Abstract
Yeast cells, when exposed to stress, can enter a protective state in which cell division, growth, and metabolism are down-regulated. They remain viable in this state until nutrients become available again. How cells enter this protective survival state and what happens at a cellular and subcellular level are largely unknown. In this study, we used electron tomography to investigate stress-induced ultrastructural changes in the cytoplasm of yeast cells. After ATP depletion, we observed significant cytosolic compaction and extensive cytoplasmic reorganization, as well as the emergence of distinct membrane-bound and membraneless organelles. Using correlative light and electron microscopy, we further demonstrated that one of these membraneless organelles was generated by the reversible polymerization of eukaryotic translation initiation factor 2B, an essential enzyme in the initiation of protein synthesis, into large bundles of filaments. The changes we observe are part of a stress-induced survival strategy, allowing yeast cells to save energy, protect proteins from degradation, and inhibit protein functionality by forming assemblies of proteins.
INTRODUCTION
To survive in a constantly changing world, cells require mechanisms to cope with environmental fluctuations. For instance, baker’s yeast, Saccharomyces cerevisiae, responds to sudden changes in nutrient abundance by adjusting its metabolism and growth rate. Under starvation conditions, yeast proliferation is efficiently shut down in favor of an energy-saving state that allows the cells to remain viable with slow metabolic activity (Choder, 1993; Werner-Washburne et al., 1993; Ireland et al., 1994; Gray et al., 2004). However, the structural changes that happen in the cells upon entry into this energy-saving state remain unclear.
The organization of yeast cytoplasm is very dynamic and changes continuously, not only following the different stages of the cell cycle, but also in response to stress conditions such as heat shock, osmotic stress, and nutrient starvation (Meaden et al., 1999; Winderickx et al., 2003; Mourão et al., 2014; Petrovska et al., 2014; Munna et al., 2015; Joyner et al., 2016; Munder et al., 2016). The transition from a growing to a dormant state, induced through a severe lack of nutrients, is coupled to various physiochemical changes, such as lowered cytosolic pH, reduced cell volume, and decreased molecule mobility in the cytoplasm (Ashe et al., 2000; Joyner et al. 2016, Munder et al., 2016), as well as physiological changes, such as reduction in protein synthesis and in enzymatic activities (Werner-Washburne et al., 1993; Gray et al., 2004). Because the majority of metabolic reactions and signaling processes take place in the cytoplasm, stress-induced changes in its physical and chemical properties might be required for the cell to transition into a survival state. However, how the cytoplasm reorganizes under sudden stress conditions is largely unclear, and comprehensive structural investigations of cytoplasmic changes occurring are missing.
Live-cell imaging has shown that several fluorescently tagged enzymes form assemblies in the cytoplasm of dormant cells (Narayanaswamy et al., 2009; Liu, 2010; Noree et al., 2010; Petrovska et al., 2014; Prouteau et al., 2017; Riback et al., 2017). Electron microscopy imaging of three of these metabolic enzymes, cytidine triphosphate synthetase (CtpS, Barry et al., 2014), glutamine synthetase (Gln1, Petrovska et al., 2014), and target of rapamycin complex 1 (TORC1, Prouteau et al., 2017), revealed that they assemble into filaments and that the filaments organize in bundles, visible in fluorescence microscopy. In all three cases, the polymerization process is reversible, and it is coupled to the inactivation of the enzymatic activities. These findings suggest that this stress-induced polymerization could be a common mechanism for down-regulating the activity of essential metabolic enzymes under adverse environmental conditions.
In this study, we used electron tomography (ET) to investigate the reorganization of the cytoplasm of energy-depleted yeast cells, and we used correlative light and electron microscopy (CLEM) to study the behavior of the essential enzyme eIF2B (eukaryotic translation Initiation Factor 2B), a guanine nucleotide exchange factor and key initiator of protein synthesis. We show that energy-depleted yeast cells undergo a dramatic reorganization of the cytoplasm that involves the formation of distinct membrane-bound and membraneless organelles, along with an almost twofold increase in macromolecular crowding. Our results show that the eIF2B decamer self-assembles into ordered bundles of filaments. We propose that eIF2B compartmentalization upon energy depletion could be a mechanism for storing and protecting the enzyme from denaturation, disassembly, or vacuolar degradation and could possibly contribute to the down-regulation of its enzymatic activity. These results are consistent with a model in which cytoplasmic reorganization and enzyme regulation via self-assembly are an important survival strategy that enable cells to cope with extreme environmental conditions and stress.
RESULTS
Energy depletion is accompanied by a reorganization of storage and membrane lipids
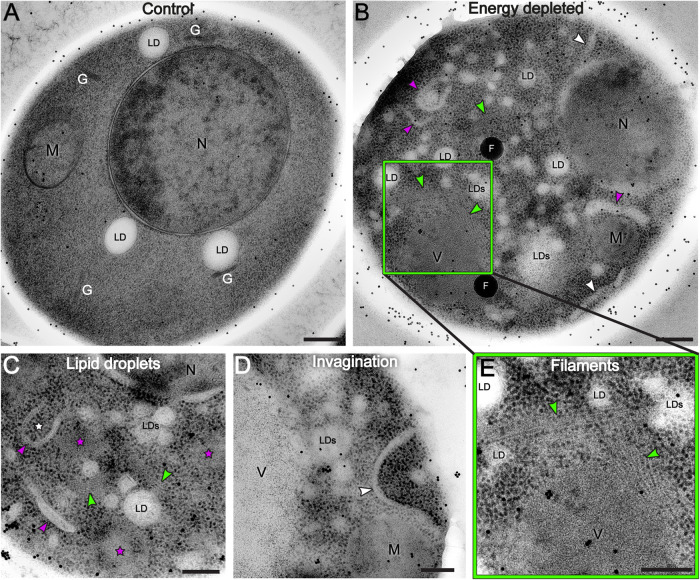
To investigate how the cytoplasm of energy-depleted yeast cells undergoes structural rearrangements, we have been mimicking sudden glucose starvation by feeding yeast cells a nonhydrolyzable analogue of glucose, 2-deoxyglucose, and by blocking mitochondrial respiration with an antimycin, de facto depriving cells of any possible source of ATP. After 15 min of treatment, we cryofixed the cells by high-pressure freezing to ensure the rapid immobilization of all macromolecular components in the cytoplasm, thus avoiding potential structural alterations that might occur during the slower process of standard chemical fixation (Frank, 2006). Subsequently, we imaged 70 to 150 nm–thick sections of control and energy-depleted yeast cells by TEM and dual-axis electron tomography. By comparing the ultrastructure of log-phase growing cells with that of energy-depleted cells, we found pronounced structural modifications between the two conditions (Figure 1; Supplemental Video 1 for controls; Supplemental Videos 2 and 3 for energy depletion), as energy depletion was characterized by extensive rearrangements of many cytoplasmic components, in particular lipids. We observed an increased number of small cytoplasmic lipid droplets (LDs), recognizable by their amorphous non-electron-dense content. They appeared fragmented and irregular (Figure 1, B and C; Supplemental Videos 2 and 3) in comparison with the fewer and larger LDs visible in control cells (Figure 1A; Supplemental Video 1). The average number of LDs in control cells was ∼3, with an average diameter of ∼160 nm. In energy-depleted cells, the number rose to ∼9 LDs per cell and the diameter was reduced to ∼120 nm (six cells were analyzed per condition—Supplemental Figure S1).
FIGURE 1:
TEM images reveal cytoplasm reorganization in dormant yeast cells. A log-phase growing yeast cell (A) is compared with energy-depleted yeast cells (B–D) showing that the cytoplasm undergoes drastic reorganization upon sudden energy depletion. The vacuole is not visible in panel (A) because it is not included in this particular section of the cell. The dispersed black dots in the images are gold beads of 15 nm diameter used as alignment fiducials during tomography reconstruction. (B–D) Energy-depleted cells contain numerous fragmented lipid droplets (LDs; smaller droplets are not labeled), nascent autophagosomes (white stars), and elongated membranous structures (magenta arrows). Ribosomes appear more densely packed in energy-depleted cells than in the control cell, and areas of ribosome exclusion are also visible (magenta stars in C). (B,C) Elongated invagination of the cell membrane (white arrows) (B,D) and filamentous structures are visible in several parts of the cell (green arrows) (B,C, inset E). N = nucleus; M = mitochondria; G = Golgi; LD(s) = lipid droplet(s); V = vacuole; F = fiducial beads. Scale bars: A and B = 300 nm; C–E = 200 nm.
Video S1.
3D tomographic reconstruction of a non‐stressed yeast cell. Scale bar= 500 nm
Video S2.
3D tomographic reconstruction of an energy‐depleted yeast cell. eIF2B is unlabelled and over‐expressed. Scale bar = 500 nm
Video S3.
3D tomographic reconstruction of an energy‐depleted yeast cell. eIF2B is unlabelled and endogenously expressed. Scale bar = 500 nm
Membrane lipids also underwent a great rearrangement through the accumulation of elongated membranous structures and the appearance of deep plasma membrane invaginations in the cytoplasm (Figure 1, B and D; Supplemental Videos 2 and 3). The invaginations extended to 1 µm in length, with diameters ranging from 20 to 100 nm.
Interestingly, in energy-depleted cells, we also observed double-layered membrane vesicles containing mostly ribosomes (Figure 1, B and C; Supplemental Figure S2 and Supplemental Video 3; see Figure 4, B–D, in The sfGFP tag does not interfere with eIF2B polymerization). These resembled autophagosome vesicles, sometimes captured in an open, pre-fusion conformation (Figure 1, B and C). These structures could indicate early steps of the autophagy process, a process that is constitutive in cells, and can also be induced by stress (Noda and Ohsumi, 1998; Jin and Klionsky, 2014).
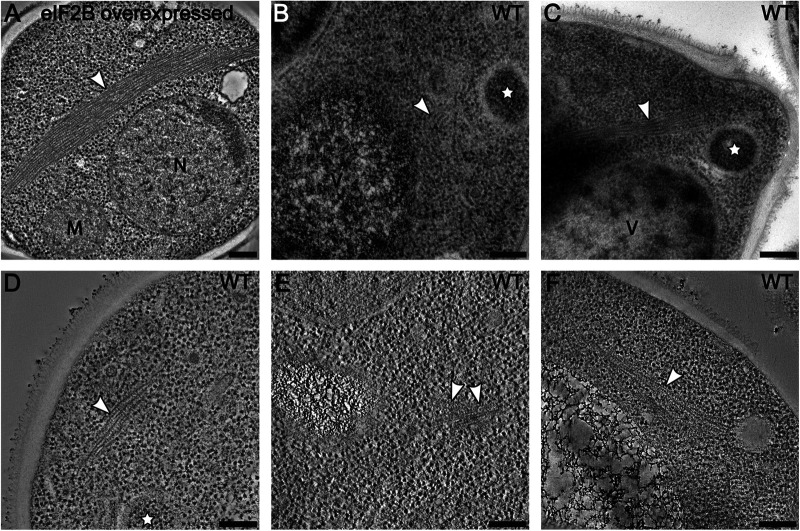
FIGURE 4:
Filament formation is independent of sfGFP tagging and overexpression of eIF2B. Average images of 20 tomographic slices of WT yeast cells showing (A) overexpressed eIF2B forming large bundles of filament in the cytoplasm and (B–F) endogenously expressed eIF2B forming smaller bundles of filaments. (B–D) White stars label autophagosomes, recognized as double-membrane vesicles containing mostly ribosomes. M = mitochondrion; N = nucleus; V = vacuole. Scale bars = 200 nm.
Ribosome density and distribution change in energy-deprived cells
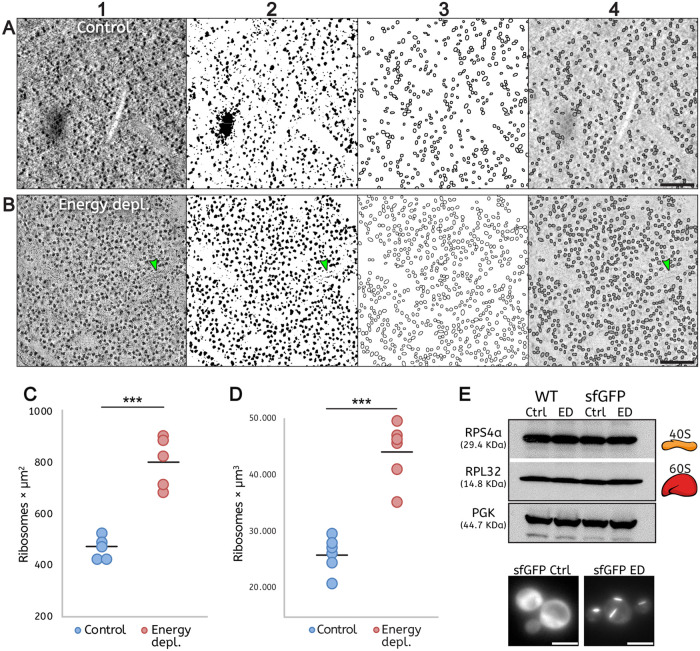
A previous study of energy-depleted yeast reported increased mechanical stability of the cytoplasm and a ∼7% reduction in cell volume (Munder et al., 2016). Based on these observations, it was proposed that macromolecular crowding increases in stressed yeast cells. However, a direct quantification of cytoplasm density was never obtained. Because ribosomes take up a significant fraction of the cellular interior (Duncan and Hershey, 1983; Warner 1999), an accurate quantification of ribosome density can be used to account for changes in cytoplasmic crowding. To use ribosome density to quantify macromolecular crowding, we first verified that ribosome number does not change significantly between log-phase growing and dormant (energy-depleted) cells. Immunoblot analysis on ribosomal proteins of both 40S and 60S subunits (RPS4a and RPL32, respectively) showed no significant change in the quantity of ribosomes between untreated and energy-depleted yeast cells (Figure 2E).
FIGURE 2:
Quantification of ribosome density demonstrates macromolecular crowding in energy-depleted cells. (A, B) Automated quantification of ribosomes involving sequential 1) filtering, 2) binarization, 3) segmentation, and 4) particle detection on single tomographic slices of cells: A, under control conditions, and B, after energy depletion. Green arrows in panels B indicate a short filament. (C, D) The ribosome number is increased almost twofold in the energy-depleted cells. (C) Ribosome density measured in tomographic slices. Five tomograms were analyzed per condition. ***p-value = 0.0001. (D) Ribosomes density is measured from tomographic volumes and assessed by manual counting. Six tomograms were analyzed per condition. ***p-value = 0.0001. (E) Western blot shows no significant change in the numbers of RPS4a (40S) and RPL32 (60S) ribosomal subunits between log-phase growing (Ctrl) and energy-depleted (ED) cells in either WT or sfGFP-tagged eIF2B strains. Cells expressing sfGFP-tagged eIF2B were imaged immediately before Western blot analysis to validate the log-phase growth: diffuse GFP signal in the control condition (sfGFP Ctrl) and condensed GFP signal in energy depletion (sfGFP ED). Scale bars: A, B = 200 nm; E = 5 μm.
In our TEM tomograms, ribosomes appeared as extremely electron-dense and were clearly distinguishable as dark dots in the cytoplasm, due to the presence of negatively charged rRNAs that are efficiently stained by contrast reagents used for sample preparation (Tzaphlidou et al., 1982; Figure 2, A1 and B1; Supplemental Figure S3, A and B). To quantify the ribosome density, we took advantage of their sharp contrast and developed a FIJI macro to automatically detect and count them in tomographic slices of six randomly selected cells per condition (Schindelin et al., 2012). The images were processed by filtering, binarization, segmentation, and particle detection (Figure 2, A and B). The accuracy of the automated quantification workflow was tested by comparison with ground truth data generated by manual counting of the ribosomes in single tomographic slices of five tomograms per condition (Supplemental Figure S3). Ribosomes were manually counted in five cytoplasmic squares in every tomogram, avoiding areas that contained mainly large organelles such as vacuoles, nuclei, or mitochondria. Both quantifications showed that ribosome density increased almost twofold in energy-depleted cells (Figure 2, C and D; Supplemental Figure S3, C).
Membraneless compartments in energy-depleted cells have two distinct morphologies
Recent studies have proposed macromolecular crowding as one of the possible physical conditions to promote condensation of fluorescently labeled enzymes in the cytoplasm of energy depleted yeast cells (Petrovska et al., 2014; Joyner et al. 2016; Munder et al., 2016). We observed that, along with the dramatic increase in ribosome density, energy-depleted cells in our study showed also cytoplasmic areas from where ribosomes and larger macromolecular complexes were clearly excluded. These areas were never observed in control cells and showed a neat separation from the rest of the cytoplasm without being delimited by membranes. In particular, two main morphologies could be distinguished: some areas appeared as amorphous aggregate-like bodies (Figure 1C; Supplemental Figure S5C—orange stars), other areas appeared as ordered self-assembly structures mostly composed of filaments organized in bundles (Figure 1, B and E; Figure 3, C, F–H—green arrows; Supplemental Videos 2 and 3—white arrows). Neither type of compartments showed any obvious specific distribution in the cytoplasm, nor any specific interaction with organelles or cellular components. The EM data suggest that these membrane-free areas of ribosome exclusion could represent specialized compartments for specific components of the cytoplasm, such as proteins linked to RNA and/or other enzymes, a possible mechanism of stress-induced segregation of such molecular components.
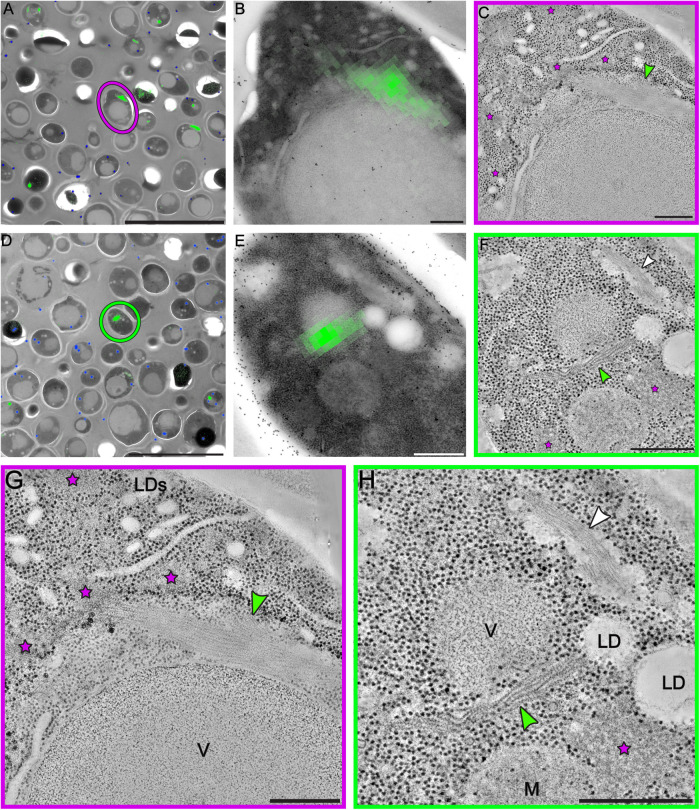
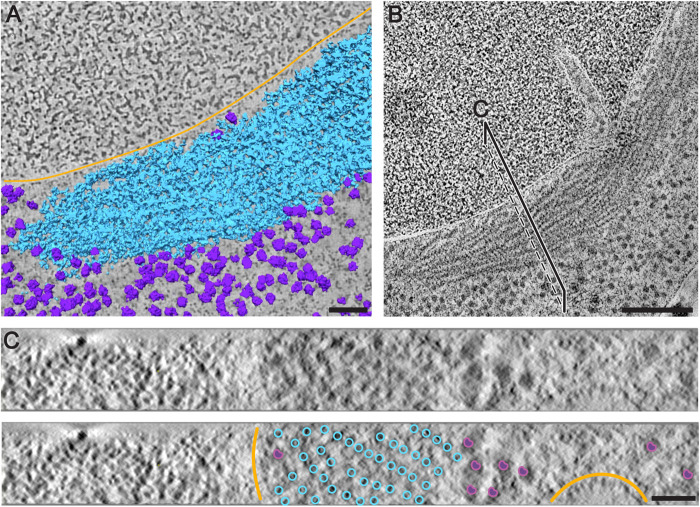
FIGURE 3:
Correlative fluorescence and electron microscopy analysis reveals organization of sfGFP-tagged eIF2B into bundles of parallel filaments. (A, D) The fluorescent signal of sfGFP-eIF2B is overlaid on low-magnification TEM images of energy-depleted yeast cells previously embedded in Lowicryl HM-20 and then sectioned. The magenta circle in A and green circle in D highlight cells that were selected for further tomographic analysis. (B, E) Close-up of the cells highlighted in A and D, respectively. (C, F, G, H) Slices through tomographic reconstructions of the same cells as in B and E. (G, H) Magnified views of the tomographic slices shown in C and F, respectively. Bundles of filamentous structures (green arrows) correspond to the fluorescence signal. eIF2B organizes in ordered, non–membrane bound arrays of filaments. Energy-depleted cells also contain other non–membrane bound compartments that do not include ribosomes. Some have an amorphous appearance (magenta stars), whereas others comprise filamentous structures, H (white arrow). These filaments, which do not display a fluorescent signal, have a different morphology than the eIF2B filaments and their protein content is unknown. LD(s) = lipid droplet(s); M = mitochondrion; V = vacuole. Scale bars: A, D = 10 μm; B, C, G = 200 nm; E, F, H = 500 nm.
eIF2B organizes rapidly into highly ordered compartments upon energy depletion
Our TEM observations are in agreement with fluorescence microscopy studies reporting that a large number of enzymes, including eIF2B, are able to form assemblies upon nutrient/energy deprivation (Campbell et al., 2005; Narayanaswamy et al., 2009; Noree et al., 2010; Liu, 2010). However, so far only Gln1, CtpS, and TORC1 enzymes have been shown to self-assemble into highly ordered polymers under such stressful conditions by TEM (Barry et al., 2014; Petrovska et al., 2014; Prouteau et al., 2017). It is not known whether other enzymes assemble into polymers and whether this is a common occurrence in the cytoplasm of energy-depleted cells.
The eIF2B translation initiation factor is a key guanine nucleotide exchange factor highly conserved throughout eukaryotes that plays a fundamental role in the initiation of protein expression (Kurat et al., 2006; Hinnebusch and Lorsch, 2012; Gordiyenko et al., 2014, 2019; Jennings and Pavitt 2014). It is known to form elongated and reversible fluorescent assemblies in yeast and mammalian cells under different stress conditions (Campbell et al., 2005; Noree et al., 2010; Nüske et al., 2018; Hodgson et al., 2019). Therefore, we tested whether eIF2B could be a constituent of one of the highly ordered membraneless compartments we described above. To test this hypothesis, we performed CLEM on energy-depleted yeast cells overexpressing a sfGFP-tagged version of the eIF2B enzyme. First, we followed the sfGFP–eIF2B signal in live-cell fluorescence microscopy (Supplemental Figure S4; Supplemental Video 4). In agreement with a previous study (Noree et al., 2010), eIF2B enzymes showed a diffuse cytoplasmic signal in log-phase growing cells (Supplemental Figure S4A). About 15 min after energy depletion, most of the eIF2B molecules were concentrated into foci and elongated structures, and cell growth and division were heavily slowed, if not arrested (Supplemental Figure S4, B and C). This condition lasted until replenishment of ATP, when the fluorescence signal promptly diffused again, and cells quickly reentered the cell cycle (∼15 min after energy depletion; Supplemental Figure S4, D and E). The reversibility of the condensation process has also been described for self-assemblies made by CtpS (Barry et al., 2014), Gln1 (Petrovska et al. 2014), and TORC1 (Prouteau et al., 2017), with clear regulatory functions for all the three enzymes.
Video S4.
The fluorescent signal of eIF2B is distributed differently between stressed yeast cells and log‐phase growing cells. According to the fluorescent signal, the sfGFP‐labeled Gcn3 (α) subunit of eIF2B present a diffuse distribution throughout the cytoplasm in log‐phase growing and dividing cells. On the contrary, the fluorescence is concentrated into condensed foci‐like or elongated structures in stressed cells that have been energy depleted for 15 minutes at pH 5.5. Scale bar = 10 µm
After imaging the fluorescent signal, we prepared yeast cells overexpressing sfGFP–eIF2B, both untreated and energy-depleted cells, for electron microscopy imaging. Then we correlated the fluorescence signals of the condensed eIF2B with the TEM images and located cell sections containing the labeled structures of our interest (Figure 3; Supplemental Figure S5). On those sections, we acquired dual-axis electron tomograms (Mastronarde 1997). The tomograms of energy-depleted cells showed that eIF2B polymerizes into a specific type of membraneless cytoplasmic compartment that is composed of long zigzagged filaments packed in big bundles (Figure 3, C, F–H). The morphology of eIF2B filaments and bundles was distinct from the morphology of other non–membrane bound compartments of stressed cells (Figure 3, F and H—green and white arrows, respectively). A single energy-depleted cell could contain several eIF2B bundles, with overexpressing cells showing larger bundles (Supplemental Figure S5). eIF2B filaments were never observed in untreated cells from the same strain and cell culture (Supplemental Video 1).
In all analyzed tomograms of stressed cells, macromolecular complexes in the size range of ribosomes were excluded from the space occupied by the eIF2B bundles. Additionally, eIF2B filaments and bundles were never observed to be associated with autophagosomes, which often engulfed cytoplasmic components in energy-depleted cells (Figure 1, B and C; Figure 4, B–D; Supplemental Figure S2—white stars; Supplemental Video 3).
The sfGFP tag does not interfere with eIF2B polymerization
It has been reported that fluorescent proteins have a natural dimerizing affinity and tend to form aggregates or even higher-order structures (Zacharias et al., 2002; Snapp et al., 2003). This might pose a problem for applications in which fluorescent proteins are used to visualize the cellular localization, dynamics, and oligomeric state of a protein. Here, we used three distinct approaches to test whether eIF2B self-assembly happens intrinsically or might be caused or enhanced by the GFP tag: 1) we used the nondimerizing version of GFP (sfGFP, Costantini et al., 2012); 2) we imaged eIF2B by immunofluorescence using the short HA polypeptide chain as a tag; 3) we acquired electron tomograms of the native untagged eIF2B in energy-depleted wild-type (WT) yeast.
For the immunofluorescence experiments, we labeled with the HA tag two distinct subunits of the eIF2B complex, the regulatory subunit Gcn3 (α) and the catalytic subunit Gcd1 (γ), and we stained the samples with antibodies against the HA tag (Supplemental Figure S6). The analysis confirmed that eIF2B also formed assemblies upon energy depletion when a different tag than sfGFP was used.
TEM analysis of energy-depleted WT cells using untagged eIF2B, either endogenously expressed or overexpressed, revealed bundles of filaments showing the same morphology observed in CLEM experiments (Figure 4). As observed for sfGFP-tagged cells, bundles of untagged eIF2B were larger in overexpressing strains (Figure 4A) than in WT cells (Figure 4, B–F). These results demonstrate that the formation of eIF2B filaments and bundles is an intrinsic property of the enzyme and that the sfGFP tag is not involved in this process. Additionally, we observed that the overexpression of eIF2B does not affect the mechanism of filament formation but leads only to bulkier bundles.
eIF2B filaments comprise repeats of decameric units
To investigate the 3D organization of the filaments in the bundle, we measured the filaments, length and the distance between filaments from the raw tomograms (Figure 5; Supplemental Video 5). The filaments’ length varied from ∼45 nm to ∼850 nm. By dividing the length of the filaments by the known length (14 nm) of the decamer from the existing cryo-EM structure of Adomavicius et al. (2019) and Gordiyenko et al. (2019), we determined that shorter filaments comprise three or four copies of eIF2B decamers. Short filaments were usually observed at the periphery of a bundle. Filaments were mostly aligned in regular rows in the bundle, with a between-row spacing of ∼13 nm and a within-row spacing of ∼26 nm (Figure 5C; Figure 6A). Diameter, periodicity, and spacing of eIF2B filaments in the bundle differed from those of previously characterized enzymatic polymers, including those made of other metabolic enzymes such as CtpS, Gln1, and TORC1 (Barry et al., 2014; Petrovska et al., 2014; Prouteau et al., 2017). In stressed cells, we also observed filaments with a morphology and subunit repeat (∼6 nm) similar to the ones described for Gln1 (Petrovska et al., 2014), with smaller diameter, shorter between-filament spacing, and smoother longitudinal striations than eIF2B filaments (Figure 3, F and H—white arrows).
FIGURE 5:
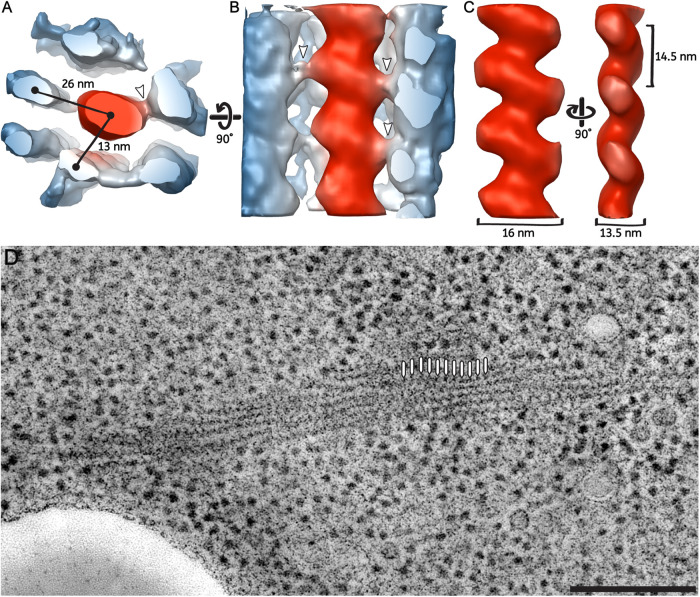
Segmentation and cross-sectional views of a eIF2B bundle showing the ordered arrangement of filaments. (A) Segmentation of membranes (orange), ribosomes (purple), and eIF2B filaments (cyan) of a cell overexpressing sfGFP-tagged eIF2B. (B) Tomographic slice through a larger field of view of the same tomogram. eIF2B filaments are densely packed and parallel to each other in the bundle. The black line indicates the cutting plane through the bundle corresponding to the cross-sectional view in C. (C) eIF2B filaments follow a regular pattern with an average center-to-center inter-filament spacing of approximately 20 nm. The positions of the filaments in the cross section have been annotated according to the segmentation. Scale bars: A = 50 nm; B = 200 nm; C = 40 nm.
FIGURE 6:
3D organization of parallel filaments in the eIF2B bundle reveals lateral connections. Subtomogram averaging of three repeating eIF2B units along a filament. (A, B) Neighboring filaments (in blue) are still visible in the average, which is indicative of their roughly consistent position around a central filament (in red). The center-to-center distance between the central filament and the surrounding ones is ∼26 nm for filaments in the same row and ∼13 nm for filaments in parallel rows. Lateral connections are visible between the central filament and the surrounding ones (white arrows). (B, C) The longitudinal view of the central filaments has a zigzag-shaped structure. The central filament shows a 14.5-nm repeat, a long diameter of 16 nm, and a short diameter of 13.5 nm. (D) Average of 10 tomographic slices showing the eIF2B bundle used for the 3D model reconstruction of the filaments and the periodicity at which particles have been picked (white lines). Scale bars: A, B, C = 10 nm; D = 200 nm.
Video S5.
Segmentation of a bundle of eIF2B filaments in a reconstructed tomogram using Super Region Volume Segmentation software [Luengo et al., 2017]. Automated segmentation of ribosomes (purple) and eIF2B filaments (cyan) in the tomographic reconstruction of a cell overexpressing sfGFP‐tagged eIF2B.
Because of their apparent repeating structure, eIF2B filaments could represent polymers of a single protein complex. We used a subtomogram-averaging approach to obtain a 3D model of the repeating unit. The 3D model showed a zigzag pattern with a 14.5-nm repeat (Figure 6C), in agreement with the shape of the pattern visible in the raw tomograms (Figures 3, 5, 6). A single filament measured ∼16 nm for the long diameter and 13.5 nm for the short diameter (Figure 6C). Contact points between central and neighboring filaments were visible in raw tomograms and in the 3D model (Figure 6, A and B—white arrows) as they protruded from both sides of the zigzag shape. The regularity of the interfilament packing, where a central filament is surrounded by six or seven equidistant neighboring ones (Figure 5; Figure 6A), suggested that the observed lateral connections could contribute to keeping distance between molecules and a regular pattern of filaments in the bundle.
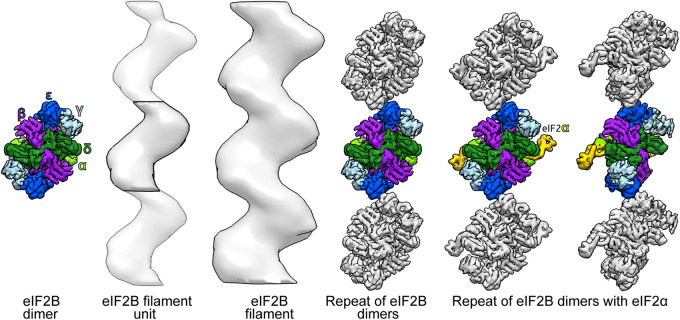
Although the sample preparation protocol used for ET in this study does not allow for the generation of high resolution 3D models by subtomogram averaging (Supplemental Figure S7), we extracted the volume of the eIF2B decamer from the cryoEM map of eIF2B/eIF2αP complex (Adomavicius et al., 2019; Gordiyenko et al., 2019), and we compared a gaussian filtered version of it with our 3D model of the filament. The comparison showed that periodicity and curvatures of three decameric eIF2B units were well accommodated in our 3D model (Figure 7). The proposed eIF2B decamers arrangement shows a similar zigzag pattern as the filaments in our tomograms. With the proposed stacking the only point of contact between units appears to be through Gcd6 ε-subunits.
FIGURE 7:
Proposed organization of eIF2B decamers in the filament. The repeating units in the eIF2B filament have a size and overall shape similar to the cryo-EM structure of the eIF2B decamer (Adomavicius et al., 2019; Gordiyenko et al., 2019). The proposed stacking of three cryo-EM structures of eIF2B is compatible with the subtomogram average of the filament, which contains three eIF2B units. This suggests that the filaments are generated by polymerization of eIF2B decamers. Light green = Gcn3 (α) subunit; purple = Gcd7 (β) subunit; light blue = Gcd1 (γ) subunit; green = Gcd2 (δ) subunit; blue = Gcd6 (ε) subunit of eIF2B; and yellow = Sui2 (α) subunit of eIF2.
DISCUSSION
In this paper, we demonstrate that entrance of yeast cells into a stress survival state is associated with a broad reorganization of molecules and the formation of structures in the cytoplasm. We reveal that the reorganization involves a massive rearrangement of membranes and lipids as well as the formation of compartments that are not surrounded by membranes.
Lipid droplets change their morphology upon energy depletion
The observed changes in lipid droplet (LD) morphology might be indicative of the dramatic effects that the scarcity of nutrients/energy has on the regulation of lipid metabolism (Rußmayer et al., 2015). In yeast cells, LDs are particularly dynamic throughout the growth phase and depending on the nutritional status of the cell (Kurat et al., 2006; Bozaquel-Morais et al., 2010). Cells that progressively reach the stationary phase and experience gradual nutrient depletion are known to accumulate and enlarge LDs, due to their shift from a lipolytic to a lipogenic metabolism, allowing them to produce basal levels of energy from fatty acids for cellular maintenance (Madeira et al., 2015). Upon replenishment of glucose, large amounts of sterols and fatty acids are mobilized and, as a result, LDs are decreased in number and size (Kurat et al., 2006). However, cells that are exposed to gradual glucose reduction seem to be unable to survive in the long term if the conditions are not reversed (Seo et al., 2017; Jarc and Petan, 2019).
In contrast, yeast cells that undergo sudden glucose starvation are known to consume lipid droplets and can survive during prolonged nutrient stress (Seo et al., 2017; Jarc and Petan, 2019). In our experiments, yeast cells were grown until early–mid log-phase and then exposed to acute energy starvation, without previous accumulation of LDs. Because starvation is known to switch yeast cell metabolism toward β-oxidation of fatty acids (Jarc and Petan, 2019), which yields more energy per gram than carbohydrates such as glucose (Gray et al. 2004; Kurat et al., 2006), LDs present in the cell are strongly mobilized and appeared smaller and fragmented, as a possible consequence of activation of lipid metabolism. In addition, LDs fragmentation could contribute to the increase in total surface area accessible to lipases, enhancing lipolytic activity and contributing to the production of energy under stress conditions, as has been shown in yeast and animal cells (Ariotti et al., 2012; Hashimoto et al., 2012; Seo et al., 2017; Thiam and Beller, 2017; Jarc and Petan, 2019).
It has been shown that lipid biosynthesis enzymes, such as fatty acid synthetase (FAS), are sequestered into distinct foci and down-regulated upon sudden glucose starvation in favor of a lipolytic metabolism (Suresh et al., 2015). Interestingly, FAS foci do not represent accumulation of misfolded proteins, as the sequestered enzymes are functional in vitro (Suresh et al., 2015). Therefore, the formation of FAS foci, similarly to what we observed for eIF2B in this study, indicates the presence of a mechanism that regulates enzymatic activity through the sequestration of enzymes without affecting their functional status. This mechanism allows the cells to return quickly to lipogenic metabolism upon glucose replenishment and release of the enzymes from the foci (Suresh et al., 2015).
Plasma membrane invaginations accumulate as a consequence of cell shrinkage
The pronounced plasma membrane invaginations present in energy-depleted cells were morphologically similar to those observed in yeast cells under severe hyperosmotic stress. This stress condition is usually associated with dehydration and rapid shrinkage of the cell volume (Morris et al., 1986; Blomberg and Adler, 1992; Gervais and Marechal, 1994; Martinez de Maranon et al., 1996; Slaninova et al., 2000; Simonin et al., 2007; Dupont et al., 2010). Concomitantly, due to the poor compressibility of biological membranes (Thiery et al., 1976), cell shrinkage is associated with wrinkling of the plasma membrane (Adya et al., 2006). In agreement with our observations, the formation of similar plasma membrane invaginations, called eisosomes, was recently described in yeast cells upon shrinkage induced by glucose starvation (Appadurai et al., 2020). Therefore, the formation of plasma membrane invaginations that we observed in energy-depleted cells can well be the consequence of the reduction in cell volume attributed to sudden water loss that follows acute energy depletion in S. cerevisiae (Munder et al., 2016).
Moreover, upon energy replenishment, the cell surface needs to increase rapidly, to allow cells to return to their normal volume. Such rapid expansion of the plasma membrane requires membrane reservoirs that provide surface area and buffer membrane tension. We speculate that the accentuated depth of plasma membrane invaginations in starved cells allows the rapid shrinkage of the membrane into reservoirs, which then provide additional surface for prompt reexpansion of the plasma membrane upon energy replenishment.
Ribosome density is a measure of increased macromolecular crowding
Munder et al. (2016) measured a dramatic decrease in particle mobility in the cytoplasm of energy-depleted cells, which was proposed to result from increased molecular crowding and condensation of the cytoplasm. However, the measured ∼7% reduction in the cell volume would hardly be sufficient to induce this pronounced effect on particle mobility. Therefore, we directly quantified ribosomes in 3D electron tomograms to verify and measure changes in molecular crowding between control and energy depleted yeast cells. While the total ribosome number remained unchanged between the two conditions, we observed an almost twofold increase in ribosome density in energy-depleted cells. Based on these data, we estimate a theoretical cell volume reduction of about 42%, which is far more pronounced than that measured by Munder et al. (2016). This discrepancy could be explained by the concomitant enlargement of the vacuole, which has previously been reported to occur in response to starvation (Desfougeres et al., 2016; Joyner et al., 2016) and also during the progression of autophagy (Noda and Ohsumi, 1998). Additionally, the exclusion of ribosomes from areas occupied by membraneless compartments formed in energy-depleted cells is likely to contribute to the increased ribosome density. On the other hand, the mere redistribution of eIF2B and other enzymes from a diffused form to the compartments should not influence the overall crowding in starved cells. Our quantification is in line with previously measured cytoplasmic volume reduction of 30% in yeast cells upon glucose starvation (Joyner et al., 2016). Therefore, we propose that a combination of cell volume reduction and vacuole enlargement is most likely responsible for the increase in molecular crowding observed in starved yeast cells. Regardless of the exact sequence of events causing the changes in the physical–chemical properties of the cytoplasm, our analysis provides a direct quantification of the contribution of molecular crowding in yeast cells.
Increased macromolecular crowding is associated with assembly and compartment formation
Changes in molecular crowding are thought to have profound effects on cytoplasm organization and on cell physiology. In vitro experiments show that the assembly of individual enzymatic species is triggered by the addition of crowding agents (Petrovska et al., 2014; Woodruff et al., 2017). Consequently, crowding has been proposed to play an important role in the formation and stabilization of assemblies and membraneless compartments in cells. The increased crowding of stressed yeast cells that we report here could indeed represent one of the physical parameters that promote the formation of protein assemblies, such as bundles of eIF2B filaments. The formation of eIF2B filaments requires the diffusion of soluble components and the incorporation of these components into a nucleation center. Filament nucleation and growth could be promoted by the increased crowding conditions in the cytoplasm, because the resulting higher protein concentration favors molecular interactions and chemical reactions (Rivas et al., 2001; Zhou et al., 2008). Conversely, excessive crowding could decrease molecular motion, especially of large particles, resulting in diminished protein assembly and binding and consequently a reduced rate of chemical reactions (Trappe et al., 2001; Miermont et al., 2013; Joyner et al., 2016; Munder et al., 2016). This suggests that filament nucleation and elongation must occur when crowding does not entirely prevent molecular mobility. Because we observed maximal crowding already after 15 min of energy depletion (Supplemental Figure S4), the recruitment of molecules into filaments must happen very rapidly at the beginning of this process. Hence, we propose that increased macromolecular crowding in the cell favors and stabilizes assemblies and membraneless compartments, but does not necessarily cause or initiate their compartmentalization in the cytoplasm.
There is increasing evidence that sudden intracellular acidification, induced by energy starvation, triggers condensation of several enzymes in yeast cells (Petrovska et al., 2014; Munder et al., 2016; Rabouille and Alberti, 2017). This pH drop is thought to change surface charges of molecules, producing new repulsive and attractive molecular interactions in the cytoplasm. Therefore, pH change in the cytoplasm may be a key initiator of enzyme-specific assemblies. Even though the chemical nature of the signal that triggers assembly of eIF2B and other enzymes in energy-depleted cells needs to be further elucidated, the process must occur at very low energy levels and before the cytoplasm becomes too crowded. Thus, stressed cells transition from a stage in which cellular components are allowed to dynamically reorganize to a stage where the cytoplasm is condensed and rather static, as reflected in the reduced mobility of organelles and foreign tracer particles (Munder et al. 2016).
Filaments and bundles form mainly via polymerization of eIF2B decamers
The rapid organization of eIF2B in highly ordered bundles of filaments in the cytoplasm of energy-depleted cells suggests that eIF2B filament formation is a specific adaptation to conditions in which energy levels are low. Using immunolabeling and tomography on WT cells, we were able to exclude that filament formation is triggered or affected by sfGFP-tagging, therefore confirming that filaments and bundles formation is an intrinsic property of eIF2B. It has been shown that energy-depleted WT yeast cells undergo translational arrest (Ashe et al., 2000; Nüske et al., 2018), whereas eIF2B mutated strains with a reduced ability to form filaments keep on translating proteins longer after energy depletion (Nüske et al., 2018). These findings seem to indicate that eIF2B filament formation promotes down-regulation of translation and suggests that eIF2B activity is inhibited upon energy depletion. This suggests that filament formation could be a mechanism to silence eIF2B enzymatic activity in cells undergoing energy depletion.
Comparison of the eIF2B cryo-EM structure from S. cerevisiae (Adomavicius et al., 2019; Gordiyenko et al., 2019) with the subtomogram average of the filament repeating unit shows similar size and overall shape. The stacking of eIF2B subunits, as we propose in Figure 7, has the same repeat as that measured for the filaments in tomograms and after averaging. This indicates that filaments are formed by polymerization of intact eIF2B decamers, rather than by aggregation of disordered condensates of eIF2B subunits (Figure 7). In more detail, the comparison between the shape of the filament and the eIF2B cryo-EM structure suggests that dimers might interact through the catalytic Gcd6 ε-subunits (Figure 7). The Gcd6 ε-subunit, together with Gcd1 γ-subunit, composes the “catalytic core” of eIF2B, which catalyzes the GDP/GTP exchange reaction (Gomez and Pavitt, 2000; Williams et al., 2001). This proposed eIF2B arrangement in the filament would partially occlude the catalytic sites of both Gcd6 ε-subunit of eIF2B decamers, suggesting a possible mechanism for enzymatic inhibition. In a different study, we have shown that mutations of the noncatalytic Gcn3 α-subunit reduce or disrupt filament formation (Nüske et al. 2018). The α-subunit is known to be located at the center of the dimer and to be important for its stabilization (Gordiyenko et al., 2014; Kashiwagi et al., 2016). Therefore, the impaired filament formation observed in the mutants might be caused by the absence of stable dimers rather than the inability of the dimers to polymerize into filaments.
In fluorescent microscopy experiments, eIF2B foci are known to colocalize with their substrate protein eIF2 (Pavitt et al., 1998; Campbell et al., 2005; Noree et al., 2010; Taylor et al., 2010; Egbe et al., 2015; Hodgson et al., 2019). Because eIF2 might be only transiently bound to eIF2B filaments (Campbell et al., 2005), it is unlikely to be resolved by averaging methods such as the one we used. However, the comparison between shapes and dimensions of the filament average with the cryo-EM structure of eIF2B (Figure 7) seems to indicate that filaments are generated by polymerization of eIF2B complexes alone.
In summary, we have described the structural rearrangements taking place in the cytoplasm of energy-depleted yeast cells (Figure 8). We propose that these changes allow cells to save energy and to protect essential cellular components, such as eIF2B, from denaturation and/or vacuolar degradation. The reversible self-assembly of eIF2B, and potentially other enzymes, aimed at storage and inhibition of their energy-demanding activities, could help cells to endure unfavorable environmental conditions. It will be interesting to investigate whether this cytoplasmic reorganization can be a general response for other proteins, in other species, and to other kind of stresses.
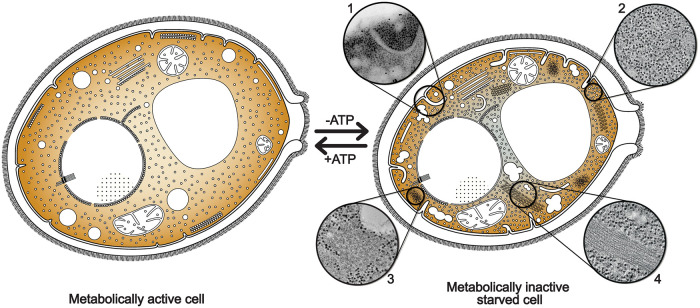
FIGURE 8:
Cytoplasm compaction and compartmentalization is a protective state for energy-depleted cells. Ultrastructural analysis shows that energy depletion induces a massive reorganization of cytoplasmic structures in yeast cells. ATP-depleted cells undergo cell volume reduction, with consequent formation of plasma membrane invaginations (1) and a pronounced increase in macromolecular crowding (2). Numerous membraneless compartments of amorphous (3) or highly ordered morphologies, such as eIF2B filaments and bundles (4), appear in the cytoplasm. Energy-depleted cells are also characterized by the fragmentation of lipid droplets, suggesting a switch to β-oxidative metabolism of fatty acids. All these cellular rearrangements result in a “solidification” of the cytoplasm that could functio¡n as a protective mechanism for molecular components, preserving them to be readily available when favorable environmental conditions are restored.
MATERIALS AND METHODS
Yeast strains, media, and energy depletion
WT Saccharomyces cerevisiae W303 cells and the two strains overexpressing untagged and sfGFP-tagged eIF2B on the C-terminus of the Gcn3 α-subunit were grown in an orbital shaker (180 rpm) at 25°C in YPD medium containing 1% (wt/vol) yeast extract, 2% peptone, and 2% glucose. Detailed information on the generation of the mutant strains is published in a related study (Nüske et al., 2018).
Stress treatment was carried out on cells grown to mid-log phase and cellular growth was monitored by optical density (OD) at 600 nm. To induce energy depletion (ATP depletion), cells at 0.5 OD600nm were washed twice and then incubated in SD (synthetic dropout) complete medium containing 20 mM 2-deoxyglucose (2-DG; Carl Roth GmbH, Karlsruhe, Germany) and 10 M antimycin-A (Sigma-Aldrich, Steinheim, Germany) for at least 15 min. These two chemicals were used to block glycolysis and mitochondrial respiration, respectively. This treatment reduces intracellular ATP levels by more than 95% (Serrano, 1977). Yeast samples were then incubated in an orbital shaker at 30°C for 1 h. Log phase yeast cells expressing only endogenous untagged eIF2B were treated in both complete and energy-depleted medium in the same way as described above.
Fluorescence microscopy
Yeast cells were imaged in concanavalin A-coated four-well Matek dishes. Fluorescence microscopy of live (time-lapse movies) and fixed cells was acquired using a Deltavision microscope system with softWoRx 4.1.2 software (Applied Precision), a 100 × 1.4 NA UPlanSApo oil immersion objective, and a CoolSnap HQ2 camera (Photometrics) at 1024 × 1024 (or 512 × 512) pixels. Exposure time was 0.3 s and the interval between consecutive frames in the time lapse was 15 min.
We determined the exact time required for yeast cells to enter the dormant state upon energy depletion by analysis of the fluorescence microscopy time-lapse movies. This information was required to select dormant cells for imaging in TEM and compare their ultrastructure with that of growing cells.
Immunostaining
For immunostaining, cells expressing Gcd1-γ-HA and Gcn3-α-HA were fixed via treatment with 3.7% paraformaldehyde (EMS, Hatfield, USA) for at least 30 min followed by 45 min incubation in spheroplasting buffer (100 mM phosphate buffer pH 7.5, 5 mM EDTA, 1.2 M Sorbitol [Sigma-Aldrich, Steinheim, Germany], Zymolyase [Zymo Research, USA]) at 30˚C with mild agitation. Spheroplasts were permeabilized with 1% Triton X-100 (Serva, Heidelberg, Germany), washed, and incubated with mouse anti-HA primary antibody (1:2000; Covance, USA) and goat anti-mouse-HRP (1:5000; Sigma, St. Louis, USA).
Western blot
Strains of WT W303 yeast and sfGFP-tagged eIF2B grown to log phase (0.5 OD600nm) were used in both control and energy-depleted experiments as described above. Cells were lysed by vortexing them for 10 min in 200 µl 10% trichloroacetic acid (TCA, Sigma) and 300 µl of glass beads. After washing the beads twice, cell extracts were pool and spun down for 10 min at 1000 × g and the supernatant was discarded to retain only the protein fraction from the extracts. Samples were then resuspended in 50 µl 3 × Laemmli buffer, vortexed, and heated to 95°C for 10 min, to denature proteins. After spinning, the supernatant was collected to measure relative protein concentration for normalization. Total protein extract (30 µg) was taken for each sample condition and loaded in each well of the polyacrylamide gel and electrophoresis was run at 130 mA for 1.5 h. Once separated, proteins were transferred to a membrane for 1 h at 150 mA. The membrane was washed in water and blocked first in PBS-Tween20 for 10 min and then in PBS-Tween20-milk for 1 h at room temperature, to reduce antibodies’ nonspecific binding. Primary antibodies against two ribosomal proteins had been developed, tested, and provided by Prof. Jonathan R. Warner (Albert Einstein College of Medicine, NY): 1) anti-ribosome protein L32, located on the large ribosomal subunit 60S; 2) anti-ribosome protein S4A/L30, which binds at the interface of the S4 protein in small ribosomal subunit 40S and L30 protein in the large subunit. A monoclonal antibody anti-3-phosphoglycerate kinase (PGK mAb, Invitrogen) was used as constitutive expression marker and loading control.
Primary antibodies were each applied to one of the membranes and incubated on a shaker overnight at 4°C. After incubation, membranes were washed three times for 5 min and the secondary antibody was applied and incubated for 1 h while shaking: horseradish-peroxidase (HRP) conjugated antibody anti-mouse, for monoclonal anti-Rpl32 and PGK, and anti-rabbit, for polyclonal anti-Rps4a (Merck). After the second incubation, membranes were washed three times for 5 min and rinsed with 2 ml of chemiluminescence buffer (Luminata Crescendo Western HRP substrate, Millipore) to activate the HRP and a Western blot was developed.
High-pressure freezing and freeze substitution of yeast cells
Yeast cells, harvested by vacuum filtration as described in Bertin and Nogales (2016), were transferred to 100 µm–deep membrane carriers and high-pressure frozen with a Leica EM PACT2 or Leica EM ICE freezer (Leica Microsystems, Wentzler, Germany). All samples were processed by freeze substitution in a Leica AFS2 temperature-controlling machine (Leica Microsystems, Wentzler, Germany) and then embedded in resin using two distinct protocols for untagged and sfGFP-tagged yeast strains.
Untagged eIF2B yeast cells.
High pressure–frozen samples were freeze-substituted using 1% osmium tetroxide, 0.1% uranyl acetate (wt/vol), and 5% H2O (vol/vol) in glass-distilled acetone. Freeze substitution was carried out at –90°C for 36 h before the temperature was raised steadily to –30°C at 4°C per hour. The samples were kept at –30°C for 5 h before they were brought to 0°C in steps of 4°C per hour. Samples were washed with acetone and infiltrated with increasing concentrations (25, 50, 75, and 100%; 2 h each) of EPON resin (Electron Microscopy Sciences). A 100% EPON solution was exchanged two times in 12-h steps. Resin-infiltrated samples were then UV-polymerized at 60°C for 48 h. Samples were cut into 150- to 200-nm sections with an ultramicrotome (Ultracut UCT; Leica) and Ultra 35° diamond knife (Diatome). The sections were mounted on Formvar-coated slot grids (Science Services).
Yeast cells sfGFP-tagged on the C-terminal of the Gcn3 α-subunit of eIF2B.
Vitrified yeast cells were freeze-substituted with 0.1% (wt/vol) uranyl acetate and 4% (vol/vol) water in acetone at –90°C as described in Kukulski et al. (2011). The samples were then embedded in Lowicryl HM-20 (Polysciences) and cut into 70, 100, and 150 nm–thick sections using an ultramicrotome (Ultracut UCT; Leica) with an Ultra 35° diamond knife (Diatome).
Correlative light and electron microscopy
For CLEM imaging, sections of yeast cells expressing the sfGFP-tagged Gcn3 (α)-subunit of eIF2B, embedded in Lowicryl HM-20 (Polysciences; Kukulski et al., 2011) were mounted on Formvar-coated finder grids (Science Services) and incubated with quenched Blue FluoSphere fiducials (diameter 200 nm) diluted 1:500 for 10 min in the dark. Grids were mounted on a glass slide with VectaShield (Vector Laboratories, Burlingame, USA) and imaged with a Zeiss Axioplan2 wide-field fluorescence CCD upright microscope to record the fluorescence signal in sections of the embedded yeast cells. Images were acquired with a 10× objective lens to generate a grid overview and a 100× objective lens (NA 1.4) to image the fluorescence in individual cells. Cells were imaged with a green GFP channel (488 nm) for the sfGFP-tagged eIF2B and a UV channel (405 nm) for the Blue FluoSphere fiducials. The correlation (overlay) between the images obtained in fluorescence microscopy and EM micrographs or tomographic slices was performed with AMIRA® 3D Visualization & Analysis Software (FEI Company, http://www.vsg3d.com/amira/; Zuse-Institut, Berlin, Germany).
Electron tomography
All sections from resin-embedded samples were stained with 1% (wt/vol) uranyl acetate in 70% (wt/vol) methanol for 5 min and 0.4% lead citrate for 3 min. Colloidal gold particles (diameter 15 nm) were added to both surfaces of the sections to serve as fiducial markers for tilt series alignment. Tomographic series were acquired in dual-axis tilt scheme (±60° and 1° increments) with SerialEM (Mastronarde, 2005), using a FEI Tecnai F30 TEM (300 kV) equipped with a Gatan US1000 CCD camera. Pixel size ranged between 7 and 12 Å/px. Tomograms reconstruction was performed using the IMOD and Etomo packages (Mastronarde, 1997).
Subtomogram averaging and tomogram segmentation
Subtomogram averaging was done with PEET software from the IMOD package (Heumann et al., 2011). Subtomograms of 54 × 54 × 54 voxels (40 × 40 × 40 nm) were automatically picked along eIF2B filaments every 15 nm. A final average was calculated from a total of ∼500 particles. A loose mask was used to refine the central filament average.
Semiautomatic segmentation of structures of interest in the tomograms was performed with the software SuRVoS (Luengo et al., 2017). The segmentation of the ribosomes, as well as the segmentation of the filamentous bundles, was done by manual segmentation using the voxel-selective brush of the SuRVoS software. The USCF Chimera package was used for 3D visualization, rendering, and animation of the reconstructed volumes (Pettersen et al., 2004).
For eIF2B fitting, eIF2(αP)/eIF2B cryo-EM maps from Adomavicius et al., 2019 (EMDB 4404) and from Gordiyenko et al., 2019 (EMDB 4544) have been segmented based on eIF2B and eIF2 subunits from their corresponding atomic models (PDB 6I3M and 6QG1, respectively, whose modeling was based on the eIF2B crystal structure of S. pombe from Kashiwagi et al., 2016). Surfaces of eIF2 subunits have been hidden and all eIF2B subunits have been gaussian filtered at 1.5 Å. The fitting in the 3D model of the filament has been performed using the “fit” command in UCSF Chimera (Pettersen et al., 2004), treating all subunits as a part of a single model.
Supplementary Material
Acknowledgments
We acknowledge J.R. Warner and S.V. Buhl from the Albert Einstein College of Medicine (NY) for kindly donating the L3 (TCM) monoclonal and the L30/S4 polyclonal ribosome antibodies. We thank T. Müller-Reichert for sharing the Amira protocol of automated filament tracing, and D. Richter for cloning HA-tagged eIF2B yeast strains. We are grateful for the technical support of P. Kiesel in G. Pigino Lab; T. Furstenhaupt, K. Gibson, J. Meissner and the EM Facility; A. Bogdanova, B. Borgonovo and the Protein Expression and Purification Facility at the Max Planck Institute of Molecular Cell Biology and Genetics (MPI-CBG). We thank O. Gonzalez for IT support and D. Diener and F. Jug for substantial advice and theoretical input. This work was supported by the Dresden International Graduate School for Biomedicine and Bioengineering (DIGS-BB), granted by the German Research Foundation (DFG) in the context of the Excellence Initiative. S.A. acknowledges funding by the Volkswagen Life Initiative and the Deutsche Forschungsgemeinschaft (AL 1061/5-1).
Abbreviations used:
- CLEM
correlative light and electron microscopy
- CtpS
cytidine triphosphate synthetase
- ED
energy depleted
- eIF2
eukaryotic translation initiation factor 2
- eIF2B
eukaryotic translation initiation factor 2B
- ET
electron tomography
- FAS
fatty acid synthetase
- GDP
guanosine diphosphate
- GTP
guanosine triphosphate
- Gln1
glutamine synthetase
- HA
human influenza hemagglutinin
- sfGFP
superfolder GFP
- TEM
transmission electron microscopy
- TORC1
target of rapamycin (TOR) complex 1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-02-0125) on April 15, 2020.
REFERENCES
- Adomavicius T, Guaita M, Zhou Y, Jennings MD, Latif Z, Roseman AM, Pavitt GD. (2019). The structural basis of translational control by eIF2 phosphorylation. Nat Commun , 2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adya AK, Canetta E, Walker GM. (2006). Atomic force microscopic study of the influence of physical stresses on Saccharomyces cerevisiae and Schizosaccharomyces pombe . FEMS Yeast Res , 120–128. [DOI] [PubMed] [Google Scholar]
- Appadurai D, Gay L, Moharir A, Lang MJ, Duncan MC, Schmidt O, Teis D, Vu TN, Silva M, Jorgensen EM, Babst M. (2020). Plasma membrane tension regulates eisosome structure and function. Mol Biol Cell , 287–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariotti N, Murphy S, Hamilton N, Wu L, Green K, Schieber NL, Li P, Parton PG. (2012). Postlipolytic insulin-dependent remodeling of micro lipid droplets in adipocytes. Mol Biol Cell ,1826–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe MP, De Long SK, Sachs AB. (2000). Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell , 833–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry RM, Bitbol AF, Lorestani A, Charles EJ, Habrian CH, Hansen JM, Li HJ, Baldwin EP, Wingreen NS, Kollmanand JM, Gitai Z. (2014). Large-scale filament formation inhibits the activity of CTP synthetase. eLife , e03638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A, Nogales E. (2016). Characterization of septin ultrastructure in budding yeast using electron tomography. In: Yeast Cytokinesis: Methods and Protocols, ed. Sanchez-Diaz A, Perez P. New York: Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg A, Adler L. (1992). Physiology of osmotolerance in fungi, Adv Microb Physiol , 145–212. [DOI] [PubMed] [Google Scholar]
- Bozaquel-Morais BL, Madeira JB, Maya-Monteiro CM, Masuda CA, Montero-Lomeli M. (2010). A new fluorescence-based method identifies protein phosphatases regulating lipid droplet metabolism. PLoS One , e13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell SG, Hoyle NP, Ashe MP. (2005). Dynamic cycling of eIF2 through a large eIF2B-containing cytoplasmic body implications for translation control. J Cell Biol , 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choder M. (1993). A growth rate-limiting process in the last growth phase of the yeast life cycle involves RPB4, a subunit of RNA polymerase. J Bacteriol , 6358–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini LM, Fossati M, Francolini M, Snapp EL. (2012). Assessing the tendency of fluorescent proteins to oligomerize under physiologic conditions. Traffic , 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desfougeres Y, Neumann H, Mayer A. (2016). Organelle size control—increasing vacuole content activates SNAREs to augment organelle volume through homotypic fusion. J Cell Sci , 2817–2828. [DOI] [PubMed] [Google Scholar]
- Duncan R, Hershey JW. (1983). Identification and quantitation of levels of protein synthesis initiation factors in crude HeLa cell lysates by two-dimensional polyacrylamide gel electrophoresis. J Biol Chem , 7228–7235. [PubMed] [Google Scholar]
- Dupont S, Beney L, Ritt JF, Lherminier J, Gervais P. (2010). Lateral reorganization of plasma membrane is involved in the yeast resistance to severe dehydration. Biochim Biophys Acta , 975–985. [DOI] [PubMed] [Google Scholar]
- Egbe NE, Paget GM, Wang H, Ashe MP. (2015). Alcohols inhibit translation to regulate morphogenesis in C. albicans . Fungal Genet Biol , 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J. (2006). Three-Dimensional Electron Microscopy of Macromolecular Assemblies: Visualization of Biological Molecules in their Native State. Oxford: Oxford University Press. [Google Scholar]
- Gervais P, Marechal PA. (1994). Yeast resistance to high levels of osmotic pressure: influence of kinetics. J Food Eng , 399–407. [Google Scholar]
- Gomez E, Pavitt GD. (2000). Identification of domains and residues within the ε subunit of eukaryotic translation initiation factor 2B (eIF2Bε) required for guanine nucleotide exchange reveals a novel activation function promoted by eIF2B complex formation. Mol Cell Biol , 3965–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordiyenko Y, Llácer JL, Ramakrishnan V. (2019). Structural basis for the inhibition of translation through eIF2α phosphorylation. Nat Commun , 2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordiyenko Y, Schmidt C, Jennings MD, Matak-Vinkovic D, Pavitt GD, Robinson CV. (2014). eIF2B is a decameric guanine nucleotide exchange factor with a gamma2epsilon2 tetrameric core. Nat Commun , 3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. (2004). “Sleeping beauty”: quiescence in Saccharomyces cerevisiae . Microbiol Mol Biol Rev , 187–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Segawa H, Okuno M, Kano H, Hamaguchi HO, Haraguchi T, Hiraoka Y, Hasui S, Yamaguchi T, Hirose F, Osumi T. (2012). Active involvement of micro-lipid droplets and lipid-droplet-associated proteins in hormone-stimulated lipolysis in adipocytes. J Cell Sci , 6127–6136. [DOI] [PubMed] [Google Scholar]
- Heumann J, Hoenger A, Mastronarde D. (2011). Clustering and variance maps for cryo-electron tomography using wedge-masked differences. J Struct Biol , 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Lorsch JR. (2012). The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol , a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson RE, Varanda BA, Ashe MP, Allen KE, Campbell CG. (2019). Cellular eIF2B subunit localization: implications for the integrated stress response and its control by small molecule drugs. Mol Biol Cell , 942–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland LS, Johnston GC, Drebot MA, Dhillon N, DeMaggio AJ, Hoekstra MF, Singer RA. (1994). A member of a novel family of yeast ‘Zn-finger’ proteins mediates the transition from stationary phase to cell proliferation. EMBO J , 3812–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarc E, Petan T. (2019) Lipid droplets and the management of cellular stress. Yale J Biol Med , 435–452. [PMC free article] [PubMed] [Google Scholar]
- Jennings MD, Pavitt GD. (2014). A new function and complexity for protein translation initiation factor eIF2B. Cell Cycle , 2660–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Klionsky DJ. (2014). Regulation of autophagy: modulation of the size and number of autophagosomes. FEBS Lett , 2457–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner RP, Tang JH, Helenius J, Dultz E, Brune C, Holt LJ, Huet S, Müller DJ Weis K. (2016). A glucose-starvation response regulates the diffusion of macromolecules. eLife , e09376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi K, Takahashi M, Nishimoto M, Hiyama TB, Higo T, Umehara T, Sakamoto K, Ito T, Yokoyama S. (2016). Crystal structure of eukaryotic translation initiation factor 2B. Nature , 4–52. [DOI] [PubMed] [Google Scholar]
- Kukulski W, Schorb M, Welsch S, Picco A, Kaksonen M, Briggs JA. (2011). Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J Cell Biol , 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurat CF, Natter K, Petschnigg J, Wolinski H, Scheuringer K, Scholz H, Zimmermann R, Leber R, Zechner R, Kohlwein SD. (2006). Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J Biol Chem , 491–500. [DOI] [PubMed] [Google Scholar]
- Liu JL. (2010). Intracellular compartmentation of CTP synthase in Drosophila . J Genet Genom , 281–296. [DOI] [PubMed] [Google Scholar]
- Luengo I, Darrow M, Spink M, Sun Y, Dai W, He C, Chiu W, Pridmore T, Ashton A, Duke E, et al. (2017). SuRVoS: super-region volume segmentation workbench. J Struct Biol , 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira JB, Masuda CA, Maya-Monteiro CM, Matos GS, Montero-Lomelí M, Bozaquel-Morais BL. (2015). TORC1 inhibition induces lipid droplet replenishment in yeast. Mol Cell Biol , 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez de Maranon I, Marechal PA, Gervais P. (1996). Passive response of Saccharomyces cerevisiae to osmotic shifts: cell volume variations depending on the physiological state. Biochem Biophys Res Commun , 519–523. [DOI] [PubMed] [Google Scholar]
- Mastronarde D. (1997). Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol , 343–352. [DOI] [PubMed] [Google Scholar]
- Mastronarde D. (2005). Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol , 36–51. [DOI] [PubMed] [Google Scholar]
- Meaden PG, Arneborg N, Guldfeldt LU, Siegumfeldt H, Jakobsen M. (1999). Endocytosis and vacuolar morphology in Saccharomyces cerevisiae are altered in response to ethanol stress or heat shock. Yeast , 1211–1222. [DOI] [PubMed] [Google Scholar]
- Miermont A, Waharte F, Hu S, McClean MN, Bottani S, Léon S, Hersen P. (2013). Severe osmotic compression triggers a slowdown of intracellular signaling, which can be explained by molecular crowding. Proc Natl Acad Sci USA , 5725–5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GJ, Winters L, Coulson GE, Clarke KJ. (1986). Effect of osmotic stress on the ultrastructure and viability of the yeast Saccharomyces cerevisiae . J Gen Microbiol , 2023–2034. [DOI] [PubMed] [Google Scholar]
- Mourão MA, Hakim JB, Schnell S. (2014). Connecting the dots: the effects of macromolecular crowding on cell physiology. Biophys J , 2761–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munder MC, Midtvedt D, Franzmann T, Nüske E, Otto O, Herbig M, Ulbricht E, Müller P, Taubenberger A, Maharana S, et al. (2016). A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. eLife , e09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munna MS, Humayun S, Noor R. (2015). Influence of heat shock and osmotic stresses on the growth and viability of Saccharomyces cerevisiae . BMC Research Notes , 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanaswamy R, Levy M, Tsechansky M, Stovall GM, O’Connell JD, Mirrielees J, Ellington AD, Marcotte EM. (2009). Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc Natl Acad Sci USA , 10147–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Ohsumi Y. (1998). Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem , 3963–3966. [DOI] [PubMed] [Google Scholar]
- Noree C, Sato BK, Broyer RM, Wilhelm JE. (2010). Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster . J Cell Biol , 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nüske E, Marini G, Richter D, Leng W, Bogdanova A, Franzmann T, Pigino G, Alberti S. (2018). Filament formation by the translation factor eIF2B regulates protein synthesis in starved cells. bioRxiv 467829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavitt GD, Ramaiah KV, Kimball SR, Hinnebusch AG. (1998). eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes dev , 514–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovska I, Nüske E, Kulasegaran G, Gibson K, Munder MC, Malinovska L, Richter D, Verbavatz JM, Alberti S. (2014). Filament formation by metabolic enzymes is a specific adaptation to the energy-depleted cellular state. eLife , e02409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E, Goddard T, Huang C, Couch G, Greenblatt D, Meng E, Ferrin T. (2004). UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem , 1605–1612. [DOI] [PubMed] [Google Scholar]
- Prouteau M, Desfosses A, Sieben C, Bourgoint C, Mozaffari LN, Demurtas D, Mitra AK, Guichard P, Manley S, Loewith R. (2017). TORC1 organized in inhibited domains (TOROIDs) regulate TORC1 activity. Nature , 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C, Alberti S. (2017). Cell adaptation upon stress: the emerging role of membrane-less compartments. Curr Opin Cell Biol , 34–42. [DOI] [PubMed] [Google Scholar]
- Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, Sosnick TR, Drummond DA. (2017). Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell , 1028–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas G, Fernández JA, Minton AP. (2001). Direct observation of the enhancement of noncooperative protein self-assembly by macromolecular crowding: indefinite linear self-association of bacterial cell division protein FtsZ. Proc Natl Acad Sci USA , 3150–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. (2003). Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol , 721–745. [DOI] [PubMed] [Google Scholar]
- Rußmayer H, Bucheics M, Gruber C, Valli M, Grillitsch K, Modarres G, Guerrasio R, Klavins K, Neubauer S, Drexler H, et al. (2015). Systems-level organization of yeast methylotrophic lifestyle. BMC Biol , 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Meth , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo AY, Lau PW, Feliciano D, Sengupta P, Gros MA, Cinquin B, Larabell CA, Lippincott-Schwartz J. (2017). AMPK and vacuole-associated Atg14p orchestrate μ-lipophagy for energy production and long-term survival under glucose starvation. eLife , e21690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano R. (1977). Energy requirements for maltose transport in yeast. Eur J Biochem , 97–102. [DOI] [PubMed] [Google Scholar]
- Simonin H, Beney L, Gervais P. (2007). Sequence of occurring damages in yeast plasma membrane during dehydration and rehydration: mechanisms of cell death. Biochim Biophys Acta , 1600–1610. [DOI] [PubMed] [Google Scholar]
- Slaninova I, Sestak S, Svoboda A, Farkas V. (2000). Cell wall and cytoskeleton reorganization as the response to hyperosmotic shock in Saccharomyces cerevisiae . Arch Microbiol , 245–252. [DOI] [PubMed] [Google Scholar]
- Snapp EL, Hegde RS, Francolini M, Lombardo F, Colombo S, Pedrazzini E, Borgese N, Lippincott-Schwartz J. (2003). Formation of stacked ER cisternae by low affinity protein interactions. J Cell Biol , 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh HG, da Silveira dos Santos AX, Kukulski W, Tyedmers J, Riezman H, Bukau B, Mogk A. (2015). Prolonged starvation drives reversible sequestration of lipid biosynthetic enzymes and organelle reorganization in Saccharomyces cerevisiae . MBoC , 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor EJ, Campbell S, Griffiths CD, Reid P, Slaven JW, Harrison R, Sims P, Pavitt GD, Delneri D, Ashe MP. (2010). Fusel alcohols regulate translation initiation by inhibiting eIF2B to reduce ternary complex in a mechanism that may involve altering the integrity and dynamics of the eIF2B body. MBoC , 2202–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam AR, Beller M. (2017). The why, when and how of lipid droplet diversity. J Cell Sci , 315–324. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Macaya G, Bernardi G. (1976). An analysis of eukaryotic genomes by density gradient centrifugation. J Mol Biol , 219–235. [DOI] [PubMed] [Google Scholar]
- Trappe V, Prasad V, Cipelletti L, Segre PN, Weitz DA. (2001). Jamming phase diagram for attractive particles. Nature , 772–775. [DOI] [PubMed] [Google Scholar]
- Tzaphlidou M, Chapman JA, Al-Samman MH. (1982). A study of positive staining for electron microscopy using collagen as a model system. In II. Staining by uranyl ions. Micron , 133–145. [Google Scholar]
- Warner JR. (1999). The economics of ribosome biosynthesis in yeast. Trends Biochem Sci , 437–440. [DOI] [PubMed] [Google Scholar]
- Werner-Washburne M, Braun E, Johnston GC, Singer RA. (1993). Stationary phase in the yeast Saccharomyces cerevisiae . Microbiol Rev , 383–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DD, Pavitt GD, Proud CG. (2001). Characterization of the initiation factor eIF2B and its regulation in Drosophila melanogaster . J Biol Chem , 3733–3742. [DOI] [PubMed] [Google Scholar]
- Winderickx J, Holsbeeks I, Lagatie O, Giots F, Thevelein J, de Winde H. (2003). Yeast stress responses—from feast to famine; adaptation to nutrient availability in yeast. In Topics in Current Genetics, , ed. Hohmann S, Mager P.W.H. Berlin/Heidelberg: Springer-Verlag, 305–386. [Google Scholar]
- Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, Hyman AA. (2017). The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell , 1066–1077. [DOI] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, Tsien RY. (2002). Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science , 913–916. [DOI] [PubMed] [Google Scholar]
- Zhou HX, Rivas G, Minton AP. (2008). Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. AnnuRev Biophys , 375–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.