Abstract
Glycosaminoglycans are sulfated polysaccharide molecules, essential for many biological processes. The 6-O sulfation of glycosaminoglycans is carried out by carbohydrate 6-O sulfotransferases (C6OSTs), previously named Gal/GalNAc/GlcNAc 6-O sulfotransferases. Here, for the first time, we present a detailed phylogenetic reconstruction, analysis of gene synteny conservation and propose an evolutionary scenario for the C6OST family in major vertebrate groups, including mammals, birds, nonavian reptiles, amphibians, lobe-finned fishes, ray-finned fishes, cartilaginous fishes, and jawless vertebrates.
The C6OST gene expansion likely started early in the chordate lineage, giving rise to four ancestral genes after the divergence of tunicates and before the emergence of extant vertebrates. The two rounds of whole-genome duplication in early vertebrate evolution (1R/2R) only contributed two additional C6OST subtype genes, increasing the vertebrate repertoire from four genes to six, divided into two branches. The first branch includes CHST1 and CHST3 as well as a previously unrecognized subtype, CHST16 that was lost in amniotes. The second branch includes CHST2, CHST7, and CHST5. Subsequently, local duplications of CHST5 gave rise to CHST4 in the ancestor of tetrapods, and to CHST6 in the ancestor of primates.
The teleost-specific gene duplicates were identified for CHST1, CHST2, and CHST3 and are result of whole-genome duplication (3R) in the teleost lineage. We could also detect multiple, more recent lineage-specific duplicates. Thus, the vertebrate repertoire of C6OST genes has been shaped by gene duplications and gene losses at several stages of vertebrate evolution, with implications for the evolution of skeleton, nervous system, and cell–cell interactions.
Keywords: carbohydrate 6-O sulfotransferases, Gal/GalNAc/GlcNAc 6-O sulfotransferases, whole-genome duplication, vertebrate
Introduction
Glycosaminoglycans (GAGs) are sulfated linear polysaccharide molecules composed of repeating disaccharides, like chondroitin sulfate (CS), dermatan sulfate (DS), keratan sulfate (KS), and heparan sulfate (HS). CS and DS are composed of N-acetylgalactosamine (GalNAc) linked to glucuronic acid or iduronic acid, respectively. HS and KS are composed of N-acetylglucosamine (GlcNAc) linked to glucuronic acid or galactose, respectively. Sulfated GAGs are found in both vertebrates and invertebrates and are important for many biological processes like cell adhesion, signal transduction, and immune response (Yamada et al. 2011; Soares da Costa et al. 2017). The polymerization of long, linear GAG chains onto core protein takes place in the Golgi apparatus and results in the formation of proteoglycans that are essential components of the extracellular matrix (Kjellén and Lindahl 1991).
Sulfation is a complex modification process that is common for most GAGs and is important for their activity (Soares da Costa et al. 2017). The 6-O sulfation of CS and KS is carried out by enzymes of the carbohydrate 6-O sulfotransferase (C6OST) family (Kusche-Gullberg and Kjellén 2003). The nomenclature of the reported members of this family is summarized in table 1. Hereafter, we will use the abbreviation C6OST to refer to the protein family in general and the symbols CHST1 through CHST7 to refer to the individual genes. The 6-O sulfation of HS is carried out by enzymes of another protein family, the heparan sulfate 6-O sulfotransferases (HS6ST1–HS6ST3) (Nagai and Kimata 2014) that will not be discussed here. The sulfation of GAGs is carried out by using 3′-phosphoadenosine 5′-phosphosulfate as a sulfonate donor. 3′-Phosphoadenosine 5′-phosphosulfate binds to C6OSTs at specific sequence motifs: the 5′-phosphosulfate binding (5′PSB) motif RxGSSF (Habuchi et al. 2003) and the 3′-phosphate binding (3′PB) motif RDPRxxxxSR (Tetsukawa et al. 2010). As an example, in the human chondroitin 6-sulfotransferase 1 protein sequence (encoded by CHST3), the 5′PSB motif corresponds to positions 142–147 (RTGSSF), and the 3′PB motif to positions 301–310 (RDPRAVLASR). Chondroitin 6-O sulfotransferase 1 is one of the most widely studied members of the C6OST family, showing activity toward both CS and KS (see references in Yusa et al. [2006]). Another enzyme, chondroitin 6-O sulfotransferase 2 (encoded by CHST7) also shows activity toward CS (Kitagawa et al. 2000). The expression of CHST3 and CHST7 has been reported in a number of tissues, including chondrocytes, immune system organs, as well as the central and peripheral nervous system (see Habuchi 2014, and references therein). Mutations in CHST3 are associated with congenital spondyloepiphyseal dysplasia, congenital joint dislocations, and hearing loss in kindred families (Waryah et al. 2016; Srivastava et al. 2017), whereas overexpression of CHST3 lowers the abundance of the proteoglycan aggrecan in the extracellular matrix of the aged brain, which is associated with the loss of neural plasticity (Miyata and Kitagawa 2016). The orthologs of CHST3 and CHST7 have been characterized in zebrafish, where chst3a, chst3b, and chst7 are expressed in pharyngeal cartilages, the notochord, and several brain regions during development (Habicher et al. 2015).
Table 1.
Nomenclature of the Carbohydrate 6-O Sulfotransferases
| Gene Name | Synonyms | Human chr. |
|---|---|---|
| CHST1 | KS6ST, KSGal6ST, GST1 | 11 |
| CHST2 | GlcNAc6ST1, Gn6ST1, GST2 | 3 |
| CHST3 | C6ST1, GST0 | 10 |
| CHST4 | GlcNAc6ST2, HEC-GlcNAc6ST, GST3 | 16 |
| CHST5 | GlcNAc6ST3, I-GlcNAc6ST, GST4-alpha | 16 |
| CHST6 | GlcNAc6ST5, C-GlcNAc6ST, GST4-beta | 16 |
| CHST7 | C6ST2, GlcNAc6ST4, GST5 | X |
Carbohydrate sulfotransferase 1 (encoded by CHST1), also called Keratan Sulfate Gal-6 Sulfotransferase 1 (KSGal6ST), is responsible for the sulfation of KS. CHST1 expression has been described in the developing mouse brain (Hoshino et al. 2014) and in human endothelial cells (Li and Tedder 1999). Another sulfotransferase showing activity toward KS is the corneal N-acetylglucosamine-6-O-sulfotransferase (C-GlcNAc6ST, encoded by CHST6). In humans, the closely related CHST5 and CHST6 genes are located ∼40 kb apart on chromosome 16. Whereas in the mouse genome, there is only one gene in the corresponding chromosomal region on chromosome 8, called CHST5. Although human CHST5 expression seems restricted to the small intestine and colon (Lee et al. 1999), mutations of CHST6 in humans cause a rare autosomal recessive macular corneal dystrophy (Akama et al. 2000; Carstens et al. 2016; Rubinstein et al. 2016). In mice, a similar condition in the form of CS/DS aggregates in the cornea is caused by the disruption of CHST5 (Parfitt et al. 2011). In addition, the similarity in enzymatic activity between the human CHST6 and mouse CHST5 gene products has been used to suggest that these genes are orthologous (Akama et al. 2001, 2002).
N-Acetylglucosamine-6-O-sulfotransferase 1 (GlcNAc6ST1, encoded by CHST2) is expressed in brain tissues and several internal organs of adult mice (Fukuta et al. 1998). It has also been reported in endothelial tissues of both humans and mice (Li and Tedder 1999). In the mouse model, CHST2 is critical for neuronal plasticity in the developing visual cortex (Takeda-Uchimura et al. 2015), and CHST2 deficiency leads to increased levels of amyloid-β phagocytosis thus modulating Alzheimer’s pathology (Zhang et al. 2017). N-Acetylglucosamine-6-O-sulfotransferase 2 (GlcNAc6ST2, encoded by CHST4) is expressed exclusively in the high endothelial venules of the lymph nodes (Bistrup et al. 2004) and its disruption in mice affects lymphocyte trafficking (Hemmerich, Bistrup, et al. 2001). CHST4 is also expressed in early-stage uterine cervical and corpus cancers (Seko et al. 2009).
Functional studies of GAGs have expanded greatly during the last 20 years. The roles of GAG modifying enzymes, including C6OSTs, in health and disease have been studied extensively in human and mouse, as well as to some extent in chicken (Fukuta et al. 1995; Yamamoto et al. 2001; Nogami et al. 2004; Kobayashi et al. 2010). However, there is a big information gap regarding which C6OST genes can be found outside mammalian vertebrates, as well as the phylogenetic relationship between them. Outside mammals and chicken, the number of studies is very limited to just a few species, including zebrafish (Danio rerio) (Habicher et al. 2015), as well as vase tunicate (Ciona intestinalis) (Tetsukawa et al. 2010), and pearl oyster (Pinctada fucata martensii) (Du et al. 2017).
Here, for the first time, we describe the evolution of the C6OST family of sulfotransferases based on detailed phylogenetic and chromosomal location analyses in major vertebrate groups, and propose a duplication scenario for the expansion of the C6OST family during vertebrate evolution. We also report a previously unrecognized KSGal6ST-like subtype of C6OST, encoded by a gene we have called CHST16 that was lost in amniotes.
Materials and Methods
Identification of C6OST Gene Sequences
C6OST amino acid sequences corresponding to the CHST1-7 genes were sought primarily in genome assemblies hosted by the National Center for Biotechnology Information (NCBI) Assembly database (www.ncbi.nlm.nih.gov/assembly) (Kitts et al. 2016; Canese et al. 2017) and the Vertebrate Genomes Project (https://vertebrategenomesproject.org) (Rhie et al. forthcoming). For some species, the Ensembl genome browser (www.ensembl.org) (Perry et al. 2018) was used. C6OST sequences were also sought in transcriptome assemblies hosted by the PhyloFish Portal (http://phylofish.sigenae.org) (Pasquier et al. 2016). Independent sources for an Eastern Newt reference transcriptome (Abdullayev et al. 2013) and the gulf pipefish genome assembly (Small et al. 2016) were also used. All the investigated species, genome/transcriptome assembly versions, and source databases are listed in supplementary table S1, Supplementary Material online. In total, 158 species were investigated. They include 18 mammalian species, 33 avian species in 16 orders, 14 nonavian reptile species, 6 amphibian species, the basal lobe-finned fish coelacanth, the holostean fishes spotted gar and bowfin, 67 teleost fish species in 35 orders, including the important model species zebrafish, 7 cartilaginous fish species, as well as 3 jawless vertebrate species, the sea lamprey (Petromyzon marinus), Arctic lamprey (Lethenteron camtschaticum, also known as Japanese lamprey), and inshore hagfish (Eptatretus burgeri). C6OST sequences from invertebrate species were also sought. Notably, the vase tunicate (Ciona intestinalis) was used to provide a relative dating point with respect to the early vertebrate whole-genome duplications (1R/2R), and the fruit fly (Drosophila melanogaster) was used as an outgroup.
The C6OST sequences were identified by searching for NCBI Entrez Gene models or Ensembl gene predictions annotated as CHST1-7, followed by extensive TBlastN searches (Altschul et al. 1990) to identify sequences with no corresponding gene annotations or for species where gene annotations were not available. In most cases (excluding transcriptome data), corresponding gene models could be found and their database IDs were recorded. In cases where no gene models could be identified, or where they included errors (Prosdocimi et al. 2012), C6OST sequences were predicted/corrected by manual inspection of their corresponding genomic regions, including flanking regions and introns. Exons were curated with respect to consensus sequences for splice donor/acceptor sites (Abril et al. 2005; Iwata and Gotoh 2011) and start of translation (Nakagawa et al. 2008), as well as sequence similarity to other C6OST family members.
All identified amino acid sequences were collected and the corresponding genomic locations (if available) were recorded. All collected sequences were verified against the Pfam database of protein families (https://pfam.xfam.org) (El-Gebali et al. 2019) to ensure they contained a sulfotransferase type 1 domain (Pfam ID: PF00685), and inspected manually to verify the characteristic 5′PSB and 3′PB motifs. All genomic locations and sequence identifiers used in this study have been verified against the latest genome assembly and database versions, including NCBI’s Reference Sequence Database (RefSeq 96, September 16, 2019) and Ensembl (version 97, July 2019).
Sequence Alignment and Phylogenetic Analyses
Sequence alignments were constructed with the MUSCLE alignment algorithm (Edgar 2004) applied through AliView 1.25 (Larsson 2014). Alignments were curated manually to adjust poorly aligned stretches with respect to conserved motifs and exon boundaries, as well as to identify faulty or incomplete sequences. Phylogenies were constructed from full-length alignments using IQ-TREE v1.6.3 which applies a stochastic maximum likelihood algorithm (Nguyen et al. 2015). The best-fit amino acid substitution model and substitution parameters were selected using IQ-TREE’s model finder with the -m TEST option (Kalyaanamoorthy et al. 2017). The proportion of invariant sites was optimized using the –opt-gamma-inv option. Branch supports were calculated using IQ-TREE’s nonparametric UltraFast Bootstrap (UFBoot) method (Minh et al. 2013) with 1,000 replicates, as well as the approximate likelihood-ratio test (aLRT) with SH-like supports (Anisimova and Gascuel 2006; Anisimova et al. 2011) over 1,000 iterations.
Conserved Synteny Analyses
The two whole-genome duplications that occurred at the base of vertebrate evolution (1R and 2R) resulted in a large number of quartets of related chromosome regions, each such quartet is called a paralogon or a paralogy group, and related chromosome regions are said to be paralogous. To investigate whether any of the vertebrate C6OST genes arose in 1R/2R, and duplicated further in the teleost whole-genome duplication 3R, we searched for patterns of conserved synteny, the conservation of gene family colocalization, across the C6OST gene-bearing chromosome regions in the human, Carolina anole lizard, spotted gar, and zebrafish genomes. The anole lizard was chosen because of the presence of a “CHST4/5-like” gene in this species. The spotted gar was chosen because of the presence of CHST16, which is missing from amniotes, because it shows a moderate degree of genome rearrangement compared with teleost fish genomes, and because its taxonomic position allows it to bridge the gap between ray-finned fishes and lobe-finned fishes (including tetrapods) (Braasch et al. 2016). The zebrafish genome was chosen to investigate the involvement of 3R. Lists of gene predictions 2.5-Mb upstream and downstream of the C6OST genes in these species were downloaded using the BioMart function in Ensembl version 83 (December 2015) (https://dec2015.archive.ensembl.org). These lists were sorted according to Ensembl protein family predictions (Ensembl 83 is the last version of the database to use these family predictions) to identify gene families with members on at least two of the C6OST gene-bearing chromosome blocks within each species. Neighboring “gene families” in the context of our study are those Ensembl protein family predictions with member genes on at least two C6OST gene-bearing chromosome regions, defined as 5-Mb around each C6OST gene. Sequence identifiers and genomic locations for each member gene were collected for the following species: human, chicken, Western clawed frog, spotted gar, zebrafish, medaka, and elephant shark. All locations and sequence identifiers from Ensembl version 83 were updated to correspond with the latest versions of the genome assemblies in the NCBI database.
Because only one CHST1 gene, chst1b, could be identified in the zebrafish (described in Results), the region of the channel catfish CHST1a gene was also used to facilitate the conserved synteny analysis: Gene models 1 Mb to each side of the channel catfish CHST1a gene were identified in the NCBI Genome Data Viewer. These were used to identify the orthologous region in the zebrafish genome which in turn was used for the conserved synteny analysis as described above.
Results
Phylogeny of the C6OST Family
C6OST amino acid sequences were collected from a large diversity of vertebrate genomes and aligned in order to produce phylogenies of the C6OST gene family across vertebrates. The first phylogeny of C6OST family in chordates is summarized in figure 1 and includes the vase tunicate along with a smaller subset of representative vertebrates. This phylogeny is rooted with two putative family members from the fruit fly, CG31637 and CG9550. These fruit fly sequences share the sulfotransferase domain (Pfam ID: PF00685) and recognizable 5′PSB and 3′PB motifs with the vertebrate C6OST sequences. In addition to this phylogeny, we produced detailed phylogenies for each C6OST subtype branch including all investigated vertebrate species. These have been included as supplementary figures S1–S5 and S7, Supplementary Material online. These additional phylogenies helped us classify all identified sequences into the correct C6OST subtypes, and improve the resolution of some phylogenetic relationships. Partial sequences shorter than 50% of the final alignment lengths were not included in any of the phylogenies. These sequences are listed in the supporting data deposited in figshare (doi:10.6084/m9.figshare.9596285).
Fig. 1.
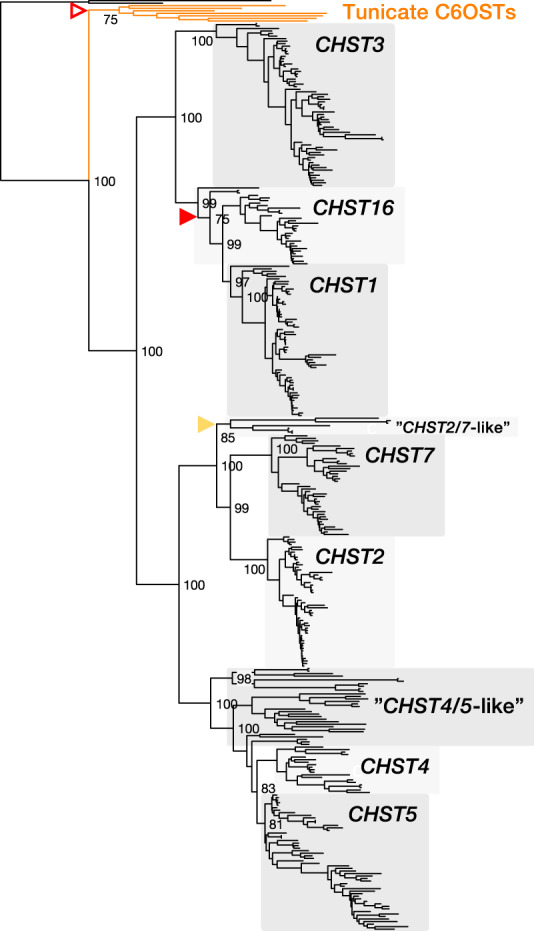
—Maximum likelihood phylogeny of chordate C6OST sequences. The phylogeny is supported by aLRT and UFBoot analyses. UFBoot supports for deep nodes are shown. Red filled arrowheads indicate unreliable nodes (≤75%) in both UFBoot and aLRT, yellow filled arrowheads indicate nodes with low aLRT support only, and red open arrowheads indicate nodes with low UFBoot support only. The phylogeny is rooted with the Drosophila melanogaster C6OST sequences CG9550 and CG31637.
Our phylogeny (fig. 1) supports the subdivision of the C6OST family into two main branches. The first branch contains three well-supported subtype clades, including CHST1 and CHST3 as well as a previously unrecognized CHST1-like subtype of genes we have named CHST16. CHST8-14 genes encode carbohydrate 4-O sulfotransferases and CHST15 encodes a GalNac4S-6-O sulfotransferase, thus CHST16 was chosen. The second C6OST branch in vertebrates contains well-supported CHST2, CHST4, CHST5, and CHST7 clades, as well as several smaller clades of “CHST4/5-like” sequences from jawless vertebrates (Agnatha), amphibians, and nonavian reptiles. We could identify putative CHST16, CHST3, as well as “CHST2/7-like” and “CHST4/5-like” sequences in lampreys (described in detail below), as well as putative CHST1, CHST16, “CHST2/7-like” and “CHST4/5-like” sequences in the inshore hagfish. Although the identity of some of these sequences remains unclear, the jawless vertebrate branches place the divergences between the C6OST subtype clades before the split between jawless and jawed vertebrates (Gnathostomata), early in vertebrate evolution. To provide an earlier dating point, we used seven C6OST-like sequences from the vase tunicate. These sequences were first identified by Tetsukawa et al. (2010), and our phylogeny (fig. 1) supports their conclusion that the tunicate C6OST genes represent an independent lineage-specific gene expansion. Thus, vertebrate C6OST genes likely diversified after the divergence of tunicates but before the divergence between jawless and jawed vertebrates. This is consistent with the time window of the two rounds of whole-genome duplication early in vertebrate evolution, 1R and 2R (Dehal and Boore 2005; Nakatani et al. 2007; Putnam et al. 2008; Sacerdot et al. 2018). We could also identify teleost-specific duplicates of CHST1, CHST2, and CHST3 (described below), which is consistent with the third round of whole-genome duplication (3R) that occurred early in the teleost lineage (Meyer and van de Peer 2005).
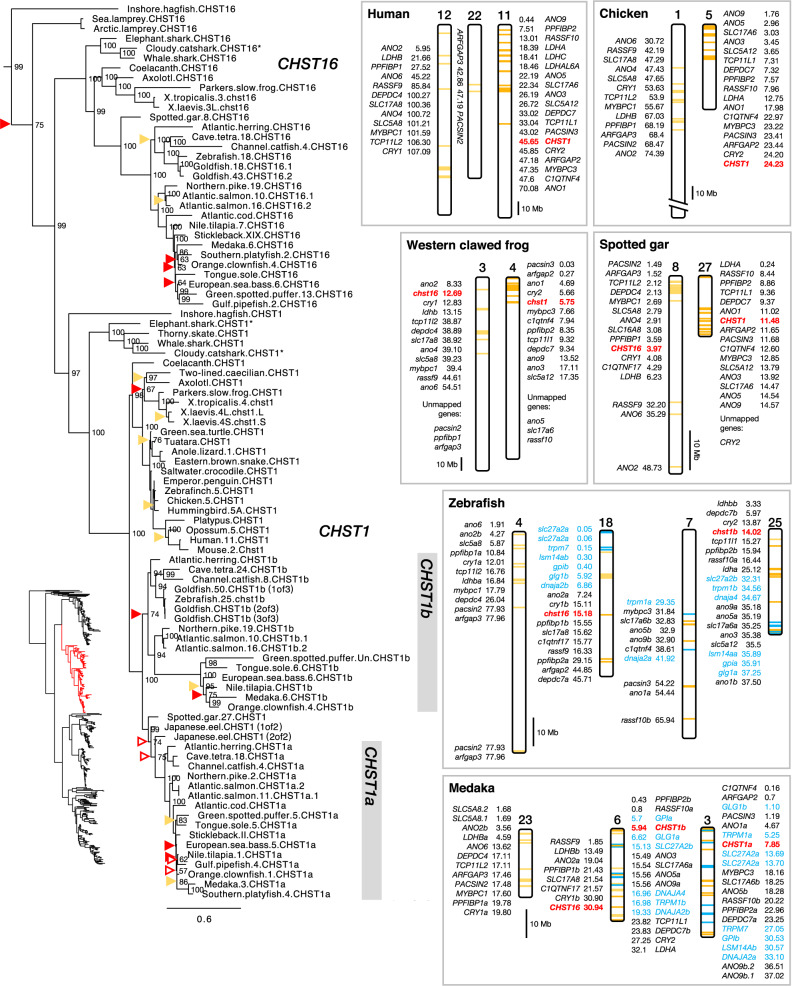
CHST1 and CHST16
The CHST1 and CHST16 branches of the C6OST phylogeny are shown in figure 2. The full-species phylogeny of CHST1 sequences is shown in supplementary figure S1, Supplementary Material online, and of CHST16 sequences in supplementary figure S2, Supplementary Material online. Both CHST1 and CHST16 are represented in cartilaginous fishes (Chondrichthyes), lobe-finned fishes (Sarcopterygii), including tetrapods and coelacanth, as well as ray-finned fishes (Actinopterygii), including the spotted gar and teleost fishes. Overall, both CHST1 and CHST16 branches follow the accepted phylogeny of vertebrate groups, and the overall topology is well supported. However, there are some notable inconsistencies: We could identify putative CHST1 and CHST16 orthologs in the inshore hagfish, as well as putative CHST16 orthologs in the Arctic and sea lampreys. However, the jawless vertebrate CHST16 sequences cluster basal to both jawed vertebrate CHST1 and CHST16 clades, and the inshore hagfish and lamprey genes do not cluster together. Within teleost fishes, we could identify duplicate CHST1 genes located on separate chromosomes, which we have named CHST1a and CHST1b. However, the spotted gar CHST1 sequence clusters together with the teleost CHST1a clade rather than basal to both CHST1a and CHST1b clades. This is true for the smaller phylogeny shown in figure 2 as well as for the phylogeny with full-species representation (supplementary fig. S1, Supplementary Material online), which also includes a CHST1 sequences from another holostean fish, the bowfin. The duplicate CHST1 genes in the investigated eel species also both cluster within the CHST1a branch. Thus, we could not determine with any certainty whether these CHST1 duplicates in eels represent CHST1a and CHST1b. These inconsistencies are likely, at least partially, caused by uneven evolutionary rates between different lineages as well as a relatively high degree of sequence conservation for CHST1 sequences. There are also duplicate CHST1, but not CHST16, genes in the allotetraploid African clawed frog, whose locations on chromosomes 4L and 4S correspond to each of the two homeologous subgenomes (Session et al. 2016).
Fig. 2.
—Phylogeny of CHST1 and CHST16, and conserved synteny between CHST1- and CHST16-bearing chromosome regions, including CHST1a- and CHST1b-bearing regions in teleost fishes. The placement of the CHST1 and CHST16 branches within the full C6OST phylogeny is indicated in the bottom left. Sequence names include species names followed by chromosome/linkage group designations (if available) and gene symbols. Asterisks indicate incomplete sequences. For node support details, see figure 1 caption. Some node support values for shallow nodes have been omitted for visual clarity. Neighboring genes identified in the vicinity of CHST1a and CHST1b genes are indicated in blue. For medaka chromosome 6, CHST16-neighboring genes are to the left and CHST1-neighboring genes are to the right.
There have been notable gene losses of both CHST1 and CHST16. CHST16 genes could not be identified in nonavian reptiles, birds, or mammals, indicating that this subtype was lost in the amniote ancestor. Within cartilaginous fishes, CHST16 was missing in all investigated skate species, indicating a loss from Rajiformes. CHST16 genes were also missing from both eel species, indicating a loss within the genus Anguilla. Not all teleost fish species preserve both CHST1 duplicates. Notably, CHST1a is missing from cypriniform fishes, including the zebrafish, and we could not identify CHST1a in one salmonid fish, the Arctic char. The CHST1b gene seems to have been lost more widely: We could not find CHST1b sequences in several basal spiny-rayed fishes (Acanthomorpha), such as opah (Lampriformes), Atlantic cod (Gadiformes), and longspine squirrelfish (Holocentriformes), as well as several basal percomorph fishes, namely bearded brotula (Ophidiiformes), mudskippers (Gobiiformes) which instead have duplicate CHST1a genes, yellowfin tuna (Scombriformes), tiger tail seahorse, and gulf pipefish (Syngnathiformes). It was also missing from turbot (Pleuronectiformes), turquoise killifish, guppy and Southern platyfish (Cyprinodontiformes), barred knifejaw (Centrarchiformes), European perch and three-spined stickleback (Perciformes), and Japanese pufferfish (Tetraodontiformes). At least some of these absences could be due to incomplete genome assemblies, as there are closely related species that do have both CHST1a and CHST1b genes, such as the giant oarfish (Lampriformes), blackbar soldierfish (Holocentriformes), tongue sole (Pleuronectiformes), Murray cod (Centrarchiformes), tiger rockfish (Perciformes), and green spotted pufferfish (Tetraodontiformes). Aside from this diversity in terms of CHST1b gene preservation or absence, CHST1b genes seem to have evolved more rapidly in neoteleost fishes, as indicated by the branch lengths within this clade in our phylogenies (fig. 2 and supplementary fig. S1, Supplementary Material online). The neoteleost CHST1b genes also have a divergent exon structure (see Exon/Intron Structures of Vertebrate C6OST Genes).
We could identify 15 gene families with members in the vicinity of both CHST1 and CHST16 (subset 1, supplementary table S2, Supplementary Material online): ANO1/2, ANO3/4/9, ANO5/6/7, ARFGAP, C1QTNF4/17, CRY, DEPDC4/7, LDH, MYBPC, PACSIN, PPFIBP, RASSF9/10, SLC5A5/6/8/12, SLC17A6/8, and TCP11. This was detected in the spotted gar genome on linkage groups 27 and 8 (fig. 2). All identified chromosome segments are shown in the supporting data (doi:10.6084/m9.figshare.9596285). In the human genome, the identified blocks of conserved synteny correspond mainly to segments of chromosomes 11 (where CHST1 is located), 12 and 22 (fig. 2), as well as 1, 6, 19, and X (not shown here). These chromosome regions have been recognized as paralogous, the result of the 1R/2R whole-genome duplications, in several large-scale reconstructions of vertebrate ancestral genomes (Nakatani et al. 2007; Putnam et al. 2008; Sacerdot et al. 2018). The CHST1- and CHST16-bearing chromosome regions correspond to the “D” paralogon in Nakatani et al. (2007), more specifically the vertebrate ancestral paralogous segments called “D1” (CHST1) and “D0” (CHST16). In the study by Sacerdot et al. (2018), they correspond to the reconstructed pre-1R ancestral chromosome 6. One of us (D.O.D.) has previously analyzed these chromosome regions extensively and could also conclude that they arose in 1R/2R (Lagman et al. 2013; Ocampo Daza and Larhammar 2018).
With respect to 3R, we could identify seven neighboring gene families in subset 1 which also had teleost-specific duplicate genes in the vicinity of CHST1a and CHST1b: ANO1/2, ANO3/4/9, ANO5/6/7, DEPDC4/7, SLC17A6/8, PPIFBP, and RASSF9/10 (fig. 2). We could also identify a further six gene families with members in the vicinity of CHST1a and CHST1b in teleost fishes (subset 2, supplementary table S2, Supplementary Material online): DNAJA1/2/4, GLG1, GPI, LSM14A, SLC27A, and TRPM1/3/6/7 (labeled with blue color in fig. 2). The identified conserved synteny blocks in the zebrafish genome correspond to segments of chromosomes 18 and 7 on one side, and 25 on the other, as well as segments of chromosomes 6 and 3 in the medaka genome. These chromosome segments have previously been identified to be the result of the teleost-specific whole-genome duplication, 3R (Kasahara et al. 2007; Nakatani et al. 2007; Nakatani and McLysaght 2017). In the reconstruction of the pre-3R genome by Nakatani and McLysaght (2017), these regions correspond to proto-chromosome 10. The chromosome segments we identified in the spotted gar, chicken, and human genome also agree with the reconstruction by Nakatani and McLysaght (2017). In the analyses of teleost fish chromosome evolution by Kasahara et al. (2007) and Nakatani et al. (2007), these regions correspond to proto-chromosome “j.” We could also detect extensive translocations between 1R/2R-generated paralogous chromosome segments in teleost fishes. See chromosome 18 in the zebrafish and chromosome 6 in the medaka, for example (fig. 2). This is also compatible with previous observations (Kasahara et al. 2007; Nakatani and McLysaght 2017; Ocampo Daza and Larhammar 2018). These rearrangements likely underlie the colocalization of CHST16 with CHST1b in many teleost genomes, as on chromosome 6 in the medaka (fig. 2), or with CHST1a within Otocephala, as on chromosome 18 in the Mexican cave tetra (see chromosome designations in the phylogeny in fig. 2). Cypriniform fishes, including the zebrafish, have lost the CHST1a gene as described above. However, CHST16 is located near the chromosome segment where CHST1a would have been located in the zebrafish genome (fig. 2).
In addition to 3R, there have been more recent whole-genome duplication events within cyprinid fishes (Cyprinidae) and salmonid fishes (Salmonidae) (Glasauer and Neuhauss 2014). We identified additional duplicates of CHST1b in the goldfish genome, however, only one of the three duplicates is mapped, to chromosome 50 (fig. 2), and the other two are identical. There are also duplicates of CHST16 on goldfish chromosomes 18 and 43. These chromosome locations correspond to homeologous chromosomes that arose in the cyprinid-specific whole-genome duplication (see Chen et al. 2019, figure 2). In salmonid fishes, there are additional duplicates of CHST1a in the Atlantic salmon and of CHST1b and CHST16 in all three investigated species, Atlantic salmon, rainbow trout, and Arctic char. Although several of these salmonid duplicates are unmapped, including one of the CHST1a duplicates in Atlantic salmon, the chromosomal locations of the syntenic CHST1b and CHST16 duplicates on chromosomes 10 and 16 in Atlantic salmon (Lien et al. 2016, figure 2), 2 and 1 in rainbow trout (Berthelot et al. 2014, figure 2), as well as 4 and 26 in Arctic char (Christensen et al. 2018, figure 1), correspond to duplicated chromosome segments from the salmonid whole-genome duplication. Note: It has recently been suggested that this Arctic char genome may instead represent a Northern dolly varden (Salvelinus malma malma) with some introgression from Arctic char (Shedko 2019). This does not affect our overall conclusions.
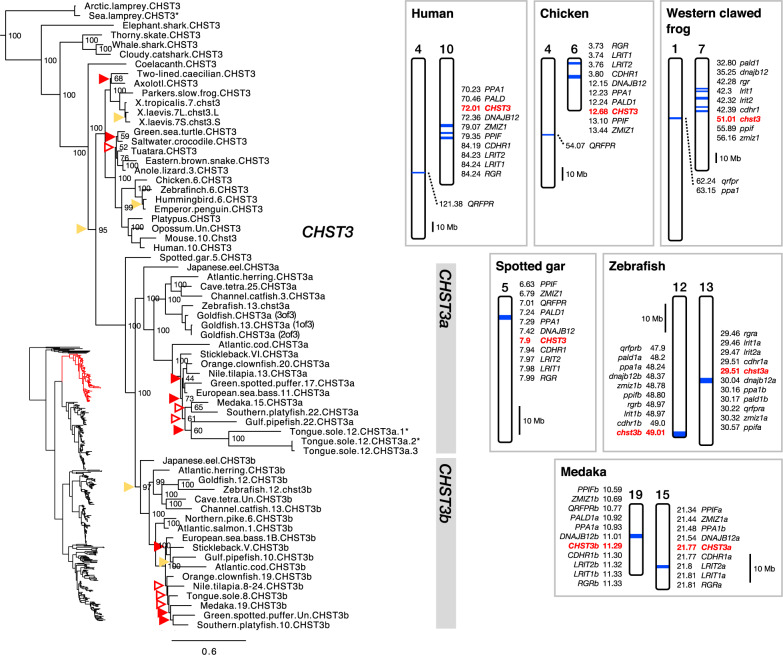
CHST3
The CHST3 branch of our C6OST phylogeny is shown in figure 3. The full-species phylogeny of CHST3 sequences is shown in supplementary figure S3, Supplementary Material online. We could identify CHST3 sequences across all major vertebrate lineages, including both lamprey species but not the inshore hagfish. The allotetraploid African clawed frog has duplicated CHST3 genes located on chromosomes 7L and 7S that correspond to each of the two homeologous subgenomes (Session et al. 2016). Overall, our phylogenies follow the accepted phylogeny of vertebrate groups. However, there are some inconsistencies in both phylogenies. The cartilaginous fish clade is not resolved, however, all cartilaginous fish CHST3 sequences cluster basal to the bony vertebrate clade. In addition, the coelacanth CHST3 sequence clusters basal to both the tetrapod and ray-finned fish clades. Nonetheless, the overall topologies of our phylogenetic analyses support the presence of a CHST3 gene before the split between jawless and jawed vertebrates and show that CHST3 is closely related to CHST1 and CHST16 (fig. 1). However, we could not identify any conserved synteny, that is, no shared genomic neighbors, between CHST3 and CHST1 or CHST16, in any of the analyzed species. In fact, the CHST3-bearing chromosome regions correspond to an entirely different vertebrate ancestral paralogon; paralogon “C” in Nakatani et al. (2007) and pre-1R ancestral chromosome 6 in Sacerdot et al. (2018).
Fig. 3.
—Phylogeny of CHST3 branch of C6OST sequences, and conserved synteny across CHST3-bearing chromosome regions, including CHST3a- and CHST3b-bearing regions in teleost fishes. See figure 2 caption for phylogeny details.
In teleost fishes, we found duplicates of CHST3 located on different chromosomes (fig. 3). These genes have been named chst3a and chst3b in the zebrafish (Habicher et al. 2015). The single CHST3 sequence from spotted gar clusters at the base of the well-supported teleost chst3a and chst3b clades, which is consistent with duplication in the time window of 3R. We could identify nine gene families with teleost-specific duplicate members in the vicinity of chst3a and chst3b: CDHR1, DNAJB12, LRIT, PALD1, PPA, PPIF, QRFPR, RGR, and ZMIZ (subset 3, supplementary table S2, Supplementary Material online). The identified paralogous segments on chromosomes 13 and 12 in zebrafish and on chromosomes 15 and 19 in medaka (fig. 3) correspond to chromosome segments that most likely arose in 3R (Kasahara et al. 2007; Nakatani et al. 2007; Nakatani and McLysaght 2017). In Kasahara et al. (2007) and Nakatani et al. (2007), they correspond to pre-3R proto-chromosome “d,” and in Nakatani and McLysaght (2017), they correspond to proto-chromosome “4.” The chromosome segments we could identify in the spotted gar, chicken, and human genomes (fig. 3) are also compatible with these previous studies, as well as with comparative genomic analyses of the spotted gar genome versus the human and zebrafish genomes (see figs. 3 and 4 and Amores et al. [2011, table S2]). Other smaller synteny blocks not shown here are shown in the supporting data (doi:10.6084/m9.figshare.9596285). We could identify three copies of CHST3a in the goldfish genome, however, only one of them is mapped, to chromosome 13. Thus, it is not possible to attribute at least one of the duplications to the cyprinid-specific whole-genome duplication. There are also three copies of the CHST3a gene in the tongue sole genome, located in tandem on chromosome 12. In salmonid fishes, no CHST3a gene could be found, indicating a loss of this gene, and no additional gene duplicates of CHST3b seem to have been preserved.
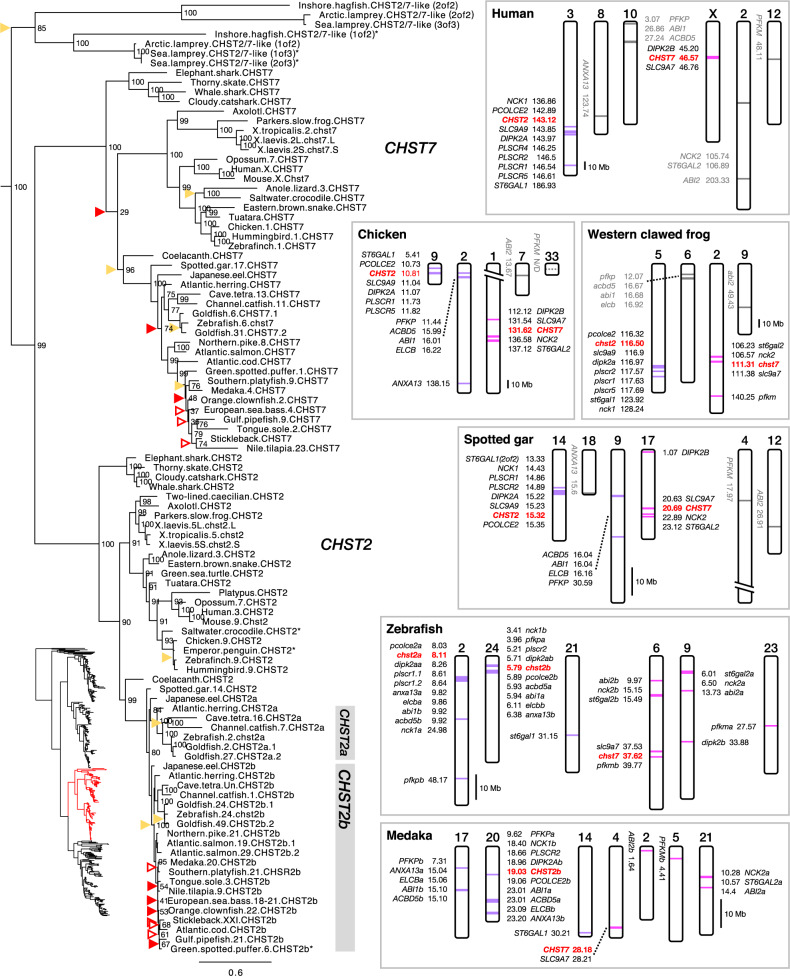
CHST2 and CHST7
The CHST2 and CHST7 branches of our C6OST phylogeny are shown in figure 4. The full-species phylogeny of CHST2 sequences is included in supplementary figure S4, Supplementary Material online, and of CHST7 sequences in supplementary figure S5, Supplementary Material online. The CHST2 and CHST7 subtype branches are well supported and follow the accepted phylogeny of vertebrate groups, overall. Both clades include orthologs from lobe-finned fishes (including coelacanth and tetrapods), ray-finned fishes (including spotted gar and teleost fishes), as well as cartilaginous fishes. However, both coelacanth CHST2 and CHST7 cluster with the respective ray-finned fish clades rather than at the base of the lobe-finned fish clades. This is likely due to the low evolutionary rate, that is, high degree of sequence conservation, of these coelacanth sequences relative to the tetrapod sequences (Amemiya et al. 2013). There are duplicate CHST2 and CHST7 genes in the allotetraploid African clawed frog (Xenopus laevis), located on the homeologous chromosomes (Session et al. 2016) 5L and 5S, 2L and 2S, respectively.
Fig. 4.
—Phylogeny of CHST2 and CHST7, and conserved synteny between CHST2- and CHST7-bearing chromosome regions, including CHST2a- and CHST2b-bearing regions in teleost fishes. Genes with uncertain synteny relationships are indicated in gray. See figure 2 caption for phylogeny details.
Notably, CHST7 could not be identified in any of the three investigated species within Testudines (turtles, tortoises, and terrapins), indicating an early deletion of this gene within the lineage. CHST7 is also missing from the two-lined caecilian (an amphibian) and several avian lineages, including penguins (Sphenisciformes, three species representing all available genera were investigated), falcons (within the genus Falco, the two available species were investigated), as well as the rock pigeon (Columbiformes), hoatzin (Ophistocomiformes), downy woodpecker (Piciformes), and hooded crow (Passeriformes). For at least some of these species, the absence of a CHST7 sequence could be due to incomplete genome assemblies, as there are other closely related species with CHST7, for example, the band-tailed pigeon (Columbiformes) and great tit (Passeriformes).
In jawless vertebrates, we could identify “CHST2/7-like” sequences from the inshore hagfish, sea lamprey, and Arctic lamprey genomes, forming two clades. These sequences cluster basal to both the jawed vertebrate CHST2 and CHST7 clades (fig. 4).
There are duplicates of CHST2 in teleost fishes located on different chromosomes (fig. 4). These have been named chst2a and chst2b in the zebrafish. We could identify both CHST2a and CHST2b only within basal teleost lineages (supplementary fig. S4, Supplementary Material online): eels (within genus Anguilla), freshwater butterflyfishes, bony-tongues, and mormyrids (Osteoglossiformes), as well as otocephalan fishes, including the Atlantic herring, European pilchard, and allis shad (Clupeiformes), zebrafish, Dracula fish, Amur ide, and goldfish (Cypriniformes), Mexican cave tetra and red-bellied piranha (Characiformes), electric eel (Gymnotiformes), and channel catfish (Siluriformes). This indicates that the CHST2a gene was lost early in the euteleost lineage, which includes the majority of the extant teleost species diversity. Both CHST2a and CHST2b clades are well supported in the phylogeny (fig. 4), and the single spotted gar CHST2 sequence clusters at the base of both branches, which is consistent with duplication within the time window of 3R. In the full-species phylogeny (supplementary fig. S4, Supplementary Material online), this topology is disrupted by the osteoglossiform CHST2a and CHST2b branches. This is likely caused by uneven evolutionary rates for teleost CHST2 sequences, as shown by the branch lengths within the phylogeny. Teleost CHST2 sequences seem to have had a very low basal amino acid substitution rate overall, however, the CHST2a sequences, as well as the CHST2b sequences in the African butterflyfish and mormyrids (Osteoglossiformes), seem to have evolved at a relatively faster rate. There are additional duplicates of both CHST2a and CHST2b, as well as of CHST7, in the goldfish genome, and their locations on chromosomes 2 and 27, 24 and 49, 6 and 31, respectively, supports their emergence through the cyprinid-specific whole-genome duplication (see Chen et al. 2019, figure 2). Salmonid fishes lack CHST2a, as all euteleost fishes do, however, there are duplicate CHST2b genes in the three species that were investigated. The CHST2b duplicates are located on chromosomes 19 and 29 in the Atlantic salmon genome, and 11 and 15 in the rainbow trout genome. These locations correspond to chromosomal segments known to have emerged in the salmonid whole-genome duplication (Berthelot et al. 2014; Lien et al. 2016). In the Arctic char, only one of the CHST2b duplicates is mapped, to chromosome 14. No CHST7 duplicates could be found in salmonids.
We could identify seven gene families with members in the vicinity of both CHST2 and CHST7: BRINP, NOS1AP, PFK, and RGS4/5/8/16 were detected in the zebrafish genome, NCK and ST6GAL in the spotted gar genome, and SLC9A6/7/9 in both the spotted gar and human genomes (subset 4, supplementary table S2, Supplementary Material online). Additionally, we could identify seven gene families with members in the vicinity of both chst2a and chst2b in the zebrafish genome: ABI, ACBD4/5, ANXA13, DIPK2, ELCB, PCOLCE, and PLSCR (subset 5, supplementary table S2, Supplementary Material online). Out of both subsets, only four gene families, NCK, SLC9A6/7/9, ST6GAL, and DIPK2, had members in the vicinity of CHST2 and CHST7 in the tetrapod and/or spotted gar genomes. This apparent deficit of conserved syntenic genes possibly reflects a low gene density in the region of CHST7. We examined the chromosome regions around CHST2 and CHST7 genes in the synteny database Genomicus version 69.10 (http://www.genomicus.biologie.ens.fr/genomicus-69.10) (Nguyen et al. 2018) and could only identify two gene pairs, in any species’ genome, in the vicinity of both CHST2 and CHST7: SLC9A9 and SLC9A7, as well as C3orf58 and CXorf36. The latter gene pair was not identified in our synteny analysis. Nevertheless, the conserved synteny blocks that we could identify (fig. 4) correspond to chromosome regions recognized to have resulted from the 1R/2R whole-genome duplications. In the reconstruction by Nakatani et al. (2007), these chromosome segments correspond to paralogon “F,” specifically the “F0” (CHST2) and “F3” (CHST7) vertebrate ancestral paralogous segments. In the analysis based around the Florida lancelet (Branchiostoma floridae) genome, they correspond to the reconstructed ancestral linkage group 10 (Putnam et al. 2008). In the more recent study by Sacerdot et al. (2018), they correspond to the reconstructed pre-1R ancestral chromosome 17. We could also identify one gene family, ITPR, with members in the vicinity of chst7 and chst16 in the zebrafish genome. However, this synteny pattern is not reproduced in any of the other investigated genomes.
With respect to 3R, seven gene families from subset 4 and subset 5 have teleost-specific duplicate members in the vicinity of both chst2a and chst2b in the zebrafish genome: PFK from subset 4, as well as ABI, ACBD4/5, ANXA13, DIPK2, ELCB, and PCOLCE from subset 5. One additional family, PLSCR, has members in the vicinity of chst2a (plscr1.1, plscr1.2) and chst2b (plscr2). However, the locations of these genes outside teleost fishes reveal that they likely arose through a local duplication in a bony fish ancestor, at the latest, rather than in teleost fishes and 3R.The identified paralogous segments on chromosomes 2 and 24 in zebrafish, and 17 and 20 in medaka (fig. 4), correspond to chromosome segments previously identified to be the result of 3R (Kasahara et al. 2007; Nakatani et al. 2007; Amores et al. 2011; Nakatani and McLysaght 2017). In Kasahara et al. (2007) and Nakatani et al. (2007), they correspond to pre-3R proto-chromosome “m,” and in Nakatani and McLysaght (2017), they correspond to proto-chromosome “13.” Our analysis also identified the possible location of the CHST2a gene in medaka on chromosome 17, had it not been lost early in the euteleost lineage. The chromosome segments for the ABI, ACBD4/5, ANXA13, ELCB, and PFK families in the spotted gar, chicken, Western clawed frog, and human genomes (fig. 4) do not correspond to the CHST2 and CHST7-bearing chromosome segments. However, several studies have identified those chromosome segments to also be part of the CHST2 and CHST7-bearing paralogon (see Kasahara et al. 2007, figure 4; Bian et al. 2016, figure 4; Nakatani and McLysaght 2017, figure 3).
In the zebrafish, the ABI, NCK, and PFK gene families have members in the vicinity of either chst2a or chst2b as well as chst7 (fig. 4), reflecting the 1R/2R-generated paralogy as described above. There are three additional families that also fulfill this criteria, BRINP, NOS1AP, and RGS4/5/8/16 (supplementary fig. S6, Supplementary Material online) but are likely not the result of 1R/2R. The single homologous paralogy blocks on human chromosome 1, chicken chromosome 8, Western clawed frog chromosome 4, and spotted gar linkage group 10 suggest that the gene duplicates BRINP2 and BRINP3, as well as RGS4, RGS5, RGS8, and RGS16 arose through ancient local duplications rather than through 1R/2R.
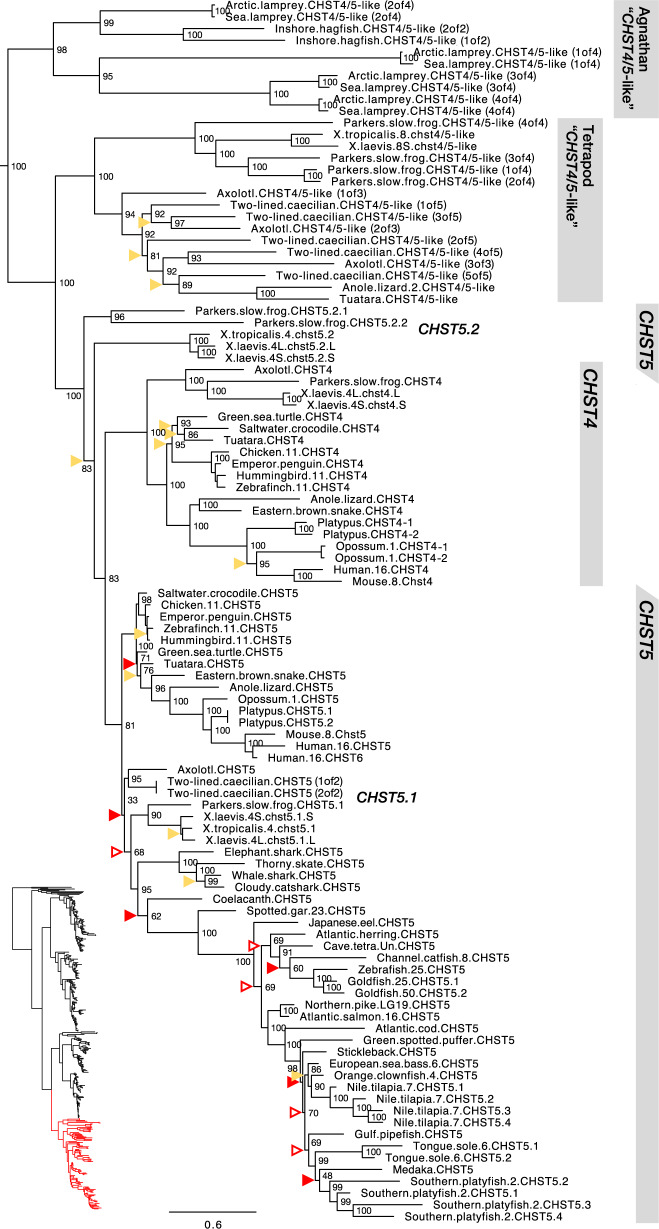
CHST4, CHST5, and Related “CHST4/5-Like” Sequences
The branch of the C6OST family that contains the known CHST4, CHST5, and CHST6 sequences is by far the most complex of the C6OST phylogeny. This branch of our C6OST phylogeny is shown in figure 5, and the full-species phylogeny is included in supplementary figure S7, Supplementary Material online. Aside from the known C6OST sequences, we can report several “CHST4/5-like” subtypes represented in amphibians and nonavian reptiles as well as jawless vertebrates.
Fig. 5.
—Phylogeny of CHST4, CHST5, and related genes, including CHST6 and “CHST4/5-like” genes. See figure 2 caption for phylogeny details.
In both our phylogenies, the known CHST4 and CHST5 sequences cluster into two well-defined and well-supported clades. This allowed us to classify a number of sequences with hitherto unclear identities. All identified sequences from cartilaginous fishes, coelacanth, and ray-finned fishes, including spotted gar and teleost fishes, cluster confidently together with tetrapod sequences within the CHST5 clade, whereas CHST4 sequences could only be identified from tetrapod species. In all tetrapod genomes with assembled chromosomes, linkage groups, or longer genomic scaffolds, CHST4 and CHST5 genes are located in tandem, and some of the “CHST4/5-like” genes described below are in turn located downstream of CHST5—see supporting data (doi:10.6084/m9.figshare.9596285). Chromosome/linkage group designations are also shown in the phylogenies. We could only identify orthologs of the human CHST6 gene in other primate species, located downstream of CHST5 and clustering confidently within the CHST5 clade. Although there are genes in other mammalian species, chicken, and Western clawed frog that have previously been identified as CHST6 (Akama and Fukuda 2014), our analyses show that they are better described as CHST5 for the nonprimate mammal and chicken genes, and as “CHST4/5-like” in the Western clawed frog. As of the publication of this article, the zebrafish chst5 gene is also erroneously annotated as chst6 in the zebrafish information network database at www.zfin.org (gene ID: ZDB-GENE-060810-74). In teleost fishes, we could identify many lineage-specific duplicates of CHST5. The largest number was found in the genome of the climbing perch (Anabantiformes) with 15 gene duplicates, some of which are likely pseudogenes. There are also CHST5 duplicates in the tongue sole (Pleuronectiformes), corkwing wrasse (Labriformes), brown dottyback (Pseudochromidae), turquoise killifish, guppy, and Southern platyfish (Cyprinodontiformes), Eastern happy, zebra mbuna, Nile tilapia, and Simochromis diagramma (Cichliformes), European pilchard (Clupeiformes), as well as in the mormyrids, Asian arowana, silver arowana, and Arapaima (Osteoglossiformes) (supplementary fig. S7, Supplementary Material online). At least some of the duplications are shared between several species within Cyprinodontiformes, Cichliformes, and Osteoglossiformes. In the genomes that have been assembled into chromosomes, linkage groups, or longer genomic scaffolds, these duplicated CHST5 genes are located in tandem. We could also identify CHST5 duplicates in the goldfish genome, located on chromosomes 25 and 50 that likely arose in the cyprinid fish whole-genome duplication (see Chen et al. 2019, figure 2). Conversely, no CHST5 duplicates from the salmonid-specific whole-genome duplication seem to have been preserved.
In addition to the CHST4 and CHST5 sequences (including CHST6), we could identify a multitude of previously unrecognized “CHST4/5-like” sequences in jawless vertebrates, amphibians, and nonavian reptiles. With some exceptions detailed below, we have used the name “CHST4/5-like” for these sequences. In jawless vertebrates, we identified four “CHST4/5-like” sequences in the Arctic lamprey and sea lamprey genomes, respectively, and two “CHST4/5-like” sequences in the inshore hagfish genome. These sequences form a well-supported clade that clusters at the base of the CHST4, CHST5, and “CHST4/5-like” branch (fig. 5). As for jawed vertebrates, we could identify a well-supported clade of “CHST4/5-like” sequences represented in amphibians as well as Lepidosauria, excluding snakes. We could identify sequences of this subtype in all investigated amphibian lineages, including the two-lined caecilian (Apoda), salamanders (Urodela), and frogs (Anura), as well as in the tuatara (Rhynchocephalia), the ocelot gecko (Gekkota), and several lizards, including the European green lizard (Laterata/Lacertoidea), the Carolina anole lizard, and the central bearded dragon (Iguania). This clade clusters basal to the main CHST4 and CHST5 branches in both phylogenies. This indicates that these sequences represent either an ancestral jawed vertebrate C6OST subtype that has not been preserved in any other lineages or more parsimoniously, “CHST4/5-like” gene duplicates with a derived mode of sequence evolution that arose in tetrapods before the split between amphibians and amniotes. A conserved synteny analysis of the Carolina anole lizard “CHST4/5-like” sequence on chromosome 2 showed no conservation of synteny with other C6OST gene-bearing regions (supplementary fig. S8, Supplementary Material online). In amphibians, there have been multiple rounds of local gene duplication within this clade. Although the Western and African clawed frogs only have one “CHST4/5-like” gene in this clade, Parker’s slow frog has four copies located in tandem on the same genomic scaffold, and the two-lined caecilian has five (supplementary fig. S9, Supplementary Material online). The phylogenetic relationships between different duplicates in different species are not resolved in either phylogeny (fig. 5 and supplementary fig. S7, Supplementary Material online). Apart from these “CHST4/5-like” sequences, we could identify CHST5-like sequences in the three frog genomes. In the phylogeny with full-species representation (supplementary fig. S7, Supplementary Material online), these sequences form a well-supported sister clade to the frog CHST5 sequences. Owing also to their arrangement downstream of CHST5 in the Western and African clawed frog genomes (supplementary fig. S9, Supplementary Material online), we have called them CHST5.2 and the frog CHST5 genes CHST5.1. The relationship between the frog CHST5.1 and CHST5.2 genes is not reproduced in the smaller phylogeny (fig. 5). Indeed, the amphibian branch of CHST5 is unresolved in both phylogenies, likely due to diverging evolutionary rates within the CHST5 clade. The amphibian CHST5 (CHST5.1 in frogs) sequences have had a lower rate of amino acid substitution, whereas the frog CHST5.2 sequences seem to have had an accelerated evolutionary rate, as shown by the branch lengths in both phylogenies. The allotetraploid African clawed frog has duplicate CHST4, CHST5.1, and CHST5.2 genes, all located on the homeologous chromosome pair 4L and 4S (supplementary fig. S9, Supplementary Material online), totaling seven genes within this branch of the C6OST phylogeny.
We could identify only two gene families showing conserved synteny between either CHST4 or CHST5 and another C6OST family member: KLF9/13/14/16 and RPGRIP1. These were detected in the zebrafish genome, with members in the vicinity of chst5 and chst2a (KLF9/13/14/16) or chst2b (RPGRIP1). However, this conserved synteny relationship is not reproduced in any of the other genomes we investigated, including the medaka. Thus, it is likely the result of chromosome rearrangements in the lineage leading to zebrafish. No other patterns of conserved synteny could be detected, and the chromosome regions bearing CHST4, CHST5, and related genes correspond to a separate vertebrate ancestral paralogon: paralogon “B” in Nakatani et al. (2007) and pre-1R ancestral chromosome “5” in Sacerdot et al. (2018).
Exon/Intron Structures of Vertebrate C6OST Genes
As has been reported previously, most mammalian C6OST genes consist of intron-less open reading frames (ORFs) (Bistrup et al. 1999; Lee et al. 1999; Kitagawa et al. 2000; Hemmerich, Lee, et al. 2001), with the exception of CHST3, whose protein-coding domain is encoded by two exons (Tsutsumi et al. 1998). Here, we can report that the protein-coding domain of CHST16 also consists of two exons, however, the positions of their respective introns are different between CHST3 and CHST16 genes (supplementary fig. S10, Supplementary Material online). These exon structures are common to jawed vertebrates and likely represent the exon/intron structures of the ancestral genes. There are several exceptions to these exon structures, all the result of several intron insertions, all within teleost fishes (supplementary fig. S10, Supplementary Material online). Notably, CHST1b genes in spiny-rayed fishes (Acanthomorpha) have acquired four introns. In jawless vertebrates, there seem to have been several independent intron insertions that make it difficult to deduce the ancestral conditions. Exon junctions for all identified C6OST genes are shown in the supporting data (doi:10.6084/m9.figshare.9596285).
Invertebrate C6OST Genes
In addition to the vase tunicate and fruit fly C6OST sequences included in our phylogeny (fig. 1), we could identify putative C6OST sequences from the Florida lancelet (Branchiostoma floridae), the hemichordate acorn worm (Saccoglossus kowalevskii), the purple sea urchin (Strongylocentrotus purpuratus) as well as the honey bee (Apis mellifera) and the silk moth (Bombyx mori). These have been deposited in the supporting data (doi:10.6084/m9.figshare.9596285). Ultimately, these sequences were not used in our final phylogenies, however, it is worth mentioning that there seem to have been extensive lineage-specific expansions of C6OST genes in all but the insect species. Seven unique C6OST sequences were previously identified in the vase tunicate by Tetsukawa et al. (2010). In addition, we could identify 16 unique C6OST sequences in the Florida lancelet, 31 in the acorn worm, and 30 in the purple sea urchin. All full-length sequences contain the characteristic sulfotransferase domain (Pfam PF00685) and have recognizable 5′PSB and 3′PB motifs.
Discussion
The Evolution of Vertebrate C6OST Genes
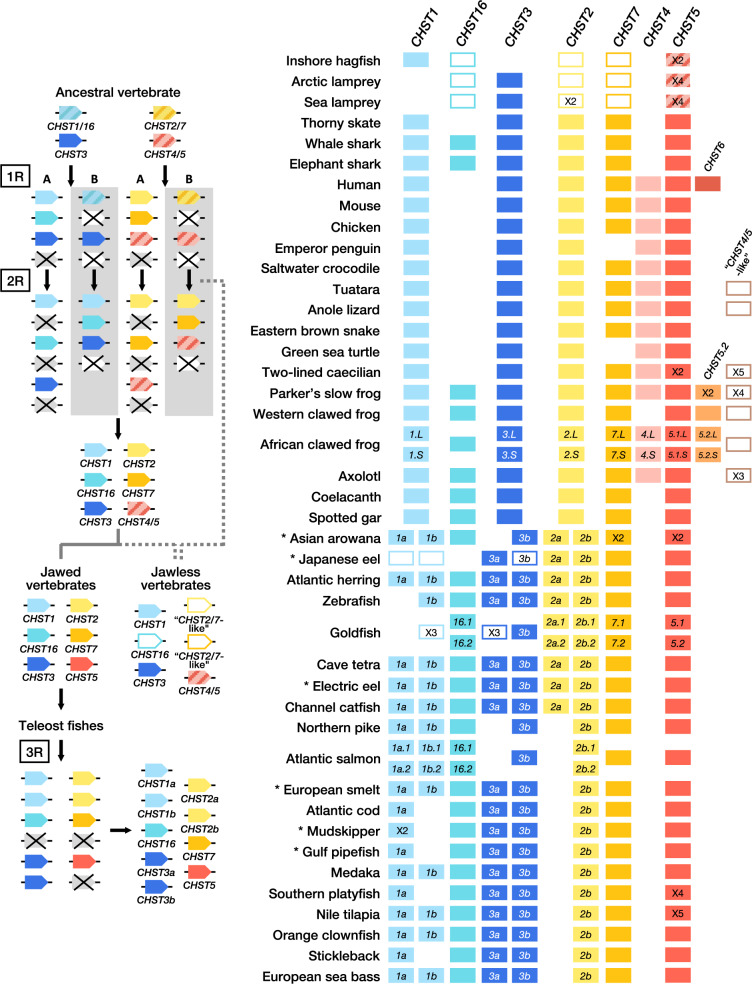
We present the first large-scale analysis and phylogenetic classification of vertebrate C6OST genes. Our analyses are based on genomic and transcriptomic data from a total of 158 species representing all major vertebrate groups, including 18 mammalian species, 33 avian species in 16 orders, 14 nonavian reptile species, 6 amphibian species, the basal lobe-finned fish coelacanth, the holostean fishes spotted gar and bowfin, 67 teleost fish species in 35 orders, 7 cartilaginous fish species, and 3 jawless vertebrate species. This allowed us to identify a previously unrecognized C6OST subtype gene that we have named CHST16, as well as several lineage-specific duplicates of the known C6OST genes. We could also identify known C6OST genes in lineages where they were previously unrecognized, like CHST7 in birds and nonavian reptiles, and CHST1a and CHST1b duplicates in teleost fishes. Our results are summarized in figure 6 (right).
Fig. 6.
—Proposed evolution of C6OST genes through the vertebrate whole-genome duplications (1R, 2R and 3R) (left) and C6OST gene repertoires in representative vertebrate species (right). A and B indicate alternative duplication scenarios through 1R/2R. The uncertain divergence of jawless vertebrates relative to 1R and 2R is indicated by dashed lines. Crossed-over boxes indicate gene losses. Open boxes indicate genes with unresolved phylogenetic positions. Some species-specific or lineage-specific duplicates are indicated by “X2” etc. within boxes. Asterisks indicate species included in the larger phylogenies shown in supplementary figures S1–S5 and S7, Supplementary Material online.
We conducted comparative analyses of conserved synteny—the conservation of gene content across several chromosomal regions that is typically the result from whole-genome duplications and found that the CHST1 and CHST16 genes, as well as the CHST2 and CHST7 genes, are located in genomic regions previously recognized to have originated in the basal vertebrate whole-genome duplications 1R and 2R (Nakatani et al. 2007; Kasahara et al. 2007; Putnam et al. 2008; Sacerdot et al. 2018). Thus, 1R/2R likely only contributed two additional genes to the ancestral vertebrate repertoire, giving rise to CHST1 and CHST16, as well as CHST2 and CHST7 (fig. 6). In this scenario, many gene duplicates were deleted after 1R/2R, and four C6OST genes were present already in a vertebrate ancestor before 1R: CHST3 as well as the ancestral CHST1/16, CHST2/7, and CHST4/5 genes (fig. 6). These four putative ancestral vertebrate C6OST genes also correspond to different ancestral chromosomes/paralogy groups from the reconstructions cited above: “D” (CHST1/16), “C” (CHST3), “F” (CHST2/7), and “B” (CHST4/5) in Nakatani et al. (2007) and “6” (CHST1/16), “14” (CHST3), “5” (CHST2/7), and “11” (CHST4/5) in Sacerdot et al. (2018). This suggests that the four vertebrate ancestral C6OST genes were located on different chromosomes before 1R. In the reconstruction by Putnam et al. (2008), which goes as far back as the divergence with cephalochordates, they correspond to five ancestral chromosomes: “14” (CHST1/6), “6” (CHST3), “10” (CHST2), “9” (CHST7), and “5” (CHST4/5). Our phylogenetic analysis places the origin of the ancestral C6OST genes between the divergence of tunicates from the main chordate branch, ∼547 Ma (Delsuc et al. 2018), and the emergence of extant vertebrate lineages. Thus, the gene family expansions that laid the ground for the vertebrate C6OST gene family likely occurred very early in chordate/vertebrate evolution, although it is not clear by which mechanism.
In jawless vertebrates, we identified putative CHST1 and CHST16 sequences from the inshore hagfish, as well as CHST16 and CHST3 sequences from the Arctic and sea lampreys (fig. 6). We could also identify several “CHST2/7-like,” and “CHST4/5-like,” sequences in all three jawless vertebrate species. The positions of the CHST16 and “CHST2/7-like” genes in our phylogeny are unresolved or ambiguous (bordered boxes in fig. 6) and raise questions about the relationship between 1R/2R and the jawless vertebrate lineage. There has been a long debate about whether jawless vertebrates diverged after 1R or whether jawless and jawed vertebrates share both rounds of whole-genome duplication (Kuraku et al. 2009). Previous studies indicate that jawless and jawed vertebrates share both whole-genome duplications (Mehta et al. 2013; Smith et al. 2013), but that the shared patterns of gene synteny are obscured by the asymmetric retention and loss of gene duplicates (Kuraku 2013). Another recent reconstruction of vertebrate genome evolution based on the retention of gene content, order and orientation, as well as gene family phylogenies, also concluded that jawless and jawed vertebrates diverged after 2R (Sacerdot et al. 2018). In contrast, an analysis of the meiotic map of the sea lamprey genome interprets the patterns of synteny conservation differently, suggesting instead one round of genome duplication at the base of vertebrates with subsequent independent segmental duplications in jawless and jawed vertebrates (Smith and Keinath 2015). Jawless and jawed vertebrates may also have preserved and lost different gene copies generated by 1R or 2R. Such differential gene losses/preservations, or “hidden paralogies,” are thought to underlie many ambiguous orthology and paralogy assignments between lamprey sequences and jawed vertebrate sequences (Kuraku 2010). Thus, any interpretation of our results in jawless vertebrates has to take several different scenarios into account. The positions of the agnathan CHST16 genes in our phylogeny (fig. 2) suggest that they may have originated before the main CHST1 and CHST16 clades, possibly in 1R. However, we identified a conserved pattern of microsynteny between the inshore hagfish and jawed vertebrate CHST16 genes, indicating their orthology (supplementary fig. S11, Supplementary Material online). Thus, their ambiguous positions in our phylogeny, and the paraphyly between the inshore hagfish and lamprey sequences, could be due to phylogenetic artifacts (see alignment in supporting data, doi:10.6084/m9.figshare.9596285). As for the two jawless vertebrate “CHST2/7-like” genes, our phylogeny suggests that they originated independently from the duplication that gave rise to CHST2 and CHST7, either through 1R or possibly through a jawless vertebrate-specific gene duplication. We could identify a conserved microsynteny pattern that seems to support the direct orthology between the “CHST2/7-like (1of2)” genes and CHST2, and the “CHST2/7-like (2of2)” genes and CHST7 (supplementary fig. S12, Supplementary Material online). However, the paucity of conserved neighboring genes precludes any definite conclusion. The jawless vertebrate “CHST4/5-like” sequences likely represent the ancestral CHST4/5 gene. In summary, our results are compatible with at least one whole-genome duplication, 1R, shared between jawless and jawed vertebrates (fig. 6).
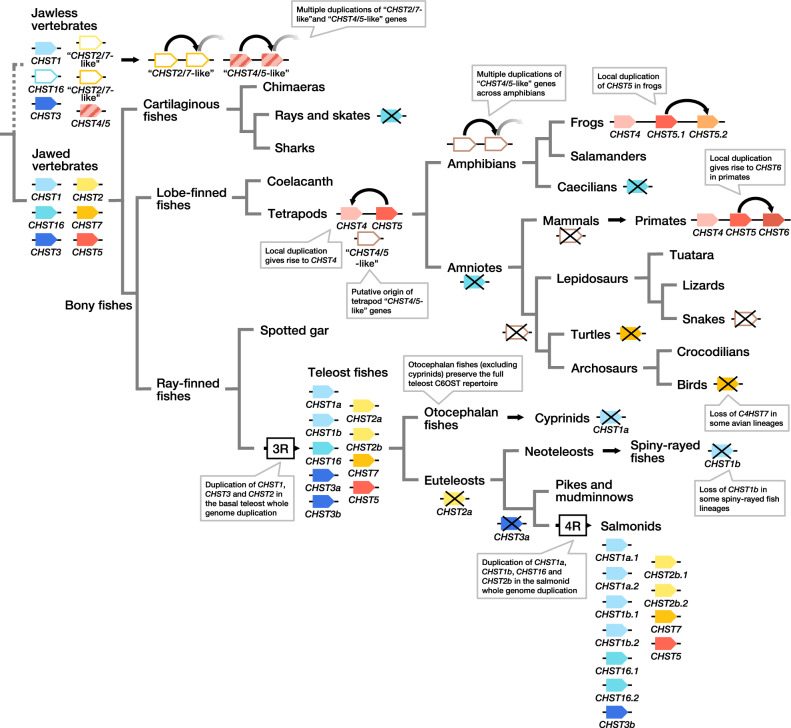
After the emergence of jawed vertebrates, and later of bony vertebrates, and the split between lobe-finned fishes and ray-finned fishes, the evolution of C6OST genes has taken different routes in different lineages, with several additional gene duplications and losses. The evolution of the CHST4/5 branch is particularly marked by local gene duplications: Local gene duplications of CHST5 generated CHST4 in a tetrapod ancestor, CHST6 in a primate ancestor, as well as a previously unrecognized gene in frogs that we called CHST5.2 (fig. 7 and supplementary fig. S9, Supplementary Material online). Only tetrapods were found to have both CHST4 and CHST5, located in tandem. The cartilaginous fish, coelacanth, spotted gar, and teleost sequences within this branch cluster confidently within the CHST5 clade, rather than basal to the tetrapod CHST4 and CHST5 clades, which indicates that CHST4 emerged later (fig. 7). Different genes have previously been identified as CHST6 in nonprimate mammals, chicken, Western clawed frog, and zebrafish. In most cases, it is CHST5 that has been misidentified, and we suggest new gene names and symbols in the results above. The origin of the “CHST4/5-like” genes found in amphibians and some lepidosaurs is uncertain, but we place its latest appearance at the base of tetrapods. It is possible that these genes represent a 1R/2R-generated duplicate of the ancestral CHST4/5 gene, however, no conserved synteny relationships could be determined with other C6OST gene-bearing chromosomes (supplementary fig. S8, Supplementary Material online).
Fig. 7.
—Proposed scenario of C6OST gene evolution after 1R and 2R. Crossed-over boxes indicate gene losses. This figure shows turtles as the sister clade to archosaurs, however, this position is still contested (Gilbert and Corfe 2013). The cyprinid-specific whole-genome duplication and the allotetraploidization in Xenopus laevis are not shown.
The newly identified CHST16 is present in cartilaginous fishes but was likely lost from skates (Rajiformes). CHST16 was also identified in all investigated ray-finned fish species, the coelacanth, and amphibians (excluding caecilians), but not in amniote species, indicating an early gene loss in this lineage. Nothing is known about the functions of CHST16, so we cannot speculate whether its deletion was concurrent with a loss of function, or whether another C6OST gene could compensate for its loss. However, it is notable that the likely emergence of CHST4 as a copy of CHST5, as well as the loss of CHST16, is associated with the time window for the transition from water to land and the emergence of the amniotic egg. Another notable loss in the tetrapod lineage is the loss of CHST7 in at least two bird lineages, penguins (Sphenisciformes) and falcons (within genus Falco), as well as in turtles (Testudines).
The C6OST gene family has had a dynamic and varied evolution within the teleost fish lineage. The teleost-specific whole-genome duplication, 3R, gave rise to duplicates of CHST1, CHST2, and CHST3, increasing the number of genes to nine in the teleost ancestor (figs. 6 and 7). This was followed by multiple differential losses; notably, the loss of CHST1a from cyprinid fishes, including zebrafish, the loss of CHST3a from salmonid fishes, and the loss of CHST2a from euteleost fishes. CHST1b seems to have been lost independently within several lineages of spiny-rayed fishes (Acanthomorpha). This apparently relaxed selection on the retention of CHST1b seems to have been preceded by a stage of rapid evolution. We found that the neoteleost branch of CHST1b sequences show an accelerated rate of amino acid substitution coupled with the insertion of four introns into the otherwise uninterrupted ORF. CHST16 is colocalized with either CHST1a or CHST1b within teleost fishes; this could be detected in all investigated teleost genomes that have been mapped to chromosomes or assembled into linkage groups (see supporting data, doi:10.6084/m9.figshare.9596285). It is still unclear whether this translocation occurred before 3R, in which case otocephalan and neoteleost fishes have preserved and lost different 3R-duplicates of CHST16, or whether the colocalization of CHST16 and CHST1a in otocephalan fishes, and of CHST16 and CHST1b in neoteleost fishes, represents two independent translocation events. Chromosome-level genome assemblies from earlier-diverging teleost lineages could help clarify this.
In addition to 3R, there have been more recent whole-genome duplications in cyprinid fishes and in salmonid fishes (Glasauer and Neuhauss 2014), occasionally called 4R. We concluded that the salmonid 4R contributed duplicates of CHST1a, CHST1b, CHST16, and CHST2b (figs. 6 and 7), raising the number of C6OST genes in this lineage to a total of 11. We identified additional duplicates of CHST16, CHST2a, CHST2b, CHST3b, CHST5, and CHST7 in the goldfish genome that likely arose in the cyprinid whole-genome duplication (not shown in fig. 7), as well as copies of CHST1b and CHST3a of an uncertain origin, raising the number of C6OST genes in this species to a total of 17. Other notable gene expansions within the teleost fishes include the local expansion of CHST5 genes in several lineages, such as killifishes and live-bearers, cichlids and osteoglossiform fishes.
Functional Considerations
The interest in the evolution of C6OSTs mainly revolves around their key roles as modulators of extracellular matrix components during the development of the brain and skeletal system. Our results suggest that four C6OST family members were already present before the divergence of jawless and jawed vertebrates, early in vertebrate evolution, and that there was only a modest expansion by two additional family members in 1R/2R. This stands in contrast to several gene families whose expansions in 1R/2R have been considered essential for the evolution of the vertebrate nervous system and skeleton (Holland and Takahashi 2005; Wada 2010), notably the Hox gene family (Sundström et al. 2008; Kuraku and Meyer 2009), and the fibrillar collagen gene family (Boot-Handford and Tuckwell 2003; Zhang and Cohn 2008). Several key gene families involved in the regulation of bone homeostasis also expanded through 1R/2R, including the parathyroid hormone gene family, the calcitonin genes (CALC), and their cognate receptor genes (Hwang et al. 2013), as well as several bone morphogenic protein genes (Marques et al. 2016; Feiner et al. 2019).
The extracellular matrix of the central nervous system forms perineuronal nets containing CS proteoglycans, which have roles in brain plasticity and memory (Foscarin et al. 2017). CS molecules are also important components of proteoglycans in cartilage, and the CS sulfotransferases, encoded by CHST3 and CHST7, have been implicated in the development and homeostasis of the skeletal system and brain. Interestingly however, CHST3 and CHST7 are not closely related, which suggests that chondroitin 6-O sulfation activity has evolved at least twice. It is also notable that CHST7 is related to CHST2, which is involved in the sulfation of KS. KS is important for the development and homeostasis of the brain as well as visual system, and KS sulfotransferases, encoded by CHST1, CHST2, and CHST5, are implicated in corneal function as well as in the developing and adult brain (Parfitt et al. 2011; Hoshino et al. 2014; Takeda-Uchimura et al. 2015; Narentuya et al. 2019). KS may also have a role in the skeletal system, in the maintenance of cartilage (Hayashi et al. 2011).
Because sulfotransferases generate the active GAGs that are subsequently combined into proteoglycans, building the scaffold for tissue formation, our results raise the question of whether extracellular matrix-modifying enzymes such as sulfotransferases were important prerequisites before genes specifically dedicated toward skeleton and brain formation could arise. As an important first step to understand the evolution of extracellular matrix enzyme genes and their cognate functions, it is essential to consider both phylogenetic and chromosomal synteny data from a wide selection of species. Together with comparative studies on gene expression and substrate specificity, as well as investigations of the extracellular matrix components in different tissues during the animals’ lifetime, these evolutionary insights may illuminate the emergence, evolution, and extant development of key vertebrate innovations, as well as inform comparative models of human disease.
Supplementary Material
Acknowledgments
This work was supported by Vetenskapsrådet (the Swedish Research Council) through an International Postdoc Grant awarded to D.O.D. (2016-00552) and a Starting Grant awarded to T.H. (621-2012-4673). We wish to thank the G10K Consortium and professor Erich D. Jarvis of Rockefeller University, New York, for access to genome assemblies from several key species, (platypus, greater horseshoe bat, Anna’s hummingbird, kakapo, zebra finch, two-lined caecilian, and thorny skate) through the Vertebrate Genomes Project (http://vertebrategenomesproject.org) (Rhie et al. forthcoming). We also acknowledge Wen Wang of the Northwestern Polytechnical University of Xi’an, China, Yong Zhang of the Institute of Zoology, Chinese Academy of Sciences, as well as Shigeru Kuratani and Juan Pascual-Anaya of the RIKEN Cluster for Pioneering Research, Japan, for access to the early draft of the inshore hagfish genome.
Data deposition: This project has been deposited in figshare with the Digital Object Identifier (DOI) 10.6084/m9.figshare.9596285.
Literature Cited
- Abdullayev I, Kirkham M, Björklund ÅK, Simon A, Sandberg R.. 2013. A reference transcriptome and inferred proteome for the salamander Notophthalmus viridescens. Exp Cell Res. 319(8):1187–1197. [DOI] [PubMed] [Google Scholar]
- Abril JF, Castelo R, Guigó R.. 2005. Comparison of splice sites in mammals and chicken. Genome Res. 15(1):111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akama TO, Misra AK, Hindsgaul O, Fukuda MN.. 2002. Enzymatic synthesis in vitro of the disulfated disaccharide unit of corneal keratan sulfate. J Biol Chem. 277(45):42505–42513. [DOI] [PubMed] [Google Scholar]
- Akama TO, et al. 2000. Macular corneal dystrophy type I and type II are caused by distinct mutations in a new sulphotransferase gene. Nat Genet. 26(2):237–241. [DOI] [PubMed] [Google Scholar]
- Akama TO, et al. 2001. Human corneal GlcNAc 6-O-sulfotransferase and mouse intestinal GlcNAc 6-O-sulfotransferase both produce keratan sulfate. J Biol Chem. 276(19):16271–16278. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Amemiya CT, et al. 2013. The African coelacanth genome provides insights into tetrapod evolution. Nature 496(7445):311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A, Catchen JM, Ferrara A, Fontenot Q, Postlethwait JH.. 2011. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics 188(4):799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O.. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 55(4):539–552. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Gil M, Dufayard J-F, Dessimoz C, Gascuel O.. 2011. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst Biol. 60(5):685–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot C, et al. 2014. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun. 5:3657.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian C, et al. 2016. The Asian arowana (Scleropages formosus) genome provides new insights into the evolution of an early lineage of teleosts. Sci Rep. 6:24501.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bistrup A, et al. 1999. Sulfotransferases of two specificities function in the reconstitution of high endothelial cell ligands for l-selectin. J Cell Biol. 145(4):899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bistrup A, et al. 2004. Detection of a sulfotransferase (HEC-GlcNAc6ST) in high endothelial venules of lymph nodes and in high endothelial venule-like vessels within ectopic lymphoid aggregates. Am J Pathol. 164(5):1635–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boot-Handford RP, Tuckwell DS.. 2003. Fibrillar collagen: the key to vertebrate evolution? A tale of molecular incest. Bioessays 25(2):142–151. [DOI] [PubMed] [Google Scholar]
- Braasch I, et al. 2016. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet. 48(4):427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canese K, et al. 2017. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 46:D8–D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens N, et al. 2016. Novel mutation in the CHST6 gene causes macular corneal dystrophy in a black South African family. BMC Med Genet. 17:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, et al. 2019. De Novo assembly of the goldfish (Carassius auratus) genome and the evolution of genes after whole genome duplication. Sci Adv. 5(6):eaav0547. [DOI] [PMC free article] [PubMed]
- Christensen KA, et al. 2018. The Arctic Char (Salvelinus alpinus) genome and transcriptome assembly. PLoS One. 13(9):e0204076. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dehal P, Boore JL.. 2005. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 3(10):e314.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, et al. 2018. A phylogenomic framework and timescale for comparative studies of tunicates. BMC Biol. 16:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, et al. 2017. The pearl oyster Pinctada fucata martensii genome and multi-omic analyses provide insights into biomineralization. GigaScience 6(8):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S, et al. 2019. The Pfam protein families database in 2019. Nucleic Acids Res. 47(D1):D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiner N, Motone F, Meyer A, Kuraku S.. 2019. Asymmetric paralog evolution between the “cryptic” gene Bmp16 and its well-studied sister genes Bmp2 and Bmp4. Sci Rep. 9:3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foscarin S, Raha-Chowdhury R, Fawcett JW, Kwok J.. 2017. Brain ageing changes proteoglycan sulfation, rendering perineuronal nets more inhibitory. Aging 9(6):1607–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuta M, Kobayashi Y, Uchimura K, Kimata K, Habuchi O.. 1998. Molecular cloning and expression of human chondroitin 6-sulfotransferase. Biochim Biophys Acta Gene Struct Expr. 1399(1):57–61. [DOI] [PubMed] [Google Scholar]
- Fukuta M, et al. 1995. Molecular cloning and expression of chick chondrocyte chondroitin 6-sulfotransferase. J Biol Chem. 270(31):18575–18580. [DOI] [PubMed] [Google Scholar]
- Gilbert SF, Corfe I.. 2013. Turtle origins: picking up speed. Dev Cell 25(4):326–328. [DOI] [PubMed] [Google Scholar]
- Glasauer SMK, Neuhauss S.. 2014. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol Genet Genomics. 289(6):1045–1060. [DOI] [PubMed] [Google Scholar]
- Habicher J, et al. 2015. Chondroitin/dermatan sulfate modification enzymes in zebrafish development. PLoS One 10(3):e0121957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habuchi H, et al. 2003. Biosynthesis of heparan sulphate with diverse structures and functions: two alternatively spliced forms of human heparan sulphate 6-O-sulphotransferase-2 having different expression patterns and properties. Biochem J. 371(1):131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habuchi O. 2014. Carbohydrate (chondroitin 6) sulfotransferase 3; carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 (CHST3, 7) In: Taniguchi N, Honke K, Fukuda M, editors. Handbook of glycosyltransferases and related genes. Tokyo (Japan: ): Springer; p. 979–987. [Google Scholar]
- Hayashi M, Kadomatsu K, Kojima T, Ishiguro N.. 2011. Keratan sulfate and related murine glycosylation can suppress murine cartilage damage in vitro and in vivo. Biochem Biophys Res Commun. 409(4):732–737. [DOI] [PubMed] [Google Scholar]
- Hemmerich S, Bistrup A, et al. 2001. Sulfation of l-selectin ligands by an HEV-restricted sulfotransferase regulates lymphocyte homing to lymph nodes. Immunity 15(2):237–247. [DOI] [PubMed] [Google Scholar]
- Hemmerich S, Lee JK, et al. 2001. Chromosomal localization and genomic organization for the galactose/N-acetylgalactosamine/N-acetylglucosamine 6-O-sulfotransferase gene family. Glycobiology 11(1):75–87. [DOI] [PubMed] [Google Scholar]
- Holland PWH, Takahashi T.. 2005. The evolution of homeobox genes: implications for the study of brain development. Brain Res Bull. 66(4–6):484–490. [DOI] [PubMed] [Google Scholar]
- Hoshino H, et al. 2014. KSGal6ST is essential for the 6-sulfation of galactose within keratan sulfate in early postnatal brain. J Histochem Cytochem. 62(2):145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J-I, et al. 2013. Expansion of secretin-like G protein-coupled receptors and their peptide ligands via local duplications before and after two rounds of whole-genome duplication. Mol Biol Evol. 30(5):1119–1130. [DOI] [PubMed] [Google Scholar]
- Iwata H, Gotoh O.. 2011. Comparative analysis of information contents relevant to recognition of introns in many species. BMC Genomics. 12(1):45.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M, et al. 2007. The medaka draft genome and insights into vertebrate genome evolution. Nature 447(7145):714–719. [DOI] [PubMed] [Google Scholar]
- Kitagawa H, Fujita M, Ito N, Sugahara K.. 2000. Molecular cloning and expression of a novel chondroitin 6-O-sulfotransferase. J Biol Chem. 275(28):21075–21080. [DOI] [PubMed] [Google Scholar]
- Kitts PA, et al. 2016. Assembly: a resource for assembled genomes at NCBI. Nucleic Acids Res. 44(D1):D73–D80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjellén L, Lindahl U.. 1991. Proteoglycans: structures and Interactions. Annu Rev Biochem. 60(1):443–475. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, et al. 2010. Functional analysis of chick heparan sulfate 6-O-sulfotransferases in limb bud development. Dev Growth Differ. 52(2):146–156. [DOI] [PubMed] [Google Scholar]
- Kuraku S. 2010. Palaeophylogenomics of the vertebrate ancestor—impact of hidden paralogy on hagfish and lamprey gene phylogeny. Integr Comp Biol. 50(1):124–129. [DOI] [PubMed] [Google Scholar]
- Kuraku S. 2013. Impact of asymmetric gene repertoire between cyclostomes and gnathostomes. Semin Cell Dev Biol. 24(2):119–127. [DOI] [PubMed] [Google Scholar]
- Kuraku S, Meyer A.. 2009. The evolution and maintenance of Hox gene clusters in vertebrates and the teleost-specific genome duplication. Int J Dev Biol. 53(5–6):765–773. [DOI] [PubMed] [Google Scholar]
- Kuraku S, Meyer A, Kuratani S.. 2009. Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol Biol Evol. 26(1):47–59. [DOI] [PubMed] [Google Scholar]
- Kusche-Gullberg M, Kjellén L.. 2003. Sulfotransferases in glycosaminoglycan biosynthesis. Curr Opin Struct Biol. 13(5):605–611. [DOI] [PubMed] [Google Scholar]
- Lagman D, et al. 2013. The vertebrate ancestral repertoire of visual opsins, transducin alpha subunits and oxytocin/vasopressin receptors was established by duplication of their shared genomic region in the two rounds of early vertebrate genome duplications. BMC Evol Biol. 13(1):238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large data sets. Bioinformatics 30(22):3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Bhakta S, Rosen SD, Hemmerich S.. 1999. Cloning and characterization of a mammalian N-acetylglucosamine-6-sulfotransferase that is highly restricted to intestinal tissue. Biochem Biophys Res Commun. 263(2):543–549. [DOI] [PubMed] [Google Scholar]
- Li X, Tedder TF.. 1999. CHST1 and CHST2 sulfotransferases expressed by human vascular endothelial cells: cDNA cloning, expression, and chromosomal localization. Genomics. 55(3):345–347. [DOI] [PubMed] [Google Scholar]
- Lien S, et al. 2016. The Atlantic salmon genome provides insights into rediploidization. Nature 533(7602):200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques CL, et al. 2016. Comparative analysis of zebrafish bone morphogenetic proteins 2, 4 and 16: molecular and evolutionary perspectives. Cell Mol Life Sci. 73(4):841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta TK, et al. 2013. Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum). Proc Natl Acad Sci U S A. 110(40):16044–16049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, van de Peer Y.. 2005. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). BioEssays 27(9):937–945. [DOI] [PubMed] [Google Scholar]
- Minh BQ, Nguyen MAT, Von Haeseler A.. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata S, Kitagawa H.. 2016. Chondroitin 6-sulfation regulates perineuronal net formation by controlling the stability of aggrecan. Neural Plast. 2016:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai N, Kimata K.. 2014. Heparan-sulfate 6-O-sulfotransferase 1-3 (HS6ST1-3) In: Taniguchi N, Honke K, Fukuda M, editors. Handbook of glycosyltransferases and related genes. Tokyo (Japan: ): Springer; p. 1067–1080. [Google Scholar]
- Nakagawa S, Niimura Y, Gojobori T, Tanaka H, Miura K-I.. 2008. Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res. 36(3):861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, McLysaght A.. 2017. Genomes as documents of evolutionary history: a probabilistic macrosynteny model for the reconstruction of ancestral genomes. Bioinformatics 33(14):i369–i378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Takeda H, Kohara Y, Morishita S.. 2007. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 17(9):1254–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narentuya, et al. 2019. GlcNAc6ST3 is a keratan sulfate sulfotransferase for the protein-tyrosine phosphatase PTPRZ in the adult brain. Sci Rep. 9:4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NTT, Vincens P, Crollius HR, Louis A.. 2018. Genomicus 2018: karyotype evolutionary trees and on-the-fly synteny computing. Nucleic Acids Res. 46(D1):D816–D822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogami K, et al. 2004. Distinctive expression patterns of heparan sulfate O-sulfotransferases and regional differences in heparan sulfate structure in chick limb bluds. J Biol Chem. 279(9):8219–8229. [DOI] [PubMed] [Google Scholar]
- Ocampo Daza D, Larhammar D.. 2018. Evolution of the growth hormone, prolactin, prolactin 2 and somatolactin family. Gen Comp Endocrinol. 264:94–112. [DOI] [PubMed] [Google Scholar]
- Parfitt GJ, et al. 2011. Electron tomography reveals multiple self-association of chondroitin sulphate/dermatan sulphate proteoglycans in Chst5-null mouse corneas. J Struct Biol. 174(3):536–541. [DOI] [PubMed] [Google Scholar]
- Pasquier J, et al. 2016. Gene evolution and gene expression after whole genome duplication in fish: the PhyloFish database. BMC Genomics. 17(1):368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry E, et al. 2018. Ensembl 2019. Nucleic Acids Res. 47:D745–D751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosdocimi F, Linard B, Pontarotti P, Poch O, Thompson JD.. 2012. Controversies in modern evolutionary biology: the imperative for error detection and quality control. BMC Genomics. 13(1):5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, et al. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature 453(7198):1064–1071. [DOI] [PubMed] [Google Scholar]
- Rhie A, et al. Forthcoming. Towards complete and error-free genome assemblies of all vertebrate species. [DOI] [PMC free article] [PubMed]
- Rubinstein Y, et al. 2016. Macular corneal dystrophy and posterior corneal abnormalities. Cornea 35(12):1605–1610. [DOI] [PubMed] [Google Scholar]
- Sacerdot C, Louis A, Bon C, Berthelot C, Roest Crollius H.. 2018. Chromosome evolution at the origin of the ancestral vertebrate genome. Genome Biol. 19(1):166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seko A, et al. 2009. N-Acetylglucosamine 6-O-sulfotransferase-2 as a tumor marker for uterine cervical and corpus cancer. Glycoconj J. 26(8):1065–1073. [DOI] [PubMed] [Google Scholar]
- Session AM, et al. 2016. Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538(7625):336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedko SV. 2019. Assembly ASM291031v2 (Genbank: GCA_002910315.2) identified as assembly of the Northern Dolly Varden (Salvelinus malma malma) genome, and not the Arctic char (S. alpinus) genome. arXiv:1912.02474.
- Small CM, et al. 2016. The genome of the Gulf pipefish enables understanding of evolutionary innovations. Genome Biol. 17(1):258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Keinath MC.. 2015. The sea lamprey meiotic map improves resolution of ancient vertebrate genome duplications. Genome Res. 25(8):1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, et al. 2013. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. 45(4):415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares da Costa D, Reis RL, Pashkuleva I.. 2017. Sulfation of glycosaminoglycans and its implications in human health and disorders. Annu Rev Biomed Eng. 19(1):1–26. [DOI] [PubMed] [Google Scholar]
- Srivastava P, Pandey H, Agarwal D, Mandal K, Phadke SR.. 2017. Spondyloepiphyseal dysplasia Omani type: CHST3 mutation spectrum and phenotypes in three Indian families. Am J Med Genet. 173(1):163–168. [DOI] [PubMed] [Google Scholar]
- Sundström G, Larsson TA, Larhammar D.. 2008. Phylogenetic and chromosomal analyses of multiple gene families syntenic with vertebrate Hox clusters. BMC Evol Biol. 8(1):254.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda-Uchimura Y, et al. 2015. Requirement of keratan sulfate proteoglycan phosphacan with a specific sulfation pattern for critical period plasticity in the visual cortex. Exp Neurol. 274:145–155. [DOI] [PubMed] [Google Scholar]
- Akama TO, Fukuda MN. 2014. Carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5 and 6 (CHST5, 6). In: Taniguchi N, Honke K, Fukuda M, editors. Handbook of glycosyltransferases and related genes. Tokyo, Japan: Springer. p 1005–1014. [Google Scholar]
- Tetsukawa A, Nakamura J, Fujiwara S.. 2010. Identification of chondroitin/dermatan sulfotransferases in the protochordate, Ciona intestinalis. Comp Biochem Physiol B Biochem Mol Biol. 157(2):205–212. [DOI] [PubMed] [Google Scholar]
- Tsutsumi K, Shimakawa H, Kitagawa H, Sugahara K.. 1998. Functional expression and genomic structure of human chondroitin 6-sulfotransferase. FEBS Lett. 441(2):235–241. [DOI] [PubMed] [Google Scholar]
- Wada H. 2010. Origin and genetic evolution of the vertebrate skeleton. Zoolog. Sci. 27(2):119–123. [DOI] [PubMed] [Google Scholar]
- Waryah AM, et al. 2016. A novel CHST3 allele associated with spondyloepiphyseal dysplasia and hearing loss in Pakistani kindred. Clin Genet. 90(1):90–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Sugahara K, Özbek S.. 2011. Evolution of glycosaminoglycans: comparative biochemical study. Commun Integr Biol. 4(2):150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Takahashi I, Ogata N, Nakazawa K.. 2001. Purification and characterization of N-acetylglucosaminyl sulfotransferase from chick corneas. Arch Biochem Biophys. 392(1):87–92. [DOI] [PubMed] [Google Scholar]
- Yusa A, Kitajima K, Habuchi O.. 2006. N-linked oligosaccharides on chondroitin 6-sulfotransferase-1 are required for production of the active enzyme, Golgi localization, and sulfotransferase activity toward keratan sulfate. J Biol Chem. 281(29):20393–20403. [DOI] [PubMed] [Google Scholar]
- Zhang G, Cohn MJ.. 2008. Genome duplication and the origin of the vertebrate skeleton. Curr Opin Genet Dev. 18(4):387–393. [DOI] [PubMed] [Google Scholar]
- Zhang Z, et al. 2017. Deficiency of a sulfotransferase for sialic acid-modified glycans mitigates Alzheimer’s pathology. Proc Natl Acad Sci U S A. 114:E2947–E2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.