Abstract
Many different enzymes in intermediate metabolism dynamically assemble filamentous polymers in cells, often in response to changes in physiological conditions. Most of the enzyme filaments known to date have only been observed in cells, but in a handful of cases structural and biochemical studies have revealed the mechanisms and consequences of assembly. In general, enzyme polymerization functions as a mechanism to allosterically tune enzyme kinetics, and it may play a physiological role in integrating metabolic signaling. Here, we highlight some principles of metabolic filaments by focusing on two well-studied examples in nucleotide biosynthesis pathways—inosine-5’-monophosphate (IMP) dehydrogenase and cytosine triphosphate (CTP) synthase.
INTRODUCTION
Classically, intermediate metabolism has been viewed as a kind of soup of enzymes and substrates, partially organized by sequestration into membrane-bound compartments and limited primarily by diffusion within those compartments. More recently, however, a significant level of physical organization of intermediate metabolism has been uncovered, where dynamic reorganization of enzymes into discreet cellular structures is linked to changes in metabolic conditions. These structures include multienzyme aggregates that colocalize different enzymes in a pathway, and filamentous polymers that form by self-assembly of a single enzyme (Chitrakar et al., 2017; Jin et al., 2017).
Several dozen different metabolic enzymes are now known to form filamentous polymers in cells (for a comprehensive recent review, see Park and Horton, 2019). Among the best described of these metabolic filaments are IMP dehydrogenase (IMPDH) and CTP synthase (CTPS), key regulatory enzymes in the purine and pyrimidine nucleotide biosynthesis pathways, respectively. Here, we focus on these two examples to highlight common aspects of metabolic filament structure, evolution, regulation, and molecular mechanisms.
Evolution of assembly
How metabolic enzyme polymerization arose during evolution is not well understood. However, theoretical and experimental models suggest that single amino acid alterations can be sufficient to drive otherwise diffuse enzymes to assemble (Garcia-Seisdedos et al., 2017). This may be especially true for metabolic enzymes, which are frequently homo-oligomeric proteins, and therefore the effect of individual mutations can be amplified by their presence in each subunit (Figure 1). Indeed, many proteins, and particularly metabolic enzymes, have been found to form large assemblies in proteomic and imaging-based screens in yeast (Noree et al., 2010, 2019; O’Connell et al., 2014; Shen et al., 2016).
FIGURE 1:
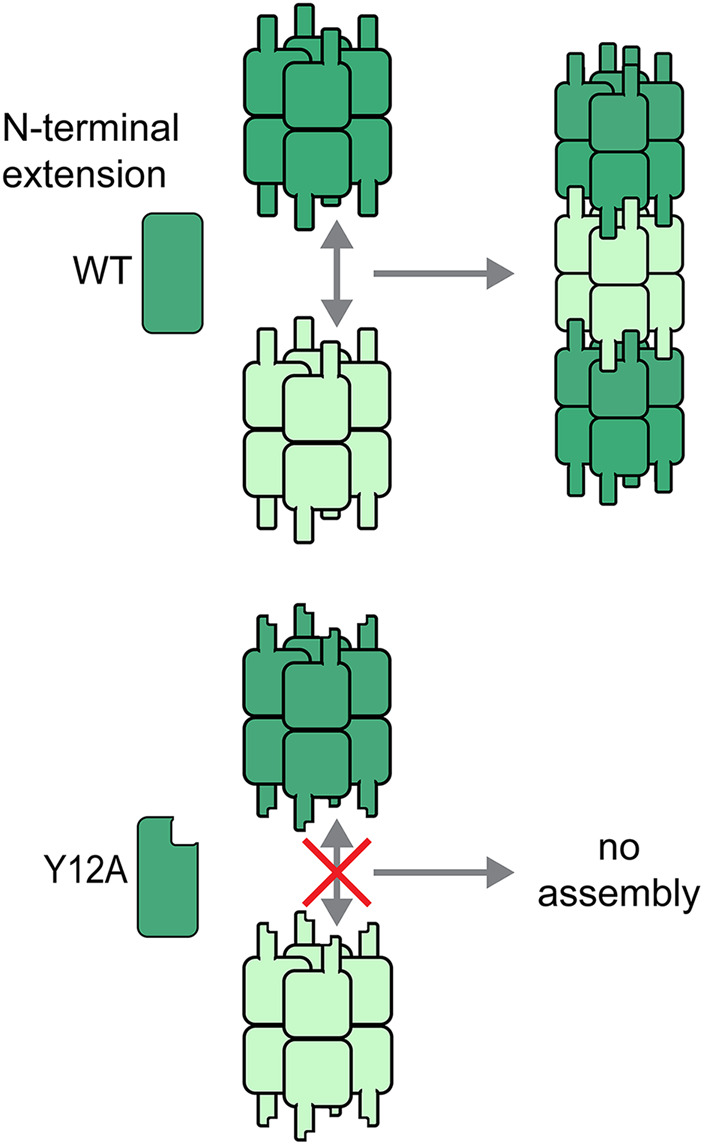
A single amino acid change in the IMPDH N-terminal extension disrupts assembly of IMPDH octamers. Intermolecular interactions between IMPDH protomers are amplified eghtfold through reciprocal interactions across the polymerization interface between octamers. The avidity effects of such multivalent interactions may facilitate evolution of polymers of oligomeric proteins. Conversely, mutation of just one amino acid, tyrosine 12 to alanine, in the N-terminal extension, completely prevents octamer polymerization.
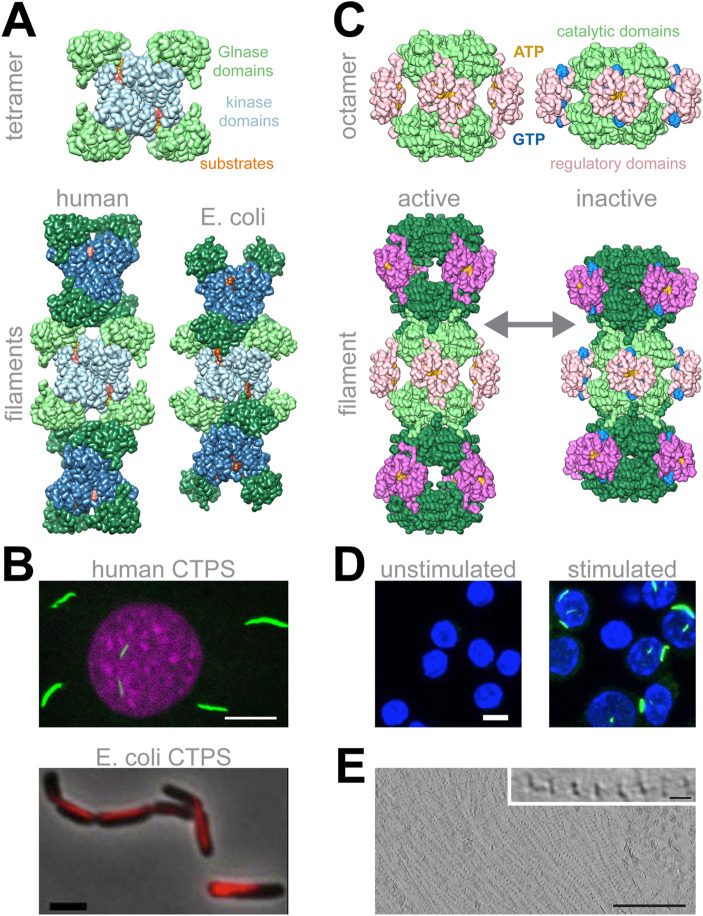
The evolution of polymerization in CTPS and IMPDH each followed a unique trajectory, but illustrate principals that may be relevant to other polymerizing enzymes. Eukaryotic and prokaryotic CTPS form a conserved tetrameric structure, but adopt very different filamentous forms mediated by different assembly contacts (Figure 2, A and B). In bacteria, filaments form from interlocking tetramers, while a unique 10 amino acid insertion mediates stacking of eukaryotic CTPS tetramers into filaments (Barry et al., 2014; Lynch et al., 2017; Lynch and Kollman, 2020). The insertion can be found in some protist and plant CTPS enzymes, suggesting that this form of CTPS polymer may have evolved ∼1.5 billion years ago. The evolution of this insert may have driven a transition in CTPS polymer morphology, with the corresponding functional consequences outlined below. Alternatively, eukaryotic and prokaryotic polymer forms may have evolved independently from a nonpolymerizing ancestor, which again would be consistent with the idea that polymerization of homo-oligomeric proteins can be readily achieved with a small number of mutations.
FIGURE 2:
Structures of CTPS and IMPDH filaments. (A) CTPS structures. CTPS monomers each have two catalytic domains (blue and green), and assemble into tetramers. Tetramers can dynamically assemble filaments with different geometries. A unique eukaryotic insert mediates assembly contacts of human CTPS, while Escherichia coli CTPS tetramers form more extensive, interlocking interactions. (B) Cellular polymerization of CTPS is broadly conserved (adapted from Barry et al. 2014; Gou et al., 2014). (C) IMPDH structures. IMPDH is an octamer, with each protomer consisting of catalytic (green) and regulatory (pink) domains. ATP binding in the regulatory domains stabilizes octamers, and GTP binding promotes a conformational change in that, in the context of the octamer and in the filament, results in a compressed, inactive conformation. (D) IMPDH polymers are observed to form in mouse lymphocytes upon TCR stimulation (adapted from Duong-Ly et al. 2018). (E) Cryo-tomography of HEp-2 cells shows IMPDH forming extensive bundles with spacing consistent with the spacing observed in in vitro reconstituted single filaments (adapted from Juda et al. 2014). Scale bars: B (top) = 10 μm, B (bottom) = 3 μm, D = 5 μm, E = 200 nm, inset in E = 10 nm.
Polymerization of IMPDH appears to have evolved more recently than for CTPS. An N-terminal sequence extension, conserved in most metazoan IMPDH enzymes, plays a key role in mediating polymerization of octamers by a mechanism related to “runaway domain coupling” (Figure 2, C and D; McPartland et al., 2018). Recent high-resolution cryo-EM structures have shown that the flexible N-terminus extends from the enzyme core to make ordered interactions across an assembly interface, and a single alanine substitution mutation in tyrosine 12 prevents the formation of IMPDH filaments (Anthony et al., 2017; Fernández-Justel et al., 2019; Johnson and Kollman, 2020). At each octamer–octamer interface, this reciprocal interaction is seen in all eight adjacent monomers, dramatically increasing the valency and buried surface area of the interaction, potentially explaining the key role of this residue. IMPDH filaments have thus far only been demonstrated in mammalian cells and despite extensive analysis are not observed in Drosophila. Consistent with this, tyrosine 12 and the surrounding amino acid sequence are not conserved in Diptera but are present in some species, such as Trichoplax adhaerens, that are more distantly related to mammals. This suggests that IMPDH assembly may have evolved during the development of multicellularity but has been lost in certain animal lineages. Consistent with their unique evolutionary trajectories, different regulatory mechanisms modulate the assembly of CTPS and IMPDH, as described next.
Regulation of assembly
In metazoans, assembly of both IMPDH and CTPS is associated with cellular conditions in which nucleotide demand exceeds supply. For example, in Drosophila ovary or salivary gland, CTPS assembly correlates with developmental periods in which cells undergo endoreplication (Liu, 2010), a process that dramatically increases nucleotide demand. CTPS filaments have also been observed in rapidly dividing cancer cells in culture (Chang et al., 2017), and mammalian T-cells transiently assemble IMPDH into filaments in response to immunological activation before cell proliferation (Calise et al., 2018; Duong-Ly et al., 2018). It remains to be seen whether assembly in these contexts is simply a response to changes in nucleotide abundance relative to demand or whether it is part of a programmed change in cell state.
Filament assembly is also observed when nucleotide supply is limited, such as in cells treated with pharmacological inhibitors of nucleotide biosynthetic enzymes (reviewed in Schiavon et al., 2018). Conversely, increasing nucleotide levels by providing them exogenously triggers filament dispersion (Gunter et al., 2008; Calise et al., 2014, 2016; Duong-Ly et al., 2018). These findings are consistent with emerging models for assembly as a means of regulating nucleotide homeostasis (Barry et al., 2014; Gou et al., 2014; Noree et al., 2014; Calise et al., 2016; Anthony et al., 2017; Lynch et al., 2017; Fernández-Justel et al., 2019).
Elevating IMPDH or CTPS protein levels in cells also promotes their assembly (Ingerson-Mahar et al., 2010; Keppeke et al., 2018; Li et al., 2018; Wu and Liu, 2019). IMPDH protein is dramatically up-regulated in primary T-cells following T-cell receptor (TCR) activation and is associated with IMPDH assembly. However, increased IMPDH expression is not sufficient for assembly, which also depends on signaling by NFAT and mTOR (Duong-Ly et al., 2018). mTOR has also been implicated in CTPS filament formation in Drosophila (Aughey et al., 2014; Sun and Liu, 2019). In Drosophila ovaries, CTPS filament assembly is regulated by Myc and activated cdc42-associated kinase (Ack; Strochlic et al., 2014; Aughey et al., 2016). It remains unknown how these pathways impact assembly but posttranslational modifications of the enzymes themselves could be involved. CTPS assembly in the Drosophila ovary is regulated by ubiquitination associated with the Cbl ubiquitin ligase (Wang et al., 2015). In cultured cells under nutrient stress, CTPS filament assembly is regulated by methylation controlled by histidine and the folate cycle (Lin et al., 2018). However, as of yet there does not appear to be a clear connection between these posttranslational modifications and known signaling pathways associated with filament assembly.
In addition to regulation by signaling pathways, small molecules including substrates, products, and allosteric regulators can influence metabolic enzyme polymerization. ATP and GTP both allosterically regulate filament assembly and activity of IMPDH (Anthony et al., 2017; Buey et al., 2017; Fernández-Justel et al., 2019) and GTP allosterically regulates CTPS activity (Habrian et al., 2016). Likewise, the presence or absence of their substrates strongly influences CTPS and IMPDH polymer assembly in vitro (Anthony et al., 2017; Lynch et al., 2017). Interestingly, in cultured cells, CTPS and IMPDH can assemble into distinct structures or can assemble together, depending on the method of induction (Carcamo et al., 2011; Chang et al., 2015, 2018). This suggests the intriguing possibility that coassembly could mediate coordination between purine and pyrimidine biosynthetic pathways. Consistent with this, pharmacological inhibitors of CTPS promote the assembly of IMPDH filaments (Carcamo et al., 2011; Chang et al., 2015, 2018).
These examples highlight the complexity of regulation of metabolic enzyme assembly. Future work must begin to connect these different levels of regulation to form a complete picture of how cells sense their metabolic state and initiate enzyme polymerization.
Assembly is an allosteric regulator
Although there are only a handful of examples of studies on the biochemical effects of polymerization, it appears that in most cases filament assembly directly tunes enzyme kinetics. Assembly-based allosteric tuning can take very different forms, including decreasing or increasing specific activity, altering the response to other allosteric regulators, or enhancing the cooperativity with which a population of enzymes transitions between activity states (Figure 3).
FIGURE 3:
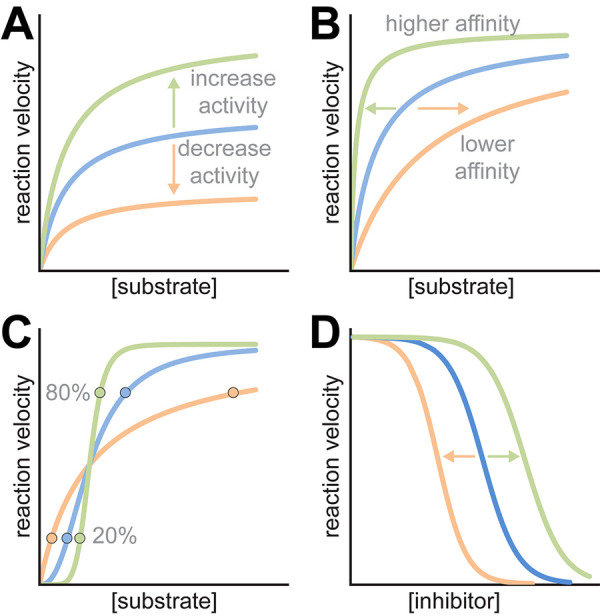
Potential effects of polymerization on enzyme kinetics. Hypothetical activity of free enzymes (blue) is compared with possible activity in filaments (green and orange). (A) Filaments can affect the catalytic rate constant, either increasing or decreasing activity across all concentrations of substrate. (B) Filaments might affect affinity for substrate (the apparent Km), resulting in different activity levels at low substrate concentrations. (C) Filaments can influence the cooperativity of reactions. Circles show the 20 and 80% activity levels, highlighting the steepening of the activity curve with increasing cooperativity. (D) Filaments can shift the sensitivity to allosteric effectors, here illustrated as an inhibitor.
Tetrameric CTPS has a conserved conformational equilibrium, with cooperative transitions between active and inactive conformations resulting from relative motions of the two catalytic domains in each protomer (Lynch et al., 2017). The inactive conformation is promoted by CTP binding, in a form of feedback inhibition. Filaments provide an additional layer of allosteric control on top of this conformational equilibrium, although these effects vary between domains of life. For example, in bacteria, filament assembly interactions sterically lock the CTPS tetramer into the inactive conformation (Figure 2A; Barry et al., 2014; Lynch et al., 2017). This suggests that for bacterial CTPS, polymerization is a mechanism to maintain homeostasis by rapidly shutting down activity in response to elevated CTP levels. A similar mechanism was recently described for filaments of budding yeast glucokinase 1, suggesting that polymerization can act as a “molecular surge protector” to prevent deleterious metabolic effects of excess flux through the initial phase of glycolysis (Stoddard et al., 2020).
Humans have two CTPS isoforms, CTPS1 and CTPS2, that both form filaments with interaction interfaces that are completely different from the bacterial interface (Figure 2A). The biochemical consequences of polymerization vary between the two human isoforms. CTPS1 is much more active in filaments, and disassembles upon inactivation (Lynch et al., 2017). CTPS2, thought to be the more “housekeeping” isoform, is able to transition between active and inactive conformations while remaining assembled; in this case, coupling conformational changes between many CTPS tetramers increases cooperativity of the structural transition between active and inactive states (Lynch and Kollman, 2020). These different regulatory outcomes likely reflect the different cellular roles of the two isoforms: shifting CTPS1 into polymers would increase flux through the enzyme to produce the higher nucleotide levels required during proliferation, while enhanced cooperativity in CTPS2 filaments provides a mechanism for ultrasensitive, switch-like control of CTP synthesis during more homeostatic conditions. Like CTPS1, other metabolic enzymes appear to be stabilized in active conformations in filaments (Webb et al., 2017; Kim et al., 2019). And like CTPS2, at least one other enzyme filament, acetyl-CoA carboxylase, can form filaments in active or inactive states depending on what allosteric ligands or regulatory partners are bound (Hunkeler et al., 2018).
Like CTPS2, IMPDH filaments can accommodate active or inactive conformations (Anthony et al., 2017), although in this case assembly does not have a direct effect on specific activity and does not enhance cooperativity (Figure 2C). Instead, IMPDH filament assembly is sensitive to the levels of both substrate and downstream product (GTP) pools. In the presence of substrate, filament assembly interactions stabilize an enzyme conformation that resists allosteric inhibition by the downstream product GTP (Johnson and Kollman, 2020). This allows IMPDH to retain activity in the presence of otherwise inhibitory GTP concentrations, consistent with the high nucleotide demands where IMPDH assembly is observed, such as lymphocyte activation (Calise et al., 2018; Duong-Ly et al., 2018).
IMPDH filaments have also been shown to form large, lateral bundles in cells; these bundles have a periodicity consistent with the spacing between octamers in in vitro reconstituted single filaments (Figure 2E; Juda et al., 2014). Given the large size of most cellular metabolic filaments, we anticipate that this kind of lateral bundling of single filaments may be a common form of higher order association, although the functional importance of bundles has not yet been explored for any enzyme filament.
Why filaments?
It may seem surprising that filament assembly has evolved as a general mechanism for tuning enzyme kinetics; after all, there are limitless examples in metabolism of allosteric control being achieved through smaller, defined oligomers. One simple explanation may be that polymer assembly is relatively easy to evolve with a small number of mutations, as suggested above (Garcia-Seisdedos et al., 2017). By this logic, filament assembly is a straightforward way to introduce an additional layer of regulation on top of existing allostery.
Nevertheless, it seems peculiarly baroque to build micron-scale cellular structures simply to regulate the activity of a single enzyme. One intriguing possibility is that large, ordered, multivalent structures like metabolic filaments may serve as platforms for signaling the state of cellular metabolism. There are hints from cell biological studies that this might be the case. For example, coassembly of IMPDH and CTPS filaments suggests the possibility that biosynthetic activity of these two separate but interrelated pathways may be coregulated through physical interaction, whether direct or mediated by an as yet unidentified scaffold. Even more intriguing, other signaling molecules colocalize with these filaments (Strochlic et al., 2014; Hayward et al., 2019), supporting the idea the filaments may be coordinating multiple activities in response to changing metabolic conditions.
Outlook
While many enzyme filaments have been observed in cells, only a few have been characterized at the level of structure and biochemical function. Given the diversity of structural and regulatory outcomes of the few cases where these details are known, it seems likely that other novel mechanisms remain to be discovered.
One pressing challenge is to uncover the physiological role of the various metabolic filaments. In vitro studies of filament structures, assembly mechanisms, and biochemical regulation are providing context and tools for studying the role of metabolic filaments in cells and tissues. Understanding how filaments specifically alter metabolite levels and/or flux through the associated pathways under varying physiological conditions is an important next step in understanding their biological function. Identifying whether other cellular factors specifically recognize and interact with metabolic filaments is another important step in understanding how they communicate with other cellular components.
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01GM-118396 to J.M.K., R01GM-083025 to J.R.P., F31EY-030732 to A.L.B., and J.C.S. was supported by T32 CA-009035).
Abbreviations used:
- ATP
adenosine triphosphate
- CTP
cytosine triphosphate
- CTPS
CTP synthase
- GTP
guanosine triphosphate
- IMP
inosine monophosphate
- IMPDH
IMP dehydrogenase
- WT
wild type
Footnotes
REFERENCES
- Anthony SA, Burrell AL, Johnson MC, Duong-Ly KC, Kuo YM, Simonet JC, Michener P, Andrews A, Kollman JM, Peterson JR. (2017). Reconstituted IMPDH polymers accommodate both catalytically active and inactive conformations. Mol Biol Cell, 10.1091/mbc.E17-04-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aughey GN, Grice SJ, Liu JL. (2016). The interplay between Myc and CTP synthase in Drosophila. PLoS Genet , e1005867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aughey GN, Grice SJ, Shen QJ, Xu Y, Chang CC, Azzam G, Wang PY, Freeman-Mills L, Pai LM, Sung LY, et al. (2014). Nucleotide synthesis is regulated by cytoophidium formation during neurodevelopment and adaptive metabolism. Biol Open , 1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry RM, Bitbol AF, Lorestani A, Charles EJ, Habrian CH, Hansen JM, Li HJ, Baldwin EP, Wingreen NS, Kollman JM, et al. (2014). Large-scale filament formation inhibits the activity of CTP synthetase. Elife , e03638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buey RM, Fernández-Justel D, Marcos-Alcalde I, Winter G, Gómez-Puertas P, de Pereda JM, Luis Revuelta J. (2017). A nucleotide-controlled conformational switch modulates the activity of eukaryotic IMP dehydrogenases. Sci Rep , 2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calise SJ, Abboud G, Kasahara H, Morel L, Chan EKL. (2018). Immune response-dependent assembly of IMP dehydrogenase filaments. Front Immunol , 2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calise SJ, Carcamo WC, Krueger C, Yin JD, Purich DL, Chan EK. (2014). Glutamine deprivation initiates reversible assembly of mammalian rods and rings. Cell Mol Life Sci , 2963–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calise SJ, Purich DL, Nguyen T, Saleem DA, Krueger C, Yin JD, Chan EK. (2016). ‘Rod and ring’ formation from IMP dehydrogenase is regulated through the one-carbon metabolic pathway. J Cell Sci , 3042–3052. [DOI] [PubMed] [Google Scholar]
- Carcamo WC, Satoh M, Kasahara H, Terada N, Hamazaki T, Chan JY, Yao B, Tamayo S, Covini G, von Muhlen CA, et al. (2011). Induction of cytoplasmic rods and rings structures by inhibition of the CTP and GTP synthetic pathway in mammalian cells. PLoS One , e29690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Jeng YM, Peng M, Keppeke GD, Sung LY, Liu JL. (2017). CTP synthase forms the cytoophidium in human hepatocellular carcinoma. Exp Cell Res , 292–299. [DOI] [PubMed] [Google Scholar]
- Chang CC, Keppeke GD, Sung LY, Liu JL. (2018). Interfilament interaction between IMPDH and CTPS cytoophidia. FEBS J , 3753–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Lin WC, Pai LM, Lee HS, Wu SC, Ding ST, Liu JL, Sung LY. (2015). Cytoophidium assembly reflects upregulation of IMPDH activity. J Cell Sci , 3550–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitrakar I, Kim-Holzapfel DM, Zhou W, French JB. (2017). Higher order structures in purine and pyrimidine metabolism. J Struct Biol , 354–364. [DOI] [PubMed] [Google Scholar]
- Duong-Ly KC, Kuo YM, Johnson MC, Kollman JM, Soboloff J, Rall GF, Andrews AJ, Peterson JR. (2018). T cell activation triggers reversible IMPDH filament assembly. J Cell Sci , jcs223289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Justel D, Núñez R, Martin-Benito J, Jimeno D, González-López A, Soriano EM, Revuelta JL, Buey RM. (2019). A nucleotide-dependent conformational switch controls the polymerization of human IMP dehydrogenases to modulate their catalytic activity. J Mol Biol , 956–969. [DOI] [PubMed] [Google Scholar]
- Garcia-Seisdedos H, Empereur-Mot C, Elad N, Levy ED. (2017). Proteins evolve on the edge of supramolecular self-assembly. Nature , 244–247. [DOI] [PubMed] [Google Scholar]
- Gou KM, Chang CC, Shen QJ, Sung LY, Liu JL. (2014). CTP synthase forms cytoophidia in the cytoplasm and nucleus. Exp Cell Res , 242–253. [DOI] [PubMed] [Google Scholar]
- Gunter JH, Thomas EC, Lengefeld N, Kruger SJ, Worton L, Gardiner EM, Jones A, Barnett NL, Whitehead JP. (2008). Characterisation of inosine monophosphate dehydrogenase expression during retinal development: differences between variants and isoforms. Int J Biochem Cell Biol , 1716–1728. [DOI] [PubMed] [Google Scholar]
- Habrian C, Chandrasekhara A, Shahrvini B, Hua B, Lee J, Jesinghaus R, Barry R, Gitai Z, Kollman J, Baldwin EP. (2016). Inhibition of Escherichia coli CTP synthetase by NADH and other nicotinamides and their mutual interactions with CTP and GTP. Biochemistry , 5554–5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward D, Kouznetsova VL, Pierson HE, Hasan NM, Guzman ER, Tsigelny IF, Lutsenko S. (2019). ANKRD9 is a metabolically-controlled regulator of IMPDH2 abundance and macro-assembly. J Biol Chem , 14454–14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunkeler M, Hagmann A, Stuttfeld E, Chami M, Guri Y, Stahlberg H, Maier T. (2018). Structural basis for regulation of human acetyl-CoA carboxylase. Nature , 470–474. [DOI] [PubMed] [Google Scholar]
- Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. (2010). The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat Cell Biol , 739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Fuller GG, Han T, Yao Y, Alessi AF, Freeberg MA, Roach NP, Moresco JJ, Karnovsky A, Baba M, et al. (2017). Glycolytic enzymes coalesce in G bodies under hypoxic stress. Cell Rep , 895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MC, Kollman JM. (2020). Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation. Elife , e53243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juda P, Smigova J, Kovacik L, Bartova E, Raska I. (2014). Ultrastructure of cytoplasmic and nuclear inosine-5′-monophosphate dehydrogenase 2 “rods and rings” inclusions. J Histochem Cytochem , 739–750. [DOI] [PubMed] [Google Scholar]
- Keppeke GD, Chang CC, Peng M, Chen LY, Lin WC, Pai LM, Andrade LEC, Sung LY, Liu JL. (2018). IMP/GTP balance modulates cytoophidium assembly and IMPDH activity. Cell Div , 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G, Azmi L, Jang S, Jung T, Hebert H, Roe AJ, Byron O, Song JJ. (2019). Aldehyde-alcohol dehydrogenase forms a high-order spirosome architecture critical for its activity. Nat Commun , 4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Xie J, Hei M, Tang J, Wang Y, Forster E, Zhao S. (2018). High level of CTP synthase induces formation of cytoophidia in cortical neurons and impairs corticogenesis. Histochem Cell Biol , 61–73. [DOI] [PubMed] [Google Scholar]
- Lin WC, Chakraborty A, Huang SC, Wang PY, Hsieh YJ, Chien KY, Lee YH, Chang CC, Tang HY, Lin YT, et al. (2018). Histidine-dependent protein methylation is required for compartmentalization of CTP synthase. Cell Rep , 2733–2745.e2737. [DOI] [PubMed] [Google Scholar]
- Liu JL. (2010). Intracellular compartmentation of CTP synthase in Drosophila. J Genet Genomics , 281–296. [DOI] [PubMed] [Google Scholar]
- Lynch EM, Hicks DR, Shepherd M, Endrizzi JA, Maker A, Hansen JM, Barry RM, Gitai Z, Baldwin EP, Kollman JM. (2017). Human CTP synthase filament structure reveals the active enzyme conformation. Nat Struct Mol Biol , 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch EM, Kollman JM. (2020). Coupled structural transitions enable highly cooperative regulation of human CTPS2 filaments. Nat Struct Mol Biol , 32–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPartland L, Heller DM, Eisenberg DS, Hochschild A, Sawaya MR. (2018). Atomic insights into the genesis of cellular filaments by globular proteins. Nat Struct Mol Biol , 705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noree C, Begovich K, Samilo D, Broyer R, Monfort E, Wilhelm JE. (2019). A quantitative screen for metabolic enzyme structures reveals patterns of assembly across the yeast metabolic network. Mol Biol Cell , 2721–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noree C, Monfort E, Shiau AK, Wilhelm JE. (2014). Common regulatory control of CTP synthase enzyme activity and filament formation. Mol Biol Cell , 2282–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noree C, Sato BK, Broyer RM, Wilhelm JE. (2010). Identification of novel filament-forming proteins in Saccharomycescerevisiae and Drosophila melanogaster. J Cell Biol , 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JD, Tsechansky M, Royall A, Boutz DR, Ellington AD, Marcotte EM. (2014). A proteomic survey of widespread protein aggregation in yeast. Mol Biosyst , 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CK, Horton NC. (2019). Structures, functions, and mechanisms of filament forming enzymes: a renaissance of enzyme filamentation. Biophys Rev , 927–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavon CR, Griffin ME, Pirozzi M, Parashuraman R, Zhou W, Jinnah HA, Reines D, Kahn RA. (2018). Compositional complexity of rods and rings. Mol Biol Cell , 2303–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QJ, Kassim H, Huang Y, Li H, Zhang J, Li G, Wang PY, Yan J, Ye F, Liu JL. (2016). Filamentation of metabolic enzymes in Saccharomycescerevisiae. J Genet Genomics , 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard PR, Lynch EM, Farrell DP, Dosey AM, DiMaio F, Williams TA, Kollman JM, Murray AW, Garner EC. (2020). Polymerization in the actin ATPase clan regulates hexokinase activity in yeast. Science , 1039–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strochlic TI, Stavrides KP, Thomas SV, Nicolas E, O’Reilly AM, Peterson JR. (2014). Ack kinase regulates CTP synthase filaments during Drosophila oogenesis. EMBO Rep , 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Liu JL. (2019). mTOR-S6K1 pathway mediates cytoophidium assembly. J Genet Genom , 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PY, Lin WC, Tsai YC, Cheng ML, Lin YH, Tseng SH, Chakraborty A, Pai LM. (2015). Regulation of CTP synthase filament formation during DNA endoreplication in Drosophila. Genetics , 1511–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb BA, Dosey AM, Wittmann T, Kollman JM, Barber DL. (2017). The glycolytic enzyme phosphofructokinase-1 assembles into filaments. J Cell Biol , 2305–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Liu JL. (2019). Cytoophidia respond to nutrient stress in Drosophila. Exp Cell Res , 159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]