Abstract
MicroRNAs (miRNAs) are endogenous, non-coding, single-stranded, tiny RNAs with 21–23 nucleotides that regulate several biological functions through binding to target mRNAs and modulating gene expression at post-transcriptional levels. Recent studies have described crucial roles for miRNAs in pathophysiology of numerous human cancers. They can act as an oncogene and promote cancer or as a tumor suppressor and alleviate the disease. Recently discovered microRNA-154 (miR-154) has been proposed to be involved in multiple physiological and pathological processes including cancer. With this aspect, aberrant expression of miR-154 has been demonstrated in variety of human malignancies, suggesting an important role for miR-154 in tumorigenesis. To be specific, it is considered as a tumor suppressor miRNA and exerts its beneficial effects by targeting several genes. This review systematically summarizes the recent advances done on the role of miR-154 in different cancers and discusses its potential prognostic, diagnostic and therapeutic values.
Keywords: microRNA-154, cancer, target gene, tumor suppressor
Introduction
Despite the condense researches done on the diagnosis and therapy of cancer, it is still one of the main health concerns of today’s society. Even in developed countries, several new cancer cases and deaths are recorded annually leading to a substantial economic burden including the financial costs of cancer for both the patient and for society as a whole. As evidence, 1,762,450 new cancer cases and 606,880 cancer deaths were recorded in the United States, in 2019.1 In a massive effort to elucidate the mechanisms of carcinogenesis, scientists have found that several genes are dysregulated in initiation and progression of cancers including the silencing of tumor suppressors, aggressive activation of oncogenes and proliferation-related signaling pathways.2 Recent studies have demonstrated that microRNAs (miRNAs) are significantly involved in the regulation of functional genes associated with oncogenesis.3,4 They can either activate or inactivate oncogenes and thereby regulate oncogenesis. MiRNAs also can target cell signaling pathways and thus control tumorigenesis.5 Considering their essential roles in pathophysiology of different malignancies, the miRNAs-related mechanisms of cancerous diseases should be fully elucidated which in return can enhance our knowledge of oncogenesis and increase therapeutic efficacy.
MiRNAs are a class of highly conserved, single stranded, endogenous, non-coding small RNAs consisting of 21–23 nucleotides. Mature miRNAs act as post-transcriptional regulators of gene expression, by base-pairing with mRNA molecules in the 3′ untranslated region (3′-UTR), controlling the translation process of the target mRNA. The level of complementarity between miRNAs and the target mRNA determines the outcomes, as this can usually result in translation blockage, or less frequently mRNA degradation.6,7 It has been claimed that miRNAs account for 1–5% of the human genome and regulate at least 30% of protein-coding genes.8 Since their discovery in 1993, miRNAs have been proposed to play a pivotal role in several biological functions such as cell growth, apoptosis, and immune responses.8 Each miRNA is able to target hundreds of genes, and each mRNA could also be targeted by several miRNAs. The remarkable point is that miRNAs can be detected not only in tissues (eg, tumor tissues), but also in the circulation, blood serum, where they are released.9 This unique feature allows miRNAs to be employed as a great tool for noninvasive and early diagnosis of several diseases, more importantly cancer. MiRNAs also possess a great therapeutic potential, as evidence, miR-208/499 and miR-195 have been proposed as a preclinical treatment for cardiovascular disease, while miR-34 and let-7 for cancer. Particularly, Miravirsen (codenamed SPC3649), antisense to miR-122, has been entered the Phase II clinical trials for treatment of hepatitis C virus infection.10–12 Within the past decade, understanding the relation between miRNAs and different malignancies has aroused considerable research interest and they are tightly being investigated for prognosis, diagnosis and therapy of human cancers.
MicroRNA-154
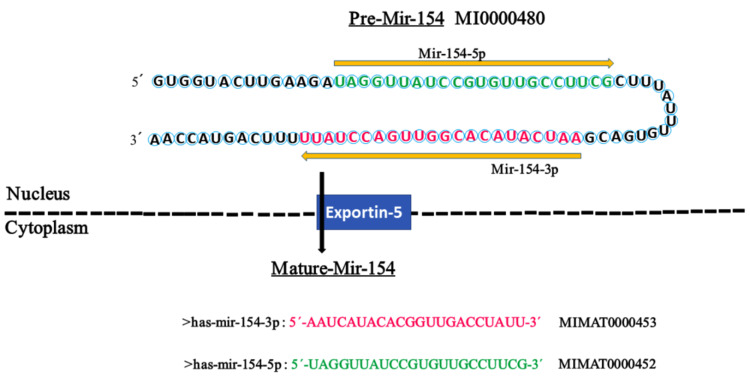
MicroRNA-154 (miR-154) is a tiny RNA particle with 22 nucleotide which is located on 14q32.31 chromosome (Figure 1). Consistent with canonical miRNA biogenesis pathway, the biosynthesis of miR-154 occurs in nucleus and cytoplasm through several steps. Initially, miR-154 is a large double-strand RNA, but following actions of several proteins and enzymes, most importantly RNA polymerases, it shrinks into a small single-strand RNA and then interacts with an Argonaute family protein, AGO2 and some additional proteins to form a complex named RISC (RNA induced silencing complex). Now, RISC can be guided to the target and inactivate one or multiple functional genes.8 The primary role and expression pattern of miR-154 were first described in idiopathic lung fibrosis by Milosevic et al in 2010.13 Then, they deeply investigated its precise role in pulmonary fibrosis in 2012.14 Despite other miRNAs (eg, miR-145), to date a little has been unraveled about the upstream regulation mechanisms of miR-154. In this regard, Milosevic et al, first identified that stimulation of normal human lung fibroblasts with transforming growth factor-β (TGF-β) could upregulate miRNAs on chr14q32. They also found 16 Smad (3 and/or 4) binding elements upstream of the up-regulated miRNAs, nine of which were detected upstream of miR-154 family members.14 Moreover, working on neuropathic pain by targeting the miR-154-5p/CXCL13 axis, just recently Chen et al, determined that a kind of long noncoding RNA (lncRNA), named small nucleolar RNA host gene 5 (SNHG5) is upstream regulator of miR-154-5p.15 Today, miR-154 is a hotspot and its roles have already been tightly investigated in numerous physiological and pathological processes such as diabetes,16 spermatogenesis,17 endometritis18 and cardiac complications.19 Currently, there is no miR-154-based treatment, however, in a clinical trial (ChiCTR1800019872), Ren et al, tried to investigate diagnostic potential of miR-154-5p in patients with type 2 diabetes mellitus. According to their results, elevated serum miR-154-5p was significantly correlated with high urinary albumin to creatinine ratio which may be a novel biomarker for early diagnosis of diabetic kidney disease.20 This review systematically summarizes the recent advances done on the role of miR-154 in different human cancers and discusses its potential prognostic, diagnostic and therapeutic values.
Figure 1.
The hairpin structure of pre-miR-154 and the sequence of mature miR-154. The miR-154 gene is transcribed into a precursor (premiR-154) with 84 nucleotides in the nucleus, which is transferred to the cytoplasm by Exportin-5 for further processing to yield two mature miRNAs with 22 nucleotides, miR-154-5p and miR154-3p. The sequence of mature miR-154-5p and miR-154-3p is colored in green and red, respectively. The arrow indicates the orientations from 5′ to 3′. The codes of miR-154 were extracted from miRBase.
MicroRNA-154 in Cancerous Diseases
As a newly-discovered miRNA, miR-154 has been demonstrated to play a key role in tumorigenesis of several human cancers including, colorectal cancer (CRC), hepatocellular carcinoma (HCC), gastric cancer (GC), non-small cell lung cancer (NSCLC), breast cancer (BC), prostate cancer (PCa), glioma and glioblastoma. It has distinct functions in specific cancers, including inhibition of proliferation, migration and invasion as well as induction of apoptosis and cell-cycle arrest. To be specific, it is generally thought that miR-154 is a tumor suppressor as evidence its expression level is frequently measured to be lower in cancerous tissues as compared to normal adjacent tissues. MiR-154 exerts its suppressive effects by targeting several genes which have been listed in Table 1. Furthermore, some of the lncRNA can target miR-154 through acting as a competitive endogenous RNA (ceRNA) leading to promotion of tumorigenesis in various cancers (Table 2). Additionally, miR-154 can interact with some cell signaling pathway, especially WNT.
Table 1.
Target Genes of miRNA-154 Responsible for Its Tumor Suppressive Action in Different Cancers
| Gene | Location | Malignancy | Cell Lines | No. of Tissue Samples (Pair) | Outcomes | References | |
|---|---|---|---|---|---|---|---|
| Normal | Cancerous | ||||||
| MTDH | 8q22.1 | Gastric cancer | GES-1 | SGC-7901, AGS, MKN-1, and BGC-823 |
36 | Inhibited the proliferation, migration and invasion |
[22] |
| DIXDC1 | 11q23.1 | Gastric cancer | GES-1 | MGC-803, MKN-87, MKN-45, and AGS |
16 | Inhibited the growth and invasion | [23] |
| TRAF6 | 11p12 | Colorectal cancer | N/A | N/A | N/A | Inhibited the proliferation, migration and invasion | [25] |
| CCND1 | 11q13.3 | Colorectal cancer | N/A | N/A | N/A | Inhibited the proliferation, migration and invasion | [25] |
| TLR2 | 4q31.3 | Colorectal cancer | HEK293 | SW480, SW620, HT29 | 41 | Inhibited the proliferation, colony formation, migration and invasion | [26] |
| CCND2 | 12p13.32 | Colorectal cancer | FHC | HCT-116, HCT-8, SW-480, SW-620, DLD-1 and HT-29 |
80 | Inhibited the proliferation and growth |
[28] |
| Prostate cancer | N/A | DU145, PC-3 | 27 | Inhibited the proliferation, colony formation, and growth | [47] | ||
| Hepatocellular carcinoma | N/A | HepG2, Hep3B | N/A | Inhibited the growth, colony formation and induced cell cycle arrest at G1/S phase | [64] | ||
| GALNT7 | 4q34.1 | laryngeal squamous cell carcinoma | 16HBE | LSCC Hep-2, TU212 | 104 | Inhibited the proliferation, colony formation, migration, and induced cell cycle arrest at G0/G1 phase | [30] |
| ZEB2 | 2q22.3 | Non-small cell lung cancer | N/A | A549 | N/A | Inhibited the migration, invasion, and suppressed EMT | [34] |
| Hepatocellular carcinoma | HL-7702 | Huh-7, SMMC7721, HepG2 HCCLM3 |
50 | Inhibited the proliferation, migration, invasion and induced apoptosis, G1cell cycle arrest at G1 phase | [65] | ||
| Non-small cell lung cancer | NBE | A549, H322, H1299, GLC-82, SPC-A1 | 42 | Inhibited the proliferation, migration invasion, and induced apoptosis | [36] | ||
| RUNX2 | 6p21.1 | Non-small cell lung cancer | NBE | A549, H322, H1299, GLC-82, SPC-A1 | 42 | ||
| RUNX2 | 6p21.1 | Bladder cancer | N/A | T24 | 86 | Inhibited the proliferation, migration, invasion, and improved prognosis | [40] |
| RSF1 | 11q14.1 | ||||||
| ATG7 | 3p25.3 | Bladder cancer | SV-HUC-1 | J82, T24, UM-UC-3 |
84 | Inhibited the proliferation, migration, invasion, and decreased advanced T stage, lymphatic invasion, distant metastasis |
[39] |
| HMGA2 | 12q14.3 | Prostate cancer | N/A | DU145, PC-3 | 4 | Inhibited the migration, invasion and suppressed EMT | [46] |
| E2F5 | 8q21.2 | Prostate cancer | N/A | DU145, PC-3 | 32 | Inhibited the migration, invasion, growth, and induced cell cycle arrest at G1 phase |
[48] |
| Osteosarcoma | N/A | MG63, HOS | 30 | Inhibited the migration, proliferation, invasion and induced cell cycle arrest at G0/G1 phase | [73] | ||
| Breast cancer | MCF-10A | MCF-7, MDA-MB-231, BT-549 MDA-MB-453 |
36 | Inhibited the proliferation, migration, invasion, and increased cell arrest at the G0/G1 phase | [59] | ||
| PIWIL1 | 12q24.33 | Glioblastoma | N/A | U251, U87, A172, LN229, SNB19, LN308 | 34a | Inhibited the proliferation, metastasis and induced apoptosis | [51] |
| Glioma | NHA | A172, SNB19, U172, U251, U87MG |
40 | Inhibited the proliferation, migration and invasion | [52] | ||
| ADAM9 | 8p11.22 | Breast cancer | MCF-10A | MCF-7, SKBR3, MDA-MB-231 | 45 | Inhibited the proliferation, migration and invasion | [60] |
| NAMPT | 7q22.3 | Breast cancer | MCF-10A, HEK-293 |
MCF-7, MDA-MB-231 | N/A | Inhibited nicotinamide adenine dinucleotide (NAD) salvage pathway, increased apoptosis, and sensitivity to doxorubicin | [61] |
| BASE (BPIFA4P) |
20q11.21 | Breast cancer | N/A | N/A | 68a | N/A | [58] |
| PCNA | 20p12.3 | Breast cancer | MCF10A | MCF7, T47D, BT474, BT549, MDA-MB-468, MDAMB231 | 30 | Inhibited the proliferation and induced cell cycle arrest at G1 phase | [62] |
| Hepatocellular carcinoma | Primary Human Hepatocytes (PHH) | HepG2, HepG2.2.15, HepAD38, HepG2-NTCP |
43a | Decreased HBV replication and growth | [66] | ||
| RHOA | 3p21.31 | Thyroid cancer | Nthy-ori3-1 | K-1, TPC-1, B-CPAP | 63 | Inhibited the proliferation, growth and induced apoptosis | [70] |
| WNT5A | 3p14.3 | Osteosarcoma | NHOst | HOS, Saos-2, U2OS, MG-63 | 44 | Inhibited the proliferation, colony formation, migration, invasion and induced cell cycle arrest at G1 phase |
[74] |
| AURKA | 20q13.2 | Melanoma | HEMa-LP | SK-MEL-2, A375, 1205Lu, UACC903, CHL-1 |
104a | Inhibited the proliferation, Invasion, migration and induced apoptosis |
[76] |
| NSD2 | 4p16.3 | Skin squamous cell carcinoma | NHEK | CRL-1555, CRL-1623 | N/A | Inhibited the growth, and induced apoptosis, G0/G1cell cycle arrest | [77] |
| CCNE1 | 19q12 | Osteosarcoma | N/A | MG63, HOS | 30 | Induced cell cycle arrest at G0/G1 phase | [73] |
| CDK2 | 12q13.2 | Osteosarcoma | N/A | MG63, HOS | 30 | Induced cell cycle arrest at G0/G1 phase | [73] |
Notes: aTissue specimens are not in pairs. The genes' names were abbreviated according to GenBank.
Abbreviation: N/A, not applicable.
Table 2.
Oncogenic Circular RNA and Long Non-Coding RNAs (lncRNA), Which Act as Competitive Endogenous RNA (ceRNA) for miR-154 (So-Called Sponging Effect) in Different Cancers
| RNA | Malignancy | Cell Lines | No. of Tissue Samples (Pair) | Outcomes | References | |
|---|---|---|---|---|---|---|
| Normal | Cancerous | |||||
| LncRNA SNHG1 | Colorectal cancer | FHC | HCT-116, HCT-8, SW-480, SW-620, DLD-1 and HT-29 |
80 | Tumor progression, poor prognosis and reduced patient survival | [28] |
| LncRNA SNHG20 | Non-small cell lung cancer | NBE | A549, H322, H1299, GLC-82, SPC-A1 | 42 | Tumor progression and poor prognosis | [36] |
| LncRNA SNHG5 |
Breast cancer | MCF10A | MCF7, T47D, BT474, BT549, MDA-MB-468, MDAMB231 | 30 | Poor prognosis, promoted proliferation and cell-cycle progression | [62] |
| LncRNA PCNAP1 |
Hepatocellular carcinoma | Primary Human Hepatocytes (PHH) | HepG2, HepG2.2.15, HepAD38, HepG2-NTCP |
43a | Increased HBV replication and cell growth | [66] |
| CircRNA Circ_101064 |
Glioma | NHA | A172, SNB19, U172, U251, U87MG |
40 | Increased tumor grading and diameter | [52] |
Note: aTissue specimens are not in pairs.
MiR-154 and Digestive System Cancers
Available evidence shows that 165,460 deaths were recorded in the United States, in 2019 due to digestive system cancers.1 The role of miR-154 in tumorigenesis of lower digestive tract cancers has been relatively well studied, however, only a single paper has been published about the upper cancers. Particularly, there is scarcity of information on the role of miR-154 in esophagus cancer. The expression level of miR-154 undergoes dramatic changes in tumors of digestive system. While some of the researchers have measured decreased expression of miR-154, others have found the increased expression. Squamous cell carcinoma (SCC) of tongue is an aggressive malignancy which is famous for its high rate of proliferation and nodal metastasis. Investigating the expression pattern of miRNAs in tongue SCC, Wong et al found that miR-154 was upregulated in the cancerous tissue specimens compared to the adjacent normal tissue.21 GC derived from gastric mucosal epithelial cells is a prevalent digestive cancer and the second leading cause of cancer-associated deaths worldwide. It has been reported that the expression of miR-154 is downregulated in GC tissue specimens and cell lines compared to the control tissue and cells. Additionally, it has been claimed that forced overexpression of miR-154 can alleviate the progression of GC by directly targeting metadherin (MTDH).22 In a similar study, Song et al measured decreased expression of miR-154 in both clinical GC tissue samples and cell lines. They also identified an inverse correlation between miR-154 expression and DIXDC1 expression, as overexpression of miR-154 using miRNA mimics could suppress GC through binding to DIXDC1. Finally, their experiments demonstrated that miR-154 could also interact with WNT signally pathway to repress GC.23 The WNT signaling pathway is an ancient and evolutionarily conserved pathway which is responsible for vital aspects of cell fate such as cell migration, cell polarity, neural patterning and organogenesis during embryonic development.24 Accumulating evidence suggests that dysregulation of miR-154 is involved in the initiation and progression of the rectal cancer (RC) and CRC. In this regard, Chung et al observed decreased expression of miR-154 in CRC cells and tissue samples. They also noted that forced overexpression of miR-154 or miR-124 could inhibit cell proliferation, migration and invasion by directly targeting 3′-UTR of TRAF6.25 The results of Xin et al’s study were supportive of the aforementioned study, as they found that miR-154-repressed CRC cell growth and motility by targeting TLR2.26 Despite the aforementioned studies which demonstrate downregulated levels of miR-154 in CRC, D’Angelo et al, measured the elevated expression of miR-154 in serum and tissue samples of patients with rectal adenocarcinoma who were not responsive to preoperative chemotherapy suggesting that miR-154 may be associated with increased resistance to chemotherapy.27 Accumulating evidence suggests that lncRNAs play a central role in both initiation and progression of different cancers. With this aspect, Xu et al exhibited that SNHG1 was involved in CRC progression. They revealed that SNHG1 was upregulated in CRC tissues which was associated with decreased patient survival. They also found that SNHG1 promoted CRC cell growth through epigenetic silencing of promoter of Kruppel like factor 2 (KLF2) and Cyclin-dependent kinase inhibitor 2B (CDKN2B) in the nucleus and increasing CCND2 expression via its sponge activity of miR-154-5p in cytoplasm.28 Although the above-mentioned studies show that miR-154 exerts tumor-suppressive effects on digestive system cancers, there are still some inconsistencies on its expression levels. Additionally, its role has not yet been studied in esophageal cancers. Therefore, further studies using larger tissue samples and different cell lines should be conducted to elucidate the role of miR-154 in digestive system tumors.
MiR-154 and Respiratory System Cancers
Respiratory diseases are one of the major threats to public health and consistently ranked among the most fatal diseases in developed countries.29 It is thought that respiratory cancers were responsible for 147,510 deaths in 2019, in the United States.1 Available evidence suggests that miR-154 is involved in the both upper and lower respiratory tract tumors. With this aspect, Niu et al reported that the expression of miR-154 was low in laryngeal SCC (LSCC) clinical tissue specimens and cell lines. Their experiments confirmed that ectopic overexpression miR-154 could repress LSCC aggressiveness by directly targeting GALNT7.30 Dysregulation of miR-154 has also been reported in various types of lung cancers which can be employed as a novel and potential biomarker for cancer diagnosis. In this regard, Huang et al studied serum miRNA profiles in nonsmokers, smokers, and lung-cancer patients and noticed that miR-154-5p was significantly downregulated in the sera of smokers and lung-cancer patients.31 Consistently, Cazzoli et al measured miRNAs expression levels in plasma samples from patients affected by lung adenocarcinomas and lung granulomas as well as healthy smokers and demonstrated that miR-154-3p could be effectively employed for discriminating between lung adenocarcinoma and granuloma.32 In another study, Lin et al assessed expression of miR-154 at the transcriptional level in 40, NSCLC tumor tissues compared to the matched adjacent normal tissues and correlated the expression level with clinicopathological features of the patients. They proved that decreased miR-154 expression was significantly associated with metastasis, larger tumor size and advanced TNM (tumor, lymph node, and metastasis) stage. According to their experiments, transfection of NSCLC A549 cells with miR-154 mimics could inhibit cell proliferation, colony formation, invasion and migration, as well as induce cell apoptosis and G0/G1 cell cycle arrest, in vitro. Additionally, enforced expression of miR-154 suppressed the growth of cancer cell xenografts, in vivo.33 In a parallel study, Lin et al noticed that miR-154 suppressed NSCLC progression by directly targeting zinc finger E-box binding homeobox 2 (ZEB2) gene.34 The studies conducted by Xue et al were supportive of the aforementioned studies, as they also proved that miR-154-3p plays tumor-suppressive role in NSCLC.35 In another study, it was determined that the expression of lncRNA SNHG20 was high in NSCLC tissues and cell lines which was associated with poor prognosis of patients. Furthermore, SNHG20 could function as a ceRNA to increase ZEB2 and RUNX2 expression by sponging miR-154.36 Conclusively, miR-154 plays an exact tumor suppressor role in lung cancers and according to the above-mentioned studies it can be employed for both diagnosis and therapy of pulmonary tumors.
MiR-154 and Genitourinary System Cancers
Tumors of the genitourinary (GU) system are one of the most common tumors encountered in clinical practice and account for 98,960 deaths in 2019, in the United States.37 Moreover, the incidence of GU cancers is higher in men than in women, and in the case of bladder cancer, it is as much as three times higher.38 Despite the recent advances in diagnostic methods particularly imaging techniques and therapeutic strategies, GU cancers are still responsible for millions of deaths worldwide. Increasing number of studies suggest that miR-154 has tumor-suppressive properties in GU cancers. Consistently, the studies conducted by Zhao et al and Zhang et al demonstrated that miR-154 was downregulated in bladder cancer tissue samples and cell lines.39,40 According to Zhao et al’s experiments, miR-154 acts as a prognostic factor and can suppress bladder cancer by directly targeting RSF1 and RUNX2.40 In agreement, Zhang et al reported that miR-154 functions as a tumor suppressor in bladder cancer by directly targeting ATG7.39 Investigating the role of miR-154 in cervical cancer, Wu et al, found that the decreased expression level of miR-154 was significantly associated with clinicopathological features including lymph node metastasis, vascular invasion, poor tumor differentiation, advanced FIGO stage, and short overall survival. They also proved that the expression level of miR-154 could independently predict prognosis of the patients.41 On the contrary, it was observed that miR-154-5p was upregulated in renal cell carcinoma (RCC) tissue specimens and cell lines which could reduce cell apoptosis and promote cell proliferation, viability, migration as well as invasion. Moreover, the expression level of miR-154 was associated with poor prognosis and decreased survival of RCC patients indicating that miR-154 acts as an oncogene.42
PCa is the second most frequent cancer diagnosed in men and the fifth leading cause of death worldwide. Initially, it may be asymptomatic and often has an indolent course that may require only active surveillance. Based on GLOBOCAN 2018 estimates, 1,276,106 new cases of PCa were reported worldwide in 2018, with higher prevalence in the developed countries.43 Several factors have been claimed to be involved in etiology of PCa. Particularly, a recent study has established a relation between active hormone dihydroxy vitamin D (1, 25(OH)2D) and miRNAs. With this aspect, Dambal et al noticed that miR-154-5p level is regulated by the active form of vitamin D and its receptors in human prostate stroma.44 Consistently, downregulation of miR-154 has been reported to be involved in PCa, as Formosa et al, observed that severe and consistent downregulation of miRNAs including miR-154 was remarkably associated with a higher incidence of metastatic events and higher prostate specific antigen levels, with similar trends observed for lymph node invasion and the Gleason score.45 High-mobility group AThook2 (HMGA2) gene has been shown to be upregulated in PCa tissues and cell lines which leads to promotion of epithelial–mesenchymal transition (EMT) and aggressive phenotype of PCa. It was proved that forced expression of miR-154 could inhibit EMT and suppress PCa progression by directly targeting HMGA2.46 In a parallel study, Zhu et al exhibited that overexpression of miR-154 using miRNA mimics could repress PCa by directly binding to 3′-UTR of CCND2.47 Similarly, Zheng et al demonstrated that miR-154-5p acted as a tumor suppressor in PCa cell lines by directly targeting E2F5.48 In spite of the aforementioned studies which emphasize on tumor suppressive roles of miR-154, Gururajan et al, have reported contrary results. They observed that miRNAs in the delta-like 1 homolog–deiodinase, iodothyronine 3 (DLK1-DIO3) including miR-154 were upregulated in bone metastatic PCa cell lines and tissues and intracardiac inoculation of miR-154 inhibitor-treated bone metastatic PCa cells in mice led to decreased bone metastasis and increased survival. According to their experiments, miR-154 could facilitate tumor growth, EMT, and bone metastasis.78 In conclusion, the roles of miR-154 in GU cancers are uncertain. Particularly, only a single study has been conducted on RCC which indicates that miR-154 is oncogene. Further studies are still required to elucidate the expression pattern and involvement of miR-154 in GU cancer specifically PCa and RCC.
MiR-154 and Nervous System Cancers
Both children and adults are affected by primary brain and CNS cancers which can be diagnosed in all anatomical regions of the CNS, mainly (>90%) occurring in the brain and the rest occurring in the meninges, spinal cord, and cranial nerves. The diagnosis and treatment of these tumors require extensive resource allocation and cutting-edge diagnostic and therapeutic technology. In 2016, there were 330,000 incident cases of CNS cancer and 227,000 deaths worldwide, and age-standardized incidence rates of CNS cancer increased globally by 17.3% between 1990 and 2016.49 These data clearly show that the incidence and mortality rate of CNS cancers are in warning state and novel diagnostic and therapeutic strategies must urgently be adopted to cut down the disease. In this regard, Wang et al measured declined expression of miR-154 in tissue specimens of patients with glioma compared to normal tissue samples and established a significant relation between the expression level and high World Health Organization (WHO) grade, large tumor size, a low Karnofsky performance status score, and a shorter overall survival. They have claimed that miR-154 might serve as a potential prognostic biomarker for patients with this disease.50 The studies conducted by Wang et al on glioblastoma tissue specimens and cell lines suggest that PIWIL1 is upregulated and there is an inverse association between PIWIL1 expression level and miR-154-5p expression. Additionally, they have shown that ectopic overexpression of miR-154 can inhibit progression of glioblastoma.51 In another study conducted on glioma tissue samples and cell lines, it was determined that PIWIL1 and circular RNA circ_101064 were upregulated and there was an inverse correlation between them and miR-154 expression. Upregulated levels of PIWIL1 and circular RNA circ_101064 were associated with aggressive phenotype of the disease and forced overexpression of miR-154 could repress the progression of glioma through binding to its downstream molecule PIWIL1.52 Contrary to the above-mentioned studies, Yang et al have claimed that miR-154 functions as an oncogene in CD133+ glioblastoma stem cells. Using T98G and U87MG cells, they showed that miR-154 was upregulated and by directly targeting PRPS1 could increase proliferation and migration of the cells.53 Although these findings are remarkable and interesting, further studies should be conducted to be able to correlate the results to clinicopathologic features of the disease. Taking together, the majority of studies suggest that miR-154 act as a tumor suppressor in CNS cancers, however, there are still some inconsistencies and the exact role of miR-154 in glioma and glioblastoma should be clarified in future studies.
MiR-154 and Breast Cancer
BC is the most common cancer diagnosed among US women (excluding skin cancers) and is the second leading cause of cancer death among women after lung cancer. Over the recent 5-year period (2012–2016), the BC incidence rate increased slightly by 0.3% per year in the United States, largely because of rising rates of local stage and hormone receptor-positive disease.54 As evidence, 271,270 new cases of BC were recorded in 2019, in the United States.1 The role of miRNA dysregulation in BC was first understood in 2005, and since then numerous studies have found dysregulated levels of miR-154 in both tissue specimens and cell lines. With this aspect, Lowery et al and Miranda et al measured significantly declined expression level of miR-154 in BC tissue specimens and cell lines, respectively.55,56 In a similar study, Li et al assessed miRNA expression in 21 ductal carcinoma in situ (DCIS), the most common type of non-invasive BC using miRNA microarray and real-time PCR and noticed that miR-154-3p was significantly downregulated in DCIS compared to the normal corresponding tissue.57 It has also been reported that the expression of miR-154 is lower in the estrogen receptor (ER) positive cell lines than ER-negative cells.58 Available evidence suggests that forced overexpression of miR-154 using miRNA mimics can suppress BC progression through targeting different genes. In this regard, Xu et al demonstrated that E2F5 was a target of miR-154, and its expression was inversely correlated with miR-154 expression in clinical BC tissues.59 Similarly, Qin et al identified ADAM9 as a novel direct target for miR-154 in BC which was upregulated in both clinical tissue specimens and cell lines.60 Working on various BC cell lines, Pour et al demonstrated that miR-154 can not only decrease BC progression but also increase its sensitivity to chemotherapy. According to their experiments, treatment of the cell lines with miR-154 mimics could suppress expression of nicotinamide phosphoribosyltransferase (NAMPT) leading to inhibition of nicotinamide adenine dinucleotide (NAD) production and decrease in intracellular energy level. Additionally, the susceptibility to doxorubicin was elevated in the miRNA mimics treated cells.61 Finally, Chi et al exhibited that the expression level of oncogene lncRNA SNHG5 was high in BC tissue specimens compared to noncancerous samples and there was a negative correlation between SNHG5 expression and miR-154-5p expression. Furthermore, they proved that by acting as a sponge for miR-154-5p, SNHG5 increases the expression of PCNA, the direct target of miR-154-5p. It was determined that upregulation of PCNA was associated with progression of BC and ectopic over expression of miR-154-5p or knockdown of SNHG5 could alleviate the disease.62 In conclusion, the aforementioned studies clearly indicate that miR-154 plays an exact tumor suppressor role in BC and it should be considered as a novel and potential therapeutic target.
MiR-154 and Liver Cancer
The incidence of liver cancer is in warning state. As evidence, it has been claimed that in 2018, 841,080 liver cancer cases were recorded globally which account for 4.7% all types of cancers.63 Novel and efficient diagnostic and therapeutic tools would significantly contribute to the reduction of liver cancer. With this aspect, in a comprehensive meta-analysis of miRNA expression microarray datasets, Wang et al, found that miR-154 was significantly downregulated in different cancerous tissues including HCC compared to adjacent normal tissues. They further investigated the role of miR-154 in HCC cell lines and revealed that miR-154 plays an antimalignance role in HCC by directly binding to 3′-UTR of CCND2.64 In another study, Pang et al proved that miR-154 functioned as a potential tumor suppressor in HCC by directly targeting ZEB2. They also showed that miR-154 expression was negatively correlated with tumor differentiation, TNM stage and lymph node metastasis.65 Hepatitis B virus (HBV) is a major risk factor for liver cancer, in which HBV covalently closed circular DNA (cccDNA) plays crucial roles. Feng et al investigated the roles of lncRNA PCNAP1 in contribution of HBV replication through modulating miR-154/PCNA/HBV cccDNA signaling in hepatocarcinogenesis and observed upregulated levels of PCNAP1 and PCNA in the liver of HBV-infectious human liver-chimeric mice. They also proved that PCNAP1 enhanced PCNA expression through functioning as a sponge for miR-154, which was associated with progression HCC.66 In summary, the above-mentioned studies illuminate that miR-154 plays pivotal role in tumor suppression of HCC suggesting that miR-154 might be an important target for both prognosis and therapy of liver cancer.
MiR-154 and Endocrine Cancers
Pancreatic cancer is ranked the seventh among the leading cause of cancer-related deaths worldwide. However, its losses are higher in more developed countries. It has not yet been clarified why the mortality rate varies tremendously among pancreatic cancer patients, but perhaps lack of appropriate diagnosis, treatment and cataloging strategies are the reasons. Considering that the patients are not really symptomatic until an advanced stage of the disease, pancreatic cancer remains one of the most lethal malignancies that caused 432,242 new deaths in 2018 (GLOBOCAN 2018 estimates). Novel diagnostic and therapeutic tools will definitely contribute to decreasing the deaths.67 Autophagy has been claimed to be one of the major contributors to occurrence and progression of pancreatic cancer. In a comprehensive study, Wei et al tried to understand differential expression of mRNAs, miRNAs, lncRNAs and circRNAs postautophagy suppression by chloroquine diphosphate in PANC-1 cells. According to their analysis, only two miRNAs, including miR-154-3p had declined expression in the cell line. Additionally, they constructed a ceRNA network containing miR-154-3p, 5 circRNAs, 2 lncRNAs and 11 genes.68 In a parallel study, Mian et al analyzed miRNA expression profile in 34 cases of sporadic medullary thyroid carcinoma (MTC), 6 cases of hereditary MTC and 2 cases of C-cell hyperplasia (CCH). Based on their experiments, miR-154 was significantly upregulated in both MTC and CCH.69 On the contrary, Fan et al measured decreased expression levels of miR-154-3p and miR-487-3p in thyroid cancer tissue specimens and cell lines which was associated with tumor size, TNM stage, histological grade, lymph node metastasis and shorter overall survival in the patients. Furthermore, they proved that both miRNA suppress progression of thyroid cancer by synergistically targeting ROHA and blocking its expression.70 Taking together, the aforementioned studies initially suggest that miR-154 may be employed for prognosis and diagnosis of endocrine cancers, however, its precise tumor suppressor role and the beyond mechanisms should be deeply investigated.
MiR-154 and Other Malignancies
Tumors arising from bones and joints are responsible for 1,660 deaths in 2019, in the United States.1 Osteosarcoma, the most common primary bone tumor occurs most frequently in adolescents but a second incidence peak among individuals over age 60 can also be observed. In young patients, it usually stem from the metaphyses of long bones, such as the distal femur, proximal tibia, and proximal humerus.71 Despite significant advancements in the diagnosis and treatment of osteosarcoma to date, overall survival has remained relatively constant for over 2 decades.72 Recent studies suggest that miR-154 plays tumor suppressor role in osteosarcoma. In this regard, Tian et al evaluated decreased expression of miR-154-5p in human osteosarcoma tissue samples. Additionally, ectopic overexpression of miR-154-5p could suppress progression of osteosarcoma and induce apoptosis via upregulating Bax and cleaved caspase 3, and downregulation of Bcl-2. Finally, they identified E2F5 as a direct target of miR-154-5p.73 In a similar study, Zhou et al demonstrated that miR-154 acted as a tumor suppressor in osteosarcoma by targeting Wnt5a.74 Melanoma is a malignant tumor that arises from uncontrolled proliferation of melanocytes-pigment-producing cells. While the most common form of melanoma is cutaneous, it can also arise in mucosal surfaces, the uveal tract, and leptomeninges. The worldwide incidence of melanoma has risen rapidly over the course of the last 50 years.75 Using melanoma tissue samples and cell lines, Wang et al measured downregulated levels of miR-154 which was associated with advanced tumor stage ulceration and shorter overall survival. Additionally, they confirmed that miR-154 through targeting AURKA could inhibit progression of melanoma.76 Cutaneous SCC is a kind of malignant tumor of keratinocytes which is correlated with recurrence, metastasis, and mortality. It was determined that the expression of miR-154 was declined in human skin SCC cell lines and WHSC1 was direct target of miR-154. Moreover, overexpression of miR-154 could silence WHSC1 expression, inhibit the activation of P53 signaling pathway and also increase expression of P73, P16 and Bax, as well as decreased expression of P53, c-myc and Bcl-2 which led to suppression of skin SCC progression.77 A quantitative expression profile analysis of 157 mature miRNAs was performed on 100 acute myeloid leukemia (AML) patients representing the spectrum of known karyotypes common in AML. It was determined that the whole set of genes, including a subset of miRNAs located in the human 14q32 imprinted domain was upregulated. Additionally, miR-154 was identified to be a potential tool for sub-classifying cancers suggesting a role in the etiology of leukemia.79
Concluding Remarks and Perspectives
Despite the improvement of lifestyle, awareness and human living environment, the incidence of different human cancers has been increased within the past decades. Several researchers are continually working to understand the detailed mechanisms of tumorigenesis and every year novel generations of anticancer drugs are being released, however, the survival rate of cancer patients has remained constant and only a little progress has been made. As a newly discovered miRNA, miR-154 has been claimed to possess considerable tumor-suppressive effects in various human cancers. Particularly, some of the studies indicate that ceRNA mechanism (so-called sponge effect) is remarkably involved in miR-154 mediated tumor suppression. The ceRNA mainly involves three kinds of RNAs, including mRNA, pseudogene transcripts and lncRNA, however, lncRNA followed by circRNA has aroused considerable research interest in the field of ceRNA family.80 Although the role of lncRNA particularly SNGH family in miR-154 mediated tumor suppression has been initially studied in a limited number of human cancers, further studies are still in demand to illuminate their oncogenic mechanisms and interactions with miR-154.
Taking together, the available evidence suggests that miR-154 is frequently downregulated in different human cancers and exerts antimalignance effects by targeting several genes. However, there are still some inconsistences particularly in tumors arising from GU and digestive systems, which should be clarified in future studies. Furthermore, the involvement of miR-154 has not yet been investigated in some of the tumors, particularly those of arising from the hematopoietic and lymphoid tissues. Additionally, the studies conducted on BC suggest that miR-154 is a novel diagnostic and therapeutic target, nevertheless, only a single study has been done on chemo-sensitivity induced by miR-154. It is a well-known concept that a combination of surgery, radiotherapy and chemotherapy are used for management of BC.81 Therefore, it is of crucial importance to deeply investigate the roles of miR-154 in chemo-sensitization and radio-sensitization. Finally, the majority of the studies summarized here have been conducted on clinical tissue specimens and cancerous cell lines, however, in vivo studies using animal models, especially mice bearing xenografts would enhance our knowledge of oncogenic mechanisms and significantly contribute to developing miR-154-based treatments. Such determinations should be covered in future researches.
Funding Statement
The authors received no financial support for the work.
Disclosure
The authors do declare that there are no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg Robert A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 3.Hesari Z, Nourbakhsh M, Hosseinkhani S, et al. Down-regulation of NAMPT expression by mir-206 reduces cell survival of breast cancer cells. Gene. 2018;673:149–158. doi: 10.1016/j.gene.2018.06.021 [DOI] [PubMed] [Google Scholar]
- 4.Yarahmadi S, Abdolvahabi Z, Hesari Z, et al. Inhibition of sirtuin 1 deacetylase by miR-211-5p provides a mechanism for the induction of cell death in breast cancer cells. Gene. 2019;711:143939. doi: 10.1016/j.gene.2019.06.029 [DOI] [PubMed] [Google Scholar]
- 5.Cloonan N, Wani S, Xu Q, et al. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol. 2011;12(12):R126. doi: 10.1186/gb-2011-12-12-r126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 7.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921 [DOI] [PubMed] [Google Scholar]
- 8.Macfarlane LA, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genomics. 2010;11(7):537–561. doi: 10.2174/138920210793175895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho Y, Baldán Á. Quest for new biomarkers in atherosclerosis. Missouri Med. 2013;110(4):325. [PMC free article] [PubMed] [Google Scholar]
- 10.Catalanotto C, Cogoni C, Zardo G. MicroRNA in control of gene expression: an overview of nuclear functions. Int J Mol Sci. 2016;17(10):1712. doi: 10.3390/ijms17101712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindow M, Kauppinen S. Discovering the first microRNA-targeted drug. J Cell Biol. 2012;199(3):407–412. doi: 10.1083/jcb.201208082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van der Ree M, Van Der Meer A, Van Nuenen A, et al. Miravirsen dosing in chronic hepatitis C patients results in decreased micro RNA‐122 levels without affecting other micro RNA s in plasma. Aliment Pharmacol Ther. 2016;43(1):102–113. doi: 10.1111/apt.13432 [DOI] [PubMed] [Google Scholar]
- 13.Milosevic J, Pandit K, Magister M, et al. The role and regulation of miR-154 microRNA family in lung fibrosis Am J Respir Crit Care Med. 2010;181:A2019. [Google Scholar]
- 14.Milosevic J, Pandit K, Magister M, et al. Profibrotic role of miR-154 in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2012;47(6):879–887. doi: 10.1165/rcmb.2011-0377OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen M, Yang Y, Zhang W, et al. Long noncoding RNA SNHG5 knockdown alleviates neuropathic pain by targeting the miR-154-5p/CXCL13 axis. Neurochem Res. 2020. doi: 10.1007/s11064-020-03021-2 [DOI] [PubMed] [Google Scholar]
- 16.Ren H, Ma X, Shao Y, Han J, Yang M, Wang Q. Correlation between serum miR-154-5p and osteocalcin in males and postmenopausal females of type 2 diabetes with different urinary albumin creatinine ratios. Front Endocrinol. 2019;10:542. doi: 10.3389/fendo.2019.00542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao Q, Ju Z, Zhang Y, et al. Association of TNP2 gene polymorphisms of the bta-miR-154 target site with the semen quality traits of Chinese Holstein bulls. PLoS One. 2014;9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pateisky P, Pils D, Szabo L, et al. hsa-miRNA-154-5p expression in plasma of endometriosis patients is a potential diagnostic marker for the disease. Reprod Biomed Online. 2018;37(4):449–466. doi: 10.1016/j.rbmo.2018.05.007 [DOI] [PubMed] [Google Scholar]
- 19.Dong P, Liu W, Wang Z. MiR-154 promotes myocardial fibrosis through β-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22(7):2052–2060. doi: 10.26355/eurrev_201804_14735 [DOI] [PubMed] [Google Scholar]
- 20.Ren H, Wu C, Shao Y, Liu S, Zhou Y, Wang Q. Correlation between serum miR-154-5p and urinary albumin excretion rates in patients with type 2 diabetes mellitus: a cross-sectional cohort study. Front Med. 2020;1–9. [DOI] [PubMed] [Google Scholar]
- 21.Wong T-S, Liu X-B, Wong BY-H, Ng RW-M, Yuen AP-W, Wei WI. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14(9):2588–2592. doi: 10.1158/1078-0432.CCR-07-0666 [DOI] [PubMed] [Google Scholar]
- 22.Qiao W, Cao N, Yang L. MicroRNA-154 inhibits the growth and metastasis of gastric cancer cells by directly targeting MTDH. Oncol Lett. 2017;14(3):3268–3274. doi: 10.3892/ol.2017.6558 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Song J, Guan Z, Li M, et al. MicroRNA-154 inhibits the growth and invasion of gastric cancer cells by targeting DIXDC1/WNT signaling. Oncol Res. 2018;26(6):847–856. doi: 10.3727/096504017X15016337254632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4(2):68–75. doi: 10.4161/org.4.2.5851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung JY, Park YR, Choi SH, Seo SY, Lee ST, Kim SW. miR-124 and miR-154 inhibit colorectal cancer cell proliferation, migration, and invasion by suppressing TRAF6. Paper presented at: Journal of Gastroenterology and Hepatology; 2018; NJ USA. [Google Scholar]
- 26.Xin C, Zhang H, Liu Z. miR-154 suppresses colorectal cancer cell growth and motility by targeting TLR2. Mol Cell Biochem. 2014;387(1–2):271–277. doi: 10.1007/s11010-013-1892-3 [DOI] [PubMed] [Google Scholar]
- 27.D’Angelo E, Fassan M, Maretto I, et al. Serum miR-125b is a non-invasive predictive biomarker of the pre-operative chemoradiotherapy responsiveness in patients with rectal adenocarcinoma. Oncotarget. 2016;7(19):28647–28657. doi: 10.18632/oncotarget.8725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu M, Chen X, Lin K, et al. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol Cancer. 2018;17(1):1–16. doi: 10.1186/s12943-018-0894-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naghavi M, Abajobir AA, Abbafati C, et al. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390(10100):1151–1210. doi: 10.1016/S0140-6736(17)32152-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niu J-T, Zhang L-J, Huang Y-W, Li C, Jiang N, Niu Y-J. MiR-154 inhibits the growth of laryngeal squamous cell carcinoma by targeting GALNT7. Biochem Cell Biol. 2018;96(6):752–760. doi: 10.1139/bcb-2018-0047 [DOI] [PubMed] [Google Scholar]
- 31.Huang J, Wu J, Li Y, et al. Deregulation of serum microRNA expression is associated with cigarette smoking and lung cancer. Biomed Res Int. 2014;2014:1–13. doi: 10.1155/2014/364316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cazzoli R, Buttitta F, Di Nicola M, et al. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol. 2013;8(9):1156–1162. doi: 10.1097/JTO.0b013e318299ac32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin X, Yang Z, Zhang P, Shao G. miR-154 suppresses non-small cell lung cancer growth in vitro and in vivo. Oncol Rep. 2015;33(6):3053–3060. doi: 10.3892/or.2015.3895 [DOI] [PubMed] [Google Scholar]
- 34.Lin X, Yang Z, Zhang P, Liu Y, Shao G. miR-154 inhibits migration and invasion of human non-small cell lung cancer by targeting ZEB2. Oncol Lett. 2016;12(1):301–306. doi: 10.3892/ol.2016.4577 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Xue YJ, Fu YJ, Wang JH. The inhibitory effect of MIR-154-3p on metastasis of non-small cell lung cancer cell lines. Tumor. 2015;35:498–507. [Google Scholar]
- 36.Lingling J, Xiangao J, Guiqing H, Jichan S, Feifei S, Haiyan Z. SNHG20 knockdown suppresses proliferation, migration and invasion, and promotes apoptosis in non-small cell lung cancer through acting as a miR-154 sponge. Biomed Pharmacother. 2019;112:108648. doi: 10.1016/j.biopha.2019.108648 [DOI] [PubMed] [Google Scholar]
- 37.Zheng W, Feng Q, Liu J, et al. Inhibition of 6-phosphogluconate dehydrogenase reverses cisplatin resistance in ovarian and lung cancer. Front Pharmacol. 2017;8:421. doi: 10.3389/fphar.2017.00421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin J, Spitz MR, Dinney CP, Etzel CJ, Grossman HB, Wu X. Bladder cancer risk as modified by family history and smoking. Cancer. 2006;107(4):705–711. doi: 10.1002/cncr.22071 [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Mao S, Wang L, et al. MicroRNA-154 functions as a tumor suppressor in bladder cancer by directly targeting ATG7. Oncol Rep. 2019;41(2):819–828. doi: 10.3892/or.2018.6879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao X, Ji Z, Xie Y, Liu G, Li H. MicroRNA-154 as a prognostic factor in bladder cancer inhibits cellular malignancy by targeting RSF1 and RUNX2. Oncol Rep. 2017;38(5):2727–2734. doi: 10.3892/or.2017.5992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu C, Xu H, Zhang M, et al. Downregulation of miR‐154 in cervical cancer and its clinicopathological and prognostic significance. Int J Clin Exp Med. 2016;9(10):19791–19796. [Google Scholar]
- 42.Lin C, Li Z, Chen P, et al. Oncogene miR-154-5p regulates cellular function and acts as a molecular marker with poor prognosis in renal cell carcinoma. Life Sci. 2018;209:481–489. doi: 10.1016/j.lfs.2018.08.044 [DOI] [PubMed] [Google Scholar]
- 43.Rawla P. Epidemiology of prostate cancer. World J Oncol. 2019;10(2):63–89. doi: 10.14740/wjon1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dambal S, Giangreco AA, Acosta AM, et al. microRNAs and DICER1 are regulated by 1, 25-dihydroxyvitamin D in prostate stroma. J Steroid Biochem Mol Biol. 2017;167:192–202. doi: 10.1016/j.jsbmb.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Formosa A, Markert E, Lena A, et al. MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p, mapped to the 14q32. 31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene. 2014;33(44):5173–5182. doi: 10.1038/onc.2013.451 [DOI] [PubMed] [Google Scholar]
- 46.Zhu C, Li J, Cheng G, et al. miR-154 inhibits EMT by targeting HMGA2 in prostate cancer cells. Mol Cell Biochem. 2013;379(2):69–75. doi: 10.1007/s11010-013-1628-4 [DOI] [PubMed] [Google Scholar]
- 47.Zhu C, Shao P, Bao M, et al. miR-154 inhibits prostate cancer cell proliferation by targeting CCND2 Urologic Oncology: Seminars and Original Investigations. 2014;32(1):31e9–16. [DOI] [PubMed] [Google Scholar]
- 48.Zheng Y, Zhu C, Ma L, et al. miRNA-154-5p inhibits proliferation, migration and invasion by targeting E2F5 in prostate cancer cell lines. Urol Int. 2017;98(1):102–110. doi: 10.1159/000445252 [DOI] [PubMed] [Google Scholar]
- 49.Patel AP, Fisher JL, Nichols E, et al. Global, regional, and national burden of brain and other CNS cancer, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019;18(4):376–393. doi: 10.1016/S1474-4422(18)30468-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, Wu L, Wu J. Downregulation of miR-154 in human glioma and its clinicopathological and prognostic significance. J Int Med Res. 2016;44(5):994–1001. doi: 10.1177/0300060516649487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X, Sun S, Tong X, et al. MiRNA-154-5p inhibits cell proliferation and metastasis by targeting PIWIL1 in glioblastoma. Brain Res. 2017;1676:69–76. doi: 10.1016/j.brainres.2017.08.014 [DOI] [PubMed] [Google Scholar]
- 52.Zhou H, Zhang Y, Lai Y, Xu C, Cheng Y. Circ_101064 regulates the proliferation, invasion and migration of glioma cells through miR-154–5p/PIWIL1 axis. Biochem Biophys Res Commun. 2020. doi: 10.1016/j.abb.2020.108366 [DOI] [PubMed] [Google Scholar]
- 53.Yang L, Yan Z, Wang Y, Ma W, Li C. Down‐expression of miR‐154 suppresses tumourigenesis in CD133+ glioblastoma stem cells. Cell Biochem Funct. 2016;34(6):404–413. doi: 10.1002/cbf.3201 [DOI] [PubMed] [Google Scholar]
- 54.DeSantis CE, Ma J, Gaudet MM, et al. Breast cancer statistics, 2019. CA Cancer J Clin. 2019;69(6):438–451. doi: 10.3322/caac.21583 [DOI] [PubMed] [Google Scholar]
- 55.Lowery A, Miller N, McNeill R, Kerin M. MicroRNA expression in primary breast tumors. Poster presented at: 30 th Annual San Antonio Breast Cancer Symposium; 2007; Texas-USA. [Google Scholar]
- 56.Miranda P, Vimalraj S, Selvamurugan N. A feedback expression of microRNA-590 and activating transcription factor-3 in human breast cancer cells. Int J Biol Macromol. 2015;72:145–150. doi: 10.1016/j.ijbiomac.2014.07.051 [DOI] [PubMed] [Google Scholar]
- 57.Li S, Meng H, Zhou F, et al. MicroRNA-132 is frequently down-regulated in ductal carcinoma in situ (DCIS) of breast and acts as a tumor suppressor by inhibiting cell proliferation. Pathol Res Pract. 2013;209(3):179–183. doi: 10.1016/j.prp.2012.12.002 [DOI] [PubMed] [Google Scholar]
- 58.Lowery A, Miller N, Kerin M. Expression of the putative breast cancer gene BASE; relationship with microRNA-154* and estrogen receptor status. Poster presented at: 30 th Annual San Antonio Breast Cancer Symposium; 2007; Texas-USA. [Google Scholar]
- 59.Xu H, Fei D, Zong S, Fan Z. MicroRNA-154 inhibits growth and invasion of breast cancer cells through targeting E2F5. Am J Transl Res. 2016;8(6):2620. [PMC free article] [PubMed] [Google Scholar]
- 60.Qin C, Zhao Y, Gong C, Yang Z. MicroRNA-154/ADAM9 axis inhibits the proliferation, migration and invasion of breast cancer cells. Oncol Lett. 2017;14(6):6969–6975. doi: 10.3892/ol.2017.7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pour ZB, Nourbakhsh M, Mousavizadeh K, et al. Suppression of nicotinamide phosphoribosyltransferase expression by miR-154 reduces the viability of breast cancer cells and increases their susceptibility to doxorubicin. BMC Cancer. 2019;19(1):1–13. doi: 10.1186/s12885-018-5219-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chi J-R, Yu Z-H, Liu B-W, et al. SNHG5 promotes breast cancer proliferation by sponging the miR-154-5p/PCNA axis. Mol Ther Nucleic Acids. 2019;17:138–149. doi: 10.1016/j.omtn.2019.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goodarzi E, Ghorat F, Jarrahi A, Adineh H, Sohrabivafa M, Khazaei Z. Global incidence and mortality of liver cancers and its relationship with the human development index (HDI): an ecology study in 2018. World J Cancer Res. 2019;6:12. [Google Scholar]
- 64.Wang W, Peng B, Wang D, et al. Human tumor microRNA signatures derived from large‐scale oligonucleotide microarray datasets. Int J Cancer. 2011;129(7):1624–1634. doi: 10.1002/ijc.25818 [DOI] [PubMed] [Google Scholar]
- 65.Pang X, Huang K, Zhang Q, Zhang Y, Niu J. miR-154 targeting ZEB2 in hepatocellular carcinoma functions as a potential tumor suppressor. Oncol Rep. 2015;34(6):3272–3279. doi: 10.3892/or.2015.4321 [DOI] [PubMed] [Google Scholar]
- 66.Feng J, Yang G, Liu Y, et al. LncRNA PCNAP1 modulates hepatitis B virus replication and enhances tumor growth of liver cancer. Theranostics. 2019;9(18):5227. doi: 10.7150/thno.34273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rawla P, Sunkara T, Gaduputi V. Epidemiology of pancreatic cancer: global trends, etiology and risk factors. World J Oncol. 2019;10(1):10. doi: 10.14740/wjon1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wei DM, Jiang MT, Lin P, et al. Potential ceRNA networks involved in autophagy suppression of pancreatic cancer caused by chloroquine diphosphate: a study based on differentially‑expressed circRNAs, lncRNAs, miRNAs and mRNAs. Int J Oncol. 2019;54(2):600–626. doi: 10.3892/ijo.2018.4660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mian C, Pennelli G, Fassan M, et al. MicroRNA profiles in familial and sporadic medullary thyroid carcinoma: preliminary relationships with RET status and outcome. Thyroid. 2012;22(9):890–896. doi: 10.1089/thy.2012.0045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.X-d F, Luo Y, Wang J, An N. miR-154-3p and miR-487-3p synergistically modulate RHOA signaling in the carcinogenesis of thyroid cancer. Biosci Rep. 2020;40(1). doi: 10.1042/BSR20193470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the surveillance, epidemiology, and end results program. Cancer. 2009;115(7):1531–1543. doi: 10.1002/cncr.24121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Geller DS, Gorlick R. Osteosarcoma: a review of diagnosis, management, and treatment strategies. Clin Adv Hematol Oncol. 2010;8(10):705–718. [PubMed] [Google Scholar]
- 73.Tian Q, Gu Y, Wang F, et al. Upregulation of miRNA-154-5p prevents the tumorigenesis of osteosarcoma. Biomed Pharmacother. 2020;124:109884. doi: 10.1016/j.biopha.2020.109884 [DOI] [PubMed] [Google Scholar]
- 74.Zhou H, Zhang M, Yuan H, Zheng W, Meng C, Zhao D. MicroRNA-154 functions as a tumor suppressor in osteosarcoma by targeting Wnt5a. Oncol Rep. 2016;35(3):1851–1858. doi: 10.3892/or.2015.4495 [DOI] [PubMed] [Google Scholar]
- 75.Matthews N, Li W, Qureshi A, Weinstock M, Cho E. Epidemiology of melanoma In: Ward W, Farma J, editors. Cutaneous Melanoma: Etiology and Therapy [Internet]. Brisbane (AU): Codon Publications; 2017. [PubMed] [Google Scholar]
- 76.Wang J, Fang Y, Liu Y, et al. MiR-154 inhibits cells proliferation and metastasis in melanoma by targeting AURKA and serves as a novel prognostic indicator. Eur Rev Med Pharmacol Sci. 2019;23(10):4275–4284. doi: 10.26355/eurrev_201905_17932 [DOI] [PubMed] [Google Scholar]
- 77.Chen H-Q, Gao D. Inhibitory effect of microRNA-154 targeting WHSC1 on cell proliferation of human skin squamous cell carcinoma through mediating the P53 signaling pathway. Int J Biochem Cell Biol. 2018;100:22–29. doi: 10.1016/j.biocel.2018.04.021 [DOI] [PubMed] [Google Scholar]
- 78.Gururajan M, Josson S, Chu GC-Y, et al. miR-154* and miR-379 in the DLK1-DIO3 microRNA mega-cluster regulate epithelial to mesenchymal transition and bone metastasis of prostate cancer. Clin Cancer Res. 2014;20(24):6559–6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dixon-McIver A, East P, Mein CA, et al. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS One. 2008;3(5):e2141. doi: 10.1371/journal.pone.0002141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mei J, Xu R, Hao L, Zhang Y. MicroRNA-613: a novel tumor suppressor in human cancers. Biomed Pharmacother. 2020;123:109799. doi: 10.1016/j.biopha.2019.109799 [DOI] [PubMed] [Google Scholar]
- 81.Jabbari N, Zarei L, Esmaeili Govarchin Galeh H, Mansori Motlagh B. Assessment of synergistic effect of combining hyperthermia with irradiation and calcium carbonate nanoparticles on proliferation of human breast adenocarcinoma cell line (MCF-7 cells). Artif Cells Nanomed Biotechnol. 2018;46(sup2):364–372. doi: 10.1080/21691401.2018.1457537 [DOI] [PubMed] [Google Scholar]