Abstract
Conditional knockout (cKO) mice are extremely valuable for biomedical research because they enable detailed analyses of gene functions in a tissue- or temporally-specific fashion. The conventional method for generating cKO mice is time consuming and labor intensive, which involves making a large gene-targeting construct, transfecting and screening many embryonic stem (ES) cell clones, injecting positive ES clones into blastocysts to produce chimeric mice, and breeding the chimeras to transmit the targeted gene through the germline. Recently developed CRISPR technology has revolutionized the way genetically engineered organisms are created. Knockout and knockin mice can now be made by directly injecting zygotes with Cas9, sgRNA, and donor DNA. In theory, cKO mice can be generated by simultaneously inserting two loxP sites using two sgRNAs and two oligonucleotides as donors, but in practice the probability of obtaining cKO mice in one step is still very low, partly because the efficiency of oligo-mediated knockin is much lower than non-homologous end joining (NHEJ) and partly because co-cutting juxtaposed sites in one allele at the same time often leads to the deletion of the entire fragment between the two cutting sites. Therefore, many laboratories prefer to insert the two loxP sites sequentially, i.e., generating mice with one loxP first and then use embryos collected from these mice to insert the second loxP site. In this chapter, we describe our procedures and timeline using this sequential method to make a Six6 cKO mouse line as a demonstration of its feasibility.
Keywords: Microinjection, CRISPR, Conditional knockout, loxP, Embryo transfer, Sequential method, Six6
1. Introduction
The knockout mouse technology has played a fundamentally important role in biomedical research, and Drs. Martin Evans [1], Oliver Smithies [2–4], and Mario Capecchi [5, 6] were recognized with a Nobel Prize in 2007 for their ground breaking work in developing this technology. The so-called conditional knockout (cKO) approach has further increased the versatility of these methods by enabling gene knockout in a tissue-specific or temporally-specific manner [7]. Traditionally, cKO mice are generated by electroporating a large gene-targeting construct into germline-competent embryonic stem (ES) cells. After screening a few hundred ES cell clones, the clones with the correctly targeted gene are microinjected into blastocyst-stage embryos for producing chimeric mice. The chimeras are then bred with wild-type mice to pass the targeted gene through the germline. Next, germline transmitted heterozygous mice are crossed with a mouse line expressing a recombinase (often Flp but sometimes Cre) for removing the selection marker gene (often neomycin-resistance gene) which was required for selecting ES cell clones earlier. Only after that, the floxed mice can be crossed with Cre-driver lines for conducting tissue-specific knockout studies.
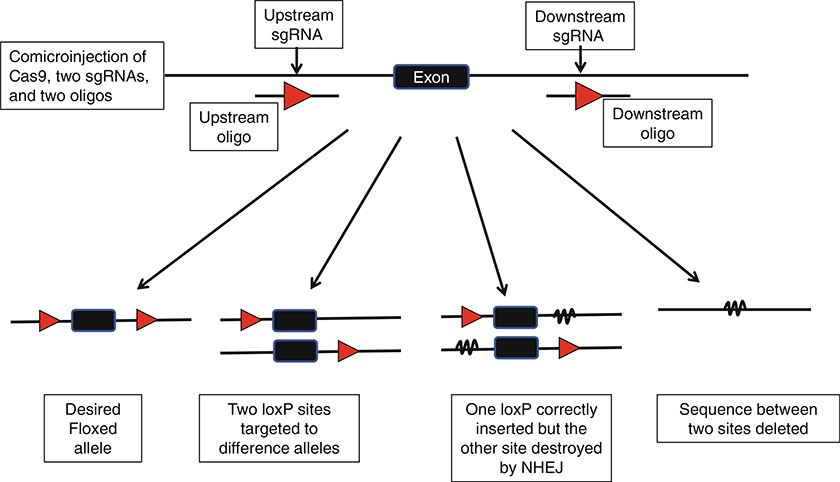
Recently developed CRISPR technology is a revolutionary method that has produced a profound impact on biomedical research [8–10]. It has dramatically speeded up the process for creating knockout and knockin mice [11], and has also been successfully used for cKO mouse generation [12]. However, in practice, it still remains challenging to insert simultaneously both loxP sites into the same allele. As depicted in Fig. 1, several possible outcomes exist when two juxtaposed sites are targeted simultaneously. At this time, the efficiency for oligonucleotide-mediated knockin is still much lower than NHEJ, so the probability of obtaining mice with both loxP insertions is still low. Most mice with one correct loxP often have indels (deletions or insertions) at the other sgRNA cutting site, which is counterproductive because this renders the mice useless for subsequent use to insert the second loxP site. In the cases when both sites are successfully knocked in, the two loxPs are inserted into different alleles in about half of founder mice. Another outcome is that the entire sequence between the two cutting sites is deleted, which occurs at a surprisingly high efficiency. If each Cas9/sgRNA complex makes the cut and the cut is immediately repaired by NHEJ or HDR as an independent event, one would expect that big deletions only happen somewhat rarely when the two CRISPR sgRNAs cut at the exact same moment. However, in reality such big deletions occur frequently, impling that Cas9/sgRNA complexes that sit in juxtaposed positions may not cut independent of each other, or that they do not repair double-strand break immediately after cutting.
Fig. 1.
Possible outcomes of co-injecting CRISPR reagents for simultaneously inserting both loxP sites
Given the challenges of simultaneously inserting both loxP sites using two sgRNAs and two oligos, various CRISPR-mediated strategies have been developed for generating tissue-specific knockout mice, including tissue-specific expression of Cas9 through Cre-activated gene expression [13] or tissue-specific promoters [14], inserting a Cre-regulated artificial small intron into the gene [15]; electroporating the two loxP sites into zygotes and 2-cell embryos on 2 consecutive days [16], and using a long single stranded DNA that contains both loxP sites as a donor [17, 18]. However, these methods are still considered at the exploratory stage, and many transgenic facilities are still using the sequential method to insert the two loxP sites in two steps, i.e., generating mice with one loxP first and then use these carrier mice as embryo donors for microinjection to insert the second loxP site. Even though, the sequential approach requires one or two (if breeding is required for expanding the colony with one loxP) generations of breeding, it is advantageous over the traditional ES cell-mediated method for the following reasons: (1) loxP sites can be inserted using single-stranded oligonucleotides and consequently eliminate the need for making a large gene-targeting construct; (2) CRISPR reagents and donor DNAs can be directly microinjected into fertilized eggs and therefore eliminate the need for using ES cells which not only saves time and labor but also removes much of the uncertainty associated with germline competency of the final ES clones; (3) CRISPR method does not require generating chimeric mice nor crossing with Flp (sometimes Cre) mice for removing the selection marker gene, and therefore save two generations of mouse breeding.
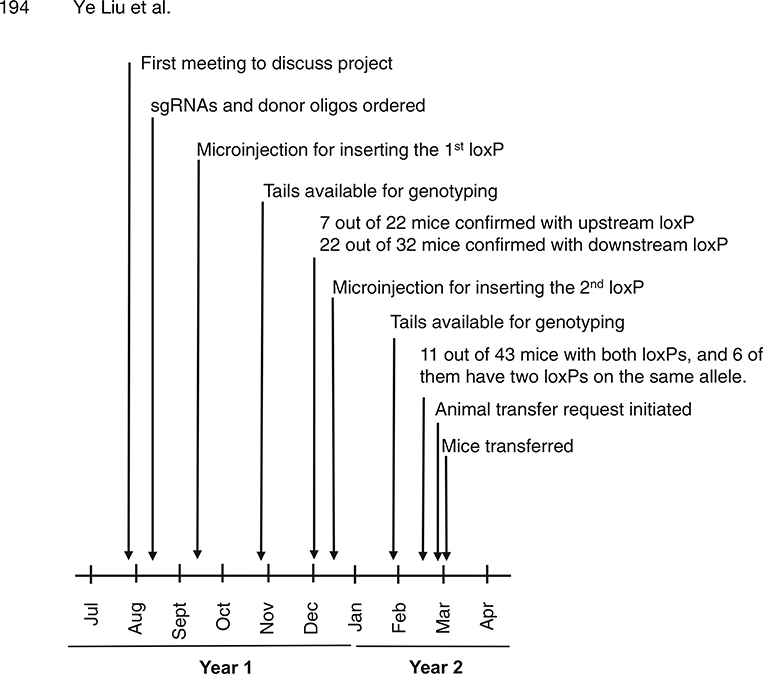
In this chapter, we describe the generation of mice carrying a floxed allele of the Six6 gene as an example of our procedures and timeline (Fig. 2) and as a demonstration of the feasibility of using the CRISPR/Cas9 technology to sequentially insert two loxP sites.
Fig. 2.
Timeline for the generation of the Six6 cKO mice by sequentially inserting the two loxP sites
2. Materials
2.1. Mice
Male and female B6D2F1 mice (The Jackson Laboratory) as embryo donors; CD-1 (Charles River) vasectomized males and CD-1 females for producing foster mothers (see Note 1 for choosing mouse strains).
2.2. Equipment and Tools for Microinjection and Mouse Surgery
Zeiss Axiovert S100 inverted microscope with 5×, 10×, 20×, and 40× objectives.
Two Eppendorf TransferMan NK-2 micromanipulators.
Eppendorf FemtoJet4i.
Eppendorf CellTram Vario.
Air table.
Sutter P-97 horizontal pipette puller.
Stereo microscopes for embryo collection and embryo transfer procedures.
CO2 incubators.
Autoclave machine for sterilizing surgical instruments.
Hot beads sterilizer for sterilizing instruments between animals when more than one animal is used during a session.
Mouth pipette for transferring embryos.
Hair clipper.
Wound clip applier.
Suturing needle holder.
Eppendorf VacuTip holding pipette.
5–0 Vicryl suture.
26–29 G disposable hypodermic needles.
T/Pump heating water blanket.
Povidone-Iodine swabsticks.
Micro dissecting scissors.
Iris forceps.
Micro dissecting tweezers #5.
Dieffenbach micro clamp.
Surgical instruments sterilization tray.
Microloader pipette tips.
Alcohol swab.
Sterile surgical gloves.
Glass capillary tubing for pulling microinjection needles: Borosil, 1.0 mm OD × 0.75 mm ID with omega dot fiber for rapid filling.
2.3. Media and Reagents for Embryo Culture and Animal Surgery
M2 culture medium.
M16 culture medium.
Ketamine/Xylazine anesthetics: mixing 0.2 mL Ketamine stock solution (100 mg/mL) and 0.1 mL Xylazine stock solution (20 mg/mL) with 1.7 mL 0.9% sodium chloride solution before ip injection (see Note 2).
Pregnant mare’s serum gonadotropin (PMSG).
Human chorionic gonadotropin (hCG).
Hyaluronidase: 10× concentrated solution is prepared by dissolving 100 mg in 20 mL M2 medium. Filter through a 0.22 μm filter, aliquot into microcentrifuge tubes, and store at −20° C in the dark.
Embryo culture-tested mineral oil.
Bupivacaine: 2.5 mg/mL solution.
Meloxicam: 5 mg/mL solution.
2.4. Reagents for CRISPR/Cas9Mediated Gene-Editing
Cas9 mRNA.
-
sgRNAs:
Six-Up (ACCTTAACTGCACGGCATTG) for inserting the upstream loxP.
Six-In (GCATAGGCCGACTAGCTTGC) for inserting the loxP in intron 1.
-
Donor single-stranded DNA oligos for using with corresponding sgRNAs to insert the upstream and downstream loxP sequences.
Six-Up: CTCTGCAGGAGGGGTCAGTGACCCGTGGCTCTGACCAATCTACCTTGCCACCTTAACTGCACGGCATAACTTCGTATAATGTATGCTATACGAAGTTATGAATTCATTGTGGGATGCAGCCTGCACTTTATCTTGCTGGCACTTCTGATCTAAAA.
Six-In: GTTTGGGCCTGAGTTGCAAGCTCCAAAAGCTGGCGAATGAAAAGCATAGGCCGACTAGCGGATCCATAACTTCGTATAGCATACATTATACGAAGTTATTTGCTGGGTGGAGACAGGATAGTTGTCATGAGGAAACACGTCTTTCTGCCTCTTAGAATC.
Microinjection buffer: 10 mM Tris–HCl (pH 7.5), 0.1 mMEDTA, 100 mM NaCl.
2.5. Genotyping Equipment and Reagents
PCR thermal cycler.
Electrophoresis power supply.
Horizontal DNA gel electrophoresis chamber.
Gel imaging system.
Agarose.
1× TBE electrophoresis buffer: 89 mM Tris–borate, 2 mM EDTA, pH 8.3.
Ethidium bromide (EtBr) stock solution: 10 mg/mL.
Alkaline lysis buffer: 50 mM NaOH.
Tail neutralization buffer: 1 M Tris–HCl, pH 7.4.
Proteinase K.
Taq DNA polymerase: usually contained in a master mix containing Taq, reaction buffer, MgCl2, dNTPs, and loading dyes.
10× standard Taq reaction buffer with 15 mM MgCl2.
100 mM dNTPs (if not included in PCR master mix).
EcoRI, BamHI, and 10× restriction reaction buffers.
1 Kb DNA ladder.
Gel loading dye (if not included in PCR master mix).
PCR DNA purification kit.
Genomic DNA purification kit.
Phenol:Chloroform:Isoamyl Alcohol 25:24:1 saturated with10 mM Tris, pH 8.0, 1 mM EDTA.
Sodium acetate solution (3 M), pH 5.2.
Isopropanol (optional).
Ethanol.
5 M Betaine.
Phusion high fidelity DNA polymerase.
-
PCR primers for genotyping the upstream loxP site (Six-Up):
Sixup-F100 (common forward), 5′-TTGGCCAGAAGGCAG AGTGC-3′;
Sixup-R26 (common reverse), 5′-AGATCAGAAGTGCCA GCAAG-3′;
Sixup-EIR (allele-specific reverse), 5′-CAGGCTGCATCCC$ ACAATGAAT-3′.
-
PCR primers for genotyping the downstream intronic loxP site
(Six-In):
Sixin-F56 (common forward), 5′-CTAAGAGGCAGAAA GACGTG-3′;
Sixin-R63 (common reverse), 5′-CTGAGGCTAGAGGAT GCTTG-3′;
Sixin-BIR (allele-specific reverse), 5′-CATAGGCCGACTA GCGGATC-3′.
For flox allele identification, Sixin-BIR is paired with Sixup-lxpF (forward): 5′-TTAACTGCACGGCATAACTTCG-3′.
3. Methods
3.1. Designing and Generating CRISPR Reagents
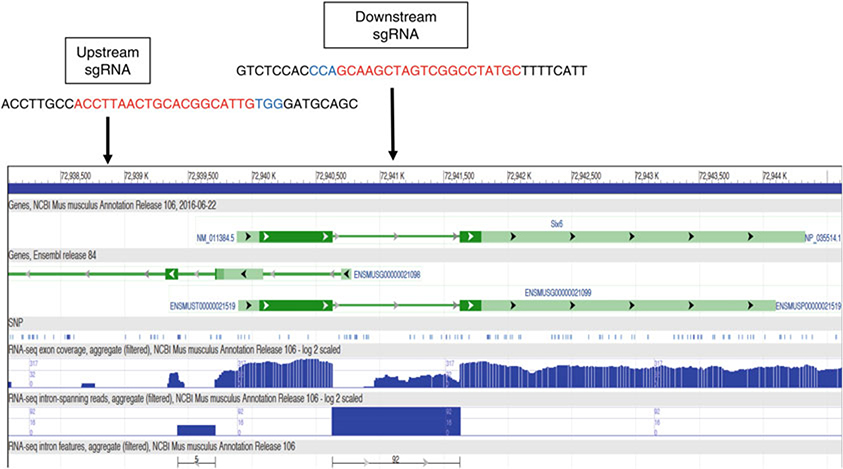
Selection of loxP insertion sites: A general strategy for creating cKO mouse lines is to flox an exon around the 5′ end/translational start codon of the gene with two loxP sites, but floxing the first exon is usually avoided because inserting a loxP site upstream of the first exon risks disrupting the promoter function and therefore affects gene expression before Cre-mediated deletion occurs (see Note 3). However, as shown in Fig. 3, the mouse Six6 gene contains only two exons and the majority of the coding sequence is contained in exon 1, therefore making it necessary to flox the first exon. In order to minimize the chance of disrupting promoter function, the upstream regions across difference vertebrate classes were aligned for avoiding inserting loxP into highly conserved regions.
After selecting the insertion regions, sequences are submitted to MIT’s CRISPR design website (http://crispr.mit.edu) for searching and analyzing possible sgRNAs. Ideal sgRNAs should have a score of >60 (higher score indicating fewer highly homologous sequences throughout the genome and consequently lowering the chance of off-target activity), a GC content of 50–70%, and should be devoid of homopolymers (a stretch of the same nucleotide).
Purchase in vitro transcribed (IVT) sgRNAs from Thermo-Fisher, and further purify the sgRNAs by pheno/chloroform extraction and ethanol precipitation.
Design donor oligos: single-stranded DNA oligos are used as donors for inserting the loxP sequences. We prefer to insert the loxP at the exact CRISPR cutting site (between 3 and 4 bp upstream of NGG (PAM) for SpCas9). We usually insert a restriction site adjacent to the loxP to facilitate genotyping. In this particular project, an EcoRI site is included in the Six-Up oligo and a BamHI site is included in the Six-In oligo. On each side of the insert (loxP + restriction site), we include a 50–60 bp homologous arm (see Note 4).
Fig. 3.
Positions of the upstream and downstream CRISPR target sites for inserting the two loxP sites flanking the Exon 1 of the mouse Six6 gene
3.2. Microinjection of CRISPR Reagents for Inserting the First loxP
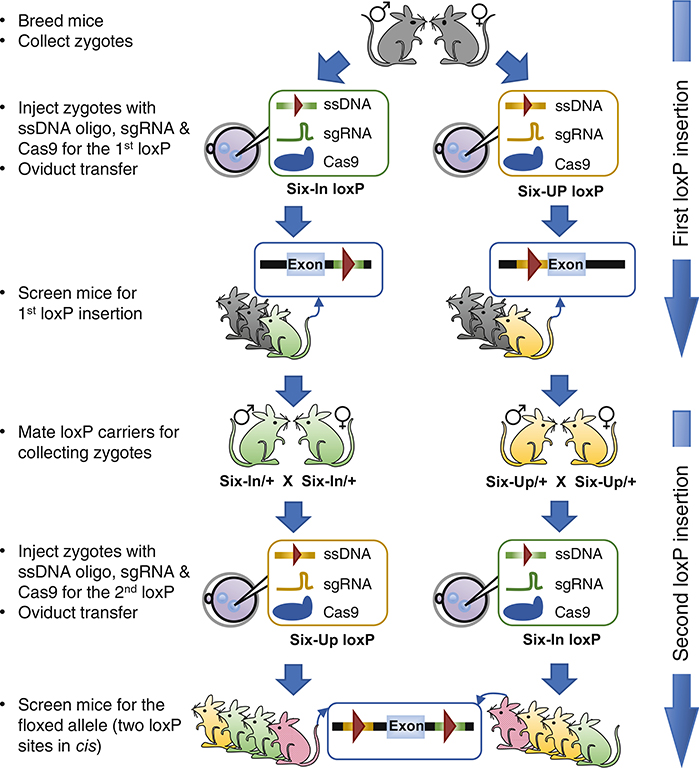
In theory, only the CRISPR reagents (Cas9 mRNA, sgRNA, and the corresponding oligos) targeting one loxP site need to be microinjected for obtaining mice with the first loxP insertion. However, in practice, it is strongly advisable to perform in parallel two separate microinjection experiments, one for each loxP site, as outlined in Fig. 4. Occasionally, some sgRNAs do not work efficiently or may be toxic to embryos, but there is no easy and reliable method to pretest the sgRNA (see Note 5) before microinjection. Injecting the two sgRNAs in parallel enables the identification of bad sgRNAs without causing a severe delay to the project, because as long as one of the two sgRNA works well, the project can proceed while the bad sgRNA is replaced in a repeat attempt for that loxP insertion. If both sgRNAs work efficiently at the first attempt, the mice with either loxP can be used for reciprocal injection of the second loxP site. Our microinjection and embryo transfer procedures are outlined below:
Three days prior to the scheduled microinjection, inject (i.p.) ten B6D2F1 female mice with 5 units of PMS. About 46 h (44–48 h) later, inject the same female mice with 5 units of hCG each. Then each female mouse is paired with an individually caged stud male.
In the next morning, check the females for vaginal plugs. Euthanize plugged females with CO2 and quickly dissect out both oviducts from each female and place all dissected oviducts in a 35 mm culture dish containing M2 medium.
Using a pair of forceps and a 29-G needle tear open the ampulla of each oviduct to allow the cumulus mass to extrude spontaneously into the media.
Add 1/10 volume of concentrated (10×) hyaluronidase solution and incubate for 2–5 min at room temperature so that individual eggs are released from the masses.
Use a mouth operated embryo transfer pipette to pick up the eggs and wash them through three drops of M2 medium. The eggs are now ready for microinjection.
Pull fresh microinjection needles using the P-97 pipette puller and load each needle with 2–3 μL of CRISPR reagents (100 ng/μL Cas9 mRNA, 50 ng/μL of sgRNA, and 100 ng/ μL of oligos dissolved in microinjection buffer) using an Eppendorf microloader tip.
Place a drop of M2 in the injection chamber and cover it under mineral oil. Load 30–40 eggs into the drop using an embryo transfer pipet. Place the injection chamber on the microscope stage.
Use the CellTram vario micromanipulator and holding pipette to grasp an egg. Insert the injection needle into its cytoplasm and quickly deliver the CRISPR reagents by pushing the foot pedal connected to the FemtoJet 4i.
After all the eggs are injected, they are washed through 2–3 M16 drops and then cultured in M16 medium overnight to allow them to develop into 2-cell stage embryos (see Note 6 for alternative embryo transfer strategies).
On the day of microinjection, mate CD-1 female mice with vasectomized males.
Check vaginal plugs in the next morning, and use pluggedCD-1 females as surrogate mothers for embryo transfer.
Anesthetize the recipient mothers by ip injection of diluted Ketamine/Xylazine solution at 10 mL/Kg of body weight. Clip hair off from a generous area on back and disinfect the clipped area by alternating applications of Betadine and 70% alcohol.
Cut open the skin (~10 mm) and muscle body wall (5–10 mm), and use a pair of iris forceps carefully take out the ovary, oviduct, and a portion of the uterine horn.
Under a dissecting microscope, carefully implant about ten injected embryos into one of the oviducts, and repeat the procedure for the oviduct on the other side.
Close the abdominal wall with a cruciate suture utilizing absorbable 5–0 Vicryl suture. Drop one or two drops of 0.25% Bupivacaine solution on the muscle at the surgical site using a 26-G needle.
Close the skin incision with stainless steel surgical wound clips. Inject the mouse with diluted (in 0.9% sodium chloride) Meloxicam (2–5 mg/kg of body weight), which serves as a long-lasting analgesic for alleviating pain.
Place the mouse in a pre-warmed mouse cage placed on a circulating water blanket. The mouse will normally awake 30–60 min after surgery.
Wound clips are removed 10–14 days post-surgery, and pups are usually born 19–20 days post implantation. At weaning (18–20 days old), young mice are marked with ear-tags and their tail tips are cut for genotyping.
Fig. 4.
Schematic outline of experimental procedures for sequentially inserting the two loxP sites in parallel
3.3. Genotyping for the First loxP Site Integration
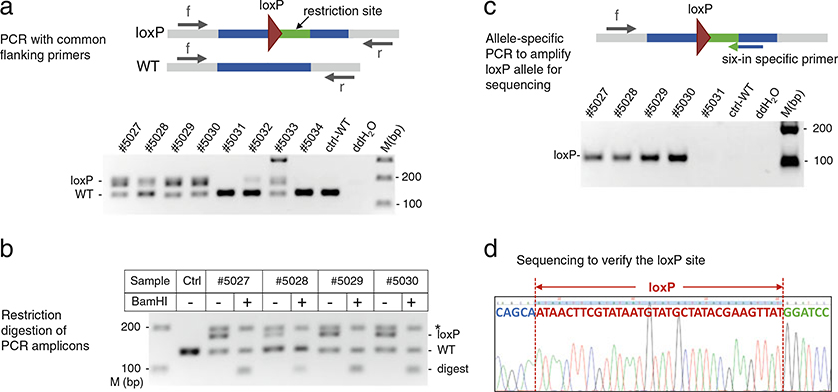
Design a primer pair targeting outside of 5′ and 3′ homology arms of the donor oligo (see Fig. 5). These primers are designated as “common flanking primers,” as they can amplify both wild-type and loxP-integrated alleles (see Note 7).
Place tail tips into 8-strip PCR tubes. Extract genomic DNA using a modified HotSHOT method [19].
Add 100 μL of alkaline lysis buffer to each sample tube and incubate at 95 ° C for 20 min. After cooling at 4 ° C for 5 min, adjust pH by adding 20 μL of tail neutralization buffer and thoroughly mix. The supernatant can be directly used for PCR as DNA template (see Note 8).
Set up a 20 μL PCR reaction for each sample by mixing 10 μL of GoTaq green master mix (2×), 1 μL each of the common primers (10 μM), 1 μL of tail DNA prep, and 7 μL of ddH2O.
Use standard parameters for PCR: initial denaturation at 94 ° C for 2 min, followed by 35 cycles of 94 ° C for 30 s, 58 ° C for 30 s, and 72 ° C for 30 s, and a final extension at 72 ° C for 10 min (see Note 9).
Make a 2.5–3% agarose gel with 1× TBE buffer and 0.5 μg/mL ethidium bromide (see Note 10). Load 6 μL PCR products per well alongside a DNA size marker. Perform electrophoresis until the wild-type (lower band) vs. loxP (upper band) amplicons of expected sizes are well separated. Visualize DNA by UV transilluminator and document gel pictures using a standard gel imaging system (see Fig. 5a lower panel).
To confirm the larger PCR amplicon indeed has loxP site integration, set up restriction digests for upper band-positive samples: mix 8 μL of PCR product, 1.2 μL of 10× Reaction Buffer, 0.5 μL of restriction enzyme (EcoRI for Six-Up; BamHI-HF for Six-In), and 10.3 μL of ddH2O in a 20 μL reaction.
After incubating at 37 ° C for 1–2 h, load each digested (20 μL) and undigested (~6 μL) sample pairwise onto a 3% agarose gel and then conduct electrophoresis. A sample carrying the loxP integrated allele displays a band shift after the restriction digest: the loxP amplicon, previously appearing as the upper band, disappears (or becomes weaker), while its digestion products, predicted as smaller than wild-type, now appear on the gel (see Fig. 5b and Note 11).
Design a loxP allele-specific reverse PCR primer that spans the junction of genomic sequence and the inserted restriction enzyme site, with the 3′ terminal 4–5 nt of the primer matching the restriction enzyme site. This primer thus only recognizes the loxP-integrated but not the wild-type allele. Pair this allele-specific primer with a common forward primer and set up PCR reactions. Amplicons should only appear in loxP-positive but not in wild-type or samples with indels (see Fig. 5c and Note 12).
Purify the loxP-specific PCR amplicon by column-based (e.g., Qiaquick PCR purification kit) or magnetic bead-based (e.g., Agencourt AMPure XP beads) method, following the manufacturer’s protocol. Sequence purified DNA using the common forward primer to confirm the loxP site (see Fig. 5d).
Analyze sequencing results and select the verified loxP-positive mice to use for the second loxP injection. As shown in Table 1 and Fig. 5, of the 140 Six-Up CRISPR injected zygotes, 75 developed to the 2-cell stage and were implanted, which result in a total of 22 live pups. Upon genotyping, eight mice were identified as potential loxP carriers, with three being heterozygous and five hemi- or homozygous for the loxP allele (see Note 13). DNA sequencing analyses showed that all but one mouse have the correct loxP site. Of 144 Six-In CRISPR injected embryos, 99 developed into 2-cell stage and were implanted, 32 pups were born, 22 were identified as loxP carriers including 16 heterozygotes and six hemi- or homozygotes. LoxP sites in all 22 mice were confirmed by sequencing.
Fig. 5.
Identification of mice with insertion of the first loxP site. (a) Upper panel: schematic presentation of the PCR strategy for identifying the integrated loxP site at the target region. Gray bar: genomic DNA; blue bar: regions complementary to oligo’s homology arms; green bar: restriction site; maroon triangle: loxP site; arrows: common forward (f) and reverse primers (r). Lower panel: gel analysis of PCR products of the Six-In locus to separate the 179 bp loxP amplicon from the 138 bp wild-type (WT) amplicon. M (bp), DNA size marker. (b) Restriction digestion (BamHI) of Six-In PCR products to confirm the integration of donor oligos. Ctrl: wild-type; digest: loxP digested fragments of 93 bp and 86 bp; a star (*) points to heteroduplexes formed between wild-type and loxP amplicons. (c) Six-In loxP allele-specific PCR (loxP, 111 bp) using common forward primer F56 and loxP-specific reverse primer Sixin-BIR. Upper panel: schematic indication of the position and direction of PCR primers relative to the inserted loxP. Lower panel: gel analysis of the PCR products. (d) DNA sequencing chromatogram for verifying perfect loxP sequence (maroon)
Table 1.
Summary of targeting experiments for the first loxP site
| Target site | No. of embryos injected/implanted | Mice weaned | No. of loxP carrier mice | Targeting efficiencya |
|---|---|---|---|---|
| Six-Up | 140/75 | 22 | 7 | 7/22 (32%) |
| Six-In | 144/99 | 32 | 22 | 22/32 (69%) |
Targeting efficiency is defined as total numbers of mice that have correctly targeted loxP over total numbers of founder mice
3.4. CRISPR-Mediated Integration of the Second loxP Site
Male mice with correct loxP sites were separated into individual cages when they reach 7 weeks of age. We normally set up 3–7 stud males for mating with females to collect zygotes for the second injection (see Note 14).
Female loxP carrier mice were superovulated (see Subheading 3.2) and mated with males carrying the same loxP allele (see steps 1–5 of Subheading 3.2). If there are insufficient female loxP carriers, wild-type females can also be used. In the latter case, the chance of obtaining mice with both loxPs integrated on the same chromosome is lower but it is usually sufficient for obtaining the desired cKO mice.
Fertilized eggs are collected and microinjected as described in steps 6–9 of Subheading 3.2, but with reciprocal CRISPR reagents, i.e., Six-Up mice injected with Six-In CRISPR and Six-In mice injected with Six-Up reagents.
Two-cell stage embryos are implanted as described in steps 10–18 of Subheading 3.2, and offspring are ear-tagged and genotyped as described in Subheading 3.5 (below).
3.5. Genotyping Analysis for the Presence of Both loxP Sites
Take a tail biopsy from each mouse at weaning. Cut 1 mm oftail tip for crude genomic DNA extraction, which is usually adequate for PCR genotyping. Save the remaining tail biopsies at −20 °C as a source of high-quality genomic DNA for long-range PCR that will be performed later.
Set up genotyping PCR for the first loxP site using the common primer pair as described in Subheading 3.3. Save only samples that contain the first loxP insertion for further analysis.
Follow the same streamlined procedures as in Subheading 3.3 to screen for correct integration of the second loxP site: perform PCR using a common primer pair for the target site, select samples positive for the second loxP amplicon of predicted size, perform the second loxP allele-specific PCR on selected samples, then purify PCR amplicons for sequencing. Mice containing both loxP sites are potential carriers for the floxed allele (two loxP sites in cis configuration) and are subject to next-step analysis (see Note 15).
Retrieve tail biopsies frozen in step 1 of Subheading 3.5 from potential floxed allele carriers. Digest each sample in 200 μL of digestion buffer with proteinase K added fresh to 0.5 mg/mL final concentration. Incubate at 55 °C with shaking until clumps of tissue disappear (3 h to overnight). Extract DNA with the QiaAmp DNA Mini Kit to obtain high-quality genomic DNA that is suitable for long-range genomic PCR (see Note 16).
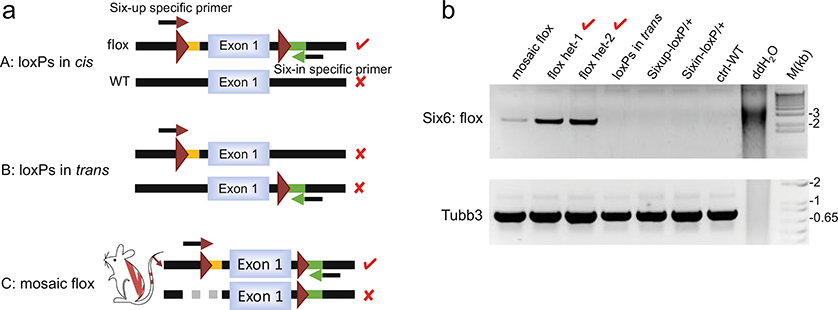
To identify alleles with two loxP sites integrated in cis, pair the Six-Up loxP-specific forward primer with the Six-In loxP-specific reverse primer to amplify regions spanning the two loxP sites (see Fig. 6a and Note 17): mix 10 μL of GoTaq green master mix (2×), 5 μL of Betaine (5 M), 0.2 μL of Phusion DNA polymerase, 1 μL of each primer (10 μM), 1 μL of high-quality genomic DNA (20 ng/μL), and 1.8 μL of ddH2O for a 20 μL reaction.
Perform PCR with an initial denaturation at 95 °C for 1 min, followed by 30 cycles of 95 °C for 25 s, 58 °C for 25 s, and 72 °C for 2 min and 40 s, and a final extension at 72 °C for 10 min.
A parallel PCR (amplicon size >500 bp) can be performed to amplify an unrelated genomic locus, serving as a control for the quality and quantity of input genomic DNA (see Fig. 6b lower panel).
PCR products are analyzed by agarose gel (1%) electrophoresis. A positive band of expected size from the allele-specific PCR indicates the presence of the floxed allele. A disproportionately faint band (taking into consideration of the intensity of control PCR bands) implies mosaicism (Fig. 6b).
Analyze PCR results and confirm by DNA sequencing: as summarized in Table 2, seven out of the 32 weaned Six-In mice injected with Six-Up CRIPSR have both loxP sites integrated. Among them, three have loxP sites in trans (unwanted) and four have loxP sites in cis (selected for breeding), including a potentially mosaic mouse. Of the 11 weaned Six-Up mice injected with Six-In CRISPR, four have both loxP sites, and two of these have loxP sites in cis (selected for breeding). Genotyping analysis of F1 offspring from the six flox-positive carrier mice indicates that three are heterozygous (het) and two are homozygous (hm) for the floxed allele of the Six6 gene, while the mosaic mouse has not transmitted the floxed allele to its offspring.
Fig. 6.
Identification of the floxed (cis-loxP sites) allele. (a) Schematic diagram of allele-specific PCR strategy to amplify DNA strand with two loxP sites integrated in cis configuration. Mouse C shows a founder mouse carrying a mosaic flox allele that does not have germline transmission. Arrow with maroon arrowhead: a forward primer with 3′ terminus complementary to the upstream loxP site; arrow with green arrowhead: a reverse primer with 3′ terminus complementary to the downstream loxP-linked restriction site. Red check mark: successful PCR amplification resulted from annealing of both primers to the DNA molecule. Red cross mark: no amplification as no primer or only one primer anneals to DNA. (b) Upper panel: representative allele-specific PCR analysis for the Six6 flox allele, with an upstream loxP-specific forward primer (Sixup-lxpF) and a downstream loxP site-specific reverse primer (Sixin-BIR) for amplification of the 2.4 Kb loxP-floxed region. Lower panel: PCR amplification of the Tubb3 gene, which serves as a control for PCR reaction
Table 2.
Summary of targeting experiments for the second loxP site
| Target site | No. of embryos injected/implanted | Mice weaned | Mice w/two loxPs | Mice w Ids loxPs | Germline transmission (het/hm) |
|---|---|---|---|---|---|
| Six-Up | 105/74 | 32 | 7 | 4 | 3(1/2) |
| Six-In | 51/36 | 11 | 4 | 2 | 2 (2/0) |
Acknowledgements
This work was supported in part by the Intramural Research Programs at the National Institute of Diabetes and Digestive and Kidney Diseases (Y.L. and D.F.), and the National Heart, Lung, and Blood Institute (Y.D., W.X., F.Z., and C.L.) at the National Institutes of Health.
Footnotes
It is strongly recommended to investigate potential strain background for known phenotypical traits or deleterious genes before initiating a long-term project. For example, for studies of visual function or retinal differentiation, a number of retinal degeneration genes (rd) are known. Some of these rd genes are common in certain mouse strains, such as the rd1 mutation (Pde6brd1) which is present in many commonly used mouse lines, including the FVB and C3H inbred strains [20]. The rd8 mutation (Crb1rd8) is present in all substrains of C57BL/6N, an NIH subline of C57BL/6 mice [21].
Ketamine is a controlled substance in the United States. It must be approved, then stored in a locked drug box and its usage clearly recorded.
For generating conditional knockout mouse lines, the most frequently floxed exon is Exon 2. However, Exon 2 should be avoided if the number of nucleotides contained in this exon is a multiple of three, which only leads to the deletion of the amino acids encoded by this exon instead of a frameshift null mutation. Under this circumstance, an alternative exon further downstream can be floxed. Multiple exons can be floxed if one is concerned with leaving too many amino acids in an open reading frame prior to the floxed exon, which can produce a truncated protein. When using conventional ES-cell-mediated gene-targeting method, the distance between the two loxP sites is often quite small because of the size limit of the targeting DNA constructs. However, for the CRISPR-mediated method, the two loxP sites are inserted independently and therefore multiple exons or even the entire gene can be floxed. This advantage comes in handy when the gene contains multiple splicing isoforms, but it is not recommended to flox a very large region without specific reasons because: 1) concerns exist that the efficiency of Cre-mediated deletions may decrease when the two loxP sites are too far apart; and 2) it is now generally believed that >80% of the noncoding genome plays some sort of functional role and therefore it is not advisable to remove unnecessarily large pieces of DNA from the chromosomes.
When using single-stranded oligos as donors for knocking in DNA sequences, one common question is whether to use the strand that is complementary or noncomplementary to the sgRNA. We sometimes prefer one strand over the other depending on whether the sgRNA and oligos can form DNA-RNA duplexes and the location of the mismatches relative to the position of CRISPR cutting site, but for the purpose of inserting loxP using the strategy we have described, we find that either strand works efficiently.
We normally do not pretest sgRNAs before microinjectingthem into zygotes, but some laboratories test guide sgRNA using in vitro (incubating Cas9 protein, template DNA, and sgRNA in test tubes) or ex vivo (transfecting into culture cells) methods. We do not believe these testing strategies can reliably predict sgRNA efficiency in a zygote after microinjection because CRISPR efficiency is also dependent on the delivery method. Culturing injected zygotes into blastocysts and genotyping them by PCR can be a reliable method for confirming the efficiency of guide RNAs, but establishing a reliable PCR protocol for genotyping blastocysts requires a significant amount of work and therefore this culturing procedure is used only in extremely important or time-sensitive projects.
We prefer to culture the injected zygotes overnight and transfer2-cell embryos the next morning because: 1) a typical microinjection day is already very busy, so culturing them overnight can alleviate some of the workload to the next day; 2) a significant portion of sgRNAs (5–10% under our standard microinjection conditions) are lethal to early embryos by blocking injected embryos from developing into 2-cell stage embryos. Culturing overnight allows earlier detection of such toxic sgRNAs and redesigning them in a timely manner.
The loxP-integrated amplicon is around 40 bp larger than the wild-type amplicon. Given this relatively small size difference, both amplicons should be designed to be less than 300 bp to allow good separation by electrophoresis on an agarose gel. It is also advisable to test the primer sets on wild-type DNA to optimize PCR conditions in advance.
In addition to the HotSHOT method, other DNA extraction methods should also work. However, the HotSHOT method is fast and cost-effective, and works well for short-amplicon PCR analysis. Caution should be exercised because DNA prepared in this way is in a crude and unstable condition. For long-term storage, freeze an aliquot at −20 °C.
Adjust the annealing temperature if necessary to achieve optimal PCR results. The upper (loxP) and lower (wild-type) band should display a relatively consistent ratio of intensity across all samples. A sample with an unusually weak loxP band may indicate mosaicism, that is only some of the cells in the mouse carry the loxP allele, and thus the germline of the mouse may or may not carry the desired mutation. Avoid using such mouse for the second targeting if possible (see Fig. 5a mouse #5032).
Gels with 2.5–3.0% agarose can be used to separate short DNA fragments (<500 bp) efficiently.
Often part of the loxP and WT amplicons form heteroduplex DNA (hDNA), which migrates as a band close to the loxP band on the gel, and is resistant to restriction digest (see bands marked with * in Fig. 5b). As a result, the digested sample will have a weaker rather than a complete absence of the “upper band.” The appearance of a smaller-than-WT digested band(s), therefore, is more reliable than loss of the upper band, for indicating successful restriction digestion.
Mixed amplicons can be problematic for sequencing. Allele-specific PCR selectively amplifies the loxP allele, eliminating the contamination of wild-type amplicon. An alternative approach is to subclone mixed PCR amplicons into a plasmid vector (e.g., TOPO-cloning kit) and subsequently sequence 6–10 individual clones for ensuring representation of all alleles. The latter method can be time-consuming if large numbers of mice are to be screened.
Some of the injected mice only display the loxP but not wild-type amplicon. This could either mean both alleles have loxP (homozygote), or one allele has loxP integration while the other allele has a big deletion such that the common PCR primers now lack a complementary genomic binding site and therefore cannot amplify that allele (hemizygote). For differentiating between these two possibilities, one can design PCR primers further away from the CRISPR cutting site which enables the detection of bigger deletions.
We prefer to set up at least three breeding pairs of mice in order to obtain enough zygotes for each microinjection session. On average, only one half to two thirds of injected zygotes can reach the 2-cell stage of development, and after implantation only about half of the embryos can lead to birth of pups at full term. When a surrogate mother is implanted with too few embryos, the litter size is small and pups often cannot survive long. In the case that only a small number of positive mice are available, it is necessary to breed them with wild-type mice to expand the colony before the second injection can take place. An alternative strategy is to use sperm collected from one loxP-positive male for in vitro fertilization (IVF) to generate a large number of zygotes. With recent improvements in IVF efficiency (see Chapter 14 of this book), this method will likely be increasingly used in coming years. Both loxP-positive and wild-type females can be used as egg donors for IVF, although the eggs from wild-type females are less efficient than those from loxP-carrier females for generating the final floxed mice. Recently developed HyperOva-stimulated ovulation method (see Chapter 13 of this volume) makes it possible for using only a few egg donors for each IVF session, and therefore greatly increases the chance of using only loxP-positive females.
The PCR protocol described here is suitable for identifying floxed mice when the two loxPs are a few kilobases apart. When the length of the DNA between the two loxPs is too big to be amplified by long-range PCR, it may be necessary to breed candidate mice with wild-type mice and then genotype their offspring. If the loxP sites are in cis (floxed), some offspring (approximately 50% or lower if mosaic) will carry both the upstream and downstream loxPs. However, if the loxP sites are in trans, they will be segregated into different offspring.
Alternatively, genomic DNA can be extracted using standard phenol-chloroform extraction method after proteinase K digestion.
The upstream loxP-specific forward primer (Sixup-lxpF) has9 nt at the 3′ terminus complementary to the loxP sequence, while the downstream loxP-specific reverse primer has 5 nt at the 3′ terminus complementary to BamHI restriction enzyme site. PCR parameters may be adjusted based on actual reactions. Betaine is a PCR enhancing agent, Phusion DNA polymerase offers high processivity. In our experience, a mixture of Phusion and Taq polymerases usually gives more robust results for long-range PCR than Phusion alone. An alternative method is to set up long-range genomic PCR using common primers that amplify the entire “floxed” region. Then subclone the PCR products into a plasmid vector (e.g., TOPO-clone kit) and sequence individual clones.
References
- 1.Evans MJ, Kaufman MH (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292:154–156 [DOI] [PubMed] [Google Scholar]
- 2.Kucherlapati RS, Eves EM, Song KY, Morse BS, Smithies O (1984) Homologous recombination between plasmids in mammalian cells can be enhanced by treatment of input DNA. Proc Natl Acad Sci U S A 81:3153–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS (1985) Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature 317:230–234 [DOI] [PubMed] [Google Scholar]
- 4.Doetschman T, Gregg RG, Maeda N, Hooper ML, Melton DW, Thompson S, Smithies O (1987) Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature 330:576–578. 10.1038/330576a0 [DOI] [PubMed] [Google Scholar]
- 5.Mansour SL, Thomas KR, Capecchi MR (1988) Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature 336:348–352. 10.1038/336348a0 [DOI] [PubMed] [Google Scholar]
- 6.Thomas KR, Capecchi MR (1987) Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51:503–512 [DOI] [PubMed] [Google Scholar]
- 7.Gu H, Zou YR, Rajewsky K (1993) Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell 73:1155–1164 [DOI] [PubMed] [Google Scholar]
- 8.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinek M, East A, Cheng A, Lin S, Ma E,Doudna J (2013) RNA-programmed genome editing in human cells. elife 2:e00471 10.7554/eLife.00471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mali P, Yang L, Esvelt KM, Aach J, Guell M,DiCarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Yang H, Shivalila CS, Dawlaty MM,Cheng AW, Zhang F, Jaenisch R (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153:910–918. 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R (2013) One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154:1370–1379. 10.1016/j.cell.2013.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Du Y, He X, Huang X, Shi YS (2017) A convenient Cas9-based conditional knockout strategy for simultaneously targeting multiple genes in mouse. Sci Rep 7:517 10.1038/s41598-017-00654-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mandasari M, Sawangarun W, Katsube K, Kayamori K, Yamaguchi A, Sakamoto K (2016) A facile one-step strategy for the generation of conditional knockout mice to explore the role of Notch1 in oroesophageal tumorigenesis. Biochem Biophys Res Commun 469:761–767. 10.1016/j.bbrc.2015.12.006 [DOI] [PubMed] [Google Scholar]
- 15.Guzzardo PM, Rashkova C, Dos Santos RL,Tehrani R, Collin P, Burckstummer T (2017) A small cassette enables conditional gene inactivation by CRISPR/Cas9. Sci Rep 7:16770 10.1038/s41598-017-16931-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horii T, Morita S, Kimura M, Terawaki N, Shibutani M, Hatada I (2017) Efficient generation of conditional knockout mice via sequential introduction of lox sites. Sci Rep 7:7891 10.1038/s41598-017-08496-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miura H, Quadros RM, Gurumurthy CB, Ohtsuka M (2018) Easi-CRISPR for creating knock-in and conditional knockout mouse models using long ssDNA donors. Nat Protoc 13:195–215. 10.1038/nprot.2017.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quadros RM, Miura H, Harms DW, Akatsuka H, Sato T, Aida T, Redder R, Richardson GP, Inagaki Y, Sakai D, Buckley SM, Seshacharyulu P, Batra SK, Behlke MA, Zeiner SA, Jacobi AM, Izu Y, Thoreson WB, Urness LD, Mansour SL, Ohtsuka M, Gurumurthy CB (2017) Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol 18:92 10.1186/s13059-017-1220-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Truett GE, Heeger P, Mynatt RL, Truett AA,Walker JA, Warman ML (2000) Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Bio-Techniques 29(52):54. [DOI] [PubMed] [Google Scholar]
- 20.Chang B, Hurd R, Wang J, Nishina P (2013) Survey of common eye diseases in laboratory mouse strains. Invest Ophthalmol Vis Sci 54:4974–4981. 10.1167/iovs.13-12289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattapallil MJ, Wawrousek EF, Chan CC, Zhao H, Roychoudhury J, Ferguson TA, Caspi RR (2012) The Rd8 mutation of the Crb1 gene is present in vendor lines of C57BL/6N mice and embryonic stem cells, and confounds ocular induced mutant phenotypes. Invest Ophthalmol Vis Sci 53:2921–2927. 10.1167/iovs.12-9662 [DOI] [PMC free article] [PubMed] [Google Scholar]