Abstract
Cell-biomaterial interactions are primarily governed by cell adhesion, which arises from the binding of cellular integrins to the extracellular matrix (ECM). Integrins drive the assembly of focal contacts that serve as mechanotransducers and signaling nexuses for stem cells, for example integrin α4β1 plays pivotal roles in regulating mesenchymal stem cell (MSC) homing, adhesion, migration and differentiation. The strategy to control the integrin-mediated cell adhesion to bioinspired, ECM-mimicking materials is essential to regulate cell functions and tissue regeneration. Previously, using one-bead one-compound (OBOC) combinatorial technology, we discovered LLP2A was a high-affinity peptidomimetic ligand (IC50 = 2 pM) against integrin α4β1. In this study, we identified LLP2A had a strong binding to human early gestation chorionic villi-derived MSCs (CV-MSCs) via integrin α4β1. To improve CV-MSC seeding, expansion and delivery for regenerative applications, we constructed artificial scaffolds simulating the structure of the native ECM by immobilizing LLP2A onto the scaffold surface as cell adhesion sites. LLP2A modification significantly enhanced CV-MSC adhesion, spreading and viability on the polymeric scaffolds via regulating outside-in signaling pathways including phosphorylation of focal adhesion kinase (FAK) and AKT, NF-kB and Caspase 9. In addition, we also demonstrated LLP2A had strong binding to MSCs of other sources, such as bone marrow-derived mesenchymal stem cells (BM-MSCs) and adipose tissue-derived mesenchymal stem cells (AT-MSCs). Therefore, LLP2A and its derivatives not only hold great promise for improving CV-MSC-mediated treatment of fetal diseases, but can also be widely applied to functionalize various biological and medical materials, which are in need of MSC recruitment, enrichment and survival, for regenerative medicine applications.
Graphical Abstract

Introduction
A number of fetal diseases may result from pathogen exposure, at any time during pregnancy, which can be devastating for afflicted children and their families1. Over the past three decades, fetal surgery has progressed from an investigational therapy to an accepted mode of therapy for selected fetal diseases including congenital diaphragmatic hernia, spina bifida and urinary tract abnormalities2–4. Fetal tissue engineering can augment existing in utero surgical techniques and has been shown to be an effective treatment option for fetal diseases5–7.
Tissue engineering is the combination of suitable stem cells, biological motifs and biomaterials to improve biological functions of tissues8. Biomaterial scaffolds provide structural support to guide cell growth and tissue regeneration9. Polyester scaffolds have received considerable attention for tissue engineering because of their appropriate mechanical properties, lower cost, and easy fabrication into complex shapes10. The electrospinning technique is a powerful tool used to make nano/microfibrous scaffolds that imitate the native tissue architecture, allow for the integration of the grafts/scaffolds with the surrounding cells, and promote tissue regeneration11. We have successfully used electrospinning techniques to fabricate microfibrous scaffolds for various tissue regeneration applications such as peripheral nerve12, 13, spinal cord14–16, and vascular tissue regeneration17, drug delivery18, and wound healing19. Though electrospun microfibrous scaffolds have a three-dimensional structure that mimics the native extracellular matrix (ECM) architecture, and they lack biological motifs and surface cytocompatibility. Furthermore, cell adhesion is an important precondition for promoting cell behavior and tissue construction, and surface modification can endow the biomaterials with special biological functions for targeting special types of cells/tissues to improve their cyto/histocompatibility20. Therefore, it is particularly pivotal to construct approaches to modify biomaterials so as to improve their biological functions. Many methods of introducing hydrophilic compounds onto the polymeric scaffold surfaces have been explored, such as plasma treatment21, surface oxidation22 and photo-induced grafting23. However, these physiochemical approaches to enhance biological interactions between cells and scaffolds are not biologically specific. Many different types of cell adhesive proteins and biofunctional motifs, such as RGD peptide24, 25, ECM-derived proteins26–28, and other bioactive factors29, have also been used to enhance cell adhesion and tissue regeneration. However, general difficulties of these include lack of high binding affinity and specificity to targeted cells, and high stability in vivo. The native ECM includes numerous ligands targeting different types of integrins that are important for cell function and cell-ECM interactions30–32. Integrin binding specificity regulates the effects of the biomaterial surface on the cell focal adhesion, signaling pathway, and functions33, 34. Therefore, integrins play critical roles in pancreatic35, cartilage36, bone37, neuronal26, and vascular11 tissue engineering. In addition, different integrin adhesion receptors on the ECM support the actin cytoskeleton and transmit biochemical signals and mechanical force to the cells38–40. Therefore, a stable and specific integrin-binding ligand is essential for biological functionalization of materials to improve cell and tissue functions in fetal tissue engineering.
In utero surgical procedures, supplemented by autologous stem cell therapy, are an ideal option for the prenatal treatment of a variety of birth defects. The placenta is a unique autologous stem cell source41, 42, and first trimester fetal stem cells possess several advantages for regenerative medicine over adult and perinatal stem cells43–47. Our previous study has shown that human early gestation chorionic villi-derived mesenchymal stem cells (CV-MSCs) possessed excellent functions, such as expansion, paracrine secretion, and neuroprotection, for fetal tissue engineering6, 7, 15, 16, 48. Therefore, an integrin-based ligand with a high binding affinity to CV-MSCs and high stability in vivo will be crucial to improve the interaction between biomaterials and fetal stem cells, and serve as a promising application for fetal tissue engineering.
One-bead one-compound (OBOC) combinatorial technology is an ultra-high throughput chemical library synthesis and screening method suitable for ligand discovery against integrins49. Previously, we employed the OBOC combinatorial technology to screen cell adhesion molecules and identified various potent ligands, such as LXY30, LXW7, and LLP2A targeting integrins α3β1, αvβ3, and α4β1, respectively50–52. In particular, LLP2A, a very high-affinity peptidomimetic ligand (IC50 = 2 pM) has a high binding affinity to integrin α4β1, which is highly expressed on mesenchymal stem cells (MSCs) and regulates MSC homing, adhesion, migration and differentiation. In addition, LLP2A contains unnatural amino acids, therefore it would be resistant to proteolysis and be more stable than other conventional peptides in vivo. LLP2A has been successfully applied in the drug field to direct MSCs for bone disease treatment53, 54. In this study, we first introduced LLP2A for tissue engineering applications. We designed a ‘Click chemistry’ strategy to functionalize the electrospun scaffold using LLP2A and evaluated the CV-MSC behaviors, including adhesion, spreading, survival and signal transduction, on the LLP2A modified scaffold to provide a new sight and evidence for fetal tissue engineering and also a promising strategy to construct MSC-dependent, integrin-based, ECM-mimicking materials for regenerative medicine.
Results and discussion
High binding affinity of LLP2A to CV-MSCs
Most of the biomolecules and strategies used to modify biomaterial-based scaffolds are limited in their translational application due to their low cell binding affinity, unstable structure, and lack of the ability to functionally interact with cells/tissue11, 55. LLP2A was discovered by OBOC technology and synthesized with D-amino acids, unnatural amino acids, and small molecule moieties so that it possessed high affinity and proteolytic stability50. Ligands with high binding affinity to cells not only support rapid and stable cell adhesion on biomaterial surfaces, but also potentially enhance cell engraftment and survival after transplantation. To confirm the high binding affinity of LLP2A to CV-MSCs, resin beads displaying LLP2A or blank resin beads (as control) were incubated with CV-MSCs. At different time points after incubation (1 min, 5 min, 10 min, or 30 min), phase contrast images were taken to determine the cell binding affinity on beads (Fig. 1A). We found that LLP2A efficiently supported CV-MSC binding, and the number of CV-MSCs attached to LLP2A beads increased over time. But very few CV-MSCs bound to the blank beads (Fig. 1A). Remarkably, A significant number of CV-MSCs already bound to the beads after only 1 min incubation (Fig. 1A), indicating that the CV-MSCs could bind to the LLP2A-modified beads rapidly. Quantification of the number of cells bound to each bead showed that the beads were almost fully covered by CV-MSCs at 10 min, and there was no significant increase in cell binding at 30 min, compared to 10 min (Fig. 1B). These results demonstrated that LLP2A possessed rapid and strong binding affinity to CV-MSCs.
Fig. 1.
Binding affinity of LLP2A to CV-MSCs. (A) Blank beads (a-d) and beads displaying with LLP2A (e-h) were incubated with CV-MSCs at 1 min, 5 min, 10 min, and 30 min. (B) The numbers of CV-MSCs bound to each bead were quantified, and statistical analyses were performed. Data were expressed as mean ± standard deviation: *p < 0.05, **p < 0.01 (n = 4).
LLP2A culture surface enhances CV-MSC attachment
The addition of tool molecules, such as biotin, to ligands will be advantageous when used in combination with other components and when used to expand the bioengineering applications of the ligands. Our previous work has shown that biotinylation of the ligand did not decrease its binding affinity and showed nearly identical binding strength to the targeted integrin11. Therefore, we conjugated LLP2A to biotin (LLP2A-biotin, Fig. 4B), as described in our previous study50. We used LLP2A-bio or D-biotin (as the negative control) to treat the culture surfaces and investigated the attachment of CV-MSCs on culture surfaces. There was more CV-MSC attachment on the LLP2A-treated surface (Fig. 2A, b), compared to the control surface (Fig. 2A, a). The number of attached CV-MSCs was quantified and showed that the LLP2A-treated surface significantly improved CV-MSC adhesion compared to the untreated surface (Fig. 2B). This finding further confirmed our previous bead-cell binding results that the LLP2A surface supported high-affinity CV-MSC binding. In order to characterize the binding affinity of LLP2A to other types of MSCs, bone marrow-derived mesenchymal stem cells (BM-MSCs) and adipose tissue-derived mesenchymal stem cells (AT-MSCs) were evaluated for cell binding. The results showed LLP2A surface also significantly elevated both BM-MSC and AT-MSC attachment (SFig. 2), which demonstrated LLP2A also supported strong binding to other sources of MSCs, indicating that LLP2A has a wide range of applications in tissue engineering.
Fig. 4.
LLP2A was modified onto electrospun scaffold via Click chemistry. Chemical structures of (A) LLP2A, (B) LLP2A-Bio, and (C) LLP2A-DBCO; (D) Schematic diagram of the chemical process involved in the LLP2A immobilization on the electrospun scaffold.
Fig. 2.
Attachment of CV-MSCs on LLP2A-treated surface. (A) Images of attached CV-MSCs on (a) untreated surface and (b) LLP2A-treated surface; (B) The number of CV-MSCs attached to different surfaces was quantified, and statistical analyses were performed. Data were expressed as mean ± standard deviation: **p < 0.01 (n = 4).
LLP2A bound to CV-MSCs via α4β1 integrin
Integrins are transmembrane receptors that facilitate cell-ECM adhesion56. Upon ligand binding, integrins activate signal transduction pathways that mediate cellular signals such as regulation of the cell cycle, organization of the intracellular cytoskeleton, and movement of new receptors to the cell membrane57–59. The presence of integrins such as integrin α4β1, allows rapid and flexible responses to events at the cell surface59, 60. Integrin α4β1 has been shown to interact with fibronectin, a major component of the ECM61, 62. Also, integrin α4β1 has been suggested to play important roles in embryogenesis, involving fetal cell adhesion and migration63. Our previous work showed that LLP2A has a high binding affinity and specificity to integrin α4β150. Integrin α4β1 is highly expressed on different types of MSCs, including placenta-derived MSCs64. To further confirm that LLP2A binds to CV-MSCs via integrin α4β1, we performed a binding/blocking test using anti-integrin α4 and anti-integrin β1 antibodies and flow cytometry. The results showed that both integrin subunits α4 and β1 were highly expressed on CV-MSCs (Fig. 3A). For the binding/blocking experiment, the results showed that LLP2A has a high binding efficiency to CV-MSCs, and the binding efficiency of LLP2A to CV-MSCs was significantly and gradually decreased after the integrin α4β1 expressed on CV-MSCs was blocked by anti-integrin α4 antibody only, anti-integrin β1 antibody only, or anti-integrin α4 and anti-integrin β1 antibodies together (Fig. 3B). These findings demonstrated that the binding of LLP2A to CV-MSCs was mediated by both integrin α4 and integrin β1, which was consistent with previous studies60, 65, 66. Integrins couple the ECM molecules outside a cell to the cytoskeleton inside the cell. The engagement of ECM and the corresponding integrins is primarily mediated by the binding of the ligands to the specific combination of the integrin α and β subunits. To further investigate the binding specificity of LLP2A to integrin α4β1, we chose human coronary artery endothelial cells (HCAECs) that do not express the integrin α4β1 combination as an additional control. Flow cytometry was performed to evaluate integrin α4 and β1 expression and the binding affinity of LLP2A to HCAECs (SFig. 3). The results showed HCAECs only expressed integrin β1 but did not express integrin α4, and LLP2A had no binding to HCAECs. The results demonstrated that LLP2A specifically bound to integrin α4β1.
Fig. 3.
Binding mechanism of LLP2A to CV-MSCs. (A) Integrin α4 and β1 were highly expressed on CV-MSCs. (B) LLP2A effectively bound to CV-MSCs (blue curve), and the binding efficiency was markedly blocked by anti-integrin α4 and anti-integrin β1 antibodies together (orange curve), anti-integrin β1 antibody only (green curve), or anti-integrin α4 antibody only (black curve).
Surface characterization after LLP2A modification
ECM provides a three-dimensional structure and native ligands for cell attachment and tissue growth and function67. To mimic the ECM structure, we employed electrospinning technology to produce microfibrous scaffolds using PLLA and PCL polymer blends. SEM images showed that the electrospun scaffolds had a porous network structure of microfibers (Fig. 6A), similar in morphology to the native ECM. To improve the biological functions of the electrospun microfibrous scaffolds, we developed a protocol to functionalize the polymer surface by the addition of LLP2A, via Click chemistry (Fig. 4D). LLP2A-DBCO, a functional derivate of LLP2A, was synthesized by adding a ring structure with an alkynyl group to LLP2A (Fig. 4C). Microfibrous scaffolds were functionalized by covalently attaching NH2-PEG-N3 to the carboxylic groups on the microfibers using EDC and Sulfo-NHS (Fig. 4D). LLP2A-DBCO was then conjugated to the NH2-PEG-N3 functionalized microfibers by Click chemistry (Fig. 4D). X-ray photoelectron spectroscopy (XPS), also known as Electron Spectroscopy for Chemical Analysis (ESCA), is generally regarded as a key technique for the surface characterization and elemental composition analysis of biomedical polymers68. The XPS results are shown in Fig. 5. There was no nitrogen peak in the untreated group, because the PLLA/PCL scaffold did not include any nitrogen. In contrast, a small nitrogen peak appeared in the linker only group that demonstrated the NH2-PEG-N3 linker had been immobilized on the PLLA/PCL scaffold successfully, due to the nitrogen included in the NH2-PEG-N3 linker. Compared to the linker only group, the nitrogen peak was significantly higher in the LLP2A with the linker group predicated LLP2A-DBCO containing additional nitrogen had been immobilized on to the scaffold successfully by adhering to the NH2-PEG-N3 linker. All these findings confirmed that LLP2A had been successfully immobilized on the electrospun microfibrous PLLA/PCL scaffold surface. The surface morphology of the scaffold after modification plays an important role for the cell attachment, growth and penetration. The scanning electron microscope (SEM, Hitachi TM-1000) was used to characterize the structure of the membrane after LLP2A modification. The results showed the significant decrement of fiber diameters after LLP2A modification compared to before modification (Fig. 6A and B), which may be because hydrolysis was performed during the process of modification to increase the density of carboxyl group on the scaffold.
Fig. 6.
Effects of LLP2A modification on surface morphology of PLLA/PCL scaffold. (A) SEM analysis was employed to evaluate the surface morphology of (a) untreated PLLA/PCL scaffold and (b) LLP2A-modified PLLA/PCL scaffold. (B) Quantification and correlative statistical analysis of the fiber diameters; Data were expressed as mean ± standard deviation: **p < 0.01 (n = 50).
Fig. 5.
XPS analysis of the untreated scaffold, the scaffold modified with linker only, and the scaffold modified with the linker and LLP2A.
LLP2A modification enhanced hydrophilic properties of the PLLA/PCL scaffold
Scaffold surface wettability in part reflects its physical and chemical properties, which play an important role in the interactions between cells and biomaterials. Hydrophilic enhancement of polymers leads to increased cell adhesion and spreading69, 70. Contact angles were employed to detect changes in the wettability of the scaffold surface. The results showed it was easier for a water droplet to come into contact with the LLP2A-modified PLLA/PCL scaffold surface (Fig. 7A, b) than the untreated PLLA/PCL scaffold surface (Fig. 7A, a). The quantification results indicated that the contact angles decreased significantly after incorporating the LLP2A onto the surface of PLLA/PCL scaffolds (Fig. 7B). This decrease in contact angle indicated that the LLP2A modification increases the hydrophilicity of the surface of the PLLA/PCL scaffold, which would further enhance its cell cytocompatibility.
Fig. 7.
Effects of LLP2A modification on hydrophilic properties of PLLA/PCL scaffold. (A) Contact angle assay was employed to evaluate the hydrophilic property of (a) untreated PLLA/PCL scaffold and (b) LLP2A-modified PLLA/PCL scaffold. (B) Quantification and correlative statistical analysis; Data were expressed as mean ± standard deviation: **p < 0.01 (n = 5).
LLP2A modification improved CV-MSC adhesion and viability on scaffold via activated related biological signals
Cell adhesion is essential in cell communication and regulation, and is of fundamental importance to the development and maintenance of tissues71. Cell viability is the most important challenge in the success of cell-based functional biomaterial implants for tissue regeneration72. To determine the effect of LLP2A modification on CV-MSC adhesion and viability on electrospun PLLA/PCL scaffolds, CV-MSCs were seeded and cultured on untreated scaffolds and LLP2A-modified scaffolds for 2 h and 7 d. CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS) results showed LLP2A modification significantly improves CV-MSC adhesion after 2 h seeding (Fig. 8A) and CV-MSC viability after 7 d culture (Fig. 8B). These results demonstrated that the function of LLP2A was well maintained after chemical modification, and LLP2A-modified biomaterials support excellent attachment and viability of CV-MSCs.
Fig. 8.
Effects of LLP2A modification on CV-MSC attachment and viability on PLLA/PCL scaffold. MTS assay was employed to determine (A) CV-MSC attachment at 2 h after cell seeding, and (B) CV-MSC viability at 7 day after cell seeding on untreated and LLP2A-modified scaffolds; Data were expressed as mean ± standard deviation: **p < 0.01 (n = 4).
Cell adhesion is involved in stimulating signals that regulate cell differentiation, cell cycle cell migration, and cell survival73. Once the integrin receptors located on the cell membrane recognize and bind to the ligand on the substrate, focal adhesions will be formed and subsequently activate focal adhesion kinase (FAK) by auto-phosphorylation. The activation of FAK is considered a marker of integrin-mediated signaling. Our results demonstrate that the activated form of FAK, phosphorylated-FAK (p-FAK), of CV-MSCs was significantly improved on the LLP2A-modified PLLA/PCL scaffolds compared with that on the untreated scaffold after 10 min cell seeding (Fig. 9A and C) or 6 h cell seeding (SFig. 4), indicating that LLP2A modification can initiate a specific integrin-mediated signal transduction between CV-MSCs and the LLP2A-modified PLLA/PCL scaffolds. The auto-phosphorylation of FAK can create a binding site that binds with phosphoinositide 3-kinase (PI-3K) and activates AKT by phosphorylation. The activated AKT plays a central role in regulating the downstream effects of the PI3K pathway. Our subsequent data showed that the activated form of AKT, phosphorylated-AKT (p-AKT), of CV-MSCs was also significantly up-regulated on the PLLA/PCL scaffold with LLP2A modification compared with that on the untreated scaffold after 10 min cell seeding (Fig. 9A and B) or 6 h cell seeding (SFig. 4). Our results also showed the p-FAK and p-AKT signals at the shorter time point 10 min were stronger than at 6 h, because it is known that phosphorylation is a very quick process. The activation of AKT does not only inhibit the pro-apoptotic factors such as caspase-9, but also activates the transcription of anti-apoptotic genes through the activation of transcription factor nuclear factor kappa B (NF-kB) related to the regulation of cell viability. Our results showed that NF-kB expression was significantly improved (Fig. 9A and D) and activation of Caspase 9 was significantly decreased (Fig. 9E) respectively of CV-MSCs on the LLP2A-modified PLLA/PCL scaffolds compared with that on the untreated scaffold after 10 min cell seeding. These findings demonstrate that LLP2A-modified PLLA/PCL scaffolds could activate FAK signaling, therefore up-regulate PI-3K/AKT pathway of CV-MSCs and regulate the downstream biological pathways related to cell viability, which are consistent with the previous studies25, 74.
Fig. 9.
Effects of LLP2A modification on CV-MSC biological functions on the PLLA/PCL scaffold. (A) Western-blot analysis of FAK, phosphorylated-FAK (pFAK), AKT, phosphorylated-AKT (pAKT), NF-kB expression in the CV-MSCs cultured on the untreated scaffold or LLP2A-modified scaffold. Quantification and the correlative statistical analysis of (B) p-AKT/AKT, (C) p-FAK/FAK and (D) NF-kB/GAPDH. Quantification and the correlative statistical analysis of Caspase 9 activity assay of CV-MSCs culture on the untreated scaffold or LLP2A-modified scaffold. Data were expressed as mean ± standard deviation: *p < 0.05 (n = 4).
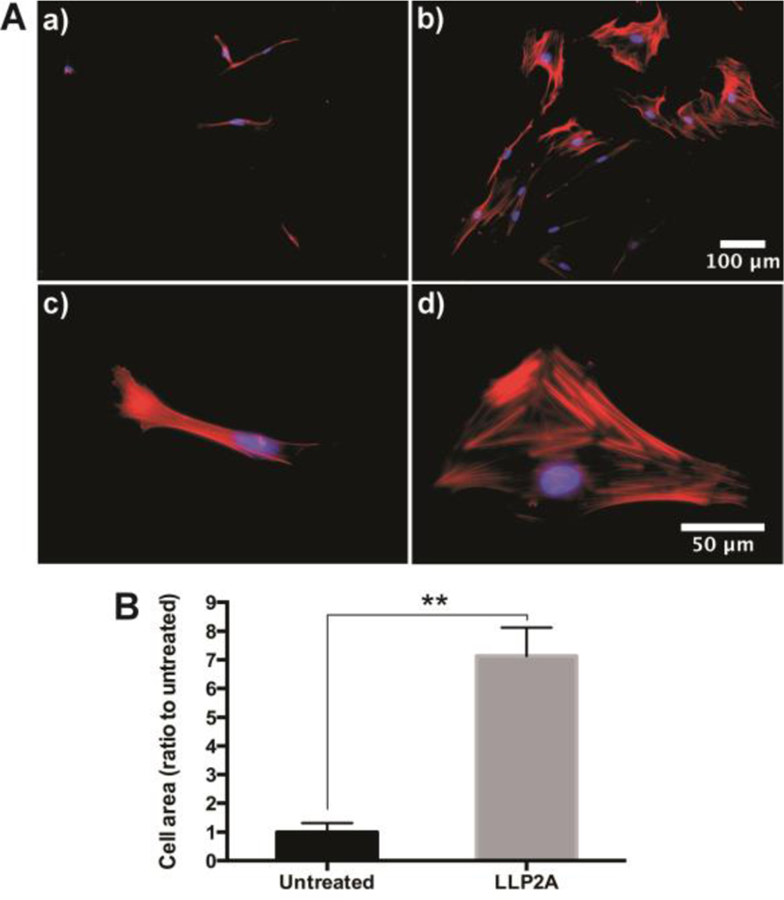
LLP2A modification enhances CV-MSC spreading and behavior on the scaffold
The degree of cell shape and spreading has important consequences for a variety of cellular behaviors, including migration, proliferation, and differentiation75–78. The cellular responses to cell spreading or shape appear to be regulated by forces generated through the cytoskeleton and cell-ECM adhesions79–81. The SEM analysis showed that CV-MSCs grow and spread much better on the LLP2A-modified scaffold (Fig. 10A, b) compared to the untreated scaffold (Fig. 10A, a). Quantification of the cell-covered area showed that there was significantly more cell-covered area on LLP2A-modified scaffolds compared to untreated scaffolds (Fig. 10B). The cytoskeleton staining showed narrow and spindly CV-MSCs on untreated scaffolds (Fig. 11A, a, c), whereas CV-MSCs on LLP2A-modified scaffolds were wide and flared (Fig. 11A, b, d). Quantification of the single cell-area showed that there was significantly more cell area on the LLP2A-modified scaffolds compared to the untreated scaffolds (Fig. 11B). These results demonstrate that LLP2A modification provides a better platform for CV-MSCs in fetal tissue regeneration.
Fig. 10.
(A) SEM images of CV-MSC coverage on an untreated scaffold surface (a) and LLP2A-modified scaffold surface (b) for 2 days; (B) Quantification and the correlative statistical analysis of cell-covered area from the SEM images. Data were expressed as mean ± standard deviation: **p < 0.01 (n = 4).
Fig. 11.
(A) Cytoskeleton of CV-MSCs cultured on (a, c) untreated scaffold and (b, d) LLP2A-modified scaffold; (B) Quantification and the correlative statistical analysis of cell area. Data were expressed as mean ± standard deviation: **p < 0.01 (n = 4).
Experimental
Cell isolation and culture
Human placental tissues (n = 4) from first trimester gestation (≤12 weeks of gestation) were collected from healthy consented patients during elective abortions at the UC Davis Medical Center, with approval from the Institutional Review Board. CV-MSCs were isolated from chorionic villus tissue using an explant culture method, previously established in our lab48. Chorionic villus tissue was washed in phosphate buffered saline (PBS, Hyclone), containing 100 U/ml penicillin and 100 µg/ml streptomycin (1% pen-strep, Thermo Fisher Scientific), and dissected into smaller pieces. Tissues were evenly spread across tissue culture-treated flasks and cultured in D5 media containing DMEM high glucose (Hyclone), 5% fetal bovine serum (FBS, Hyclone), 20 ng/mL recombinant human bFGF (R&D systems), 20 ng/mL recombinant human EGF (R&D systems), and incubated at 37°C. Cells were allowed to migrate from the tissue and grow to 80–90% confluency, before the first passage. The media was changed every 3–4 days. CV-MSCs were used between P3 to P5 for all experiments. HCAECs were purchased from ATCC. BM-MSCs were obtained from Dr. Arnold I. Caplan (Case Western Reserve University). AT-MSCs were obtained from Dr. David E. Sahar (University of California, Davis).
Synthesis and screening of the OBOC peptide library
In our previous study, we synthesized and screened the OBOC peptide library and discovered a high-affinity peptidomimetic ligand, LLP2A, targeting integrin α4β1 (half maximal inhibitory concentration (IC50) = 2 pM)50. Integrin subunits α4 and β1 are expressed on the surface of MSCs64. In this study, we synthesized LLP2A on TentaGel resin beads and used them as a convenient platform for cell−bead binding assay to investigate the ligand−cell binding affinity.
CV-MSC-bead binding assay
For the cell bead-binding assay, 6 × 105 CV-MSCs in 2 mL of D5 media were added to an ultralow attachment 35 mm Petri dish (Corning Incorporated), followed by blank resin beads or resin beads displaying with LLP2A49. The dishes were incubated in a shaking incubator at 37 °C for various time points, at 40 rpm. Phase contrast images were taken at different time points using an Olympus IX81 microscope.
CV-MSC attachment assay on LLP2A surface
We synthesized biotinylated LLP2A (LLP2A-bio, Fig. 4B) using established solid phase peptide synthesis protocols50. Peptide-bio was designed to have biotin attached to the side chain of lysine, and two hydrophilic linkers between the peptide and Lys (biotin). To modify the culture surface with ligands, target culture wells in a 24-well plate were coated with 500 μL of 20 μg/mL Avidin (Thermo Fisher Scientific) and incubated for 1 h at 37 °C. Avidin coated wells were rinsed three times with DPBS and were treated with 500 μL of 2 μM D-biotin (Thermo Fisher Scientific) or LLP2A-bio. After 1 h, the wells were washed three times with DPBS and blocked with 1% BSA (Thermo Fisher Scientific) for 1 h. After the wells were rinsed three times with DPBS, for the cell attachment assay, 5 × 104 CV-MSCs, BM-MSCs or AT-MSCs suspended in the D5 media were added to the wells and incubated for 10 min at 37 °C. The wells were washed three times with DPBS, and the adhered cells were fixed in 10% formalin (Azer Scientific) for 20 min. Phase contrast images were taken using an Olympus IX81microscope for evaluation of the CV-MSC attachment. Quantification of images was performed using the ImageJ software (NIH).
Flow cytometry analysis of ligand−cell binding affinity
To quantitatively compare cell-ligand binding affinity, flow cytometry analysis of LLP2A-bio and CV-MSCs was performed following our previous protocol11. Specifically, 5 × 105 CV-MSCs or HCAECs were incubated with 1 μM LLP2A-bio in binding buffer (1× HEPES containing 10% FBS) on ice for 30 min. The samples were washed three times with wash buffer (DPBS containing 1% FBS) and incubated with streptavidin-phycoerythrin (Life Technologies) on ice for 30 min, and then washed with DPBS. To test the expression of the α4 and β1 integrin subunits on CV-MSCs and HCAECs, samples were stained with mouse anti-human α4 or β1 integrin antibodies (Millipore) on ice for 30 min, washed three times with wash buffer, incubated with goat anti-mouse 546 conjugate (Life Technologies) in DPBS on ice for 30 min, and then washed with DPBS. To confirm that LLP2A binds to CV-MSCs mainly via integrin α4β1, we performed a binding/blocking experiment using monoclonal anti-α4 and anti-β1 antibodies. To block α4 or β1 integrin subunits, cells were first incubated with excess mouse anti-human α4 or β1 integrin antibodies on ice for 30 min, washed three times with wash buffer, and then incubated with 1 μM LLP2A-bio, for another 30 min. The samples were washed three times with wash buffer and incubated with streptavidin PECy7 conjugated in DPBS on ice for 30 min, and then washed with DPBS. Samples were analyzed on a BD Fortessa LSR Cell Analyzer, and further data analysis and gating were performed using FlowJo software (Treestar, Inc.).
Preparation and characterization of LLP2A-modified biomaterial scaffolds
Fabrication of electrospun microfibrous scaffolds was performed as previously reported11. Poly (L-lactic acid; PLLA; MW 67 400, Sigma-Aldrich) and polycaprolactone (PCL, MW 2000, Polysciences) were used to fabricate the microfibrous membranes. The polymer blends (e.g., 19% PLLA and 5% PCL; w/v) were completely dissolved in 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP, Aladdin). Microfibrous membranes with a thickness of about 200 μm were prepared by electrospinning polymer fibers onto the rotating drum collector. The negative voltage of 4.5 kV was applied to the mandrel, and a positive voltage of 4 kV was applied to the spinneret, by using a high voltage generator (Gamma High Voltage).
LLP2A was grafted onto the PLLA/PCL membrane surface in three steps (Fig. 4D). First, membranes were incubated in 0.01 M sodium hydroxide for 10 min to expose the carboxyl groups on the surface. Second, the membranes were further incubated in a solution of 1-ethyl-3-(3-(dimethylamino)propyl) carbodiimide hydrochloride (EDC, Thermo Fisher Scientific) and N-hydroxysulfosuccinimide (sulfo-NHS, Thermo Fisher Scientific) in 0.5 M morpholino ethanesulfonic acid (MES) buffer (pH 5.5, Thermo Fisher Scientific) for 30 min. After brief washing with DPBS, the membranes were incubated in a solution of amine-PEG11-azido (NH2-PEG-N3, MW 570.7, BROADPHARM) in alkalescent DPBS (pH≈7.8) for 2 h on a shaker. Third, 20 μM LLP2A-DBCO (Fig. 4C), synthesized via a similar approach to that for LLP2A-bio, but with a 5- azidopentanoic acid attached to the side chain of lysine (Supporting Information Figure S1), was conjugated to azido-decorated membranes via Click chemistry in the water system for 16 h82. The PEG linker and LLP2A modification of the membranes was characterized by XPS using a PHI 5400 spectrometer (Physical Electronics). The structure of the membranes was characterized with SEM. Fiber diameters were measured using ImageJ software. 50 fiber diameters were measured and the average was reported.
Contact angle measurement
The contact angles of ultrapure water over the surface of untreated PLLA/PCL scaffolds and LLP2A-treated PLLA/PCL scaffolds were measured with a VCA2000 video contact angle system (Advanced Surface Technology Inc.). For this measurement, a drop of ultrapure water was placed over the surface of the scaffold. Subsequently, an image of the drop of water over the surface was immediately obtained. The computer processed the pictures and calculated the angle of contact for the pictures. Five independent measurements were performed per treatment.
CV-MSC attachment and viability
The LLP2A-modified electrospun microfibrous membranes and untreated membranes were placed in 35 mm tissue culture dishes. The membranes were rinsed with DPBS and incubated with CV-MSCs in D5 media, at a density of 1 × 105 cells/cm2. For the cell attachment test, after 2 h of incubation, the media was aspirated and unattached cells were washed off with DPBS three times. The adhered cells were characterized using MTS. For the cell viability test, after 2 h of incubation, new D5 media was added into the dishes, and the cells were sequentially cultured for 7 d. The cell viability was determined by an MTS assay. The amount of soluble formazan product produced by the reduction of MTS by metabolically active cells was measured by a 96-well spectrophotometer, at a 490 nm absorbance. The cell morphologies on the membranes were characterized using SEM. The cell-covered area was quantified using ImageJ software.
Western-blot and Caspase 9 analysis
Immunoblotting of FAK, p-FAK, AKT, p-AKT and NF-kB was performed after 10 min or 6 h cell seeding. The detached cells were collected first. The attached cells were washed twice with ice cold DPBS, and membranes were cut into small pieces using a blade and pooled with the unattached cells in 1.5 ml centrifuge tubes. After centrifugation, lysis buffer containing RIPA lysis buffer, sodium metavanadate (NaVO3), PMSF and complete protease inhibitor cocktail tablet (all from Santa Cruz Biotechnology) were added to the cell pellets and the remaining materials, and then they were incubated for 1 h at 4 °C. The protein concentrations were determined using a BCA protein assay (Thermo Fisher Scientific). A total of 15 μg of each sample was loaded, separated using a 4−12% Bis-Tris NuPAGE gel (Thermo Fisher Scientific), and transferred to a nitrocellulose membrane. The membrane was blocked in 5% BSA in TBST (Trisbuffered saline with 0.5% Tween-20) and subsequently incubated with primary antibodies anti-FAK, anti-phospho-FAK, anti-AKT, anti-phospho-AKT, anti-NF-kB and GAPDH (all purchased from Cell Signaling Technologies) overnight at 4 °C. After washing three times with TBST, the membranes were incubated with respective conjugated secondary antibodies in 5% nonfat dry milk in TBST (BioRad) for 1 h at room temperature. After washing, the protein bands were visualized using a West Dura Substrate (Thermo Fisher Scientific) under a Gel Documentation System (Bio-Rad) and quantified using ImageJ software. Sensitive measurement of Caspase 9 activity in CV-MSCs was performed by Caspase 9 Assay Kit (Abcam) according to the instruction. The colorimetric was measured by a 96-well spectrophotometer, at a 405 nm absorbance.
Cytoskeleton staining
Cells were incubated on untreated and LLP2A-modified electrospun microfibrous membranes for 3 days after seeding. Cells were washed three times with DPBS before fixation in 4% paraformaldehyde for 20 mins. The cells were incubated with TRITC-conjugated Phalloidin and DAPI (Millipore), according to the manufacturer’s instructions. After three washes with DPBS, the cells were imaged using an Olympus IX81 microscope. The cell area was quantified using ImageJ software.
Statistical analysis
For two-sample comparison, student’s t test was used. For multiple-sample comparison, analysis of variance (ANOVA) was performed to detect whether a significant difference existed between groups with different treatments. A p-value of 0.05 or less indicates a significant difference between samples in comparison.
Conclusions
In this study, we described the use of LLP2A, a ligand with a high binding affinity to integrin α4β1, in engineering the surface of polymeric material scaffolds for MSC transplantation, and investigated the interactions between the LLP2A-modified surface and different types of MSCs, such as CV-MSCs, BM-MSCs and AT-MSCs. In the current proof of concept study, we chose a representative type of MSCs, CV-MSCs, which hold significant promise for fetal tissue engineering, but do not seem to be able to survive well after transplantation. LLP2A has an exceptionally high binding affinity to CV-MSCs, via integrin α4β1, and a high stability in vivo, which is why it is able to overcome the key obstacles that other biomolecules have previously encountered when used to improve CV-MSC loading on polymeric scaffolds for fetal tissue engineering applications. Furthermore, we can also use LLP2A as an excellent lead ligand, for further optimization and modification, to develop LLP2A-based bioactive ligands with an even higher binding affinity, more specific binding capability and superior functionality for MSC-based tissue regeneration applications.
Supplementary Material
Acknowledgements
This work was in part supported by the Shriners Hospitals for Children Postdoctoral Fellowship (84705-NCA-19 to DH) and the UC Davis School of Medicine Dean’s Fellowship (to AW) awards, NIH grants (5R01NS100761-02, R03HD091601-01), Shriners Hospitals for Children research grants (87200-NCA-19, 85108-NCA-19), and the March of Dimes Foundation Basil O’Connor Starter Scholar Research Award (5FY1682). We acknowledge Nicole Kreutzberg and Alexandra Maria Iavorovschi for their help with manuscript editing and submission.
Footnotes
Conflicts of interest
There are no conflicts of interest to declare.
References
- 1.Barbero S and Ponte PL, Arch Sci Med (Torino), 1977, 134, 413–435. [PubMed] [Google Scholar]
- 2.Adzick NS, Thom EA, Spong CY, Brock JW 3rd, Burrows PK, Johnson MP, Howell LJ, Farrell JA, Dabrowiak ME, Sutton LN, Gupta N, Tulipan NB, D’Alton ME, Farmer DL and Investigators M, The New England journal of medicine, 2011, 364, 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deprest JA, Flake AW, Gratacos E, Ville Y, Hecher K, Nicolaides K, Johnson MP, Luks FI, Adzick NS and Harrison MR, Prenatal diagnosis, 2010, 30, 653–667. [DOI] [PubMed] [Google Scholar]
- 4.Harrison MR, Fetal diagnosis and therapy, 2004, 19, 513–524. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe M, Kim AG and Flake AW, Fetal diagnosis and therapy, 2015, 37, 197–205. [DOI] [PubMed] [Google Scholar]
- 6.Chen YJ, Chung K, Pivetti C, Lankford L, Kabagambe SK, Vanover M, Becker J, Lee C, Tsang J, Wang A and Farmer DL, Journal of pediatric surgery, 2017, DOI: 10.1016/j.jpedsurg.2017.10.040. [DOI] [PubMed] [Google Scholar]
- 7.Wang A, Brown EG, Lankford L, Keller BA, Pivetti CD, Sitkin NA, Beattie MS, Bresnahan JC and Farmer DL, Stem Cells Transl Med, 2015, 4, 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langer R and Vacanti JP, Science, 1993, 260, 920–926. [DOI] [PubMed] [Google Scholar]
- 9.Tabata Y, J R Soc Interface, 2009, 6 Suppl 3, S311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edgar L, McNamara K, Wong T, Tamburrini R, Katari R and Orlando G, Materials (Basel), 2016, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hao D, Xiao W, Liu R, Kumar P, Li Y, Zhou P, Guo F, Farmer DL, Lam KS, Wang F and Wang A, ACS chemical biology, 2017, 12, 1075–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang A, Tang Z, Park IH, Zhu Y, Patel S, Daley GQ and Li S, Biomaterials, 2011, 32, 5023–5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Y, Wang A, Patel S, Kurpinski K, Diao E, Bao X, Kwong G, Young WL and Li S, Tissue engineering. Part C, Methods, 2011, 17, 705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Downing TL, Wang A, Yan ZQ, Nout Y, Lee AL, Beattie MS, Bresnahan JC, Farmer DL and Li S, Journal of controlled release : official journal of the Controlled Release Society, 2012, 161, 910–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saadai P, Nout YS, Encinas J, Wang A, Downing TL, Beattie MS, Bresnahan JC, Li S and Farmer DL, Journal of pediatric surgery, 2011, 46, 2279–2283. [DOI] [PubMed] [Google Scholar]
- 16.Saadai P, Wang AJ, Nout YS, Downing TL, Lofberg K, Beattie MS, Bresnahan JC, Li S and Farmer DL, Journal of pediatric surgery, 2013, 48, 158–163. [DOI] [PubMed] [Google Scholar]
- 17.Yu J, Wang AJ, Tang ZY, Henry J, Lee BLP, Zhu YQ, Yuan FL, Huang FP and Li S, Biomaterials, 2012, 33, 8062–8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi HX, Hu P, Xu J and Wang AJ, Biomacromolecules, 2006, 7, 2327–2330. [DOI] [PubMed] [Google Scholar]
- 19.Lee BLP, Jeon H, Wang AJ, Yan ZQ, Yu J, Grigoropoulos C and Li S, Acta biomaterialia, 2012, 8, 2648–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu GS, Li PH, Feng HQ, Zhang XM and Chu PK, J Mater Chem B, 2015, 3, 2024–2042. [DOI] [PubMed] [Google Scholar]
- 21.Biederman H and Slavinska D, Surf Coat Tech, 2000, 125, 371–376. [Google Scholar]
- 22.Ko YG, Kim YH, Park KD, Lee HJ, Lee WK, Park HD, Kim SH, Lee GS and Ahn DJ, Biomaterials, 2001, 22, 2115–2123. [DOI] [PubMed] [Google Scholar]
- 23.Guan JJ, Gao CY, Feng LX and Shen JC, J Appl Polym Sci, 2000, 77, 2505–2512. [Google Scholar]
- 24.Zhang HF, Li ZJ, Fu X, Ma JX and Ma XL, Arch Med Res, 2013, 44, 69–74. [DOI] [PubMed] [Google Scholar]
- 25.Zhang HN, Lin CY and Hollister SJ, Biomaterials, 2009, 30, 4063–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen WS, Guo LY, Tang CC, Tsai CK, Huang HH, Chin TY, Yang ML and Chen-Yang YW, Nanomaterials (Basel), 2018, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He W, Ma Z, Yong T, Teo WE and Ramakrishna S, Biomaterials, 2005, 26, 7606–7615. [DOI] [PubMed] [Google Scholar]
- 28.Mardilovich A, Craig JA, McCammon MQ, Garg A and Kokkoli E, Langmuir, 2006, 22, 3259–3264. [DOI] [PubMed] [Google Scholar]
- 29.Monteiro N, Martins A, Pires R, Faria S, Fonseca NA, Moreira JN, Reisa RL and Neves NM, Biomater Sci-Uk, 2014, 2, 1195–1209. [DOI] [PubMed] [Google Scholar]
- 30.Shi YB, Cell Biochem Biophys, 1995, 27, 179–202. [DOI] [PubMed] [Google Scholar]
- 31.Streuli CH, Mol Biol Cell, 2016, 27, 2885–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teti A, J Am Soc Nephrol, 1992, 2, S83–87. [DOI] [PubMed] [Google Scholar]
- 33.Keselowsky BG, Collard DM and Garcia AJ, Biomaterials, 2004, 25, 5947–5954. [DOI] [PubMed] [Google Scholar]
- 34.Keselowsky BG, Collard DM and Garcia AJ, P Natl Acad Sci USA, 2005, 102, 5953–5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krishnamurthy M and Wang R, Front Biosci (Schol Ed), 2009, 1, 477–491. [DOI] [PubMed] [Google Scholar]
- 36.Liang W, Zhu C, Liu F, Cui W, Wang Q, Chen Z, Zhou Q, Xu S, Zhai C and Fan W, Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology, 2015, 37, 1301–1314. [DOI] [PubMed] [Google Scholar]
- 37.El-Amin SF, Attawia M, Lu HH, Shah AK, Chang R, Hickok NJ, Tuan RS and Laurencin CT, J Orthop Res, 2002, 20, 20–28. [DOI] [PubMed] [Google Scholar]
- 38.Calderwood DA, Shattil SJ and Ginsberg MH, The Journal of biological chemistry, 2000, 275, 22607–22610. [DOI] [PubMed] [Google Scholar]
- 39.Choquet D, Felsenfeld DP and Sheetz MP, Cell, 1997, 88, 39–48. [DOI] [PubMed] [Google Scholar]
- 40.Parsons JT, Horwitz AR and Schwartz MA, Nature reviews. Molecular cell biology, 2010, 11, 633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy SV and Atala A, Semin Reprod Med, 2013, 31, 62–68. [DOI] [PubMed] [Google Scholar]
- 42.Portmann-Lanz CB, Schoebedein A, Huber A, Sager R, Malek A, Holzgreve W and Surbek DV, American journal of obstetrics and gynecology, 2006, 194, 664–673. [DOI] [PubMed] [Google Scholar]
- 43.Guillot PV, Gotherstrom C, Chan J, Kurata H and Fisk NM, Stem cells, 2007, 25, 646–654. [DOI] [PubMed] [Google Scholar]
- 44.Izumi M, Pazin BJ, Minervini CF, Gerlach J, Ross MA, Stolz DB, Turner ME, Thompson RL and Miki T, Journal of reproductive immunology, 2009, 81, 39–43. [DOI] [PubMed] [Google Scholar]
- 45.Poloni A, Maurizi G, Serrani F, Mancini S, Discepoli G, Tranquilli AL, Bencivenga R and Leoni P, Cell proliferation, 2012, 45, 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang ZY, Teoh SH, Chong MSK, Schantz JT, Fisk NM, Choolani MA and Chan J, Stem cells, 2009, 27, 126–137. [DOI] [PubMed] [Google Scholar]
- 47.Jones GN, Moschidou D, Puga-Iglesias TI, Kuleszewicz K, Vanleene M, Shefelbine SJ, Bou-Gharios G, Fisk NM, David AL, De Coppi P and Guillot PV, PloS one, 2012, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lankford L, Selby T, Becker J, Ryzhuk V, Long C, Farmer D and Wang A, World journal of stem cells, 2015, 7, 195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM and Knapp RJ, Nature, 1991, 354, 82–84. [DOI] [PubMed] [Google Scholar]
- 50.Peng L, Liu R, Marik J, Wang X, Takada Y and Lam KS, Nature chemical biology, 2006, 2, 381–389. [DOI] [PubMed] [Google Scholar]
- 51.Xiao WW, Wang Y, Lau EY, Luo JT, Yao NH, Shi CY, Meza L, Tseng H, Maeda Y, Kumaresan P, Liu RW, Lightstone FC, Takada Y and Lam KS, Molecular cancer therapeutics, 2010, 9, 2714–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao NH, Xiao WW, Wang XB, Marik J, Park SH, Takada Y and Lam KS, Journal of medicinal chemistry, 2009, 52, 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guan M, Yao W, Liu R, Lam KS, Nolta J, Jia J, Panganiban B, Meng L, Zhou P, Shahnazari M, Ritchie RO and Lane NE, Nat Med, 2012, 18, 456–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yao W, Guan M, Jia J, Dai W, Lay YA, Amugongo S, Liu R, Olivos D, Saunders M, Lam KS, Nolta J, Olvera D, Ritchie RO and Lane NE, Stem cells, 2013, 31, 2003–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vasita R, Shanmugam IK and Katt DS, Curr Top Med Chem, 2008, 8, 341–353. [DOI] [PubMed] [Google Scholar]
- 56.Giancotti FG and Ruoslahti E, Science, 1999, 285, 1028–1032. [DOI] [PubMed] [Google Scholar]
- 57.Petrie TA, Raynor JE, Dumbauld DW, Lee TT, Jagtap S, Templeman KL, Collard DM and Garcia AJ, Science translational medicine, 2010, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hynes RO, Cell, 2002, 110, 673–687. [DOI] [PubMed] [Google Scholar]
- 59.Olivares-Navarrete R, Rodil SE, Hyzy SL, Dunn GR, Almaguer-Flores A, Schwartz Z and Boyan BD, Biomaterials, 2015, 51, 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ip JE, Wu YJ, Huang J, Zhang LN, Pratt RE and Dzau VJ, Mol Biol Cell, 2007, 18, 2873–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Globus RK, Doty SB, Lull JC, Holmuhamedov E, Humphries MJ and Damsky CH, Journal of cell science, 1998, 111, 1385–1393. [DOI] [PubMed] [Google Scholar]
- 62.Moursi AM, Globus RK and Damsky CH, Journal of cell science, 1997, 110, 2187–2196. [DOI] [PubMed] [Google Scholar]
- 63.Wu CY, Fields AJ, Kapteijn BAE and Mcdonald JA, Journal of cell science, 1995, 108, 821–829. [DOI] [PubMed] [Google Scholar]
- 64.Brooke G, Tong H, Levesque JP and Atkinson K, Stem Cells Dev, 2008, 17, 929–940. [DOI] [PubMed] [Google Scholar]
- 65.Zwolanek D, Flicker M, Kirstatter E, Zaucke F, van Osch GJVM and Erben RG, Bioresearch Open Acc, 2015, 4, 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Semon JA, Nagy LH, Llamas CB, Tucker HA, Lee RH and Prockop DJ, Cell Tissue Res, 2010, 341, 147–158. [DOI] [PubMed] [Google Scholar]
- 67.Wang N, Tytell JD and Ingber DE, Nature reviews. Molecular cell biology, 2009, 10, 75–82. [DOI] [PubMed] [Google Scholar]
- 68.van der Marel D, van Elp J, Sawatzky GA and Heitmann D, Phys Rev B Condens Matter, 1988, 37, 5136–5141. [DOI] [PubMed] [Google Scholar]
- 69.Allen LT, Tosetto M, Miller IS, O’Connor DP, Penney SC, Lynch I, Keenan AK, Pennington SR, Dawson KA and Gallagher WM, Biomaterials, 2006, 27, 3096–3108. [DOI] [PubMed] [Google Scholar]
- 70.Lee JN, Jiang X, Ryan D and Whitesides GM, Langmuir, 2004, 20, 11684–11691. [DOI] [PubMed] [Google Scholar]
- 71.Khalili AA and Ahmad MR, Int J Mol Sci, 2015, 16, 18149–18184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cooke MJ, Vulic K and Shoichet MS, Soft Matter, 2010, 6, 4988–4998. [Google Scholar]
- 73.Huang S and Ingber DE, Nat Cell Biol, 1999, 1, E131–138. [DOI] [PubMed] [Google Scholar]
- 74.Huang Y, Hu J, Zheng J, Li J, Wei T, Zheng Z and Chen Y, J Exp Clin Cancer Res, 2012, 31, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen CS, Mrksich M, Huang S, Whitesides GM and Ingber DE, Science, 1997, 276, 1425–1428. [DOI] [PubMed] [Google Scholar]
- 76.McBeath R, Pirone DM, Nelson CM, Bhadriraju K and Chen CS, Developmental cell, 2004, 6, 483–495. [DOI] [PubMed] [Google Scholar]
- 77.Brock A, Chang E, Ho CC, LeDuc P, Jiang XY, Whitesides GM and Ingber DE, Langmuir, 2003, 19, 1611–1617. [DOI] [PubMed] [Google Scholar]
- 78.Jiang XY, Bruzewicz DA, Wong AP, Piel M and Whitesides GM, P Natl Acad Sci USA, 2005, 102, 975–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen CS, Alonso JL, Ostuni E, Whitesides GM and Ingber DE, Biochem Bioph Res Co, 2003, 307, 355–361. [DOI] [PubMed] [Google Scholar]
- 80.Liu WF and Chen CS, Mater Today, 2005, 8, 28–35. [Google Scholar]
- 81.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K and Chen CS, P Natl Acad Sci USA, 2003, 100, 1484–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nandivada H, Chen HY, Bondarenko L and Lahann J, Angew Chem Int Edit, 2006, 45, 3360–3363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.