids.
pDH22 and pDH25 were generously provided by the Hinnebusch lab in strains YDH209 and YDH226, respectively (Dong et al., 2005). The p426Met25-FFL-GFP (luciferase-GFP) plasmid was generously provided by the Morano lab (Tkach and Glover, 2008). Plasmid p416-103Q GPD is from the Lindquist collection (Addgene 1180). Our sequence analysis revealed that a more appropriate nomenclature for this plasmid is UASTDH3-Htt-Q97-GFP (Krobitsch and Lindquist, 2000). See below and Table 3 for details concerning these and other plasmids.
TABLE 3.
Plasmids.
| Plasmid Name | Description | Source |
|---|---|---|
| pDH22 | ARB1 in pRS316 | Dong et al. (2005) |
| pDH25.1 | UASG-UBI-R-FH-ARB1 in YCp111; LEU2, CEN | Dong et al. (2005) |
| p426Met25-FFL-GFP | UASMET25-FFL-GFP in pRS426; URA3, 2μ | Tkach and Glover (2008) |
| P416/Q103Addgene 1179 | UASTDH3-Htt-Q103-GFP in pRS416; URA3, CEN (actually Q97) | Krobitsch and Lindquist (2000) |
| P416/Q46(GC391)Addgene 1178 | UASTDH3-Htt-Q46-GFP in pRS416; URA3, CEN | Krobitsch and Lindquist (2000) |
| P416/Q25(GC390)Addgene 1177 | UASTDH3-Htt-Q25-GFP in pRS416; URA3, CEN | Krobitsch and Lindquist (2000) |
| isolated from Dharmacon strain | UASG PCYC1 GST-6xHis-ARB1 in pURA3; LEU2d, 2μ | Dharmacon Zhu et al. (2001) |
| pDH129 | FLAG-His6-ARB1 in YC1ac111; LEU2, CEN | Dong et al. (2005) |
| pRS425; pRS315 | LEU2, 2μ; LEU2, CEN | Christianson et al., (1992); Sikorski and Hieter (1989) |
| pRS313 Hsp104 K218T; K620TAddgene 1192 | HSP104 Double Walker A mutant; ATPase mutant (K218T and K620T); HIS3, CEN | Patino et al. (1996) |
| pRS416GAL-Hsp104A503V | HSP104 (A503V) in pRS416; URA3, CEN | Jackrel and Shorter (2014) |
| pADH1pr-MCS-ter | Sequences upstream and downstream of ADH1 (–371 to –12 and +1048–1236) separated by a polylinker in PCR2.1 | This study (S.S.) |
| pJF2140 | ADH1 fragment isolated from GC99 in pRS315; LEU2, CEN | This study (S.S.) |
| pSS2148 & pSS2175 | ADH1 fragment isolated from GC99 in pRS425; LEU2, 2μ using two different strategies | This study (S.S.) |
| pSOC4ΔSmaI | ADH1 (–371 to –12 and +1048–1236) fragment; URA3, 2μ | This study (D.L.W.) derivative of pSOC4 (gift of Williams and Cyert) |
| pXL-ABCF2-URA3 | PADH1 X. laevis ABCF2 cDNA in pSOC4ΔSmaI; URA3, 2μ | This study (D.L.W.) |
| pSS2182 | PADH1 X. laevis ABCF2; Kl LEU2, 2μ | This study (S.S.); derivative of pXL-ABCF2-URA3 |
| pSS2198, pSS2199 | PADH1 X.laevis ABCF2-K431W in pJF2182; LEU2 2μ (two clones constructed differently) | This study (SS) |
| pSS2125 | UASGC. elegans abcf-2 cDNA in pESC-LEU2; LEU2, 2μ | This study (S.S.); backbone from Agilent Technologies |
| pSS2174 | UASG C. elegans abcf-2-FLAG; LEU2, 2μ | This study (S.S.); derivative of pSS2125 |
| pSS2183 | PADH1 C. elegans abcf-2-FLAG; LEU2, 2μ | This study (S.S.); derivative of pSS2174 |
| pHS-ABCF3(high) | PADH1 human ABCF3 cDNA in pJF2148; LEU2 2μ | This study (A.G. and D.L.W.) |
| pHS-ABCF2(low) | PADH1 human ABCF2 cDNA in pJF2140; LEU2, CEN | This study (A.G. and D.L.W.) |
| pHS-ABCF3(low) | PADH1 human ABCF3 cDNA in pJF2140; LEU2, CEN | This study (A.G. and D.L.W.) |
| pHS-ABCF1(high) | PADH1 human ABCF1 in pJF2148; LEU2 2μ | This study (A.G. and D.L.W.) |
| pHS-ABCF1(low) | PADH1 human ABCF1 in pJF2140; LEU2, CEN | This study (A.G. and D.L.W.) |
| pBTP78 | C. elegans abcf-2 in PCR2.1 | This study (A.C. and B.Z.) |
| pBTP79 | C. elegans abcf-2 in pL4440 | This study (A.C. and B.Z.) |
ADH1 promoter vectors.
A 0.644-kb yeast ADH1 promoter fragment was amplified with Taq polymerase (New England Biolabs) from pre-existing plasmid, pSOC4 (gift from M. Cyert), and then ligated into PCR2.1-TOPO. The promoter fragment was subsequently isolated via ApaI–SacI double digest and ligated into ApaI–SacI-digested pRS425 or pRS315 to generate pSS2148 and pSS2140, respectively. pSS2175 is a variant of pSS2148 in which fewer restriction sites were carried over from the PCR2.1-TOPO starting plasmid (GC99).
Human ABCF expression plasmids.
The human ABCF cDNAs were purchased from Transomic Technology and introduced into high (pJF2148) and low copy (pJF2140) pRS vectors containing sequences (–312 to –12) upstream of the ADH1 ATG using HiFi DNA Assembly cloning (New England Biolabs). All constructs were verified by sequencing. ABCF2 constructs were verified by Western blot analysis. Human ABCF2 was also cloned into the plasmid pRN3P (a gift from John Gurdon, Cambridge University) by substitution of the eGFP in pRN3P with the open reading frame encoding human ABCF2.
X. laevis ABCF2 expression plasmids.
cDNA was isolated from Xenopus J strain oocyte mRNA using reverse transcriptase and ligated into pSOC4 from which the SOC4 gene had been deleted to create plasmid pXL-PADH1ABCF2-URA3. The K431W mutation was generated in the Walker A-2 domain using oligonucleotide-directed PCR mutagenesis. The mutation was verified by Sanger sequencing. A fragment containing the mutation was then introduced into pSS2182 by gap repair to generate pSS2198 and pSS2199. ABCF2 constructs were verified by Western blot analysis (Supplemental Figure S4).
C. elegans expression plasmids.
abcf-2 cDNA was amplified from N2 worms, cloned into pCR2.1-TOPO, isolated as a NotI–SpeI fragment, and ligated into pESC-LEU2 (Agilent Technologies) to generate pSS2125. A FLAG tag was added to the C-terminus by gap repair to generate plasmid pSS2174. The UASG sequence in pSS2174 was replaced by upstream regulatory sequences (coordinates –421 to –1) from ADH1 to generate plasmid pSS2183. The plasmid was verified by sequencing and abcf-2 expression was confirmed by α-FLAG Western blot analysis.
Yeast GCN20 plasmid.
A 2.345-kb GCN20 fragment was amplified with Q5 polymerase (New England Biolabs) from genomic DNA. The PCR product was A-tailed with Taq polymerase and ligated into PCR2.1-TOPO. The fragment was subsequently isolated as a BamHI–NotI fragment which was ligated into pSS2175 to generate pSS2181. The construct was verified by restriction digest and complementation of the cycloheximide sensitive phenotype of a gcn20Δ mutant.
C. elegans methods
C. elegans plasmids.
The following C. elegans plasmids were used: pBTP78, which consists of C. elegans abcf-2 cDNA inserted into PCR2.1 using forward primer ATGCCGTCTGATGCGAAGAAAG and reverse primer TTAACGCTCCTTGACAACTCCC; pBTP79, which consists of C. elegans abcf-1 inserted in pL4440 using forward primer GAAGATACGGTTTGGTCGGA and reverse primer GAGCTCCAGCATCATCATCA.
Antibody staining.
DAPI and immunostaining of C. elegans germlines and embryos were performed as in MacQueen and Villeneuve (MacQueen and Villeneuve, 2001) with the exceptions that worms were dissected in PBST instead of EGG buffer and fixed in 1% formaldehyde instead of 4% and that the BSA block was doubled from 0.5 to 1%. All antibodies were diluted with PBS containing Tween 0.1%. Antibodies include: A11 (1:400) (AB9234 Millipore Sigma), OC (1:250) (AB2286 Millipore Sigma), PGL-1 (1:25) (OIC1D4 DSHB), GFP (1:500) (ab13970 Abcam), Mcherry (1:500) (ab125096 Abcam), Donkey anti-Mouse IgG Alexa Fluor 488 ab150105 (1:500) (Abcam), Goat anti-Rabbit IgG Alexa Fluor 594 A11037 (1:500) (Invitrogen), Goat anti Chicken IgY Alexa Fluor 488 A-11039 (1:500) (Invitrogen), Donkey anti-Mouse IgG2a Alexa Fluor 594 A21135 (1:500) (Invitrogen), and Donkey anti-Rabbit IgG Alexa Fluor 488 A21206 (1:500) (Invitrogen).
X-34 staining.
A total of 500 ml of a 100-µM solution of the amyloid stain X-34 (1,4-bis(3-carboxy-4-hydroxyphenylethenyl)-benzene) was synthesized following the procedure outlined by Ikonomovic et al. (2006). Prior to synthesis, both Ikonomovic et al. (2006) and Styren et al. (2000) should be consulted to understand details and safety measures required. Many of the reagents are flammable, toxic, and/or volatile. In brief, 32 mg of tetraethyl p-xylylenediphosphonate (TCI America, Portland, OR, cat #T1582) were dissolved in 2 ml of fresh anhydrous dimethylformamide (DMF) (Aldrich, cat #22,705–6). After the tetraethyl p-xylylenediphosphonate had dissolved, 95 mg dry potassium tert-butoxide (0.85 mmol) (Aldrich, cat #15,667–1) were added. The resulting mixture was red and opaque.
In a second bottle, 23 mg 5-formylsalicylic acid (Aldrich, cat #F1,760–1) were dissolved in 1.0 ml anhydrous DMF; 250-µl aliquots of the 5-formylsalicylic acid solution were sequentially added to the tetraethyl p-xylylenediphosphonate/tert-butoxide mixture, capped, and incubated at room temperature overnight with mixing. The solution thickened and turned a burnt-orange color; 2 ml of the reaction were added to 500 ml of 40% ethanol and mixed well. The pH was adjusted to 9.8 to 10.2 using NaOH or HCl as needed.
Adult C. elegans hermaphrodites were injected with 1 mM X-34 diluted in 10 mM Tris, pH 7.5, recovered for 6 h at room temperature, and then mounted on slides for imaging. Samples were visualized using Zeiss Filter set 18, excitation 390–420 nm with a longpass emission filter at 450 nm.
ThioT staining.
C. elegans hermaphrodites were raised on NGM plates containing 0.018 mg/ml ThioT, made from a 50× ThioT stock (0.8 mg/ml ThioT in PBS). For slide mounting, the 50× stock was diluted in water to 10×, 1 µl of which was then added to a solution of 2 µl of dissected germlines in M9 plus 7 µl mount solution (Vectashield) for a final concentration of 0.016 mg/ml ThioT. Slides were aspirated to remove excess fluid and sealed for imaging.
RNAi.
RNAi was performed using standard techniques using the pL4440 feeding vector (Timmons et al., 2001).
HSP104 misexpression methods.
Two different HSP104 mRNAs were used in this study. One was derived from the coding region of pRS416GAL-HSP104A503V (Jackrel and Shorter, 2014), which was subcloned into pRN3P (a gift from John Gurdon; Zernicka-Goetz et al., 1996) where the original GFP was substituted for mRed to make C-terminal RFP fusion, and contains 5′ and 3′ untranslated regions from ß-globin. The mutant Hsp104 was derived from pRS313- HSP104K218;K620T (Patino et al. 1996; Addgene), which was cloned into the modified pRN3P vector. Transcription templates were generated using PCR, and following purification (Qiagen), mRNA was generated from the PCR product using mMESSAGE mMACHINE T3 Transcription Kit (AM1348, Ambion), precipitated with LiCl, and diluted to 0.1 µg/µl in DEPC water for injection into adult C. elegans hermaphrodite germlines. Recombinant Hsp104 protein (LifeSpan BioSciences) was injected following dilution to 0.1 mg/ml in a buffer containing 20 mM Tris HCl, 100 mM NaCl, and 2 mM EDTA, pH 8.
Xenopus methods
General Xenopus methodology.
Trials using Xenopus were carried out using J strain X. laevis obtained from the Xenopus National Resource at Woods Hole Oceanographic Institute. Embryos were obtained after hCG (human chorionic gonadotropin)-induced egg laying and in vitro fertilization using standard techniques and staged by the method of Nieuwkoop and Faber (1967). All animal protocols were reviewed and approved by the University of Iowa Animal Care and Use Committee.
In situ hybridization.
In situ hybridization was carried out essentially as described by Harland (1991). Embryos were fixed at the appropriate stage of development using MEMFA (0.1 M Mops, 2 mM EGTA, 1 mM MgSO4, and 0.4% paraformaldehyde) and rehydrated in TTW (200 mM NaCl, 50 mM Tris, pH 7.4, and 0.1% Tween 20). Digoxigenin (DIG)-labeled probes for ABCF2 were generated from plasmids containing its cDNAs using an Ambion Megascript kit with DIG RNA labeling mix. Probes were hybridized with fixed embryos using hybridization buffer (50% formamide, 5 SSC [1× SSC is 0.15 M NaCl, 0.015 M sodium citrate], 1 mg/ml total yeast RNA, 1% Denhardt’s, 0.1% Tween 20, and 5 mM EDTA) and, after extensive washing in 0.2× SSC, were incubated with alkaline phosphatase–conjugated anti-DIG antibody (1:5000; Roche) in MAB (100 mM maleic acid, 150 mM NaCl, pH 7.5, and 0.02 g/ml BMBR [Boehringer Mannheim] blocking reagent [Roche, Mannheim, Germany]) overnight at 4°C. After extensive washing at room temperature with 100 mM Tris (pH 9.5), 50 mM MgCl2, 100 mM NaCl, 0.1% Tween 20, and 1 mM levamisol to remove unbound antibody, colorimetric detection of hybrids was accomplished using 0.3 mg/ml Nitro BlueTetrazolium and 0.2 mg/ml BCIP (5-bromo-4-chloroindol-3-yl phosphate) in the washing solution. Color reactions were stopped using 100% methanol.
Oligonucleotide-mediated degradation of ABCF2.
Modified oligonucleotides were synthesized using H-phosphonate chemistry on an ExpediteTM synthesizer (Applied Biosystems, Foster City, CA) (Dagle et al., 2000; Dagle and Weeks, 2000). All reagents used for automated DNA synthesis were obtained from Glen Research (Sterling, VA). To generate unmodified phosphodiester bonds, hydrogen phosphonate diesters were oxidized for 4 min with freshly prepared 5% iodine in THF (tetrahydrofuran)/pyridine/water (15:2:2) and then for 3 min with the same solution diluted 1:1 with 8% triethylamine in THF/water (43:3). Oxidative amidation of hydrogen phosphonate diesters was performed manually by using a 10% solution of dimethyl ethylene diamine (DMED, Aldrich, Milwaukee, WI) in anhydrous carbon tetrachloride (Sigma, St. Louis, MO). Further processing and purification of oligonucleotides using reverse-phase HPLC and gradient elution with acetyl nitrile as the mobile phase were performed as previously described (Dagle et al., 2000; Dagle and Weeks, 2000). After Sephadex G-25 column chromatography, oligonucleotides were dissolved in sterile water and quantified by UV spectroscopy. The sequence of the oligonucleotide used to degrade endogenous ABCF2 mRNA was T+T+C+T+TCTTGGCC+A+G+A+T+C with the sequence complementary to the 17 nucleotides starting 9 nucleotides downstream of the start of the open reading frame of the oligonucleotide of the ABCF2 mRNA (XM_018224093.1). Rescue of oligonucleotide inhibition was accomplished using mRNA transcribed from the human ABCF2 cDNA.
Oligonucleotides and rescue mRNAs were injected using a Singer MK-1 (Somerset, England) micromanipulator and Inject+Matic injector (Geneva, Switzerland). Oligonucleotide effectiveness and dose response was tested using Trizol (Invitrogen) isolated RNA from injected embryos as a substrate for RT–PCR. Protoscript II first strand cDNA synthesis reaction (New England Biolabs) was used to generate cDNA from 1 µg of total embryonic RNA. The levels of ABCF2 and Histone H4 (used as a reaction control) were determined using PCR reactions with 12% of the cDNA generated, the appropriate primers, and Q5 polymerase (New England Biolabs).
Data and reagent availability
The authors affirm that all data necessary for confirming the conclusions of this article are represented fully within the article and its tables and figures. Primer sequences are available on request.
Supplementary Material
Acknowledgments
We thank Albert Erives for the initial discussions on the potential role of Abcf2 in amyloid processing. We also acknowledge contributions to the early stages of this project by undergraduate and high school students, Arida Dhanaswar, Priya Kanolkar, and Rhea Nagpal. Gifts of plasmids and strains are gratefully acknowledged from John Gurdon, Alan Hinnebusch, Kevin Morano, the Lindquist lab, James Shorter, Judith Kimble, Addgene, and the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs [P40 OD01440]. We are grateful for helpful discussions by members of the Fassler, Phillips, and Weeks labs as well as UI colleague, Daniel Summers. This work was supported by a University of Iowa Office of the Vice President for Research grant (D.L.W. and J.S.F.), a University of Iowa Center for Biocatalysis and Biotechnology grant (D.L.W., B.T.P., J.S.F.), NIH award GM124063 (D.L.W.) NSF IOS-1456941, NIH GM114007 (B.T.P.), and NSF award 1917169 (J.S.F. and B.T.P.). S.S. acknowledges support from a University of Iowa Graduate College fellowship award; M.H. was a trainee in the Medical Scientist Training Program at the University of Iowa.
Abbreviations used:
- aa
amino acid
- ABC
ATP binding cassette
- BSA
bovine serum albumin
- DAmP
Decreased Abundance by mRNA Perturbation
- DIG
digoxigenin
- DMED
dimethyl ethylene diamine
- DMF
dimethylformamide
- HB
Hygromycin B
- PBS
phosphate-buffered saline
- PGC
primordial germ cell
- SDD-AGE
semidenaturing detergent agarose gel electrophoresis
- ThioT
Thioflavin T
- THF
tetrahydrofuran.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-08-0443) on April 22, 2020.
REFERENCES
- Abrams JL, Morano KA. (2013). Coupled assays for monitoring protein refolding in Saccharomyces cerevisiae. J Vis Exp e50432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ader C, Frey S, Maas W, Schmidt HB, Gorlich D, Baldus M. (2010). Amyloid-like interactions within nucleoporin FG hydrogels. Proc Natl Acad Sci USA , 6281–6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanese V, Reissmann S, Frydman J. (2010). A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis. J Cell Biol , 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando-Akatsuka Y, Shimizu T, Numata T, Okada Y. (2012). Involvements of the ABC protein ABCF2 and alpha-actinin-4 in regulation of cell volume and anion channels in human epithelial cells. J Cell Physiol , 3498–3510. [DOI] [PubMed] [Google Scholar]
- Aron R, Higurashi T, Sahi C, Craig EA. (2007). J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. EMBO J , 3794–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balguerie A, Dos Reis S, Ritter C, Chaignepain S, Coulary-Salin B, Forge V, Bathany K, Lascu I, Schmitter JM, Riek R, Saupe SJ. (2003). Domain organization and structure-function relationship of the HET-s prion protein of Podospora anserina. EMBO J , 2071–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardill JP, Dulle JE, Fisher JR, True HL. (2009). Requirements of Hsp104p activity and Sis1p binding for propagation of the [RNQ(+)] prion. Prion , 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardill JP, True HL. (2009). Heterologous prion interactions are altered by mutations in the prion protein Rnq1p. J Mol Biol , 583–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biancalana M, Koide S. (2010). Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim Biophys Acta , 1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom MS, Koshland D, Guacci V. (2018). Cohesin function in cohesion, condensation, and DNA Repair is regulated by Wpl1p via a common mechanism in Saccharomyces cerevisiae. Genetics , 111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boel G, Smith PC, Ning W, Englander MT, Chen B, Hashem Y, Testa AJ, Fischer JJ, Wieden HJ, Frank J, et al. (2014). The ABC-F protein EttA gates ribosome entry into the translation elongation cycle. Nat Struct Mol Biol , 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast , 115–132. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA. (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science , 1729–1732. [DOI] [PubMed] [Google Scholar]
- Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, Braun S, Madhani HD, Krogan NJ, Weissman JS. (2008). A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods , 711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JC. (2014). Optimization of SDD-AGE as a Method to Study Amyloid Conversion of Human Recombinant Prion Protein. In Biochemistry. Vol. M.S. Texas State University. 42 Unpublished thesis; https://digital.library.txstate.edu/handle/10877/5272.
- Chen Y, Pan FC, Brandes N, Afelik S, Solter M, Pieler T. (2004). Retinoic acid signaling is essential for pancreas development and promotes endocrine at the expense of exocrine cell differentiation in Xenopus. Dev Biol , 144–160. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. (1995). Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science , 880–884. [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. (1992). Multifunctional yeast high-copy-number shuttle vectors. Gene , 119–122. [DOI] [PubMed] [Google Scholar]
- Collart C, Owens ND, Bhaw-Rosun L, Cooper B, De Domenico E, Patrushev I, Sesay AK, Smith JN, Smith JC, Gilchrist MJ. (2014). High-resolution analysis of gene activity during the Xenopus mid-blastula transition. Development , 1927–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagle JM, Littig JL, Sutherland LB, Weeks DL. (2000). Targeted elimination of zygotic messages in Xenopus laevis embryos by modified oligonucleotides possessing terminal cationic linkages. Nucleic Acids Res , 2153–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagle JM, Weeks DL. (2000). Selective degradation of targeted mRNAs using partially modified oligonucleotides. Methods Enzymol , 420–436. [DOI] [PubMed] [Google Scholar]
- Dichmann DS, Harland RM. (2012). fus/TLS orchestrates splicing of developmental regulators during gastrulation. Genes Dev , 1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmic MW, Rest JS, Mindell DP, Goldstein RA. (2002). rtREV: an amino acid substitution matrix for inference of retrovirus and reverse transcriptase phylogeny. J Mol Evol , 65–73. [DOI] [PubMed] [Google Scholar]
- Dong J, Lai R, Jennings JL, Link AJ, Hinnebusch AG. (2005). The novel ATP-binding cassette protein ARB1 is a shuttling factor that stimulates 40S and 60S ribosome biogenesis. Mol Cell Biol , 9859–9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duennwald ML, Echeverria A, Shorter J. (2012). Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol , e1001346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erives AJ, Fassler JS. (2015). Metabolic and chaperone gene loss marks the origin of animals: evidence for Hsp104 and Hsp78 chaperones sharing mitochondrial enzymes as clients. PLoS One , e0117192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. (2006). Functional amyloid formation within mammalian tissue. PLoS Biol , e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Carroni M, Nussbaum-Krammer C, Mogk A, Nillegoda NB, Szlachcic A, Guilbride DL, Saibil HR, Mayer MP, Bukau B. (2015). Human Hsp70 Disaggregase Reverses Parkinson’s-Linked alpha-Synuclein Amyloid Fibrils. Mol Cell , 781–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi M, Lilie H, Funfschilling U, Mun A, Ross S, Lithgow T, Rucknagel P, Rospert S. (2001). RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc Natl Acad Sci USA , 3762–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenning M. (2010). Binding mode of Thioflavin T and other molecular probes in the context of amyloid fibrils-current status. J Chem Biol , 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. (2012). Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature , 363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Lindquist S. (2010). Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science , 629–632. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW. (2005). AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol , 519–529. [DOI] [PubMed] [Google Scholar]
- Harland RM. (1991). In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol , 685–695. [DOI] [PubMed] [Google Scholar]
- Hashimshony T, Wagner F, Sher N, Yanai I. (2012). CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep , 666–673. [DOI] [PubMed] [Google Scholar]
- Hayes MH, Peuchen EH, Dovichi NJ, Weeks DL. (2018). Dual roles for ATP in the regulation of phase separated protein aggregates in Xenopus oocyte nucleoli. eLife . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes MH, Weeks DL. (2016). Amyloids assemble as part of recognizable structures during oogenesis in Xenopus. Biol Open , 801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iconomidou VA, Cordopatis P, Hoenger A, Hamodrakas SJ. (2011). The silkmoth eggshell as a natural amyloid shield for the safe development of insect oocyte and embryo: insights from studies of silkmoth chorion protein peptide-analogues of the B family. Biopolymers , 723–733. [DOI] [PubMed] [Google Scholar]
- Ikonomovic MD, Abrahamson EE, Isanski BA, Debnath ML, Mathis CA, Dekosky ST, Klunk WE. (2006). X-34 labeling of abnormal protein aggregates during the progression of Alzheimer’s disease. Methods Enzymol , 123–144. [DOI] [PubMed] [Google Scholar]
- Inoue Y, Kawai-Noma S, Koike-Takeshita A, Taguchi H, Yoshida M. (2011). Yeast prion protein New1 can break Sup35 amyloid fibrils into fragments in an ATP-dependent manner. Genes Cells , 545–556. [DOI] [PubMed] [Google Scholar]
- Jackrel ME, DeSantis ME, Martinez BA, Castellano LM, Stewart RM, Caldwell KA, Caldwell GA, Shorter J. (2014). Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell , 170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackrel ME, Shorter J. (2014). Potentiated Hsp104 variants suppress toxicity of diverse neurodegenerative disease-linked proteins. Dis Model Mech , 1175–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. (2012). Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell , 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, McKnight SL. (2018). A solid-state conceptualization of information transfer from gene to message to protein. Annu Rev Biochem , 351–390. [DOI] [PubMed] [Google Scholar]
- Kayed R, Head E, Sarsoza F, Saing T, Cotman CW, Necula M, Margol L, Wu J, Breydo L, Thompson JL, et al. (2007). Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol Neurodegener , 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr ID. (2004). Sequence analysis of twin ATP binding cassette proteins involved in translational control, antibiotic resistance, and ribonuclease L inhibition. Biochem Biophys Res Commun. , 166–173. [DOI] [PubMed] [Google Scholar]
- Kirstein J, Arnsburg K, Scior A, Szlachcic A, Guilbride DL, Morimoto RI, Bukau B, Nillegoda NB. (2017). In vivo properties of the disaggregase function of J-proteins and Hsc70 in Caenorhabditis elegans stress and aging. Aging Cell , 1414–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles TP, Vendruscolo M, Dobson CM. (2014). The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol , 384–396. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI, Aravind L. (2000). Protein fold recognition using sequence profiles and its application in structural genomics. Adv Protein Chem , 245–275. [DOI] [PubMed] [Google Scholar]
- Koplin A, Preissler S, Ilina Y, Koch M, Scior A, Erhardt M, Deuerling E. (2010). A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J Cell Biol , 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg PA, Melton DA. (1985). Developmental regulation of a gastrula-specific gene injected into fertilized Xenopus eggs. EMBO J , 3463–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krobitsch S, Lindquist S. (2000). Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc Natl Acad Sci USA , 1589–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. (2003). Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem , 49636–49643. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol , 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackie RE, Maciejewski A, Ostapchenko VG, Marques-Lopes J, Choy WY, Duennwald ML, Prado VF, Prado MAM. (2017). The Hsp70/Hsp90 chaperone machinery in neurodegenerative diseases. Front Neurosci , 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link CD, Johnson CJ, Fonte V, Paupard M, Hall DH, Styren S, Mathis CA, Klunk WE. (2001). Visualization of fibrillar amyloid deposits in living, transgenic Caenorhabditis elegans animals using the sensitive amyloid dye, X-34. Neurobiol Aging , 217–226. [DOI] [PubMed] [Google Scholar]
- MacQueen AJ, Villeneuve AM. (2001). Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev , 1674–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, Rissman RA, Singru PS, Nilsson KP, Simon R, Schubert D, et al. (2009). Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science , 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton MJ, Vazquez de Aldana CR, Qiu H, Chakraburtty K, Hinnebusch AG. (1997). Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF2alpha kinase GCN2. Mol Cell Biol, 4474–4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell , 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR. (1986). Genealogy of principal strains of the yeast genetic stock center. Genetics , 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AN, Kelly JW. (2012). Hsp104 gives clients the individual attention they need. Cell , 695–697. [DOI] [PubMed] [Google Scholar]
- Nathan DF, Vos MH, Lindquist S. (1997). In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc Natl Acad Sci USA , 12949–12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby GA, Lindquist S. (2013). Blessings in disguise: biological benefits of prion-like mechanisms. Trends Cell Biol , 251–259. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. (1982). A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell , 675–686. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. (1967). Normal Table of Xenopus laevis (Daudin). A Systematical And Chronological Survey of the Development from the Fertilized Egg Till the End of Metamorphosis, Amsterdam: North-Holland Publishing Company, 260 p. with illus. with 210 fold. l. pp. [Google Scholar]
- Nillegoda NB, Bukau B. (2015). Metazoan Hsp70-based protein disaggregases: emergence and mechanisms. Front Mol Biosci , 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nillegoda NB, Kirstein J, Szlachcic A, Berynskyy M, Stank A, Stengel F, Arnsburg K, Gao X, Scior A, Aebersold R, et al. (2015). Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature , 247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nillegoda NB, Wentink AS, Bukau B. (2018). Protein Disaggregation in Multicellular Organisms. Trends Biochem Sci , 285–300. [DOI] [PubMed] [Google Scholar]
- Otzen D, Riek R. (2019). Functional amyloids. Cold Spring Harb Perspect Biol , a033860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino MM, Liu JJ, Glover JR, Lindquist S. (1996). Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science , 622–626. [DOI] [PubMed] [Google Scholar]
- Penna TC, Ishii M, Junior AP, Cholewa O. (2004). Thermal stability of recombinant green fluorescent protein (GFPuv) at various pH values. Appl Biochem Biotechnol , 469–483. [DOI] [PubMed] [Google Scholar]
- Peshkin L, Wuhr M, Pearl E, Haas W, Freeman RM, Jr, Gerhart JC, Klein AM, Horb M, Gygi SP, Kirschner MW. (2015). On the relationship of protein and mRNA dynamics in vertebrate embryonic development. Dev Cell , 383–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissler S, Deuerling E. (2012). Ribosome-associated chaperones as key players in proteostasis. Trends Biochem Sci , 274–283. [DOI] [PubMed] [Google Scholar]
- Putnam A, Cassani M, Smith J, Seydoux G. (2020). A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat Struct Mol Biol , 220–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaran RN, Serpell LC. (2008). Amyloid fibrils: abnormal protein assembly. Prion , 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampelt H, Kirstein-Miles J, Nillegoda NB, Chi K, Scholz SR, Morimoto RI, Bukau B. (2012). Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J , 4221–4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova NV, Chernoff YO. (2009). Hsp104 and prion propagation. Protein Pept Lett , 598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW. (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics , 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M, Sibbald PR, Wittinghofer A. (1990). The P-loop–a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci , 430–434. [DOI] [PubMed] [Google Scholar]
- Saupe SJ. (2000). Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol Mol Biol Rev , 489–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods , 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J. (2011). The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS One , e26319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J. (2017). Designer protein disaggregases to counter neurodegenerative disease. Curr Opin Genet Dev , 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. (2004). Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science , 1793–1797. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. (2006). Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell , 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. (2010). Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell , 421–435. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics , 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider J, Houry WA. (2008). AAA+ proteins: diversity in function, similarity in structure. Biochem Soc Trans , 72–77. [DOI] [PubMed] [Google Scholar]
- Stroo E, Koopman M, Nollen EA, Mata-Cabana A. (2017). Cellular regulation of amyloid formation in aging and disease. Front Neurosci , 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styren SD, Hamilton RL, Styren GC, Klunk WE. (2000). X-34, a fluorescent derivative of Congo red: a novel histochemical stain for Alzheimer’s disease pathology. J Histochem Cytochem , 1223–1232. [DOI] [PubMed] [Google Scholar]
- Sweeny EA, Shorter J. (2016). Mechanistic and structural insights into the prion-disaggregase activity of Hsp104. J Mol Biol , 1870–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol , 1596–1599. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. (2001). Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene , 103–112. [DOI] [PubMed] [Google Scholar]
- Tkach JM, Glover JR. (2008). Nucleocytoplasmic trafficking of the molecular chaperone Hsp104 in unstressed and heat-shocked cells. Traffic , 39–56. [DOI] [PubMed] [Google Scholar]
- Tompa P. (2012). Intrinsically disordered proteins: a 10-year recap. Trends Biochem Sci , 509–516. [DOI] [PubMed] [Google Scholar]
- Torrente MP, Chuang E, Noll MM, Jackrel ME, Go MS, Shorter J. (2016). Mechanistic insights into Hsp104 potentiation. J Biol Chem , 5101–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrente MP, Shorter J. (2013). The metazoan protein disaggregase and amyloid depolymerase system: Hsp110, Hsp70, Hsp40, and small heat shock proteins. Prion , 457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey JW. (1949). Comparing individual means in the analysis of variance. Biometrics , 99–114. [PubMed] [Google Scholar]
- Vassar PS, Culling CF. (1959). Fluorescent stains, with special reference to amyloid and connective tissues. Arch Pathol , 487–498. [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. (1982). Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J , 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Fuxreiter M. (2016). The structure and dynamics of higher-order assemblies: amyloids, signalosomes, and granules. Cell , 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Costanzo M, Heisler LE, Paw J, Kaper F, Andrews BJ, Boone C, Giaever G, Nislow C. (2008). Yeast Barcoders: a chemogenomic application of a universal donor-strain collection carrying bar-code identifiers. Nat Methods , 719–725. [DOI] [PubMed] [Google Scholar]
- Yoshiike Y, Minai R, Matsuo Y, Chen YR, Kimura T, Takashima A. (2008). Amyloid oligomer conformation in a group of natively folded proteins. PLoS One , e3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernicka-Goetz M, Pines J, Ryan K, Siemering KR, Haseloff J, Evans MJ, Gurdon JB. (1996). An indelible lineage marker for Xenopus using a mutated green fluorescent protein. Development , 3719–3724. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang S, Zhang L, Lu J, Zhao C, Luo F, Li D, Li X, Liu C. (2019). Heat shock protein 104 (HSP104) chaperones soluble Tau via a mechanism distinct from its disaggregase activity. J Biol Chem , 4956–4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, et al. (2001). Global analysis of protein activities using proteome chips. Science , 2101–2105. [DOI] [PubMed] [Google Scholar]
- Zhu H, Snyder M. (2001). Protein arrays and microarrays. Curr Opin Chem Biol , 40–45. [DOI] [PubMed] [Google Scholar]
- Zwicker D, Decker M, Jaensch S, Hyman AA, Julicher F. (2014). Centrosomes are autocatalytic droplets of pericentriolar material organized by centrioles. Proc Natl Acad Sci USA , E2636–E2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.