Abstract
Actin-based tubular connections between cells have been observed in many cell types. Termed “tunneling nanotubes (TNTs),” “membrane nanotubes,” “tumor microtubes (TMTs),” or “cytonemes,” these protrusions interconnect cells in dynamic networks. Structural features in these protrusions vary between cellular systems, including tubule diameter and the presence of microtubules. We find tubular protrusions, which we classify as TMTs, in a pancreatic cancer cell line, Dartmouth-Hitchcock Pancreatic Cancer (DHPC)-018. TMTs are present in DHPC–018-derived tumors in mice, as well as in a mouse model of pancreatic cancer and a subset of primary human tumors. DHPC-018 TMTs have heterogeneous diameter (0.39–5.85 µm, median 1.92 µm) and contain actin filaments, microtubules, and cytokeratin 19-based intermediate filaments. TMTs do not allow intercellular transfer of cytoplasmic GFP. Actin filaments are cortical within the protrusion, as opposed to TNTs, in which filaments run down the center. TMTs are dynamic in length, but are long lived (median >60 min). Inhibition of actin polymerization, but not microtubules, results in TMT loss. Extracellular calcium is necessary for TMT maintenance. A second class of tubular protrusion, which we term cell-substrate protrusion, has similar width range and cytoskeletal features but makes contact with the substratum as opposed to another cell. Similar to previous work on TNTs, we find two assembly mechanisms for TMTs, which we term “pull-away” and “search-and-capture.” Inhibition of Arp2/3 complex inhibits TMT assembly by both mechanisms. This work demonstrates that the actin architecture of TMTs in pancreatic cancer cells is fundamentally different from that of TNTs and demonstrates the role of Arp2/3 complex in TMT assembly.
INTRODUCTION
Cells possess a variety of mechanisms for exchange of materials and information, including soluble growth factors/chemokines, exosomes, adherens junctions, and gap junctions (Ribeiro-Rodrigues et al., 2017; Mathieu et al., 2019). Over 15 years ago, it was revealed that tubular connections can exist between cells. Originally termed cytonemes (Ramírez-Weber and Kornberg, 1999) or tunneling nanotubes (TNTs) (Önfelt and Davis, 2004; Rustom et al., 2004), these protrusions are thin (<500 nm width) and can extend over 100 µm. Given their dimensions, and the fact that they are not on the basal surface but are extended between cells like a tightrope, the protrusions are easily destroyed by certain fixation procedures (Rustom et al., 2004; Sartori-Rupp et al., 2019), perhaps contributing to their relatively late identification.
Since these initial discoveries, a variety of similar structures have been described in multiple systems. With this wider spread has come a wider variety of structural features. One difference between the structures concerns cytoskeletal composition. While the initially identified structures were shown to contain actin but not microtubules (Ramírez-Weber and Kornberg, 1999; Rustom et al., 2004; Sowinski et al., 2008; Sartori-Rupp et al., 2019), a number of subsequently identified structures contain microtubules (Önfelt et al., 2006; Gerdes et al., 2013; Osswald et al., 2015; Kumar et al., 2017; Resnik et al., 2018; Kim et al., 2019). Where examined, intermediate filaments (IFs) have also been identified (Iglicˇ et al., 2007; Ady et al., 2014; Sáenz-de-Santa-María et al., 2017; Resnik et al., 2018). A second difference concerns the size of the structures. TNTs are generally defined as having a diameter less than 500 nm (Rustom et al., 2004; Ariazi et al., 2017; Sartori-Rupp et al., 2019), but other structures can be several microns wide (Vidulescu et al., 2004; Osswald et al., 2015; Kumar et al., 2017). These variations have led to use of additional names such as “microtubes” to reflect the structural differences. To avoid a diameter specification, we will refer to the structures collectively as intercellular membrane tubules (IMTs).
IMTs have been identified in vivo in both Drosophila (Ramírez-Weber and Kornberg, 1999; Roy et al., 2011, 2014; Huang et al., 2019) and mammals (Chinnery et al., 2008), suggesting physiological significance. A number of communication functions have been attributed to IMTs, including targeted growth factor transmission (Roy et al., 2011, 2014), neurotransmitter signaling from epithelial cells (Huang et al., 2019), and direct exchange of cytoplasmic materials including: mitochondria (Kumar et al., 2017; Kretschmer et al., 2019), endosomes/lysosomes (Rustom et al., 2004; Kumar et al., 2017; Sartori-Rupp et al., 2019), plasma membrane proteins (Önfelt and Davis, 2004), mRNA (Haimovich et al., 2017), and microRNAs (Thayanithy et al., 2014).
Three pathogenic roles have also been linked to IMTs. First, pathogens such as viruses (Sowinski et al., 2008; Hashimoto et al., 2016; Kumar et al., 2017; Panasiuk et al., 2018) and bacteria (Önfelt et al., 2006; Kim et al., 2019) can use IMTs as a means of cell-to-cell transfer. Second, misfolded protein aggregates including huntingtin (Costanzo et al., 2013), prion protein (Zhu et al., 2015), and α-synuclein (Abounit et al., 2016) have also been shown to transfer between cells through IMTs. Finally, IMTs have been linked to increased treatment resistance of several cancers. In a glioblastoma model, IMTs of >100 µm length extended at the invasive edge of the tumor into peripheral tissue. These IMTs had a diameter ∼1.6 µm, contained both actin and microtubules, and correlated with increased resistance to radiation and chemotherapy (Osswald et al., 2015; Weil et al., 2017). Similarly, IMTs that contribute to treatment resistance have been identified in pancreatic cancer (Desir et al., 2018), prostate cancer (Kretschmer et al., 2019), mesothelioma (Lou et al., 2012), and leukemias (Polak et al., 2015).
In terms of IMT assembly, there is strong consensus that the process is actin dependent (Ariazi et al., 2017). Interestingly, two distinct mechanisms for assembly have been observed (Sowinski et al., 2008; Veranicˇ et al., 2008; Gerdes et al., 2013; Kumar et al., 2017), which we term “pull-away” and “search-and-capture.” In pull-away assembly, an IMT is created between two cells that are closely associated when one of the cells migrates away from the other. In search-and-capture, one cell extends a protrusion that makes stable contact with another cell. The factors that nucleate the actin filaments involved in IMT assembly by either mechanism have not been defined. Due to their thin, tubular nature and their actin dependence, it has been tempting to view IMTs as specialized filopodia. However, evidence from a neuronal cell line suggests distinct and even opposite molecular characteristics between IMTs and filopodia (Delage et al., 2016), although IMTs in this system still contain parallel actin filaments that run the length of the IMT, similar to filopodia (Sartori-Rupp et al., 2019).
In this study, we examine IMTs of a variety of diameters emanating from a low-passage pancreatic ductal adenocarcinoma (PDAC) cell line, which we define as TMTs based on their presence in tumors and morphological characteristics. Actin filaments are present in all IMTs, with microtubules and IFs also present in > 90% of the structures. In IMTs of median width (∼2 µm), actin filaments are enriched along the IMT membrane, whereas microtubules and IFs run down the center. A second structure, which we refer to as a cell surface protrusion (CSP), does not contact another cell but interacts with the substratum. IMT assembly occurs equally by pull-away and search-and-capture mechanisms. Inhibition of Arp2/3 complex results in a decrease in IMT assembly by both mechanisms. We also find IMTs in PDAC tumors in several contexts, including a subset of primary human tumors. These results suggest that IMTs can represent a heterogeneous population of structures, even within a single cell type.
RESULTS
DHPC-018 cells possess two types of lateral fingerlike protrusions
The DHPC-018 (Dartmouth-Hitchcock Pancreatic Cancer) cell line was established from a human peritoneal metastasis that had been surgically removed after neo-adjuvant treatment. Examination of live DHPC-018 cells by differential interference contrast (DIC) microscopy revealed two clear features. First, the cells contain frequent intercellular connections, in the form of thin protrusions (Figure 1A). The mean frequency of these protrusions is 0.315 ± 0.06 protrusions/cell (Figure 1B). A wide variety of protrusion lengths is present, with a range from 5 to 244 µm, and a median of 26.7 µm (Figure 1C). The width of the protrusions is also variable, with a range from 0.39 to 5.85 µm and a median of 1.92 µm (Figure 1D). Due to their width (generally larger than traditionally defined TNTs) and their presence in a tumor-derived cell line, we choose to refer to these structures as tumor microtubes (TMTs).
FIGURE 1:
DHPC-018 cells possess TMTs and cell–substratum protrusions (CSPs) of variable length and width. (A) DIC microscopy of live DHPC-018 cells. Focal plane starts 1 µm above basal surface, 20×, 0.75 NA objective. Arrows and arrowheads denote examples of TMTs and CSPs respectively. Scale bar, 25 µm. (B) TMT and CSP frequency; 21 fields, 4963 cells, 1487 TMTs, 182 CSPs. Bars are medians: 0.315 ± 0.01 TMT/cell (SEM), 0.041 ± 0.01 CSP/cell. (C) TMT and CSP length; 544 TMTs, 63 CSPs. Bars are medians: 26.7 ± 1.2 µm for TMT (SEM), 37.7 ± 3.8 µm for CSPs. (D) TMT and CSP width; 82 TMTs, 22 CSPs. Bars are medians: 1.90 ± 0.14 µm for TMT (SEM), 2.68 ± 0.25 µm for CSPs. (E) Comparative Airyscan images of fixed U2OS (left) and DHPC-018 (right) cells, stained with anti-Tom20 (green, mitochondria), anti-tubulin (white), TRITC-phalloidin (red, actin), and DAPI (blue, nucleus); 0.2-µm Z slices, 100 × 1.4 NA objective. Top: max intensity plan view. Bottom: side view of Z-stack rotated 50° from the orientation of the plan view to show the TMT clearly. Arrow denotes TMT, with the receiving cell being out of the field. Scale bars, 10 µm. Movie 1 shows a rotating view of the 3D reconstruction. (F) TMT height above the substratum; 82 TMTs. Bar is median, 5.2 ± 0.27 µm (SEM). (G) Example of a CSP. Max intensity plan view of a field of DHPC-018 cells (Airyscan, 0.2-µm Z slices). Cells were stained with anti-CK19 (green), anti-tubulin (magenta), TRITC-phalloidin (red), and DAPI (blue). Scale bar, 25 µm. (H) Zoom of the boxed CSP from G, rotated 70° counterclockwise from the orientation of the plan view. Scale bar, 10 µm. (I) Side view of Z-stack of boxed CSP from G. Dashed line indicates the substratum. Scale bar, 10 µm.
A second interesting feature is that most DHPC-018 cells do not spread extensively on the fibronectin-coated substratum, but rather remain extended in the Z direction. This feature is best appreciated in comparison to an established culture cell line such as U2OS osteosarcoma cells (Figure 1E; Supplemental Figure S1A; Supplemental Movie S1). Examination of Z-stacks of DHPC-018 cells shows that TMTs typically protrude from the lateral cell surface, at a median height of 5.2 ± 2.5 μm above the base of the cell (Figure 1F).
Movie S1.
Rotating view of 3D reconstruction of DHPC-018 cell, to go with Figure 1E. Cell stained with anti-Tom20 (green, mitochondria), anti-tubulin (white), TRITC-phalloidin (red, actin), and DAPI (blue, nucleus). Arrow indicates TMT. Scale bar, 10 μm.
We also observed a second population of protrusions from DHPC-018 cells that did not connect with another cell but to the substratum (Figure 1, A and G–I), and which we term CSP. Similar to TMTs, CSPs generally originate from the lateral cell surface, contacting the substratum at their distal tips (Figure 1I; Supplemental Movie S2). While morphologically akin to TMTs, CSPs are slightly longer (Figure 1C) and wider (Figure 1D), although there is considerable heterogeneity in both parameters. The distal tips of CSPs tend to display an expanded width compared with the CSP shaft (Figure 1H). CSPs are 7.7-fold less abundant than TMTs (Figure 1B, mean 0.041 ± 0.01 CSP/cell).
Movie S2.
Rotating view of 3D reconstruction of a CSP, to go with Figure 1I. Cell stained with anti-CK19 (green), anti-tubulin (magenta), TRITC-phalloidin (red), and DAPI (blue). Scale bar, 10 μm.
These experiments were conducted on cells plated onto fibronectin. We asked whether the nature of the substratum influenced the size or abundance of TMTs or CSPs. Plating onto the following coated surfaces results in minor variation of these parameters from fibronectin: uncoated glass, laminin, type-1 collagen, and concanavalin A (Supplemental Figure S1, B–E).
Cytoskeletal organization in DHPC-018 TMTs
We next examined the cytoskeletal components present in TMTs, initially focusing on actin filaments and microtubules. Using low-magnification fixed-cell imaging of actin filaments (TRITC-phalloidin), microtubules (anti-alpha-tubulin), and cell membranes (wheat germ agglutinin, WGA), we find that all TMTs contain actin filaments and ∼93% contain microtubules (Figure 2, A and B). We used higher resolution Airyscan microscopy to examine cytoskeletal organization in more detail. For TMTs of median thickness (∼2 µm), actin filament staining exists mainly along the cortex of the TMT, whereas microtubules enrich in the central region (Figure 2C; Supplemental Figure S2A). This organization of actin at the cortex is also observed on 3D reconstruction of Z-stacks (Figure 2D). For thinner TMTs (<1 µm), the spatial organization is more difficult to determine at this resolution (Figure 2E; Supplemental Figure S2B).
FIGURE 2:
Cytoskeletal organization in DHPC-018 TMTs. Airyscan confocal images of cells were plated on fibronectin, fixed, and stained with anti-CK19 (green), anti-tubulin (white), TRITC-phalloidin (red), and WGA (magenta). (A) Max projection of a field of cells. Arrow indicates a TMT with actin, tubulin, and CK19; arrowhead indicates a TMT with only actin and CK19. Scale bar, 20 µm. (B) Percentage of TMTs containing CK19 and/or microtubules. All TMTs contain actin filaments. 611 TMTs. (C) High-resolution image of a TMT of median thickness (∼2 μm). Scale bars, 5 µm. (D) 3D reconstruction of a TMT interior. Top: plan view. Scale bar, 1 µm. Bottom: XZ cross-section of TMT, taken at the point indicated by the dashed line in the top panel. Actin filaments trace the TMT periphery, while CK19 and microtubules localize to the interior. (E) A TMT of thinner width (<1 μm). Scale bars, 5 µm. (F) Two examples of CSP cytoskeletal distribution. Left: CSP of intermediate width (average 3.00 μm) and length (44.0 μm). Right: CSP of larger width (average 4.84) and length (70.5 μm). Scale bars, 5 µm.
We also examined DHPC-018 cells for the presence of cytokeratin 19 (CK19), since this IF protein has been used as a marker for PDAC (Jain et al., 2010; Cen et al., 2017). CK19 staining is enriched in ∼94% of TMTs (Figure 2, A and B). Similar to microtubules, CK19-containing IFs run through the interior of the TMT (Figure 2C; Supplemental Figure S2A). However, while IFs and microtubules are often found in close proximity in the TMT, they are not completely overlapping in localization, suggesting that they are not obligatorily associated.
Finally, we examined the cytoskeletal organization in CSPs. CSPs of two different widths are shown in Figure 2F, with additional examples in Supplemental Figure S2C. In the CSP shaft, organization is largely similar to TMTs, with actin enriched along the cortex, while microtubules and CK19 are largely in the central region. For wider CSPs, there is some evidence for long filaments/bundles in the central region (Figure 2F, right). The CSP tip is more variable. In some cases, the tip is considerably wider than the shaft, with leading edge actin staining reminiscent of a lamellipodium or growth cone (Figure 2F). In other cases, the CSP tip is less broad, with greatly reduced leading edge actin staining (Supplemental Figure S2C).
Assembly mechanisms and dynamics for DHPC-018 TMTs and CSPs
We used live-cell DIC microscopy to evaluate TMT assembly mechanisms and dynamics in DHPC-018 cells, allowing cells to adhere to fibronectin-coated coverslips for 4 h before acquiring images at multiple Z planes at 15- or 30-min intervals over the next 12–20 h. From these examinations, both TMTs and CSPs display a range of lifetimes, from one frame to over 12 h (Figure 3A).
FIGURE 3:
Assembly and dynamics of TMTs and CSPs. (A) Graph of protrusion lifetimes. Bars are medians, 75 ± 21.2 min for TMTs (40 events); 60 ± 22.4 min CSPs (52 events). (B) Graph of total TMT assemblies per field as a function of assembly mechanism. Bars are medians, 122 ± 9.29 for pull-away (710 events); 59.5 ± 5.51 for search-and-capture (352 events). (C) Graph of TMT lifetimes as a function of assembly mechanism. Bars are medians, 75 ± 17.5 min pull-away (35 events); 67.5 ± 17.2 min search-and-capture (35 events). (D) Time-lapse DIC montage (30-min frame interval) of pull-away assembly of a TMT. Asterisk depicts the cell that pulls away from the adjacent cells, creating three TMTs and a CSP (arrowheads) that appear to condense together. Z plane 3–4 µm above basal surface. Scale bar, 20 µm. From Supplemental Movie S3. (E) Time-lapse DIC montage (15-min frame interval) of search-and-capture TMT assembly. The cell marked with the asterisk extends a CSP (arrowhead) by moving away from the CSP tip. Another cell (marked by a hashtag) contacts the CSP, then pulls away. The resulting TMT is denoted by an arrow. Scale bar, 20 µm. From Supplemental Movie S5. (F) Time-lapse DIC montage (30-s frame interval) of an individual TMT, which undergoes periodic ruffling (asterisks), filopodial assembly (arrowhead), and translocation of a bulged region (arrows). Scale bar, 10 µm. Z plane 3–4 µm above basal surface. From Supplemental Movie S6. (G) Time-lapse DIC montage (30-s frame interval) of an individual CSP whose dynamic activity includes a filopodial assembly (arrowhead), ruffling (asterisks), and blebbing. Scale bar, 10 µm. Z plane on basal surface. From Supplemental Movie S7. (H) Time-lapse DIC montage (30-s frame interval) of an individual CSP that undergoes retraction, preceded by termination of ruffling at the tip. Scale bar, 10 µm. Z plane on basal surface. From Supplemental Movie S8. Cells were plated on glass coverslips in C and D and on fibronectin-coated coverslips in E–G. All error calculations are SEM.
Similar to past studies (Sowinski et al., 2008; Wang et al., 2010; Gerdes et al., 2013; Kumar et al., 2017), we observe two main mechanisms of TMT assembly, which we term pull-away and search-and-capture. Pull-away represents ∼2/3 of the events under the plating conditions used here (Figure 3B). The lifetimes of TMTs created by either mechanism are similar (Figure 3C). For both mechanisms, a number of features suggest considerable flexibility in the process.
In pull-away assembly, one cell migrates away from another, leaving a TMT tether that can persist for multiple hours. Intriguingly, in some cases, several TMTs can assemble between two cells in the early stages of pull-away, and subsequently condense into an apparent single TMT (Figure 3D; Supplemental Movie S3). In other cases, a single TMT is pulled between cells (Supplemental Figure S3; Supplemental Movie S4).
Movie S3.
Pull-away TMT assembly in which multiple TMTs are created initially, then condense to one TMT, to go with Figure 3D. Scale bar, 20 μm.
In search-and-capture assembly, one cell extends a CSP and eventually makes contact with another cell (the “receiving” cell). An interesting feature of search-and-capture in DHPC-018 cells is that it is not the CSP-containing cell that is doing the searching. CSPs are generally not elongating from their tips, but by the cell body moving away from the stationary CSP tip. In the context of search-and-capture, the receiving cell is the one that contacts the stationary CSP (Figure 3E; Supplemental Movie S5). In the example shown, the receiving cell then pulls away from the CSP, creating its own CSP in the process and resulting in a TMT containing segments from both cells. We cannot determine from these movies (Z-resolution 1.5 µm) whether the tip of the CSP lifts off from the substratum after stable contact is made.
After assembly, the TMT can undergo extensive changes in length, as well as withstand considerable deformation by intervening cells (Figure 3, D and E; Supplemental Movies S4 and S5). We also examined TMT dynamics on a shorter timescale (60 min, 2 frames/min). In this context, additional dynamics are apparent including: protrusion of filopodia-like structures from the TMT, lamellipodia-like ruffling from the TMT/cell body interface, and directional movement of material along the TMT, suggestive of organellar contents (Figure 3F; Supplemental Movie S6). For CSPs, the tip does not generally translocate but displays considerable dynamics in the form of blebbing, filopodia, or ruffling (Figure 3, G and H; Supplemental Movies S7 and S8). CSPs can undergo retraction, which is accompanied by a decrease in dynamics at the tip (Figure 3H; Supplemental Movie S8).
Movie S4.
Pull-away TMT assembly in which one TMT is assembled initially, to go with Supplementary Figure 3A. Scale bar, 50 μm.
Movie S5.
Search-and-capture TMT assembly to go with Figure 3E. Scale bar, 20 μm.
Movie S6.
TMT dynamics, including ruffling, filopodium formation, and particle movement, to go with Figure 3F. Scale bar, 10 μm.
Movie S7.
CSP dynamics, including ruffling at CSP tip, to go with Figure 3G. Scale bar, 10 μm.
Movie S8.
CSP ruffling, filopodia, and retraction, to go with Figure 3H. Scale bar, 10 μm.
In some systems, TMTs can serve as bridges for transfer of cytoplasmic contents between cells (Önfelt and Davis, 2004; Rustom et al., 2004; Thayanithy et al., 2014; Haimovich et al., 2017; Kumar et al., 2017; Sartori-Rupp et al., 2019). In some instances, even bulk cytoplasm has been shown to transit through TMTs (Biran et al., 2014). We tested the possibility of cytoplasmic transfer by mixing GFP+ cells and GFP– cells at a 1:9 ratio, then examining the percentage of GFP+ cells after 24 h of plating. No significant change in the GFP+:GFP– ratio occurred over this time period (Supplemental Figure S4A). When individual GFP+ pairings with GFP– cells are examined, there is no evidence of faint GFP staining in the GFP– cells (Supplemental Figure S4B). By live-cell microscopy, there is no evidence of GFP transfer over a 30-min period after a pull-away event of a GFP– cell from a GFP+ cell (Supplemental Figure S4C; Supplemental Movie S9). These results suggest that TMTs do not serve as open conduits for the transfer of cytoplasmic materials in DHPC-018 cells.
Movie S9.
DHPC-018 TMT produced during pull-away from a GFP-PDAC cell, to go with Supplementary Figure 4C. Scale bar, 20 μm.
One possible mechanism for TMT assembly may be during the cytokinetic process, in which the divided cells maintain a stable bridge. In the case of DHPC-018 cells, the division process results in two tightly apposed cells (Supplemental Figure S4D; Supplemental Movie S10). Therefore, this mechanism is unlikely here.
Movie S10.
GFP-positive cell division, to go with Supplementary Figure 4D. Scale bar, 25 μm.
TMT assembly is Arp2/3 complex dependent
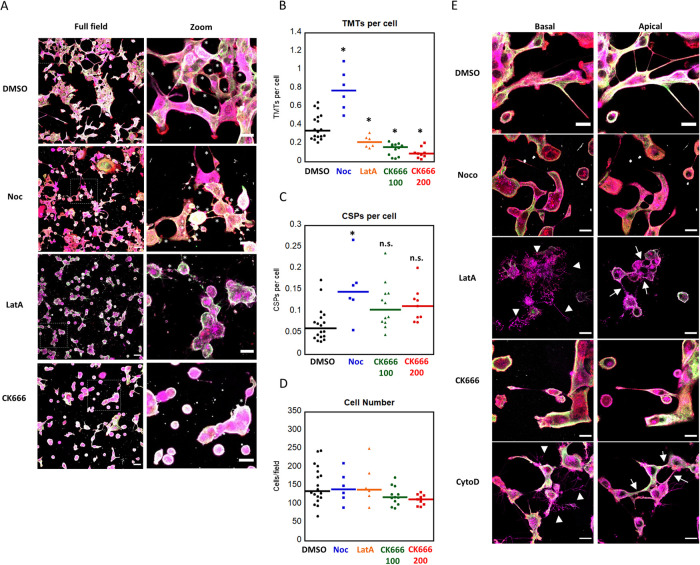
We next examined the roles of actin and microtubules in TMT assembly and maintenance using latrunculin A (LatA, actin depolymerization) and nocodazole (Noc, microtubule depolymerization). Similar to past studies (Vallabhaneni et al., 2012; Wittig et al., 2012; Takahashi et al., 2013; Han et al., 2016), treatment with LatA for 24 h causes a significant decrease in TMTs, whereas TMT and CSP numbers increase with Noc treatment (Figure 4, A–C). These differences are not due to the effects on cell number, since neither LatA nor Noc affect this parameter significantly (Figure 4D). An additional effect occurring on LatA treatment is the elaboration of numerous basal protrusions (Figure 4E). These protrusions are clearly distinct from TMTs, in that they are on the basal surface and are largely devoid of actin filaments, microtubules, and CK19. Similar basal protrusions occur on use of another drug that affects actin depolymerization, cytochalasin D (Figure 4E). Since these protrusions occupy the basal plane, they preclude determination of CSP number with LatA treatment.
FIGURE 4:
Actin depolymerization and Arp2/3 complex inhibition reduce TMT number. (A) Airyscan confocal max projections of fixed DHPC-018 cells following 24 h treatment with DMSO, 50 µM Noc, 5 µM LatA, or 200 µM CK666. Boxed areas in the fields on the left correspond to the zooms on the right. Cells stained with anti-CK19 (green), anti-tubulin (white), TRITC-phalloidin (red), and WGA (magenta). Scale bars, 50 µm for full fields, 20 µm for zooms. (B) Number of TMTs per cell after 24 h treatment; 5991 cells, 3606 TMTs total; 2725 cells, 1128 TMTs for DMSO; 872 cells, 720 TMTs for Noc; 927 cells, 210 TMTs for LatA; 1467 cells, 209 TMTs for 100 µM CK666; 1005 cells, 94 TMTs for 200 µM CK666. Bars are medians, 0.47 ± 0.06 DMSO, 0.77 ± 0.10 Noc, 0.21 ± 0.03 LatA, 0.16 ± 0.02 100 µM CK666, 0.09 ± 0.02 200 µM CK666. *p ≤ 0.05 by ANOVA with Tukey’s Honest Significant Difference. (C) Number of CSPs per cell after 24 h treatment with DMSO, 50 µM Noc, or 200 µM CK666; 182 CSPs DMSO, 119 Noc, 156 100 µM CK666, 115 200 µM CK666. Bars are medians, 0.07 ± 0.01 DMSO, 0.15 ± 0.03 Noc, 0.10 ± 0.02 100 µM CK666, 0.11 ± 0.01 200 µM CK666. CSPs in LatA-treated cells were not quantified due to the extensive basal protrusions formed. *p ≤ 0.05 by ANOVA with Tukey’s Honest Significant Difference; n.s. indicates no statistical significance. (D) Number of cells per field after the indicated 24 h treatment. Bars are medians, 135.5 ± 12.06 DMSO, 140 ± 17.46 Noc, 139.5 ± 22.96 LatA, 118.5 ± 7.31 100 µM CK666, 113 ± 4.92 200 µM CK666. (E) Airyscan confocal images of DHPC-018 cells fixed after DMSO, Noc, LatA, CK666, or cytochalasin D treatments. Left: single 0.4-µm Z slice basal images, right: single Z slice apical images. Arrowheads indicate the basal retraction fibers following LatA and CytoD treatments, while arrows indicate apical TMTs. Staining as in A. Scale bars, 20 µm. All error calculations are SEM.
To probe the role of actin in more detail, we utilized an inhibitor of Arp2/3 complex, CK666. Arp2/3 complex is a major actin nucleation factor and is required for a wide range of cellular actin-based structures (Campellone and Welch, 2010). Treatment with 100 or 200 µM CK666 for 24 h causes a significant decrease in TMTs (Figure 4, A and B) without causing a significant drop in cell number (Figure 4D). Unlike LatA, CK666 treatment does not result in basal surface protrusions (Figure 4E).
We also examined the effects of CK666 treatment on live cells, in order to determine the mechanism leading to TMT loss. Over a 3-h treatment period, CK666-treated cells display a 66% decrease in TMT assembly events (Figure 5A). This decrease is consistent over the experiment time course (Figure 5B), suggesting that Arp2/3 complex is acutely required for TMT assembly. Both pull-away and search-and-capture events are reduced by this treatment (Figure 5C). Both CK666 and LatA cause a change in cell–substratum adhesion, with LatA being much more dramatic. On minutes after LatA treatment, cells retract to leave the basal protrusions, indicating that these apparent protrusions are actually retraction fibers (Figure 5D). CK666 does cause a milder change in overall cell shape, with treated cells becoming less spread than control cells, suggesting an additional effect on cell–substratum adhesion. Neither cell number nor cell viability is affected by either treatment, however.
FIGURE 5:
Arp2/3 acts in assembly of new TMTs. (A) Number of total TMT assembly events quantified from live DIC imaging over 3 h of treatment with DMSO or 200 µM CK666; 350 DMSO TMT assemblies, 120 CK666 TMT assemblies. Bars are medians, 39 ± 6.97 DMSO, 15 ± 4.69 CK666. *p < 0.005 by Wilcoxon Rank Sum test. (B) TMT assemblies as a function of time during DMSO or 200 µM CK666 treatments. (C) TMT assemblies by type following treatment with DMSO or 200 µM CK666. Bars are medians, 37 ± 6.36 pull-away and 3 ± 0.83 search-and-capture for DMSO, 15 ± 2.04 pull-away, and 0 ± 0.30 search-and-capture for CK666. *p < 0.005 by unpaired Student’s t test. (D) Live DIC montages of DHPC-018 cells over 3-h treatments with DMSO (top), 200 µM CK666 (middle), or 5 µM LatA (bottom). Red asterisks denote new TMT assembly. Blue arrowheads denote basal protrusions. Note that LatA montage is taken over a different time period. Scale bars, 30 µm. (E) Examples of TMTs that persist following 1 h of treatment with DMSO, 5 µM LatA, 50 µM Noc, or 100 µM CK666. Cells stained with anti-CK19 (green), anti-tubulin (white), TRITC-phalloidin (red), and WGA (magenta). Scale bars, 5 µm. All error calculations are SEM.
Finally, we examined cytoskeletal distribution in TMTs after inhibitor treatments. Actin filament staining is largely eliminated from TMTs after a 1-h treatment with LatA (Figure 5E). In contrast, actin filaments persist in CK666-treated cells over this time period (Figure 5E). Noc treatment results in loss of microtubules, but actin filaments stay intact (Figure 5E). Interestingly, both CK666 and LatA treatments result in decreased microtubule staining in TMTs (Figure 5E), which could suggest actin- and Arp2/3 complex dependence for microtubule maintenance.
These results suggest that Arp2/3 complex is not required for continuous maintenance of actin filaments in existing TMTs. Interestingly, both CK666 and LatA treatment results in bright WGA punctae in the TMT lumen, suggesting an increase in transport vesicles or endosomal compartments in the TMT.
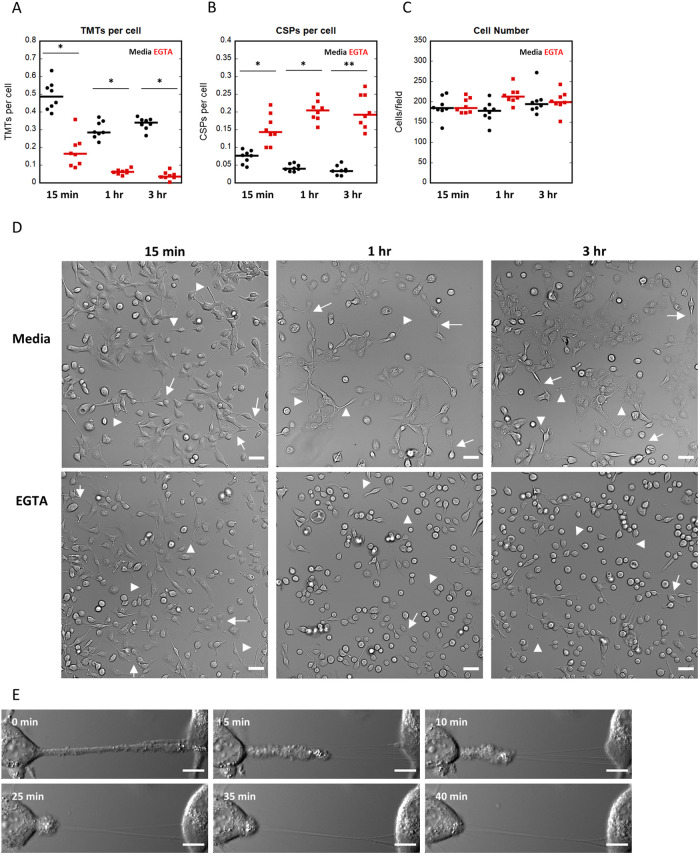
TMT stability requires extracellular calcium
Our data suggest that TMTs in DHPC-018 cells are not open cytoplasmic tubes between the two cells (Supplemental Figure S4). We asked whether the adhesion between the TMT and receiving cell could be disrupted by extracellular calcium chelation, which might suggest adhesion by cadherins or other calcium-dependent mechanisms. Addition of EGTA to the medium results in a significant drop in TMT number within 15 min, with a further drop to near baseline in 1 h (Figure 6A). In contrast, CSP number increases on a similar time course (Figure 6B). While the majority of cells round up during this time, they remain attached to the substratum (Figure 6, C and D), suggesting that TMT loss is not due to loss of cells. Live-cell imaging of cells after EGTA addition shows that TMTs detach at the interface between the TMT and the cell body (Figure 6E; Supplemental Movie S11), suggesting that the connection between TMT and receiving cell is disrupted. In many cases, thin membrane tubes persist (Figure 6E; Supplemental Movie S11). These data suggest that TMTs are attached to receiving cells through calcium-dependent adhesions and can be rapidly detached, but with retention of some continued connection.
FIGURE 6:
TMT stability requires extracellular calcium. DHPC-018 cells were treated with 3 mM EGTA, then monitored by fixed-cell (A–D) or live-cell (E) DIC microscopy. (A) Number of TMTs per cell after 15 min, 1 h, and 3 h treatment with EGTA or media (negative control). Eight fields (20× objective) monitored. 9368 cells, 3275 TMTs total. Bars are medians, 0.49 ± 0.03 15 min media (1497 cells, 735 TMTs), 0.16 ± 0.03 15 min EGTA (1523 cells, 274 TMTs), 0.29 ± 0.02 1 h media (1404 cells, 421 TMTs), 0.06 ± 0.01 1 h EGTA (1727 cells, 108 TMTs), 0.34 ± 0.01 3 h media (1604 cells, 539 TMTs), 0.04 ± 0.01 3 h EGTA (1613 cells, 68 TMTs). *p ≤ 0.0001 by Student’s t test. (B) Number of CSPs per cell after the indicated treatment. 1130 CSPs total. Bars are medians, 0.08 ± 0.01 15 min media (107 CSPs), 0.14 .10.02 15 min EGTA (226 CSPs), 0.04 .00.01 1 h media (60 CSPs), 0.20 ± 0.01 1 h EGTA (353 CSPs), 0.03 ± 0.01 3 h media (57 CSPs), 0.19 ± 0.02 3 h EGTA (141 CSPs). *p ≤ 0.0002, **p ≤ 0.0001 by Student’s t test, respectively. (C) Number of cells per field after the indicated treatment. Bars are medians, 184.5 ± 9.44 15 min media, 184.5 846.44 15 min EGTA, 178 s 9.03 1 h media, 213 ± 7.31 1 h EGTA, 194.5 ± 11.02 3 h media, 199.5 ± 9.12 3 h EGTA. (D) Fixed-cell DIC images of 15 min, 1 h, and 3 h treatments with media or EGTA. Arrows and arrowheads denote TMTs and CSPs, respectively. Scale bars, 50 µm. (E) Live DIC imaging of a TMT retracting from the cell on the right on treatment with EGTA. From Supplemental Movie S11. Scale bar, 10 µm. All error calculations are SEM.
Movie S11.
TMT retraction upon EGTA treatment, to go with Figure 6E. Scale bar, 10 μm.
TMTs in PDAC tumors
To test whether TMTs are present in a tumorlike environment, we injected DHPC-018 cells into the flanks of immune-compromised mice, resulting in tumors of approximately 1 cm diameter in 5 wk. Formalin-fixed, paraffin-embedded (FFPE) sections of the excised tumors were stained with anti-CK19 and DAPI. This approach resulted in a dense CK19 staining pattern due to the fact that DHPC-018 cells represent close to 100% of the cells in the tumor (Supplemental Figure S5A). The dense cell packing precluded detection of TMTs connecting individual DHPC-018 cells.
As an alternative approach, we created a DHPC-018 cell line stably expressing GFP and raised flank tumors in which 10% of the injected cells were GFP-expressing. Low-resolution tile-scans reveal GFP throughout the tumor (Figure 7A). High-resolution imaging with 3D reconstruction reveals readily discernable TMTs, regardless of the fixation/preparation techniques used (Figure 7B; Supplemental Movie S12). These results suggest that TMTs are maintained in DHPC-018 cells in a tumorlike environment. One qualification here is that we cannot definitively distinguish between TMTs and CSPs in these images and those shown below. For simplicity, we will refer to them as TMTs.
FIGURE 7:
TMTs in the PDAC tumor environment. (A) Low magnification (10× objective) tile-scan image of a section from a DHPC-018 flank tumor, in which 10% of the cells express GFP. Section stained with DAPI (blue). Left: overlay, right: GFP alone, enhanced contrast. Scale bars, 500 µm. (B) Representative higher magnification (63× objective) 3D reconstruction images of four DHPC-018 flank tumors (10% GFP). Left to right: formaldehyde perfusion fixation by cardiac puncture, excision, and fixation by formaldehyde, excision and fixation by glutaraldehyde, excision, and fixation by combination of formaldehyde and glutaraldehyde (from Supplemental Movie S12). Arrowheads indicate GFP-positive TMTs. Scale bars, 10 μm for excision fixations, 5 μm for perfusion fixation. (C) Low magnification (10× objective) tile-scan images of FFPE pancreas sections from C57BL/6 mice. Left: pancreas after tumor formation by orthotopically injected KPC4662 acinar cells. Right: control pancreas. Sections stained with DAPI (blue) and anti-CK19 (green). Scale bars, 500 µm. (D) Higher magnification (63× objective) 3D reconstruction image of the KPC4662 tumor from C. Arrowheads/outlines indicate CK19+ TMTs. From Supplemental Movie S13. Scale bars, 5 µm. (E) Representative low magnification tile-scans of sections from FFPE samples of primary tumors from two patients. Sections stained with DAPI (blue) and anti-CK19 (green). Scale bars, 500 µm. (F) Higher magnification (63× objective) 3D reconstruction images of the tumors from E, focused on CK19+ regions. Arrowheads/outlines indicate CK19+ TMTs in tumor D-038, while tumor D-046 appears negative for CK19+ TMTs. Bottom left: XZ view of TMT boxed in D-038; arrowhead indicates TMT. Bottom right: closer view of CK19+ region of D-046, devoid of TMTs. Sections stained with DAPI (blue) and anti-CK19 (green). From Supplemental Movie S14. Scale bars, 10 µm.
Movie S12.
Rotating view of 3D reconstruction of section from a DHPC-018 flank tumor in which 10% of the injected cells express GFP (green), to go with Figure 7B. Cells stained with DAPI (blue).
We next extended our study to a mouse model system in which PDAC is established in an immune-competent environment by injection of cultured KPC 4662 (KrasG12D/+, Trp53R172H/+, Pdx1-Cre) pancreatic acinar cells into the pancreas of C57BL/6 mice (Winograd et al., 2015). The low cellularity of these tumors allows evaluation of TMTs by CK19 staining. Tile-scans reveal elevated and localized CK19 staining compared with control mouse pancreas (Figure 7C). High-resolution imaging with 3D reconstruction reveals CK19-positive TMT connections between cells (Figure 7D; Supplemental Movie S13). An assumption here is that the linear CK19 staining denotes TMTs and not IF bundles within the cell body. The combination of our cell culture results on DHPC-018 cells (showing that long CK19-rich extensions are invariably TMTs or CSPs) and our results with GFP-expressing flank tumors (showing TMT extensions) support the identity of these CK19-rich linear structures as TMTs.
Movie S13.
Rotating view of 3D reconstruction of KPC 4662-derived tumor, to go with Figure 7D. Cells stained with DAPI (blue) and anti-CK19 (green).
To assess the presence of TMTs in a clinically relevant situation, we imaged CK19-stained FFPE sections of six PDAC tumors from Dartmouth-Hitchcock Medical Center (Table 1; Figure 7, E and F; Supplemental Figure S5, B–D). Five samples were resected tumors (four from pancreas, one from a liver metastasis), and one was a needle biopsy core. All patients had undergone neoadjuvant treatment prior to resection/biopsy. Low-resolution tile-scans revealed sporadic pockets of CK19-positive cells (Figure 7E; Supplemental Figure S5B), which were more closely examined at higher resolution for TMTs (Figure 7F; Supplemental Figure S5, C and D). We identified TMTs in three tumors, while two tumors displayed no detectable TMTs by our analysis (Figure 7F; Supplemental Figure S5, C and D; Supplemental Movie S14).
Movie S14.
Rotating view of 3D reconstruction of primary human PDAC tumor D-038, to go with Figure 7F. Cells stained with DAPI (blue) and anti-CK19 (green).
DISCUSSION
In this study, we identify a population of intercellular linear protrusions in a pancreatic cancer (PDAC) cell line, which we define as TMTs. In addition to these structures, a second type of protrusion of similar morphology makes contact with the substratum rather than with another cell, and we define these as CSPs. Both TMTs and CSPs emanate from the lateral plasma membrane as opposed to the basal surface, a property similar to TNTs (Rustom et al., 2004; Gurke et al., 2008; Abounit and Zurzolo, 2012). The majority of TMTs and CSPs contain three cytoskeletal elements: actin filaments, microtubules, and IFs, with actin filaments being enriched along the membrane while microtubules and IFs run along the center. Live-cell microscopy shows that two assembly mechanisms are employed for TMTs, which we term pull-away and search-and-capture, and are similar to previously elucidated TNT assembly mechanisms (Sowinski et al., 2008; Veranicˇ et al., 2008; Gerdes et al., 2013; Kumar et al., 2017). Actin polymerization through Arp2/3 complex is necessary for TMT assembly, and extracellular calcium is necessary for TMT maintenance. We observe TMTs in PDAC tumors in a number of contexts, including a subset of primary human tumors.
An important finding of our work is that the structures we identified are quite heterogeneous in a number of characteristics, including width and assembly mechanism. For this reason, we struggled with which name to assign to the structures. We ultimately chose TMTs for the following reasons: the majority are in the larger width range (>1 µm), they are present in a tumor-derived cell line, and they contain microtubules. However, 21% of the structures are <1 µm and 9% are <0.5 µm in width, which is in the range broadly defined for TNTs. The heterogeneity in this population suggests the possibility of fundamentally distinct functions for these structures. A second point to consider is that the width of an individual TMT can vary over time, which is apparent from our live-cell DIC imaging. A previous study of TNTs from neuronal cell lines showed that multiple TNTs could bundle together into a larger structure (Sartori-Rupp et al., 2019). This scenario is unlikely here, due to the absence of plasma membrane staining within the thicker TMTs we observe. In addition, the structures observed in neuronal cell lines were devoid of microtubules, which is not the case here.
Another heterogeneous aspect of this TMT population is in assembly mechanism. While we broadly use two assembly categories that have been previously defined for TNTs (Sowinski et al., 2008; Veranicˇ et al., 2008; Gerdes et al., 2013; Kumar et al., 2017), our live-cell imaging results suggest heterogeneity within both mechanisms. For the pull-away mechanism, there is a subset of cases in which multiple TMTs assemble initially and then zipper into one. In the search-and-capture mechanism, the receiving cell can pull away from the ‘donor’ cell (the cell extending the CSP), creating a hybrid TMT containing material from both cells. Such hybrid structures have been observed previously for TNTs (Sowinski et al., 2008). Another interesting feature of search-and-capture is that the receiving cell appears to be the one to initiate contact, in that it migrates to the comparatively immobile CSP. This process is fundamentally different from TNT search-and-capture observed in PC12 (Rustom et al., 2004), immune cells (Önfelt and Davis, 2004), or cytonemes (Ramírez-Weber and Kornberg, 1999), in which filopodia protrude to make contact with the receiving cell.
The organization of cytoskeletal elements within TMTs in our system was surprising to us. The most detailed examination of actin organization in TNTs shows long, parallel filaments akin to those in filopodia and microvilli (Sartori-Rupp et al., 2019), although the actin-binding proteins involved appear to be different for filopodia and TNTs (Delage et al., 2016). In the case of PDAC TMTs, actin clearly enriches along the plasma membrane, with little evidence of filaments running down the center of the structure. Instead, both microtubules and IFs occupy the central region. These findings suggest that neither TMTs nor CSPs are an elaboration of filopodia, but rather are more similar to neuronal-like processes, as suggested for cytonemes (Roy et al., 2014; Huang et al., 2019).
Actin filament localization near the plasma membrane is reminiscent of the cortical actin found in the cell body of many cell types (Chugh et al., 2017). Cortical actin consists of a meshwork of actin filaments and myosin II, and the contractile force exerted by the myosin resists cellular turgor pressure. Arp2/3 complex has been shown to be an important assembly factor for cortical actin (Bovellan et al., 2014). It is, therefore, surprising that the actin filament staining in TMTs is maintained after a 1-h treatment with the Arp2/3 complex inhibitor CK666, whereas a 1-h treatment with the actin sequestering molecule LatA results in loss of actin staining.
Our results show, however, that Arp2/3 complex is important in TMT assembly. The exact mechanism by which Arp2/3 complex participates in TMT assembly remains to be defined. Both pull-away and search-and-capture involve translocation of the entire cell, generally considered to involve Arp2/3 complex (Blanchoin et al., 2014). However, cell–cell adhesion also involves Arp2/3 complex (Collins et al., 2017), so TMT assembly could also be affected at this level.
TMTs in our DHPC-018 cells also contain microtubules and IFs. While microtubules are absent from some TNTs (Rustom et al., 2004; Sowinski et al., 2008; Wang et al., 2010; Desir et al., 2018; Kretschmer et al., 2019), they are present in others (Önfelt et al., 2006; Osswald et al., 2015; Jansens et al., 2017; Sáenz-de-Santa-María et al., 2017; Resnik et al., 2018; Zhang et al., 2019). Where examined, the microtubule-containing structures are wider than those lacking microtubules (Önfelt et al., 2006). Microtubules could play clear roles in organelle trafficking within TMTs, but also may be important in maintaining the wider diameter of TMTs (Zhang et al., 2019). It is intriguing in this respect that Noc treatment results in an increase in both TMTs and CSPs for DHPC-018 cells. This result suggests that microtubules are not essential for TMT assembly or maintenance, and even that microtubules might even inhibit these processes, at least in the context of the in vitro system employed here.
A number of IF proteins have been identified in TNTs, including cytokeratins and vimentin (Veranicˇ et al., 2008; Ady et al., 2014; Sáenz-de-Santa-María et al., 2017; Resnik et al., 2018). We find abundant CK19 in TMTs from pancreatic cancer cells both in culture and in a tumor environment. Given the role of keratin-containing IFs in resisting mechanical tension in epithelial cells (Coulombe and Omary, 2002), CK19 may serve to resist the significant tension exerted on TMTs during their lifetime (see, for example, Supplemental Movie S3). It is possible that the presence of CK19 and other IF proteins allows TMTs to resist fixation by formaldehyde (this paper and unpublished observations), while thinner TNTs are much more labile to fixation (Rustom et al., 2004; Koyanagi et al., 2005; Watkins and Salter, 2005).
An interesting question relates to the specific composition of the IFs in TMTs, given the ∼70 filament-forming IF proteins found in mammals (Chung et al., 2013). Thus far, CK7 (Veranicˇ et al., 2008; Resnik et al., 2018), CK19 (this study), and vimentin (Ady et al., 2014; Sáenz-de-Santa-María et al., 2017) have been identified in this context. In view of the growing appreciation for the pleiotropic roles of IF in cancer (Karantza, 2011), their function in TNTs and TMTs assumes greater importance.
Two results suggest that TMTs are not open tubes between two cells, but are rather adhesions between a cell extension and a cell body. First, cytoplasmic GFP does not transfer between cells (Biran et al., 2014). Second, TMTs retract rapidly on chelation of extracellular calcium, seemingly due to detachment of the TMT tip from the cell body of the receiving cell. It is unclear whether this specific mechanism occurs for all TMTs, but the fact that TMTs are almost completely abolished by the treatment suggests the general need for extracellular calcium to maintain connection. The calcium requirement could suggest cadherin-mediated adhesion, and it will be interesting to elucidate the specific cadherin(s) involved.
Our finding that TMTs are present in the pancreatic tumor environment agrees with previous observations in several tumor types (Lou et al., 2012; Osswald et al., 2015; Griessinger et al., 2017; Desir et al., 2018). One function of TMTs is to provide resistance to radiation, chemotherapy, or other assaults (Osswald et al., 2015; Weil et al., 2017; Desir et al., 2018). Interestingly, we can only identify clear TMTs in a subset of primary tumor samples. Expansion of this study to a larger number of tumors, and development of additional TMT markers, is needed to determine the features of TMT-containing tumors in terms of TMT initiation and role in tumor progression. The differential presence of TMTs could provide an additional means of stratification of pancreatic cancer, which would inform treatment and eventual use of TMT-disrupting therapies.
MATERIALS AND METHODS
Culture cells
The DHPC-018 cell line was developed from a peritoneal metastasis that had been surgically removed from a 59-year old male (D11129-008) at the Dartmouth-Hitchcock Medical Center. Prior to surgery, the patient had received three cycles of gemcitabine/docetaxel/capecitabine treatment. Postsurgical testing revealed the tumor to contain the K-Ras G12R mutation. The tumor was minced, then cultured in DMEM +10% fetal bovine serum (FBS) +1% penicillin/streptomycin (all from Hyclone) on tissue culture plastic. Cells were passaged every 3 d by 0.05% trypsinization with 0.53 mM EDTA (Corning, 25-052-Cl). After 10 passages, cells were frozen. On thawing, cells were maintained in DMEM (Corning, 10-013-CV) + 10% FBS (Sigma-Aldrich, F4135) for a maximum of 35 passages. KPC4662 cells (KrasG12D/+, Trp53R172H/+, Pdx1-Cre) pancreatic acinar cells were obtained from Robert Vonderheide (Winograd et al., 2015) and cultured in DMEM/F12 (Corning, 15-090-CV) +10% FBS +1× Glutamax (Life Technologies, 35050-061) +1× penicillin/streptomycin (Sigma, P4333) to a maximum of 35 passages. All cultures were tested every 3 mo for mycoplasma contamination using the LookOut kit (Sigma, MP0035).
To generate a stable GFP-expressing line, DHPC-018 cells were infected at passage 20 with lentivirus expressing FUGW-eGFP (Addgene, #14883), then sorted on a FACS Aria II cell sorter for GFP fluorescence. Following cell sorting, the GFP-DHPC-018 cells were cultured as the DHPC-018 wild-type cells.
Antibodies and dyes
The following primary antibodies and dyes were used: anti-alpha tubulin (Sigma, T9026, used at 1:10000), anti-Tom20 (Santa Cruz, 11415, used at 1:300), anti-CK19 (Abcam, 52625, used at 1:1000 for IF and 1:100 for IHC), phalloidin-tetramethylrhodamine B isothiocyanate (Sigma, P1951, used at 1:500), DAPI (Sigma, D9542, used at 1:500), and WGA-647 (Molecular probes, W32466, used at 0.1 mg/ml). For immunofluorescence, the following secondary antibodies were used: anti-rabbit FITC (Invitrogen, F2765, used at 1/500), anti-mouse AF405 (Invitrogen, A31553, used at 1/500), and anti-mouse AF647 (Life technologies, A21236, used at 1/500). Tissue sections were mounted in VectaShield containing added DAPI (Vector Laboratories, H-1200).
Cell plating and inhibitor treatments
Unless stated otherwise, for all cell experiments, cells were seeded at 3 × 10^5 cells in 1.5 ml of media onto 10 µg/ml fibronectin (Sigma-Aldrich, F1141) coated MatTek dishes (MatTek Corporation, P35G-1.5-14-C) and left to incubate 16–24 h before treatments, fixation, or live imaging. For inhibitor treatments, cells were treated with the following: DMSO (Life technologies, D12345) at 0.5%, LatA (EMD Millipore, 428021) at a 5 µM dilution from a 1 mM stock in DMSO, Noc (Sigma-Aldrich, M1404) at a 50 µM dilution from a 33 mM stock in DMSO, CK666 (Sigma-Aldrich, SML0006) at a 100 or 200 µM dilution from a 20 mM stock in DMSO, and cytochalasin D (Sigma-Aldrich, C8273) at a 1 µM dilution from a 2 mM stock in DMSO. For EGTA treatment, cells were incubated for 3 h with 3 mM EGTA (Sigma-Aldrich, E3889) in media from a 0.95 M stock. Trypan blue (Bio-Rad, 145-0013) exclusion assay showed that cell viability was at least 93% for all inhibitor treatment conditions, conducted at the longest time point.
Immunofluorescence
Cells were fixed on MatTek dishes with 1% glutaraldehyde (Electron Microscopy Sciences, 16019) in BRB80 (80 mM PIPES, pH 6.9; 1 mM MgCl2; 1 mM EGTA) at 23°C, quenched with 3 × 10-min treatments of 1 mg/ml sodium borohydride in phosphate-buffered saline (PBS), permeabilized with 0.25% triton in PBS for 15 min, blocked with 10% normal calf serum (HyClone, SH30118.03) in PBS with 0.02% sodium azide, and stained with antibodies and dyes in 250 µl primary or 300 µl secondary antibodies/dye incubations for 60 min each. When used, WGA-647 was administered for 5 min after quenching and before permeabilization. Dishes were stored in PBS at 4°C and imaged in PBS. Imaging was conducted on an LSM 880 equipped with a 63 × 1.4 NA Apochromat oil objective using the Airyscan detector (Carl Zeiss Microscopy). For DAPI and AF405 secondary antibodies, the 405 nm diode laser and band pass (BP) 420–480 + BP 495–550 filter were used. For FITC and AF488 secondary antibodies the 488-nm Ar-ion laser and BP 420–480 + BP 495–550 filter were used. For rhodamine phalloidin the 561-nm diode-pumped solid state laser and BP 420–480 + 495–620 filter were used. For WGA-647 the 633-nm HeNe laser and BP 570–620 + long pass 645 filter were used. Images were acquired using Zen Black 2012 software and processed with Imaris 8.3.1 software.
Live-cell imaging
MatTek dishes were imaged on a Nikon Ti inverted epifluorescence microscope utilizing a 20×/0.75 NA dry plan apo and 63×/1.40 NA oil plan apo objectives. DIC images were acquired with NIS Elements v5.11.02 and an Andor Zyla sCMOS FLASH 4.0 v3 camera while cells were in a TokaiHit stage-top incubator (37°C and 5% CO2) in DMEM + 10% FBS culture media. To maintain proper humidity over extended imaging times, a MatTek dish lid was modified by replacing a cut square region with a glass coverslip, allowing the dish to be imaged with the lid on.
Tumor samples and immunohistochemistry
Flank tumors from the DHPC-018 cell line were generated in NOD/SCID mice following IACUC protocol #2072, allowing 6 wk for tumor growth. For most preparations, mice were killed by CO2 asphyxiation, then tumors were excised and fixed in 10% formalin, 4% paraformaldehyde (Electron Microscopy Sciences, 15710), 1% glutaraldehyde (Electron Microscopy Sciences, 16000), or a combination of 1% glutaraldehyde and 4% paraformaldehyde in PBS. Fixed tumors were either sectioned immediately on a vibratome (50-µm sections) and stored in PBS with 4% sucrose until staining or paraffin-embedded and sectioned later (FFPE, 20-µm sections). For one mouse, fixation was performed by cardiac puncture using 4% PFA (Acros Organics, AC416780250) and 4% sucrose (Fisher, BP220-1) in PBS (National Diagnostics, CL-253) fixative (following IACUC protocol #0002073[m15]), then the excised tumor was sectioned immediately (50-µm sections). For FFPE samples, deparaffinization was conducted in xylene (EMD Millipore, XX0055), followed by hydration in graded ethanol/water, then the tumor underwent antigen retrieval in boiling Tris-EDTA buffer (10 mM Tris, 1 mM EDTA, pH 9) for 15 min before blocking in 4% Triton-X100 with 10% normal calf serum (Hyclone, SH30118.03) in PBS, staining with anti-CK19 (where stated) and mounting in DAPI-containing VectaShield. Nonembedded samples were subject to the same procedures, omitting the deparaffinization, hydration, and antigen retrieval steps. For assessment of tumors directly in mouse pancreas, tumors were created by orthotopic injection of KPC4662 cells. Briefly, 5 × 10^5-1 × 10^6 cultured KPC4662 cells (KrasG12D/+, Trp53R172H/+, Pdx1-Cre) pancreatic acinar cells (Winograd et al., 2015) are injected into the pancreatic parenchyma (below the capsule) of C57BL/6 mice. At 4 wk postinjection, the mouse is killed by CO2 asphyxiation, and the pancreas is excised and subject to FFPE preparation, sectioning, and staining as described above. Primary human tumors were obtained under IACUC protocol 2177 (see Table 1) as FFPE blocks; 20-µm tumor sectioning and staining were prepared as for mouse FFPE sections above.
Tumors were imaged on an LSM880 Airyscan confocal microscope. Tile-scans were performed using either a 10 × 0.3 NA or 20 × 0.8 NA dry objective (depending on the size of the tissue section), acquiring a series of 3-µm Z-steps and 16–50+ individual fields which were stitched together using Zen Black image processing prior to Airyscan processing. Higher resolution images of tumor TMTs were performed using a 63×/0.75 NA oil objective, acquiring a series of 0.2-µm Z-steps at regions with cells staining positive for CK19.
Quantification of TMT and CSP dimensions and frequency
TMT/CSP frequency and length were determined from 20× Z-stacks acquired on the Dragonfly microscope (10–25 slices at 1 µm). Fields were acquired randomly throughout the dish. For frequency, the number of cells, TMTs, and CSPs were counted in the entire imaging field (Andor Zyla sCMOS FLASH 4.0 v3 camera, 660 × 660 µm area). Length was measured for all TMTs and CSPs throughout the field. TMTs were counted if they were at least two slices above the basal surface, and CSPs were counted if they originated at least two slices above the basal surface. TMT and CSP width and height above basal surface were measured from 63× Z-stacks acquired on the Airyscan microscope (15-40 slices at 0.4 µm). Width was determined using WGA staining, at the z-slice containing the widest TMT section. Line-scans were taken across the TMT, and the exterior position at half height of each side of the TMT was used to mark the position of the plasma membrane.
Quantification of TMT assembly frequency
For Figure 3B, 3.5 × 10^5 cells in 1.5 ml of media were seeded onto 10 µg/ml fibronectin-coated MatTek dishes and left in the incubator for 4 h. After incubation, five fields of DIC images were acquired on a Nikon Eclipse Ti microscope, 20×/0.75 NA objective, with a time interval of 30 min for 24 h. During the first 3 h, every new TMT assembly was counted and categorized as either a pull-away or a search-and-capture event. For Figure 5C, cells were seeded at 3 × 10^5 cells in 1.5 ml of media onto 10 µg/ml fibronectin-coated MatTek dishes and left to incubate 16–24 h before treatment with DMSO or 200 µM CK666. Dishes were imaged with the same microscope and objective as above, with the following changes: three fields were acquired per experiment with 15-min intervals for 3 h.
Supplementary Material
Acknowledgments
We thank the PDX Mouse Service at the Norris Cotton Cancer Center, Robert Vonderheide (University of Pennsylvania) for supplying the KPC4662 cell line, Tim Bruceos for forming connections, and Shivaprasad Sathyanarayana for help with perfusion fixation and vibratome sectioning. This work was supported by National Institutes of Health (NIH) R35 GM122545, a seed grant from the Hirshberg Foundation for Pancreatic Cancer Research, and a Prouty Pilot Project grant to H.N.H.; NIH R01 CA204228-01 to S.D.L.; a Steven B. Currier Clinical Oncology Scholar Award to K.D.S.; NCI Cancer Center Support Grant 5P30CA023108-37 to the Norris Cotton Cancer Center; and NIH P20 GM113132.
Abbreviations used:
- BP
band pass
- CK19
cytokeratin 19
- CSP
cell surface protrusion
- DHPC
Dartmouth-Hitchcock Pancreatic Cancer
- DIC
differential interference contrast
- FBS
fetal bovine serum
- FFPE
formalin-fixed, paraffin-embedded
- IF
intermediate filament
- IMT
intercellular membrane tubule
- LatA
latrunculin A
- Noc
nocodazole
- PBS
phosphate-buffered saline
- PDAC
pancreatic ductal adenocarcinoma
- TMT
tumor microtube
- TNT
tunneling nanotube
- WGA
wheat germ agglutinin.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-11-0605) on April 8, 2020.
REFERENCES
- Abounit S, Bousset L, Loria F, Zhu S, Chaumont F, Pieri L, Olivo-Marin J, Melki R, Zurzolo C. (2016). Tunneling nanotubes spread fibrillar α-synuclein by intercellular trafficking of lysosomes. EMBO J , 2120–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abounit S, Zurzolo C. (2012). Wiring through tunneling nanotubes - from electrical signals to organelle transfer. J Cell Sci , 1089–1098. [DOI] [PubMed] [Google Scholar]
- Ady JW, Desir S, Thayanithy V, Vogel RI, Moreira AL, Downey RJ, Fong Y, Manova-Todorova K, Moore MAS, Lou E. (2014). Intercellular communication in malignant pleural mesothelioma: properties of tunneling nanotubes. Front Physiol , 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariazi J, Benowitz A, De Biasi V, Den Boer M, Cherqui S, Cui H, Douillet N, Eugenin E, Favre D, Goodman S, et al. (2017). Tunneling nanotubes and gap junctions–their role in long-range intercellular communication during development, health, and disease conditions. Front Mol Neurosci , 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biran A, Perelmutter M, Gal H, Burton DGA, Ovadya Y, Vadai E, Geiger T, Krizhanovsky V. (2014). Senescent cells communicate via intercellular protein transfer. Genes Dev , 791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. (2014). Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev , 235–263. [DOI] [PubMed] [Google Scholar]
- Bovellan M, Romeo Y, Biro M, Boden A, Chugh P, Yonis A, Vaghela M, Fritzsche M, Moulding D, Thorogate R, et al. (2014). Cellular control of cortical actin nucleation. Curr Biol , 1628–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. (2010). A nucleator arms race: Cellular control of actin assembly. Nat Rev Mol Cell Biol , 237–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen D, Chen J, Li Z, Zhao J, Cai X. (2017). Prognostic significance of cytokeratin 19 expression in pancreatic neuroendocrine tumor: A meta-analysis. PLoS One , e0187588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery HR, Pearlman E, McMenamin PG. (2008). Cutting edge: membrane nanotubes in vivo: a feature of MHC class II + cells in the mouse cornea. J Immunol , 5779–5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh P, Clark AG, Smith MB, Cassani DAD, Dierkes K, Ragab A, Roux PP, Charras G, Salbreux G, Paluch EK. (2017). Actin cortex architecture regulates cell surface tension. Nat Cell Biol , 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BM, Rotty JD, Coulombe PA. (2013). Networking galore: Intermediate filaments and cell migration. Curr Opin Cell Biol , 600–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C, Denisin AK, Pruitt BL, Nelson WJ. (2017). Changes in E-cadherin rigidity sensing regulate cell adhesion. Proc Natl Acad Sci USA , E5835–E5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Abounit S, Marzo L, Danckaert A, Chamoun Z, Roux P, Zurzolo C. (2013). Transfer of polyglutamine aggregates in neuronal cells occurs in tunneling nanotubes. J Cell Sci , 3678–3685. [DOI] [PubMed] [Google Scholar]
- Coulombe PA, Omary MB. (2002). “Hard” and “soft” principles defining the structure, function and regulation of keratin intermediate filaments. Curr Opin Cell Biol , 3678–3685. [DOI] [PubMed] [Google Scholar]
- Delage E, Cervantes DC, Pénard E, Schmitt C, Syan S, Disanza A, Scita G, Zurzolo C. (2016). Differential identity of Filopodia and Tunneling Nanotubes revealed by the opposite functions of actin regulatory complexes. Sci Rep , 39632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desir S, O’Hare P, Vogel R, Sperduto W, Sarkari A, Dickson E, Wong P, Nelson A, Fong Y, Steer C, et al. (2018). Chemotherapy-induced tunneling nanotubes mediate intercellular drug efflux in pancreatic cancer. Sci Rep , 9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes HH, Rustom A, Wang X. (2013). Tunneling nanotubes, an emerging intercellular communication route in development. Mech Dev , 381–387. [DOI] [PubMed] [Google Scholar]
- Griessinger E, Moschoi R, Biondani G, Peyron JF. (2017). Mitochondrial Transfer in the Leukemia Microenvironment. Trends Cancer , 828–839. [DOI] [PubMed] [Google Scholar]
- Gurke S, Barroso JFV, Hodneland E, Bukoreshtliev NV, Schlicker O, Gerdes HH. (2008). Tunneling nanotube (TNT)-like structures facilitate a constitutive, actomyosin-dependent exchange of endocytic organelles between normal rat kidney cells. Exp Cell Res , 3669–3683. [DOI] [PubMed] [Google Scholar]
- Haimovich G, Ecker CM, Dunagin MC, Eggan E, Raj A, Gerst JE, Singer RH. (2017). Intercellular mRNA trafficking via membrane nanotube-like extensions in mammalian cells. Proc Natl Acad Sci USA , E9873–E9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Hu J, Yan Q, Zhu J, Zhu Z, Chen Y, Sun J, Zhang R. (2016). Bone marrow-derived mesenchymal stem cells rescue injured H9c2 cells via transferring intact mitochondria through tunneling nanotubes in an in vitro simulated ischemia/reperfusion model. Mol Med Rep , 1517–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Bhuyan F, Hiyoshi M, Noyori O, Nasser H, Miyazaki M, Saito T, Kondo Y, Osada H, Kimura S, et al. (2016). Potential Role of the Formation of Tunneling Nanotubes in HIV-1 Spread in Macrophages. J Immunol , 1832–1841. [DOI] [PubMed] [Google Scholar]
- Huang H, Liu S, Kornberg TB. (2019). Glutamate signaling at cytoneme synapses. Science , 948–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglicˇ A, Lokar M, Babnik B, Slivnik T, Veranicˇ P, Hägerstrand H, Kralj-Iglicˇ V. (2007). Possible role of flexible red blood cell membrane nanodomains in the growth and stability of membrane nanotubes. Blood Cells Mol Dis , 14–23. [DOI] [PubMed] [Google Scholar]
- Jain R, Fischer S, Serra S, Chetty R. (2010). The use of cytokeratin 19 (CK19) immunohistochemistry in lesions of the pancreas, gastrointestinal tract, and liver. Appl Immunohistochem Mol Morphol , 9–15. [DOI] [PubMed] [Google Scholar]
- Jansens RJJ, Van den Broeck W, De Pelsmaeker S, Lamote JAS, Van Waesberghe C, Couck L, Favoreel HW. (2017). Pseudorabies virus US3-induced tunneling nanotubes contain stabilized microtubules, interact with neighboring cells via cadherins, and allow intercellular molecular communication. J Virol , e00749-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantza V. (2011). Keratins in health and cancer: More than mere epithelial cell markers. Oncogene ,127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B-W, Lee J-S, Ko Y-G. (2019). Mycoplasma exploits mammalian tunneling nanotubes for cell-to-cell dissemination. BMB Rep , 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi M, Brandes RP, Haendeler J, Zeiher AM, Dimmeler S. (2005). Cell-to-cell connection of endothelial progenitor cells with cardiac myocytes by nanotubes: A novel mechanism for cell fate changes? Circ Res , 1039–1041. [DOI] [PubMed] [Google Scholar]
- Kretschmer A, Zhang F, Somasekharan S, Tse C, Leachman L, Gleave A, Li B, Asmaro I, Huang T, Kotula L, et al. (2019). Stress-induced tunneling nanotubes support treatment adaptation in prostate cancer. Sci Rep , 7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Kim JH, Ranjan P, Metcalfe MG, Cao W, Mishina M, Gangappa S, Guo Z, Boyden ES, Zaki S, et al. (2017). Influenza virus exploits tunneling nanotubes for cell-to-cell spread. Sci Rep , 40360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou E, Fujisawa S, Morozov A, Barlas A, Romin Y, Dogan Y, Gholami S, Moreira AL, Manova-Todorova K, Moore MAS. (2012). Tunneling nanotubes provide a unique conduit for intercellular transfer of cellular contents in human malignant pleural mesothelioma. PLoS One , e33093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu M, Martin-Jaular L, Lavieu G, Théry C. (2019). Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol , 9–17. [DOI] [PubMed] [Google Scholar]
- Önfelt B, Davis DM. (2004). Can membrane nanotubes facilitate communication between immune cells? Biochem Soc Trans , 676–678. [DOI] [PubMed] [Google Scholar]
- Önfelt B, Nedvetzki S, Benninger RKP, Purbhoo MA, Sowinski S, Hume AN, Seabra MC, Neil MAA, French PMW, Davis DM. (2006). Structurally distinct membrane nanotubes between human macrophages support long-distance vesicular traffic or surfing of bacteria. J Immunol , 8476–8483. [DOI] [PubMed] [Google Scholar]
- Osswald M, Jung E, Sahm F, Solecki G, Venkataramani V, Blaes J, Weil S, Horstmann H, Wiestler B, Syed M, et al. (2015). Brain tumour cells interconnect to a functional and resistant network. Nature , 93–98. [DOI] [PubMed] [Google Scholar]
- Panasiuk M, Rychłowski M, Derewon´ko N, Bien´kowska-Szewczyk K. (2018). Tunneling nanotubes as a novel route of cell-to-cell spread of herpesviruses. J Virol . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak R, De Rooij B, Pieters R, Den Boer ML. (2015). B-cell precursor acute lymphoblastic leukemia cells use tunneling nanotubes to orchestrate their microenvironment. Blood , 2404–2414. [DOI] [PubMed] [Google Scholar]
- Ramírez-Weber FA, Kornberg TB. (1999). Cytonemes: Cellular processes that project to the principal signaling center in Drosophila imaginal discs. Cell , 599–607. [DOI] [PubMed] [Google Scholar]
- Resnik N, Prezelj T, De Luca GMR, Manders E, Polishchuk R, Veranicˇ P, Kreft ME. (2018). Helical organization of microtubules occurs in a minority of tunneling membrane nanotubes in normal and cancer urothelial cells. Sci Rep , 17133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro-Rodrigues TM, Laundos TL, Pereira-Carvalho R, Batista-Almeida D, Pereira R, Coelho-Santos V, Silva AP, Fernandes R, Zuzarte M, Enguita FJ, et al. (2017). Exosomes secreted by cardiomyocytes subjected to ischaemia promote cardiac angiogenesis. Cardiovasc Res , 1338–1350. [DOI] [PubMed] [Google Scholar]
- Roy S, Hsiung F, Kornberg TB. (2011). Specificity of Drosophila cytonemes for distinct signaling pathways. Science , 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Huang H, Liu S, Kornberg TB. (2014). Cytoneme-mediated contact-dependent transport of the Drosophila decapentaplegic signaling protein. Science , 1244624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustom A, Saffrich R, Markovic I, Walther P, Gerdes HH. (2004). Nanotubular highways for intercellular organelle transport. Science , 1007–1010. [DOI] [PubMed] [Google Scholar]
- Sáenz-de-Santa-María I, Bernardo-Castiñeira C, Enciso E, García-Moreno I, Chiara JL, Suarez C, Chiara MD. (2017). Control of long-distance cell-to-cell communication and autophagosome transfer in squamous cell carcinoma via tunneling nanotubes. Oncotarget , 20939–20960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori-Rupp A, Cordero Cervantes D, Pepe A, Gousset K, Delage E, Corroyer-Dulmont S, Schmitt C, Krijnse-Locker J, Zurzolo C. (2019). Correlative cryo-electron microscopy reveals the structure of TNTs in neuronal cells. Nat Commun , 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowinski S, Jolly C, Berninghausen O, Purbhoo MA, Chauveau A, Köhler K, Oddos S, Eissmann P, Brodsky FM, Hopkins C, et al. (2008). Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat Cell Biol , 211–219. [DOI] [PubMed] [Google Scholar]
- Takahashi A, Kukita A, Li YJ, Zhang JQ, Nomiyama H, Yamaza T, Ayukawa Y, Koyano K, Kukita T. (2013). Tunneling nanotube formation is essential for the regulation of osteoclastogenesis. J Cell Biochem , 1238–1247. [DOI] [PubMed] [Google Scholar]
- Thayanithy V, Dickson EL, Steer C, Subramanian S, Lou E. (2014). Tumor-stromal cross talk: Direct cell-to-cell transfer of oncogenic microRNAs via tunneling nanotubes. Transl Res , 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhaneni KC, Haller H, Dumler I. (2012). Vascular smooth muscle cells initiate proliferation of mesenchymal stem cells by mitochondrial transfer via tunneling nanotubes. Stem Cells Dev , 3104–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veranicˇ P, Lokar M, Schütz GJ, Weghuber J, Wieser S, Hägerstrand H, Kralj-Iglicˇ V, Iglicˇ A. (2008). Different types of cell-to-cell connections mediated by nanotubular structures. Biophys J , 4416–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidulescu C, Clejan S, O’Connor KC. (2004). Vesicle traffic through intercellular bridges in DU 145 human prostate cancer cells. J Cell Mol Med , 388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Veruki ML, Bukoreshtliev NV, Hartveit E, Gerdes HH. (2010). Animal cells connected by nanotubes can be electrically coupled through interposed gap-junction channels. Proc Natl Acad Sci USA , 17194–17199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins SC, Salter RD. (2005). Functional connectivity between immune cells mediated by tunneling nanotubules. Immunity , 309–318. [DOI] [PubMed] [Google Scholar]
- Weil S, Osswald M, Solecki G, Grosch J, Jung E, Lemke D, Ratliff M, Hänggi D, Wick W, Winkler F. (2017). Tumor microtubes convey resistance to surgical lesions and chemotherapy in gliomas. Neuro Oncol , 1316–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winograd R, Byrne KT, Evans RA, Odorizzi PM, Meyer AR, Bajor DL, Clendenin C, Stanger BZ, Furth EE, Wherry EJ, et al. (2015). Induction of T-cell immunity overcomes complete resistance to PD-1 and CTLA-4 blockade and improves survival in pancreatic carcinoma. Cancer Immunol Res , 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig D, Wang X, Walter C, Gerdes HH, Funk RHW, Roehlecke C. (2012). Multi-level communication of human retinal pigment epithelial cells via tunneling nanotubes. PLoS One , e33195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhang J, Zhao L, Xin Y, Liu S, Cui W. (2019). Differential roles of microtubules in the two formation stages of membrane nanotubes between human mesenchymal stem cells and neonatal mouse cardiomyocytes. Biochem Biophys Res Commun , 441–447. [DOI] [PubMed] [Google Scholar]
- Zhu S, Victoria GS, Marzo L, Ghosh R, Zurzolo C. (2015). Prion aggregates transfer through tunneling nanotubes in endocytic vesicles. Prion , 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.