Abstract
Background
Liquid-based cytology (LBC) is now a widely used method for cytologic screening and cancer diagnosis. Since the cells are fixed with alcohol-based fixatives, and the specimens are stored in a liquid condition, LBC specimens are suitable for genetic analyses.
Methods
Here, we established a small cancer gene panel, including 60 genes and 17 microsatellite markers for next-generation sequencing, and applied to residual LBC specimens obtained by endometrial cancer screening to compare with corresponding formalin-fixed paraffin-embedded (FFPE) tissues.
Results
A total of 49 FFPE and LBC specimens (n = 24) were analyzed, revealing characteristic mutations for endometrial cancer, including PTEN, CTNNB1, PIK3CA, and PIK3R1 mutations. Eight cases had higher scores for both tumor mutation burden (TMB) and microsatellite instability (MSI), which agree with defective mismatch repair (MMR) protein expression. Paired endometrial LBC, and biopsied and/or resected FFPE tissues from 7 cases, presented almost identical mutations, TMB, and MSI profiles in all cases.
Conclusion
These findings demonstrate that our ad hoc cancer gene panel enabled the detection of therapeutically actionable gene mutations in endometrial LBC and FFPE specimens. Endometrial cancer LBC specimens offer an alternative and affordable source of molecular testing materials.
Keywords: Endometrial cytology, LBC, NGS, Endometrial cancer, MSI, TMB
Background
Endometrial cancer is the most common gynecologic cancer worldwide, affecting over 500,000 women every year [1]. Since lifestyle changes can affect the risk of developing endometrial cancer, an epidemiological approach is essential to manage the disease [2]. Moreover, genome-wide association studies have identified new susceptibility loci for endometrial cancer in the human genome, providing candidate genes for further studies aiming to unravel the mechanisms driving carcinogenesis, which could offer new opportunities for early screening and identification of therapeutic targets [3]. Unlike squamous cell carcinoma of the uterine cervix, which is characterized by pathogenesis closely related to that of human papilloma virus infection [4], endometrial carcinogenesis is linked to common cancer gene mutations and genome instability [5, 6].
Cytology specimens, including conventional smear, cytospin, liquid-based cytology (LBC), and cell block, are valuable sources of routine molecular diagnostics [7]. Extensive studies have been reported that a variety of cytology materials, containing LBC specimens suitable for next-generation sequencing (NSG) testing, especially in non-small cell lung cancer [7–12]. Meanwhile, the endometrial cytology is not only a safe and easy clinical procedure but also comparable to suction endometrial biopsy for detecting atypical endometrial hyperplasia and cancers [13–16]. Since 1987, in Japan, a cytologic examination for endometrial cancer screening has been established by the National Health Insurance law. After that, endometrial cytology, using a specific device, was introduced as a less invasive and less painful tool [17, 18] to screen high-risk population and perimenopausal women with abnormal genital bleeding. Conventional endometrial cytology preparation, however, has some disadvantages such as bloody background, cellular overlapping, and thick cell clusters; therefore, the resulting cytological screening would not be widely accepted in endometrial cancer management except in Japan [19]. After the introduction of LBC in endometrial cytology in combination with transvaginal sonography, LBC-based endometrial cytology, a less invasive and expensive procedure, was proposed to screen cancer also in asymptomatic postmenopausal women [20, 21].
To date, LBC represents a routine endometrial cytology method for cancer diagnosis and gene analysis [22]. We also previously reported that residual LBC samples could better preserve the genome quality for downstream NSG, as compared to formalin-fixed paraffin-embedded (FFPE) tissues, even after several years of storage [23]. In the present study, we established a small custom cancer gene panel with full exon coverage of 60 cancer-related genes and 17 microsatellite foci. We then used this custom panel to comparatively analyze the genetic profiles obtained from residual endometrial LBC specimens and corresponding FFPE tissues with respect to the presence of somatic gene mutations, tumor mutation burden (TMB), and microsatellite instability (MSI). We further conducted immunohistochemistry for detecting the expression of mismatch repair (MMR) proteins in the FFPE sections to compare the results obtained by genomic analysis. The detection of MMR protein expression is known as a reliable proxy of the MSI status [24]. This study demonstrates the feasibility of using a small NGS-based cancer gene panel to screen for endometrial cancer in residual LBC specimens. The reliability of the results was compared with those obtained from biopsied and resected FFPE tissue analysis.
Methods
Histological, cytological and blood specimens
In this study, 53 tissue and cytology samples from 24 patients with endometrial cancer registered in the Clinical Research of Cancer Gene Panel Analysis of Gynecologic Cancers study from January to August 2019 at Kagoshima University Hospital were used. The specimens included 2 mL of whole blood as a reference, FFPE tissues from 10 biopsied, 29 resected tissues (39 FFPE tissues in total), and 10 residual LBC samples of endometrial cytology. The tissues were fixed with phosphate-buffered neutral 10% formalin within 24 h and routinely processed for paraffin embedding, followed by sectioning for hematoxylin and eosin (H&E) staining. The FFPE blocks were stored in the dark at room temperature (25 ± 2 °C). The endometrial cytology samples were obtained using a specific device (Endocyte® sampler, Laboratoire CCD, Paris, France) [17, 18], and collected cells were fixed with CytoRich Red solution (Becton Dickinson, Franklin Lakes, NJ, USA). The cytology samples were routinely processed for LBC using BD SurePath liquid-based Pap Test System (Becton Dickinson). The residual LBC specimens were stored at 4 °C.
The pathological diagnosis was made by two board-certified pathologists (IK and AT), according to the World Health Organization classification [25], and the tumor fraction in the sections (% area) was evaluated in 10% increments. Similarly, the cytological diagnosis was made by two board-certified cytopathologists (IK and TH), according to the classification system proposed by the Japanese Society of Clinical Cytology [26]. For cytology specimens, atypical or tumor cell numbers were counted in five high-power fields (HPF) at × 400 magnification. The average counts were calculated and the total tumor cells (T) subjected to NGS were estimated. Similarly, non-tumor cell counts were calculated as the average of five HPF, and total non-tumor cells (N) were estimated, and then the tumor cell ratio (T/N + T) was calculated. The cytological and pathological studies were performed independently from the molecular studies described below.
DNA extraction and quality check
Whole blood DNA was extracted using the QIAamp DNA Blood Mini Kit (Qiagen). DNA from FFPE tissues was obtained from 3 to 6 sections with 10-μm thickness, including more than 30% of the cancer area, or after microdissection using a microdissection laser (Leica Biosystems, Nussloch GmbH, Germany). One mL of residual LBC samples was centrifuged (× 12,000 g), and the pellets were resuspended in 95% ethanol and air-dried for 5 min. Both the FFPE sections and LBC pellets were then incubated with proteinase K (Promega, Madison, WI, USA) for 15 h at 70 °C, followed by a 1-h incubation at 98 °C in lysis buffer (Promega). After centrifugation (× 12,000 g), the supernatants were applied to the Maxwell RSC DNA FFPE kit and Maxwell 16 system (Promega). After measuring the extracted DNA concentration using the Qubit 3.0 dsDNA BR assay kit (Life Technologies, Grand Island, NY, USA), DNA qualities were monitored using the QIAseq DNA quantimize kit (Qiagen). A quality check score (QC score) less than 0.04 was considered as a high-quality DNA, according to our previous report [23].
Design of the custom gene panel
A total of 60 cancer-related genes and 17 microsatellite foci [27] were selected from QIAseq Targeted DNA Custom Panel (Qiagen [28]), including 2615 primers for the regions of interest, with an average exon coverage of 99.87%. The cancer-related genes were used for the construction of a custom panel (detailed in Table 1). The selection of these 60 genes was validated according to the Catalogue Of Somatic Mutations In Cancer (COSMIC ver. 90, https://cancer.sanger.ac.uk/cosmic) to include target genes of molecular targeting drugs for solid cancers, and genes frequently expressed in ovarian and endometrial cancers.
Table 1.
Region of interests of custom panel
| 60 genes | 17 MSI regions | |||||
|---|---|---|---|---|---|---|
| Target | Loci | Gene | ||||
| AKT1 | CTNNB1 | MAP2K1 | PIK3CA | Bat-25 | chr4:55598208–55,598,241 | KIT |
| APC | DDR2 | MAP2K2 | PIK3R1 | Bat-26 | chr2:47641403–47,641,591 | MSH2 |
| ATM | EGFR | MAP2K4 | PMS2 | MONO-27 | chr2:39564890–39,564,926 | MAP4K3 |
| AR | ERBB2 | MED12 | PTEN | NR-21 | chr14:23652343–23,652,372 | SLC7A8 |
| ARID1A | ERBB3 | MET | RB1 | NR-24 | chr2:95849358–95,849,389 | ZNF2 |
| BARD1 | ERBB4 | MDM2 | RAD51 | MSI-1 | chr1:201754407–201,754,432 | NAV1 |
| BRAF | ESR1 | MLH1 | STK11 | MSI-3 | chr2:62063090–62,063,115 | FAN161A |
| BRCA1 | FGFR1 | MSH2 | TP53 | MSI-4 | chr2:108479619–108,479,675 | RGPD4 |
| BRCA2 | FGFR2 | MSH6 | RET | MSI-6 | chr5:172421757–172,421,780 | ATP6V0E1 |
| BRIP1 | FGFR3 | MTOR | MSI-7 | chr6:142691947–142,691,972 | GPR126 | |
| CCND1 | FLT3 | MUTYH | MSI-8 | chr7:1787516–1,787,541 | ELFN1 | |
| CD274 | HRAS | MYC | MSI-11 | chr11:106695511–106,695,531 | GUCY1A2 | |
| CDK4 | IDH1 | NF1 | HSPH1-T17 | chr13:31722617–31,722,642 | HSPH1 | |
| CDK6 | IDH2 | NRAS | MSI-12 | chr15:45897768–45,897,790 | BLOC1S6 | |
| CDKN2A | KDR | PALB2 | MSI-13 | chr16:18882656–18,882,679 | SMG1 | |
| CDH1 | KIT | PDCD1 | MSI-14 | chr17:19314914–19,314,940 | RNF112 | |
| CTLA4 | KRAS | PDGFRA | EWSR1 | chr22:29696465–29,696,489 | EWSR1 | |
NGS analysis
Forty ng of DNA from whole blood cells and LBC specimens and 100–200 ng of DNA from FFPE tissue sections were used to construct the NGS library based on QIAseq Targeted DNA Custom Panel (Qiagen). The quality of the libraries was monitored using an Agilent high-sensitivity DNA kit (Agilent Technologies, Santa Clara, CA, USA) to confirm the successful generation of 300-bp PCR products. The libraries were then applied to MiSeq sequencer (Illumina, San Diego, CA, USA) after dilution with a hybridization buffer to a final concentration of 20 pM. The obtained sequencing data were analyzed by the Qiagen Web Portal service (https://www.qiagen.com/us/shop/genes-and-pathways/data-analysis-center-overview-page/ ). Qiagen Web Portal service, equipped with a smCounter analyzing pipeline [29], indicates that a mean unique molecular index (UMI) sequence depth of 2,000 is necessary for detection of 0.5% variant allele frequency (VAF), and the cut-off value of VAF varies, inversely proportional to the mean sequence depth. Our NGS, with a condition of tumor fraction ≥20% and cluster passing filter rate ≥ 90%, yielding a mean UMI depth of 500, at which the SNP detection noise was low when VAF was ≥10%. Therefore, the cut-off value of VAF was set to 10%, also considering the sample loading size in one NGS run and its cost. Human genome reference GRCh37 hg19 (https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13/) and COSMIC database were used as the analytical references. For validation of the NSG panel, all the sequence data were analyzed and annotated by Qiagen Web Portal service and Mitsubishi Space Software (Amagasaki, Hyogo, Japan, https://www.mss.co.jp/business/life-science/). Human cell lines (HEK293T [catalogue # ATCC CRL-3216], HCT166 [catalogue # ATCC CCL-247], and MDA-MB453 cells [catalogue # ATCC HTB-131]) obtained from American Type Culture Collection (Rockville, MD) were analyzed as controls, and the genomic sequence data were compared to those deposited in the COSMIC database. In this study, DNA sequence data obtained from whole blood were used only for reference, whereas germline analysis was not performed.
Calculation of TMB and MSI scores
The numbers of missense mutations, including non-synonymous mutations and internal deletions, with more than 10% VAF, were counted as somatic mutations. TMB was calculated as the number of single nucleotide variants/Mbp of DNA sequence [30, 31]. MSI scores were determined by MSIsensor (ver. 1.0) [32, 33].
Immunohistochemistry for MMR proteins
To evaluate the expression of MMR proteins, FFPE tissue sections were applied to immunohistochemistry (IHC) using antibodies against MLH1 (clone ES05 M3640), MSH2 (clone FE11 M3639), MSH6 (clone EP49 M3646) and PMS2 (clone EP51 M3647) purchased from DAKO (Tokyo, Japan) [34, 35]. Staining was performed on representative 5-μm-thick FFPE sections with Envision FLEX High pH K8000 system (DAKO) according to the manufacturer’s protocol. Positive nuclear staining of lymphocytes in the tissue sections was monitored as a positive control. MMR deficiency was defined as complete loss of nuclear staining for both MLH1 and PMS2, both MSH2 and MSH6, MSH6 only, or PMS2 only. When partial loss of each protein was observed, the case was defined as showing heterogeneous expression.
Statistical analyses
All values are expressed as the mean ± standard deviation. Significant differences were analyzed using Welch’s t-test and Mann-Whitney U test. Values of p < 0.05 were considered statistically significant. The cut-off values for the evaluation of TMB-high and MSI-high conditions were determined by receiver operator characteristic (ROC) curves.
Ethical approval for the genome studies
The studies using clinical samples were approved by the Ethics Committees for Clinical and Epidemiologic Research at Kagoshima University, and written-informed consent was obtained from each participant. The studies did not include participants younger than the age of 20.
Results
DNA quality and quantity obtained from FFPE and LBC specimens
The results of the pathological diagnosis, DNA quality, and input DNA for NGS of the 24 endometrial cancer cases are summarized in Table 2. The storage period of FFPE and LBC specimens ranged from 2 weeks to 3 years. The tumor fraction of the FFPE sections ranged from 30 to 90%. In endometrial LBC specimens, estimated tumor cell count (T) subjected to NGS varied from 1500 to 140,000, and the tumor cell ratio varied from 27 to 95% (T/N + T). All of the genomic DNA extracted from the FFPE and LBC samples demonstrated high-quality and sufficient quantity for library construction and successful sequencing.
Table 2.
Pathological and DNA sample information for NGS analysis
| Case no. | Sample no. | Samples | Pathological diagnosis | Storage time | DNA yield (ng/μL) | QC score | Input DNA (ng) | Tumor fraction in FFPE (%) | Tumor cell count and tumor cell ratio in LBC | Insufficient VAF call | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tumor cells (T) | Non-tumor cells (N) | Tumor cell ratio (T/N+T)% | ||||||||||
| 1 | 1 | SR(Ut) | EC G1 | 1 m | 1.655 | 0.012 | 200 | 30 | ||||
| 2 | 2 | SR(Ut) | EC G1 | 2 m | 5.510 | 0.003 | 100 | 80 | ||||
| 3 | 3 | SR(Ut) | EC G2 | 1 m | 2.980 | 0.008 | 100 | 60 | ||||
| 4 | 4 | SR(Ut) | EC G1 | 1 m | 1.521 | 0.016 | 200 | 50 | ||||
| 5 | 5 | SR(Ut) | EC G1 | 3 m | 4.033 | 0.010 | 119 | 80 | ||||
| 6 | 6 | SR(Ut) | EC G1 | 3 m | 6.636 | 0.003 | 72 | 30 | ||||
| 7 | SR(Ut) | EC G2 | 3 m | 6.071 | 0.004 | 79 | 60 | |||||
| 7 | 8 | SR(Ut) | EC G1 + Sq | 1 m | 5.912 | 0.003 | 200 | 70 | ||||
| 8 | 9 | SR(Ut) | Mixed EC/SC | 2 m | 3.487 | 0.010 | 138 | 50 | ||||
| 10 | SR(Om) | Mixed EC/SC | 2 m | 4.897 | 0.007 | 98 | 30 | |||||
| 9 | 11 | Bx | SC | 3 m | 6.425 | 0.004 | 75 | 80 | ||||
| 12 | SR(Ut) | CCC | 3 m | 1.930 | 0.015 | 168 | 90 | |||||
| 13 | End LBC | AC | 3 m | 28.879 | 0.000 | 20 | 6661 | 10,059 | 39.8 | |||
| 10 | 14 | SR(Ut) | DC | 3 m | 3.337 | 0.007 | 144 | 70 | ||||
| 15 | SR(Ut) | EC G1 | 3 m | 5.743 | 0.004 | 84 | 70 | |||||
| 11 | 16 | SR(Ut) | AC | 1 m | 2.327 | 0.011 | 168 | 60 | ||||
| 17 | SR(Ut) | DC | 1 m | 0.939 | 0.016 | 168 | 50 | |||||
| 18 | SR(Ov) | EC G3 | 1 m | 1.778 | 0.012 | 168 | 40 | |||||
| 12 | 19 | Bx | AC | 1 m | 2.264 | 0.010 | 168 | 60 | ||||
| 20 | SR(Ut) | EC G2 | 1 m | 3.025 | 0.006 | 168 | 70 | |||||
| 21 | End LBC | Malig | 2 m | 21.861 | 0.002 | 22 | 19,588 | 4087 | 82.7 | |||
| 13 | 22 | SR(Ut) | EC G1 | 1 m | 2.338 | 0.010 | 168 | 50 | ||||
| 14 | 23 | Bx | EC G1 | 1 m | 5.664 | 0.001 | 85 | 70 | ||||
| 24 | End LBC | AGC | 2 m | 23.839 | −0.003 | 10 | 1450 | 136 | 91.4 | |||
| 15 | 25 | SR(Ut) | DC | 1 m | 6.249 | 0.008 | 77 | 90 | ||||
| 26 | SR(Ut) | AC | 1 m | 5.830 | 0.010 | 85 | 90 | |||||
| 16 | 27 | Bx | AC | 1 m | 5.029 | 0.010 | 95 | 80 | ||||
| 28 | SR(Ut) | Serous | 1 m | 0.872 | 0.017 | 168 | 40 | |||||
| 29 | End LBC | AC | 2 m | 14.257 | 0.005 | 31 | 22,261 | 3928 | 85.0 | |||
| 17 | 30 | SR(Ut) | AH | 1 m | 3.443 | 0.007 | 140 | 70 | ||||
| 31 | SR(Ut) | EC G1 | 1 m | 4.137 | 0.006 | 116 | 60 | |||||
| 18 | 32 | Bx | EC G1 | 1 m | 3.794 | 0.009 | 127 | 90 | ||||
| 33 | SR(Ut) | EC G1 | 1 m | 3.025 | 0.012 | 168 | 80 | |||||
| 34 | End LBC | EC | 2 m | 3.392 | 0.008 | 11 | 8043 | 20,911 | 27.8 | * | ||
| 19 | 35 | SR(Ut) | EC G1 + Sq | 2 m | 4.585 | 0.004 | 105 | 30 | ||||
| 36 | End LBC | EC | 4 m | 16.984 | 0.000 | 26 | 7739 | 9261 | 45.5 | |||
| 20 | 37 | Bx | EC G2 | 3y | 0.492 | 0.022 | 168 | 60 | ||||
| 21 | 38 | Bx | EC G1 | 3 m | 2.612 | 0.012 | 168 | 90 | ||||
| 39 | SR(Ut) | EC G1 | 1 m | 1.321 | 0.025 | 154 | 90 | |||||
| 40 | End LBC | EC | 3 m | 16.114 | 0.005 | 36 | 16,855 | 10,739 | 61.1 | |||
| 22 | 41 | Bx | EC G3 | 2y | 1.024 | 0.022 | 168 | 40 | ||||
| 42 | End LBC | AGC | 1 m | 16.208 | 0.004 | 30 | 14,816 | 1019 | 93.6 | |||
| 23 | 43 | Bx | AC | 2 m | 3.479 | 0.010 | 138 | 90 | ||||
| 44 | SR(Ut) | EC G2 | 2w | 2.443 | 0.012 | 168 | 90 | |||||
| 45 | End LBC | EC | 2 m | 8.576 | 0.004 | 54 | 45,220 | 2537 | 94.7 | |||
| 24 | 46 | Bx | EC | 4 m | 2.443 | 0.009 | 168 | 80 | ||||
| 47 | SR(Ut) | EC G1 | 2 m | 5.058 | 0.005 | 95 | 80 | |||||
| 48 | SR(Ov) | EC G1 | 2 m | 1.694 | 0.011 | 168 | 70 | |||||
| 49 | End LBC | AC | 4 m | 24.088 | −0.001 | 31 | 21,930 | 6661 | 76.7 | |||
SRsurgical resection,BxEndometrial biopsy,LBCliquid-based cytology,EndEndometrial,UtUterus,OmOmentumOvOvary,ECEndometrioid carcinoma,SCSerous carcinoma,CCCClear cell carcinoma,ACAdenocarcinoma,SqSquamous differentiation,DCDedifferentiated carcinoma,MaligMalignant cell,AGCAtypical glandular cell,AHAtypical hyperplasia,w, week,mmonthy, year
Mutations detected in FFPE specimens
The detected mutations in endometrial cancers and genomic information of the variants are summarized in supplemental Tables S1 and S2. Among the 24 cases, 18 were finally diagnosed as endometrioid carcinoma (EC) by biopsy or surgical resection, including 12 cases of G1, 5 cases of G2, and 1 case of G3 EC. The other 6 cases consisted of 3 dedifferentiated carcinomas (DC), and 1 case each of mixed EC/serous carcinoma (SC), SC, and clear cell carcinoma (CCC). These endometrial cancers showed common mutation profiles, including PTEN, CTNNB1, PIK3CA, and PIK3R1 mutations.
The cases of mixed EC/SC, CCC, and SC had additional TP53 mutations. Two different FFPE sections relative to case no. 6 were analyzed, revealing G1 and G2 EC. While NGS analysis unveiled common mutations in PTEN, CTNNB1, and ARID1A, different PIK3CA and PIK3R1 mutations were also detected, suggesting the existence of at least two cancer clones. The three DC cases harbored PTEN, CTNNB1, PIK3CA, or PIK3R1 mutations along with multiple mutations in receptor-type tyrosine kinase genes, such as FGFR, ERBB, RET, and FLT. The mutation profiles of the EC and DC sections were not completely comparable but exhibited similarity with respect to PTEN mutation.
Mutations detected in LBC specimens
Endometrial LBC specimens contained abundant atypical cells, resulting in a higher frequency of mutation detection in the endometrial LBC specimens (9 out of 10 cases; supplemental Table S1). Mutations in PTEN, CTNNB1, and PIK3CA were identified in 1 case of atypical cell cytology (case no. 14), in which the diagnosis of G1 EC was confirmed by endometrial curettage biopsy.
Relations between MMR protein expression, TMB, and MSI
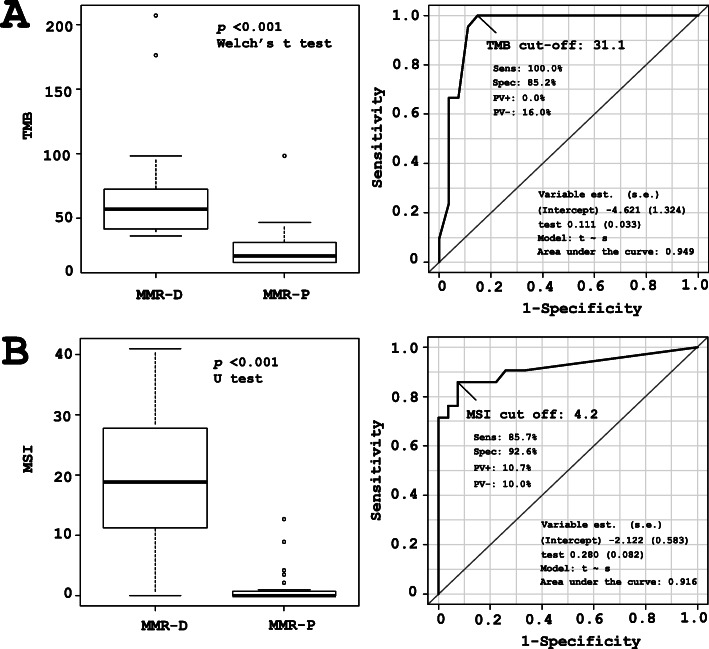
The overall relationships between MMR protein deficiency (MMR-D), TMB, and MSI status are shown (Fig. 1). In cases of MMR-D (9 cases, 21 samples), the TMB score was significantly higher than in cases of MMR protein proficiency (MMR-P) (15 cases, 27 samples) (p < 0.001; Fig. 2a, left). The MSI score of MMR-D cases was also significantly higher than that in MMR-P cases (p < 0.001; Fig. 2b, left). Estimated cut-off values for the evaluation of TMB-high (TMB-H) and MSI-high (MSI-H) were > 31.1 and > 4.2, respectively, as determined by the ROC curve (Fig. 2a and b).
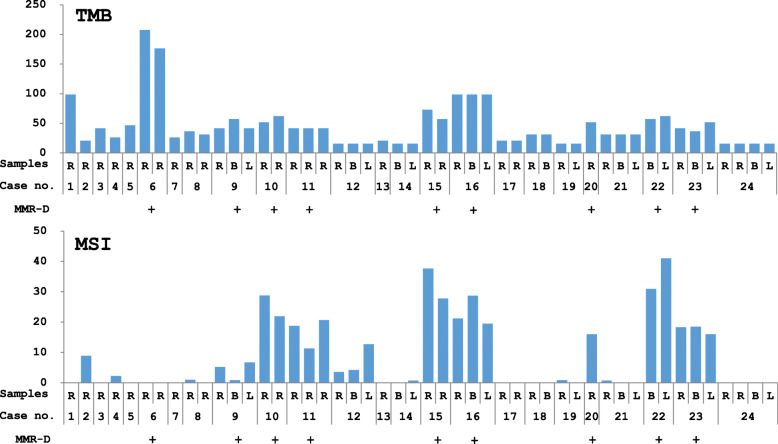
Fig. 1.
TMB and MSI scores in each case of endometrial cancer. Scores for TMB and MSI were calculated from the NGS analysis of FFPE tissues obtained by surgical resection (R), biopsy (B), and by LBC specimens (L). Most of the MMR protein-deficient cases (MMR-D: +), screened by IHC, had both higher TMB and MSI scores. In case nos. 1, 3, 5, and 12, no MMR-D was detected despite higher TMB or MSI
Fig. 2.
Relation of TMB, MSI scores, and MMR protein expression. a TMB scores and ROC curve. b, c, d MSI score and ROC curve. Cases with positive MMR protein expression (MMR-P) exhibited lower TMB and MSI scores, but the MMR protein-deficient cases (MMR-D) significantly demonstrated higher scores for TMB (p < 0.001) and MSI (p < 0.001). The ROC curve was used to determine the cut-off scores for TMB-H and MSI-H as 31.1 and 4.2, respectively
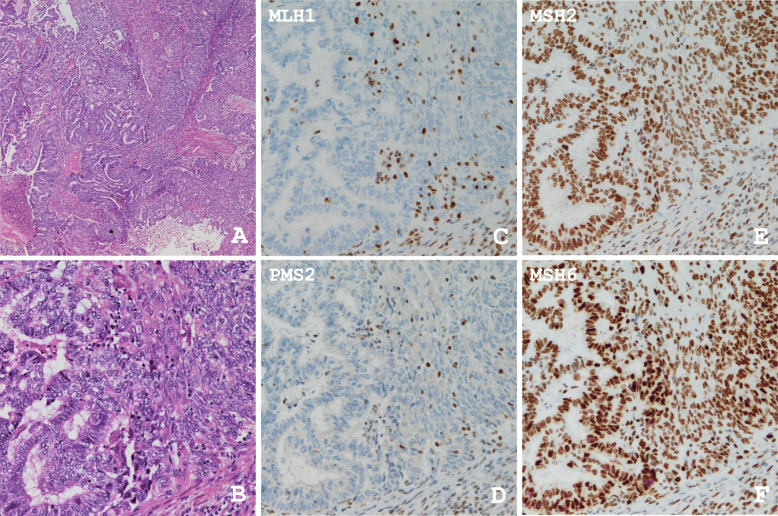
The IHC results, TMB, and MSI status are summarized in supplemental Table S3. The majority of MMR-D cases (8 of 9 cases), but not MMR-P cases, showed both TMB-H and MSI-H. The exception was the case no. 6, exhibiting heterogeneous loss of expression in MLH1, PMS2, and MSH6, which was only classified as TMB-H. One case with high TMB and MSI scores (case no. 16) presented a pathogenic mutation in MSH6 and the corresponding loss of MSH6 expression. Three cases of DC with MMR-D were found to be both TMB-H and MSI-H, among which 2 cases harbored pathogenic MLH1 mutations and loss of MLH1 and PMS2 protein expression. In 3 cases (case nos. 1, 3 and 5), no loss of MMR protein was detected despite a TMB-H status. In 1 case (case no. 12), despite being MSI-H positive, no MMR protein loss was detected. The photomicrographs of H&E staining and IHC from a representative MMR-D case of G2 EC (case no. 23) with loss of MLH1 and PMS2 expression are shown in Fig. 3. Very similar findings were obtained from cases no. 20 and 22, and both the TMB and MSI scores were over the cut-off values in these cases.
Fig. 3.
Representative H&E sections and IHC for MMR protein expression. a Scanning view of endometrioid carcinoma G2 (H&E, original magnification: 40×). b Higher power view of the endometrioid carcinoma G2 arranged in solid and glandular patterns (H&E, original magnification: 200×). c Absent expression of MLH1 in both the glandular and solid components (IHC, original magnification 200×). d Absent expression of PMS2 (IHC, original magnification 200×). e MSH2 expression was noted in both components (IHC, original magnification 200×). f MSH6 expression was also observed in the glandular and solid parts (IHC, original magnification 200×). Note the expression of these four proteins in stromal lymphocytes as an internal control
Correlation of the genetic diagnosis from LBC and FFPE specimens
In 10 cases (case nos. 9, 12, 14, 16, 18, 19, 21–24), the FFPE tissues from biopsy and/or resection along with endometrial LBC, were subjected to gene panel analysis. For cases no. 14 and 22, biopsied and endometrial LBC samples were analyzed together, revealing the same mutations. In 8 cases (case nos. 9, 12, 16, 18, 21, 23, and 24), 3 paired endometrial LBC, biopsied, and resection FFPE specimens were available for genetic studies. Seven of these 8 cases (except for case no. 18) exhibited almost identical mutation profiles and a similar TMB/MSI status.
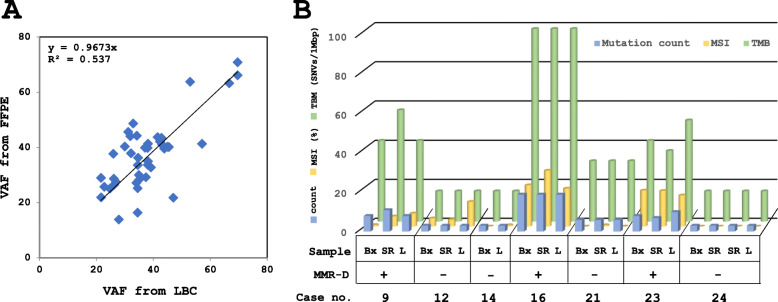
The VAF detected from 7 endometrial LBC specimens (case nos. 9, 12, 14, 16, 21, 23, and 24) showed a remarkably similar VAF to that of the same gene mutations from the corresponding biopsied FFPE (Fig. 4a). In case no. 22, VAF of APC markedly changed from 85.6% in LBC to 58.9% in biopsied FFPE, possibly because chemotherapy resulted in tumor clonal selection after LBC sampling and before the biopsy, and, therefore, the data were excluded from the Fig. 4a. All together with the results obtained from these 7 cases, a fine concordance was observed between endometrial LBC, and biopsied and/or resected FFPE tissue specimens in numbers of mutations, TMB and MSI scores, and MMR protein expression (Fig. 4b).
Fig. 4.
Correlation between corresponding LBC and FFPE specimens in VAF, Mutation counts, TMB, and MSI. a The VAF of mutated genes obtained from 7 endometrial LBC and that of the same gene mutations from corresponding FFPE were plotted, showing a good correlation between the selected LBC and FFPE specimens. The data consisted of VAF of 42 genes detected by NGS analysis in 7 cases with paired LBC and biopsy specimens. b Concordance of mutation counts, TMB, and MSI scores in paired LBC and FFPE (biopsy and resection) specimens from the 7 cases. Bx, biopsy; SR, surgical resection; L, LBC; MMR-D, mismatch repair protein deficiency
Discussion
We showed that LBC specimens offered sufficient DNA quality and quantity for NGS analyses. Endometrial LBC samples contained a much higher tumor cell ratio, allowing for NGS analysis to be efficiently performed. The endometrial LBC specimen containing at least 1450 tumor cells, and 10 ng of input DNA were successfully analyzed with the present protocol. A previous report suggested that 5000 viable cells are necessary for successful NGS using a 50-cancer gene hotspot panel from conventional cytology samples [36]. Therefore, appropriate cytology specimens containing more than several thousand cells with a higher tumor ratio could be an alternative source for genetic analysis to FFPE and conventional cytology specimens. However, the use of LBC specimens for NGS analysis faces a few disadvantages [37, 38]. Unlike NGS in scraped cells from smears, in which the same cells observed by cytopathologists are subjected to molecular testing, NGS analysis from LBC specimens is started with pooled residual samples. Therefore, although we checked for the presence of tumor cells in the residual LBC specimens and then applied to DNA extraction, false-negative materials may be applied to NGS. A rapid on-site evaluation to ensure the sufficient preservation of tumor cells are unavailable in LBC specimens, either. To the best of our knowledge, another disadvantage is that an efficient method to enrich tumor cell fraction in LBC specimens has not yet been developed. In this regard, the LBC specimens, such as inflammatory cell-rich ascites, may not be always suitable for NGS. Conventional smears and cell blocks are superior to LBC for increasing tumor cell fraction by trimming techniques [38, 39]. Conversely, one of the advantages of LBC specimens is the well-preserved DNA quality and yield even after long-term storage [23]. The absence of diagnostic material loss due to cell scraping and less complicated procedures represent additional advantage of LBC specimens.
Analysis with our custom-built panel identified mutations in PTEN (79%), CTNNB1 (54%), PIK3R1 (42%), PIK3CA (58%), ARID1A (67%), and TP53 (21%) in the FFPE specimens, and these variant frequencies agree with previous reports [40–42]. In 9 of 10 endometrial LBC samples, genetic analysis was successfully completed with PTEN (56%), CTNNB1 (44%), PIK3R1 (33%), PIK3CA (56%), ARID1A (56%), and TP53 (33%) mutation frequencies comparable to those previously reported based on amplicon sequencing for 5 genes (PETN, PIK3CA, CTNNB1, KRAS, and TP53), in which at least one pathogenic variant was identified in 17 of 20 cases (85%) [43]. Of note, a few differences in genomic profiles among different specimens from the same patients were observed. One possible explanation would be the presence of cancer heterogeneity harboring distinct gene mutations [44]. Furthermore, in endometrial neoplastic lesions, a complex non-linear molecular evolution happens between atypical endometrial hyperplasia (AH)/endometrial intraepithelial neoplasia (EIN) and EC [45]. The presence of minor AH/EIN components might result in genomic heterogeneity. In the present study, we did not analyze AH/EIN cases. Since a significant overlap of gene mutations is reported between AH, EIN, and EC [45, 46], a combination study with cytology and pathology would further improve the diagnostic accuracy.
Since the loss of MMR protein expression can occur by methylation and mutations in the promoter region, in addition to loss-of-function mutations in the coding region, a validation of the exon sequence would not be sufficient to evaluate the MMR expression and function [47–49]. Actually, in the present study, only 3 of 9 MMR-D cases presented pathogenic mutations in MLH1, MSH6, or PMS2. Although IHC is considered a practical tool for speculation of TMB and MSI conditions as a first screening step [50, 51], the results of MMR protein IHC were not always comparable to those of MMR gene mutation identification. Thus, evaluation of TMB and MSI using NGS-based genome analysis along with IHC would be a practical strategy for clinical testing [52–54]. NGS analysis with endometrial LBC specimens, which were not usually used for immunocytochemistry of MMR proteins, consistently detected TMB-H and MSI-H. Therefore, our custom-made panel would possibly be beneficial to determine TMB-H and MSI-H status. However, the sample size was relatively small, and the NGS panel is also insufficient to comprehensively detect TMB and MSI at the whole-exome sequencing level, in which TMB calculation algorithm often weights down mutation data from hotspot to prevent TMB over-estimation [32, 33, 55, 56].
Conclusion
The NGS-based panel with coverage of 60 cancer genes and 17 microsatellite foci demonstrated highly recurrent somatic mutations, TMB, and MSI in FFPE and endometrial LBC specimens. In addition, the endometrial cytology can efficiently collect cells from the entire part of the endometrial cavity to predict histological subtypes [16] and also is available to detect asymptomatic endometrial cancer by the less invasive cytologic screening [14]. Therefore, the small size cancer gene panel and endometrial LBC specimens would be an alternative tool for genetic testing as a diagnostic or therapeutic strategy. As in the case of NGS analysis from FFPE tissues, it is vital that cytopathologists correctly estimate the amounts of tumor cells, to select suitable LBC specimens for molecular diagnosis appropriately [7].
Supplementary information
Additional file 1: Table S1. Somatic mutation prolife detected in endometrial cancers.
Additional file 2: Table S2. Genomic information of variants detected in endometrial cancers.
Additional file 3: Table S3. TMB, MSI and MMR protein deficiency.
Acknowledgements
The authors greatly appreciate the excellent technical assistance of Ms. Orie Iwaya, Ms. Mai Tokudome, and Mr. Kei Matsuo (Department of Pathology, Kagoshima University Graduate School of Medical and Dental Sciences), and Ms. Yukari Nishida (Department of Surgical Pathology, Kagoshima University Hospital). We would like to thank Editage (www.editage.com) for English language editing.
Abbreviations
- LBC
Liquid-based cytology
- FFPE
Formalin-fixed paraffin-embedded
- TMB
Tumor mutation burden
- MSI
Microsatellite instability
- MMR
Mismatch repair
- NGS
Next-generation sequencing
- H&E
Hematoxylin and eosin
- HPF
High-power fields
- VAF
Variant allele frequency
- IHC
Immunohistochemistry
- ROC
Receiver operator characteristic
- EC
Endometrioid carcinoma
- DC
Dedifferentiated carcinoma
- SC
Serous carcinoma
- CCC
Clear cell carcinoma
Authors’ contributions
AT, SU, and HK designed the study. SY1, MK, ST, and SS interpreted the patient data. IK and TH contributed to interpretation of pathological data. TA, SY2, IS, and SN performed the genetic analyses. AT wrote the manuscript with the assistance of TA, IK and SN. All authors read and approved the final manuscript.
Funding
No funding for the present study.
Availability of data and materials
The datasets generated and analyzed during the current study are made available as supplementary Tables (Supplementary Tables S1, S2, and S3) that lists all qualified variants. Database used in this study were Catalogue Of Somatic Mutations In Cancer (COSMIC ver. 90, https://cancer.sanger.ac.uk/cosmic), Qiagen Web Portal service (https://www.qiagen.com/us/shop/genes-and-pathways/data-analysis-center-overview-page/), and Human genome reference GRCh37 hg19 (https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13/).
Ethics approval and consent to participate
The studies using clinical samples were approved by the Ethics Committees for Clinical and Epidemiologic Research at Kagoshima University, and written-informed consent was obtained from each participant. The studies did not include participants younger than the age of 20.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12920-020-00753-6.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. 10.3322/caac.21262. [DOI] [PubMed]
- 2.Moore K, Brewer MA. Endometrial Cancer: is this a new disease? Am Soc Clin Oncol Educ Book. 2017;37:435–442. doi: 10.14694/EDBK_175666. [DOI] [PubMed] [Google Scholar]
- 3.O’Mara TA, Glubb DM, Amant F, Annibali DA, Shton K, Attia J, Auer PL, Beckmann MW, Black A, Bolla MK, et al. Identification of nine new susceptibility loci for endometrial cancer. Nat Commun. 2018;9:3166. doi: 10.1038/s41467-018-05427-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev. 2003;16:1–17. doi: 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rabban JT, Gilks CB, Malpica A, Matias-Guiu X, Mittal K, Mutter GL, Oliva E, Parkash V, Ronnett BM, Staats P, et al. Issues in the differential diagnosis of uterine low-grade endometrioid carcinoma, including mixed endometrial carcinomas: recommendations from the international society of gynecological pathologists. Int J Gynecol Pathol. 2019;38:S25–S39. doi: 10.1097/PGP.0000000000000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soslow RA, Tornos C, Park KJ, Malpica A, Matias-Guiu X, Oliva E, Parkash V, Carlson J, McCluggage WG, Gilks CB. Endometrial carcinoma diagnosis: use of FIGO grading and genomic subcategories in clinical practice: recommendations of the international society of gynecological pathologist. Int J Gynecol Pathol. 2019;38:S64–S74. doi: 10.1097/PGP.0000000000000518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roy-Chowdhuri S, Pisapia P, Salto-Tellez M, Savic S, Nacchio M, de Biase D, Tallini G, Troncone G, Schmitt F. Invited review-next-generation sequencing: a modern tool in cytopathology. Virchows Achiv. 2019;475:3–11. doi: 10.1007/s00428-019-02559-z. [DOI] [PubMed] [Google Scholar]
- 8.Karnes HE, Duncavage EJ, Bernadt CT. Targeted next-generation sequencing using fine-needle aspirates from adenocarcinomas of the lung. Cancer Cytopathol. 2014;122:104–113. doi: 10.1002/cncy.21361. [DOI] [PubMed] [Google Scholar]
- 9.Velizheva NP, Rechsteiner MP, Wong CE, Zhong Q, Rossle M, Bode B, Moch H, Soltermann A, Wind PJ, Tischler V. Cytology smears as excellent starting materials for next-generation sequencing-based molecular testing of patients with adenocarcinoma of the lung. Cancer Cytopathol. 2017;125:30–40. doi: 10.1002/cncy.21771. [DOI] [PubMed] [Google Scholar]
- 10.Padmanabhan V, Steinmetz HB, Rozzo EJ, Erskine AJ, Fairbank TL, de Abreu FB, Tsongalis GJ, Tafe LJ. Improving adequacy of small biopsy and fine-needle aspiration specimens for molecular testing by next-generation sequencing in patients with lung cancer. Arch Pathol Lab Med. 2017;141:402–409. doi: 10.5858/arpa.2016-0096-OA. [DOI] [PubMed] [Google Scholar]
- 11.Turner SR, Buonocore D, Desmeules P, Rekhtman N, Dogan S, Lin O, Arcila ME, Jones DR, Huang J. Feasibility of endobronchial ultrasound transbronchial needle aspiration for massively parallel next-generation sequencing in thoracic cancer patients. Lung Cancer. 2018;119:85–90. doi: 10.1016/j.lungcan.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jain D, Roy-Chowdhuri S. Molecular pathology of lung cancer cytology specimens a concise review. Arch Pathol Lab Med. 2018;142:1127–1133. doi: 10.5858/arpa.2017-0444-RA. [DOI] [PubMed] [Google Scholar]
- 13.Dijkhuizen FP, Mol BW, Brölmann HA, Heintz AP. The accuracy of endometrial sampling in the diagnosis of patients with endometrial carcinoma and hyperplasia: a meta-analysis. Cancer. 2000;89:1765–1772. doi: 10.1002/1097-0142(20001015)89:8<1765::AID-CNCR17>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 14.Jobo T, Arai T, Sato R, Kuramoto H. Clinicopathologic relevance of asymptomatic endometrial carcinoma. Acta Cytol. 2003;47:611–615. doi: 10.1159/000326577. [DOI] [PubMed] [Google Scholar]
- 15.Nishimura Y, Watanabe J, Jodo T, Hattori M, Arai T, Kuramoto H. Cytologic scoring of endometrioid adenocarcinoma of the endometrium. Cacner. 2005;105:8–12. doi: 10.1002/cncr.20787. [DOI] [PubMed] [Google Scholar]
- 16.Okadome M, Saito T, Nishiyama N, Ariyoshi K, Shimamoto K, Shimada T, Kodama K, Imamura S, Nishiyama K, Taguchi K. Prediction of histological types of endometrial cancer by endometrial cytology. J Obstet Gynaecol Res. 2014;40:1931–1939. doi: 10.1111/jog.12436. [DOI] [PubMed] [Google Scholar]
- 17.Byrne AJ. Endocyte endometrial smears in the cytodiagnosis of endometrial carcinoma. Acta Cytol. 1990;34:373–381. [PubMed] [Google Scholar]
- 18.Norimatsu Y, Shimizu K, Kobayashi TK, Moriya T, Tsukayama C, Miyake Y, Ohno E. Cellular features of endometrial hyperplasia and well differentiated adenocarcinoma using the endocyte sampler. Diagnostic criteria based on the cytoarchitecture of tissue fragments. Cancer Cytoplathol. 2006;108:77–85. doi: 10.1002/cncr.21734. [DOI] [PubMed] [Google Scholar]
- 19.Norimatus Y, Yanoh K, Hirai Y, Kurokawa T, Kobayashi TK, Fulciniti F. A diagnostic approach to endometrial cytology by means of liquid-based preparations. Acta Cytol. 2019;30:1–13. doi: 10.1159/000502108. [DOI] [PubMed] [Google Scholar]
- 20.Buccoliero AM, Castiglione F, Gheri CF, Garbini F, Fambrini M, Bargelli G, Pappalardo S, Scarselli G, Taddei GL. Liquid-based endometrial cytology: its possible value in postmenopausal asymptomatic women. Int J Gynecol Cancer. 2007;17:182–187. doi: 10.1111/j.1525-1438.2006.00757.x. [DOI] [PubMed] [Google Scholar]
- 21.Buccoliero AM, Gheri CF, Castiglione F, Garbini F, Fambrini M, Bargelli G, Barbetti A, Pappalardo S, Tadei A, Boddi V, et al. Liquid-based endometrial cytology in the management of sonographically thickened endometrium. Diagn Cytopathol. 2007;35:398–402. doi: 10.1002/dc.20655. [DOI] [PubMed] [Google Scholar]
- 22.Di Lorito A, Rosini S, Falò E, Gustapane S, Gomes M, Costa JL, Schmitt FC. Molecular alterations in endometrial archived liquid-based cytology. Diagn Cytopathol. 2013;41:492–496. doi: 10.1002/dc.22869. [DOI] [PubMed] [Google Scholar]
- 23.Akahane T, Yamaguchi T, Kato Y, Yokoyama S, Hamada T, Nishida Y, Higashi M, Nishihara H, Suzuki S, Ueno S, et al. Comprehensive validation of liquid-based cytology specimens for next-generation sequencing in cancer genome analysis. PLoS One. 2019;14:e0217724. doi: 10.1371/journal.pone.0217724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stelloo E, Jansen AML, Osse EM, Nout RA, Creutzberg CL, Ruano D, Church DN, Morreau H, Smit VTHBM, van Wezel T, et al. Practical guidance for mismatch repair-deficiency testing in endometrial cancer. Ann Oncol. 2017;28:96–102. doi: 10.1093/annonc/mdw542. [DOI] [PubMed] [Google Scholar]
- 25.Zaino R, Carinelli SG, Ellenson LH, Eng C, Katabuchi H, Konishi I, et al. Tumours of the uterine corpus. In: Kurman RJ, Carcangiu ML, Herrington CS, Young RH, et al., editors. WHO classification of tumours of female reproductive organs. 4. Lyon: IARC; 2014. pp. 125–135. [Google Scholar]
- 26.Yanoh K, Hirai Y, Sakamoto A, Aoki D, Moriya T, Hiura M, Yamawaki T, Shimizu K, Nakayama H, Sasaki H, et al. New terminology for intrauterine endometrial samples: a group study by the Japanese Society of Clinical Cytology. Acta Cytol. 2012;56:233–241. doi: 10.1159/000336258. [DOI] [PubMed] [Google Scholar]
- 27.Hempelmann JA, Lockwood CM, Konnick EQ, Schweizer MT, Antonarakis ES, Lotan TL, Montgomery B, Nelson PS, Klemfuss N, Salipante SJ, et al. Microsatellite instability in prostate cancer by PCR or next-generation sequencing. J Immunother Cancer. 2018;6:29. doi: 10.1186/s40425-018-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu C, Nezami Ranjbar MR, Wu Z, Dicarlo J, Wang Y. Detecting very low allele fraction variants using targeted DNA sequencing and a novel molecular barcode-aware variant caller. BMC Genomics. 2017;18:5. doi: 10.1186/s12864-016-3425-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu C, Gu X, Padmanabhan R, Wu Z, Pneg Q, DiCarlo J, Wang Y. smCounter2: an accurate low-frequency variant caller for targeted sequencing data with unique molecular identifiers. Bioinformatics. 2019;35:1299–1309. doi: 10.1093/bioinformatics/bty7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galuppini F, Dal Pozzo CA, Deckert J, Loupakis F, Fassan M, Baffa R. Tumor mutation burden: from comprehensive mutational screening to the clinic. Cancer Cell Int. 2019;19:209. doi: 10.1186/s12935-019-0929-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, Wendl MC, Ding L. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2014;30:1015–1016. doi: 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boll Johansen AF, Kassentoft CG, Knudsen M, Laursen MB, Madsen AH, Iversen LH, Sunesen KG, Rasmussen MH, Andersen CL. Validation of computational determination of microsatellite status using whole exome sequencing data from colorectal cancer patients. BMC Cancer. 2019;19:971. doi: 10.1186/s12885-019-6227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chapel DB, Patil SA, Plagov A, Puranik R, Mendybaeva A, Steinhardt G, Wanjari P, Lastra RR, Kadri S, Segal JP, Ritterhouse LL. Quantitative next-generation sequencing-based analysis indicates progressive accumulation of microsatellite instability between atypical hyperplasia/endometrial intraepithelial neoplasia and paired endometrioid endometrial carcinoma. Mod Pathol. 2019;32:1508–1520. doi: 10.1038/s41379-019-0298-5. [DOI] [PubMed] [Google Scholar]
- 35.Haruma T, Nagasaka T, Nakamura K, Haraga J, Nyuya A, Nishida T, Goel A, Masuyama H, Hiramatsu Y. Clinical impact of endometrial cancer stratified by genetic mutational profiles, POLE mutation, and microsatellite instability. PLoS One. 2018;13:e0195655. doi: 10.1371/journal.pone.0195655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gleeson FC, Kipp BR, Kerr SE, Voss JS, Lazaridis KN, Katzka DA, Levy MJ. Characterization of endoscopic ultrasound fine-needle aspiration cytology by targeted next-generation sequencing and theranostic potential. Clin Gastroenterol Hepatol. 2015;13:37–41. doi: 10.1016/j.cgh.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 37.Bellevicine C, Malapella U, Vigliar E, Pisapia P, Vita G, Troncone G. How to prepare cytological samples for molecular testing. J Clin Pathol. 2017;70:819–826. doi: 10.1136/jclinpath-2017-204561. [DOI] [PubMed] [Google Scholar]
- 38.Roy-Chowdhuri S, Stewart J. Preanalytic variables in cytology: lessons learned from next-generation sequencing – the MD Anderson experience. Arch Pathol Lab Med. 2016;140:1191–1199. doi: 10.5858/arpa.2016-0117-RA. [DOI] [PubMed] [Google Scholar]
- 39.Tian SK, Killian JK, Rekhtman N, Benayed R, Middha S, Ladanyi M, Lin O, Arcila ME. Optimizing workflows and processing of cytologic samples for comprehensive analysis by next-generation sequencing. Memorial Sloan Kettering Cancer Centre experience. Arch Pathol Lab Med. 2016;140:1200–1205. doi: 10.5858/arpa.2016-0108-RA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Network CGAR, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iijima M, Banno K, Okawa R, Yanokura M, Iida M, Takeda T, Kunitomi-Irie H, Adachi M, Nakamura K, Umene K, et al. Genome-wide analysis of gynecologic cancer: The Cancer Genome Atlas in ovarian and endometrial cancer. Oncol Lett. 2017;13:1063–1070. doi: 10.3892/ol.2017.5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Hara AJ, Bell DW. The genomics and genetics of endometrial cancer. Adv Genomics Genet. 2012;2:33–47. doi: 10.2147/AGG.S28953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsuura M, Yamaguchi K, Tamate M, Satohisa S, Teramoto M, Iwasaki M, Iwasaki M, Sugita S, Hasegawa T, Koubo R, et al. Efficacy of liquid-based genetic diagnosis of endometrial cancer. Cancer Sci. 2018;109:4025–4032. doi: 10.1111/cas.13819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Späth SS, Marjani SL, Zhang W, Pan X. Characterization of cancer genomic heterogeneity by next-generation sequencing advances precision medicine in cancer treatment. Precis Clin Med. 2018;1:29–48. doi: 10.1093/pcmedi/pby007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russo M, Broach J, Sheldon K, Houser KR, Liu DJ, Kesterson J, Phaeton R, Hossler C, Hempel N, Baker M, et al. Clonal evolution in paired endometrial intraepithelial neoplasia/atypical hyperplasia and endometrioid adenocarcinoma. Hum Pathol. 2017;67:69–77. doi: 10.1016/j.humpath.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 46.Russo M, Newell JM, Budyrlean L, Houser KR, Sheldon K, Kesterson J, Phaeton R, Hossler C, Rosenberg J, DeGraff D, et al. Mutational profile of endometrial hyperplasia and risk of progression to endometrioid adenocarcinoma. Cancer. 2020;126:2775–2783. doi: 10.1002/cncr.32822. [DOI] [PubMed] [Google Scholar]
- 47.Esteller M. Epigenetic lesions causing genetic lesions in human cancer: promoter hypermethylation of DNA repair genes. Eur J Cancer. 2000;36:2294–2300. doi: 10.1016/s0959-8049(00)00303-8. [DOI] [PubMed] [Google Scholar]
- 48.Banno K, Yanokura M, Iida M, Masuda K, Aoki D. Carcinogenic mechanisms of endometrial cancer: involvement of genetics and epigenetics. J Obstet Gynaecol Res. 2014;40:1957–1967. doi: 10.1111/jog.12442. [DOI] [PubMed] [Google Scholar]
- 49.Yokoyama T, Takehara K, Sugimoto N, Kaneko K, Fujimoto E, Okazawa-Sakai M, Okame S, Shiroyama Y, Yokoyama T, Teramoto N, et al. Lynch syndrome-associated endometrial carcinoma with MLH1 germline mutation and MLH1 promoter hypermethylation: a case report and literature review. BMC Cancer. 2018;18:576. doi: 10.1186/s12885-018-4489-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goodfellow PJ, Billingsley CC, Lankes HA, Ali S, Cohn DE, Broaddus RJ, Ramirez N, Pritchard CC, Hampel H, Chassen AS, et al. Combined Microsatellite Instability, MLH1 Methylation Analysis, and Immunohistochemistry for Lynch Syndrome Screening in Endometrial Cancers From GOG210: An NRG Oncology and Gynecologic Oncology Group Study. J Clin Oncol. 2015;33:4301–4308. doi: 10.1200/JCO.2015.63.9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adar T, Rodgers LH, Shannon KM, Yoshida M, Ma T, Mattia A, Lauwers GY, Iafrate AJ, Hartford NM, Oliva E, et al. Universal screening of both endometrial and colon cancers increases the detection of Lynch syndrome. Cancer. 2018;124:3145–3153. doi: 10.1002/cncr.31534. [DOI] [PubMed] [Google Scholar]
- 52.Bruegl AS, Kernberg A, Broaddus RR. Adv Anat Pathol. 2017;24:372–378. doi: 10.1097/PAP.0000000000000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salipante SJ, Scroggins SM, Hampel HL, Turner EH, Pritchard CC. Microsatellite instability detection by next generation sequencing. Clin Chem. 2014;60:1192–1199. doi: 10.1373/clinchem.2014.223677. [DOI] [PubMed] [Google Scholar]
- 54.Dong F, Costigan DC, Howitt BE. Targeted next-generation sequencing in the detection of mismatch repair deficiency in endometrial cancers. Mod Pathol. 2019;32:252–257. doi: 10.1038/s41379-018-0125-4. [DOI] [PubMed] [Google Scholar]
- 55.Campbell BB, Light N, Fabrizio D, Zatzman M, Fuligni F, de Borja R, Davidson S, Edwards M, Elvin JA, Hodel KP, et al. Comprehensive analysis of hypermutation in human cancer. Cell. 2017;171:1042–1056. doi: 10.1016/j.cell.2017.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, Schrock A, Campbell B, Shlien A, Chmielecki J, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Somatic mutation prolife detected in endometrial cancers.
Additional file 2: Table S2. Genomic information of variants detected in endometrial cancers.
Additional file 3: Table S3. TMB, MSI and MMR protein deficiency.
Data Availability Statement
The datasets generated and analyzed during the current study are made available as supplementary Tables (Supplementary Tables S1, S2, and S3) that lists all qualified variants. Database used in this study were Catalogue Of Somatic Mutations In Cancer (COSMIC ver. 90, https://cancer.sanger.ac.uk/cosmic), Qiagen Web Portal service (https://www.qiagen.com/us/shop/genes-and-pathways/data-analysis-center-overview-page/), and Human genome reference GRCh37 hg19 (https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13/).