Abstract
Background
Epithelial-to-mesenchymal transition (EMT) and mesenchymal-to-epithelial transition (MET) are both reversible processes, and regulation of phenotypical transition is very important for progression of several cancers including hepatocellular carcinoma (HCC). Recently, it is defined that cancer cells can attain a hybrid epithelial/mesenchymal (hybrid E/M) phenotype. Cells with hybrid E/M phenotype comprise mixed epithelial and mesenchymal properties, they can be more resistant to therapeutics and also more capable of initiating metastatic lesions. However, the mechanisms regulating hybrid E/M in HCC are not well described yet. In this study, we investigated the role of the potential crosstalk between lncRNA HOTAIR and c-Met receptor tyrosine kinase, which are two essential regulators of EMT and MET, in acquiring of hybrid E/M phenotype in HCC.
Methods
Expression of c-Met and lncRNA HOTAIR were defined in HCC cell lines and patient tissues through HCC progression. lncRNA HOTAIR was overexpressed in SNU-449 cells and its effects on c-Met signaling were analyzed. c-Met was overexpressed in SNU-398 cells and its effect on HOTAIR expression was analyzed. Biological significance of HOTAIR/c-Met interplay was defined in means of adhesion, proliferation, motility behavior, invasion, spheroid formation and metastatic ability. Effect of ectopic lncRNA HOTAIR expression on phenotype was defined with investigation of molecular epithelial and mesenchymal traits.
Results
In vitro and in vivo experiments verified the pivotal role of lncRNA HOTAIR in acquisition of hybrid E/M phenotype through modulating expression and activation of c-Met and its membrane co-localizing partner Caveolin-1, and membrane organization to cope with the rate limiting steps of metastasis such as survival in adhesion independent microenvironment, escaping from anoikis and resisting to fluidic shear stress (FSS) in HCC.
Conclusions
Our work provides the first evidence suggesting a role for lncRNA HOTAIR in the modulation of c-Met to promote hybrid E/M phenotype. The balance between lncRNA HOTAIR and c-Met might be critical for cell fate decision and metastatic potential of HCC cells.
Video Abstract
Keywords: HCC, long non-coding RNA, HOTAIR, c-Met, Caveolin-1, Hybrid E/M
Background
Liver cancer is ranked as 7th cancer type in the list of cancers with the highest number of cases incident between 2007 and 2017 by Global Burden of Disease (GBD) 2017 study. Hepatocellular Carcinoma (HCC) accounts more than 80% of liver cancer cases and it is estimated to be the 4th most common cause of cancer related deaths worldwide [1]. HCC is reported to have complex and heterogeneous molecular features which is previously stressed out by integrative studies combining genomic characterization, exome sequencing, transcriptome analysis and clinical trials. Molecular heterogeneity and aetiological complexity of HCC makes it unlikely for one treatment or agent to effectively target all or most HCCs [2, 3].
c-Met, a receptor tyrosine kinase, is known to be upregulated in liver diseases favoring hepatocyte proliferation. Besides potential benefits in chronic liver diseases, c-Met contributes to initiation, development and progression of HCC. c-Met activation in HCC is mostly driven by molecular networks instead of activating mutations and it is activated by non-canonical signaling mechanisms as well as canonical activation by its ligand, hepatocyte growth factor (HGF). c-Met is regarded as one of the most promising targets for HCC treatment and c-Met targeted clinical trials are being conducted, currently [4]. In addition to its contribution to HCC development and progression, c-Met is also considered to be a key player in drug resistance [5].
HOX transcript antisense intergenic RNA, HOTAIR, is a 2148-nt-long spliced and polyadenylated long non-coding RNA (lncRNA) encoded within HOXC cluster. HOTAIR is first defined to take role in epidermal tissue development by recruiting chromatin remodeling complexes to its epigenetic targets [6, 7]. Overexpression of HOTAIR has been associated with poor prognosis, invasiveness and aggressiveness of various cancer types [8–11]. Complex secondary structure and ability to form independent structural domains ensure the multi-acting nature of HOTAIR and it has been defined to contribute to various cellular mechanisms via different molecular interactions such as scaffolding protein complexes, decoying microRNAs, epigenetically targeting genes and enabling RNA-protein/DNA-protein interactions [7, 12–14].
Transcription factors, non-coding RNAs and epigenetic regulators play critical roles in cell-fate determination such as transition between epithelial and mesenchymal phenotypes. Starting from the origin of a tumor, cancer cells go through complex and dynamic phenotypical changes -epithelial to mesenchymal transition (EMT), or its reverse mesenchymal to epithelial transition (MET)- to cope with metastasis rate-limiting steps. It has been reported that, some cancer cells display both epithelial and mesenchymal markers and this phenotype is referred as hybrid, intermediate, partial, metastable or incomplete EMT phenotype [15–18]. The tumor cells simultaneously expressing both mesenchymal and epithelial markers may be more plastic and most likely to contribute to metastatic outgrowth. Cells bearing this hybrid phenotype are first defined in circulating tumor cells (CTCs) of patients with both epithelial and mesenchymal traits [19, 20]. Hybrid cells that co-express mesenchymal and epithelial markers migrate collectively, more resistant to fluidic shear stress damage in circulation, exit from anoikis and have enhanced metastatic ability than cells with complete epithelial or mesenchymal phenotypes [21]. That is why, cancer cells that attain a hybrid E/M (epithelial/mesenchymal) phenotype pose greater risk for metastasis in cancer patients [22].
c-Met is reported to play an essential and complex role in modulation of transitional states within the broad spectrum of cellular phenotypes [23]. Although HOTAIR is also reported to contribute attainment of cellular phenotypes, its role is generally defined in means of correlations with aggressiveness traits of cancer cells [10, 11, 24, 25]. As the number of the studies reporting induced expression and critical roles of these two molecules in HCC are increasing, the likelihood of their direct or indirect molecular interaction becomes non-negligible. In this study, we aimed to investigate the nature of their interaction and here, we describe the interplay between HOTAIR and c-Met in HCC context. Our data present strong evidence that HOTAIR ensures hybrid E/M phenotype and its downregulation is required for c-Met induced complete mesenchymal phenotype in HCC cells.
Materials and methods
Cell culture
All Human HCC cell lines were kindly provided by Prof. Dr. Mehmet Öztürk (Izmir Biomedicine and Genome Center, Izmir, Turkey). FOCUS, SNU-449, SK-HEP-1, SNU-475, SNU-387, SNU-423, SNU-398, MAHLAVU, HEP-3B and HuH-7 cell lines were cultured as described previously [26]. All cell lines were tested for mycoplasma infection and confirmed as negative before the experiments. SU11274 (Calbiochem, 448101) is solubilized with DMSO and used with 2500 nM concentration. c-Met kinase activity was inhibited with SU11274 (2500 nM in 2% FBS complete medium). Ligand dependent c-Met activation was induced by 10 ng/ml Hepatocyte Growth Factor (HGF) in 2% FBS complete medium (R&D system, USA). Sorafenib treatment was continuously applied with the amount of their growth inhibition 50 (GI50) values (6 μM of Sorafenib) to MAHLAVU sorafenib resistant cell clones which have been established in our previous studies [5].
Cells were sheared with 205 U/CA multichannel peristaltic pump and Marprene tube elements of 2.79 bore for tubing (Watson Marlow, UK). Adherent cells were detached with 0.25% Trypsin /EDTA (Biological Industries, #03–050-1B, Israel) and 3 million cells in 8 ml of culture media were cultured in circulation under 0.5 dyn/ cm2 shear stress at 37 °C, in humidified incubator supplied with 5% CO2, for 1 hour. After FSS application, cells were stained with trypan blue and counted with hemocytometer to evaluate cell viability under shear stress.
mRNA isolation, reverse transcription and RT-qPCR
NucleoSpin RNA Isolation kit (Macherey-Nagel, Germany) has been used for mRNA isolation. mRNA isolation performed according to the manufacturer’s instructions. mRNA eluted with DNAse/RNAse free distilled water and its concentration is measured via NanoDrop.
SensiFast™ cDNA Synthesis Kit (Bioline Meridian Bioscience, USA) were used to transcribe cDNA from mRNA. qPCR reactions were performed with primers designed specific for cDNA sequences of gene of interests (CDS) from NCBI-Gene Bank. RT-qPCR analysis performed using SensiFast™ SYBR No-ROX kit (Bioline Meridian Bioscience, USA) with ABI 7500 Fast Real-Time PCR System (Thermo Scientific, USA) according to the manufacturer’s instructions. RPL41 and/or GAPDH gene expressions were used as internal controls to normalize relative expression of the genes. Primers targeting mRNAs of interest were listed in Supplementary Document 1.
siRNA gene silencing experiments
HuH-7 cells were transfected with 10 nM non-targeting (untargeted, negative control-NC, Dharmacon, Lincode non-targeting SMARTpool, D-001320-10-50) and HOTAIR targeting siRNAs (Dharmacon, Lincode Human HOTAIR SMARTpool, R-187951-00-0050) by Roche X-tremeGENE™ HP DNA Transfection Reagent (6366244001, Roche). Cells were harvested after 48 h post-transfection and processed for further protein and mRNA investigations.
Fluorescent In-Situ Hybridization (FISH)
HOTAIR mRNA was hybridized with Stellaris® FISH Probes, Human HOTAIR with Quasar® 570 Dye (LGS Biosearch Technologies, VSMF-2178-5). HCC cells were grown on glass coverslips and fixed with 3:1 methanol-acetic acid (MeOH-AcOH) fixation solution for 10 min. Hybridization was performed according to the manufacturer’s instructions with 500 nM HOTAIR targeting probes for 4 h at 37 °C. Cell nucleus was stained with DAPI.
Labeling with c-Met antibody and hybridizing HOTAIR mRNA with fluorescent-labeled probes were performed sequentially. For sequential labeling, HCC cells were firstly fixed with 3.7% (vol./vol.) formaldehyde in 1X-PBS for 10 min at room temperature. Fixed cells were incubated with primary antibody against c-Met protein (Cell Signaling, #3127) for 2 h at room temperature. After couple of washes with 1X-PBS, cells were incubated with secondary antibody (Alexa488-conjugated goat anti-mouse secondary antibody, Invitrogen, A-11001) for 1 h at RT. After immunofluorescent labeling, cells were fixed again with 3:1 methanol-acetic acid (MeOH-AcOH) fixation solution for 10 min. Following hybridization was performed according to the manufacturer’s instructions with 500 nM HOTAIR targeting probes for 4 h at 37 °C. Cell nucleus was stained with DAPI and cells were analyzed with Zeiss LSM 880-Confocal Laser Scanning Microscopy with Airyscan in IBG Optic Imaging Core Facility.
Immunoblotting
The cells were lysed using RIPA lysis buffer containing 1 mM Na3VO2, 1 mM NaF, and 1% protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN, USA), and the lysates were subjected to Western blot analysis as described previously [26]. Primary antibodies are listed in Supplementary Document 1.
Producing retroviral and lentiviral virus particles and generation of stable cell lines
SNU-449 cells were infected with retroviral vector bearing viruses. Retroviral vector bearing virus particles produced by transfection of HEK-293T cells with VSV-G envelope expressing plasmid (5 μg), gag-pol expressing plasmid (5 μg) and retroviral target vectors (5 μg) together by Roche X-tremeGENE™ HP DNA Transfection Reagent (6366244001, Roche). After 24 h of transfection, cell culture media refreshed and collected 48 h post-transfection. HCC cells were infected with 1:1 titration of retrovirus containing media. After 48 h, infected SNU-449 cells were selected with 13 μg/ml and SNU-398 cells were selected with 0.5 μg/ml puromycine (Life Technologies, #A1113803, USA) for at least 3 passages.
HEK-293T cells were transfected with vectors bearing Rev (1 μg), gag-pol (6 μg) and VSV-G envelope (3 μg) genes with lentiviral target vectors (2.5 μg) by transfection reagent. After 48 h, cell culture media collected and SNU-398 cells were infected with 1:5 titration of lentivirus containing media. After 48 h of infection, infected cells were selected with 0.5 μg/ml puromycine for at least 3 passages. Lentiviral and retroviral vectors and their origins are listed in Supplementary Document 1.
Immunofluorescent analysis of the cells
HCC cells were grown on glass cover-slips and after experimental conditions set, cells were immunofluorescently labeled as described in previous study [27]. Imaging was performed with upright fluorescence microscope (Olympus - BX61) and Zeiss LSM 880-Confocal Laser Scanning Microscopy with Airyscan at IBG Optic Imaging Core Facility.
Trans-well motility and invasion assays
Trans-well inserts with 8 μm pore size (SPL Life Sciences, #37224, Korea) were used to analyze motility and invasion ability of HCC cells. The mix of 0.25 mg/ml basal membrane extract (Corning Matrigel® Growth Factor Reduced (GFR) Basement Membrane Matrix, Phenol Red-Free, #356231) was coated on invasion inserts. Trans-well motility and invasion experiments performed as described in previous study [26]. Motile and invasive cells located at the exterior surface of the inserts were counted with bright field inverted microscope and statistical analysis was performed via Prism 8.
Real-time adhesion, proliferation, motility and invasion assays
Real-time cell growth monitoring was performed with the Real-Time Cell Analyzer, xCELLigence System (Roche, Germany) as described previously [28]. 5000 cell/well was plated to analyze proliferation and adhesion to cell-culture treated wells of E-Plates. For motility and invasion assays, CIM-plates (ACEA Biosciences, # 0566581701, USA) were used with xCELLigence System. CIM-plate wells were coated with 0.25 mg/ml basal membrane extract (Corning Matrigel® Growth Factor Reduced (GFR) Basement Membrane Matrix, Phenol Red-Free, #356231) for invasion assay. For motility and invasion assays, 30.000 cells/well were plated to CIM-Plate wells and incubated for 24 h by monitoring with xCELLigence RTCA system.
Hanging drop spheroid formation assay
2500 HCC cells were incubated within 20 μl culture medium drops which were hanging from the inverted lid of 10 cm cell culture plates. The reservoir part of 10 cm of tissue culture dish was filled with 3–5 ml of 1X PBS to provide a humidified environment. The culture dish was incubated at 37 °C, 5% CO2, 95% humidified incubator for 72 h or 96 h until the spheroids formation and spheroids were imaged under Olympus SZX10 stereo microscope and SC50 camera system.
2D Colony formation assay
SNU-449 HOTAIR OE and SNU-449 MOCK cells seeded on 24-well plates as 70 cells/well. The cells were cultured until single-cell colony formation. Cell culture media was refreshed in every three days and cells were incubated for one week. The day of analysis, cell culture media was aspirated and colonies were fixed with 1:1 cold Aceton:Methanol solution at -20 °C for 10 min. Colonies stained with 20% Giemsa solution for 20 min. After washing with distilled water, plate was incubated at room temperature for air dry. Plates were imaged under BIO-RAD Gel Doc™ XR+ Gel Documentation System and the images were analyzed via ImageJ - ColonyArea plug-in [29].
Phalloidin staining
HCC cells were seeded on glass cover slips (22 × 22 cm) and incubated overnight for adhesion. Phalloidin (Abcam, abID#176756, USA)staining was performed as previously described [30]. Imaging performed at IBG Optic Imaging Core Facility with fluorescent (Olympus - BX61) and Zeiss LSM 880-Confocal Laser Scanning Microscopy with Airyscan.
MTT assay
SNU-449 HOTAIR OE and SNU-449 MOCK 1000 cells per well were seeded on 96-well plate and incubated for 24, 48 and 72 h. MTT analysis was performed as described previously [31]. Measured absorbance was analyzed by Microsoft Excel 2016 and plotted by Prism 8.
Analysis of TCGA and GEO microarray datasets
Normalized gene expression data from dataset GSE89377, GSE98091 and GSE109483 were analyzed with GEO2R and GREIN [32]. Expression and correlation analysis of HOTAIR and MET performed with LncTarD [33]. Expression values plotted and statistically analyzed with Prism 8.
Zebrafish xenograft assay
Zebrafish xenograft assay was performed with SNU-449 MOCK and SNU-449 HOTAIR cell clones. The experiment was performed as described in a previous study [30]. Briefly, 48hpf embryos were anesthetized and DiI labeled cells were injected to the yolk sack.100 cells per embryo were injected, successful injections to yolk with localized cells were selected 4 h after injection. Xenografts were analyzed with Olympus SZX16 Fluorescent stereomicroscope. Three independent experiments were performed and each group consisted average number of 50 embryos with intact tumorospheres. All experiments were performed in compliance with local ethics regulations and EU Directive 2006 and approved by IBG Animal Ethics Committee with the protocol number of 03/2019.
Statistical analysis
Statistical analysis performed using the GraphPad Prism 8 software. Statistical methods included Analysis of variance (ANOVA), Student’s t-test and linear regression. Zebrafish xenograft data was statistically analyzed with chi-square with Yates correction as described in previous study [34]. Results with p < 0.05 were considered as statistically significant.
Results
Expression of HOTAIR is low in HCC cells with high c-Met expression and activation
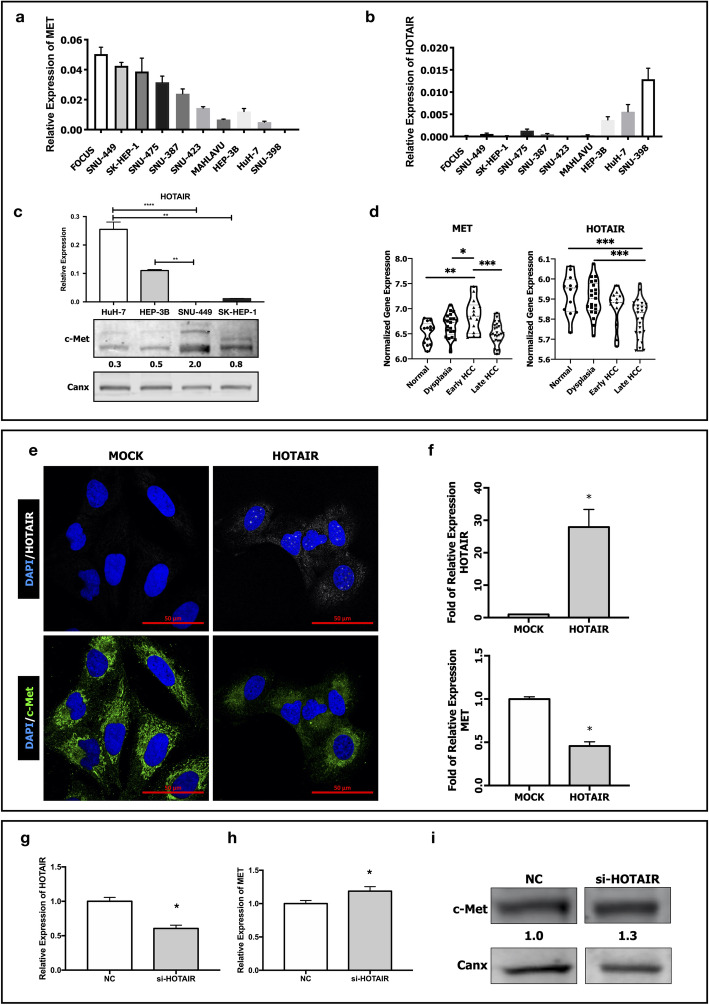
To examine expression levels of HOTAIR and c-Met, we analyzed their mRNA expression in 10 HCC cell lines (FOCUS, SNU-449, SK-HEP-1, SNU-475, SNU-387, SNU-423, MAHLAVU, Hep-3B, HuH-7 and SNU-398). HOTAIR expression was only abundant in HuH-7 and SNU-398 cell lines which are also known to have low or no c-Met expression, respectively (Fig. 1a-b). To evaluate the potential relation between HOTAIR and c-Met protein expressions, we selected two cell lines with constitutive c-Met activation (SNU-449 and SK-HEP-1) and two with low c-Met expression (HuH-7 and Hep-3B). RT-qPCR analysis of HOTAIR mRNA expression levels and western blot analysis of c-Met protein expression revealed that HOTAIR expression is low in HCC cell lines with high c-Met protein expression (Fig. 1c). To understand the relationship between HOTAIR and c-Met expression in HCC, we analyzed HOTAIR and MET gene expression in a microarray dataset, GSE89377, which comprise gene expression analysis in patient tissues from normal liver, dysplasia, early and late HCC stages [35]. Expression analysis of the dataset showed that HOTAIR expression decreases through HCC progression whereas MET gene expression increases (Fig. 1d).
Fig. 1.
Expression of HOTAIR and c-Met in HCC cell lines and patient tissues through HCC progression. RT-qPCR analysis of (a) MET and (b) HOTAIR expressions in HCC cell lines FOCUS, SNU-449, SK-HEP-1, SNU-475, SNU-387, SNU-423, MAHLAVU, Hep-3B, HuH-7 and SNU-398. (c) RT-qPCR analysis of HOTAIR expression and immunoblotting of c-Met protein in HuH-7, Hep-3B, SNU-449 and SK-HEP-1 cells. (d) Normalized expression levels of MET and HOTAIR in normal, dysplasia, early and late HCC tissues in microarray dataset GSE89377. (e) Confocal microscopy image of fluorescent-in-situ hybridized HOTAIR mRNA and immunofluorescent labeled c-Met protein expression in control (MOCK) and HOTAIR over-expressing (HOTAIR OE) SNU-449 cells. RT-qPCR analysis of (f) MET and HOTAIR mRNA expression in MOCK and HOTAIR OE clones. RT-qPCR analysis of (g) HOTAIR and (h) c-Met mRNA expression in control si-RNA (NC) and si-HOTAIR transfected HuH-7 cells. Western blot analysis of (i) c-Met protein expression in control (NC) and si-HOTAIR transfected HuH-7 cells. Densitometric analysis of band intensities in immunoblots were analyzed with ImageJ and normalized with internal control protein band intensities. All graphs of experiments are presented as the mean ± SEM of at least 3 independent experiments. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001
To evaluate the effect of ectopic HOTAIR expression on c-Met expressing HCC cells, we overexpressed HOTAIR in SNU-449 cell line which has constitutive c-Met activation. Overexpression of HOTAIR (HOTAIR OE, presented in figures as HOTAIR clone) and its effect on c-Met expression were analyzed and confirmed with confocal microscopy imaging of fluorescent in-situ hybridization (FISH) of HOTAIR, immunofluorescent labeling of c-Met (Fig. 1e), and also with RT-qPCR (Fig. 1f). HOTAIR overexpression suppressed both mRNA and protein levels of c-Met, dramatically.
To understand the possible interplay between two molecules in detail, we knocked down HOTAIR expression with siRNA transfection in HuH-7 cell line and then analyzed mRNA and protein expression of c-Met. Analysis of three independent experiments showed that average of 45% knock-down of HOTAIR (Fig. 1g) induced c-Met mRNA (Fig. 1h) and protein expression (Fig. 1i), significantly. Uncut membrane images and expression levels are presented in Supplementary Fig. 1a).
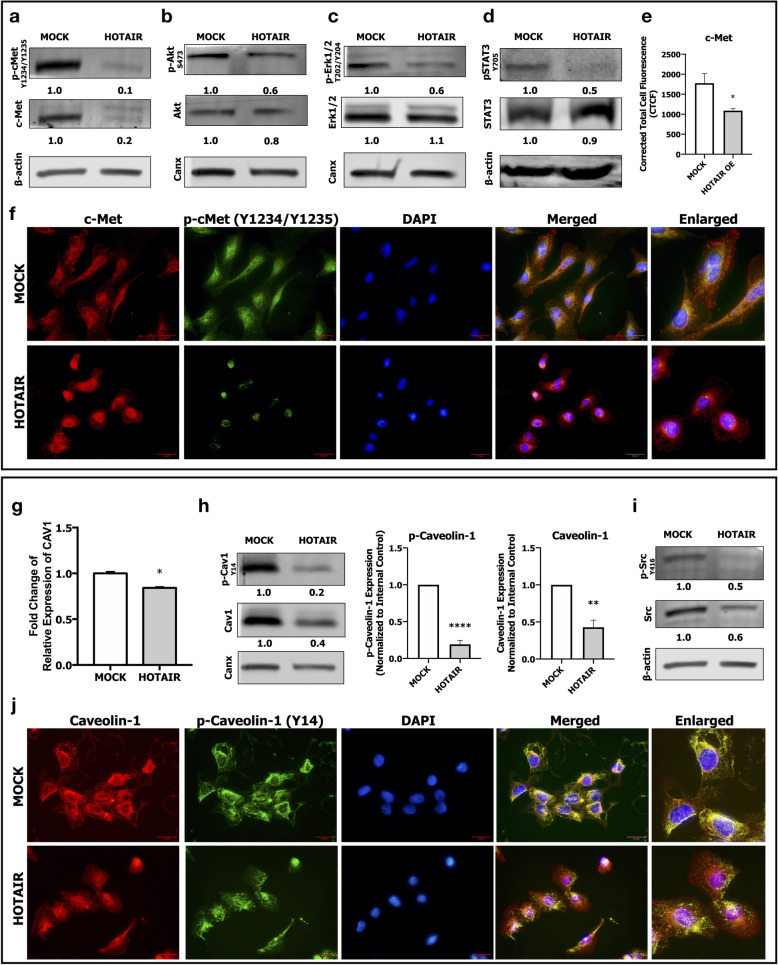
lncRNA HOTAIR overexpression suppresses c-Met downstream signaling and modulates its cellular localization via suppressing Caveolin-1 expression
To understand the effects of HOTAIR on c-Met signaling, we analyzed activation of c-Met and its downstream signaling pathways Akt, MAPK and STAT3. Western blot analysis showed that HOTAIR OE suppressed c-Met protein expression and activation (Fig. 2a). Repression of c-Met activation was a result of diminished c-Met protein expression and there was not a significant inhibition on c-Met phosphorylation (p-cMet) when normalized to total c-Met protein expression (Supplementary Fig. 1b). HOTAIR OE caused a modest decrease in Akt activation (Fig. 2b), and suppressed MAPK (Fig. 2c) and STAT3 (Fig. 2d) activations, dramatically. We confirmed these results by analyzing expression and activation of c-Met by immunofluorescent microscopy (Fig. 2e and f). Macroscopic analysis of the cells revealed that HOTAIR OE cells were smaller with roundish morphology and even the immunofluorescence staining intensities of c-Met seem to be higher in HOTAIR OE cells, analysis of immunofluorescence intensity per cells showed that is an optical illusion caused by size (smaller) and morphological (rounder) attributes of HOTAIR OE cells (Fig. 2e). Furthermore, immunofluorescent labeling revealed a differential organization and localization of c-Met in HOTAIR OE cells (Fig. 2f). In our previous studies, we reported that Caveolin-1 (CAV1), a membrane protein enriched in lipid-rafts, enhances c-Met activation and modulates its membrane localization in HCC cells [27]. To investigate the possible role of CAV1 in HOTAIR/c-Met interaction, we analyzed expression and activation of CAV1 and its activating kinase, Src, in HOTAIR OE cells. HOTAIR OE suppressed CAV1 mRNA level (Fig. 2g), protein expression and activation (Fig. 2, Supplementary Fig. 1c) as well as Src kinase expression and activation (Fig. 2i). Moreover, immunofluorescent staining revealed that membrane organization of Caveolin-1 was disrupted in HOTAIR OE cells which was compatible with disorganization of c-Met (Fig. 2j).
Fig. 2.
Effect of HOTAIR overexpression in c-Met downstream signaling. Analysis of c-Met and downstream signaling in MOCK and HOTAIR OE cells. Immunoblot analysis of (a) c-Met protein expression, c-Met activation phosphorylation (Tyr-1234/Tyr-1235); (b) Akt1 protein expression, Akt1 activation phosphorylation (Ser-473); (c) Erk1/2 protein expression, Erk1/2 activation phosphorylation (Thr-202/Tyr-204); (d) STAT3 protein expression and activation phosphorylation (Tyr-705). Column graph of corrected total cell fluorescence of c-Met immunofluorescent staining (e). Immunofluorescence imaging of (f) total c-Met protein (red) and its activation (green). RT- qPCR analysis of (g) CAV1 mRNA expression, immunoblotting of (h) Caveolin-1 protein and its activation phosphorylation (Tyr-14) and column graph of densitometric analysis of Caveolin-1 activation and total protein expressions. Immunoblotting of (i) Src kinase expression and its activation phosphorylation (Tyr-416). Immunofluorescence imaging of (j) total Caveolin-1 protein (red) and its activation (green). DAPI (blue) was used as a nuclear marker in immunofluorescent images. Densitometric analysis of band intensities in immunoblots were analyzed with ImageJ and normalized with internal control protein band intensities. All graphs of experiments are presented as the mean ± SEM of at least 3 independent experiments. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001
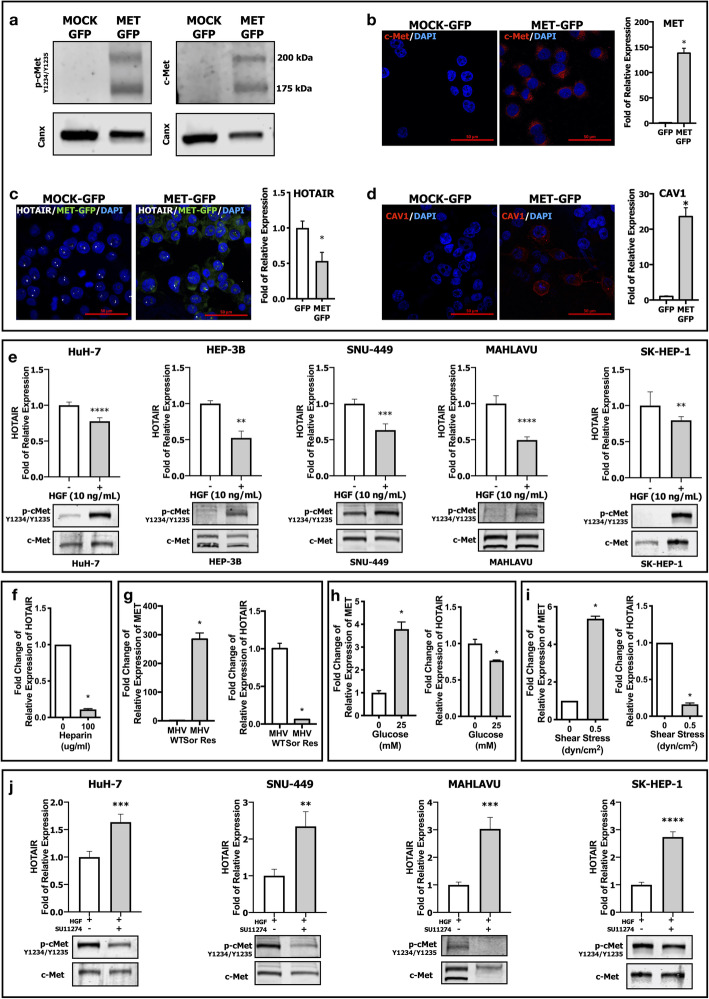
Overexpression and/or activation of c-Met decreases HOTAIR expression in HCC cell lines
To understand the interplay between HOTAIR and c-Met further, we applied an opposite approach by overexpressing c-Met in SNU-398 HCC cell line which lacks c-Met protein expression and activation in wild-type (Fig. 1a). Overexpression and activation phosphorylation of c-Met were confirmed by western blot (Fig. 3a), immunofluorescent labeling and RT-qPCR analysis (Fig. 3b). Analysis of HOTAIR expression in MET-OE-SNU 398 (presented in figures as MET-GFP) cells by FISH and RT-qPCR showed that HOTAIR mRNA expression was significantly suppressed by c-Met overexpression (Fig. 3c). To investigate HOTAIR/c-Met/Caveolin-1 axis further, we analyzed Caveolin-1 expression in c-MET OE cell lines and found that Caveolin-1 expression was significantly upregulated, consistent with HOTAIR down-regulation (Fig. 3d).
Fig. 3.
Analysis of HOTAIR-c-Met interaction in MET overexpressed, activated and inhibited conditions. Immunoblotting of (a) c-Met activation phosphorylation (Tyr-1234/Tyr-1235), and total c-Met protein expression. Confocal microscopy imaging of (b) c-Met protein and RT-qPCR analysis of c-Met mRNA expression in MET OE SNU-398 clones. (c) Confocal imaging of FISH labeling (white dots) and RT-qPCR analysis of HOTAIR mRNA expression in MET OE clones. (d) Confocal imaging of Caveolin-1 protein and qPCR analysis of CAV1 mRNA expression in MET OE cells. (e) Immunoblot of c-Met activation and qPCR analysis of HOTAIR expression in ligand-dependent activation (HGF 10 ng/ml) of c-Met in HuH-7, HEP-3B, SNU-449, MAHLAVU and SK-HEP-1 cells. RT-qPCR analysis of HOTAIR expression (f), in response to heparin (100 μg/mL) induced c-Met activation in SK-HEP-1 cells. RT-qPCR analysis of MET and HOTAIR expressions in sorafenib resistance (MAHLAVU cell line) (g) and high-glucose (25 mM, HuH-7 cell line) (h) and fluidic shear stress (0.5 dyn/cm2, HuH-7 cell line) (i) induced c-Met activations. (j) Immunoblot of c-Met activation and qPCR analysis of HOTAIR expression in tyrosine kinase activity inhibition of c-Met by SU11274 (2500 nM) HuH-7, SNU-449, MAHLAVU and SK-HEP-1 cells. All graphs of experiments are presented as the mean ± SEM of at least 3 independent experiments. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001
To understand the role of c-Met activation in HOTAIR expression, we analyzed the effect of HGF induced c-Met activation on HCC cell lines with different c-Met and HOTAIR expression levels such as HuH-7, HEP-3B, SNU-449, MAHLAVU and SK-HEP-1. Ligand-dependent c-Met activation by 10 ng/ml HGF for 1 h suppressed HOTAIR expression significantly in all tested cell lines (Fig. 3e). In addition to ligand-dependent activation, we examined the role of ligand-independent c-Met activation on HOTAIR transcription. In our previous studies, we showed that heparin treatment [36], acquired sorafenib resistance [5], high glucose induction [37] or fluidic shear stress treatment [38] induces ligand independent c-Met activation in HCC cell lines. Similar to HGF, heparin (Fig. 3f), sorafenib (Fig. 3g), high-glucose (Fig. 3h) and fluidic shear stress (Fig. 3i) treatments induced c-Met activation resulted in a substantial decrease in HOTAIR transcription. The maintenance of the interplay between HOTAIR and c-Met in different contexts supports the hypothesis that there is a reciprocal crosstalk between these two molecules in HCC cell lines. We further confirmed the rescue of HGF-suppressed HOTAIR expression after c-Met tyrosine kinase inhibition by SU11274 in HCC cell lines HuH-7, SNU-449, MAHLAVU and SK-HEP-1. c-Met tyrosine kinase inhibition rescued HGF-induced HOTAIR suppression in HCC cell lines (Fig. 3j).
To understand the biological significance of this interaction, we investigated the phenotypic and biological results of HOTAIR overexpression in SNU-449 HCC cell line.
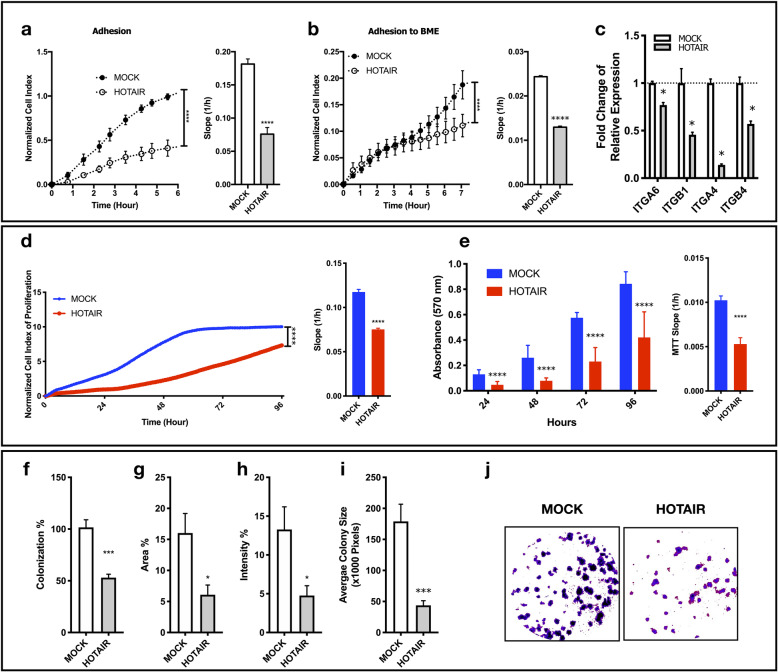
HOTAIR OE induced c-Met inhibition decreases adhesion, proliferation and colony formation of SNU-449 cells
HOTAIR OE cells showed reduced adhesion capacity to cell culture-treated (Fig. 4a) and basal membrane extract coated cell culture surfaces (Fig. 4b) in xCELLigence real time cell analysis system (RTCA). To understand the cause of decreased extracellular matrix (ECM) attachment capacity, we analyzed the expression of cell surface integrins that contribute to matrix attachment. ITGA6, ITGB1, ITGA4 and ITGB4 expressions were down-regulated in HOTAIR OE cells, significantly (Fig. 4c).
Fig. 4.
HOTAIR OE induced c-Met inhibition decreases adhesion, proliferation and colony formation. Normalized cell index and slope analysis of adhesion to (a) cell culture treated and (b) basal membrane extract-coated surfaces analyzed with xCELLigence RTCA. (c) RT-qPCR analysis of ITGA6, ITGB1, ITGA4 and ITGB4 expressions. (d) Analysis of normalized cell index and rate (slope) of proliferation by xCELLigence RTCA system. Normalized (e) MTT absorbance (570 nm) and proliferation rate (slope) calculated by MTT absorbance. 2D colony formation analysis of MOCK and HOTAIR clones presented as (f) percentage of colonization, (g) percentage of area covered by colonies, (h) intensity of Giemsa-staining (i) average colony size in pixels and (j) representative images of clones generated with ImageJ plugin ColonyArea. 2D colony formation is analyzed by ImageJ plugin ColonyArea. All graphs of experiments are presented as the mean ± SEM of at least 3 independent experiments. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001
Proliferation of the cells were evaluated with both xCELLigence RTCA system (Fig. 4d) and MTT analysis (Fig. 4e). HOTAIR OE cells showed significant reduction in proliferation rate in both techniques. Compatible with proliferation assays, 2D colony forming potential of HOTAIR OE cells were dramatically low. Colony counts are normalized to number of cells plated for evaluation of colonization percentage. HOTAIR OE cells showed significantly lower colonization ability in comparison to MOCK cells (Fig. 4f). Coverage area of colonies in well-plates, staining intensity of colonies and average colony sizes were also significantly lower in HOTAIR OE cells (Fig. 4g-j). These results reflect lower capacities of adhesion and proliferation.
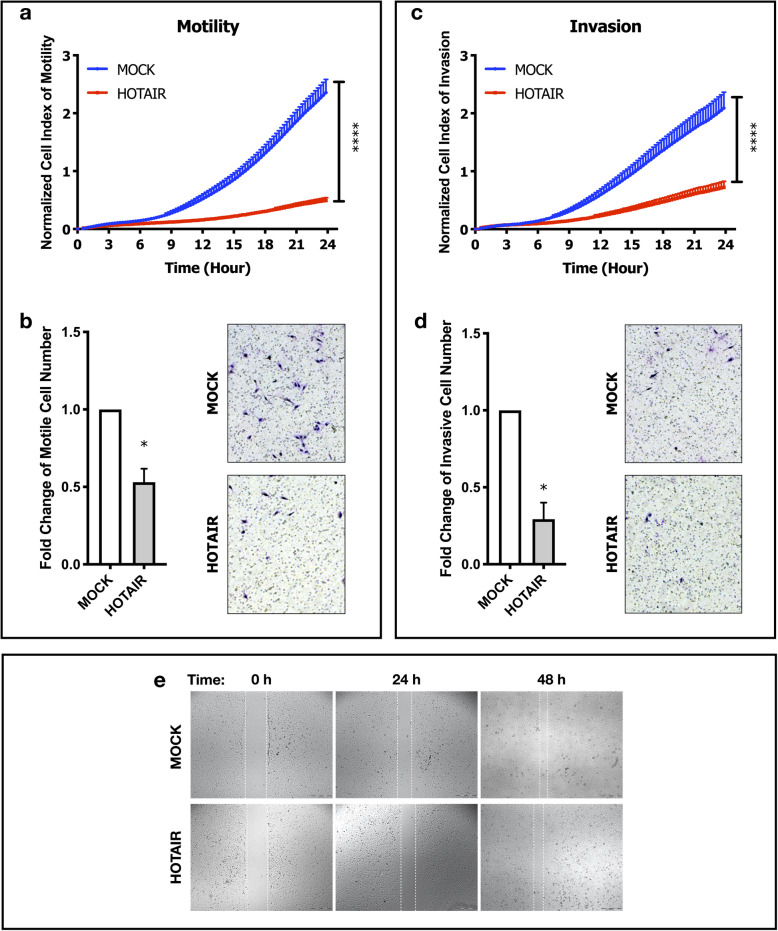
HOTAIR OE induced c-Met inhibition decreases motility and invasion capacity of SNU-449 cells
Motility and invasion experiments were performed in parallel by both xCELLigence RTCA system and trans-well cell culture systems. Cell index of motile (Fig. 5a) and invasive (Fig. 5c) cells in HOTAIR OE clones were significantly lower in xCELLigence RTCA (p ≤ 0.001). Consistent with real-time motility and invasion analysis; the number of motile and invasive cells migrated through trans-well cell culture inserts were reduced in HOTAIR OE cells (Fig. 5b, d), significantly (p ≤ 0.05).
Fig. 5.
HOTAIR OE induced c-Met inhibition decreases individual motility and invasion ability of SNU-449 cells. Real-time (by xCELLigence RTCA) and end-point (cell culture inserts) analysis of motility and invasion of MOCK and HOTAIR OE cell clones. Normalized cell index of (a) motility and (c) invasion analyzed by xCELLigence RTCA system. Fold change graphs and brightfield images of (b) motile and (d) invaded cells in trans-well cell culture inserts. (e) Brightfield microscopy images of 0-, 24- and 48-h scratch (wound-healing) assay of MOCK and HOTAIR OE clones. All graphs of experiments are presented as the mean ± SE of at least 3 independent experiments. * p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001
Since trans-well systems especially tests individual cell movement ability, we further analyzed collective migration ability of clones in scratch (wound-healing) assay. Scratch assay enables cells to move collectively, without losing cell-cell attachments. Even though HOTAIR OE cells have reduced ability to migrate individually, they were still motile. Rather than migrating individually, HOTAIR OE cells migrated without losing cell-cell contacts, collectively (Fig. 5e).
lncRNA HOTAIR inhibits c-Met induced cell scattering and promotes metastasis in zebrafish embryos
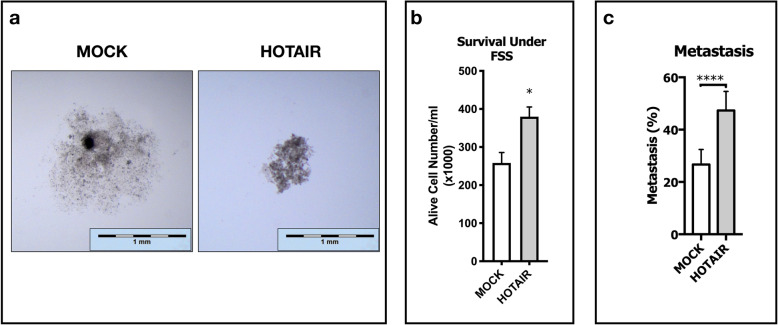
Decrease in attachment capacity to ECM led us to investigate spheroid formation capacity of HOTAIR OE cells. We analyzed spheroid formation ability of those cells with hanging-drop spheroid formation assay. SNU-449 cells have a complete mesenchymal phenotype, as a result of scattering effect of constitutive c-Met activation, and they are not inclined to form spheroids in hanging-drops. Due to their reduced ability to form cell-cell contacts maintained by constitutive c-Met activation; MOCK cells formed loose, scattered and wide clusters whereas HOTAIR OE cells formed more tight, stacked and small spheroids (Fig. 6a). Significant ability to form intact spheroids led us to investigate their survival capacity under fluidic shear stress (FSS). To test their survival ability, cells sheared for 1 h under FSS of 0.5 dyn/cm2, mimicking the sinusoidal shear stress in liver [39]. HOTAIR OE improved survival ability under sinusoidal FSS. Alive cell numbers after one hour of FSS was significantly higher (p ≤ 0.05) in HOTAIR OE clones (Fig. 6b).
Fig. 6.
lncRNA HOTAIR suppresses c-Met induced cell scattering, enhances survival under FSS and promotes metastasis in zebrafish embryos. Spheroid formation, survival under fluidic shear stress and metastatic capacity of MOCK and HOTAIR OE cells. (a) Spheroid images were collected with stereo-microscopy and magnified equally with scale-bars to generate more clarified presentation of data. (b) Graphical presentation of alive cell numbers of MOCK and HOTAIR OE clones cultured under 0.5 dyn/cm2 fluidic shear stress for 1 h. Embryonic zebrafish xenograft experiments with MOCK and HOTAIR OE SNU-449 cell clones presented as (c) percentage of metastasis in groups that were analyzed in total of three biological experiments. Statistical significance was analyzed by chi-square with Yates correction. All graphs of experiments are presented as the mean ± SE of at least 3 independent experiments. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001
Finally, embryonic zebrafish xenograft model was used to test in vivo metastatic capacity of HOTAIR OE, further. Average number of 100 HOTAIR OE and MOCK cells were injected to the yolk sac of 48 h zebrafish embryos and embryos which bear an intact tumorosphere after 4 h of injection were raised for 4 days to analyze metastatic capacity. Three independent experiments were performed and in every experiment each group consisted an average number of 50 embryos with intact tumorospheres. Metastatic capacity was calculated by number of xenograft hosts that have metastasized cells at 4 days post injection (dpi). Even though wild-type SNU-449 cells are known to be metastatic in embryonic zebrafish model [30], HOTAIR OE dramatically increased their metastatic capacity when compared to MOCK cells (Fig. 6c).
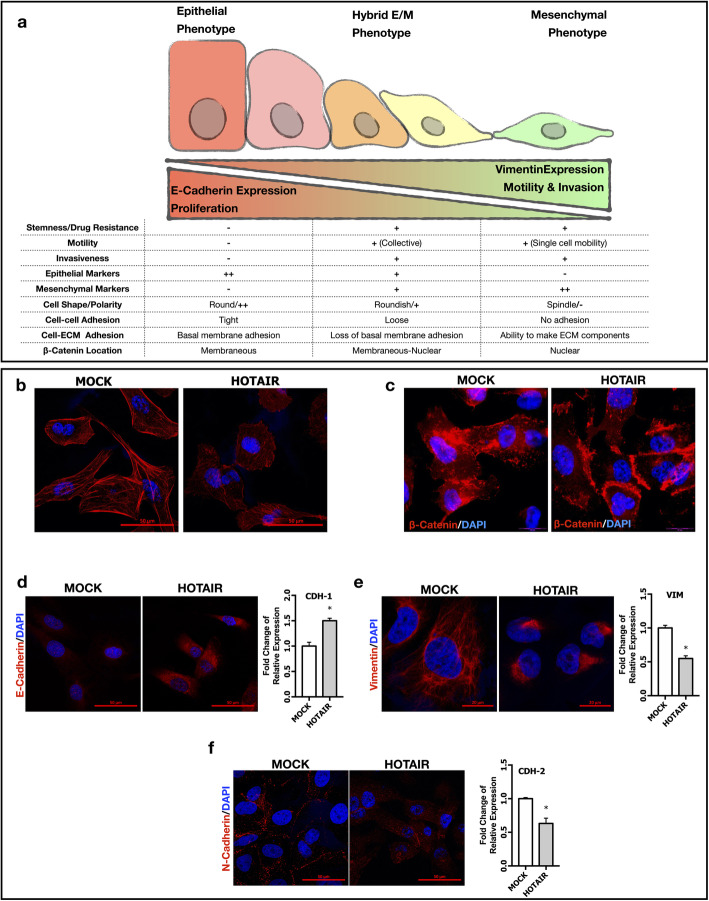
lncRNA HOTAIR maintains hybrid E/M phenotype to promote metastasis
Our experiments with HOTAIR OE cells show morphological and biological properties that are consistent with the hybrid E/M phenotype which can be defined as a fluidic state between epithelial and mesenchymal phenotypes (Fig. 7a). To test this hypothesis, we examined previously defined EMT markers in HOTAIR OE SNU-449 cells [40–42]. First, we found that in contrast to the mesenchymal and elongated phenotype, HOTAIR OE cells were round-shaped with thinner F-actin stress fibrils indicated with phalloidin staining (Fig. 7b). Then, we performed a detailed investigation of molecular biomarkers of hybrid E/M phenotype. In HOTAIR OE cells, beta-catenin was enriched in the plasma membrane (Fig. 7c), expression of epithelial biomarker E-cadherin was upregulated (Fig. 7d) whereas mesenchymal biomarker Vimentin expression was significantly down-regulated and showed polarity (Fig. 7e). Consistent with epithelial phenotype, N-Cadherin expression was down-regulated and mostly located in the plasma membrane of HOTAIR OE cells (Fig. 7f). Immunofluorescent analysis of cytoskeletal organization of F-actin and Vimentin in SNU-398 MET OE clones showed that ectopic MET expression and its suppressive effect on HOTAIR expression was also resulted in thickening of F-actin stress fibrils, and increase in Vimentin expression and polarity (Supplementary Fig. 1d).
Fig. 7.
lncRNA HOTAIR maintains hybrid E/M phenotype to promote metastasis. (a) Schematic presentation of morphological, molecular and biological biomarkers of epithelial, mesenchymal and hybrid E/M phenotypes. Confocal imaging of (b) Alexa-555 labeled phalloidin staining of F-actin filaments (red). Immunofluorescence imaging of (c) beta-Catenin protein (red) and its cellular localization. Confocal imaging and RT-qPCR analysis of (d) E-Cadherin (e) Vimentin and (f) N-Cadherin expressions. Alexa-594 conjugated secondary antibodies were used in all immunolabeling and confocal imaging experiments. DAPI (blue) was used as a nuclear marker in immunofluorescence and confocal images. All graphs of experiments are presented as the mean ± SEM of at least 3 independent experiments. * p ≤ 0.05
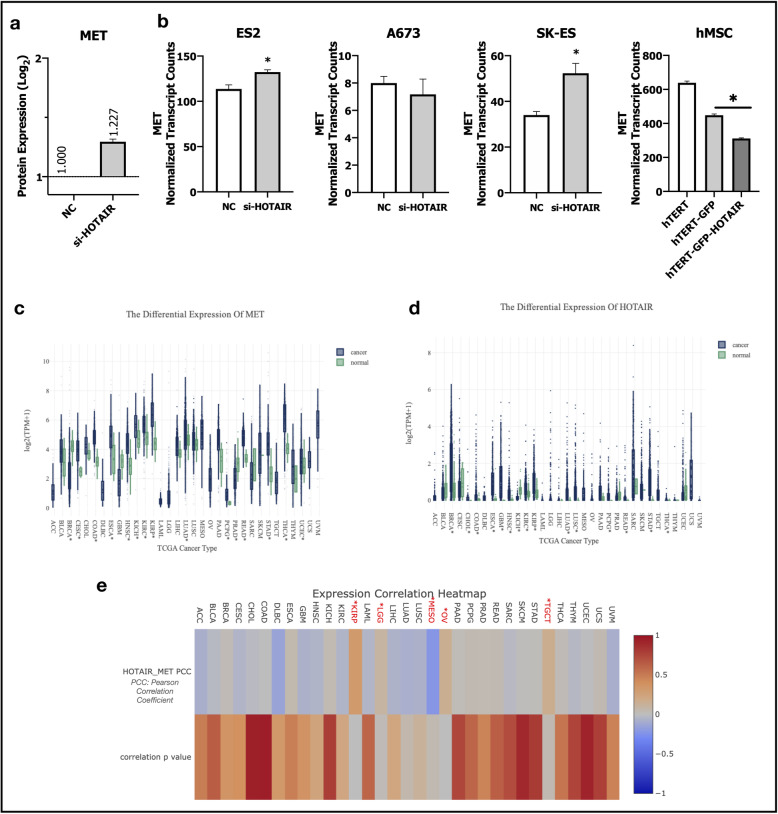
In addition to in-vitro experiments, we performed bioinformatic analysis of available data-sets GSE98091 and GSE109483. GSE98091 dataset has an integrated RNA sequencing transcriptomic and quantitative proteomic analysis those performed to explore the regulatory role of HOTAIR in HCC [43]. Researchers inhibited HOTAIR expression by siRNA silencing in HepG2 cells and analyzed differentially expressed genes and proteins between control-siRNA and HOTAIR-siRNA HepG2 clones. Even though transcriptional dysregulation of MET was not significant in this study, protein expression of c-Met was significantly higher in HepG2 cells transfected with HOTAIR siRNA (Fig. 8a). In GSE109483 dataset, researchers repressed HOTAIR expression in three Ewing sarcoma cell lines (ES2, A673, SK-ES) and overexpressed HOTAIR in hTERT-immortalized human mesenchymal stem cells to evaluate its effects on gene expression [44]. They performed RNA-Seq in Ewing sarcoma cell lines (control-GapmeR and HOTAIR repressing GapmeR clones), and hTERT-immortalized hMSCs with expression of control GFP and HOTAIR. MET gene expression was significantly upregulated by HOTAIR repression in ES2 and SK-ES Ewing Sarcoma cell lines and its expression was significantly repressed by HOTAIR over-expression in hTERT-immortalized human mesenchymal stem cells (Fig. 8b). The data of these datasets supports proposed interaction of HOTAIR and MET and they are very important to show similar results with HCC in different cancer and cellular contexts.
Fig. 8.
Bioinformatic analyses of GEO and TCGA datasets supports HOTAIR and c-Met interplay. (a) Column graph of c-Met protein expression in control-siRNA and HOTAIR-siRNA transfected HepG2 cells in GEO dataset GSE98091. (b) MET gene expression in experimental groups of GEO dataset GSE109483. Column graphs of control-GapMeR and HOTAIR-GapmeR transfected Ewing sarcoma cell lines ES2, A673 and SK-ES (1st, 2nd and 3rd graphs). Last graph belongs to hTERT-immortalized human mesenchymal stem cells (hMSC), hMSCs transfected with GFP (hTERT-GFP) and HOTAIR-GFP (hTERT-GFP-HOTAIR) overexpression vectors. Analysis of (c) MET (d) HOTAIR gene expressions in TCGA datasets. (e) Heatmap of gene expression correlation analysis of HOTAIR and MET in TCGA datasets. GEO dataset analysis are performed with GREIN and TCGA gene expression and correlation analysis are performed with LncTarD. * p ≤ 0.05
Finally, we analyzed TCGA datasets of various cancers to have a foresight about the possible relationship of two molecules [3]. HOTAIR and MET gene transcripts in TCGA datasets were analyzed by GREIN and LncTarD [32, 33]. Despite the limitations in RNAseq sample counts among TCGA datasets that comprise RNA sequencing of long non-coding RNAs, HOTAIR and c-Met were differentially expressed between normal and primary tumor tissues in various cancer types (Fig. 8c-d). While MET expression was significantly suppressed in primary tumor tissues in breast invasive carcinoma (BRCA) and upregulated in kidney chromophobe (KIHC), HOTAIR expression was significantly higher and lower, respectively (Fig. 8c-d). Analysis of the correlation of these two molecules among TCGA datasets showed that and they were reversely correlated with each other in mesothelioma (MESO) but positively correlated in kidney renal papillary cell carcinoma (KIRP), brain lower grade glioma (LGG), ovarian serous cystadenocarcinoma (OV) and testicular germ cell tumors (TGCT) with statistical significance. Even lack of significance, tendency of reverse regulation was significance in adrenocortical carcinoma (ACC), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), lymphoid neoplasm diffuse large b-cell lymphoma (DLBC), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), acute myeloid leukemia (LAML), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD) and ulveal melanoma (UVM), (Fig. 8e). Although lncRNA HOTAIR, one of the most studied long non-coding RNAs, is reported to be elevated in patients with metastasis and poor prognosis in many cancer types, analysis of HOTAIR in TCGA datasets revealed that HOTAIR has a context-dependent expression pattern which is not always upregulated through the progression of the disease (Fig. 8d-e).
Discussion
Our results suggest c-Met and HOTAIR axis as a modulator of epithelial/mesenchymal hybrid state in hepatocellular carcinoma cells. Mechanistically, we demonstrated that the interplay between c-Met receptor tyrosine kinase and HOTAIR is critical for maintenance of hybrid E/M state in HCC cells to cope with rate-limiting steps of tumor metastasis, such as survival in adhesion independent microenvironment, escaping from anoikis and resisting to fluidic shear stress (FSS) in HCC.
HCC cell lines with high c-Met expression and activation are defined to have mesenchymal phenotype in previous definitive studies [45, 46]. In our study, we report that HCC cells can be divided into two clusters which show differential expression tendency for HOTAIR and c-MET genes. Expression of these two molecules among 10 HCC cell lines demonstrated that most HCC cell lines show a reverse tendency in levels of c-Met and HOTAIR expression. To further confirm the possible reverse interaction between HOTAIR and c-Met, we overexpressed HOTAIR in SNU-449 cell line which has ligand-independent constitutive c-Met activation and silenced HOTAIR in HuH-7 cell line which expresses HOTAIR, abundantly. Our results show that constitutive c-Met expression is suppressed by HOTAIR over-expression and HOTAIR silencing induced c-Met expression in HCC cells. It is very important to note that inhibition of expression with siRNAs is limited mostly with cytoplasmic HOTAIR mRNA targeting and further studies should be done to address the detailed mechanism of HOTAIR action in means of molecular interactions and/or epigenetic function in HCC cells.
Analysis of microarray dataset comprised of tissues from normal liver, dysplastic liver, early and late HCC of patients (GSE89377) showed that HOTAIR was downregulated through hepatocarcinogenesis whereas c-Met expression was upregulated. This analysis supports our data showing HOTAIR downregulation is a requirement for c-Met activation and c-Met triggered complete mesenchymal phenotype in HCC cell lines. Contrary to the reports of melanoma, retinoblastoma and colon cancer studies [47–49] and expectations on the parallel expression tendencies of c-Met and HOTAIR in HCC cell lines; comparative analysis of HCC cell lines and cancer datasets shows the opposite, clearly. Indicated studies suggested HOTAIR as a competing long non-coding RNA decoying microRNAs targeting c-Met in different cancer contexts, and those microRNAs are reported to have different molecular targets in HCC rather than c-Met [50, 51].
In addition to suppression of c-Met protein expression and activation as a consequence, HOTAIR weakens downstream Akt1, MAPK and STAT3 signaling pathways not only by decreasing c-Met expression but also by modulating its membrane organization. In our previous studies, we showed that Caveolin-1 enhances c-Met signaling by co-localizing in plasma membrane [27]. Src kinase, a well-identified downstream effector of c-Met, phosphorylates Caveolin-1 from Tyr-14 residue to induce lipid-raft enrichment of c-Met with Caveolin-1 and formation of caveola structures [27, 52]. HOTAIR over-expression reduced Caveolin-1 expression and activation via suppressing activation of c-Met and its downstream effector Src kinase. In parallel to c-Met, Caveolin-1 organization in plasma membrane was also disrupted in HOTAIR OE cells. Tsai et al. defined function of HOTAIR in epigenetic regulation of its target genes and they reported Caveolin-1 as one of the most differentially expressed genes in response to HOTAIR knock-down in foreskin fibroblasts [12]. In addition to the HOTAIR OE data, we showed that HOTAIR knock-down elevated CAV1 expression in HuH-7 cells (Supplementary Fig. 1e). We confirmed the interaction axis of c-Met, HOTAIR and Caveolin-1 is conserved in c-Met over-expression model of SNU-398 cell line. Its wild type is lack of c-Met expression but expresses HOTAIR, abundantly. In addition to the lack of c-Met expression, SNU-398 cells also do not express Caveolin-1 [26]. As a consequence of c-Met overexpression and HOTAIR suppression, Caveolin-1 expression was induced in those cells. In addition to the over-expression studies of c-Met and HOTAIR, we showed that c-Met and HOTAIR reverse interaction is conserved in both ligand-dependent and independent c-Met activation contexts and also c-Met kinase inhibition. These results support the defined interplay between two molecules.
To understand the biological significance of HOTAIR/c-Met interaction, we analyzed the results of HOTAIR overexpression on c-Met related biological responses which are clearly defined in the literature as contributing to aggressive phenotype of HCC cells [53]. HOTAIR over-expressing cells showed delayed adhesion to surfaces and suppressed expression of adhesion-related integrins [54]. In addition to decreased attachment to surface, proliferation and colony formation ability of HOTAIR OE cells were also decreased. Suppression of individual motility and invasion of HOTAIR OE cells were consistent with suppressed scattering effect of c-Met signaling. HOTAIR OE cells were migrating collectively rather than individually which is compatible with previous studies explaining migration behavior of cells with hybrid E/M phenotype [15]. Increased expression of cell adhesion molecules such as CDH-1 and JAM-2 (Supplementary Fig. 1f) were consistent with the evidence of collective migration that requires preservation of cell-to-cell interactions [55, 56].
Surprisingly, HOTAIR over-expressing cells showed increased survival ability in adhesion independent culture and under FSS. Escaping from anoikis and overcoming damage incurred by shear forces are important necessities of survival in circulation to achieve metastasis [57]. Response to fluidic shear-stress or adhesion-independent conditions are context-dependent processes those depend on the molecular pool of the cell; HOTAIR over-expressing cells resisted those unfavorable conditions via formation of spheres [39]. Aggregation into spheroids to overcome the stress of unfavorable microenvironment, enhancing cell-cell adhesion and surviving under damage of shear forces are well-defined requirements of metastasis [39, 57]. As expected, by attainment of those abilities, HOTAIR over-expression increased metastatic ability of SNU-449 cells which was defined to be highly metastatic in our previous studies in zebrafish xenograft model [30]. Our study is the first in the literature defining the molecular basis of HOTAIR overexpression mediated increase in the metastatic ability of HCC cells.
Since the definition of EMT has been changed and broadened, transitional states between epithelial and mesenchymal phenotypes are called “partial EMT”. Existence of intermediate hybrid phenotypes have been defined in different molecular and biological processes such as fibrosis, development, wound healing and cancer [15]. In our study, we found that HOTAIR acts as a fine tuner of maintenance of hybrid E/M phenotype by modulating c-Met signaling. Compatibly with the literature, we analyzed defined morphological and molecular markers of epithelial and mesenchymal phenotypes in HOTAIR over-expressing cells [15, 16, 41, 42]. HOTAIR OE cells were round-shaped and had thinner F-actin stress fibrils. Beta-Catenin was enriched in membrane compatible with increased E-cadherin expression and hybrid E/M phenotype characteristics [18]. In comparison with MOCK cells, mesenchymal biomarkers Vimentin and N-cadherin expression was suppressed in HOTAIR OE cells (Fig. 7). Consistent with HOTAIR over-expression data in SNU-449 cells, c-Met overexpression in SNU-398 cells increased F-actin stress fibrils and Vimentin expression and, HOTAIR knock-down induced Vimentin expression in HuH-7 cells (Supplementary Fig. 1 g). While over-expression of c-Met in SNU-398 cells suppressed HOTAIR expression and led acquirement of mesenchymal attributes; inhibition of HOTAIR expression in HuH-7 cells enhanced mesenchymal phenotype. Changes in cytoskeletal components and dis−/re-assembling of cell-cell contacts regulates gene expression, motility and cell-cycle. Cells placed in spectrum between epithelial and mesenchymal phenotypes are reported to arrest cell cycle and decrease their proliferative activity [15, 17]. Consistent with the literature, proliferation rate of HOTAIR OE cells were decreased (Fig. 4) and cell cycle was arrested (Supplementary Fig. 1 h-i) in these cells.
Decrease in biological responses such as individual cell motility and invasion are consistent with suppressed scattering effect of c-Met signaling activity. Even individual cell motility was decreased, HOTAIR OE clones have ability to enclose the scratch via collective migration as fast as MOCK clones in wound-healing assay (Fig. 5). Collective migration is not only important for maintenance of invading front of tumors but also it is an emerging mechanism for seeding of secondary tumors [58]. Taken together, our data shows that decrease in HOTAIR expression and its effects on c-Met and Caveolin-1 activation is crucial through the completion of mesenchymal phenotype.
Concordant with our data, we report that c-Met and HOTAIR expression profiles show inverse correlation in mesothelioma but positively correlated in kidney renal papillary cell carcinoma, brain lower grade glioma, ovarian serous cystadenocarcinoma and testicular germ cell tumors with statistical significance. With the fact that all samples in TCGA cancer datasets do not compromise long non-coding gene expression data, analysis of expression correlation with HOTAIR has its limitations. HOTAIR and MET gene expression showed a remarkable tendency of reverse regulation in adrenocortical carcinoma, cervical squamous cell carcinoma and endocervical adenocarcinoma, lymphoid neoplasm diffuse large b-cell lymphoma, glioblastoma multiforme, head and neck squamous cell carcinoma, kidney renal clear cell carcinoma, acute myeloid leukemia, liver hepatocellular carcinoma, lung adenocarcinoma and uveal melanoma (Fig. 8e). The power of correlation analysis would be more enhanced by expansion of the sample sizes with non-coding gene expression data of public cancer datasets. Besides TCGA, we further analyzed available public research datasets in GEO database. Analysis of available data generated by siRNA knock-down of HOTAIR in an HCC cell line HepG2 [43], (Fig. 8a) and human sarcoma cell lines ES2, A673 and SK-ES (Fig. 8b) supported our in-vitro HOTAIR knock-down data in HuH-7 cells [44]. Moreover, data generated by over-expression of HOTAIR in human mesenchymal stem cells was consistent with our data in SNU-449 cells [44], (Fig. 8b, hMSC).
Our study clearly defines the fine tuning of c-Met signaling by HOTAIR to maintain hybrid E/M phenotype which ensures flexibility of commitment and enhances metastatic ability under unfavorable conditions through rate-limiting steps of metastasis. Hybrid phenotype contributes metastatic colonization and may play a crucial role in intra-and extrahepatic metastasis [59]. On the other way around, c-Met activation proceeds E/M spectrum through complete mesenchymal phenotype by suppressing HOTAIR expression. Even this study defines the interplay between two molecules, the detailed molecular interactions are still needed to be investigated. Considering the complex structure and multifunctional behavior, HOTAIR might be regulating c-Met signaling through physically interacting and epigenetic or transcriptional suppression of c-Met and/or its regulators. Further studies are needed to address the molecular cascade between HOTAIR, c-Met and Caveolin-1 in detail.
Conclusions
In conclusion, HOTAIR over-expression suppresses c-Met expression, activation and also disrupts its organization on plasma membrane by modulating Caveolin-1 expression and activation. HOTAIR over-expression provides an advantage to HCC cells via maintaining hybrid E/M phenotype thereby enhancing survival in adhesion-independent and shear-stressed conditions. HOTAIR overexpression decreases adhesion dependency and proliferation while increasing spheroid formation and improving collective migration abilities and overall improving metastatic potential of HCC cells. Further studies are needed to brighten detailed regulatory signaling cascades and mapping potential targets for therapeutic applications.
Supplementary information
Additional file 1: Figure S1. Includes supplementary data indicated in the text.
Additional file 2: Supplementary document 1. Includes primer sequences used in gene expression analysis and catalog numbers of antibodies used in immunoblotting and/or immunofluorescent labeling experiments.
Acknowledgments
We thank Prof. Dr. Mehmet Öztürk for providing HCC cells, Izmir Biomedicine and Genome Center for establishment of required technical and instrumental infrastructure for this study, IBG Vivarium Zebrafish facility for housing and providing the animals, Melek Ucuncu and Didem Cimtay from IBG Optical Imaging Core Facility for their supports for imaging processes of samples.
Materials & Correspondence:
Requests should be addressed to Nese Atabey (nese.atabey@ibg.edu.tr).
Abbreviations
- ACC
Adrenocortical carcinoma
- BRCA
Breast invasive carcinoma
- CESC
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- COAD
Colon adenocarcinoma
- CTCs
Circulating tumor cells
- DLBC
Lymphoid neoplasm diffuse large B-cell lymphoma
- EMT
Epithelial to mesenchymal transition
- FSS
Fluidic shear stress
- GBD
Global Burden of Disease
- GBM
Glioblastoma multiforme
- HCC
Hepatocellular carcinoma
- HNSC
Head and neck squamous cell carcinoma
- HOTAIR
HOX Transcript Antisense Intergenic RNA
- KICH
Kidney chromophobe
- KIRC
Kidney renal clear cell carcinoma
- KIRP
Kidney renal papillary cell carcinoma
- LAML
Acute myeloid leukemia
- LGG
Brain lower grade glioma
- LIHC
Liver hepatocellular carcinoma
- lncRNA
long non-coding RNA
- LUAD
Lung adenocarcinoma
- MESO
Mesothelioma
- MET
Mesenchymal to epithelial transition
- OE
Over-expression
- OV
Ovarian serous cystadenocarcinoma
- RT-qPCR
Reverse transcriptase-quantitative polymerase chain reaction
- RT
Room temperature
- RTCA
Real time cell analysis system
- TCGA
The Cancer Genome Atlas
- UVM
Uveal melanoma
Authors’ contributions
HT, EB, DC and GB performed the experiments. HT performed data analysis and curation. GCA supervised and trained HT for Zebrafish xenograft model establishment and performance. HT and NA conceived the experiments and study plan. Data was mainly interpreted by HT and NA. EB, DC and GB contributed experiment discussions. HT prepared the original draft and NA performed review edit of the manuscript. NA performed supervision, project administration and funding acquisitions. The authors critically read and approved the manuscript.
Funding
This research was funded by “Department of Scientific Research Projects of Dokuz Eylul University (BAP), grant number 2016.KB.SAG.005” and “Scientific and Technological Research Council of Turkey (TUBITAK), grant number 114S359“. HT was awarded with a Short-Term Fellowship from the European Molecular Biology Organization (EMBO) between 01.06.17–01.07.17.
Availability of data and materials
Eun et al., 2016, performed expression profiling array to identify novel drivers of hepatocellular carcinoma and reveal clinical relevance as early diagnostic and prognostic biomarkers (data accessible at NCBI GEO database (Eun et al., 2016), accession GSE89377). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE89377
Wu et al., 2018, performed an integrated proteomic and transcriptomic analysis to examine the overall transcriptomic changes in HepG2 cells after HOTAIR knockdown by RNA sequencing (data accessible at NCBI GEO database (Wu et al., 2018), accession GSE98091).
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE98091
Shah et al., generated a next-generation RNA-sequencing data of models of Ewing sarcoma with modulated expression (repression with locked nucleic acid antisense oligonucleotides and over-expression with lentiviral transduction) of the lncRNA HOTAIR (data accessible at NCBI GEO database (Shah et al., 2018), accession GSE109483).
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE109483
The results reported here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Ethics approval and consent to participate
In vitro experiments of this study were approved by Non-Interventional Research Ethics Board of Dokuz Eylül University with the protocol number of 1597-GOA. Animal experiments of this study was approved by IBG Animal Ethics Committee with the protocol number of 03/2019.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12964-020-00602-0.
References
- 1.Global Burden of Disease Cancer C. Fitzmaurice C, Abate D, Abbasi N, Abbastabar H, Abd-Allah F, et al. Global, regional, and National Cancer Incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2017: a systematic analysis for the global burden of disease study. JAMA Oncol. 2019;5:1749–1768. doi: 10.1001/jamaoncol.2019.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Electronic address wbe, Cancer Genome Atlas Research N Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169(7):1327–1341. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouattour M, Raymond E, Qin S, Cheng AL, Stammberger U, Locatelli G, et al. Recent developments of c-met as a therapeutic target in hepatocellular carcinoma. Hepatology. 2018;67(3):1132–1149. doi: 10.1002/hep.29496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Firtina Karagonlar Z, Koc D, Iscan E, Erdal E, Atabey N. Elevated hepatocyte growth factor expression as an autocrine c-met activation mechanism in acquired resistance to sorafenib in hepatocellular carcinoma cells. Cancer Sci. 2016;107(4):407–416. doi: 10.1111/cas.12891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng J, Yang M, Jiang R, An N, Wang X, Liu B. Long non-coding RNA HOTAIR regulates the proliferation, self-renewal capacity, tumor formation and migration of the cancer stem-like cell (CSC) subpopulation enriched from breast cancer cells. PLoS One. 2017;12(1):e0170860. doi: 10.1371/journal.pone.0170860. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan SK, Pastori C, Penas C, Komotar RJ, Ivan ME, Wahlestedt C, et al. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol Cancer. 2018;17(1):74. doi: 10.1186/s12943-018-0822-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W, He X, Zheng Z, Ma X, Hu X, Wu D, et al. Serum HOTAIR as a novel diagnostic biomarker for esophageal squamous cell carcinoma. Mol Cancer. 2017;16(1):75. doi: 10.1186/s12943-017-0643-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang X, Feng D, Li M, Zhou J, Li X, Zhao D, et al. Transcriptomic analysis of mRNA-lncRNA-miRNA interactions in hepatocellular carcinoma. Sci Rep. 2019;9(1):16096. doi: 10.1038/s41598-019-52559-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Somarowthu S, Legiewicz M, Chillon I, Marcia M, Liu F, Pyle AM. HOTAIR forms an intricate and modular secondary structure. Mol Cell. 2015;58(2):353–361. doi: 10.1016/j.molcel.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166(1):21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 16.Denisov EV, Perelmuter VM. A fixed partial epithelial-mesenchymal transition (EMT) triggers carcinogenesis, whereas asymmetrical division of hybrid EMT cells drives cancer progression. Hepatology. 2018;68(3):807–810. doi: 10.1002/hep.29784. [DOI] [PubMed] [Google Scholar]
- 17.Lovisa S, LeBleu VS, Tampe B, Sugimoto H, Vadnagara K, Carstens JL, et al. Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat Med. 2015;21(9):998–1009. doi: 10.1038/nm.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basu S, Cheriyamundath S, Ben-Ze'ev A. Cell-cell adhesion: linking Wnt/beta-catenin signaling with partial EMT and stemness traits in tumorigenesis. F1000Res. 2018;7:F1000 Faculty Rev-1488. doi: 10.12688/f1000research.15782.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lecharpentier A, Vielh P, Perez-Moreno P, Planchard D, Soria JC, Farace F. Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. Br J Cancer. 2011;105(9):1338–1341. doi: 10.1038/bjc.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Armstrong AJ, Marengo MS, Oltean S, Kemeny G, Bitting RL, Turnbull JD, et al. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol Cancer Res. 2011;9(8):997–1007. doi: 10.1158/1541-7786.MCR-10-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Revenu C, Gilmour D. EMT 2.0: shaping epithelia through collective migration. Curr Opin Genet Dev. 2009;19(4):338–342. doi: 10.1016/j.gde.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Grosse-Wilde A, Fouquier d'Herouel A, McIntosh E, Ertaylan G, Skupin A, Kuestner RE, et al. Stemness of the hybrid epithelial/mesenchymal state in breast cancer and its association with poor survival. PLoS One. 2015;10(5):e0126522. doi: 10.1371/journal.pone.0126522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Almale L, Garcia-Alvaro M, Martinez-Palacian A, Garcia-Bravo M, Lazcanoiturburu N, Addante A, et al. C-met signaling is essential for mouse adult liver progenitor cells expansion after transforming growth factor-beta-induced epithelial-mesenchymal transition and regulates cell phenotypic switch. Stem Cells. 2019;37(8):1108–1118. doi: 10.1002/stem.3038. [DOI] [PubMed] [Google Scholar]
- 24.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18(5):1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 25.Ishibashi M, Kogo R, Shibata K, Sawada G, Takahashi Y, Kurashige J, et al. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 2013;29(3):946–950. doi: 10.3892/or.2012.2219. [DOI] [PubMed] [Google Scholar]
- 26.Cokakli M, Erdal E, Nart D, Yilmaz F, Sagol O, Kilic M, et al. Differential expression of Caveolin-1 in hepatocellular carcinoma: correlation with differentiation state, motility and invasion. BMC Cancer. 2009;9:65. doi: 10.1186/1471-2407-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korhan P, Erdal E, Kandemis E, Cokakli M, Nart D, Yilmaz F, et al. Reciprocal activating crosstalk between c-met and caveolin 1 promotes invasive phenotype in hepatocellular carcinoma. PLoS One. 2014;9(8):e105278. doi: 10.1371/journal.pone.0105278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gunes A, Iscan E, Topel H, Avci ST, Gumustekin M, Erdal E, et al. Heparin treatment increases thioredoxin interacting protein expression in hepatocellular carcinoma cells. Int J Biochem Cell Biol. 2015;65:169–181. doi: 10.1016/j.biocel.2015.05.025. [DOI] [PubMed] [Google Scholar]
- 29.Guzman C, Bagga M, Kaur A, Westermarck J, Abankwa D. ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS One. 2014;9(3):e92444. doi: 10.1371/journal.pone.0092444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunes A, Bagirsakci E, Iscan E, Cakan-Akdogan G, Aykutlu U, Senturk S, et al. Thioredoxin interacting protein promotes invasion in hepatocellular carcinoma. Oncotarget. 2018;9(96):36849–36866. doi: 10.18632/oncotarget.26402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bagirsakci E, Sahin E, Atabey N, Erdal E, Guerra V, Carr BI. Role of albumin in growth inhibition in hepatocellular carcinoma. Oncology. 2017;93(2):136–142. doi: 10.1159/000471807. [DOI] [PubMed] [Google Scholar]
- 32.Mahi NA, Najafabadi MF, Pilarczyk M, Kouril M, Medvedovic M. GREIN: An interactive web platform for re-analyzing GEO RNA-seq data. Sci Rep. 2019;9(1):7580. doi: 10.1038/s41598-019-43935-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao H, Shi J, Zhang Y, Xie A, Yu L, Zhang C, et al. LncTarD: a manually-curated database of experimentally-supported functional lncRNA-target regulations in human diseases. Nucleic Acids Res. 2020;48(D1):D118–DD26. doi: 10.1093/nar/gkz985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avci ME, Keskus AG, Targen S, Isilak ME, Ozturk M, Atalay RC, et al. Development of a novel zebrafish xenograft model in ache mutants using liver cancer cell lines. Sci Rep. 2018;8(1):1570. doi: 10.1038/s41598-018-19817-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eun J, Nam S. Identifying novel drivers of human hepatocellular carcinoma and revealing clinical relevance as early diagnostic and prognostic biomarker. 2017. [Google Scholar]
- 36.Iscan E, Gunes A, Korhan P, Yilmaz Y, Erdal E, Atabey N. The regulatory role of heparin on c-met signaling in hepatocellular carcinoma cells. J Cell Commun Signal. 2017;11(2):155–166. doi: 10.1007/s12079-016-0368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yilmaz Y, Topel H, Atabey N. 25th biennial congress of the European Association for Cancer Research; 2018/7/1. Amsterdam: ESMO Open: BMJ Publishing Group Limited; 2018. Identification of C-met and insulin receptor heterodimer as a mediator for hepatocellular carcinoma; p. A297-A. [Google Scholar]
- 38.Topel H, Comez D, Atabey N. 25th biennial congress of the European Association for Cancer Research; 2018/7/1. Amsterdam: BMJ Publishing Group Limited; 2018. HGF/c-met signalling pathway downregulates lncRNA HOTAIR to induce adhesion independent growth in HCC by increasing caveolin-1 expression; pp. A282–A2A3. [Google Scholar]
- 39.Huang Q, Hu X, He W, Zhao Y, Hao S, Wu Q, et al. Fluid shear stress and tumor metastasis. Am J Cancer Res. 2018;8(5):763–777. [PMC free article] [PubMed] [Google Scholar]
- 40.Jolly MK, Somarelli JA, Sheth M, Biddle A, Tripathi SC, Armstrong AJ, et al. Hybrid epithelial/mesenchymal phenotypes promote metastasis and therapy resistance across carcinomas. Pharmacol Ther. 2019;194:161–184. doi: 10.1016/j.pharmthera.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 41.Kroger C, Afeyan A, Mraz J, Eaton EN, Reinhardt F, Khodor YL, et al. Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. Proc Natl Acad Sci U S A. 2019;116(15):7353–7362. doi: 10.1073/pnas.1812876116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jolly MK, Boareto M, Huang B, Jia D, Lu M, Ben-Jacob E, et al. Implications of the hybrid epithelial/mesenchymal phenotype in metastasis. Front Oncol. 2015;5:155. doi: 10.3389/fonc.2015.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu Y, Xiong Q, Li S, Yang X, Ge F. Integrated proteomic and transcriptomic analysis reveals long noncoding RNA HOX transcript antisense intergenic RNA (HOTAIR) promotes hepatocellular carcinoma cell proliferation by regulating opioid growth factor receptor (OGFr) Mol Cell Proteomics. 2018;17(1):146–159. doi: 10.1074/mcp.RA117.000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shah N. In: Next-generation RNA-sequencing data of models of Ewing sarcoma with modulated expression of the lncRNA HOTAIR. Geo N, editor. 2018. [Google Scholar]
- 45.Hirschfield H, Bian CB, Higashi T, Nakagawa S, Zeleke TZ, Nair VD, et al. In vitro modeling of hepatocellular carcinoma molecular subtypes for anti-cancer drug assessment. Exp Mol Med. 2018;50(1):e419. doi: 10.1038/emm.2017.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuzugullu H, Benhaj K, Ozturk N, Senturk S, Celik E, Toylu A, et al. Canonical Wnt signaling is antagonized by noncanonical Wnt5a in hepatocellular carcinoma cells. Mol Cancer. 2009;8:90. doi: 10.1186/1476-4598-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luan W, Li R, Liu L, Ni X, Shi Y, Xia Y, et al. Long non-coding RNA HOTAIR acts as a competing endogenous RNA to promote malignant melanoma progression by sponging miR-152-3p. Oncotarget. 2017;8(49):85401–85414. doi: 10.18632/oncotarget.19910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang G, Fu Y, Lu X, Wang M, Dong H, Li Q. LncRNA HOTAIR/miR-613/c-met axis modulated epithelial-mesenchymal transition of retinoblastoma cells. J Cell Mol Med. 2018;22(10):5083–5096. doi: 10.1111/jcmm.13796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu B, Liu Q, Pan S, Huang Y, Qi Y, Li S, et al. The HOTAIR/miR-214/ST6GAL1 crosstalk modulates colorectal cancer procession through mediating sialylated c-met via JAK2/STAT3 cascade. J Exp Clin Cancer Res. 2019;38(1):455. doi: 10.1186/s13046-019-1468-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang X, Wu J, Zhang Y, Wang S, Yu X, Li R, et al. MiR-613 functions as tumor suppressor in hepatocellular carcinoma by targeting YWHAZ. Gene. 2018;659:168–174. doi: 10.1016/j.gene.2018.03.036. [DOI] [PubMed] [Google Scholar]
- 51.Wang W, Zhang H, Wang L, Zhang S, Tang M. miR-613 inhibits the growth and invasiveness of human hepatocellular carcinoma via targeting DCLK1. Biochem Biophys Res Commun. 2016;473(4):987–992. doi: 10.1016/j.bbrc.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 52.Gottlieb-Abraham E, Shvartsman DE, Donaldson JC, Ehrlich M, Gutman O, Martin GS, et al. Src-mediated caveolin-1 phosphorylation affects the targeting of active Src to specific membrane sites. Mol Biol Cell. 2013;24(24):3881–3895. doi: 10.1091/mbc.E13-03-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia-Vilas JA, Medina MA. Updates on the hepatocyte growth factor/c-met axis in hepatocellular carcinoma and its therapeutic implications. World J Gastroenterol. 2018;24(33):3695–3708. doi: 10.3748/wjg.v24.i33.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sokeland G, Schumacher U. The functional role of integrins during intra- and extravasation within the metastatic cascade. Mol Cancer. 2019;18(1):12. doi: 10.1186/s12943-018-0937-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aurrand-Lions M, Johnson-Leger C, Wong C, Du Pasquier L, Imhof BA. Heterogeneity of endothelial junctions is reflected by differential expression and specific subcellular localization of the three JAM family members. Blood. 2001;98(13):3699–3707. doi: 10.1182/blood.v98.13.3699. [DOI] [PubMed] [Google Scholar]
- 56.Zhao H, Yu H, Martin TA, Zhang Y, Chen G, Jiang WG. Effect of junctional adhesion molecule-2 expression on cell growth, invasion and migration in human colorectal cancer. Int J Oncol. 2016;48(3):929–936. doi: 10.3892/ijo.2016.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Campbell K, Casanova J. A common framework for EMT and collective cell migration. Development. 2016;143(23):4291–4300. doi: 10.1242/dev.139071. [DOI] [PubMed] [Google Scholar]
- 59.Giannelli G, Koudelkova P, Dituri F, Mikulits W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J Hepatol. 2016;65(4):798–808. doi: 10.1016/j.jhep.2016.05.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Includes supplementary data indicated in the text.
Additional file 2: Supplementary document 1. Includes primer sequences used in gene expression analysis and catalog numbers of antibodies used in immunoblotting and/or immunofluorescent labeling experiments.
Data Availability Statement
Eun et al., 2016, performed expression profiling array to identify novel drivers of hepatocellular carcinoma and reveal clinical relevance as early diagnostic and prognostic biomarkers (data accessible at NCBI GEO database (Eun et al., 2016), accession GSE89377). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE89377
Wu et al., 2018, performed an integrated proteomic and transcriptomic analysis to examine the overall transcriptomic changes in HepG2 cells after HOTAIR knockdown by RNA sequencing (data accessible at NCBI GEO database (Wu et al., 2018), accession GSE98091).
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE98091
Shah et al., generated a next-generation RNA-sequencing data of models of Ewing sarcoma with modulated expression (repression with locked nucleic acid antisense oligonucleotides and over-expression with lentiviral transduction) of the lncRNA HOTAIR (data accessible at NCBI GEO database (Shah et al., 2018), accession GSE109483).
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE109483
The results reported here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.