Abstract
The intestinal microbiota is crucial to intestinal homeostasis. Porcine epidemic diarrhea virus (PEDV) is high pathogenic to intestines, causing diarrhea, even death in piglets. To investigate the detailed relationship between PEDV infection and intestinal microbiota, the composition and distribution of intestinal microbiota from pigs were first analyzed using 16S rRNA sequencing technology. The results demonstrated that the composition and distribution of microbes in different intestinal segments were quite similar between 1-week-old and 2-week-old piglets but different from 4-week-old (weaned) piglets. Then piglets at different ages were inoculated with PEDV. The results showed that the 1-week-old piglets exhibited the most severe pathogenicity comparing to the other age groups. Further investigations indicated that Lactobacillus, Escherichia coli, and Lactococcus in the intestinal microbiota of piglets were significantly changed by PEDV infection. These results strengthen our understanding of viruses influencing intestinal microbes and remind us of the potential association between PEDV and intestinal microbes.
Keywords: Porcine epidemic diarrhea virus, Intestinal microbiota, Pathogenicity
1. Introduction
Porcine epidemic diarrhea (PED) is an infectious enteric disease, causing significant economic loss to the pig industry across the world (Lee and Lee, 2018). The pathogen responsible for PED is the porcine epidemic diarrhea virus (PEDV), a member of the coronaviruses (Brian and Baric, 2005). The first PEDV strain was isolated by a research group from Gent University, Belgium, in 1978 and named CV777(Sun et al., 2008). They found that the PEDV shared a similar structure with transmissible gastroenteritis virus (TGEV), a known swine enteric coronavirus at that time. However, PEDV cannot be neutralized by antiserum of TGEV, although both viruses are from the same category and caused the same clinical symptom of watery diarrhea in pigs (Debouck and Pensaert, 1980). Lately, the PEDV was found in Asian pig farms in 1982 and caused mild diarrhea and limited mortality in pig herds. During the early 2010s, virulent PEDV strains emerged and spread promptly between the pig farms from Asia, North America, and the European Union, causing enormous economic loss (Li et al., 2019; Mole, 2013). Currently, the virulent strain of PEDV is a continuous threat to the pig industry.
PEDV-infected piglets suffer severe clinical symptoms like vomiting, watery diarrhea, dehydration, and high mortality (up to 100% mortality in 1-day to 3-day old piglets) (Lee, 2015; Lee et al., 2015). Generally, virus shedding in feces is detectable within 48 h and may last for four weeks (Shibata et al., 2000). Compared to fattening pigs, the piglets are easier to get infected by PEDV due to lower turnover of their intestinal epithelium cells (Moon et al., 1973; Norman et al., 1973). Kwonil et al. found that the pigs with intestinal damages, like some destructed junction proteins like adherin in villous and crypt epithelium of intestines, were more susceptible to secondary infection (Jung et al., 2015).
The intestinal microbiota is a stable microenvironment within the whole intestines and plays a critical role in the development of the intestinal mucosal immune system. It can compete with other pathogens for nutrients and binding sites, inhibiting their invasion into intestinal mucosa (Goulet, 2015). When the homeostasis of the intestinal microbiota is destroyed, the number of conditioned pathogenic bacteria can increase while the number of beneficial bacteria will decrease, resulting in inflammation or diarrhea. PEDV infection was reported to alter intestinal microbiota distribution. Huang et al. found that the taxon composition profile of infected suckling piglets was significantly different from that of uninfected suckling piglets (three to eight days old). The abundance of Firmicutes was higher in infected suckling piglets, whereas the abundance of Bacteroidetes was lower in infected piglets (Huang et al., 2018b). The intestinal microbiota was also changed after PEDV infection, whether in pigs around 4–21 days (Liu et al., 2015) or pigs less than three months old (Koh et al., 2015). Liu found that Verrucomicrobia whose abundance was 20% in control piglets was approximately zero in diarrheal piglets while the abundance of Fusobacteria in diarrheal piglets (36%) was seven times higher than that in control piglets (5%). Koh found that eight genera were more abundant in the gut after PEDV infection, including Fusobacterium and Bacteroides. However, these studies did not tell that the different intestinal microbiota distribution with changed intestinal segments and ages of healthy pigs. Besides, they did not tell how PEDV infection influences intestinal microbiota in pigs of different ages.
In this study, we investigated the distribution of swine intestinal microbiota in different segments of small intestines and different age groups. We also analyzed the differential distribution of intestinal microbes affected by PEDV infection. The results that originated from this study will contribute to the understanding of age-dependent pathogenicity of PEDV infection and associations between PEDV infection and intestinal microbes.
2. Results
2.1. Intestinal microbial community in different segments of the small intestine
A total of 54 intestinal content samples were collected from different segments of the small intestine of piglets and subjected to 16S rRNA sequencing. After screening of the original sequence reads, 2,671,688 valid reads for the 16S rRNA V3–V4 region was obtained, with an average of 49,476 reads per sample (range from 33,155 to 12,0788). The average length of the amplicon was 462 bp. The effective sequence lengths were distributed between 430 bp and 470 bp (Fig. S1), with average length at 462 bp. The detailed information of each sample is listed in DATASET S1. A total of 2809 OTUs classified into different taxonomies. Twelve phyla, 21 classes, 38 orders, 74 families, and 215 genera were identified from these samples based on sequence clustering. A phylogenetic tree for the top 30 bacteria was generated by the maximum likelihood method and showed in Fig. S2.
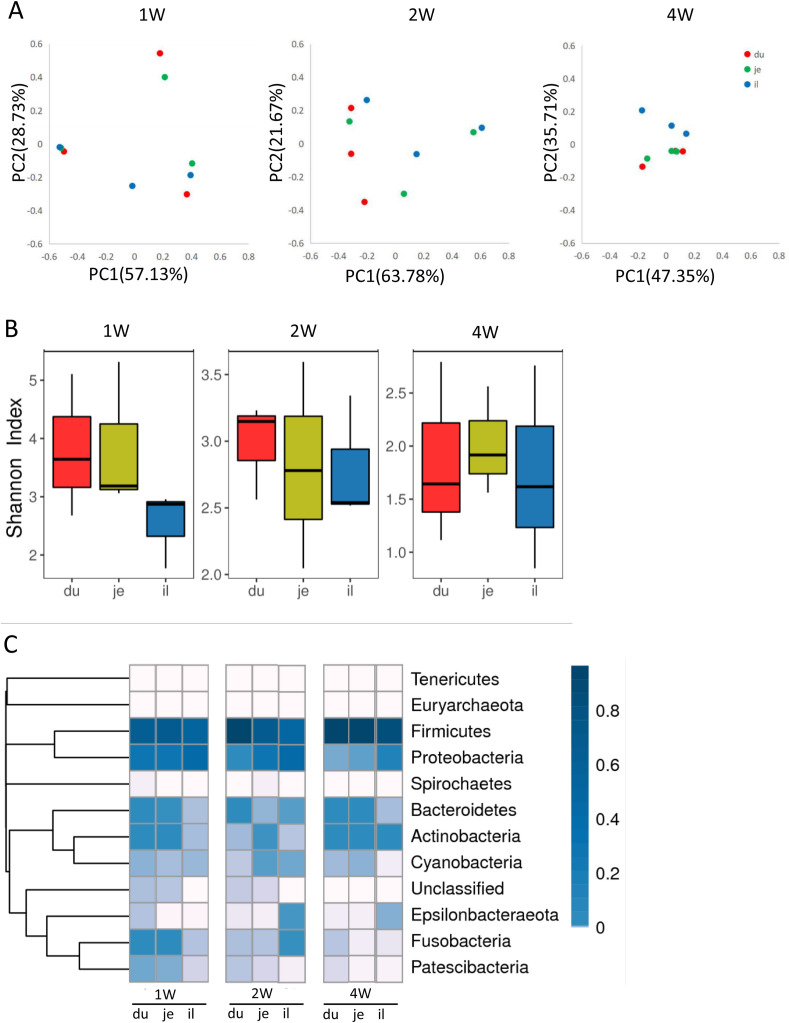
According to the relative genus abundance, PCoA analysis revealed that no significant microbiota difference was observed from duodenum, jejunum, and ileum of the piglets at the same age (Fig. 1 A). The P-value from Anosim analysis of the difference between different intestinal segments in every pig's age was 0.832, 0.513, and 0.313, respectively. The further analysis illustrated that the duodenum and jejunum had higher microbial diversity than that of the ileum of 1-week-old piglets, while no diversity difference was shown between the different segments in 2-week-old and 4-week-old piglets (Fig. 1B). The relative abundance analysis at the phylum level demonstrated that Firmicutes, Bacteroidetes, and Proteobacteria were the main intestinal phyla in all age groups (Fig. 1C). The relative abundances of Bacteroidetes, Actinobacteria, and Fusobacteria were lower in the ileum of 1-week-old piglets than in the other two segments. Besides, the ileum contained a higher abundance of Fusobacteria in 2-week-old piglets but lower Bacteroidetes in 4-week-old piglets, compared to the abundances in duodenum and jejunum (Fig. 1C). The LEfSe analysis showed that the Flavobacteriales in the duodenum of 1-week-old piglets, the Streptosorangiales and Nocardiopsaceae in the jejunum of 2-week-old piglets, the Enterobacteriales, Escherichia Shigella, Enterobacteriaceae in the ileum, and the Oceanospirillales, Halomonadaceae in the jejunum of 4-week-old piglets were the discriminatory species between each segment and age group (Fig. S3).
Fig. 1.
Analysis of intestinal microbe in different segments in the small intestine of healthy piglets. Intestinal contents from duodenum, jejunum, and ileum of 1-week-old, 2-week-old, and 4-week-old healthy piglets were collected and subjected to 16S rRNA sequencing. (A) NMDS analysis of the relative abundance of microbe in duodenum, jejunum, and ileum from a different age of piglets. (B) Shannon index of microbial diversity in duodenum, jejunum, and ileum from a different age of piglets. (C) Heatmap of bacterial distribution in the level of phylum.
2. Temporal development and maturation of swine intestinal microbiota
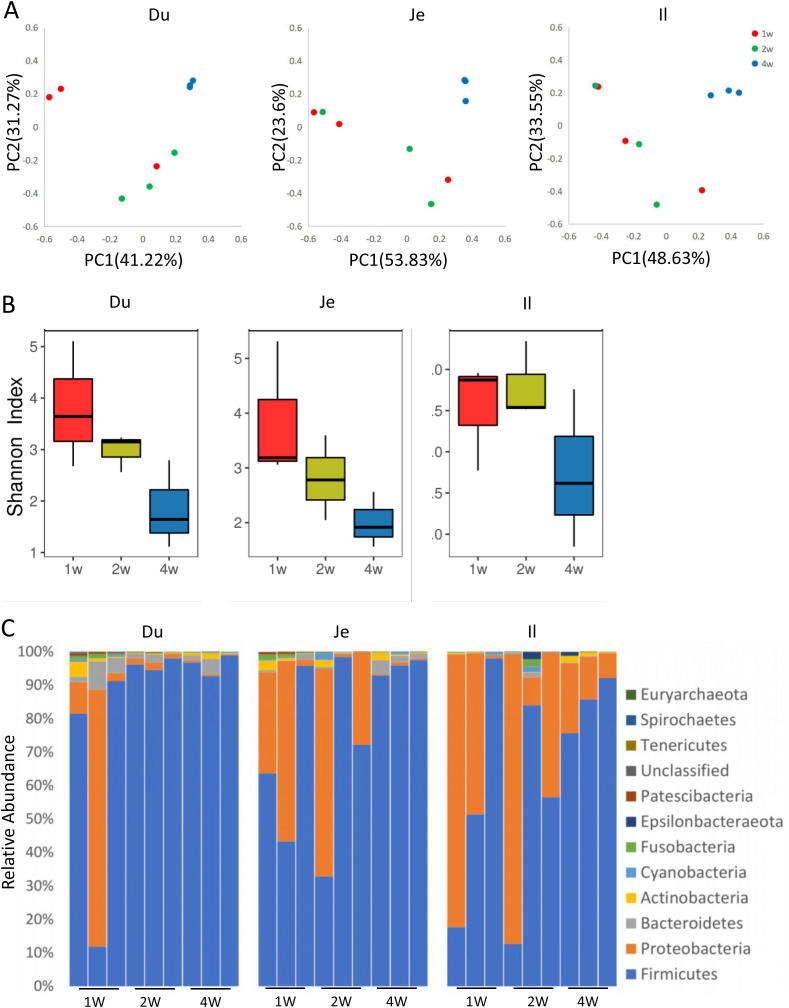
The intestinal microbiota of piglets alters with the change of feeds during different growth stages. PCoA analysis showed that 4-week-old piglets have different microbial diversity comparing to the 1-week-old and 2-week-old piglets, and the latter two age groups shared similar microbial diversity (Fig. 2 A). The P-value from Anosim analysis of the difference between different ages in every intestinal segment of pigs was 0.014, 0.063, 0.043, respectively. The 1-week-old and 2-week-old piglets' microbial diversity was higher than that in 4-week-old piglets (Fig. 2B). Generally, Firmicutes and Proteobacteria were mainly present in all piglets, with a substantial decrease of Proteobacteria in the duodenum of 2-week-old and 4-week-old piglets (Fig. 2C). The abundance of Proteobacteria was reduced while Firmicutes was increased with the growth of piglets (Fig. 2C). The LFfSe analysis revealed that the Oceanospirillales, Moraxella of Proteobacteria, were the central discriminative microbial populations, with higher abundance in the duodenum of 1-week-old piglets than that of 2-week-old and 4-week-old piglets (Fig. S4). The low abundance of Gemella and high abundance of Wohlfahrtimorias were presented in the ileum of 1-week-old piglets and the jejunum of 4-week-old piglets, respectively (Fig. S4).
Fig. 2.
Changes of intestinal microbe at different age of healthy piglets. (A) The changes in the relative abundance of intestinal bacteria at different age of piglets were analyzed by NMDS. (B) Shannon index of microbial diversity in a different segment of the small intestine from a different age of piglets. (C) The bacterial distribution diagram at the level of phylum.
3. Pathogenicity of PEDV infection
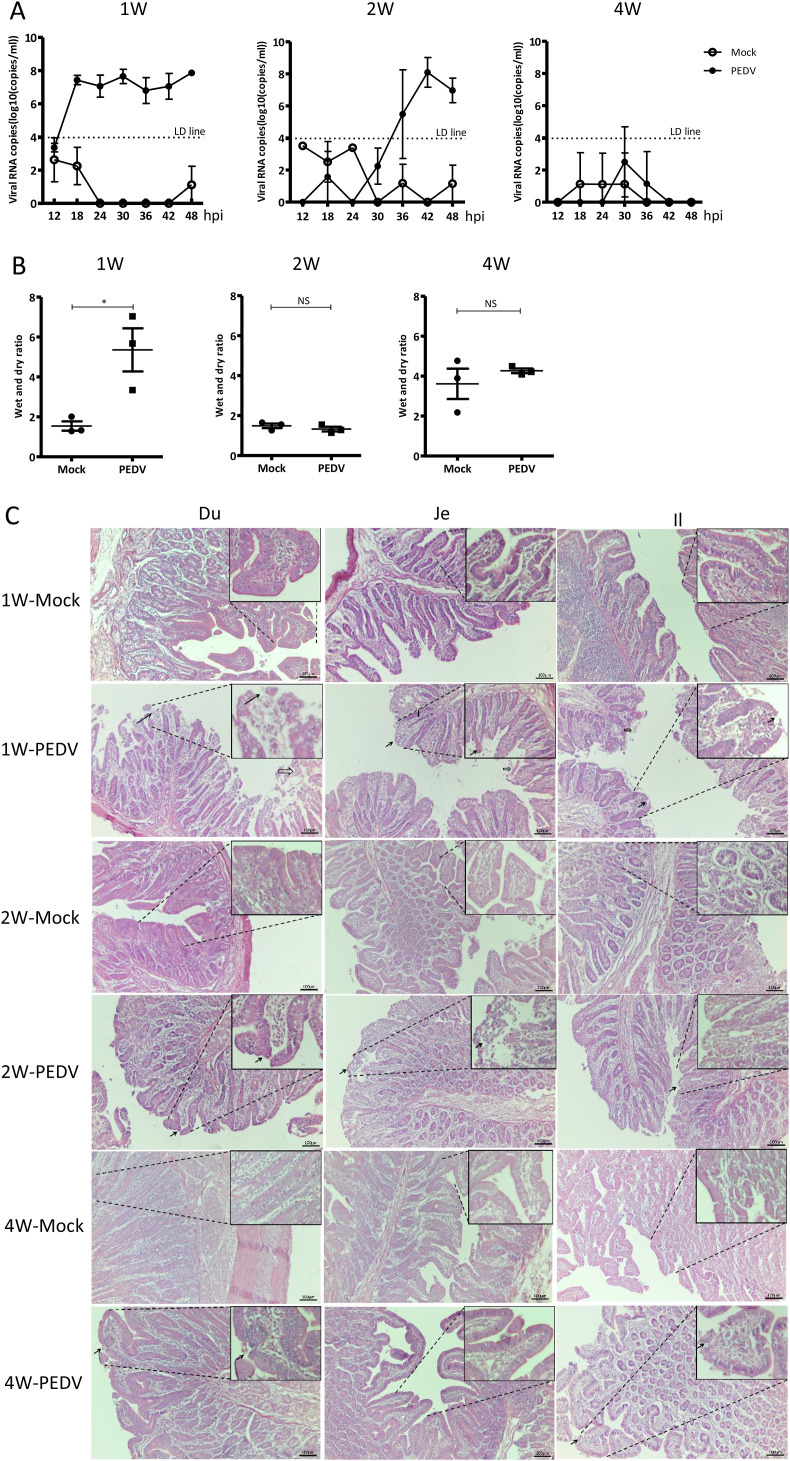
After PEDV infection, anal swabs were collected every 6 hpi and subjected to viral loads analysis by real-time RT-qPCR. The results showed that the virus was detectable in PEDV-infected 1-week-old and 2-week-old piglets but not from 4-week-old piglets (Fig. 3 A). In the 1-week-old group, the viral titers increased dramatically after challenge, peaked at 18 hpi, and remained stable after approximately 108 gene copies/ml until being sacrificed at 48 hpi (Fig. 3A), with one piglet succumbing to the infection at this time point. However, the 2-week-old group exhibited a delayed viral load peak at 42 hpi (Fig. 3A). PEDV infection did not cause a significant change in their body temperatures for all tested groups (Fig. S5A). Fecal consistency, wet and dry ratio were employed to evaluate the level of diarrhea. Both results illustrated that the 1-week-old piglets suffered more severe diarrhea after PEDV inoculation than that 4-week-old piglets did (Fig. 3B and Fig. S5B). The severe lesion, showing thin intestinal wall and yellowish watery intestinal contents, was observed from PEDV-infected 1-week-old piglets while the 2-week-old and 4-week-old piglets only showed mild clinic symptom, compared to the mock-infected piglets (Fig. S5C). The histological lesions were characterized by intestinal epithelial cell detachment, intestinal villus atrophy, cellulosic exudation, lymphocyte infiltration, and vascular congestion in PEDV-infected 1-week-old piglets (Fig. 3C). These results imply that the sensitivity of piglets to PEDV infection was reversely correlated to their ages.
Fig. 3.
Pathogenicity induced by PEDV infection. (A) Kinetics of viral shedding in a different age of piglets. The viral loads in anal swabs at indicated time points were analyzed by real-time RT-qPCR. LD line was the maximum value from blank control detected by RT-qPCR. (B) Fecal wet/dry ratios for each age group were calculated to evaluate the severity of diarrhea. (C) H&E staining of different segments of the small intestine from different age groups. Solid arrows indicate the detachment of intestinal epithelial cells. Empty arrows represent cellulosic exudation. Triangle means vascular congestion.
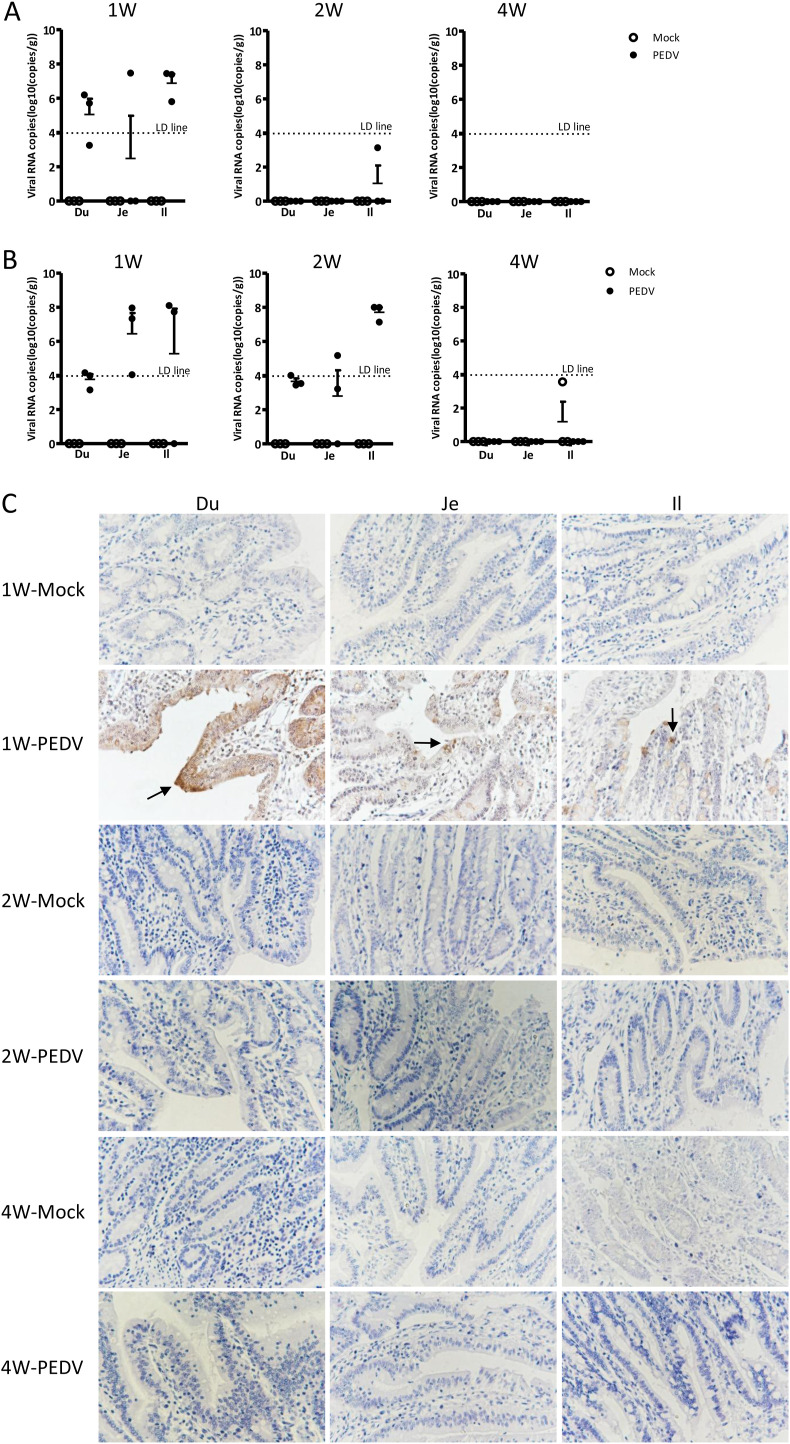
4. Virus detection in small intestines
Different segments of small intestines and associated intestinal contents were harvested and subjected to PEDV viral loads analysis by a TaqMan-probe-based real-time RT-qPCR developed in our laboratory (Huang et al., 2019). The results demonstrated that the virus was detectable in all segments of small intestines in 1-week-old challenged piglets, but no virus was found in any 2-week-old and 4-week-old age groups (Fig. 4 A). PEDV was detected in the intestinal contents derived from different small intestine segments in both 1-week-old and 2-week-old piglets but absent in 4-week-old piglets (Fig. 4B). The immunohistochemistry staining against PEDV N protein showed that viral antigen was presented on the villi of duodenum, jejunum, and ileum in 1-week-old challenged, comparing to mock-infected piglets. However, no detectable viral genome was found from any segments of small intestines in both 2-week-old and 4-week-old piglets (Fig. 4C). Taken together, the PEDV antigen detection results were precisely identical to the genome detection results.
Fig. 4.
Detection of PEDV in different segments of the small intestine and intestinal contents. Different segments of the small intestine and associated intestinal contents were collected at the time of sacrificing. Viral loads in intestines (A) and intestinal content (B) were measured by real-time RT-qPCR. (C) Immunohistochemical analysis for detection of PEDV N protein. The arrows indicate PEDV-positive cells.
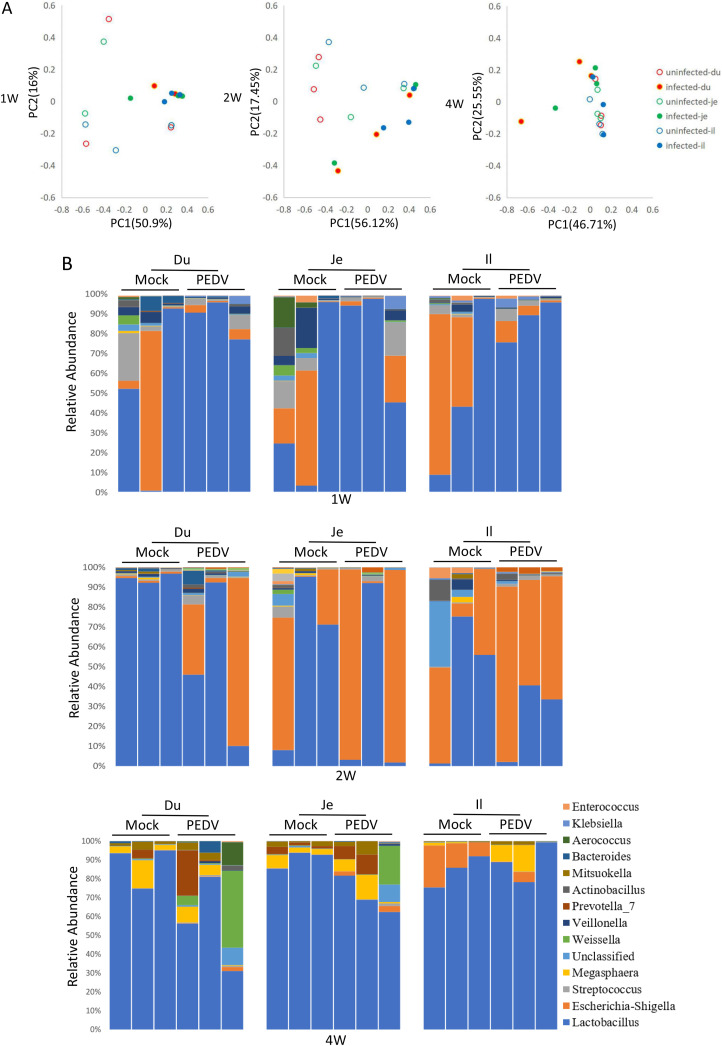
5. PEDV reshape the pig microbiota
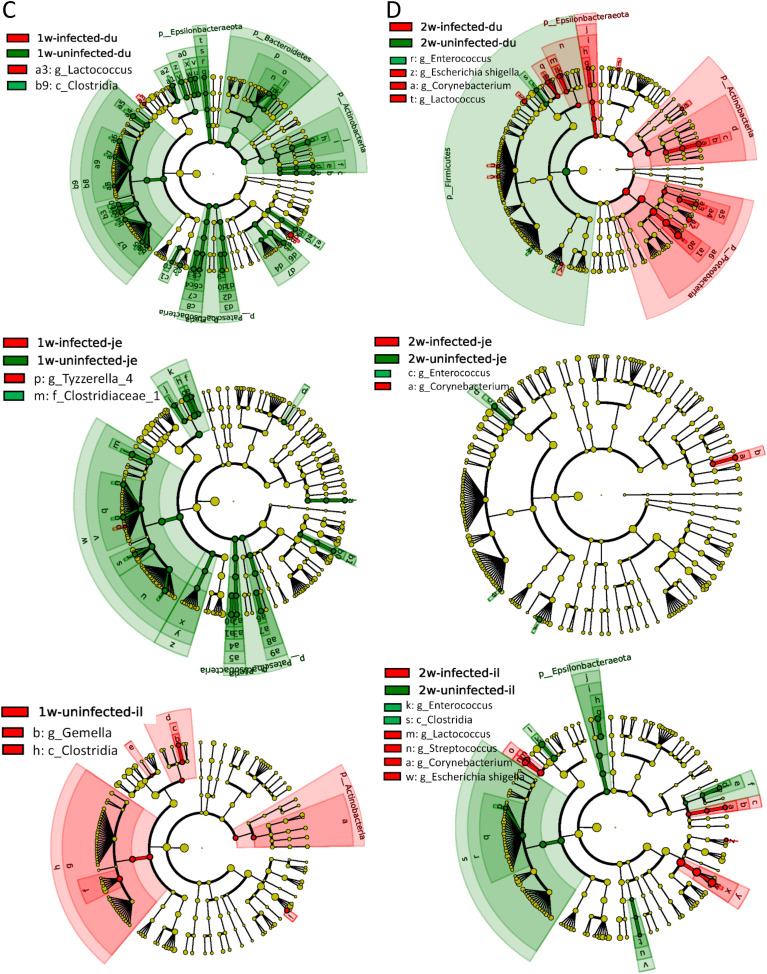
The intestinal contents from PEDV-infected piglets were collected and subjected to 16S rRNA analysis. The results indicated that PEDV infection reshaped the intestinal microbiota. The PCoA analysis revealed that the intestinal microbiota in the PEDV-infected piglets was distinct significantly from mock-infected piglets. The P-value from Anoism analysis between uninfected and infected group were 0.004, 0.003, 0.029 in 1-week, 2-week, and 4-week pigs, respectively. (Fig. 5 A). The relative abundance of the microbiota was analyzed at phylum (Fig. S6) and genus (Fig. 5B) levels. The results demonstrated that Proteobacteria was decreased while Firmicutes was increased in 1-week-old PEDV-infected piglets. On the contrary, the tendency was reversed in 2-week-old piglets. The Firmicutes and Proteobacteria in the 4-week-old group were slightly changed, but the Bacteroidetes were significantly increased in duodenum and jejunum, not ileum. (Fig. S6A). The genus-level analysis results indicated that the upregulation of Lactobacillus and downregulation of Escherichia-shigella were induced by PEDV infection in duodenum, jejunum, and ileum of 1-week-old piglets. Again, an opposite pattern was observed in 2-week piglets. In the 4-week-old group, PEDV infection downregulated the abundance of Lactobacillus in duodenum and jejunum, but the abundances of Prevotella and Weissella were higher than in mock-infected piglets (Fig. 5B).
Fig. 5.
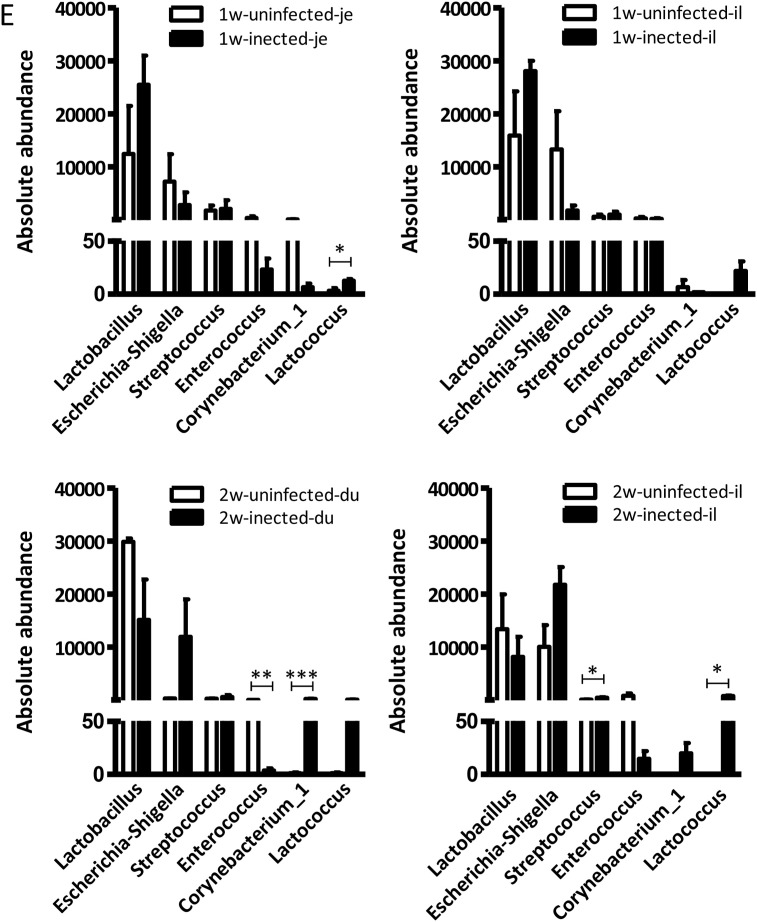
PEDV infection affects the intestinal microbiota of piglets. (A) PCoA analysis of the relative abundance of intestinal bacterial distribution after PEDV infection. (The 16S rRNA data from uninfected pigs is the same as in Fig. 1, Fig. 2.) (B) The bacterial distribution diagram at the level of the genus. The changes of intestinal microbe in 1-week-old (C) and 2-week-old piglets (D) were analyzed by LEfSe. (E) Absolute numbers of intestinal microbe in 1-week-old and 2-week-old piglets.
The representative LEfSe results showed that PEDV infection significantly decreased the abundance of Clostridia in duodenum, jejunum, and ileum of 1-week-old piglets. PEDV infection also induced upregulations of Lactococcus in the duodenum and Tyzzerellain, the jejunum (Fig. 5C). In 2-week-old piglets, the relative abundance of Enterococcus in all intestines and Clostridia in the ileum was significantly decreased by PEDV infection. In contrast, the Escherichia shigella, Corynebacterium in the duodenum and the Escherichia-shigella, Lactococcus, Streptococcus, and Corynebacterium in ileum were increased (Fig. 5D). The number of Escherichia shigella was increased in the duodenum but decreased in ileum after infection in 4-week-old piglets (Fig. S6B).
The abundance distribution of the top five differential bacterial strains between PEDV-infected and mock-infected piglets was analyzed by Metastats and presented in the supplemental DATASET S3. Absolute abundance of intestinal bacteria was analyzed by SPSS software (significantly changed data were shown as follows), and the results demonstrated that Lactococcus was significantly increased not only in the jejunum (p = 0.031) and the ileum of infected 1-week-old piglets but also in duodenum and ileum (p = 0.025) of 2-week-old infected piglets (Figs. 5E and S6C). Besides, the 2-week-old PEDV-infected piglets had a large number of Corynebacterium (p = 0.001) but decreased the amount of Enterococcus (p = 0.005) in their duodenum (Figs. 5E and S6C). For 4-week-old PEDV-infected piglets, the number of Escherichia Shigella (p = 0.044) was reduced in ileum while Streptococcus (p = 0.015) was increased in the duodenum (Figs. 5E and S6C). This analysis revealed that PEDV infection reshaped the intestinal microbiota in an age-dependent manner, characterized by significant changes in Lactobacillus and Escherichia Shigella.
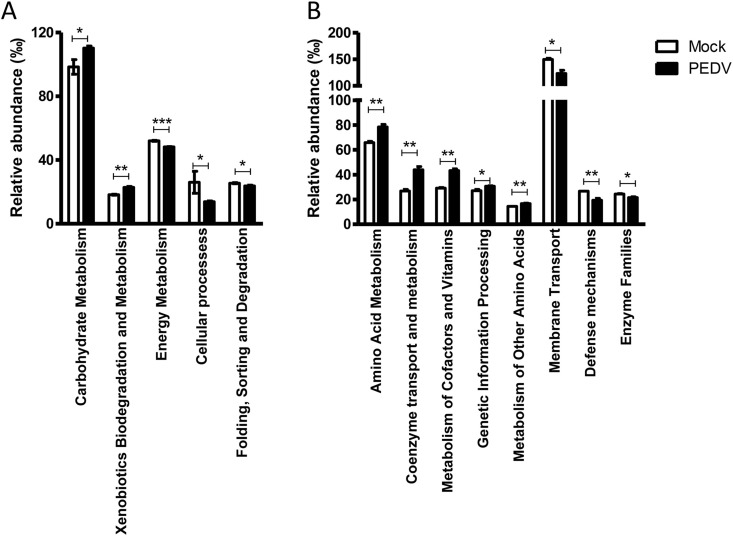
6. PEDV infection potentially influences microbial functions
We next analyzed the change of predicted microbial functions caused by PEDV infection in 1-week-old and 2-week-old piglets. In 1-week-old piglets, PEDV infection significantly upregulated the relative abundance of bacteria being responsible for carbohydrate metabolism and xenobiotics biodegradation but downregulated the abilities of energy metabolism, cellular processes, and folding, sorting and degradation (Fig. 6 A). In 2-week-old piglets, the bacterial RNA profiling of amino acid metabolism, coenzyme transport, and metabolism, metabolism of cofactors and vitamins, genetic information processing, metabolism of other amino acids were significantly induced by PEDV infection. However, the predicted functions of membrane transport, defense mechanisms, enzyme families, etc. were decreased by PEDV infection (Fig. 6B). Although the changes are small, these results might indicate that PEDV infection also affects the biological functions of intestinal microbiota in piglets.
Fig. 6.
The predicted functions of intestinal microbes in 1-week-old (A) and 2-week-old (B) piglets were analyzed by KEGG or COG.
7. Discussion
Microbiota is a critical player in the microenvironment of intestines, where the PEDV invades and replicates. The complex microbe in the gastrointestinal tract plays a critical role in maintaining the gut health of humans, animals, and birds (Ivanov and Honda, 2012). Since 2010, high virulent PEDV strains emerged in pig farms, causing severe diarrhea, intestinal damage, and up to 100% mortality in piglets younger than 10-day-old (Li et al., 2012). Thus, we hypothesized that PEDV infection changes the diversity and abundance of intestinal microbiota in young piglets. In this study, we investigated the distribution and abundance of intestinal bacteria residing in different small intestine segments from healthy piglets at different ages. The alterations of distribution, abundance, and potential functions caused by PEDV infection were also analyzed. To our knowledge, this is the first comprehensive investigation to report differential intestinal microbiota from a different age of piglets and influenced by PEDV infection.
A previous investigation in pigs showed that the bacteria in the ileum, cecum, and colon were differentially distributed (Kim and Isaacson, 2015). However, there was no such report to compare the composition of microbiota in different segments of swine small intestines in pigs. The small intestine, susceptible to PEDV invasion, plays an essential role in sustaining intestinal homeostasis via a wide variety of microbes. Our results indicated that Bacteroidetes, Firmicutes, and Proteobacteria were the predominant phyla of intestinal microbiota. Song et al. reported that PEDV infection decreased the number of Bacteroidetes (Song et al., 2017), which produces a short fatty acid to protect the intestines (Onderdonk et al., 1982). The reduced number of Bacteroidetes might influence intestinal homeostasis, making it easier for PEDV invasion.
In this study, the influence of PEDV infection on the intestinal microbial abundance in different intestine segments from a different age of piglets was investigated. The results demonstrated that 1-week-old and 2-week-old healthy piglets shared high similarity in the microbiota distribution and abundance but differed from the 4-week-old piglets. Their diet pattern may be responsible for this difference since younger piglets were fed with milk substitutes while the 4-week-old piglets were fed with pellet feed. Regardless of age, diet, and breeds of piglets, the Bacteroidetes and Firmicutes were still the predominant phyla in the small intestine, consistent with a previous report (Kim et al., 2015). The higher abundance of Proteobacteria in 1-week-old and 2-week-old piglets rather than 4-week-old piglets may result in higher viral loads in intestinal contents of 1-week-old and 2-week-old piglets because the γ-Proteobacteria were reported to cause sustained intestinal inflammation and increased susceptibility to neonatal and adult models of intestinal injury (Mirpuri et al., 2014).
Fecal consistency, a visual scoring system, is widely used to evaluate the severity of diarrhea, which is inherently subjective. To avoid inaccuracy caused by the individual investigator, both blind scoring and wet/dry ratios (an objective measure of fecal consistency) (Liu et al., 2009) were applied to assess diarrhea in this study. High coherence was observed between the visual assessment of fecal samples and their wet/dry ratios (Figs. 3B and S5B). Besides, our results demonstrated that the viral loads in intestinal contents were higher than those in intestinal tissues. We speculated that intestinal microbiota and/or their metabolite might improve the invasiveness and replication of viruses. For example, bacterial polysaccharides enhance poliovirus stability and cell attachment so that viruses can easily invade intestinal tissue and replicate robustly (Robinson et al., 2014). Our research illustrated that more detectable PEDV was found from the jejunum and ileum than from the duodenum (Fig. 4A and B). We hypothesize that bacteria facilitate PEDV invasion via microfold cells (M cells) since M cells are widely distributed in the jejunum and ileum and are the main entry sites for many bacteria like Salmonella typhimurium, Shigella Flexner, Yersinia entericolitica and Brucella abortus (Sansonetti and Phalipon, 1999). Further investigations are required to reveal the detailed relationships between the PEDV invasion, intestinal microbiota, and M cells.
Our results demonstrated that Lactobacillus and Escherichia-shigella were affected by PEDV infection. Lactobacillus plays an essential role in maintaining the homeostasis of intestinal microbiota and enteric immunity. Wang et al. reported that Lactobacillus casei modulates enteritis by regulating the differentiation of Treg/Th17 cells (Wang et al., 2017), critical players in inflammatory diseases (Velasquez-Lopera et al., 2008). Lactobacillus prevents pathogenic bacteria from invading intestines and inhibits inflammation (Alexandre et al., 2014). Our results showed that the PEDV infection induced severe pathogenicity and increased Lactobacillus's numbers in the small intestine of 1-week-old piglets, whose adaptive immune system is immature. The increase of Lactobacillus can inhibit inflammation and the growth of other pathogenic bacteria by stimulating innate immunity. More studies are needed to illuminate the relationship between Lactobacillus and intestinal pathogenicity. Escherichia-shigella was increased after PEDV infection (Koh et al., 2015) and could invade the basolateral side of epithelial cells (Mounier et al., 1992), and is one of the most common bacteria causing diarrhea (Zhu et al., 2011). The toxin of Escherichia-shigella secreted by type III secretion system is capable of changing cellular metabolism, lysing cellular membranes, and facilitating bacterial invasion (Snapper et al., 2001). The absence of detectable PEDV in the intestines of 2-week-old and older piglets (Fig. 4A and C) may have been caused by the increase of Escherichia-shigella and associated with upregulation of IFN III expression, which is essential to defend intestinal viral entry (Odendall et al., 2017). Detailed investigations are required to address this hypothesis. Besides, microbial changes influence cellular metabolism (3). Intestinal microbe imbalances caused by infection lead to metabolic disorders, consistent with our predicted microbial functions. Experimental data should be generated to verify and validate the predicted functions.
In conclusion, this study investigated the distribution of intestinal microbiota in different segments of the small intestine and different ages of piglets using 16S rRNA sequencing technology. Our results show that PEDV infection reshapes the intestinal microbiota, especially in younger piglets. Future studies will investigate whether the absence or presence of specific microbes in the context of PEDV infection changes disease susceptibility in piglets. Understanding the effect of intestinal microbes on the infectivity and severity of PEDV will facilitate the development of new prevention strategies for viral diarrhea.
8. Materials and methods
8.1. Piglets and virus
To obtain PEDV-free piglets, piglets of different ages and sows were screened by real-time RT-qPCR and ELISA, specific for both PEDV genome and antibodies. After screening, six PEDV-free piglets (Duroc-Landrace-Yorkshire) for each age group (1-week old, 2-week old, and 4-week old) were purchased from a pig farm located in Dingxi, Gansu province and housed in isolated animal rooms. Pigs at the same age were from the same sow, and the sows were feed in the same environment. The gender of pigs was chosen randomly. The 1-week and 2-week old piglets were fed with a milk substitute (Anyou, China) while the 4-week old piglets were fed with regular feed (typically weaned at 21 days).
The virulent PEDV field strain LJX01/2014 was obtained from diarrheic piglets from a pig farm in LiuJiaXia, Gansu, China. To get rid of all the intestinal bacteria, the intestinal contents from the diarrheal piglets were resuspended in sterile PBS and centrifuged at 4 °C for 20 min. The supernatant was then collected, subjected to filtration through 0.45 μm filters, and quantified for PEDV loads by a probe-based real-time RT-qPCR. No other enteric viruses were found in the viral stock, and the PEDV viral stock was then preserved at −80 °C until use.
8.2. Animal experiments
Six piglets of each age group were randomly divided into two groups, challenge group and mock control group (n = 3/group). For three challenge groups, the piglets were orally inoculated with 2 × 107 copies PEDV LJX01/2014 strain. The mock-infected piglets were orally inoculated with an equal volume of sterile PBS. Their rectal temperature was measured every 12 h after inoculation while the anal swabs of each piglet were collected every 6 h for detection of viral shedding by real-time RT-qPCR. In addition, fecal samples were collected directly from the rectum of each piglet and scored (0, normal feces; 1, mixed stool samples containing both solid and pasty feces; 2, pasty feces; 3, semiliquid feces; 4, liquid feces) independently and blindly by two investigators to evaluate the severity of diarrhea. The feces were immediately weighed after scoring, then dried at 70 °C overnight, and reweighed. Wet/dry ratios were determined by dividing the initial weight by the dry weight to assess diarrhea. At 48 hpi, all piglets were sacrificed and subjected to necropsy to harvest intestinal contents in duodenum, jejunum, and ileum separately for both viral loads detection and intestinal microbiota analysis by 16S rRNA sequencing. The tissues of duodenum, jejunum, and ileum were harvested for viral load detection and evaluation of pathological changes. All experimental procedures and animal care protocols were approved by the guidelines for Care and Use of Laboratory Animals of Lanzhou Veterinary Research Institute (LVRI), Chinese Academy of Agricultural Sciences, China.
8.3. Viral loads detection
Anal swabs and intestinal contents in sterile PBS were vortexed thoroughly and subjected to centrifugation at 12,000 rpm for 10 min at 4 °C. Intestines were homogenized in pre-chilled sterile PBS containing 0.5 mm silicon beads with a bead-beater (Eppendorf) and centrifuged at 12,000 rpm for 10 min at 4 °C. The total RNAs from each sample were extracted from supernatants using Trizol reagent (Invitrogen) and reverse-transcribed into cDNAs using random primers. The cDNAs were applied to a Taqman probe-based real-time qPCR analysis for detection PEDV genome using the ABI 7500 system with TransStart Probe qPCR Supermix (Transgen Biotech). The detailed information of the primer set and probe targeting PEDV M gene were as follows: forward primer: 5′-GAT ACT TTG GCC TCT TGT GT-3′, reverse primer: 5′-CAC AAC CGA ATG CTA TTG ACG-3′, and Taqman probe: 5′-FAM-TTC AGC ATC CTT ATG GCT TGC ATC-TAMRA-3'. The amplification was carried out with denaturing at 95 °C for 30s, followed by 40 cycles at 94 °C for 5s and 60 °C for 30s. The genome copies were calculated based on a standard curve.
8.4. Histological analysis and immunohistochemistry
Different intestinal tissue segments were collected, fixed for 24 h in 10% formalin, dehydrated according to the standard protocol, and then embedded in paraffin. The sections were cut from each tissue block, deparaffinized in xylene, stained with Hematoxylin and Eosin (H&E), and observed under a microscope to evaluate pathological changes.
After antigen retrieval, the paraffin sections were incubated with a monoclonal antibody against PEDV nucleoprotein, then HRP-conjugated goat anti-mouse (1:500) secondary antibody and followed by staining with Mayer's hematoxylin as the nuclear dye. The slides were then observed under a microscope to detect viral particles, showing brown precipitation in the cells.
8.5. Intestinal microbiota analysis
Genomic DNA was extracted from intestinal contents within duodenum, jejunum, and ileum by utilizing the PowerSoil® DNA Isolation Kit (MO BIO Laboratories Inc., Carlsbad, CA, USA), according to the manufacturer's protocol. Furthermore, DNA quality was assessed with NanoDrop ND-2000 Spectrophotometer (NanoDrop Technologies Inc., Wilmington, USA) and a spectrophotometer (Qubit 2.0 Fluorometer, ThermoScientific, USA). Then the high pure genomic DNA was used for library preparations and Illumina high-throughput sequencing by GENEWIZ (Suzhou, China). Details regarding Illumina sequencing, experimental steps, and data analyses are described elsewhere (You et al., 2016). Briefly, V3 and V4 hypervariable regions of prokaryotic 16S rRNA were selected for generating amplicons and following taxonomy analysis. The V3 and V4 regions were amplified using forward primers “CCTACGGRRBGCASCAGKVRVGAAT” and reverse primers “GGACTACNVGGGTWTCTAATCC."
Meanwhile, indexed adapters were added to the ends of the 16S rRNA amplicons to generate indexed libraries ready for downstream NGS sequencing on Illumina Miseq. The DNA library concentration was validated by Qubit3.0 Fluorometer. After quantification, the library was diluted to 10 nM, DNA libraries were multiplexed and loaded on an Illumina MiSeq instrument according to manufacturer's instructions (Illumina, San Diego, CA, USA).
The forward and reverse reads obtained by two-terminal sequencing were joined based on barcode and truncated by cutting off the barcode and primer sequence (Cutadapt v1.9.1). After removing the chimeric sequence, sequences with high quality were retained and compared to the reference database (Silva 132) for operational taxonomic unit (OTU) analysis. VSEARCH (1.9.6) was used for sequence clustering (sequence similarity was set to 97%). Then, the RDP classifier (Ribosomal Database Program) Bayesian algorithm was used to analyze the representative sequence of OTU, and the community composition of each sample was counted at different taxonomic levels.
Based on the results of OTU analysis, alpha diversity index was calculated by random sampling of sample sequences. Based on the Bray-Curtis distance matrix between samples, the visualization maps of PCoA (principal coordinates analysis) and NMDS (non-metric multidimensional scaling analysis) were used to show beta diversity. Advanced analysis, such as Metastats and LefSe (LDA Effect Size), was employed to understand genomic differences between groups better to understand the kinetics of intestinal microbiota and PEDV infection. Besides, after the OTU abundance being normalized, the COG family/KO information corresponding to each OTU through the Greengene ID corresponding was obtained, and the abundance of each COG was calculated. Finally, their predicted biological functions were annotated.
8.6. Data analysis
All data were analyzed by SPSS. The significant difference analysis was performed by one-way ANOVA (”***” means p < 0.001, “**” means p < 0.01, “*” means p < 0.05). Data presented as Mean ± SEM. All figures were generated by “R” or GraphPad prism software. The P-value from Anosim analysis was used to describe the PCoA difference in different groups. “p < 0.05” means a significant difference.
CRediT authorship contribution statement
Shanshan Yang: Conceptualization, Methodology, Validation, Formal analysis, Data curation, Writing - original draft, Project administration. Yang Li: Investigation. Bin Wang: Investigation. Ning Yang: Investigation. Xin Huang: Investigation. Qingbo Chen: Investigation. Shuxian Geng: Investigation. Yawei Zhou: Investigation. Han Shi: Investigation. Leyi Wang: Formal analysis, Data curation. Sylvia Brugman: Supervision. Huub Savelkoul: Writing - review & editing, Supervision. Guangliang Liu: Conceptualization, Methodology, Validation, Resources, Data curation, Writing - review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Key R&D Program of China (2016YFD0500103), the National Natural Science Foundation of China (31972689), the Elite Youth Program of CAAS, and partly by China Central Public-interest Scientific Institution Basal Research Fund (1610312020020), and by WUR-CAAS joint Ph.D. Program.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2020.07.001.
Availability of data and materials
The raw data for this article were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database under BioProject No. PRJNA526581.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Alexandre Y., Le Blay G., Boisrame-Gastrin S., Le Gall F., Hery-Arnaud G., Gouriou S., Vallet S., Le Berre R. Probiotics: a new way to fight bacterial pulmonary infections? Med. Maladies Infect. 2014;44(1):9–17. doi: 10.1016/j.medmal.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debouck P., Pensaert M. Experimental infection of pigs with a new porcine enteric coronavirus, CV 777. Am. J. Vet. Res. 1980;41(2):219–223. [PubMed] [Google Scholar]
- Goulet O. Potential role of the intestinal microbiota in programming health and disease. Nutr. Rev. 2015;73(Suppl. 1):32–40. doi: 10.1093/nutrit/nuv039. [DOI] [PubMed] [Google Scholar]
- Huang M.Z., Wang S.Y., Wang H., Cui D.A., Yang Y.J., Liu X.W., Kong X.J., Li J.Y. Differences in the intestinal microbiota between uninfected piglets and piglets infected with porcine epidemic diarrhea virus. PLoS One. 2018;13(2) doi: 10.1371/journal.pone.0192992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Chen J., Yao G., Guo Q., Wang J., Liu G. A TaqMan-probe-based multiplex real-time RT-qPCR for simultaneous detection of porcine enteric coronaviruses. Appl. Microbiol. Biotechnol. 2019;103(12):4943–4952. doi: 10.1007/s00253-019-09835-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.I., Honda K. Intestinal commensal microbes as immune modulators. Cell Host Microbe. 2012;12(4):496–508. doi: 10.1016/j.chom.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Eyerly B., Annamalai T., Lu Z., Saif L.J. Structural alteration of tight and adherens junctions in villous and crypt epithelium of the small and large intestine of conventional nursing piglets infected with porcine epidemic diarrhea virus. Vet. Microbiol. 2015;177(3–4):373–378. doi: 10.1016/j.vetmic.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.B., Isaacson R.E. The pig gut microbial diversity: understanding the pig gut microbial ecology through the next generation high throughput sequencing. Vet. Microbiol. 2015;177(3–4):242–251. doi: 10.1016/j.vetmic.2015.03.014. [DOI] [PubMed] [Google Scholar]
- Kim J., Nguyen S.G., Guevarra R.B., Lee I., Unno T. Analysis of swine fecal microbiota at various growth stages. Arch. Microbiol. 2015;197(6):753–759. doi: 10.1007/s00203-015-1108-1. [DOI] [PubMed] [Google Scholar]
- Koh H.W., Kim M.S., Lee J.S., Kim H., Park S.J. Changes in the swine gut microbiota in response to porcine epidemic diarrhea infection. Microb. Environ. 2015;30(3):284–287. doi: 10.1264/jsme2.ME15046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol. J. 2015;12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Kim Y., Lee C. Isolation and characterization of a Korean porcine epidemic diarrhea virus strain KNU-141112. Virus Res. 2015;208:215–224. doi: 10.1016/j.virusres.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee C. Genomic and antigenic characterization of porcine epidemic diarrhoea virus strains isolated from South Korea, 2017. Transbound Emerg Dis. 2018;65(4):949–956. doi: 10.1111/tbed.12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Su M., Yin B., Guo D., Wei S., Kong F., Feng L., Wu R., Sun D. Integrin alphavbeta3 enhances replication of porcine epidemic diarrhea virus on Vero E6 and porcine intestinal epithelial cells. Vet. Microbiol. 2019;237:108400. doi: 10.1016/j.vetmic.2019.108400. [DOI] [PubMed] [Google Scholar]
- Li W., Li H., Liu Y., Pan Y., Deng F., Song Y., Tang X., He Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg. Infect. Dis. 2012;18(8):1350–1353. doi: 10.3201/eid1808.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Kahan S.M., Jia Y., Karst S.M. Primary high-dose murine norovirus 1 infection fails to protect from secondary challenge with homologous virus. J. Virol. 2009;83(13):6963–6968. doi: 10.1128/JVI.00284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Zhao L., Zhai Z., Zhao W., Ding J., Dai R., Sun T., Meng H. Porcine epidemic diarrhea virus infection induced the unbalance of gut microbiota in piglets. Curr. Microbiol. 2015;71(6):643–649. doi: 10.1007/s00284-015-0895-6. [DOI] [PubMed] [Google Scholar]
- Mirpuri J., Raetz M., Sturge C.R., Wilhelm C.L., Benson A., Savani R.C., Hooper L.V., Yarovinsky F. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microb. 2014;5(1):28–39. doi: 10.4161/gmic.26489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mole B. Deadly pig virus slips through US borders. Nature. 2013;499(7459):388. doi: 10.1038/499388a. [DOI] [PubMed] [Google Scholar]
- Moon H.W., Norman J.O., Lambert G. Age dependent resistance to transmissible gastroenteritis of swine (TGE). I. Clinical signs and some mucosal dimensions in small intestine. Can. J. Comp. Med. 1973;37(2):157–166. [PMC free article] [PubMed] [Google Scholar]
- Mounier J., Vasselon T., Hellio R., Lesourd M., Sansonetti P.J. Shigella flexneri enters human colonic Caco-2 epithelial cells through the basolateral pole. Infect. Immun. 1992;60(1):237–248. doi: 10.1128/iai.60.1.237-248.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman J.O., Lambert G., Moon H.W., Stark S.L. Age dependent resistance to transmissible gastroenteritis of swine (TGE). II. Coronavirus titer in tissues of pigs after exposure. Can. J. Comp. Med. 1973;37(2):167–170. [PMC free article] [PubMed] [Google Scholar]
- Odendall C., Voak A.A., Kagan J.C. Type III IFNs are commonly induced by bacteria-sensing TLRs and reinforce epithelial barriers during infection. J. Immunol. 2017;199(9):3270–3279. doi: 10.4049/jimmunol.1700250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onderdonk A.B., Markham R.B., Zaleznik D.F., Cisneros R.L., Kasper D.L. Evidence for T cell-dependent immunity to Bacteroides fragilis in an intraabdominal abscess model. J. Clin. Invest. 1982;69(1):9–16. doi: 10.1172/JCI110445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C.M., Jesudhasan P.R., Pfeiffer J.K. Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe. 2014;15(1):36–46. doi: 10.1016/j.chom.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansonetti P.J., Phalipon A. M cells as ports of entry for enteroinvasive pathogens: mechanisms of interaction, consequences for the disease process. Semin. Immunol. 1999;11(3):193–203. doi: 10.1006/smim.1999.0175. [DOI] [PubMed] [Google Scholar]
- Shibata I., Tsuda T., Mori M., Ono M., Sueyoshi M., Uruno K. Isolation of porcine epidemic diarrhea virus in porcine cell cultures and experimental infection of pigs of different ages. Vet. Microbiol. 2000;72(3–4):173–182. doi: 10.1016/S0378-1135(99)00199-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapper S.B., Takeshima F., Anton I., Liu C.H., Thomas S.M., Nguyen D., Dudley D., Fraser H., Purich D., Lopez-Ilasaca M., Klein C., Davidson L., Bronson R., Mulligan R.C., Southwick F., Geha R., Goldberg M.B., Rosen F.S., Hartwig J.H., Alt F.W. N-WASP deficiency reveals distinct pathways for cell surface projections and microbial actin-based motility. Nat. Cell Biol. 2001;3(10):897–904. doi: 10.1038/ncb1001-897. [DOI] [PubMed] [Google Scholar]
- Song D., Peng Q., Chen Y., Zhou X., Zhang F., Li A., Huang D., Wu Q., Ye Y., He H., Wang L., Tang Y. Altered gut microbiota profiles in sows and neonatal piglets associated with porcine epidemic diarrhea virus infection. Sci. Rep. 2017;7(1):17439. doi: 10.1038/s41598-017-17830-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Feng L., Shi H., Chen J., Cui X., Chen H., Liu S., Tong Y., Wang Y., Tong G. Identification of two novel B cell epitopes on porcine epidemic diarrhea virus spike protein. Vet. Microbiol. 2008;131(1–2):73–81. doi: 10.1016/j.vetmic.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez-Lopera M.M., Eaton V.L., Lerret N.M., Correa L.A., Decresce R.P., Garcia L.F., Jaramillo A. Induction of transplantation tolerance by allogeneic donor-derived CD4(+)CD25(+)Foxp3(+) regulatory T cells. Transpl. Immunol. 2008;19(2):127–135. doi: 10.1016/j.trim.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Wang K., Dong H., Qi Y., Pei Z., Yi S., Yang X., Zhao Y., Meng F., Yu S., Zhou T., Hu G. Lactobacillus casei regulates differentiation of Th17/Treg cells to reduce intestinal inflammation in mice. Can. J. Vet. Res. 2017;81(2):122–128. [PMC free article] [PubMed] [Google Scholar]
- You J., Wu G., Ren F., Chang Q., Yu B., Xue Y., Mu B. Microbial community dynamics in Baolige oilfield during MEOR treatment, revealed by Illumina MiSeq sequencing. Appl. Microbiol. Biotechnol. 2016;100(3):1469–1478. doi: 10.1007/s00253-015-7073-4. [DOI] [PubMed] [Google Scholar]
- Zhu J., Yin X., Yu H., Zhao L., Sabour P., Gong J. Involvement of quorum sensing and heat-stable enterotoxin a in cell damage caused by a porcine enterotoxigenic Escherichia coli strain. Infect. Immun. 2011;79(4):1688–1695. doi: 10.1128/IAI.01281-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data for this article were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database under BioProject No. PRJNA526581.