Abstract
The multi-component cytoplasmic dynein transports cellular cargoes with the help of another multi-component complex dynactin, but we do not know enough about factors that may affect the assembly and functions of these proteins. From a genetic screen for mutations affecting early-endosome distribution in Aspergillus nidulans, we identified the prp40AL438* mutation in Prp40A, a homologue of Prp40, an essential RNA-splicing factor in the budding yeast. Prp40A is not essential for splicing, although it associates with the nuclear splicing machinery. The prp40AL438* mutant is much healthier than the ∆prp40A mutant, but both mutants exhibit similar defects in dynein-mediated early-endosome transport and nuclear distribution. In the prp40AL438* mutant, the frequency but not the speed of dynein-mediated early-endosome transport is decreased, which correlates with a decrease in the microtubule plus-end accumulations of dynein and dynactin. Within the dynactin complex, the actin-related protein Arp1 forms a mini-filament. In a pull-down assay, the amount of Arp1 pulled down with its pointed-end protein Arp11 is lowered in the prp40AL438* mutant. In addition, we found from published interactome data that a mammalian Prp40 homologue PRPF40A interacts with Arp1. Thus, Prp40 homologues may regulate the assembly or function of dynein–dynactin and their mechanisms deserve to be further studied.
INTRODUCTION
Genes in eukaryotic cells contain introns, which are transcribed into precursor mRNA (pre-mRNA) and later removed by the RNA-splicing machinery, the spliceosomes (Wahl et al., 2009; Shi, 2017). The spliceosomal complexes contain both RNA and proteins. Specifically, five uridine-rich small nuclear RNAs (snRNAs), U1, U2, U4, U5, and U6, together with various proteins, assemble into large ribonucleoprotein (snRNP) complexes that function at various stages of splicing (Wahl et al., 2009; Shi, 2017). Among the many proteins associated with snRNPs, Prp40 (pre-mRNA-processing protein 40) was initially identified in the budding yeast Saccharomyces cerevisiae as a splicing factor and is essential for cell viability (Kao and Siliciano, 1996). Prp40 has been implicated in the early stages of spliceosome assembly when it associates with the U1 snRNP and is involved in the recognition of the 5′ splicing site (Becerra et al., 2016). In mammalian cells, there are two homologues of Prp40, PRPF40A and PRPF40B: PRPF40A is also known as HYPA (Huntingtin-interacting protein A) or FBP11 (formin-binding protein 11), and PRPF40B is also known as HYPC (Huntingtin-interacting protein C) or FBP21 (formin-binding protein 21) (Chan et al., 1996; Bedford et al., 1998; Faber et al., 1998; Huang et al., 2009). In general, these Prp40 homologues play roles in splicing but may also play nonsplicing-related roles (Diaz Casas et al., 2017; Lorenzini et al., 2019). Prp40 and its mammalian homologues all contain two tyrosine-rich WW domains at the N-terminus followed by several FF (phenylalanine-phenylalanine) domains (Bedford and Leder, 1999). A WW domain normally contains 35 to 40 amino acids (aa) and binds to motifs with core proline-rich sequences, and a FF domain is an unusual phosphopeptide-binding module with about 60 aa and often follows a WW domain in the same protein (Bedford and Leder, 1999; Allen et al., 2002; Ingham et al., 2005). The WW domains in human PRPF40A and PRPF40B interact with proteins such as huntingtin involved in Huntington’s disease and MeCP2 involved in Rett syndrome (Faber et al., 1998; Passani et al., 2000; Buschdorf and Stratling, 2004). Although Prp40 in S. cerevisiae has been studied in detail, its homologues in other systems have not been well studied.
We have been using the filamentous fungus Aspergillus nidulans as a genetic model organism for studying the function of cytoplasmic dynein, a minus-end-directed microtubule motor that transports various cargoes in eukaryotic cells (Reck-Peterson et al., 2018; Olenick and Holzbaur, 2019). Cytoplasmic dynein is a multi-component complex with the two heavy chains (HCs) containing motor domains for ATP-dependent motility along microtubules (Bhabha et al., 2016; Schmidt and Carter, 2016; Grotjahn and Lander, 2019), and its function requires dynactin, another multi-component complex important for dynein–cargo interaction and dynein activation (Schroer, 2004; Reck-Peterson et al., 2018; Olenick and Holzbaur, 2019). In A. nidulans and other filamentous fungi, dynein is responsible for distributing multiple nuclei along hyphae and for transporting a variety of cargoes including early endosomes and cargoes that hitchhike on early endosomes (Wedlich-Soldner et al., 2002; Baumann et al., 2012; Etxebeste et al., 2013; Xiang et al., 2015, 2018; Peñalva et al., 2017; Salogiannis and Reck-Peterson, 2017; Steinberg et al., 2017). To identify proteins that are required for dynein-mediated early-endosome transport, we took a genetic approach to screen for mutants defective in early-endosome distribution (eed) (Yao et al., 2014, 2015; Zhang et al., 2014). From the eed mutants, we have identified genes encoding for the HookA, FhipA, and VezA proteins (Yao et al., 2014, 2015; Zhang et al., 2014). HookA and FhipA are components of the Fts-Hook-Fhip (FHF) complex that link early endosomes to dynein–dynactin (Xu et al., 2008; Bielska et al., 2014; Yao et al., 2014; Zhang et al., 2014; Guo et al., 2016; Reck-Peterson et al., 2018; Olenick and Holzbaur, 2019), and VezA is homologous to human vezatin whose higher eukaryotic homologues have just been found to participate in dynein functions (Spinner et al., 2020).
Here we report the study of a new eed mutant, eedD5, which exhibited an obvious defect in dynein-mediated transport of early endosomes as well as a mild defect in nuclear distribution. We identified the eedD gene as the Prp40 homologue in A. nidulans (Prp40A), and the eedD5 mutation is the nonsense Prp40AL438* mutation.
RESULTS
The eedD5 mutation is identified as prp40A L438*
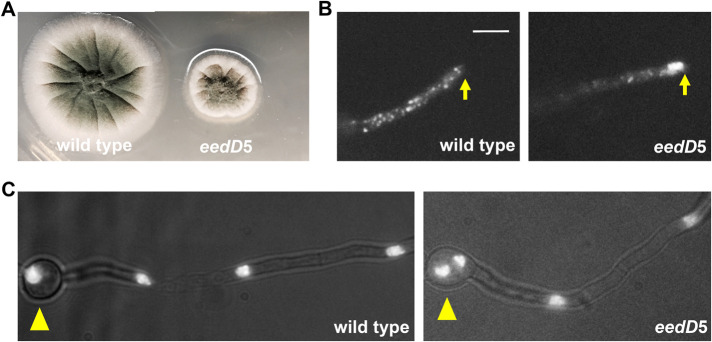
The eedD5 mutant was isolated after UV mutagenesis of a strain containing mCherry-labeled RabA and GFP-labeled Histone-H1, which allowed observation of early endosomes and nuclei, respectively (Abenza et al., 2009; Xiong and Oakley, 2009; Zhang et al., 2010; Abenza et al., 2012; Egan et al., 2012). The eedD5 mutant formed a colony obviously smaller than that of a wild type (Figure 1A). In A. nidulans, the plus ends of microtubules face the hyphal tip and minus ends are either at the spindle-pole body or at the septum (Oakley et al., 1990; Han et al., 2001; Konzack et al., 2005; Efimov et al., 2006; Xiong and Oakley, 2009; Egan et al., 2012; Zeng et al., 2014; Zhang et al., 2017b; Gao et al., 2019). An abnormal accumulation of early endosomes at the hyphal tip was obvious in every single hypha of the mutant (Figure 1B), indicating a defect in dynein-mediated early-endosome transport (Lenz et al., 2006; Abenza et al., 2009; Zekert and Fischer, 2009; Zhang et al., 2010, 2011; Egan et al., 2012). Nuclear distribution was mildly affected as about half of the spore heads in the mutant contained two nuclei instead of a single nucleus in a vast majority of wild-type hyphae (Figure 1C), whereas a nuclear-distribution (nud) mutant in the dynein pathway typically shows four or more nuclei inside the spore head (Osmani et al., 1990; Xiang et al., 1994, 1995).
FIGURE 1:
Phenotype of the eedD5 mutant. (A) Colony phenotypes of the eedD5 mutant and a wild-type strain. (B) Microscopic images showing the distributions of mCherry–RabA-labeled early endosomes (mCherry-RabA) in wild type and the eedD5 mutant. Bar, 5 μm. Although bidirectional movements of mCherry–RabA-labeled early endosomes are not completely abolished, most of the eedD5 hyphal tips (∼80%) show an abnormal accumulation of mCherry-RabA signals (n = 50). Hyphal tip is indicated by a yellow arrow. (C) Images of nuclei labeled with GFP-tagged Histone H1 in wild type and the eedD5 mutant. Spore head is indicated by a yellow arrowhead.
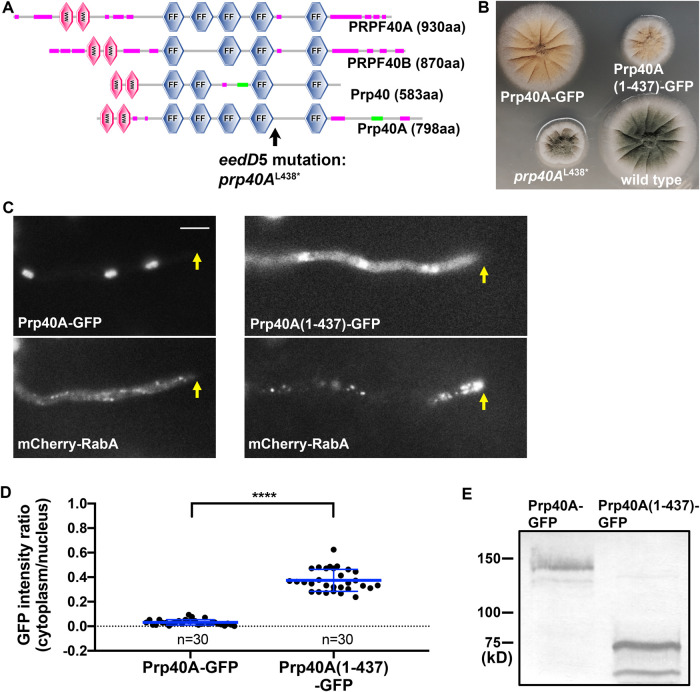
To determine whether the defect in early-endosome transport and the small-colony phenotype are genetically linked or caused by the same mutation, we crossed the eedD5 mutant with a wild-type strain. We found that five progeny with the small-colony phenotype all showed the abnormal accumulation of early endosomes at the hyphal tip, while five progeny with normal colony size all showed normal distribution of early endosomes. This result indicated that the colony phenotype and the early-endosome-distribution phenotype are causally linked to the same locus. To identify the eedD5 mutation, we combined the spores from the five mutant strains and prepared genomic DNA from the mutant sample for whole genome sequencing. As a control, we also prepared genomic DNA from five wild-type strains, and whole genome sequencing of the wild-type sample was performed together with the mutant sample. Whole-genome sequencing was done using the genome sequencing and bioinformatic service of Otogenetics (www.otogenetics.com). We used the software Integrative Genomics Viewer (IGV 2.4.3) to visualize the genomic sequencing data to identify the mutation. The eedD5 mutation was found in An1249 (www.AspGD.org), which encodes a protein with 798 aa, and the mutation substituted a T with A that changed the codon for leucine (TTA) to a stop codon (TAA) at residue 438. The protein encoded by An1249 is homologous to the budding yeast Prp40 (Supplemental Figure S1), a previously studied mRNA splicing factor associated with U1 snRNP (Becerra et al., 2016). We named this protein Prp40A (Prp40 in A. nidulans). Prp40A (798 aa) is much bigger than Prp40 (583 aa). Similar to the yeast Prp40, which contains two WW domains followed by four FF domains (SMART domain analysis), Prp40A contains two WW domains followed by five FF domains (Figure 2A). The eedD5 mutation (the only eedD mutant allele so far), identified as prp40AL438*, should result in a C-terminal truncated protein missing the last FF domain.
FIGURE 2:
Phenotypic analysis of the strain containing the Prp40A(1-437)-GFP fusion gene. (A) Domain structures of Prp40 proteins in different organisms including PRPF40A (human), PRPF40B (human), Prp40 (budding yeast), and Prp40A (A. nidulans). Position of the eedD5 mutation, prp40AL438*, is indicated by a black arrow. (B) Colony phenotypes of the strains containing different Prp40A alleles. (C) Localization of Prp40A-GFP or Prp40A(1-437)-GFP and mCherry–RabA-labeled early endosomes in strains containing one of the GFP fusions. Hyphal tip is indicated by a yellow arrow. Bar, 5 μm. (D) A quantitative analysis on the ratio of GFP signal intensity in the cytoplasm to that in the nucleus (Prp40A-GFP: n = 30; Prp40A(1-437)-GFP: n = 30). Scatter plots with mean and SD values were generated by Prism 8. The Mann–Whitney test (unpaired, two-tailed) was used for analyzing the two data sets without assuming normal distribution of the data. ****p < 0.0001. (E) A Western blot detecting the Prp40A(1-437)-GFP fusion, suggesting that the fusion protein is stable. The polyclonal anti-GFP antibody (Clontech) was used to probe the blot.
The Prp40A(1-437)-GFP fusion is defective in localization and function
To characterize the putative truncated Prp40A in the eedD5 mutant, we constructed a strain containing the Prp40A(1-437)-GFP fusion gene at the prp40A locus, and we also constructed a strain containing the full-length (FL) Prp40A-GFP at the prp40A locus as a control. While the strain containing the FL Prp40A-GFP formed a normal colony, the strain containing the Prp40A(1-437)-GFP fusion formed a small colony just like that formed by the prp40AL438* mutant (Figure 2B). In the strain containing the FL Prp40A-GFP, GFP signals labeled the nuclei, and mCherry–RabA-labeled early endosomes distributed normally along hyphae (Figure 2C). However, in the strain containing Prp40A(1-437)-GFP, the GFP signals appeared in both nuclei and cytoplasm, and mCherry–RabA-labeled early endosomes abnormally accumulated at the hyphal tip (Figure 2C). These data not only confirmed the eedD5 causal mutation being Prp40AL438* but also showed that the C-terminus of Prp40A is important but not essential for the nuclear localization of Prp40A, a point further supported by our quantitative analysis of the nuclear and cytoplasmic intensity of GFP signals (Figure 2D). The C-terminal truncated Prp40A(1-437)-GFP protein of expected size was clearly detected on a Western blot probed with an anti-GFP antibody, suggesting that the truncated protein is stably expressed but functionally defective (Figure 2E).
The Prp40A(1-437)-GFP and Prp40A-GFP fusion proteins associate with RNA-splicing factors and proteins involved in actin- and/or microtubule-based functions
To verify that Prp40A is associated with the splicing machinery in A. nidulans, we used a Prp40A-GFP-containing strain for a proteomic analysis on proteins pulled down with Prp40A-GFP. In the same experiment, we also included the Prp40A(1-437)-GFP-containing strain for comparison. Our proteomic analysis indicates that the FL Prp40A-GFP is associated with many splicing factors, including the top hit Prp8, the largest spliceosomal protein, and proteins in the Prp19 complex (Table 1). Many of these proteins were also pulled down with the truncated (TR) Prp40A(1-437)-GFP (Table 1). While some splicing factors appear to associate preferentially with the FL Prp40A-GFP, others seem to associate preferentially with the truncated Prp40A(1-437)-GFP (Table 1). The result that the truncated protein still associates with many splicing factors suggests that its splicing function may at least be partially retained.
TABLE 1:
Some pre-mRNA splicing proteins pulled down with the FL Prp40A-GFP (FL) and the truncated Prp40A(1-437)-GFP (TR).
| FL (total) | TR (total) | FL (unique) | TR (unique) | FL (coverage) | TR (coverage) | |
|---|---|---|---|---|---|---|
| Prp8 (An4523, 2940aa) | 270 | 142 | 72 | 54 | 25.40% | 21.30% |
| Brr2 (An10194, 2208aa) | 264 | 127 | 67 | 59 | 31.90% | 28.80% |
| Prp19 (An6906, 475aa) | 42 | 4 | 5 | 3 | 17.10% | 11.80% |
| Syf1 (An0111, 867aa) | 38 | 20 | 17 | 12 | 22.60% | 17.40% |
| Cus1 (An5098, 549aa) | 37 | 24 | 14 | 13 | 37.20% | 34.20% |
| Clf1 (An1259, 673aa) | 32 | 11 | 14 | 9 | 23.80% | 18.10% |
| Prp21 (An4760, 530aa) | 29 | 8 | 10 | 5 | 24.40% | 17.60% |
| Msl5 (An10615, 554aa) | 27 | 14 | 6 | 6 | 18.40% | 15.00% |
| Prp9 (An8015, 502aa) | 25 | 8 | 11 | 6 | 23.10% | 15.10% |
| Prp4 (An1468, 520aa) | 19 | 4 | 8 | 3 | 20.50% | 7.70% |
| Prp17 (An5196, 531aa) | 18 | 2 | 7 | 2 | 18.60% | 3.40% |
| Prp39 (An1635, 588aa) | 17 | 3 | 9 | 3 | 16.50% | 5.60% |
| Prp45 (An8180, 583aa) | 17 | 8 | 8 | 5 | 19.20% | 13.20% |
| Prp3 (An0727, 520aa) | 15 | 6 | 6 | 6 | 16.20% | 16.80% |
| Prp31 (An1260, 521aa) | 14 | 5 | 7 | 4 | 21.90% | 12.70% |
| Snp1 (An6903, 373aa) | 13 | 6 | 6 | 5 | 19.80% | 18.20% |
| Cwc22 (An0289, 760aa) | 13 | 4 | 6 | 3 | 7.60% | 4.30% |
| Smb1 (An10561, 196aa) | 12 | 4 | 5 | 2 | 27.60% | 11.70% |
| Prp46 (An1208, 452aa) | 12 | 2 | 6 | 2 | 21.00% | 4.90% |
| Smd2 (An1802, 114aa) | 10 | 5 | 4 | 2 | 43.40% | 17.70% |
| Syf2 (An1861, 297aa) | 9 | 0 | 4 | 0 | 19.90% | 0% |
| Slt11 (An7145, 377aa) | 8 | 0 | 5 | 0 | 20.40% | 0% |
| Slu7 (An4788, 466aa) | 8 | 0 | 6 | 0 | 16.70% | 0% |
| Cwf7 (An4244, 215aa) | 7 | 0 | 4 | 0 | 21.90% | 0% |
| Usp107 (An6036, 762aa) | 95 | 327 | 24 | 40 | 28.50% | 42.40% |
| Prp10 (An2420, 1241aa) | 13 | 85 | 9 | 31 | 9.90% | 31.40% |
| Cwf10 (An1408, 985aa) | 23 | 52 | 13 | 20 | 14.80% | 26.80% |
| Prp6 (An7447, 941aa) | 12 | 41 | 7 | 21 | 11.20% | 26.00% |
| Cwf11 (An7014, 1422aa) | 2 | 24 | 2 | 15 | 1.90% | 15.50% |
| Prp5 (An1266, 1173aa) | 2 | 23 | 2 | 17 | 3.00% | 21.10% |
| Prp28 (An1634, 782aa) | 9 | 23 | 4 | 10 | 7.90% | 19.20% |
| Luc7 (An1776, 279aa) | 13 | 20 | 5 | 7 | 20.10% | 22.80% |
| Prp22 (An4721, 1241aa) | 0 | 6 | 0 | 5 | 0% | 7.00% |
Proteins preferentially pulled down with FL are listed in the top part, and those preferentially pulled down with TR are listed in the bottom part. S. cerevisiae names are used unless there is no S. cerevisiae homologue or if the name of the S. pombe homologue indicates more clearly a role in RNA splicing than that of the S. cerevisiae homologue.
Interestingly, some proteins required for the actin cytoskeleton function and/or endocytosis were preferentially pulled down with the truncated protein Prp40A(1-437)-GFP (Table 2) (Goode et al., 2015). The top hit is SepA, the only formin protein in A. nidulans (Harris et al., 1997; Breitsprecher and Goode, 2013). This was not entirely unexpected given that the Prp40 homologues FBP11 and FBP21 were identified as FBPs because their WW domains bind to formin’s proline-rich motifs (Chan et al., 1996; Bedford et al., 1998). Homologues of Pan1 and Las17/WASP (Goode et al., 2015), both containing proline-rich motifs, were also pulled down (Table 2). Other proteins involved in the actin cytoskeleton and endocytosis were pulled down presumably because they are associated with these proteins (Goode et al., 2015).
TABLE 2:
Some proteins involved in functions of the actin cytoskeleton and/or endocytosis pulled down with the truncated Prp40A(1-437)-GFP (TR) or the FL Prp40A-GFP (FL).
| TR (total) | FL (total) | TR (unique) | FL (unique) | TR (coverage) | FL (coverage) | |
|---|---|---|---|---|---|---|
| *SepA formin (An6523,1762aa) | 225 | 23 | 69 | 14 | 39.70% | 11.30% |
| SlaB (An2756, 1022aa) | 85 | 0 | 34 | 0 | 44.70% | 0% |
| *Pan1 homologue (An4270, 1484aa)) | 76 | 0 | 30 | 0 | 28.20% | 0% |
| Clathrin HC (An4463, 1676aa) | 67 | 0 | 36 | 0 | 25.80% | 0% |
| Myosin V (An8862, 1569aa) | 51 | 0 | 28 | 0 | 25.00% | 0% |
| RGD2 homologue (An5677, 883aa) | 21 | 0 | 11 | 0 | 14.50% | 0% |
| AmpA Amphiphysin (An2516, 413aa) | 10 | 0 | 7 | 0 | 27.90% | 0% |
| *Las17/WASP homologue (An11104, 657aa) | 8 | 0 | 5 | 0 | 9.40% | 0% |
| Sla1 homologue (An1462, 1113aa) | 7 | 0 | 6 | 0 | 11.60% | 0% |
| AmpB Amphiphysin (An8831, 261aa) | 6 | 0 | 4 | 0 | 24.10% | 0% |
| Myosin I or MyoA (An1558, 1249aa) | 5 | 0 | 3 | 0 | 3.80% | 0% |
| Ent2 homologue (An3696, 561aa) | 5 | 0 | 2 | 0 | 8.60% | 0% |
*Containing proline-rich motifs that could possibly bind the WW domains of Prp40A directly.
Importantly, several proteins involved in dynein- and microtubule-based functions were also pulled down, including the dynein HC, the LC8 dynein light chain, Arp1 of dynactin, and p50 of dynactin (Table 3) (King 2000; Schroer 2004; Chowdhury et al., 2015; Urnavicius et al., 2015).
TABLE 3:
Proteins involved in dynein- or microtubule-based functions pulled down with the FL Prp40A-GFP and the truncated Prp40A(1-437)-GFP (TR).
| FL (total) | TR (total) | FL (unique) | TR (unique) | FL (coverage) | TR (coverage) | |
|---|---|---|---|---|---|---|
| NudA/dynein HC (An0118, 4345aa) | 6 | 7 | 6 | 6 | 2.00% | 2.00% |
| p50 of dynactin (An3589, 467aa) | 6 | 3 | 5 | 3 | 11.60% | 6.40% |
| NudK/Arp1 of dynactin (An1953, 380aa) | 2 | 5 | 1 | 1 | 3.90% | 3.90% |
| NudG/LC8 dynein light chain (An0420, 94aa) | 1 | 0 | 1 | 0 | 23.40% | 0% |
| NudF/LIS1 (An6197, 444aa) | 1 | 0 | 1 | 0 | 3.20% | 0% |
| NudM/p150 of dynactin (An6323, 1342aa) | 0 | 1 | 0 | 1 | 0% | 1.30% |
| ClipA/CLIP-170 (An1475, 672aa) | 1 | 0 | 1 | 0 | 2.20% | 0% |
| KinA/kinesin-1 (An5343, 927aa) | 0 | 1 | 0 | 1 | 0% | 1.40% |
| KipB/Kip3/kinesin-8 (An3970, 464aa) | 1 | 8 | 1 | 5 | 2.70% | 11.80% |
| UncA/kinesin-3 (An7547, 1630aa) | 1 | 0 | 1 | 0 | 0.90% | 0% |
| BenA beta tubulin (An1182, 447aa) | 16 | 22 | 7 | 7 | 20.40% | 20.40% |
| TubB alpha tubulin (An7570, 451aa) | 2 | 10 | 1 | 7 | 2.40% | 22.00% |
| MipA gamma tubulin (An0676, 454aa) | 1 | 0 | 1 | 0 | 3.30% | 0% |
| TubC beta tubulin (An6838, 449aa) | 0 | 4 | 0 | 3 | 0% | 10.00% |
The ∆ prp40A mutant exhibits a partial defect in dynein-mediated processes
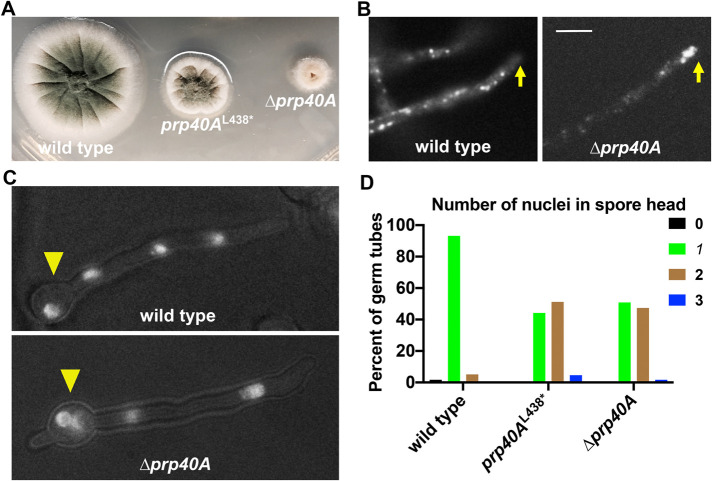
To determine if the phenotype of the original prp40A truncation mutant prp40AL438* is caused by a loss-of-function of the protein or a gain-of-function of the truncated protein Prp40A(1-437), we constructed the deletion mutant of prp40A, ∆prp40A, in which the whole open reading frame of Prp40A is deleted. The ∆prp40A mutant exhibited a much more severe growth defect than that exhibited by the prp40AL438* mutant, but it still formed a tiny colony (Figure 3A). Thus, Prp40A is not an essential protein in A. nidulans. To determine if RNA splicing in general is essential for A. nidulans growth, we created deletion constructs for genes encoding Prp5 (Abu Dayyeh et al., 2002) and Clf1 (Chung et al., 1999; Gasch et al., 2006) homologues in A. nidulans (prp5A and clf1A) (Supplemental Figures S2 and S3). Neither the prp5A-null mutant nor the clf1A-null mutant in A. nidulans is viable as shown by heterokaryon analyses (Supplemental Figure S4) (Osmani et al., 2006). Specifically, transformants that grew on nonselective medium were not able to grow on the medium selective for the AfpyrG marker linked to the deletion constructs (Supplemental Figure S4) (Osmani et al., 2006). Together, these results suggest that while splicing in general is essential for the viability of A. nidulans strains, Prp40A is not essential, which differs from the situation in budding yeast where Prp40 is essential for viability (Kao and Siliciano, 1996).
FIGURE 3:
Phenotype of the ∆prp40A mutant. (A) Colony phenotypes of a wild-type strain, the prp40AL438* mutant and the ∆prp40A mutant. (B) Microscopic images showing the distributions of mCherry–RabA-labeled early endosomes (mCherry-RabA) in wild type and the ∆prp40A mutant. Hyphal tip is indicated by a yellow arrow. Bar, 5 μm. (C) Images of nuclei labeled with GFP-tagged Histone H1 in wild type and the ∆prp40A mutant. Spore head is indicated by a yellow arrowhead. (D) A quantitative analysis on the percentage of germ tubes containing 0, 1, 2, or 3 nuclei in the spore head (wild type: n = 59; prp40AL438*: n = 43; ∆prp40A: n = 57). The number of nuclei in the spore head of the prp40AL438* mutant or the ∆prp40A mutant is higher than that in wild type (p < 0.0001 in both cases, Kruskal–Wallis ANOVA test with Dunn’s multiple comparisons test, unpaired). However, the number of nuclei in the spore head of the prp40AL438* mutant is not significantly different from that in the ∆prp40A mutant (p > 0.05, Kruskal–Wallis ANOVA test with Dunn’s multiple comparisons test, unpaired).
Importantly, the ∆prp40A mutant exhibited an abnormal accumulation of early endosomes (mCherry-RabA) at the hyphal tip (Figure 3B), similar to that exhibited by the prp40AL438* mutant. In addition, the ∆prp40A mutant exhibited a mild defect in nuclear distribution. In the ∆prp40A mutant, about half of the germ tubes have two nuclei inside the spore head, just like that in the prp40AL438* mutant but unlike wild-type germ tubes, which usually have one nucleus in the spore head (Figure 3, C and D). Thus, the prp40AL438* and ∆prp40A mutants exhibited similar defects in dynein-mediated processes, suggesting that the prp40AL438* mutation is a loss-of-function rather than a gain-of-function mutation. In addition, combined with the fact that the prp40AL438* mutant grows much better than the ∆prp40A mutant, this result also suggests that prp40AL438* represents a function-separating mutation that partially separates Prp40A’s splicing and dynein-related functions. Because the prp40AL438* mutant grows much better than the ∆prp40A mutant (Figure 3A), we used only the prp40AL438* mutant for further analyses.
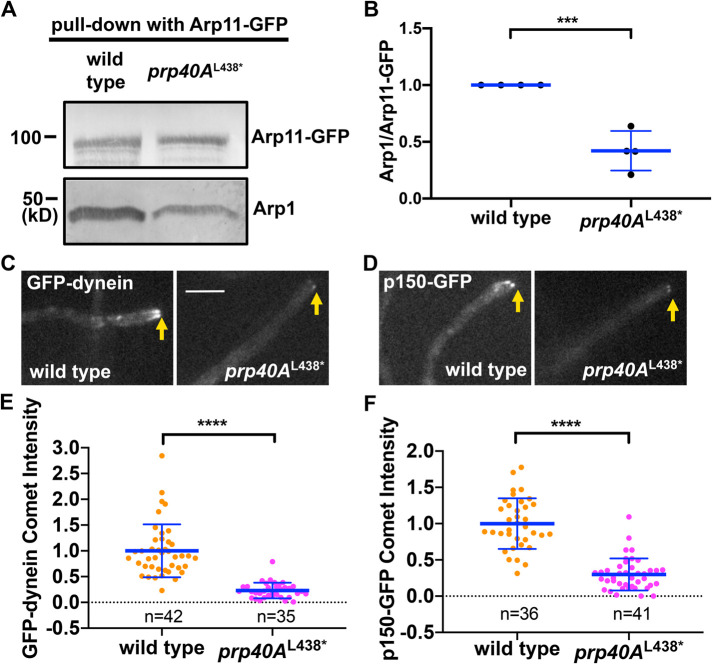
The prp40A L438* mutation decreases the amount of Arp1 associated with Arp11
The in vivo function of dynein requires dynactin, a complex containing multiple components including p150 and the actin-related protein Arp1, which forms a mini-filament whose pointed end binds directly to another actin-related protein Arp11 (Eckley et al., 1999; Schroer, 2004; Chowdhury et al., 2015; Urnavicius et al., 2015; Reck-Peterson et al., 2018; Olenick and Holzbaur, 2019). In A. nidulans, the Arp11 protein is encoded by a gene (An3185) (Zhang et al., 2008) without any intron. From a previous study on mammalian dynactin, we also know that Arp11 can stably exist within the pointed-end subcomplex without Arp1 (Eckley et al., 1999). Thus, we made an Arp11-GFP strain and used it for protein pull-down experiments. As shown by Western analyses, the amount of Arp1 pulled down with Arp11-GFP is lowered in the prp40AL438* mutant (Figure 4, A and B). This could be related to splicing as Arp1 is encoded by a gene (An1953, nudK) (Xiang et al., 1999) with two introns, but this could also be related to a moonlighting function of Prp40A on dynactin assembly.
FIGURE 4:
Decreased amounts of Arp1 pulled down with Arp11-GFP and decreased microtubule plus-end accumulation of dynein–dynactin in the prp40AL438* mutant. (A) Western blots showing that the amount of Arp1 pulled down with Arp11-GFP is lower in the prp40AL438* mutant than in the wild-type control. (B) A quantitative analysis on the ratio of pulled-down Arp1 to Arp11-GFP (Arp1/Arp11-GFP). The values were generated from Western analyses of four independent pull-down experiments (n = 4 for all). The wild-type values are set as 1. Scatter plots with mean and SD values were generated by Prism 8. ***p < 0.001 (Student’s t test, two-tailed, unpaired, normal distribution was assumed). (C) Images of GFP-dynein in wild type and the prp40AL438* mutant. Hyphal tip is indicated by a light brown arrow. Bar, 5 μm. (D) Images of dynactin p150-GFP in wild type and the prp40AL438* mutant. Hyphal tip is indicated by a light brown arrow. (E) A quantitative analysis on GFP-dynein comet intensity in wild type (n = 42) and the prp40AL438* mutant (n = 35). (F) A quantitative analysis on p150-GFP comet intensity in wild type (n = 36) and the prp40AL438* mutant (n = 41). For both E and F, all values are relative to the average value for wild type, which is set as 1. Scatter plots with mean and SD values were generated by Prism 8. The Mann–Whitney test (unpaired, two-tailed) was used for analyzing the two data sets without assuming normal distribution of the data. ****p < 0.0001.
The prp40A L438* mutation decreases the microtubule plus-end accumulation of dynein–dynactin
In fungi and higher eukaryotic cells, GFP-labeled dynein and dynactin accumulate at the dynamic microtubule plus ends, which were represented by the dynamic comet-like structures (Wu et al., 2006). In filamentous fungi, dynein and dynactin form plus-end comets near the hyphal tip (Han et al., 2001; Zhang et al., 2003), which is important for dynein-mediated early-endosome transport (Lenz et al., 2006). To test if the plus-end accumulation of dynein or dynactin is defective in the prp40AL438* mutant, we examined GFP-dynein HC and p150-dynactin-GFP signals in the prp40AL438* background. We found that the comet signal intensity of either GFP-dynein HC or p150-dynactin-GFP was significantly reduced in the mutant (Figure 4, C–F). This is consistent with Arp1 being affected by the mutation, because Arp1 is important for the stability of dynactin p150 (Minke et al., 1999; Haghnia et al., 2007; Zhang et al., 2008), both Arp1 and p150 are required for dynein–dynactin interaction (Karki and Holzbaur, 1995; Vaughan and Vallee, 1995; Chowdhury et al., 2015; Urnavicius et al., 2015, 2018; Grotjahn et al., 2018), and both are required for the plus-end localization of A. nidulans dynein (Xiang et al., 2000; Han et al., 2001; Zhang et al., 2003, 2008; Egan et al., 2012; Yao et al., 2012). However, it is hard to totally exclude the possibility that this reduction in the plus-end accumulation of dynein is due to a general decrease in the amounts of dynein–dynactin components, as most of them are encoded by intron-containing genes. For example, the genes encoding dynein intermediate chain (NudI, An1454), dynein light intermediate chain (NudN, An4664), and the 8-kD dynein light chain (NudG, An0420) all contain two or more introns and all of these proteins are important for the plus-end accumulation of dynein in A. nidulans (Beckwith et al., 1998; Zhang et al., 2002, 2009; Liu et al., 2003).
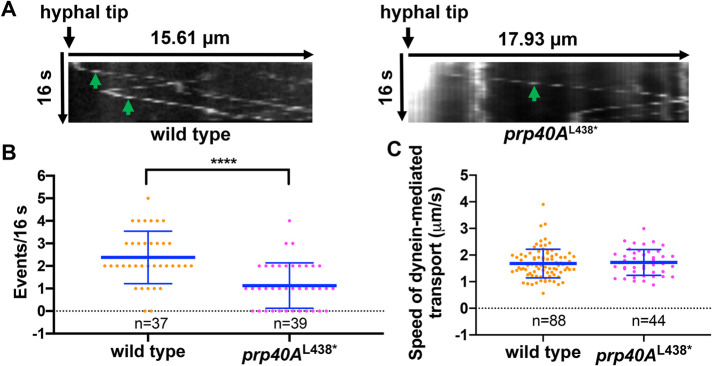
The prp40A L438* mutation decreases the frequency but not the speed of dynein-mediated early-endosome transport
To examine in more detail how the defects detected in the prp40AL438* mutant affect dynein-mediated early-endosome transport, we measured the frequency and speed of early-endosome transport away from the hyphal tip in wild type and the prp40AL438* mutant (Figure 5, A–C). While the frequency of dynein-mediated transport is significantly decreased in the mutant, the speed is not significantly affected (Figure 5, A–C).
FIGURE 5:
A quantitative analysis on dynein-mediated early-endosome transport in the prp40AL438* mutant showing a decrease in the frequency but not the speed of the transport. (A) Kymographs showing the movements of early endosomes in wild type and the prp40AL438* mutant. Green arrows indicate dynein-mediated movements away from the hyphal tip. (B) A quantitative analysis on the frequency of dynein-mediated transport in wild type (n = 37 hyphal tips) and the prp40AL438* mutant (n = 39 hyphal tips). Scatter plots with mean and SD values were generated by Prism 8. ****p < 0.0001 (unpaired, Mann–Whitney test, Prism 8). (C) A quantitative analysis on the speed of dynein-mediated early-endosome movement in wild type (n = 88 movements) and the prp40AL438* mutant (n = 44 movements). The difference between wild type and the mutant is insignificant at p = 0.05 (unpaired, Mann–Whitney test, Prism 8).
DISCUSSION
Via a screen for mutants defective in dynein-mediated early-endosome transport, we identified the prp40AL438* mutation that causes a C-terminal truncation of Prp40A, the A. nidulans homologue of Prp40, which is an essential splicing factor in budding yeast. In A. nidulans, many genes essential for colony growth contain introns. For example, a never-in-mitosis gene nimA (An9504) contains two introns, and NimA is essential for colony formation (Morris, 1975; Osmani et al., 1987, 1988). The fact that the ∆prp40A mutant still formed a tiny colony indicates that Prp40A is not essential for splicing. Interestingly, the prp40AL438* mutant formed a colony that is much healthier than that of the ∆prp40A mutant, suggesting that the splicing function of Prp40A may only be partially lost in the prp40AL438* mutant. Given this and the result that the prp40AL438* and ∆prp40A mutations cause similar defects in dynein-mediated processes, we postulate that Prp40A may have a moonlighting function that affects dynein-mediated processes.
It has been reported that another splicing factor has microtubule-related function in the filamentous fungus Ustilago maydis, where the Num1 protein in the Prp19 complex interacts with kinesin-1, thereby affecting dynein localization and function (Kellner et al., 2014; Zhou et al., 2018). Kinesin-1 is indeed important for the plus-end accumulation of dynein in filamentous fungi and neurons (note that in higher eukaryotes, kinesin light chains interact with dynein intermediate chains directly) (Zhang et al., 2003; Ligon et al., 2004; Lenz et al., 2006; Arimoto et al., 2011; Twelvetrees et al., 2016), and it is important for dynein-mediated early-endosome transport (Lenz et al., 2006). In the A. nidulans kinesin-1 mutant, both dynein and dynactin decorate along microtubules (Zhang et al., 2010; Yao et al., 2012). However, this was not seen in the prp40AL438* mutant. The A. nidulans homologue (An4244) of U. maydis Num1 (fission yeast Cwf7) is associated with Prp40A-GFP (Table 1). However, the number of total peptide from kinesin-1 (a 927-aa protein) detected in the Prp40A(1-437)-GFP pull down is only one whereas the total peptide number was five for Arp1 (a much smaller 380-aa protein) (Table 3), suggesting that Arp1 is more likely a true interactor than kinesin-1. It is also interesting to note that the mammalian Prp40 homologues interact with Huntingtin (Faber et al., 1998; Passani et al., 2000), a protein interact with dynein–dynactin and affects dynein function (Caviston and Holzbaur, 2009; Liot et al., 2013; Wong and Holzbaur, 2014). However, no Huntingtin homologue is found in A. nidulans genome.
In the prp40AL438* mutant, the amount of Arp1 pulled down with Arp11-GFP was decreased. This could be splicing-related, but we would not exclude the possibility that Prp40A affects Arp1 stability or Arp1 mini-filament assembly in a splicing-independent manner. Notably, we recently found that a mammalian Prp40 homologue PRPF40A was identified as one of the 32 total proteins interacting with Arp1 (ACTR1B) (https://thebiogrid.org/230579/summary/mus-musculus/actr1b.html) (Table 4) (Hein et al., 2015). Moreover, PRPF40A was also identified as one of the 74 total proteins interacting with p24 (DCTN3) (https://thebiogrid.org/207318/summary/mus-musculus/dctn3.html?sort=evidence) that is located on the Arp1 mini-filament with p50 (Echeverri et al., 1996; Karki et al., 1998; Pfister et al., 1998; Eckley et al., 1999; Schroer, 2004; Melkonian et al., 2007; Cheong et al., 2014; Hein et al., 2015; Urnavicius et al., 2015). Among the 32 Arp1 interactors, many are components of the dynactin complex and chaperonins that could be involved in the assembly of the complex (Table 4) (Hein et al., 2015). Importantly, both PRPF40A and vezatin are among the interactors (Table 4). The A. nidulans vezatin homologue VezA was identified from a mutant screen similar to the one performed in this study (Yao et al., 2015). Although mammalian vezatin was initially identified as a protein involved in cell adhesion (Kussel-Andermann et al., 2000), vezatin homologues in Drosophila and zebrafish have recently been found to play roles in dynein-mediated axonal transport (Spinner et al., 2020). Thus, our genetic approach has continued to identify conserved dynein regulators. Our current data combined with the mammalian interactome data suggest the possibility that Prp40 homologues may affect Arp1 either directly or indirectly, which may promote either the stability of Arp1 or its function. The Arp1 mini-filament is the backbone of the dynactin complex (Schafer et al., 1994; Chowdhury et al., 2015; Urnavicius et al., 2015), and this filament provides the binding sites for not only the dynein tails but also the coiled-coiled domains of several dynein-activating cargo adapters (Chowdhury et al., 2015; Urnavicius et al., 2015, 2018; Grotjahn et al., 2018; Reck-Peterson et al., 2018; Olenick and Holzbaur, 2019). In the prp40AL438* mutant, there is an obvious decrease in the frequency but not the speed of dynein-mediated early-endosome transport. This could be resulted from a decrease in the number of functional dynein–dynactin molecules at the plus end to support the dynein–dynactin–cargo interaction, but it is also possible that some dynein–dynactin–cargo complexes assembled at the plus end may be too abnormal to support cargo-activated initiation of dynein movement (Lenz et al., 2006; McKenney et al., 2014; Schlager et al., 2014; Zhang et al., 2017a; Qiu et al., 2019). While Prp40A’s mechanism of action needs to be further studied, we report our genetic finding and suggest that it may be worthwhile to examine the possible roles of Prp40 homologues in dynein functions in higher eukaryotes.
TABLE 4:
A list of the 32 unique interactors of ACTR1B (Arp1 actin-related protein 1B).
| Gene | Protein |
|---|---|
| MTNR1A | melatonin receptor 1A |
| SNCA | synuclein, alpha (non A4 component of amyloid precursor |
| CCT8 | chaperonin-containing TCP1, subunit 8 (theta) |
| ATXN1 | ataxin-1 |
| SYNPO2 | synaptopodin 2 |
| DCTN5 | dynactin-5 (p25) |
| OBSCN | obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| PDCL3 | phosducin-like 3 |
| ACTR10 | dynactin Arp11 or actin-related protein 10 homologue (S. cerevisiae) |
| POLR3B | RNA polymerase III polypeptide B |
| PRPF40A | PRP40 pre-mRNA processing factor 40 homologue A (S. cerevisiae) |
| VEZT | Vezatin, adherens junctions transmembrane protein |
| DCTN4 | dynactin 4 (p62) |
| CCT5 | Chaperonin-containing TCP1, subunit 5 (epsilon) |
| DCTN3 | dynactin-3 (p22 or p24) |
| DCTN6 | dynactin 6 (p27) |
| KCNMA1 | Potassium large conductance calcium activated channel, subfamily M, alpha member 1 |
| CCT2 | chaperonin-containing TCP1, subunit 2 (beta) |
| CCT4 | chaperonin-containing TCP1, subunit 4 (delta) |
| DCTN2 | dynactin 2 (p50) |
| ACTR1A | Arp1 actin related protein 1 homologue A, centractin alpha |
| MTA1 | metastasis associated 1 |
| YWHAH | Tyrosine 3-monoxygenase/tryptophan 5-monoxygenase activation protein |
| CCT3 | chaperonin-containing TCP1, subunit 3 (gamma) |
| TCP1 | t-complex 1, CCT-alpha |
| SVIL | Supervillain |
| DCTN1 | dynactin 1 (p150) |
| CCT6A | chaperonin-containing TCP1, subunit 6A (zeta 1) |
| CAPZB | capping protein (actin filament), beta |
| CAPZA2 | capping protein (actin filament), alpha 2 |
| CAPZA1 | capping protein (actin filament), alpha 1 |
| MAPT | Microtubule-associated protein tau |
MATERIALS AND METHODS
A. nidulans strains and media
A. nidulans strains used in this study are listed in Table 5. UV mutagenesis on spores of A. nidulans strains was done as previously described (Willins et al., 1995; Xiang et al., 1999). Solid rich medium was made of either YAG (0.5% yeast extract and 2% glucose with 2% agar) or YAG+UU (YAG plus 0.12% uridine and 0.12% uracial). Genetic crosses were done by standard methods. Solid minimal medium containing 1% glucose was used for selecting progeny from a cross. For live-cell imaging experiments, cells were cultured in liquid minimal medium containing 1% glycerol overnight at 32°C. All the biochemical analyses were done using cells grown overnight at 32°C in liquid YG-rich medium (0.5% yeast extract and 2% glucose).
TABLE 5:
A. nidulans strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| XX222 | GFP-nudAHC; argB2::[argB*-alcAp::mCherry-RabA];∆nkuA::argB; pantoB100; yA2 | ( Zhang et al., 2011) |
| XY13 | p150-GFP-AfpyrG;∆nkuA::argB; pyrG89; pyroA4 | ( Yao et al., 2012) |
| XY42 | argB2::[argB*-alcAp::mCherry-RabA];∆nkuA::argB; pantoB100; yA2 | ( Qiu et al., 2018) |
| eedD5 | eedD5 (prp40AL438*); hhoA-GFP-AfriboB; argB2::[argB*-alcAp::mCherry-rabA]; pantoB100; ∆nkuA::argB | This work |
| JZ788 | Arp11-GFP-AfpyrG; argB2::[argB*-alcAp::mCherry-rabA]; pantoB100; ∆nkuA::argB; yA2 | This work |
| JZ901 | prp40AL438*; Arp11-GFP-AfpyrG; argB2::[argB*-alcAp::mCherry-rabA]; ∆nkuA::argB; yA2 | This work |
| RQ246 | Prp40A(1-437)-GFP; argB2::[argB*-alcAp::mCherry-rabA];∆nkuA::argB; pantoB100; yA2 | This work |
| RQ249 | Prp40A-GFP; argB2::[argB*-alcAp::mCherry-rabA];∆nkuA::argB; pantoB100; yA2 | This work |
| RQ255 | ∆prp40A-AfpyrG; argB2::[argB*-alcAp::mCherry-rabA]; pantoB100; ∆nkuA::argB; yA2 | This work |
| XX357 | hhoA-GFP-AfriboB; argB2::[argB*-alcAp::mCherry-rabA]; pyrG89; pantoB100; ∆nkuA::argB | This work |
| XX402 | hhoA-GFP-AfriboB; argB2::[argB*-alcAp::mCherry-rabA]; pantoB100; ∆nkuA::argB | This work |
| XX403 | hhoA-GFP-AfriboB; argB2::[argB*-alcAp::mCherry-rabA]; pyrG89; ∆nkuA::argB | This work |
| XX470 | prp40AL438*; GFP-nudAHC; argB2::[argB*-alcAp::mCherry-rabA]; possibly ∆nkuA::argB | This work |
| XX471 | prp40AL438*; p150-GFP-AfpyrG | This work |
| XX498 | ∆prp40A-AfpyrG; hhoA-GFP-AfriboB; possibly ∆nkuA::argB; possibly yA2 | This work |
| XX512 | ∆prp40A-AfpyrG; GFP-nudAHC; argB2::[argB*-alcAp::mCherry-rabA];∆nkuA::argB; pyrG89; possibly yA2 | This work |
| XX513 | ∆prp40A-AfpyrG; p150-GFP-AfpyrG; argB2::[argB*-alcAp::mCherry-rabA];∆nkuA::argB; possibly pyrG89; possibly yA2 | This work |
Live-cell imaging and analyses
All images were captured using an Olympus IX73 inverted fluorescence microscope linked to a PCO/Cooke Corporation Sensicam QE cooled CCD camera. An UPlanSApo 100× objective lens (oil) with a 1.40 numerical aperture was used. A filter wheel system with GFP/mCherry-ET Sputtered series with high transmission (Biovision Technologies) was used. The IPLab software was used for image acquisition and analysis. For all images, cells were grown in the LabTek Chambered #1.0 borosilicate coverglass system (Nalge Nunc International, Rochester, NY). For the analysis on the frequency and velocity of early-endosome transport, cells were grown for overnight at 32°C and time-lapse sequences were captured at room temperature. All the images were taken with a 0.1-s exposure time (binning: 2 × 2). The hyphal-tip region was defined as a region within ∼2 μm to the hyphal tip. For measuring early-endosome movement, we focused on movements within ∼7 μm to the hyphal tip within the hyphal-tip cell (“hyphal-tip cell” refers to the hyphal segment between the hyphal tip and the most proximal septum). Retrograde movements initiating from the hyphal tip region are defined as “dynein-mediated.” Thirty frames were taken for each sequence with a 0.1-s exposure time and a 0.3-s interval between frames, and the “generate-kymograph” and “measure-kymograph functions” of IPLab were used for analysis. For measuring the signal intensity of the individual GFP-dynein or p150-GFP comets, we followed a previously described method (Zhang et al., 2011). Specifically, an area containing the whole comet was selected as a region of interest (ROI), and the Max/Min tool of the IPLab program was used to measure the maximal intensity within the ROI. Then the ROI box was dragged outside of the cell to take the background value, which was then subtracted from the value of the comet.
Construction of the strain containing the Prp40A-GFP allele at the PrpA40 locus
Constructions of all strains are done by using standard procedures used in A. nidulans (Yang et al., 2004; Nayak et al., 2006; Szewczyk et al., 2006). For constructing the Prp40A-GFP fusion, we used the following six oligos to amplify genomic DNA and the GFP-AfpyrG fusion (Yang et al., 2004): AN1249-51 (5′-CGGCGGTGTTAGTTCACTC-3′), AN1249-34 (5′-CGCCTGCACCAGCTCCATCCTCCTCAATCTCTCCTTCC-3′), GA4F (5′-GGAGCTGGTGCAGGCG-3′), AN1249-35 (5′-TCCCATATTCACCGTACACCTCTACTGTCTGAGAGGAGGCACTGAT-3′), AN1249-53 (5′-TAGAGGTGTACGGTGAATATGGGA-3′), and AN1249-36 (5′-GAATTGTGAATGACCCGCCAC-3′). Specifically, AN1249-51 and AN1249-34 were used to amplify the 2.1-kb fragment in the coding region, and AN1249-53 and AN1249-36 were used to amplify the 1-kb fragment in the 3′ untranslated region using wild-type genomic DNA as template, and GA4F and AN1249-35 were used to amplify the 2.7-kb GFP-AfpyrG fragment using genomic DNA from the RQ69 strain (Qiu et al., 2018) containing p25-GFP-AfpyrG as template. We then used two oligos, AN1249-52 (5′-TTACGTGGACCCCGTAATGAT-3′) and AN1249-36, for a fusion PCR of the three fragments to generate the 4.7-kb PRP40-GFP-AfpyrG fragment that we used to transform into XY42 containing ∆nkuA (Nayak et al., 2006) and mCherry-RabA (Abenza et al., 2009; Zhang et al., 2010; Qiu et al., 2018). The transformants were screened by microscopically observing the GFP signals in cell nuclei and further confirmed by Western blot with a polyclonal anti-GFP antibody from Clontech.
Construction of the strain containing the Prp40A(1-437)-GFP allele at the PrpA40 locus
For constructing the Prp40A(1-437)-GFP strain, we made the Prp40A(1-437)-GFP-AfpyrG fragment by inserting the GFP-AfpyrG fragment into the Prp40ALeu438* mutation site right before the stop codon. The following oligos were used to make the Prp40A(1-437)-GFP-AfpyrG construct: AN1249-51, GA4F, AN1249-52, AN1249-36, AN1249-31 (5′-CGCCTGCACCAGCTCCCGACCTTTCTTCTTCTTCGAC-3′), AN1249-32 (5′-ATCATTACGGGGTCCACGTAACTGTCTGAGAGGAGGCACTG -3′), and AN1249-33 (5′-CAGGAACAGCAGATGTAGGC-3′). Specifically, AN1249-51 and AN1249-31 were used to amplify a 1-kb fragment of the 5′ coding region, and AN1249-52 and AN1249-36 were used to amplify a 2.1-kb fragment containing the 3′ coding region and 3′ untranslated region using wild-type genomic DNA as template, and GA4F and AN1249-32 were used to amplify the 2.7-kb GFP-AfpyrG fragment using genomic DNA from the RQ69 (Qiu et al., 2018) strain as template. We then used two oligos, AN1249-51 and AN1249-33, for a fusion PCR to fuse the three fragments to generate the 4.7-kb Prp40A(1-437)-GFP-AfpyrG fragment that we used to transform the XY42 strain. The transformants were screened by microscopically observing the GFP signals in nuclei and further confirmed by Western blotting analysis with a polyclonal anti-GFP antibody from Clontech.
Construction of the ∆ prp40A mutant
For constructing the ∆prp40A mutant, we used fusion PCR to make the ∆prp40A (An1249) construct with the selective marker pyrG from Aspergillus fumigatus, AfpyrG, in the middle of the sequences flanking the Prp40A open reading frame. Specifically, AN1249-55 (5′-TCACGAGTCGGTGTTGGATTC-3′) and AN1249-39 (5′-TTTGCACCTTGATGACAAGTTTTCAA-3′) were used to amplify a ∼0.7-kb 5′ untranslated region from the strain containing PrpA40-GFP, and AN1249-58 (5′-TTGAAAACTTGTCATCAAGGTGCAAAACCGGTCGCCTCAAACAAT-3′) and AN1249-38 (5′-CAAATCACAGGCTGAAGACACG-3′) were used to amplify a 2.5-kb fragment containing AfpyrG and 3′ untranslated region from the Prp40A-GFP strain, and the two fragments were fused together by fusion PCR using AN1249-54 (5′-GATTCTGGAGGTGGGCTTTTAG-3′) and AN1249-37 (5′-CGTATTGAAGAAACTGCCAGGT-3′) as primers. The product (∼3.0 kb) was transformed into the XY42 strain. The transformants were first screened by the obvious small-colony phenotype. Site-specific integration in the ∆prp40A mutant was verified by PCR amplification using one oligo pair of AN1249-55 and AFPyrG3 (5′-GTTGCCAGGTGAGGGTATTT-3′), which should generate a ∼700 bp product from the ∆prp40A mutant but no product from a wild-type control, and another pair of AN1249-37 and AN1249-52 (5′-TTACGTGGACCCCGTAATGAT-3′), which should generate no product from the ∆prp40A mutant but a 1.7-kb product from the wild type control.
Construction of the ∆ prp5A and ∆ clf1A mutants
For constructing the ∆prp5A mutant, we first made the ∆prp5A (An1266, www.AspGD.org) construct with the selective marker AfpyrG, in the middle of the linear construct. Specifically, we used APYRGF (5′-TGCTCTTCACCCTCTTCGCG-3′), and APYRGR (5′-CTGTCTGAGAGGAGGCACTGA-3′) as primers and the pFNO3 plasmid (deposited in the FGSC by Stephen Osmani) as a template to amplify a 1.9-kb AfpyrG fragment. We used PRP5NN (5′-ACTTATCCTTCATAGCATTGACCC-3′), PRP5NC (5′-CGCGAAGAGGGAGAAGAGCATGTTGCAGGCCAGCAATGAG-3′), PRP5CN (5′-ATCAGTGCCTCCTCTCAGACAGAAATGCGCGCCTGTAACATAC-3′), and PRP5CC (5′-CGCCAAGTCCTATCTCTTATCG-3′) as primers and wild-type genomic DNA as a template to amplify 1-kb fragments upstream and downstream of prp5A’s coding sequence. We then used PRP5NN1 (5′-GGCAAGGTAGGCAGTAACTTC-3′) and PRP5CC1 (5′-ACGACGAAGCATAGGCAGA-3′) to perform a fusion PCR to fuse the three fragments and obtained a 3.9-kb fragment, which we transformed into the A. nidulans strain XY42 (Qiu et al., 2018).
For constructing the ∆clf1A mutant, we first made the ∆clf1A (An1259, www.AspGD.org) construct with the selective marker AfpyrG. We used CLF1NN (5′-TGCAGACACAATGCGAATTGC-3′), CLF1NC (5′-CGCGAAGAGGGAGAAGAGCAATCTGAGAGAGCTGGTGCAATAG-3′), CLF1CN (5′-ATCAGTGCCTCCTCTCAGACAGTACATACAAGCGCCCGCTG-3′), and CLF1CC (5′-GTGAGAGGGTGATTAATTAATTTGGAC-3′) as primers and wild-type genomic DNA as a template to amplify 1-kb fragments upstream and downstream of clf1A’s coding sequence. We then used two primers, CLF1NN1 (5′-TGTCTCTTCAGGTCATTGGG-3′) and CLF1CC1 (5′-GGATAGGCTGGCACTGAAAC-3′), to perform a fusion PCR to fuse these fragments with AfpyrG and obtained a 3.9-kb fragment, which we transformed into the XY42 strain.
The transformants obtained were streaked out on both YAG and YUU plates as a standard procedure for analyzing essential genes (Osmani et al., 2006). Many colonies were nonviable on YAG but viable on YUU, consistent with the lethality of the ∆prp5A and ∆clf1A mutants (Osmani et al., 2006).
Construction of the Arp11-GFP strain
For constructing the Arp11-GFP fusion, we used the following six oligos to amplify genomic DNA and the GFP-AfpyrG fusion from the plasmid pFNO3: A11GNC (5′-GGCTCCAGCGCCTGCACCAGCTCCCCCCCACCCTGCCAACGTCC-3′), A11GNN (5′-TGTTTACGCGGCTTTTGGGC-3′), A11GCN (5′-ATCAGTGCCTCCTCTCAGACAGTTAGCCGGGAGGCTTTTCTC-3′), A11GCC (5′-TTCCCTGGCGACGTCCTCAC-3′), GAGAF (5′-GGAGCTGGTGCAGGCGCTG-3′), and pyrG3 (5′-CTGTCTGAGAGGAGGCACTGAT-3′). A fusion PCR was performed using A11GNN and A11GCC as primers to generate the 4.7 kb Arp11-GFP-AfpyrG fragment that we used to transform into XY42. Transformants were screened for GFP signals under microscope and positive strains were confirmed by PCR using these two oligos: AfpyrG5 (5′-AGCAAAGTGGACTGATAGC-3′) and A11GCC2 (5′-CATGCGCTACAGGGCTTCC-3′), which should generate a 1.1-kb product.
Biochemical pull-down assays, Western analysis, and mass spectrometry analysis
The μMACS GFP-tagged protein isolation kit (Miltenyi Biotec) was used to pull down GFP-tagged proteins and proteins associated with them. About 0.4 g hyphal mass was harvested from overnight culture for each sample, and cell extracts were prepared using a lysis buffer containing 50 mM Tris-HCl, pH 8.0, and 10 μg/ml protease inhibitor cocktail (Sigma-Aldrich). Cell extracts were centrifuged at 8000 × g for 15 min and then 16,000 × g for 15 min at 4°C, and supernatant was used for the pull-down experiment. To pull down GFP-tagged proteins, 25 μl anti-GFP MicroBeads were added into the cell extracts for each sample and incubated at 4°C for 30 min. The MicroBeads/cell extracts mixture was then applied to the μColumn followed by gentle wash with the lysis buffer used above for protein extraction (Miltenyi Biotec). Preheated (95°C) SDS–PAGE sample buffer was used as elution buffer. Western analyses were performed using the alkaline phosphatase system and blots were developed using the AP color development reagents from Bio-Rad. The antibody against GFP was from Clontech (polyclonal). A polyclonal antibody against Arp1 was generated in a previous study by injecting proteins produced in bacteria into rabbits followed by affinity purification of the antibody (Zhang et al., 2008). Quantitation of the protein band intensity was done using the IPLab software as described previously (Qiu et al., 2013). Specifically, an area containing the whole band was selected as a ROI, and the intensity sum within the ROI was measured. Then, the ROI box was dragged to the equivalent region of the negative control lane or a blank region without any band on the same blot to take the background value, which was then subtracted from the intensity sum. For proteomic analysis, eluted protein samples were run on an SDS–PAGE gel until proteins had reached the interphase between the stacking and the separating gels as previously described (Liu et al., 2010). A single gel slice containing the proteins was sliced out for mass spectrometry analysis of each sample, which was done using the Taplin Mass Spectrometry Facility at Harvard Medical School.
Supplementary Material
Acknowledgments
We thank Berl Oakley, David Horowitz, and Tharun Sundaresan for helpful discussions; Berl Oakley, Miguel Peñalva, Martin Egan, Samara Reck-Peterson, and Xuanli Yao for Aspergillus strains; the Fungal Genetic Stock Center for the pFNO3 plasmid; and Stephen Osmani for depositing it. This work was funded by the National Institutes of Health RO1 GM121850-A1 (to X.X.).
Abbreviations used:
- aa
amino acid
- eed
early-endosome distribution
- FBP
formin-binding protein
- FL
full length
- HC
heavy chain
- nud
nuclear-distribution
- pre-RNA
precursor mRNA
- Prp40
pre-mRNA-processing protein 40
- RNP
ribonucleoprotein
- ROI
region of interest.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-03-0166) on April 8, 2020.
REFERENCES
- Abenza JF, Galindo A, Pinar M, Pantazopoulou A, de los Rios V, Peñalva MA. (2012). Endosomal maturation by Rab conversion in Aspergillus nidulans is coupled to dynein-mediated basipetal movement. Mol Biol Cell , 1889–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abenza JF, Pantazopoulou A, Rodriguez JM, Galindo A, Peñalva MA. (2009). Long-distance movement of Aspergillus nidulans early endosomes on microtubule tracks. Traffic , 57–75. [DOI] [PubMed] [Google Scholar]
- Abu Dayyeh BK, Quan TK, Castro M, Ruby SW. (2002). Probing interactions between the U2 small nuclear ribonucleoprotein and the DEAD-box protein, Prp5. J Biol Chem , 20221–20233. [DOI] [PubMed] [Google Scholar]
- Allen M, Friedler A, Schon O, Bycroft M. (2002). The structure of an FF domain from human HYPA/FBP11. J Mol Biol , 411–416. [DOI] [PubMed] [Google Scholar]
- Arimoto M, Koushika SP, Choudhary BC, Li C, Matsumoto K, Hisamoto N. (2011). The Caenorhabditis elegans JIP3 protein UNC-16 functions as an adaptor to link kinesin-1 with cytoplasmic dynein. J Neurosci , 2216–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann S, Pohlmann T, Jungbluth M, Brachmann A, Feldbrugge M. (2012). Kinesin-3 and dynein mediate microtubule-dependent co-transport of mRNPs and endosomes. J Cell Sci , 2740–2752. [DOI] [PubMed] [Google Scholar]
- Becerra S, Andres-Leon E, Prieto-Sanchez S, Hernandez-Munain C, Sune C. (2016). Prp40 and early events in splice site definition. Wiley Interdiscip Rev RNA , 17–32. [DOI] [PubMed] [Google Scholar]
- Beckwith SM, Roghi CH, Liu B, Ronald Morris N. (1998). The “8-kD” cytoplasmic dynein light chain is required for nuclear migration and for dynein heavy chain localization in Aspergillus nidulans. J Cell Biol , 1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Leder P. (1999). The FF domain: a novel motif that often accompanies WW domains. Trends Biochem Sci , 264–265. [DOI] [PubMed] [Google Scholar]
- Bedford MT, Reed R, Leder P. (1998). WW domain-mediated interactions reveal a spliceosome-associated protein that binds a third class of proline-rich motif: the proline glycine and methionine-rich motif. Proc Natl Acad Sci USA , 10602–10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhabha G, Johnson GT, Schroeder CM, Vale RD. (2016). How dynein moves along microtubules. Trends Biochem Sci , 94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielska E, Schuster M, Roger Y, Berepiki A, Soanes DM, Talbot NJ, Steinberg G. (2014). Hook is an adapter that coordinates kinesin-3 and dynein cargo attachment on early endosomes. J Cell Biol , 989–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Goode BL. (2013). Formins at a glance. J Cell Sci , 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschdorf JP, Stratling WH. (2004). A WW domain binding region in methyl-CpG-binding protein MeCP2: impact on Rett syndrome. J Mol Med (Berl) , 135–143. [DOI] [PubMed] [Google Scholar]
- Caviston JP, Holzbaur EL. (2009). Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol , 147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC, Bedford MT, Leder P. (1996). Formin binding proteins bear WWP/WW domains that bind proline-rich peptides and functionally resemble SH3 domains. EMBO J , 1045–1054. [PMC free article] [PubMed] [Google Scholar]
- Cheong FK, Feng L, Sarkeshik A, Yates JR, 3rd, Schroer TA. (2014). Dynactin integrity depends upon direct binding of dynamitin to Arp1. Mol Biol Cell , 2171–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, Ketcham SA, Schroer TA, Lander GC. (2015). Structural organization of the dynein-dynactin complex bound to microtubules. Nat Struct Mol Biol , 345–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, McLean MR, Rymond BC. (1999). Yeast ortholog of the Drosophila crooked neck protein promotes spliceosome assembly through stable U4/U6.U5 snRNP addition. Rna , 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Casas A, Chazin WJ, Pastrana-Rios B. (2017). Prp40 homolog a is a novel centrin target. Biophys J , 2529–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri CJ, Paschal BM, Vaughan KT, Vallee RB. (1996). Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J Cell Biol , 617–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckley DM, Gill SR, Melkonian KA, Bingham JB, Goodson HV, Heuser JE, Schroer TA. (1999). Analysis of dynactin subcomplexes reveals a novel actin-related protein associated with the arp1 minifilament pointed end. J Cell Biol , 307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov VP, Zhang J, Xiang X. (2006). CLIP-170 homologue and NUDE play overlapping roles in NUDF localization in Aspergillus nidulans. Mol Biol Cell , 2021–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MJ, Tan K, Reck-Peterson SL. (2012). Lis1 is an initiation factor for dynein-driven organelle transport. J Cell Biol , 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etxebeste O, Villarino M, Markina-Inarrairaegui A, Araujo-Bazan L, Espeso EA. (2013). Cytoplasmic dynamics of the general nuclear import machinery in apically growing syncytial cells. PLoS One , e85076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber PW, Barnes GT, Srinidhi J, Chen J, Gusella JF, MacDonald ME. (1998). Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet , 1463–1474. [DOI] [PubMed] [Google Scholar]
- Gao X, Schmid M, Zhang Y, Fukuda S, Takeshita N, Fischer R. (2019). The spindle pole body of Aspergillus nidulans is asymmetrical and contains changing numbers of gamma-tubulin complexes. J Cell Sci , jcs234799. [DOI] [PubMed] [Google Scholar]
- Gasch A, Wiesner S, Martin-Malpartida P, Ramirez-Espain X, Ruiz L, Macias MJ. (2006). The structure of Prp40 FF1 domain and its interaction with the crn-TPR1 motif of Clf1 gives a new insight into the binding mode of FF domains. J Biol Chem , 356–364. [DOI] [PubMed] [Google Scholar]
- Goode BL, Eskin JA, Wendland B. (2015). Actin and endocytosis in budding yeast. Genetics , 315–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotjahn DA, Chowdhury S, Xu Y, McKenney RJ, Schroer TA, Lander GC. (2018). Cryo-electron tomography reveals that dynactin recruits a team of dyneins for processive motility. Nat Struct Mol Biol , 203–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotjahn DA, Lander GC. (2019). Setting the dynein motor in motion: New insights from electron tomography. J Biol Chem , 13202–13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Farias GG, Mattera R, Bonifacino JS. (2016). Rab5 and its effector FHF contribute to neuronal polarity through dynein-dependent retrieval of somatodendritic proteins from the axon. Proc Natl Acad Sci USA , E5318–E5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghnia M, Cavalli V, Shah SB, Schimmelpfeng K, Brusch R, Yang G, Herrera C, Pilling A, Goldstein LS. (2007). Dynactin is required for coordinated bidirectional motility, but not for dynein membrane attachment. Mol Biol Cell , 2081–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G, Liu B, Zhang J, Zuo W, Morris NR, Xiang X. (2001). The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr Biol , 719–724. [DOI] [PubMed] [Google Scholar]
- Harris SD, Hamer L, Sharpless KE, Hamer JE. (1997). The Aspergillus nidulans sepA gene encodes an FH1/2 protein involved in cytokinesis and the maintenance of cellular polarity. EMBO J , 3474–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, et al. (2015). A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell , 712–723. [DOI] [PubMed] [Google Scholar]
- Huang X, Beullens M, Zhang J, Zhou Y, Nicolaescu E, Lesage B, Hu Q, Wu J, Bollen M, Shi Y. (2009). Structure and function of the two tandem WW domains of the pre-mRNA splicing factor FBP21 (formin-binding protein 21). J Biol Chem , 25375–25387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham RJ, Colwill K, Howard C, Dettwiler S, Lim CS, Yu J, Hersi K, Raaijmakers J, Gish G, Mbamalu G, et al. (2005). WW domains provide a platform for the assembly of multiprotein networks. Mol Cell Biol , 7092–7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Siliciano PG. (1996). Identification of Prp40, a novel essential yeast splicing factor associated with the U1 small nuclear ribonucleoprotein particle. Mol Cell Biol , 960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki S, Holzbaur EL. (1995). Affinity chromatography demonstrates a direct binding between cytoplasmic dynein and the dynactin complex. J Biol Chem , 28806–28811. [DOI] [PubMed] [Google Scholar]
- Karki S, LaMonte B, Holzbaur EL. (1998). Characterization of the p22 subunit of dynactin reveals the localization of cytoplasmic dynein and dynactin to the midbody of dividing cells. J Cell Biol , 1023–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner N, Heimel K, Obhof T, Finkernagel F, Kamper J. (2014). The SPF27 homologue Num1 connects splicing and kinesin 1-dependent cytoplasmic trafficking in Ustilago maydis. PLoS Genet , e1004046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM. (2000) The dynein molecular motor. Biochim Biophys Acta , 60–75. [DOI] [PubMed] [Google Scholar]
- Konzack S, Rischitor PE, Enke C, Fischer R. (2005). The role of the kinesin motor KipA in microtubule organization and polarized growth of Aspergillus nidulans. Mol Biol Cell , 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussel-Andermann P, El-Amraoui A, Safieddine S, Nouaille S, Perfettini I, Lecuit M, Cossart P, Wolfrum U, Petit C. (2000). Vezatin, a novel transmembrane protein, bridges myosin VIIA to the cadherin-catenins complex. EMBO J , 6020–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz JH, Schuchardt I, Straube A, Steinberg G. (2006). A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J , 2275–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon LA, Tokito M, Finklestein JM, Grossman FE, Holzbaur EL. (2004). A direct interaction between cytoplasmic dynein and kinesin I may coordinate motor activity. J Biol Chem , 19201–19208. [DOI] [PubMed] [Google Scholar]
- Liot G, Zala D, Pla P, Mottet G, Piel M, Saudou F. (2013). Mutant Huntingtin alters retrograde transport of TrkB receptors in striatal dendrites. J Neurosci , 6298–6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HL, Osmani AH, Ukil L, Son S, Markossian S, Shen KF, Govindaraghavan M, Varadaraj A, Hashmi SB, De Souza CP, Osmani SA. (2010). Single-step affinity purification for fungal proteomics. Eukaryot Cell , 831–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Xiang X, Lee YR. (2003). The requirement of the LC8 dynein light chain for nuclear migration and septum positioning is temperature dependent in Aspergillus nidulans. Mol Microbiol , 291–301. [DOI] [PubMed] [Google Scholar]
- Lorenzini PA, Chew RSE, Tan CW, Yong JY, Zhang F, Zheng J, Roca X. (2019). Human PRPF40B regulates hundreds of alternative splicing targets and represses a hypoxia expression signature. Rna , 905–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD. (2014). Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science , 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkonian KA, Maier KC, Godfrey JE, Rodgers M, Schroer TA. (2007). Mechanism of dynamitin-mediated disruption of dynactin. J Biol Chem , 19355–19364. [DOI] [PubMed] [Google Scholar]
- Minke PF, Lee IH, Tinsley JH, Bruno KS, Plamann M. (1999). Neurospora crassa ro-10 and ro-11 genes encode novel proteins required for nuclear distribution. Mol Microbiol , 1065–1076. [DOI] [PubMed] [Google Scholar]
- Morris NR. (1975). Mitotic mutants of Aspergillus nidulans. Genet Res , 237–254. [DOI] [PubMed] [Google Scholar]
- Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, Hynes MJ, Osmani SA, Oakley BR. (2006). A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics , 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley BR, Oakley CE, Yoon Y, Jung MK. (1990). Gamma-tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell , 1289–1301. [DOI] [PubMed] [Google Scholar]
- Olenick MA, Holzbaur ELF. (2019). Dynein activators and adaptors at a glance. J Cell Sci , jcs227132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani SA, May GS, Morris NR. (1987). Regulation of the mRNA levels of nimA, a gene required for the G2-M transition in Aspergillus nidulans. J Cell Biol , 1495–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani SA, Pu RT, Morris NR. (1988). Mitotic induction and maintenance by overexpression of a G2-specific gene that encodes a potential protein kinase. Cell , 237–244. [DOI] [PubMed] [Google Scholar]
- Osmani AH, Oakley BR, Osmani SA. (2006). Identification and analysis of essential Aspergillus nidulans genes using the heterokaryon rescue technique. Nat Protoc , 2517–2526. [DOI] [PubMed] [Google Scholar]
- Osmani AH, Osmani SA, Morris NR. (1990). The molecular cloning and identification of a gene product specifically required for nuclear movement in Aspergillus nidulans. J Cell Biol , 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passani LA, Bedford MT, Faber PW, McGinnis KM, Sharp AH, Gusella JF, Vonsattel JP, MacDonald ME. (2000). Huntingtin’s WW domain partners in Huntington’s disease post-mortem brain fulfill genetic criteria for direct involvement in Huntington’s disease pathogenesis. Hum Mol Genet , 2175–2182. [DOI] [PubMed] [Google Scholar]
- Peñalva MA, Zhang J, Xiang X, Pantazopoulou A. (2017). Transport of fungal RAB11 secretory vesicles involves myosin-5, dynein/dynactin/p25 and kinesin-1 and is independent of kinesin-3. Mol Biol Cell , 947–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister KK, Benashski SE, Dillman JF, 3rd, Patel-King RS, King SM. (1998). Identification and molecular characterization of the p24 dynactin light chain. Cell Motil Cytoskeleton , 154–167. [DOI] [PubMed] [Google Scholar]
- Qiu R, Zhang J, Xiang X. (2013). Identification of a novel site in the tail of dynein heavy chain important for dynein function in vivo. J Biol Chem , 2271–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu R, Zhang J, Xiang X. (2018). p25 of the dynactin complex plays a dual role in cargo binding and dynactin regulation. J Biol Chem , 15606–15619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu R, Zhang J, Xiang X. (2019). LIS1 regulates cargo-adapter-mediated activation of dynein by overcoming its autoinhibition in vivo. J Cell Biol , 3630–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson SL, Redwine WB, Vale RD, Carter AP. (2018). The cytoplasmic dynein transport machinery and its many cargoes. Nat Rev Mol Cell Biol , 382–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salogiannis J, Reck-Peterson SL. (2017). Hitchhiking: a non-canonical mode of microtubule-based transport. Trends Cell Biol , 141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Gill SR, Cooper JA, Heuser JE, Schroer TA. (1994). Ultrastructural analysis of the dynactin complex: an actin-related protein is a component of a filament that resembles F-actin. J Cell Biol , 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager MA, Hoang HT, Urnavicius L, Bullock SL, Carter AP. (2014). In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J , 1855–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Carter AP. (2016). Review: Structure and mechanism of the dynein motor ATPase. Biopolymers , 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer TA. (2004). Dynactin. Annu Rev Cell Dev Biol , 759–779. [DOI] [PubMed] [Google Scholar]
- Shi Y. (2017). Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat Rev Mol Cell Biol , 655–670. [DOI] [PubMed] [Google Scholar]
- Spinner MA, Pinter K, Drerup CM, Herman TG. (2020). Vezatin is required for the retrograde axonal transport of endosomes in Drosophila and zebrafish, bioRxiv, 10.1101/2020.02.09.940890. [DOI] [Google Scholar]
- Steinberg G, Peñalva MA, Riquelme M, Wosten HA, Harris SD. (2017). Cell biology of hyphal growth. Microbiol Spectr , doi: 10.1128/microbiolspec.FUNK-0034-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. (2006). Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc , 3111–3120. [DOI] [PubMed] [Google Scholar]
- Twelvetrees AE, Pernigo S, Sanger A, Guedes-Dias P, Schiavo G, Steiner RA, Dodding MP, Holzbaur EL. (2016). The dynamic localization of cytoplasmic dynein in neurons is driven by kinesin-1. Neuron , 1000–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnavicius L, Lau CK, Elshenawy MM, Morales-Rios E, Motz C, Yildiz A, Carter AP. (2018). Cryo-EM shows how dynactin recruits two dyneins for faster movement. Nature , 202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnavicius L, Zhang K, Diamant AG, Motz C, Schlager MA, Yu M, Patel NA, Robinson CV, Carter AP. (2015). The structure of the dynactin complex and its interaction with dynein. Science , 1441–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan KT, Vallee RB. (1995). Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J Cell Biol , 1507–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. (2009). The spliceosome: design principles of a dynamic RNP machine. Cell , 701–718. [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner R, Straube A, Friedrich MW, Steinberg G. (2002). A balance of KIF1A-like kinesin and dynein organizes early endosomes in the fungus Ustilago maydis. EMBO J , 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willins DA, Xiang X, Morris NR. (1995). An alpha tubulin mutation suppresses nuclear migration mutations in Aspergillus nidulans. Genetics , 1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YC, Holzbaur EL. (2014). The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J Neurosci , 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Xiang X, Hammer JA., 3rd (2006). Motor proteins at the microtubule plus-end. Trends Cell Biol , 135–143. [DOI] [PubMed] [Google Scholar]
- Xiang X. (2018). Nuclear movement in fungi. Semin Cell Dev Biol , 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Beckwith SM, Morris NR. (1994). Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc Natl Acad Sci USA , 2100–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Han G, Winkelmann DA, Zuo W, Morris NR. (2000). Dynamics of cytoplasmic dynein in living cells and the effect of a mutation in the dynactin complex actin-related protein Arp1. Curr Biol , 603–606. [DOI] [PubMed] [Google Scholar]
- Xiang X, Osmani AH, Osmani SA, Xin M, Morris NR. (1995). NudF, a nuclear migration gene in Aspergillus nidulans, is similar to the human LIS-1 gene required for neuronal migration. Mol Biol Cell , 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Qiu R, Yao X, Arst HN, Jr, Peñalva MA, Zhang J. (2015). Cytoplasmic dynein and early endosome transport. Cell Mol Life Sci , 3267–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Zuo W, Efimov VP, Morris NR. (1999). Isolation of a new set of Aspergillus nidulans mutants defective in nuclear migration. Curr Genet , 626–630. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Oakley BR. (2009). In vivo analysis of the functions of gamma-tubulin-complex proteins. J Cell Sci , 4218–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Sowa ME, Chen J, Li X, Gygi SP, Harper JW. (2008). An FTS/Hook/p107(FHIP) complex interacts with and promotes endosomal clustering by the homotypic vacuolar protein sorting complex. Mol Biol Cell , 5059–5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Ukil L, Osmani A, Nahm F, Davies J, De Souza CP, Dou X, Perez-Balaguer A, Osmani SA. (2004). Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot Cell , 1359–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Arst HN, Jr, Wang X, Xiang X. (2015). Discovery of a vezatin-like protein for dynein-mediated early endosome transport. Mol Biol Cell , 3816–3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Wang X, Xiang X. (2014). FHIP and FTS proteins are critical for dynein-mediated transport of early endosomes in Aspergillus. Mol Biol Cell , 2181–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Zhang J, Zhou H, Wang E, Xiang X. (2012). In vivo roles of the basic domain of dynactin p150 in microtubule plus-end tracking and dynein function. Traffic , 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zekert N, Fischer R. (2009). The Aspergillus nidulans kinesin-3 UncA motor moves vesicles along a subpopulation of microtubules. Mol Biol Cell , 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng CJ, Kim HR, Vargas Arispuro I, Kim JM, Huang AC, Liu B. (2014). Microtubule plus end-tracking proteins play critical roles in directional growth of hyphae by regulating the dynamics of cytoplasmic microtubules in Aspergillus nidulans. Mol Microbiol , 506–521. [DOI] [PubMed] [Google Scholar]
- Zhang K, Foster HE, Rondelet A, Lacey SE, Bahi-Buisson N, Bird AW, Carter AP. (2017a). Cryo-EM Reveals How Human Cytoplasmic Dynein Is Auto-inhibited and Activated. Cell , 1303–1314.e1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gao X, Manck R, Schmid M, Osmani AH, Osmani SA, Takeshita N, Fischer R. (2017b). Microtubule-organizing centers of Aspergillus nidulans are anchored at septa by a disordered protein. Mol Microbiol , 285–303. [DOI] [PubMed] [Google Scholar]
- Zhang J, Han G, Xiang X. (2002). Cytoplasmic dynein intermediate chain and heavy chain are dependent upon each other for microtubule end localization in Aspergillus nidulans. Mol Microbiol , 381–392. [DOI] [PubMed] [Google Scholar]
- Zhang J, Li S, Fischer R, Xiang X. (2003). Accumulation of cytoplasmic dynein and dynactin at microtubule plus ends in Aspergillus nidulans is kinesin dependent. Mol Biol Cell , 1479–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li S, Musa S, Zhou H, Xiang X. (2009). Dynein light intermediate chain in Aspergillus nidulans is essential for the interaction between heavy and intermediate chains. J Biol Chem , 34760–34768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Qiu R, Arst HN, Jr, Peñalva MA, Xiang X. (2014). HookA is a novel dynein-early endosome linker critical for cargo movement in vivo. J Cell Biol , 1009–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wang L, Zhuang L, Huo L, Musa S, Li S, Xiang X. (2008). Arp11 affects dynein-dynactin interaction and is essential for dynein function in Aspergillus nidulans. Traffic , 1073–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yao X, Fischer L, Abenza JF, Peñalva MA, Xiang X. (2011). The p25 subunit of the dynactin complex is required for dynein-early endosome interaction. J Cell Biol , 1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhuang L, Lee Y, Abenza JF, Peñalva MA, Xiang X. (2010). The microtubule plus-end localization of Aspergillus dynein is important for dynein-early-endosome interaction but not for dynein ATPase activation. J Cell Sci , 3596–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Obhof T, Schneider K, Feldbrugge M, Nienhaus GU, Kamper J. (2018). Cytoplasmic transport machinery of the SPF27 homologue Num1 in Ustilago maydis. Sci Rep , 3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.