Abstract
Ferroptosis is widely involved in degenerative diseases in various tissues including kidney, liver and brain, and is a targetable vulnerability in clear-cell carcinomas and therapy-resistant cancers. Accumulation of phospholipid hydroperoxides in cellular membranes is recognized as the hallmark and rate-limiting step of ferroptosis; however, the enzymes contributing to lipid peroxidation remain poorly characterized. Using genome-wide, CRISPR/Cas9-mediated suppressor screens, we identify cytochrome P450 oxidoreductase (POR) as a contributor to ferroptotic cell death in cancer cells exhibiting inherent and induced susceptibility to ferroptosis. By genetic depletion of POR in cancer cells, we reveal that POR contributes to ferroptosis across a wide range of lineages and cell-states, and in response to distinct mechanisms of ferroptosis induction. Using systematic lipidomic profiling, we further map POR’s activity to the lipid peroxidation step in ferroptosis. Hence, our work suggests POR as a key mediator of ferroptosis and a potential druggable target for developing anti-ferroptosis therapeutics.
Introduction
Ferroptosis is an iron-dependent and non-apoptotic form of cell death that is widely implicated in human pathological conditions including cancer therapy-resistance and brain injury1-4. Modulating ferroptosis activities holds great promise for therapeutic development against malignant and non-malignant diseases4-6. In addition to mammalian cells, ferroptosis is also observed in unicellular parasites such as Trypanosoma brucei and higher plants7,8, suggesting that mechanisms causing ferroptotic cell death might be conserved during evolution.
The chemical nature of ferroptosis can be defined by an imbalance between the production and dissipation of polyunsaturated phospholipid hydroperoxides (LOOH)9. Aberrant accumulation of LOOH leads to rapid membrane rupture and subsequent irreversible cell death10. Recently, the selenoenzyme glutathione peroxidase 4 (GPX4) was identified as the key regulator that protects cells from undergoing ferroptosis by using glutathione to reduce lipid hydroperoxides to lipid alcohols6,11,12. Distinct from other GPXs, GPX4 is the only enzyme identified thus far that acts on phospholipid hydroperoxides6,13. Hence, inhibitors of GPX4, including ML210, ML162 and 1S,3R-RSL3 (RSL3), are used as specific inducers of ferroptosis in cells with high levels of polyunsaturated lipids - lipids that are highly susceptible to oxidation1,14,15.
The mechanisms mediating the production of lipid hydroperoxides, however, remain poorly characterized. Lipid peroxidation can occur non-enzymatically via spontaneous autoxidation, or be facilitated by cellular enzymes16-19. Prior research suggested that enzymatic lipid peroxidation is primarily catalyzed by arachidonate lipoxygenases (ALOXs)2,20,21. Nonetheless, the necessity for ALOXs’ involvement in ferroptosis was recently questioned by previously unappreciated additional radical-trapping activities of ALOX-inhibiting small molecules used to establish the ALOX-ferroptosis connection22. On the other hand, the possibility that other proteins may be involved in lipid peroxidation remains largely unexplored.
Using a series of genome-wide, CRISPR/Cas9 suppressor screens, we identified cytochrome P450 oxidoreductase (POR) as a key mediator of ferroptotic cell death in cells that exhibit intrinsic as well as induced susceptibility to ferroptosis. The pro-ferroptosis role of POR applies to diverse cancer lineages, and to the effects of various ferroptosis-inducing agents with distinct structures and mechanism of action (MoA). We further revealed that POR contributes to ferroptosis by enabling membrane polyunsaturated phospholipid peroxidation. Our work suggests that targeting POR could have therapeutic value in disease contexts where blocking ferroptosis is desired.
RESULTS
CRISPR screens identify POR as a pro-ferroptotic gene
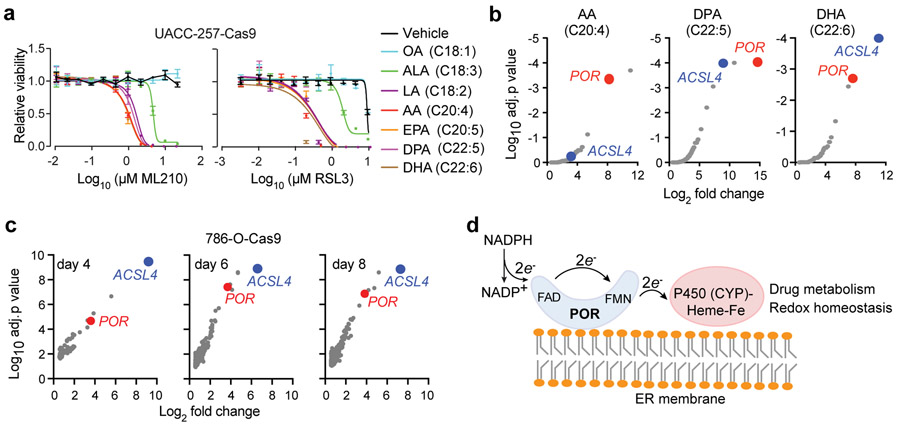
To identify genes mediating lipid peroxidation in the ferroptosis cascade, we devised two complementary groups of genome-wide, CRISPR/Cas9 screens: one in the 786-O clear-cell renal cell carcinoma (ccRCC) cell line that exhibits strong intrinsic sensitivity to ferroptosis14, and the other in UACC-257, a melanoma cell line that is inherently resistant to ferroptosis, yet was induced to ferroptosis-susceptible states by supplementing with polyunsaturated fatty acids (PUFA) in the culture media (Fig. 1a). The fatty acids that induced strong sensitization to ferroptosis triggered by ML210 (1) or RSL3 (2) treatment in UACC-257 cells included ω−6 PUFAs linoleic acid (LA, C18:2) and arachidonic acid (AA, C20:4), and ω−3 PUFAs ɑ-linolenic acid (ALA, C18:3), eicosapentaenoic acid (EPA, C20:5), docosapentaenoic acid (DPA, C22:5), and docosahexaenoic acid (DHA, C22:6), but not monounsaturated fatty acid (MUFA) oleic acid (OA, C18:1) (Fig. 1a, Supplementary Figs. 1a-b), supporting that lipid peroxidation and ferroptosis is specifically driven by PUFA-lipids.
Figure 1. Genome-wide CRISPR screens identify POR as a mediator of ferroptosis.
a. Viability curves for UACC-257-Cas9 melanoma cells pre-treated with bovine serum albumin (BSA)-containing vehicle or indicated fatty acids for 3 days, then treated with indicated concentrations of GPX4 inhibitors ML210/RSL3. Abbreviations: OA, oleic acid (C18:1); LA, linoleic acid (C18:2); ALA, ɑ-linolenic acid (C18:3); AA, arachidonic acid (C20:4); EPA, eicosapentaenoic acid (C20:5); DPA, docosapentaenoic acid (C22:5); DHA, docosahexaenoic acid (C22:6). n=4, error bars: mean±s.d. Representative plot of experiments performed in triplicate.
b. Partial volcano plots showing the top hits in AA, DPA or DHA treated screening conditions in UACC-257-Cas9 cells, highlighting POR and ACSL4. Target genes of sgRNAs enriched in the ML210-treated condition are presented.
c. Partial volcano plots showing the top hits in the 786-O-Cas9 screen, highlighting POR and ACSL4. The top left corner indicates how many days of ML210 treatment was used in each condition. Target genes of sgRNAs enriched in the ML210-treated condition are presented.
d. Brief scheme summarizing the structure and functions of the cytochrome P450 enzyme (POR / CYP) system. NADPH, nicotinamide adenine dinucleotide phosphate; FAD, flavin adenine dinucleotide; FMN, flavin mononucleotide.
In all genetic screens, we used ML210 to trigger ferroptotic cell death, and uncovered the sgRNAs that confer resistance to death by sequencing14 (Supplementary Fig. 1c). Top shared hits included acyl-CoA synthetase long-chain family member 4 (ACSL4) (Figs. 1b-c, Supplementary Fig. 1d and Supplementary Dataset 1), which catalyzes the esterification of PUFAs to PUFA-CoAs for their subsequent incorporation into membrane phospholipids and is implicated in ferroptosis across multiple cell types23,24, thus supporting the validity of our screens.
In search of mechanisms regulating the catalytic reaction of lipid hydroperoxide formation, we focused on characterizing hits that are directly involved in redox regulation. This criterion nominated cytochrome P450 oxidoreductase (POR, also known as CYPOR) as the most relevant candidate gene that is shared by both groups of CRISPR screens (Figs. 1b-c, Supplementary Fig. 1d and Supplementary Dataset 1). The most common cytochrome P450 enzyme system is comprised of a shared NADPH-dependent oxidoreductase (POR) unit and typically a cytochrome P450 (CYP) isoenzyme that contains a protoporphyrin IX heme-iron prosthetic group25 (Fig. 1d). Together, POR-CYPs play key roles in cellular redox homeostasis and detoxification of xenobiotic chemicals26,27. In addition to CYPs, POR also donates electrons to other redox partners including heme oxygenase, cytochrome b5, and squalene monooxygenase28. Importantly, POR is also a hit in our recently reported genome-wide, CRISPR/Cas9, ferroptosis screen in the pancreatic carcinoma cell line KP-4 (ref. 1). Together, these screens nominate POR as a mediator of ferroptotic cell death in cells that exhibit intrinsic, as well as PUFA-induced, ferroptosis susceptibility.
POR mediates ferroptosis in multiple cancer lineages
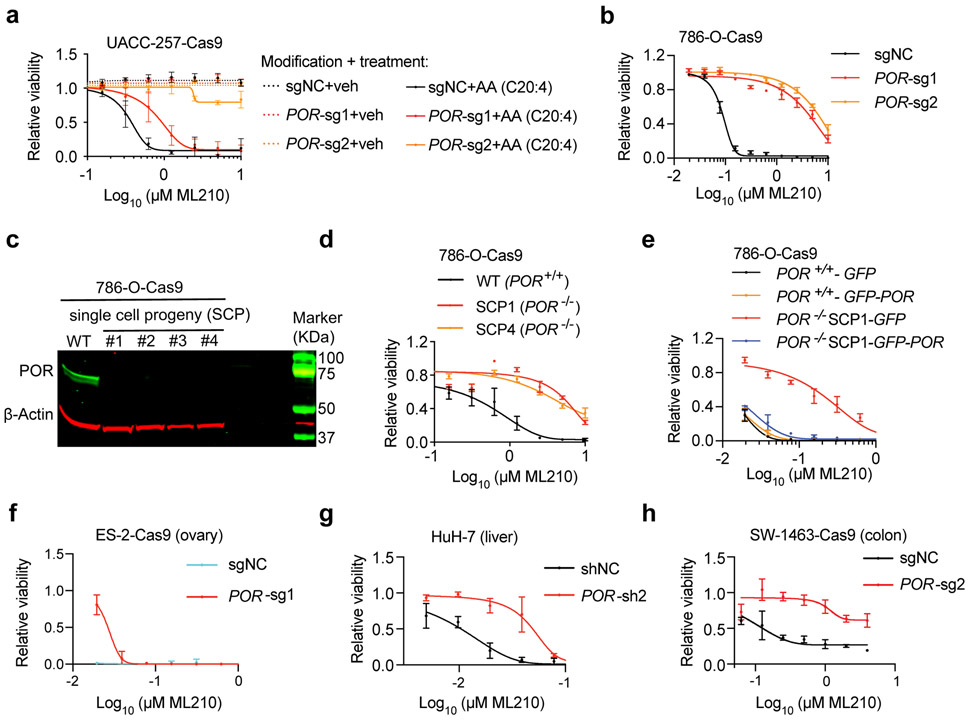
To validate the pro-ferroptotic role of POR, we used three sequence-independent sgRNAs to deplete POR proteins in Cas9-expressing UACC-257 cells (Supplementary Fig. 2a). POR-depletion suppressed arachidonic acid-induced sensitivity to ML210/RSL3 in a dose-dependent manner (Fig. 2a, Supplementary Fig. 2b). In addition to suppressing PUFA-induced ferroptosis susceptibility, POR-depletion by constitutive or inducible knockout also compromised the intrinsic ferroptosis sensitivity in ccRCC cells 786-O and 769-P (Fig. 2b, Supplementary Figs. 2c-f). The knockout effects were recapitulated by shRNA-mediated POR mRNA knockdown in UACC-257, 786-O, 769-P cells and a third ccRCC cell line OS-RC2 (Supplementary Figs. 2g-m).
Figure 2. POR contributes to ferroptotic cell death in multiple cancer lineages.
a. Viability curves for sgNC or sgPOR-expressing UACC-257-Cas9 melanoma cells pre-treated with vehicle (veh) or arachidonic acid (AA, C20:4) for 3 days, then treated with indicated concentrations of ML210.
b. Viability curves of 786-O-Cas9 cells expressing sgNC or POR-targeting sgRNAs treated with indicated concentrations of ML210.
c. Immunoblot analysis of POR protein levels in wildtype 786-O-Cas9 cells, and POR−/− single cell progenies (SCP) 1-4. β-Actin was used as a loading control. Representative plot of experiment performed twice. See Supplementary Figure 10 for the original blots.
d. Viability curves of 786-O-Cas9 cells expressing control sgRNA (WT), or POR−/− SCP1 and SCP4 treated with indicated concentrations of ML210.
e. Viability curves of POR+/+ and POR−/− 786-O cells expressing GFP or POR-GFP cDNAs treated with indicated concentrations of ML210.
f. Viability curves of ES-2-Cas9 ovarian carcinoma cells expressing sgNC or POR-targeting sgRNA treated with indicated concentrations of ML210.
g. Viability curves of HuH-7 hepatocellular carcinoma cells expressing shNC or POR-targeting shRNA treated with indicated concentrations of ML210.
h. Viability curves of SW-1463-Cas9 colorectal adenocarcinoma cells expressing sgNC or POR-targeting sgRNA treated with indicated concentrations of ML210.
Viability experiments were independently performed three times and representative plots are presented. In each plot, n=4 for each data point, data and error bars indicate mean±s.d.
To demonstrate further the specific role of POR in ferroptosis, we generated four POR−/− 786-O single cell progenies (SCP1-4) using sgRNA-2 (Fig. 2c). Both SCP1 and SCP4 – two clones that we analyzed, are resistant to ML210 and RSL3 compared with the parental POR+/+ cells (Fig. 2d, Supplementary Figs. 2n-o). Sensitivity to ferroptosis in POR−/− cells was restored by POR re-expression (Fig. 2e, Supplementary Figs. 2p-q), supporting the on-target effects of CRISPR/Cas9-mediated POR gene disruption. Notably, the ferroptosis-protective effect mediated by complete POR knockout is approaching that provided by liproxstatin-1 (Lip-1)12,29, a lipophilic radical-trapping antioxidant (RTA), in the ML210/RSL3 concentrations we tested (Supplementary Fig. 2o). These data indicate that POR is a key enzyme involved in the execution of ML210/RSL3-induced ferroptotic cell death in melanoma and ccRCC cells.
By analyzing the Genotype-Tissue Expression database (GTEx)30 and the cBioportal31 for The Cancer Genome Atlas (TCGA) datasets, we found that POR mRNA is ubiquitously expressed in most human normal and cancerous tissues, respectively (Supplementary Figs. 3a-b). To test the generality of POR’s ferroptosis-mediating role in different tissue contexts, we applied the POR sgRNAs and shRNAs to a collection of cancer cell lines from various lineages including ovary, endometrium, liver, colorectum, lung and pancreas. While the eight cell lines we tested exhibited a wide range of inherent ferroptosis susceptibility, POR-depletion reduced the sensitivity to ML210/RSL3 in all cell line models (Figs. 2f-h, Supplementary Figs. 4a-k). Additionally, shRNA-mediated POR-depletion also suppressed arachidonic acid-induced sensitivity to ferroptosis in immortalized HEK-293T cells (Supplementary Figs. 4l-m). Together, these data imply that POR contributes to ferroptosis regardless of the cell types and tissue contexts. Of notice, POR-inhibition by CRISPR/Cas9 or shRNA does not significantly impair the cellular viability in majority of the cell lines analyzed in the Cancer Dependency Map (DepMap)32 (Supplementary Fig. 4n), suggesting that POR is a non-essential gene and inhibiting POR might represent a low-cytotoxicity approach to attenuate ferroptosis in different disease contexts.
POR depletion does not alter GPX4-ML210/RSL3 binding
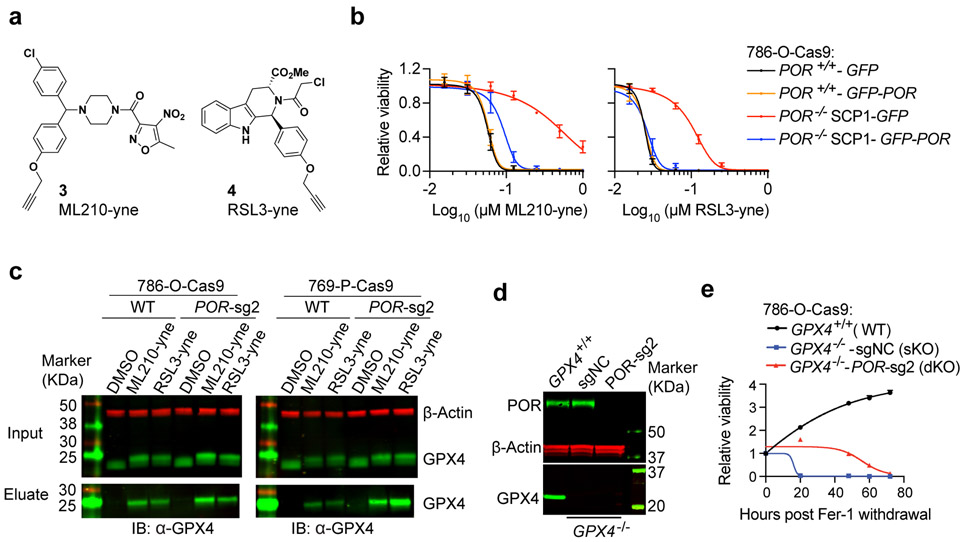
One of POR’s main functions is to provide electrons to facilitate the oxidation and detoxification of foreign chemicals25-27; hence, a caveat for POR’s involvement in GPX4 inhibition-induced ferroptosis could be that it alters the reactivity of ML210/RSL3 molecules. We employed two approaches to rule out potential POR-dependent ML210/RSL3 processing events. First, we synthesized alkyne analogs of ML210 and RSL3 – namely ML210-yne (3) and RSL3-yne (4)15, both of which preserved their ferroptosis-inducing activities in a POR-dependent manner (Figs. 3a-b, Supplementary Fig. 5a). We next determined the interactions between ML210-yne / RSL3-yne and GPX4 protein in wildtype and POR-depleted cells. Click chemistry-mediated pull-down of small-molecule-GPX4 complexes followed by anti-GPX4 immunoblotting showed that the interactions between GPX4 and ML210-yne / RSL3-yne are not altered by POR depletion in both 786-O and 769-P cells (Fig. 3c, Supplementary Fig. 5b), indicating that POR is not involved in ML210/RSL3 and GPX4 engagement.
Figure 3. POR is not required for GPX4-ML210/RSL3 binding.
a.Chemical structure of RSL3/ML210 alkyne analogs and ML210-yne (compound 3) and RSL3-yne (compound 4).
b. Viability curves of POR+/+ and POR−/− 786-O cells expressing GFP or POR-GFP cDNAs treated with indicated concentrations of ML210-yne or RSL3-yne. n=4, error bar: mean±s.d.
c. Immunoblot analysis showing the interaction between GPX4 and ML210-yne/RSL3-yne in 786-O-Cas9 or 769-P-Cas9 cells expressing sgNC (WT) or POR-sg2. Representative plot of experiment performed twice.
d. Immunoblot analysis of POR protein levels in 786-O-Cas9 cells that are GPX4+/+, GPX4−/−-sgNC or GPX4−/−-POR-sg2. Representative plot of experiment performed three times.
e. Viability time course of 786-O-Cas9 cells that are GPX4+/+ (wildtype, WT), GPX4−/−-sgNC (single knockout, sKO) or GPX4−/−-POR-sg2 (double knockout, dKO) after withdrawing ferrostatin-1 (Fer-1). n=4, error bar: mean±s.d.
β-Actin was used as a loading control in immunoblotting analyses. See Supplementary Figure 10 for the original blots.
In parallel, we genetically depleted GPX4 to bypass the use of GPX4 inhibitors for ferroptosis induction. GPX4 single knockout (GPX4−/− sKO) 786-O-Cas9 cells were previously generated and maintained in the presence of lipophilic RTA ferrostatin-1 (Fer-1)29,33, and GPX4/POR double knockout (dKO) cells were further produced by expressing POR-targeting sgRNAs in the sKO cells14 (Fig. 3d). Upon Fer-1 withdrawal, sKO cells underwent cell death rapidly (Fig. 3e); in contrast, the dKO cells exhibited significantly longer survival than their sKO counterparts (Fig. 3e). Together, POR-depleted cells are resistant to ferroptosis induced by both genetic and chemical inhibition of GPX4, underscoring the important role of POR in mediating ferroptotic cell death.
POR mediates ferroptosis induced by distinct mechanisms
We next explored whether POR also mediates ferroptosis induced by other mechanisms besides ML210/RSL3-mediated GPX4 inhibition. We selected a series of small molecules targeting different ferroptosis-related factors with distinct structures and MoA (Supplementary Figs. 5c-d). These chemicals include erastin (5) – an inhibitor of system xc− (SLC7A11) cystine/glutamate antiporter29,33, FIN56 (6), which promotes GPX4 protein degradation and CoQ10 depletion34, L-buthionine-[S,R]-sulfoximine (BSO) (7) – an inhibitor of γ-glutamylcysteine synthetase35, and FINO2 (8), a 1,2-dioxolane that directly oxidizes iron and indirectly inhibits GPX4 enzymatic function36. Importantly, POR-depletion suppressed ferroptosis induced by all four compounds (Figs. 4a-b, Supplementary Fig. 5e), highlighting the importance of POR in mediating ferroptosis induced by different mechanisms.
Figure 4. POR contributes to ferroptosis induced by distinct mechanisms.
a. Viability curves of 786-O-Cas9 cells expressing control sgRNA (WT) or POR-sg2, and POR−/− single cell clones SCP1 and SCP4 cells treated with the indicated concentrations of ferroptosis inducers including erastin, FIN56, FINO2 or BSO.
b. Viability curves of 769-P-Cas9 cells expressing control sgRNA (WT, POR+/+) or POR-sg2 treated with the indicated concentrations of ferroptosis inducers.
Viability experiments were independently performed three times and representative plots are presented. In each plot, n=4 for each data point, data and error bars indicate mean±s.d.
POR mediates ferroptosis by promoting lipid peroxidation
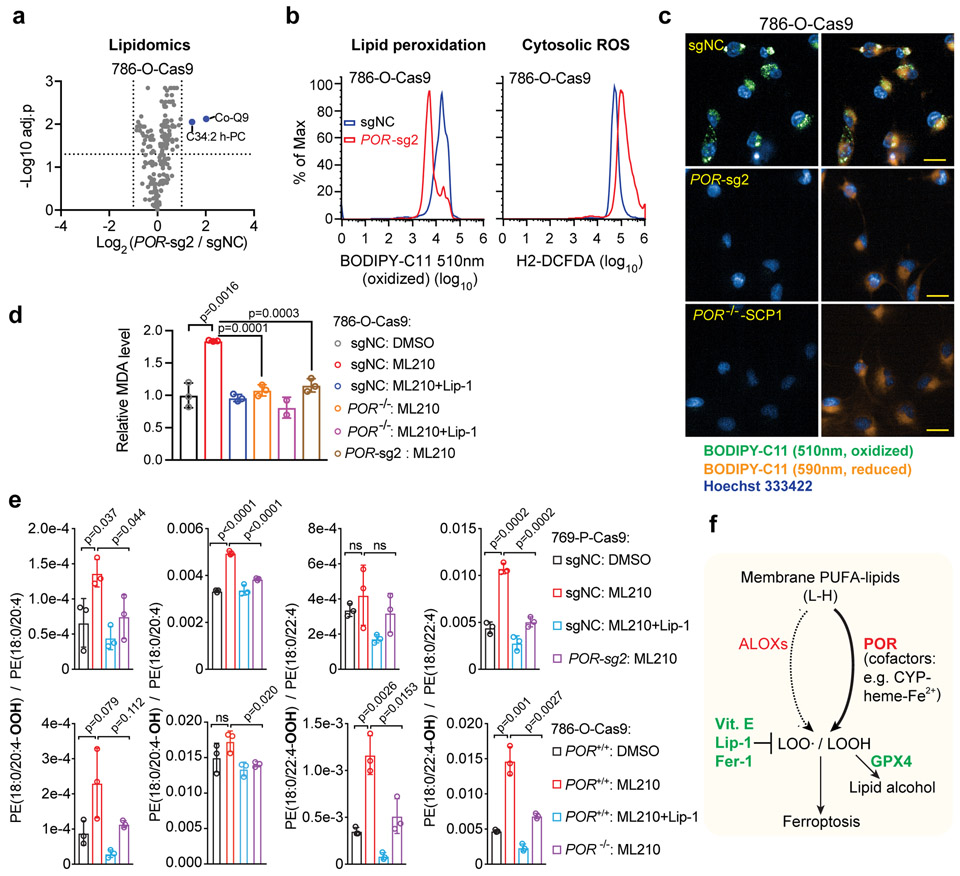
We next investigated the potential mechanisms by which POR modulates ferroptosis susceptibility. Since POR is an NADPH-dependent, FMN-containing oxidoreductase that was previously implicated in maintaining cellular redox homeostasis as well as regulating activities of lipid elongases37 and desaturases38, we proposed two major possibilities: first, POR may regulate ferroptosis susceptibility by reshaping the polyunsaturated lipidome, similar to activities of ACSL4 or lysophosphatidylcholine acyltransferase 3, LPCAT3)23. By lipidomic profiling, nonetheless, we found that POR-deletion did not significantly alter the global lipidome in 786-O and 769-P cells (Fig. 5a, Supplementary Fig. 6a and Supplementary Dataset 2), supporting that POR regulates ferroptosis through distinct mechanisms.
Figure 5. POR mediates ferroptosis by facilitating lipid peroxidation.
a. Volcano plot showing the lipidomics analysis of 786-O-Cas9 cells expressing control sgRNA (sgNC) or POR-sg2. n=3, two-tailed T-test, p values were adjusted for multi-testing using the Benjamini-Hochberg method.
b. Flow cytometry analyses of lipid peroxidation levels and cytosolic reactive oxygen species (ROS) levels reported respectively by BODIPY-C11 and H2-DCFDA in 786-O-Cas9 cells expressing sgNC or POR-sg2 and treated with ML210. Representative plots of experiments repeated three times.
c. Fluorescent microscope images of BODIPY-C11 (oxidized: green, reduced, orange) in control or POR-depleted 786-O-Cas9 cells. Scale bar, 25μm. Representative images of experiments repeated three times.
d. Bar plot showing the malondialdehyde (MDA) levels in control or POR-depleted 786-O-Cas9 cells treated with DMSO, ML210 or ML210+Lip-1. n=3, error bar: mean±s.d. Two-tailed T-tests. Representative plot of experiments repeated twice.
e. Bar plots showing the hydroperoxyl (−OOH) or hydroxyl (−OH) derivatives of indicated polyunsaturated phosphatidylethanolamine (PUFA-PE) species in WT (sgNC, POR+/+) or POR-depleted 786-O-Cas9 and 769-P-Cas9 cells treated with DMSO, ML210 or ML210+Lip-1 and analyzed by redox-lipidomics. n=3, error bar: mean±s.d. Two-tailed T-test; ns, not significant.
f. Schematic diagram summarizing the role of POR in lipid peroxidation and ferroptosis. Abbreviations: L-H, symbol for polyunsaturated- (PUFA-) phospholipid; CYP, cytochrome P450 proteins; POR, cytochrome P450 oxidoreductase; LOOH, lipid hydroperoxides; GPX4, glutathione peroxidase 4; Vit. E, vitamin E; Lip-1, liproxstatin-1; Fer-1, ferrostatin-1.
Alternatively, POR might be directly involved in lipid peroxidation via donating electrons to downstream effectors such as CYPs. Indeed, certain CYPs were shown to exhibit lipid peroxidation activity in contexts including liver microsomes and lymphocytes39-42. We hence assessed membrane lipid hydroperoxide levels in POR-depleted cells by BODIPY-C11 oxidation. Indeed, POR-depleted cells exhibited markedly reduced lipid peroxidation in UACC-257, 786-O and 769-P cells treated with ML210, as revealed by flow cytometry (Fig. 5b, Supplementary Figs. 6b-d) and fluorescent microscope analyses (Fig. 5c, Supplementary Figs. 6e-f). In contrast, POR-depletion only modestly altered cytosolic ROS levels, which was reported by the fluorescent 2’,7’-dichlorofluorescein (DCF) dye (Fig. 5b, Supplementary Fig. 6c). Consistently, POR-depletion significantly inhibited the production of malondialdehyde (MDA), a degradation product of polyunsaturated-lipid hydroperoxides and a commonly used biomarker for lipid peroxidation (Fig. 5d, Supplementary Fig. 6g).
To verify the role of POR in lipid peroxidation further, we performed redox-lipidomics in 786-O and 769-P cells. A short-term ML210 treatment induced the accumulation of hydroperoxyl- and hydroxyl-phosphatidylethanolamines (PE) that contain arachidonic acid (C20:4) or docosatetraenoic acid (also known as adrenic acid, C22:4) side chains (Fig. 5e and Supplementary Dataset 3). Lip-1 treatment effectively suppressed the accumulation of these oxidized PUFA-PEs (Fig. 5e). Importantly, POR-depletion significantly reduced the levels of both hydroperoxyl- and hydroxyl-PUFA-PEs induced by ML210 (Fig. 5e). Additionally, hydroperoxyl-PUFA-phosphatidylcholines (PC) were not detected in our experiments, while hydroxyl-PUFA-PCs were detected yet did not appear to be significantly induced by ML210 treatment (Supplementary Fig. 6h and Supplementary Dataset 3). These results are consistent with previously reported redox-lipidomics analysis using RSL3 to induce ferroptosis in Pfa1 mouse embryonic fibroblasts9,23. Collectively, these data suggest that POR facilitates lipid peroxidation in cells under ferroptotic stress.
ALOX mRNAs exhibit limited tissue expression
Previous research has highlighted the involvement of lipoxygenases, largely the ALOX5/12/15 subfamily, in mediating lipid peroxidation2,20. To explore the relative contribution of POR and lipoxygenases in ferroptosis, we interrogated the Cancer Cell Line Encyclopedia RNA-Seq data to assess their expression patterns43. Notably, ALOX mRNA expression levels are below the detection limit in majority of the CCLE cancer cells, including the aforementioned 11 cell lines we validated for POR’s ferroptosis contribution (Supplementary Figs. 7a and 8a). Additionally, sgRNAs designed to deplete the trace amount of ALOX12 protein, though the validation of which was prevented by the lack of target protein expression on immunoblots, did not provide significant protection from ferroptosis in 786-O, ES-2, OVISE or SNU-685 cells (Supplementary Figs. 8b-e). In contrast to the limited tissue expression of ALOX family members, POR is highly expressed by most cancer cells (Supplementary Figs. 3a-b, and 7a) and may therefore contribute to PUFA-lipid peroxidation in various tissue contexts.
Discussion
Ferroptosis is a cell death modality mediated by oxidative modifications of membrane polyunsaturated phospholipids10. Here, by combining small molecule-mediated induction of ferroptosis and genome-wide CRISPR/Cas9 screens, we identified POR as a contributor of ferroptotic cell death. We revealed that POR promotes ferroptosis by up-regulating the peroxidation of membrane polyunsaturated phospholipids (Fig. 5f). Moreover, we showed that POR mediates ferroptotic cell death in various cellular lineages and in response to distinct mechanisms of ferroptosis induction. Thus, our work reveals an important oxidoreductase involved in lipid peroxidation and ferroptotic cell death.
While the MoA for how POR promotes lipid peroxidation remains to be fully demonstrated, a plausible mechanism could be depicted based on our findings and insights from prior studies using liver microsomes44-46 (Supplementary Fig. 9a), which hinted that POR may facilitate lipid peroxidation by accelerating the cycling between Fe(II) and Fe(III) in the heme component of CYPs. Specifically, this process can be initiated by a putative iron-oxo species-mediated hydrogen abstraction of a polyunsaturated-lipid that leads to a bis-allylic radical45. Lipid peroxyl radicals can then be generated by the insertion of an additional di-oxygen molecule to the bis-allylic radical. The resultant peroxyl radical can react with a neighboring lipid, forming a lipid hydroperoxide and propagating the radical chain reaction22,45. In the presence of reactive iron species, lipid hydroperoxides can be converted into oxy- and peroxy-radicals (LO· and LOO·), which are highly reactive and induce significant damage to membrane structures47 (Supplementary Fig. 9a), and together can trigger ferroptotic cell death.
Under physiological conditions, GPX4 reduces lipid hydroperoxides at the expense of the reduced form of its cofactor glutathione6. Additionally, lipophilic radical-trapping antioxidants, including Vitamin E and recently developed ferrostatin-1 (Fer-1) and liproxstatin-1 (Lip-1) are able to trap chain-propagating lipid peroxyl radicals, providing a pharmacological approach to prevent ferroptosis from entering the irreversible phase29,33 (Supplementary Fig. 9a). Recent reports suggested that cancer cells could suppress ferroptotic cell death by expressing apoptosis-inducing factor, mitochondrion-associated, 2 (AIFM2, recently renamed as ferroptosis suppressor protein 1, or FSP1)18,19. FSP1 uses NADPH to regenerate ubiquinol, the reduced form of CoQ10, which acts as a cellular intrinsic lipophilic radical-trapping antioxidant and prevents oxidative lipid damage18,19. These findings indicate that certain cancer cells can boost their antioxidant capacity to counteract the lipid peroxidation stress produced by cellular intrinsic mechanisms.
While our genetic study strongly supports POR’s participation in lipid peroxidation, the above model suggests there is an enzymatic partner/electron acceptor of POR in this reaction. Prior studies suggested that CYPs could accept electrons from POR, and catalyze the peroxidation of PUFA lipids39-42. However, potentially due to functional redundancy between various CYP members, our CRISPR/Cas9 loss-of-function screens were not able to nominate specific CYP isozymes as POR partners in promoting ferroptosis. On the other hand, our screening analysis did not rule out the involvement of non-CYP electron acceptors such as heme oxygenases and squalene monooxygenase28. Given the prevalence of POR’s contribution to ferroptosis, identifying the electron acceptor downstream of POR in lipid peroxidation merits further investigation.
In contrast to prior evidence suggesting a dominant role of lipoxygenases in mediating lipid peroxidation and ferroptosis, we found that many cancer cells capable of undergoing ferroptosis lack significant expression of ALOX enzymes, at least at the mRNA level. Recently, several general or isoform-specific ALOX inhibitors including zileuton, PD146176, and nordihydroguaiaretic acid (NDGA)22 were reported to exhibit intrinsic radical-trapping activities and were sufficient to block lipid peroxidation without ALOX inhibition. Genetic depletion of ALOXs also failed to alleviate ferroptotic cell death induced by GPX4-inhibition in several cell lines and mouse models12,20,48. This evidence argues against a prevalent contribution by lipoxygenases in promoting ferroptosis. Considering that ALOX overexpression potently sensitized 786-O and HEK-293 cells to ferroptosis induced by GPX4 inhibition14,22, lipoxygenases apparently can play a positive role in ferroptotic cell death, although this role may not be necessary for ferroptosis to occur in many contexts. Our study suggests that other mechanisms, exemplified by POR-dependent lipid peroxidation, could play a substantial role in ferroptosis. Further characterizations on the POR pathway, as well as other potential non-ALOX mediated ferroptosis mechanisms, warrant further investigation.
Ferroptosis was recently revealed to be involved in severe pathological conditions including ischemia/reperfusion-induced kidney injury, stroke, traumatic brain injury, cardiomyopathy and Alzheimer’s Disease4,10,49. In such contexts, ferroptosis prevention represents attractive therapeutic avenues. Considering that POR appears to be a non-essential gene in most cancer cell lines in the DepMap analyses, our findings on the direct connection between POR and lipid peroxidation nominate POR as a potential druggable target for the development of anti-ferroptosis therapeutics that may exhibit low toxicity. Additionally, the wide tissue spectrum for POR’s ferroptosis contribution suggests that POR-targeted therapies may be effective in diseases arising from many tissues. The development of such therapies is worth future investigation.
In addition to nucleated cell types, ferroptosis was recently observed in non-nucleated platelets as well, the death of which appeared to be triggered by lipid peroxidation induced by hemoglobin released heme during hemolysis50. While lipoxygenases promote ferroptosis largely using non-heme iron, the CYP/POR enzyme system contains high-levels of heme-iron that chemically and functionally resemble that in hemoglobin, further supporting a prominent role of heme-chelated iron in promoting lipid peroxidation and ferroptosis under physio-pathological conditions.
Overall, our study highlights an important role of POR in membrane phospholipid peroxidation and consequent ferroptotic cell death, and suggests that targeting POR could have therapeutic potential in protecting cells from ferroptosis in pathological conditions.
Online Methods
Cell lines and culture conditions.
UACC-257, 786-O, 769-P, OS-RC2, ES-2, OVISE, SW-1463, and SNU-182 cells were cultured in RPMI-1640 (Gibco) media; HuH-7, Hs-766T and HEK-293T cells were cultured in DMEM (Gibco) media. All media were supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin (Corning). All cells were cultured in a humidified incubator at 37 °C with 5% CO2. All cancer cell lines were obtained from the Cancer Cell Line Encyclopedia (CCLE) distributed by the Broad Institute Biological Samples Platform, except HEK-293T cells that were obtained from American Type Culture Collection (ATCC) and HuH-7 cells were a gift from Michael Deran (Broad Institute), who originally obtained the cells from the Broad Institute Biological Samples Platform. All cells were regularly tested for mycoplasma contamination and cells used in experiments were negative for mycoplasma.
Chemicals.
The following compounds were obtained from Sigma-Aldrich: ML210 (SML0521, purity ≥ 98%, used at wide concentration range), 1S, 3R-RSL3 (RSL3, SML2234, purity ≥ 98%, used at wide concentration range), ferrostatin-1 (Fer-1, SML0583, purity ≥ 95%, used at 5 μM), erastin (E7781, purity ≥ 98%, used at wide concentration range) and L-buthionine-[S,R]-sulfoximine (BSO, B2515, purity ≥ 97%, used at wide concentration range). Free fatty acids, including oleic acid (OA, C18:1), linoleic acid (LA, C18:2), a-linolenic acid (ALA, C18:3), arachidonic acid (AA, C20:4), eicosapentaenoic acid (EPA, C20:5), docosapentaenoic acid (DPA, C22:5), and docosahexaenoic acid (DHA, C22:6) were purchased from Cayman Chemicals and conjugated with fatty-acid free BSA (Sigma-Aldrich) using previously described protocols51, and treated cells at 20 μM for 3 days in the viability assays. The following compounds were prepared according to known literature procedures: FINO236 and FIN5634. Alkyne analogs of ML210 and RSL3 (ML210-yne and RSL3-yne, respectively) were synthesized as previously described15.
Viability assays.
For cellular viability assays, cells were seeded in 384-well opaque white tissue culture and assay plates (Corning, 8867C) at 1000 cells/well. 18-24 hours after seeding, cells were treated with compounds at the indicated concentrations for 48-72h. Cellular ATP levels were quantified using CellTiter-Glo (Promega) on an Envision multi-plate reader (Perkin Elmer). Relative viability was normalized to the respective DMSO-treated condition using RStudio unless otherwise indicated. For data presentation, the mean and standard deviation for the four biological replicates of each data point in a representative experiment is plotted in Prism 8 (Graphpad) and presented. Sigmoidal non-linear regression models were used to compute the regression fit curves.
Crystal violet staining.
200,000 UACC-257 cells were seeded per well in 6 well plates. Fatty acids were added 3 hours after seeding. After 3 days of incubation, cells were then treated with ML210 or DMSO for 2 days. After media removal, cells were stained with crystal violet staining solution (0.05% w/v crystal violet, 1% formaldehyde, 1% methanol in 1x PBS) for 1 hour before washing and air-drying.
Genome-wide CRISPR screen.
For sgRNA library infection, 360 ×10e6 UACC-257 cells were resuspended in 240 ml culture media, mixed with the Brunello sgRNA library52 in the presence of 8 μg/ml of polybrene, aliquoted to 12-well plates, centrifuged at 1000 g (RCF) for 2 hours at 30 °C and incubated overnight. Infected cells were selected by 2 μg/ml of puromycin for 2 days, with an estimated infection rate at 40 %. Positively infected cells were allowed to expand and undergo genome-editing for another 10 days, before being treated with BSA-containing vehicle or 20 μM of BSA-conjugated fatty acids for 3 days. 5 μM of ML210 was then added to the fatty acid treated cells for two days, before elevating the ML210 concentration to 10 μM for another 9 days of treatment. Genomic DNA was extracted from cell pellets in each condition using the QIAamp DNA Blood Maxi/Midi/Mini kits (Qiagen) according to the manufacturer’s protocols and quantified using a Nanodrop 2000 (Thermo Fisher Scientific).
The library was prepared, sequenced, and analyzed as previously described53-55. Briefly, sgRNA cassettes were PCR-amplified and barcoded with sequencing adaptors utilizing ExTaq DNA Polymerase (Clontech). PCR products were purified with Agencourt AMPure XP SPRI beads (Beckman Coulter A63880) according to the manufacturer’s instructions, quantified using a Nanodrop 2000, pooled into a master sequencing pool, and sequenced on a HiSeq 2500 sequencer (Illumina), loaded at 60% with a 5% spike-in of PhiX DNA.
For CRISPR screening data analysis, the sgRNA sequences were mapped to a reference file containing all SpCas9 sgRNAs in the Brunello library (available from [https://media.addgene.org/cms/filer_public/8b/4c/8b4c89d9-eac1-44b2-bb2f-8fea95672705/broadgpp-brunello-library-contents.txt]), and the sgRNA-associated barcodes were counted and mapped to the barcode reference file. After obtaining the read counts, log-normalized data were obtained by using log2(reads from an individual perturbation ÷ total reads in PCR well * 10e6 + 1) and gene level data were obtained by using a hypergeometric distribution model (https://github.com/mhegde/volcano_plots). Genes represented by 3-10 sgRNAs were included in the differential enrichment analysis. Genes with average log2 fold change (LFC) > 1.5 (indicating fold of enrichment >2.83) and average -log10 p-value > 2 (indicating p <0.01) are called as top hits and used for analyzing the overlapped genes with hits from the ccRCC CRISPR screen14.
Genome-editing and shRNA-mediated gene silencing.
For CRISPR/Cas9-mediated genome-editing, cells were engineered for Cas9 expression with the pLX-311-Cas9 vector (Addgene 96924), which contains the blasticidin S-resistance gene driven by the SV40 promoter and the SpCas9 gene driven by the EF1ɑ promoter. sgRNA sequences were cloned into the pXPR_BRD050 constitutive sgRNA expression vector. Alternatively, for inducible knockout experiments, sgRNA sequences were cloned into the pLV709 doxycycline-inducible sgRNA expression vector. Doxycycline (Sigma-Aldrich) was applied at 1 μg/ml for 7-14 days to induce sgRNA expression and genome-editing. For shRNA-mediated RNA interference, shRNAs targeting the genes of interest were pre-cloned into constitutive shRNA expression vectors pLKO.1 or pLKO-TRC005 by the Broad Institute Genetic Perturbation Platform.
Lentiviruses were generated from sgRNA/shRNA constructs in HEK-293T packaging cells in 96-well plate format using FUGENE6 (Promega) as the transfection reagent and infected the target cells in the presence of 4 μg/ml of polybrene (Sigma-Aldrich Millipore). Infected cells were selected with puromycin at 2 μg/mL starting 48 hours post-infection and propagated for further analysis. Cells transduced with inducible sgRNA constructs were treated with 1 μg/ml doxycycline (Sigma-Aldrich) for 7-14 days prior to gene-knockout validation by immunoblot analysis. Sequences for POR sgRNA #1: TCGTACAGCACGTTGGTACG, sgRNA #2: GTGGTCCCCAGATTCATACC, sgRNA #3: ACATGCCTCGCATCCCGTAG. Target sequences for POR shRNA #1: GTCCAACAAGAAGCACCCATT, shRNA #2: TGGCCGAAGAAGTATCTCTTT, shRNA #3: CCTAACCTACTGGTTCCTCTT, shRNA #4: CACCTGTGGAAGTTGATCGAA. Sequences for ALOX12 sgRNA #1: CCCCCATATCCGCTACACCA, sgRNA #2: TCCAAATATGAGATTCCATG, sgRNA #3: TCCATCTTCAGCATAACGAG. POR cDNA was obtained from commercial source (Sino Biological, HG10365-ACGLN, encoding NM_000941.2).
Generation of GPX4/POR double knockout clones.
Lentiviruses with control sgRNA (sgNC) or POR-sg2 expressing vectors were used to infect previously generated GPX4−/− 786-O-Cas9 single-cell clones14 in the presence of 4 μg/ml of polybrene (Sigma-Aldrich Millipore). Infected cells were selected with puromycin at 2 μg/ml, expanded and validated for POR and GPX4 protein depletion by immunoblotting.
Immunoblot analysis.
Adherent cells were briefly washed twice with PBS and lysed with 1% SDS lysis buffer containing 10 mM EDTA and 50 mM Tris-HCl, pH 8.0. Lysates were collected, briefly sonicated, then incubated at 95°C for 10 min and the protein concentrations were determined by BCA Protein Assay kit (Pierce). Calibrated samples were diluted with 4x LDS sample buffer (Novus), separated by SDS-PAGE using NuPAGE 4-12% Bis-Tris protein gels (Novus), and transferred to nitrocellulose or PVDF membranes by iBlot2 protein-transfer system (Thermo Fisher Scientific). Membranes were blocked with 50% Odyssey blocking buffer (LiCor) diluted with 0.1% Tween-20-containing TBS (TBST) and immunoblotted with antibodies against POR (Abcam, ab133303, rabbit monoclonal antibody, clone number UOTR1B493, Lot #GR290350-7, used at 1:1000 dilution), GPX4 (Abcam, ab41787, rabbit polyclonal antibody, Lot #GR56784-1, used at 1:1000 dilution), and β-Actin (8H10D10, #3700 and 13E5, #4970, Cell Signaling Technologies, used at 1:5000 dilution). Membranes were then washed with TBST and incubated with IRDye 800CW goat-anti-Rabbit or 680RD donkey-anti-Mouse secondary antibodies (LiCor). Immunoblotting images were acquired on an Odyssey equipment (LiCor) according to the manufacturer’s instructions, and analyzed in the ImageStudio software (LiCor). β-Actin was used as loading controls.
GPX4 pulldown assay.
GPX4 affinity enrichment experiments were performed as described previously15. Briefly, 786-O or 769-P cells were treated with the indicated alkyne probes (10 mM stocks in DMSO, final alkyne concentration = 10 μM) or DMSO for 1 h at 37 °C. After removal of the media, cells were washed once with ice-cold PBS (pH 7.4). Cells were lysed in PBS containing 1% Triton X-100 and protease inhibitors (Roche Complete) for 20 min on ice. Lysates were cleared by centrifugation (20 min, 20,000 g, 4 °C) and the total protein concentration was adjusted to 1.5 mg/mL. Copper-catalyzed azide-alkyne cycloaddition (CuAAC) reactions were then performed with azide-PEG3-biotin conjugate (Sigma Aldrich). Typical reactions were performed with a final volume of 120 μL consisting of 100 μL lysate (2 mg/mL; final concentration = 1.67 mg/mL), 2.4 μL SDS (10%; final concentration = 0.2%), 2.4 μL azide-PEG3-biotin conjugate (5 mM in DMSO; final concentration = 100 μM), and 15.2 μL of catalyst mix (final concentration = 1.3 mM Cu2SO4, 1.3 mM TCEP, and 75 μM TBTA). The catalyst mix stock was prepared by mixing 3 parts TBTA (1 mM in 1:4 DMSO/tBuOH), 1 part 50 mM Cu2SO4 in water, and 1 part TCEP (50 mM in water, pH 7.0). After addition of all components, CuAAC reactions were vortexed and allowed to react for 1 h at ambient temperature and then diluted with 120 μL 0.2% SDS in PBS. A 40 μL aliquot was removed and quenched with 6x SDS sample buffer (8 μL) as an input control. Pierce high-capacity streptavidin agarose beads (Thermo Fisher) were added to the remaining sample and rotated overnight at 4 °C. The beads were then separated by centrifugation and washed sequentially with 1% SDS (3 × 1 mL) and PBS (2 × 1 mL). Proteins were eluted by boiling the beads in 75 μL of 2x SDS sample buffer for 10 minutes. After centrifugation of the samples, the supernatant was removed and analyzed by SDS-PAGE and immunoblotting.
Flow cytometry analysis.
UACC-257, 786-O, 769-P and derivative cells were treated with DMSO or ML210 (5 μM) for 90 min, while for the last 45 min cells were also incubated with 5 μM of BODIPY-C11 dye or 2 μM of H2-DCFDA (Abcam). BODIPY-C11 was used to detect reactive oxygen species (ROS) in membrane lipids via a shift of fluorescence emission peak upon oxidation of its polyunsaturated butadienyl group56. For H2-DCFDA, fluorescent 2’,7’-dichlorofluorescein (DCF) was produced from oxidation of 2’,7’-dichlorodihydrofluorescein diacetate (H2-DCF-DA)57. Treatment with other reagents including BSA and polyunsaturated fatty acids were indicated in the figure legends of each experiment. Before flow cytometry, cells were washed with PBS twice, stained with Hoechst 33342 for 5 min, trypsinized and filtered into single cell suspensions. Flow cytometry analysis was performed on a SONY SH800 cell sorter with standard settings, using PE-Texas Red filter for reduced BODIPY-C11 and the FITC filter for oxidized BODIPY-C11, and the FITC filter for H2-DCFDA. A minimum of 10,000 cells were analyzed for each condition, and each experiment was independently performed at least twice. Data analysis was performed using the FlowJo 10 software. An example gating strategy is demonstrated in Supplementary Figure 6.
BODIPY-C11 imaging assay.
Wildtype or POR-depleted 786-O and 769-P cells were plated at 5,000 cells per well in 96-well plates with standard culture media. Before imaging, cells were treated with 10 μM ML210 for 3h. During the last 30 min of ML210 treatment, cells were co-treated with 5 μM of BODIPY-C11 dye, and Hoechst 33342 was added (1 μg/mL) 10 minutes before imaging. Imaging were acquired on an Operetta Imaging equipment (Perkin Elmer) at 563 nm for the reduced form BODIPY-C11, 488 nm for the oxidized form, and 405 nm for Hoechst.
Malondialdehyde (MDA) assay.
The measurement of MDA levels was performed using the lipid peroxidation (MDA) assay kit according to manufacturer’s instructions (Abcam, ab118970). Briefly, 2 million cells were seeded in a 10 cm culture dish one day prior to the experiment. The cells were treated with DMSO, ML210 or ML210+Lip-1 for 90 min as indicated, and collected with 600 μl MDA lysis buffer supplemented with BHT (butylated hydroxytoulene) using a cell scraper. Cell lysates were homogenized using a Dounce Homogenizer for 20 strokes, and subsequently centrifuged for 10 min at 13,000 g (RCF). The supernatants were collected and aliquoted into three 180 μl replicates, with the remaining 50 μl used for protein concentration measurement using Qubit (Life Technologies) . 600 μl of 10 mg/ml TBA solution was added to each replicate, and incubated at 95 ℃ for 60 min. Samples were immediately aliquoted to a microplate for reading at Ex/Em = 532/553 nm for fluorometric assay. MDA signals from each sample were normalized to the protein concentration.
Lipidomics profiling.
Lipidomics analysis is performed as previously described14. Briefly, analyses of polar and non-polar lipids were conducted using an LC-MS system comprising a Shimadzu Nexera X2 U-HPLC (Shimadzu Corp.) coupled to an Exactive Plus orbitrap mass spectrometer (Thermo Fisher Scientific). Lipids were extracted from cells with 0.8 mL isopropanol (HPLC Grade; Honeywell). Three replicates were analyzed for each cell line or condition. Cell extracts were centrifuged at 10,000 g (RCF) for 10 minutes to remove residual cellular debris prior to injecting 10 μL onto an ACQUITY BEH C8 column (100 × 2.1 mm, 1.7 μm; Waters). The column was eluted isocratically with 80% mobile phase A (95:5:0.1 vol/vol/vol 10 mM ammonium acetate/methanol/formic acid) for 1 minute followed by a linear gradient to 80% mobile-phase B (99.9:0.1 vol/vol methanol/formic acid) over 2 min, a linear gradient to 100% mobile phase B over 7 min, then 3 min at 100% mobile-phase B. MS data were acquired using electrospray ionization in the positive-ion mode over 200–1100 m/z and at 70,000 resolutions. Other MS settings were: sheath gas 50, in source CID 5 eV, sweep gas 5, spray voltage 3 kV, capillary temperature 300 °C, S-lens RF 60, heater temperature 300 °C, microscans 1, automatic gain control target 1e6, and maximum ion time 100 ms. Raw data were processed using TraceFinder 3.3 (Thermo Fisher Scientific) and Progenesis QI (Nonlinear Dynamics) software for detection and integration of LC-MS peaks. Lipid identities were determined based on comparison to reference standards and reference plasma extracts and were denoted by total number of carbons in the lipid acyl chain(s) and total number of double bonds in the lipid acyl chain(s).
For lipidomics data analysis, median normalization was performed between each sample in the same experiment. Median-normalized lipidomic dataset is presented in Supplementary Dataset 2. Differential-abundance analysis was performed between previously annotated lipid species (about 200 lipids were previously annotated) using two-tailed Student’s T-test. For fold-change analysis, each dataset was normalized to the mean of the wildtype cell condition for each lipid species, and the ratio between Test/WT was log2 transformed and presented as volcano plots. P values are adjusted for multiple-test correction using Benjamini-Hochberg correction method and presented as −log10 p adj.
Redox-lipidomics analysis.
Redox-lipidomic profiling was performed as previously described with minor modifications58. Briefly, on average 10 million cells per sample were treated with DMSO, ML210 or ML210+Lip-1 for 90 min in 15 cm culture plates. For sample collection, cells were washed with cold PBS once, collected in cold PBS using a cell scraper, and centrifuged at 200 g for 5min. After completely removing the PBS, cell pellets were immediately snap frozen in liquid nitrogen and then stored at minus 80 °C or on dry ice until further sample processing. For sample analysis, upon removal from the freezer, 300 μL water and 500 μL methanol were added to each sample, which was then transferred to glass a test tube (12 × 75 mm) containing 500 μL methanol and the internal standards: 100 ng PE (12:0/12:0 and 200 ng PC(18:0/20:4-d8). After adding 500 μL chloroform, samples were vigorously vortexed. After further addition of 500 μL chloroform and 500 μL water, samples were vortexed and centrifuged at 1,300 g for 10 min. The lower organic phase was transferred using a Pasteur pipette to a clean glass tube, and the solvent was evaporated under vacuum in a SpeedVac centrifuge. For LC-MS/MS, each lipid extract was resuspended in 100 μL of HPLC solvent mix, and an aliquot of 10 μL was injected into the LC-MS/MS system for analysis. The chromatographic profile of each analyte-specific m/z transition was monitored and the area under the peak (ion intensity vs. elution time) was integrated using commercial software (MultiQuant, Sciex). For data analysis, a ratio between the “area ratio analyte /internal standard” value of each oxidized phospholipid and the corresponding non-oxidized phospholipid species was calculated and plotted. The raw “area ratio analyte /internal standard” values for each lipid species are presented in Supplementary Dataset 3.
Public database queries.
For gene-expression analysis of The Cancer Genome Atlas (TCGA) tumor samples, normalized RNA-Seq results for each gene of interest was downloaded through cBioPortal (www.cbioportal.org) and replotted by ranking the mean of log2-transformed expression levels (RPKM) in each cancer type using RStudio.
For gene-expression analysis from Genotype-Tissue Expression (GTEx, https://gtexportal.org/home/) database, box-whisker plots were generated and downloaded from the GTEx online portal using the dataset GTEx Analysis Release V8 (dbGaP Accession phs000424.v8.p2), and annotated in Adobe Illustrator CC 2019. The number of samples for each tissue is listed as a table in the metadata associated with this public dataset (https://www.gtexportal.org/home/tissueSummaryPage).
For Cancer Dependency Map (DepMap) datasets, dependency scores for each gene of interest, including CERES scores from CRISPR/Cas9 screening experiments and DEMETER2 scores for RNA interference (RNAi) screening experiments, were downloaded from the DepMap web portal (https://depmap.org/portal), and replotted using GraphPad Prism 8.
Software and statistical analysis.
Data are generally expressed as mean±s.d. unless otherwise indicated. No statistical methods were used to predetermine sample sizes. Statistical significance was determined using a two-tailed student’s T-test using Prism 8 software (GraphPad Software) unless otherwise indicated. The Benjamini-Hochberg correction method was used to adjust the p values where multi-testing corrections were involved. Statistical significance was set at p <0.05 unless otherwise indicated.
Supplementary Material
Acknowledgements
The authors thank R. Aoyagi, M. Arita, P. Kennedy, M. Gijón, and T. Zhao for assisting the redox-lipidomics analyses. This work was supported in part by the NCI’s Cancer Target Discovery and Development (CTD2) Network (grant number U01CA217848, awarded to S.L. Schreiber), and in part by the National Institute of General Medical Sciences (grant number R35GM127045, awarded to S.L. Schreiber). G. Sandoval-Gomez is supported by a fellowship from the Howard Hughes Medical Institute Exceptional Research Opportunities Program.
Footnotes
Data Availability and Code Availability Statement
Raw sequencing data for CRISPR/Cas9 screens in UACC-257 melanoma cells are deposited in Gene Expression Omnibus (accession number GSE130982). Processed CRISPR screening data are supplied as Supplementary Dataset 1. The global lipidomics analysis results are presented in Supplementary Dataset 2, and the redox-lipidomics analysis results are presented in Supplementary Dataset 3. All original data and computational code that support the findings of this study are available upon request.
Competing Financial Interests Statement
S.L.S. serves on the Board of Directors of the Genomics Institute of the Novartis Research Foundation (“GNF”); is a shareholder and serves on the Board of Directors of Jnana Therapeutics; is a shareholder of Forma Therapeutics; is a shareholder and advises Decibel Therapeutics and Eikonizo Therapeutics; serves on the Scientific Advisory Boards of Eisai Co., Ltd., Ono Pharma Foundation, Exo Therapeutics, and F-Prime Capital Partners; and is a Novartis Faculty Scholar. The remaining authors declare no competing interests.
References
- 1.Viswanathan VS et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 547, 453–457 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wenzel SE et al. PEBP1 Wardens Ferroptosis by Enabling Lipoxygenase Generation of Lipid Death Signals. Cell 171, 628–641.e26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hangauer MJ et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 551, 247–250 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alim I et al. Selenium Drives a Transcriptional Adaptive Program to Block Ferroptosis and Treat Stroke. Cell 177, 1262–1279.e25 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Li Q et al. Inhibition of neuronal ferroptosis protects hemorrhagic brain. JCI Insight 2, e90777 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang WS et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogacz M & Krauth-Siegel RL Tryparedoxin peroxidase-deficiency commits trypanosomes to ferroptosis-type cell death. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Distéfano AM et al. Heat stress induces ferroptosis-like cell death in plants. J. Cell Biol 216, 463–476 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kagan VE et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol 13, 81–90 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stockwell BR et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 171, 273–285 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingold I et al. Selenium Utilization by GPX4 Is Required to Prevent Hydroperoxide-Induced Ferroptosis. Cell 172, 409–422.e21 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Friedmann Angeli JP et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol 16, 1180–1191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ursini F, Maiorino M, Valente M, Ferri L & Gregolin C Purification from pig liver of a protein which protects liposomes and biomembranes from peroxidative degradation and exhibits glutathione peroxidase activity on phosphatidylcholine hydroperoxides. Biochim. Biophys. Acta 710, 197–211 (1982). [DOI] [PubMed] [Google Scholar]
- 14.Zou Y et al. A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat. Commun 10, 1617 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eaton JK et al. Targeting a Therapy-Resistant Cancer Cell State Using Masked Electrophiles as GPX4 Inhibitors. bioRxiv. doi: 10.1101/376764. [DOI] [Google Scholar]
- 16.Conrad M & Pratt DA The chemical basis of ferroptosis. Nat. Chem. Biol 15, 1137–1147 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Yin H, Xu L & Porter NA Free Radical Lipid Peroxidation: Mechanisms and Analysis. Chemical Reviews vol. 111 5944–5972 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Doll S et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature 575, 693–698 (2019) [DOI] [PubMed] [Google Scholar]
- 19.Bersuker K et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 575, 688–692 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang WS et al. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proceedings of the National Academy of Sciences vol. 113 E4966–E4975 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn H, Banthiya S & van Leyen K Mammalian lipoxygenases and their biological relevance. Biochim. Biophys. Acta 1851, 308–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shah R, Shchepinov MS & Pratt DA Resolving the Role of Lipoxygenases in the Initiation and Execution of Ferroptosis. ACS Cent Sci 4, 387–396 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doll S et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol 13, 91–98 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dixon SJ et al. Human Haploid Cell Genetics Reveals Roles for Lipid Metabolism Genes in Nonapoptotic Cell Death. ACS Chem. Biol 10, 1604–1609 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandey AV & Flück CE NADPH P450 oxidoreductase: structure, function, and pathology of diseases. Pharmacol. Ther 138, 229–254 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Foti RS & Dalvie DK Cytochrome P450 and Non-Cytochrome P450 Oxidative Metabolism: Contributions to the Pharmacokinetics, Safety, and Efficacy of Xenobiotics. Drug Metab. Dispos 44, 1229–1245 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Veith A & Moorthy B ROLE OF CYTOCHROME P450S IN THE GENERATION AND METABOLISM OF REACTIVE OXYGEN SPECIES. Curr Opin Toxicol 7, 44–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riddick DS et al. NADPH–Cytochrome P450 Oxidoreductase: Roles in Physiology, Pharmacology, and Toxicology. Drug Metabolism and Disposition vol. 41 12–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zilka O et al. On the Mechanism of Cytoprotection by Ferrostatin-1 and Liproxstatin-1 and the Role of Lipid Peroxidation in Ferroptotic Cell Death. ACS Cent Sci 3, 232–243 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carithers LJ & Moore HM The Genotype-Tissue Expression (GTEx) Project. Biopreservation and Biobanking vol. 13 307–308 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao J et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 6, l1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsherniak A et al. Defining a Cancer Dependency Map. Cell 170, 564–576.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dixon SJ et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimada K et al. Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nat. Chem. Biol 12, 497–503 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffith OW & Meister A Potent and specific inhibition of glutathione synthesis by buthionine sulfoximine (S-n-butyl homocysteine sulfoximine). J. Biol. Chem 254, 7558–7560 (1979). [PubMed] [Google Scholar]
- 36.Gaschler MM et al. FINO initiates ferroptosis through GPX4 inactivation and iron oxidation. Nat. Chem. Biol 14, 507–515 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ilan Z, Ilan R & Cinti DL Evidence for a new physiological role of hepatic NADPH:ferricytochrome (P-450) oxidoreductase. Direct electron input to the microsomal fatty acid chain elongation system. J. Biol. Chem 256, 10066–10072 (1981). [PubMed] [Google Scholar]
- 38.Lamb DC, Warrilow AG, Venkateswarlu K, Kelly DE & Kelly SL Activities and kinetic mechanisms of native and soluble NADPH-cytochrome P450 reductase. Biochem. Biophys. Res. Commun 286, 48–54 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Huang B, Bao J, Cao Y-R, Gao H-F & Jin Y Cytochrome P450 1A1 (CYP1A1) Catalyzes Lipid Peroxidation of Oleic Acid-Induced HepG2 Cells. Biochemistry 83, 595–602 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Minoda Y & Kharasch ED Halothane-dependent lipid peroxidation in human liver microsomes is catalyzed by cytochrome P4502A6 (CYP2A6). Anesthesiology 95, 509–514 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Ghosh MK, Mukhopadhyay M & Chatterjee IB NADPH-initiated cytochrome P450-dependent free iron-independent microsomal lipid peroxidation: specific prevention by ascorbic acid. Mol. Cell. Biochem 166, 35–44 (1997). [DOI] [PubMed] [Google Scholar]
- 42.Dey A, Parmar D, Dhawan A, Dash D & Seth PK Cytochrome P450 2E1 dependent catalytic activity and lipid peroxidation in rat blood lymphocytes. Life Sci. 71, 2509–2519 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Ghandi M et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bast A, Brenninkmeijer JW, Maria Savenije-Chapel E & Noordhoek J Cytochrome P450 oxidase activity and its role in NADPH dependent lipid peroxidation. FEBS Letters vol. 151 185–188 (1983). [DOI] [PubMed] [Google Scholar]
- 45.Sevanian A, Nordenbrand K, Kim E, Ernster L & Hochstein P Microsomal lipid peroxidation: the role of NADPH--cytochrome P450 reductase and cytochrome P450. Free Radic. Biol. Med 8, 145–152 (1990). [DOI] [PubMed] [Google Scholar]
- 46.Ursini F, Maiorino M, Hochstein P & Ernster L Microsomal lipid peroxidation: mechanisms of initiation. The role of iron and iron chelators. Free Radic. Biol. Med 6, 31–36 (1989). [DOI] [PubMed] [Google Scholar]
- 47.Girotti AW Lipid hydroperoxide generation, turnover, and effector action in biological systems. J. Lipid Res 39, 1529–1542 (1998). [PubMed] [Google Scholar]
- 48.Brütsch SH et al. Expression of Inactive Glutathione Peroxidase 4 Leads to Embryonic Lethality, and Inactivation of the Alox15 Gene Does Not Rescue Such Knock-In Mice. Antioxidants & Redox Signaling vol. 22 281–293 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Bai Y-T et al. ENPP2 protects cardiomyocytes from erastin-induced ferroptosis. Biochem. Biophys. Res. Commun 499, 44–51 (2018). [DOI] [PubMed] [Google Scholar]
- 50.NaveenKumar SK et al. The Role of Reactive Oxygen Species and Ferroptosis in Heme-Mediated Activation of Human Platelets. ACS Chem. Biol 13, 1996–2002 (2018). [DOI] [PubMed] [Google Scholar]
Methods-only References
- 51.Hannah VC, Ou J, Luong A, Goldstein JL & Brown MS Unsaturated fatty acids down-regulate srebp isoforms 1a and 1c by two mechanisms in HEK-293 cells. J. Biol. Chem 276, 4365–4372 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Doench JG et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hegde M, Strand C, Hanna RE & Doench JG Uncoupling of sgRNAs from their associated barcodes during PCR amplification of combinatorial CRISPR screens. PLoS One 13, e0197547 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Najm FJ et al. Orthologous CRISPR–Cas9 enzymes for combinatorial genetic screens. Nat. Biotechnol 36, 179–189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanson KR et al. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat. Commun 9, 5416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naguib YM A fluorometric method for measurement of peroxyl radical scavenging activities of lipophilic antioxidants. Anal. Biochem 265, 290–298 (1998). [DOI] [PubMed] [Google Scholar]
- 57.Cathcart R, Schwiers E & Ames BN Detection of picomole levels of hydroperoxides using a fluorescent dichlorofluorescein assay. Anal. Biochem 134, 111–116 (1983). [DOI] [PubMed] [Google Scholar]
- 58.Wang W et al. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 569, 270–274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.