Abstract
Background
Intratumoral heterogeneity is a crucial factor to the outcome of patients and resistance to therapies, in which structural variants play an indispensable but undiscovered role.
Methods
We performed an integrated analysis of optical mapping and whole-genome sequencing on a primary tumor (PT) and matched metastases including lymph node metastasis (LNM) and tumor thrombus in the pulmonary vein (TPV). Single nucleotide variants, indels and structural variants were analyzed to reveal intratumoral genetic heterogeneity among tumor cells in different sites.
Results
Our results demonstrated there were less nonsynonymous somatic variants shared with PT in LNM than in TPV, while there were more structural variants shared with PT in LNM than in TPV. More private variants and its affected genes associated with tumorigenesis and progression were identified in TPV than in LNM. It should be noticed that optical mapping detected an average of 77.1% (74.5–78.5%) large structural variants (>5,000 bp) not detected by whole-genome sequencing and identified several structural variants private to metastases.
Conclusions
Our study does demonstrate structural variants, especially large structural variants play a crucial role in intratumoral genetic heterogeneity and optical mapping could make up for the deficiency of whole-genome sequencing to identify structural variants.
Keywords: Heterogeneity, lung squamous cell carcinoma (LUSC), metastasis, optical mapping, structural variants
Introduction
Lung cancer is the leading cause of cancer-related death worldwide (1). The two major histological types are non-small-cell lung cancer (NSCLC) and small-cell lung cancer (SCLC) (2). Lung squamous cell carcinoma (LUSC), one of the common histological types of NSCLC, remains poor prognosis despite of development in therapeutic strategies (3-5). Meanwhile, intratumoral heterogeneity, which refers to heterogeneity among tumor cells of a single patient, is crucial for the clinical outcome of patients with lung cancer, impacting the curative effect of chemotherapy, radiotherapy and immunotherapy (6,7).
Next-generation sequencing (NGS), a method relying on short reads, has been performed on multiregional tumors to explore intratumoral genetic heterogeneity (ITGH) in NSCLC (8-10). Previous studies focused more on ITGH involving mutations that distinguish different tumor cells in a single or multiple primary NSCLC (7-9,11). A previous study explored the ITGH based on analysis of single nucleotide variants (SNVs) and copy number variants (CNVs) using whole-genome sequencing (WGS) on primary tumors, metastatic lymph nodes and tumor cells in the pleura (10). Because of the challenge in detecting technology, structural variants (SVs) increasingly appears to have an indispensable but undiscovered role in ITGH (12,13). However, ITGH which manifests uneven distribution of genetic alterations among lung tumor cells in primary tumor and associated metastases is not comprehensively characterized due to the lack of studies focusing on distant metastasis and SVs. Recently, optical mapping, a newly non-sequencing method, shed a light to dig large SVs (14,15).
In this study, we combined optical mapping and WGS to reveal the ITGH in various forms of SNVs, indels and SVs, especially large SVs (>5 kb) within primary tumor and associated metastases in a LUSC patient. We also compared SVs detected by optical mapping and those detected by WGS. Furthermore, after comparing the genes affected by variants with those associated with tumorigenesis and progression, we inferred the functional consequence of distinct genomic alterations among tumor cells within the primary site and paired metastatic sites.
Methods
Tissue collection
Surgical specimens of primary tumor (PT), lymph node metastases (LNM), tumor thrombus in the pulmonary vein (TPV) and adjacent normal lung tissue (at least 2cm away from tumor) were obtained from a patient who diagnosed with pathologically confirmed lung squamous cell carcinoma. This study was approved by the Committee for Ethical Review of Research. Informed consent was obtained.
Whole-genome sequencing
DNA extraction and sequencing: After fragmented by sonication to a size of 350 bp, genomic DNA fragments were end-polished, A-tailed, and ligated with adapter for Illumina sequencing. Then after further PCR amplification and purification, libraries were analyzed for size distribution by Agilent 2100 Bioanalyzer and quantified for concentration (2 nM) by flurogenic-quantitative PCR (Qubit 2.0). Then DNA libraries were sequenced on Illumina Novaseq 6000 sequencing platform with 30X sequencing depth. 150 bp paired-end reads were generated. Contaminated reads including adaptors, low quality reads and those with more “N” was extracted based on chastity score and quality score.
Variants detection and filtration: Paired-end reads in FastQ format were aligned to the reference human genome (UCSC Genome Browser, version hg19) by Burrows-Wheeler Aligner (BWA) (16). Subsequent BAM files were processed by SAMtools (17), Picard tool (http://picard.sourceforge.net/), and the Genome Analysis Toolkit (GATK) (18) to sort and remove duplication, local realignment, and base quality recalibration.
SNVs and indels detection: Mutect (19) was used to detect the somatic SNVs and indel with tumor-normal paired BAM files. ANNOVAR was used to further annotate for VCF (Variant Call Format) (20). Somatic SNVs were further filtered for analysis of mutational spectrum and signatures with the following criteria: SNVs which has no record in 1000 Genomes project, dbsnp or Berry4000 (Berry Genomics) were filtered (21,22).
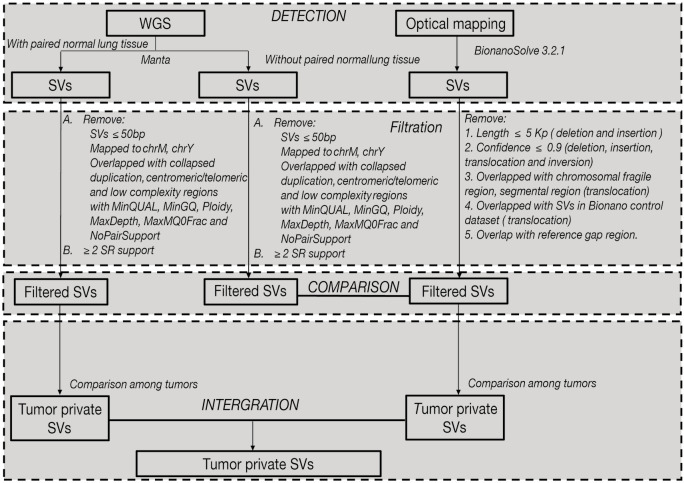
SVs detection, filtration and classification: Manta was applied for SVs detection (23), SVs were reported as INS (insertion), DEL (deletion), DUP (duplication), INV (inversion), and BND (further identified as inter-chromosomal translocation). Somatic SVs in PT, LNM and TPV were identified with the data of adjacent normal lung sample as control. ANNOVAR was applied for annotation (20). SVs were filtered if: SVs <50 bp; mapped to the mitochondrial genome or chromosome Y; overlapped with gap region, telomere, centromere or low complexity regions; with MinQUAL, MinGQ, Ploidy, MaxDepth, MaxMQ0Frac and NoPairSupport in VCF FILTER fields; and supported by <2 split reads (SR).
Optical mapping
DNA preparation: High Molecular Weight (HMW) DNA were extracted using Bionano Prep Animal Tissue DNA Isolation Fibrous Tissue Protocol (https://bionanogenomics.com/support-page/animal-tissue-dna-isolation-kit/) from the tissue of frozen PT, LNM and TPV. Firstly, approximately 10 mg of tissue were fixed, disrupted with a rotor-stator, embedded in 2% agarose, and digested with proteinase K and RNase. After multiple stabilization and recovery followed by digestion with Agarase (Thermo Fisher) enzyme, HMW DNA were released, cleaned by drop dialysis and homogenized. HMW DNA were quantitated using Qubit dsDNA BR Assay Kit.
Direct labeling: HMW DNA were extracted using Bionano Prep Direct Label and Stain (DLS) Protocol (https://bionanogenomics.com/support-page/dna-labeling-kit-dls/). Firstly, 750 ng HMW DNA were nicked by DLE-1 enzyme, recovered, labled with fluorophore and stained. Then labled and stained DNA were quantitated using modified Qubit dsDNA HS (High Sensitivity) Assay Kit. Each labeled sample was added to a BioNano Saphyr Chip (Bionano Genomics) and run on the Bionano Saphyr instrument, targeting 100× human genome coverage. The raw data were filtered by Bionano Access (v1.2.1) with the following criteria: molecule length >150 kb with average label density of 10–25/100 kb.
SVs detection and filtration: De novo assembly of long molecules into genome map and SVs detection by comparing with Hg19 were performed with software Bionano Solve (version 3.2.1). SVs were annotated by Enliven (Berry Genomics). Then SVs were filtered if: for translocation and inversion, (I) confidence value <0.9, (II) breakpoints were located in the chromosome fragile site, (III) breakpoints were located in the segmental region of the chromosome, (IV) breakpoints were within these previously identified SVs (24); For insertion and deletion, (I) confidence value <0.9, (II) length of variation <5 kb, (III) breakpoints were in the gap region of reference genome.
Comparison of SVs from optical mapping and WGS
WGS provide SVs breakpoints (start and end) with base pair resolution, while optical mapping provides only the nearest labeling site to the interval of SVs. We determined whether SVs from optical mapping overlap with SVs from WGS with the following criteria: (I) Deletions, insertions and duplications detected by WGS must overlap with the interval of SVs detected by optical mapping. (II) The breakpoints of Inversions detected by WGS must lie within 500 kb to the interval of SVs detected by optical mapping.
Comparison of SVs from WGS among PT, LNM and TPV
Somatic SVs from WGS in PT, LNM and TPV were classified as shared SVs or private SVs among tumors with the following criteria: SVs has the same breakpoints (start and end), consistent type with SVs in another tumor were identified as identical and classified as shared SVs.
Comparison of SVs from optical mapping among PT, LNM and TPV
SVs from optical mapping in PT, LNM and TPV were classified as shared or private SVs among tumors with the following criteria: SVs have overlapped interval, consistent type with SVs in another tumor were identified as shared SVs. We further filtered the shared SVs in all tumors due to the shared somatic SVs and germline SVs could not be distinguished.
Identification of genes affected by SVs
For variants from WGS, we inferred a gene affected by variants if (I) a protein coding gene is annotated with an exon-annotated deletion, insertion and duplication; (II) the breakpoint (start or end) of inversion or inter-chromosome translocation lies within one or more exon of the genes; (III) the genes carried an nonsynonymous variants (nonsynonymous SNVs or frameshifting indels).
For SVs from optical mapping, we inferred a gene affected by variants if the gene was annotated with an exon-annotated SVs.
Functional consequence analysis
For genes affected by variants, we inferred whether these genes are associated with tumorigenesis and progression based on data of lung cancer driver genes (25-27), pan-cancer driver genes (28), COSMIC (https://cancer.sanger.ac.uk/census) (29), DNA repair genes (30) and hallmark genes of epithelial-mesenchymal transition (EMT) (31-38). Based on the data of The Human Protein Atlas (www.proteinatlas.org) (39-41), we further examined whether RNA expression of these genes correlate with the outcome of lung cancer and its protein expression and classified them as unprognostic, prognostic favorable and prognostic unfavorable genes.
KEGG enrichment
Genes only affected by variants in LNM and TPV were used to KEGG enrichment analysis by The Database for Annotation, Visualization and Integrated Discovery (DIVID) (42) and KOBAS 3.0 (http://kobas.cbi.pku.edu.cn/index.php).
Statistical analysis
We used R (version 3.3.3, version 3.6.1) software. “SomaticSignatures”, “ggplot2”, “ggrepel”, “ggthemes” were used in the analyses (43,44).
Results
Patients’ characterization
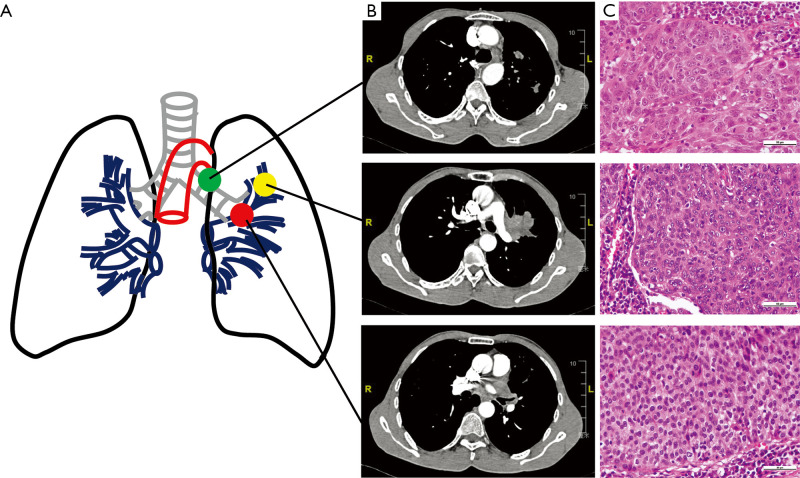
A 50-year-old East Asian male with 20 pack year history of smoking for 20 years, was diagnosed with lung squamous cell carcinoma with histopathological confirmation (Figure 1). Before systematic treatment, primary tumor (PT) located in the left upper lobe of lung, metastasis of left lower paratracheal (4L) lymph node (LNM) and tumor thrombus of the left Superior pulmonary vein (TPV) were sampled by surgical section. Furthermore, there is no reported family history of lung cancer. No significant difference in Tumor grade heterogeneity among tumor cells in primary and metastatic sites were identified by hematoxylin and eosin staining (Figure 1C, Figure S1).
Figure 1.
Clinical and histological diagnostic results of a patient with LUSC. (A) Schematic diagram of the primary tumors (PT) and lymph node metastases (LNM) and tumor thrombus in pulmonary vein (TPV). (B) Preoperative enhanced computerized tomography (enhanced-CT) scanning showed the PT (upper), LNM (middle) and TPV (lower). (C) Postoperative paraffin section and hematoxylin and eosin (H&E) staining image based on 400× magnification. Tumor cells in PT, LNM and TPV were moderately or poorly differentiated. PT, primary tumor; LNM, lymph node metastases; TPV, tumor thrombus in pulmonary vein.
Figure S1.
Postoperative paraffin section and hematoxylin and eosin (H&E) staining image for PT (A and B), LNM (C) and TPV (D) based on 40–100× magnification. PT, primary tumor; LNM, lymph node metastases; TPV, tumor thrombus in pulmonary vein.
ITGH in the form of SNVs and indels
To gain an insight into alterations of different mutational characteristics between the primary tumor and the metastases, we performed WGS on PT, LNM, TPV and adjacent normal lung tissue at an average depth of 30X.
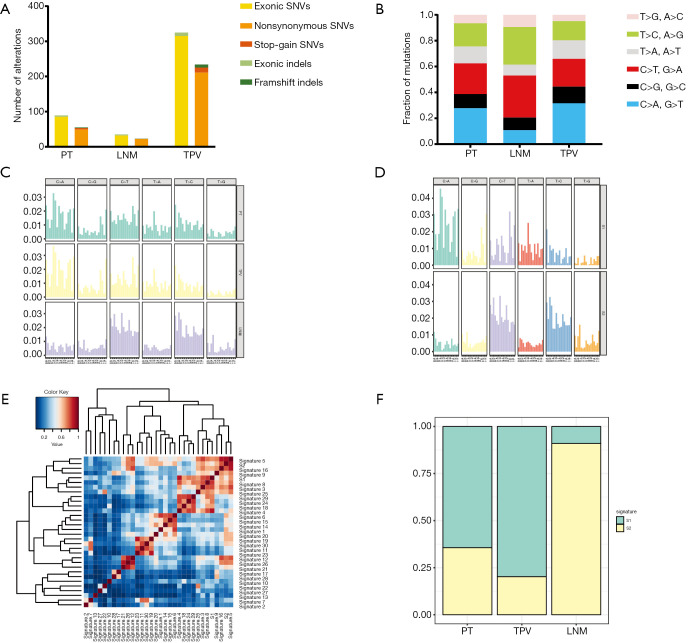
A total of 268 nonsynonymous somatic variants (including nonsynonymous SNVs and frameshifting indels) in 252 genes were identified in at least one tumor (Table S1), and 14.2% (38) of these variants were shared between PT and either one of the two metastases (Figure 2 and Figure 3A). Among them, 3 mutations were common in all tumors, while compared with LNM (5), a larger number of mutations (36) in TPV were shared with PT. 17, 15 and 195 mutations were uniquely seen in PT, LNM and TPV, respectively. Specifically, nonsynonymous SNV in TP53 which is one of the most commonly mutated gene in LUCC (45) were only detected in TPV. We further analyzed the mutation spectrum of SNVs (Figure 3A,B,C), trying to identify significant discordance between LNM and TPV. To be specific, we identified that TPV and PT both displayed a predominance of cytosine-adenine (C > A) nucleotide transversions which implied a correlation with tobacco exposure (46), consistent with the long-term smoking history of this patient. Meanwhile, the LNM exhibited a distinct preponderance of guanine-adenine (G > A) and adenine-guanine (A > G). Moreover, the detailed analysis of mutational signature showed S1 and S2 were extracted (Figure 3D). Compared with the previously known mutational signatures shown in COSMIC (29), S1 had the most similarity with signature 4 likely due to direct damage by mutagens in tobacco, and S2 exhibits the thymine-cytosine (T > C) as same as the signature 5 increased in many cancer types due to tobacco smoking (Figure 3E). Primary tumor and metastasis shared identical mutational signatures, but the proportion is different (Figure 3F). These results demonstrated patient have primary tumor and metastasis in different sites has high ITGH in the form of SNVs and indels.
Table S1. Somatic nonsynonymous SNVs and indels detected in PT, LNM and TPV.
| Start | End | Ref | Alt | Exonicfunc | Sample | Gene |
|---|---|---|---|---|---|---|
| 8399673 | 8399673 | C | A | Stopgain | PT, TPV | SLC45A1 |
| 13183833 | 13183833 | C | T | Nonsynonymous SNV | PT, LNM, TPV | HNRNPCL2 |
| 33385852 | 33385852 | C | T | Nonsynonymous SNV | PT | AQP7 |
| 79403883 | 79403883 | T | C | Nonsynonymous SNV | PT, TPV | ADGRL4 |
| 33385863 | 33385863 | G | T | Nonsynonymous SNV | PT | AQP7;AQP7 |
| 146057344 | 146057344 | T | C | Nonsynonymous SNV | PT, LNM | NBPF11 |
| 144061414 | 144061414 | G | A | Nonsynonymous SNV | PT | ARHGEF5 |
| 242121845 | 242121845 | G | T | Nonsynonymous SNV | PT, TPV | BECN2 |
| 69034420 | 69034420 | G | T | Nonsynonymous SNV | PT, TPV | ARHGAP25 |
| 84822875 | 84822875 | C | G | Nonsynonymous SNV | PT, TPV | DNAH6 |
| 88478308 | 88478308 | G | A | Nonsynonymous SNV | PT, TPV | THNSL2 |
| 98127921 | 98127921 | T | C | Nonsynonymous SNV | PT, LNM | ANKRD36B |
| 143713839 | 143713839 | A | T | Nonsynonymous SNV | PT, TPV | KYNU |
| 40523437 | 40523437 | C | G | Nonsynonymous SNV | PT, TPV | ZNF619 |
| 42956494 | 42956494 | G | T | Nonsynonymous SNV | PT, TPV | ZNF662 |
| 1201932 | 1201932 | G | T | Nonsynonymous SNV | PT, TPV | SLC6A19 |
| 38972028 | 38972028 | C | G | Nonsynonymous SNV | PT, TPV | RICTOR |
| 75427978 | 75427978 | G | A | Nonsynonymous SNV | PT, TPV | SV2C |
| 26056229 | 26056229 | C | A | Nonsynonymous SNV | PT, TPV | HIST1H1C |
| 32713598 | 32713598 | T | C | Nonsynonymous SNV | PT, TPV | HLA-DQA2 |
| 34949727 | 34949727 | G | C | Nonsynonymous SNV | PT, TPV | ANKS1A |
| 51656112 | 51656112 | T | G | Nonsynonymous SNV | PT, TPV | PKHD1 |
| 143269952 | 143269952 | A | T | Nonsynonymous SNV | PT | CTAGE15 |
| 48545953 | 48545953 | C | T | Nonsynonymous SNV | PT, TPV | ABCA13 |
| 161487805 | 161487805 | T | C | Nonsynonymous SNV | PT | FCGR2A |
| 82934997 | 82934997 | T | C | Nonsynonymous SNV | PT | GOLGA6L10 |
| 118922882 | 118922882 | A | C | Nonsynonymous SNV | PT | HYOU1 |
| 45994014 | 45994014 | C | T | Nonsynonymous SNV | PT | KRTAP10-4 |
| 150269712 | 150269712 | G | A | Nonsynonymous SNV | PT, TPV | GIMAP4 |
| 100642828 | 100642828 | C | T | Nonsynonymous SNV | PT | MUC12 |
| 100643427 | 100643427 | G | A | Nonsynonymous SNV | PT | MUC12 |
| 70918964 | 70918964 | G | A | Nonsynonymous SNV | PT, TPV | FOXD4L3 |
| 90502176 | 90502176 | C | A | Nonsynonymous SNV | PT, TPV | SPATA31E1 |
| 107266990 | 107266990 | G | A | Stopgain | PT, TPV | OR13F1 |
| 112189256 | 112189256 | C | T | Stopgain | PT, TPV | PTPN3 |
| 4967678 | 4967678 | G | A | Nonsynonymous SNV | PT, LNM, TPV | OR51A4 |
| 145326106 | 145326106 | A | T | Nonsynonymous SNV | PT | NBPF10 |
| 248616705 | 248616711 | TGCTGCG | – | Frameshift deletion | PT | OR2T2 |
| 78591144 | 78591144 | A | G | Nonsynonymous SNV | PT, TPV | NAV3 |
| 24523931 | 24523931 | G | C | Nonsynonymous SNV | PT, TPV | CARMIL3 |
| 6797520 | 6797520 | G | C | Nonsynonymous SNV | PT | RSPH10B;RSPH10B2 |
| 68475842 | 68475842 | T | G | Nonsynonymous SNV | PT | TESMIN |
| 50830413 | 50830413 | C | G | Stopgain | PT, TPV | CYLD |
| 60050130 | 60050130 | T | A | Nonsynonymous SNV | PT, TPV | MED13 |
| 72341009 | 72341009 | G | T | Nonsynonymous SNV | PT, TPV | KIF19 |
| 18534948 | 18534948 | G | C | Nonsynonymous SNV | PT, TPV | ROCK1 |
| 3150255 | 3150255 | G | C | Nonsynonymous SNV | PT, TPV | GNA15 |
| 1306817 | 1306817 | G | A | Nonsynonymous SNV | PT | TPSD1 |
| 39111054 | 39111054 | C | G | Nonsynonymous SNV | PT, TPV | EIF3K |
| 40399430 | 40399430 | T | C | Nonsynonymous SNV | PT, TPV | FCGBP |
| 55100038 | 55100038 | C | A | Nonsynonymous SNV | PT, TPV | FAM209A |
| 32647032 | 32647032 | A | C | Nonsynonymous SNV | PT | TXLNA |
| 16277757 | 16277757 | C | T | Nonsynonymous SNV | PT, LNM, TPV | POTEH |
| 10472843 | 10472843 | T | G | Nonsynonymous SNV | PT | TYK2 |
| 104379506 | 104379506 | – | TT | Frameshift insertion | PT, TPV | TDG;TDG |
| 12942047 | 12942047 | C | T | Nonsynonymous SNV | LNM | PRAMEF4 |
| 145302775 | 145302775 | T | G | Nonsynonymous SNV | LNM | NBPF10 |
| 195509939 | 195509939 | G | T | Nonsynonymous SNV | LNM | MUC4 |
| 195509941 | 195509941 | A | C | Nonsynonymous SNV | LNM | MUC4 |
| 140574103 | 140574103 | T | G | Nonsynonymous SNV | LNM | PCDHB10 |
| 56499000 | 56499000 | A | G | Nonsynonymous SNV | LNM | DST |
| 74159167 | 74159167 | G | C | Nonsynonymous SNV | LNM, TPV | GTF2I |
| 100644127 | 100644127 | C | T | Nonsynonymous SNV | LNM | MUC12 |
| 100644211 | 100644211 | C | T | Nonsynonymous SNV | LNM, TPV | MUC12 |
| 100644793 | 100644793 | C | T | Nonsynonymous SNV | LNM | MUC12 |
| 128471007 | 128471007 | T | G | Nonsynonymous SNV | LNM | FLNC |
| 135440222 | 135440222 | C | T | Nonsynonymous SNV | LNM | FRG2B |
| 89819380 | 89819380 | A | G | Nonsynonymous SNV | LNM | UBTFL1 |
| 74363307 | 74363307 | C | T | Nonsynonymous SNV | LNM, TPV | GOLGA6A |
| 54745682 | 54745682 | C | T | Nonsynonymous SNV | LNM | LILRA6;LILRB3 |
| 56274086 | 56274086 | G | A | Nonsynonymous SNV | LNM | RFPL4A |
| 24579049 | 24579049 | G | A | Nonsynonymous SNV | LNM | SUSD2 |
| 23653975 | 23653975 | - | CCGG | Frameshift insertion | LNM | BCR |
| 2523380 | 2523380 | G | T | Nonsynonymous SNV | TPV | MMEL1 |
| 55545264 | 55545264 | C | T | Nonsynonymous SNV | TPV | USP24 |
| 91403621 | 91403621 | C | G | Nonsynonymous SNV | TPV | ZNF644 |
| 108771623 | 108771623 | C | A | Nonsynonymous SNV | TPV | NBPF4 |
| 117158857 | 117158857 | C | T | Nonsynonymous SNV | TPV | IGSF3 |
| 145356733 | 145356733 | C | G | Nonsynonymous SNV | TPV | NBPF19 |
| 156531719 | 156531719 | C | T | Nonsynonymous SNV | TPV | IQGAP3 |
| 157514189 | 157514189 | C | T | Nonsynonymous SNV | TPV | FCRL5 |
| 179562624 | 179562624 | G | A | Nonsynonymous SNV | TPV | TDRD5 |
| 204438869 | 204438869 | C | A | Nonsynonymous SNV | TPV | PIK3C2B |
| 214184949 | 214184949 | G | T | Nonsynonymous SNV | TPV | PROX1 |
| 247769320 | 247769320 | G | A | Nonsynonymous SNV | TPV | OR2G3 |
| 248737734 | 248737734 | G | A | Nonsynonymous SNV | TPV | OR2T34 |
| 11337731 | 11337731 | T | A | Nonsynonymous SNV | TPV | ROCK2 |
| 71795319 | 71795319 | G | C | Nonsynonymous SNV | TPV | DYSF |
| 108487966 | 108487966 | A | G | Nonsynonymous SNV | TPV | RGPD4 |
| 121729586 | 121729586 | G | T | Nonsynonymous SNV | TPV | GLI2 |
| 128364989 | 128364989 | G | T | Nonsynonymous SNV | TPV | MYO7B |
| 128615641 | 128615641 | C | T | Nonsynonymous SNV | TPV | POLR2D |
| 141946102 | 141946102 | C | A | Nonsynonymous SNV | TPV | LRP1B |
| 178098960 | 178098960 | C | G | Nonsynonymous SNV | TPV | NFE2L2 |
| 179398041 | 179398041 | T | C | Nonsynonymous SNV | TPV | TTN |
| 179456813 | 179456813 | G | T | Nonsynonymous SNV | TPV | TTN |
| 196599665 | 196599665 | G | T | Nonsynonymous SNV | TPV | SLC39A10 |
| 225422494 | 225422494 | T | C | Nonsynonymous SNV | TPV | CUL3 |
| 228137779 | 228137779 | G | T | Nonsynonymous SNV | TPV | COL4A3 |
| 238672406 | 238672406 | G | T | Nonsynonymous SNV | TPV | LRRFIP1 |
| 4829646 | 4829646 | C | T | Stopgain | TPV | ITPR1 |
| 12458381 | 12458381 | G | A | Nonsynonymous SNV | TPV | PPARG |
| 37670790 | 37670790 | G | A | Nonsynonymous SNV | TPV | ITGA9 |
| 49721811 | 49721811 | C | T | Nonsynonymous SNV | TPV | MST1 |
| 121350823 | 121350823 | C | T | Nonsynonymous SNV | TPV | HCLS1 |
| 165547837 | 165547837 | C | A | Nonsynonymous SNV | TPV | BCHE |
| 169565951 | 169565951 | C | A | Nonsynonymous SNV | TPV | LRRC31 |
| 193028470 | 193028470 | G | C | Nonsynonymous SNV | TPV | ATP13A5 |
| 194118528 | 194118528 | G | T | Nonsynonymous SNV | TPV | GP5 |
| 1231985 | 1231985 | C | A | Stopgain | TPV | CTBP1 |
| 1920144 | 1920144 | A | G | Nonsynonymous SNV | TPV | NSD2 |
| 98902467 | 98902467 | T | G | Nonsynonymous SNV | TPV | STPG2 |
| 118005739 | 118005739 | T | A | Nonsynonymous SNV | TPV | TRAM1L1 |
| 123236706 | 123236706 | C | G | Nonsynonymous SNV | TPV | KIAA1109 |
| 162577500 | 162577500 | A | T | Nonsynonymous SNV | TPV | FSTL5 |
| 177071237 | 177071237 | A | T | Nonsynonymous SNV | TPV | WDR17 |
| 187549886 | 187549886 | T | A | Nonsynonymous SNV | TPV | FAT1 |
| 24505347 | 24505347 | C | G | Nonsynonymous SNV | TPV | CDH10 |
| 41911175 | 41911175 | T | C | Nonsynonymous SNV | TPV | C5orf51 |
| 75858298 | 75858298 | T | A | Nonsynonymous SNV | TPV | IQGAP2 |
| 90024685 | 90024685 | C | A | Nonsynonymous SNV | TPV | ADGRV1 |
| 113740318 | 113740318 | A | G | Nonsynonymous SNV | TPV | KCNN2 |
| 114860009 | 114860009 | C | T | Nonsynonymous SNV | TPV | FEM1C |
| 131007333 | 131007333 | C | T | Nonsynonymous SNV | TPV | FNIP1 |
| 131931309 | 131931309 | C | T | Stopgain | TPV | RAD50 |
| 140307748 | 140307748 | C | A | Nonsynonymous SNV | TPV | PCDHAC1 |
| 140554795 | 140554795 | C | G | Nonsynonymous SNV | TPV | PCDHB7 |
| 27222843 | 27222843 | G | T | Nonsynonymous SNV | TPV | PRSS16 |
| 32713784 | 32713784 | C | A | Nonsynonymous SNV | TPV | HLA-DQA2 |
| 41899529 | 41899529 | G | C | Nonsynonymous SNV | TPV | BYSL |
| 64422909 | 64422909 | A | C | Nonsynonymous SNV | TPV | PHF3 |
| 66005999 | 66005999 | G | C | Nonsynonymous SNV | TPV | EYS |
| 90402365 | 90402365 | C | A | Nonsynonymous SNV | TPV | MDN1 |
| 126196041 | 126196041 | A | T | Nonsynonymous SNV | TPV | NCOA7 |
| 136599115 | 136599115 | C | A | Nonsynonymous SNV | TPV | BCLAF1 |
| 150343262 | 150343262 | T | C | Nonsynonymous SNV | TPV | RAET1L |
| 152614857 | 152614857 | C | T | Nonsynonymous SNV | TPV | SYNE1 |
| 158538843 | 158538843 | G | T | Nonsynonymous SNV | TPV | SERAC1 |
| 168708765 | 168708765 | C | G | Nonsynonymous SNV | TPV | DACT2 |
| 7622874 | 7622874 | G | C | Nonsynonymous SNV | TPV | MIOS |
| 29915496 | 29915496 | T | A | Nonsynonymous SNV | TPV | WIPF3 |
| 37951827 | 37951827 | G | T | Nonsynonymous SNV | TPV | SFRP4 |
| 39379482 | 39379482 | C | A | Nonsynonymous SNV | TPV | POU6F2 |
| 49815575 | 49815575 | G | A | Nonsynonymous SNV | TPV | VWC2 |
| 107720188 | 107720188 | A | G | Nonsynonymous SNV | TPV | LAMB4 |
| 128478472 | 128478472 | T | A | Nonsynonymous SNV | TPV | FLNC |
| 140051918 | 140051918 | T | C | Nonsynonymous SNV | TPV | SLC37A3 |
| 140179090 | 140179090 | C | A | Nonsynonymous SNV | TPV | MKRN1 |
| 150778698 | 150778698 | G | T | Nonsynonymous SNV | TPV | TMUB1 |
| 150835349 | 150835349 | G | T | Nonsynonymous SNV | TPV | AGAP3 |
| 151856028 | 151856028 | G | T | Nonsynonymous SNV | TPV | KMT2C |
| 154863275 | 154863275 | G | T | Nonsynonymous SNV | TPV | HTR5A |
| 24324457 | 24324457 | A | C | Nonsynonymous SNV | TPV | ADAM7 |
| 70591803 | 70591803 | G | T | Nonsynonymous SNV | TPV | SLCO5A1 |
| 92988192 | 92988192 | C | G | Nonsynonymous SNV | TPV | RUNX1T1 |
| 107715182 | 107715182 | G | A | Nonsynonymous SNV | TPV | OXR1 |
| 113275870 | 113275870 | A | T | Stopgain | TPV | CSMD3 |
| 145193975 | 145193975 | G | A | Nonsynonymous SNV | TPV | HGH1 |
| 21187197 | 21187197 | G | T | Nonsynonymous SNV | TPV | IFNA4 |
| 21974676 | 21974676 | C | T | Nonsynonymous SNV | TPV | CDKN2A;CDKN2A |
| 27558545 | 27558545 | C | T | Nonsynonymous SNV | TPV | C9orf72 |
| 69423770 | 69423770 | C | T | Nonsynonymous SNV | TPV | ANKRD20A4 |
| 85597659 | 85597659 | G | A | Nonsynonymous SNV | TPV | RASEF |
| 23622026 | 23622026 | T | C | Nonsynonymous SNV | TPV | C10orf67 |
| 28030395 | 28030395 | T | G | Nonsynonymous SNV | TPV | MKX |
| 68526048 | 68526048 | G | T | Nonsynonymous SNV | TPV | CTNNA3 |
| 86133479 | 86133479 | G | C | Nonsynonymous SNV | TPV | CCSER2 |
| 93702292 | 93702292 | G | A | Nonsynonymous SNV | TPV | BTAF1 |
| 116247751 | 116247751 | C | T | Nonsynonymous SNV | TPV | ABLIM1 |
| 116605214 | 116605214 | G | A | Nonsynonymous SNV | TPV | FAM160B1 |
| 134942632 | 134942632 | C | A | Nonsynonymous SNV | TPV | ADGRA1 |
| 4929407 | 4929407 | C | A | Nonsynonymous SNV | TPV | OR51A7 |
| 5068137 | 5068137 | G | A | Nonsynonymous SNV | TPV | OR52J3 |
| 6291913 | 6291913 | G | C | Nonsynonymous SNV | TPV | CCKBR |
| 6341448 | 6341448 | G | T | Nonsynonymous SNV | TPV | CAVIN3 |
| 44296961 | 44296961 | G | C | Stopgain | TPV | ALX4 |
| 64084615 | 64084615 | C | A | Nonsynonymous SNV | TPV | TRMT112 |
| 64877317 | 64877317 | G | A | Stopgain | TPV | VPS51 |
| 68845988 | 68845988 | G | C | Nonsynonymous SNV | TPV | TPCN2 |
| 68846022 | 68846022 | G | C | Nonsynonymous SNV | TPV | TPCN2 |
| 68846223 | 68846223 | G | C | Nonsynonymous SNV | TPV | TPCN2 |
| 70118395 | 70118395 | G | C | Nonsynonymous SNV | TPV | PPFIA1 |
| 100211220 | 100211220 | A | G | Nonsynonymous SNV | TPV | CNTN5 |
| 120329909 | 120329909 | G | T | Nonsynonymous SNV | TPV | ARHGEF12 |
| 2711117 | 2711117 | T | C | Nonsynonymous SNV | TPV | CACNA1C |
| 3788238 | 3788238 | G | C | Nonsynonymous SNV | TPV | CRACR2A |
| 15747894 | 15747894 | G | T | Nonsynonymous SNV | TPV | PTPRO |
| 88482957 | 88482957 | T | A | Nonsynonymous SNV | TPV | CEP290 |
| 122745983 | 122745983 | C | A | Nonsynonymous SNV | TPV | VPS33A |
| 128899361 | 128899361 | G | T | Nonsynonymous SNV | TPV | TMEM132C |
| 21563012 | 21563012 | C | A | Nonsynonymous SNV | TPV | LATS2 |
| 24243249 | 24243249 | C | G | Nonsynonymous SNV | TPV | TNFRSF19 |
| 32757165 | 32757165 | A | T | Nonsynonymous SNV | TPV | FRY |
| 33017514 | 33017514 | C | A | Nonsynonymous SNV | TPV | N4BP2L2 |
| 33247368 | 33247368 | C | G | Nonsynonymous SNV | TPV | PDS5B |
| 35683531 | 35683531 | T | A | Stopgain | TPV | NBEA |
| 61103338 | 61103338 | G | T | Nonsynonymous SNV | TPV | TDRD3 |
| 107822979 | 107822979 | T | G | Nonsynonymous SNV | TPV | FAM155A |
| 19553478 | 19553478 | G | A | Nonsynonymous SNV | TPV | POTEG |
| 22138850 | 22138850 | A | G | Nonsynonymous SNV | TPV | OR4E1 |
| 79432646 | 79432646 | T | A | Nonsynonymous SNV | TPV | NRXN3 |
| 93581417 | 93581417 | C | A | Nonsynonymous SNV | TPV | ITPK1 |
| 95582849 | 95582849 | C | T | Nonsynonymous SNV | TPV | DICER1 |
| 23811612 | 23811612 | C | T | Nonsynonymous SNV | TPV | MKRN3 |
| 24922008 | 24922008 | A | T | Nonsynonymous SNV | TPV | NPAP1 |
| 33941414 | 33941414 | G | A | Nonsynonymous SNV | TPV | RYR3 |
| 33954985 | 33954985 | C | A | Nonsynonymous SNV | TPV | RYR3 |
| 42289384 | 42289384 | C | T | Nonsynonymous SNV | TPV | PLA2G4E |
| 43572000 | 43572000 | C | A | Stopgain | TPV | TGM7 |
| 76136822 | 76136822 | G | T | Nonsynonymous SNV | TPV | UBE2Q2 |
| 93015599 | 93015599 | A | G | Nonsynonymous SNV | TPV | C15orf32 |
| 94841718 | 94841718 | A | G | Nonsynonymous SNV | TPV | MCTP2 |
| 23711953 | 23711953 | C | T | Nonsynonymous SNV | TPV | ERN2 |
| 51172691 | 51172691 | C | T | Nonsynonymous SNV | TPV | SALL1 |
| 74419248 | 74419248 | C | G | Nonsynonymous SNV | TPV | NPIPB15 |
| 3101635 | 3101635 | A | T | Nonsynonymous SNV | TPV | OR1A2 |
| 4720319 | 4720319 | G | A | Nonsynonymous SNV | TPV | PLD2 |
| 6381356 | 6381356 | G | A | Nonsynonymous SNV | TPV | PITPNM3 |
| 7574003 | 7574003 | G | A | Stopgain | TPV | TP53 |
| 12620686 | 12620686 | A | T | Nonsynonymous SNV | TPV | MYOCD |
| 18539842 | 18539842 | C | T | Nonsynonymous SNV | TPV | TBC1D28 |
| 28782467 | 28782467 | T | C | Nonsynonymous SNV | TPV | CPD |
| 29123323 | 29123323 | G | A | Nonsynonymous SNV | TPV | CRLF3 |
| 32953362 | 32953362 | G | A | Nonsynonymous SNV | TPV | TMEM132E |
| 47121429 | 47121429 | T | G | Nonsynonymous SNV | TPV | IGF2BP1 |
| 47121430 | 47121430 | T | G | Nonsynonymous SNV | TPV | IGF2BP1 |
| 48542697 | 48542697 | A | C | Nonsynonymous SNV | TPV | CHAD |
| 71281726 | 71281726 | C | T | Nonsynonymous SNV | TPV | CDC42EP4 |
| 11610531 | 11610531 | G | A | Nonsynonymous SNV | TPV | SLC35G4 |
| 19395677 | 19395677 | A | T | Nonsynonymous SNV | TPV | MIB1 |
| 2115396 | 2115396 | T | A | Nonsynonymous SNV | TPV | AP3D1 |
| 3623954 | 3623954 | T | C | Nonsynonymous SNV | TPV | CACTIN |
| 9086220 | 9086220 | C | G | Nonsynonymous SNV | TPV | MUC16 |
| 10469852 | 10469852 | A | T | Nonsynonymous SNV | TPV | TYK2;TYK2 |
| 12739889 | 12739889 | A | G | Nonsynonymous SNV | TPV | ZNF791 |
| 15756539 | 15756539 | C | T | Nonsynonymous SNV | TPV | CYP4F3 |
| 18375446 | 18375446 | C | A | Nonsynonymous SNV | TPV | KIAA1683 |
| 22941567 | 22941567 | A | G | Nonsynonymous SNV | TPV | ZNF99 |
| 23040922 | 23040922 | C | G | Stopgain | TPV | ZNF723 |
| 47870310 | 47870310 | A | G | Nonsynonymous SNV | TPV | DHX34 |
| 51984886 | 51984886 | C | A | Nonsynonymous SNV | TPV | CEACAM18 |
| 54515274 | 54515274 | C | A | Nonsynonymous SNV | TPV | CACNG6 |
| 57293327 | 57293327 | A | G | Nonsynonymous SNV | TPV | ZIM2 |
| 21330036 | 21330036 | A | G | Nonsynonymous SNV | TPV | XRN2 |
| 25655939 | 25655939 | C | T | Nonsynonymous SNV | TPV | ZNF337 |
| 50286574 | 50286574 | C | T | Nonsynonymous SNV | TPV | ATP9A |
| 55206742 | 55206742 | T | C | Nonsynonymous SNV | TPV | TFAP2C |
| 37618419 | 37618419 | T | C | Nonsynonymous SNV | TPV | DOPEY2 |
| 45953704 | 45953704 | C | G | Nonsynonymous SNV | TPV | TSPEAR |
| 45993666 | 45993666 | A | G | Nonsynonymous SNV | TPV | KRTAP10-4 |
| 47320917 | 47320917 | G | A | Nonsynonymous SNV | TPV | PCBP3 |
| 19883067 | 19883067 | T | G | Nonsynonymous SNV | TPV | TXNRD2 |
| 29957800 | 29957800 | T | C | Nonsynonymous SNV | TPV | NIPSNAP1 |
| 50518810 | 50518810 | A | G | Nonsynonymous SNV | TPV | MLC1 |
| 50704016 | 50704016 | G | A | Nonsynonymous SNV | TPV | MAPK11 |
| 31792183 | 31792183 | C | A | Nonsynonymous SNV | TPV | DMD |
| 32382707 | 32382707 | C | A | Nonsynonymous SNV | TPV | DMD |
| 35938079 | 35938079 | G | C | Nonsynonymous SNV | TPV | CFAP47 |
| 110491848 | 110491848 | C | A | Nonsynonymous SNV | TPV | CAPN6 |
| 148577938 | 148577938 | C | A | Nonsynonymous SNV | TPV | IDS |
| 157803028 | 157803028 | C | – | Frameshift deletion | TPV | CD5L |
| 171627269 | 171627269 | – | A | Frameshift insertion | TPV | ERICH2 |
| 6574049 | 6574052 | TACT | – | Frameshift deletion | TPV | VAMP1 |
| 63970153 | 63970153 | – | T | Frameshift insertion | TPV | HERC1 |
| 63970155 | 63970155 | – | AACT | Frameshift insertion | TPV | HERC1 |
| 38969124 | 38969124 | C | – | Frameshift deletion | TPV | RYR1 |
| 58570657 | 58570657 | C | – | Frameshift deletion | TPV | ZNF135 |
| 19420859 | 19420868 | TCATTCCCAT | – | Frameshift deletion | TPV | MRPL40 |
PT, primary tumor; LNM, lymph node metastases; TPV, tumor thrombus in pulmonary vein.
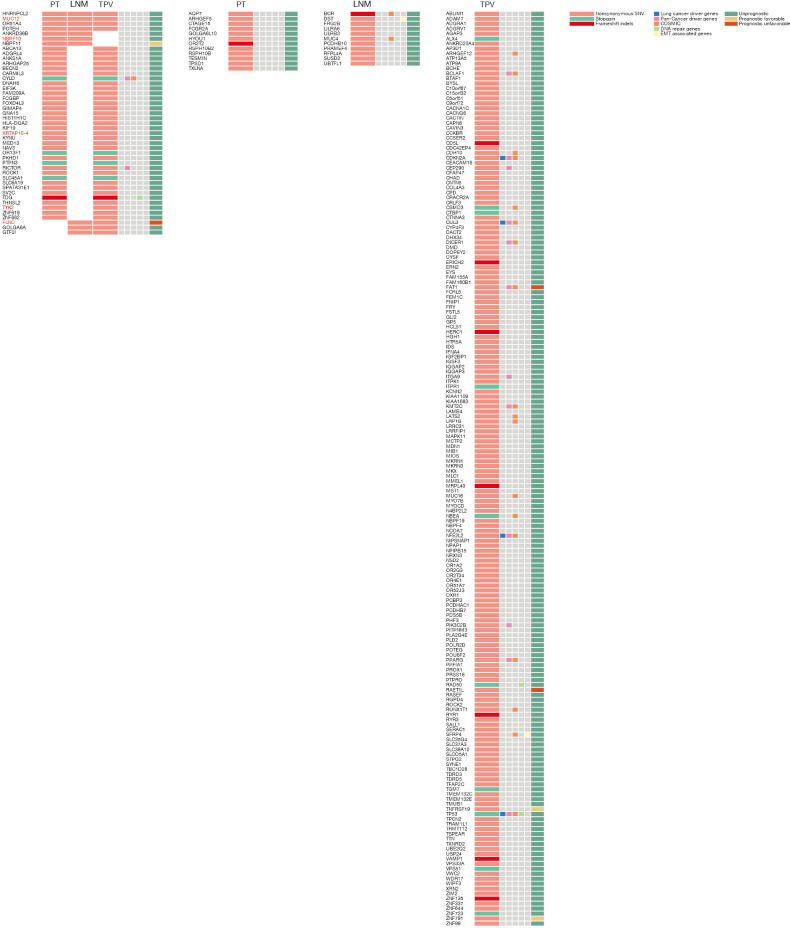
Figure 2.
Exonic somatic variants identified in PT, LNM and TPV. The exonic somatic variants were classified as shared or private variants. Red color represent genes contain different variants among different tumors. PT, primary tumor; LNM, lymph node metastases; TPV, tumor thrombus in pulmonary vein.
Figure 3.
Intratumoral genetic heterogeneity in form of SNVs and indels. (A) The number of exonic somatic variants (SNVs and indels) and nonsynonymous somatic variants in each of tumors. (B) The mutation spectrum of SNVs in PT, LNM and TPV. (C) Mutational signatures of all tumor sample. (D) Two mutational signatures (S1, S2) extracted from all tumors. (E) Cluster analysis of S1, S2 and 30 COSMIC mutational signature based on the cosine similarity. (F) The proportion of S1 and S2 in PT, LNM and TPV. PT, primary tumor; LNM, lymph node metastases; TPV, tumor thrombus in pulmonary vein.
Comparison of structural variants detected by WGS and optical mapping
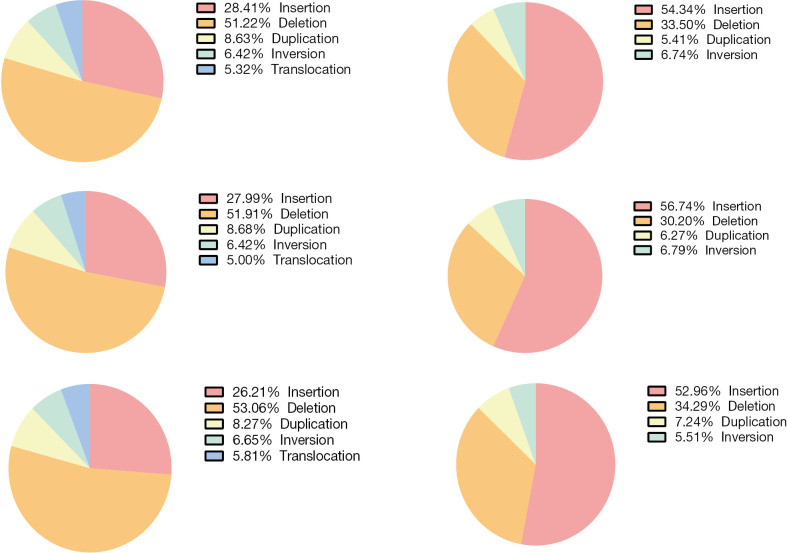
We utilized WGS data and performed optical mapping on PT, LNM and TPV at 100X coverage. SVs were called and filtered as presented in Figure 4. There were a mean of 3,617 SVs detected by WGS (3,907, 3,580, and 3,365 in PT, LNM, and TPV, respectively), of which deletions were most commonly detected type of SV (Figure S2). While SVs detected by optical mapping was 1,026 on average (979, 1,118, 980 in PT, LNM, TPV, respectively), Insertions account for the most (Figure S2).
Figure 4.
Workflow for detection of structural variants. The workflow for extracting structural variants from a combination of whole-genome sequencing and optical mapping. Detail explanation seen in Methods.
Figure S2.
The proportions of different types of SVs detected by whole-genome sequencing (left) or optical mapping (right) in PT (upper), LNM (middle) and TPV (lower). PT, primary tumor; LNM, lymph node metastases; TPV, tumor thrombus in pulmonary vein.
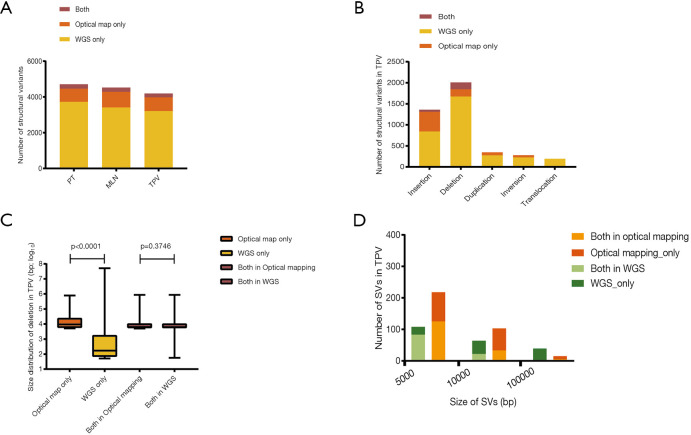
By comparing the SVs detected by WGS and optical mapping, we observed an average of 22.9 percent of SVs detected by optical mapping overlapped with those detected by WGS (25.1%, 21.4% and 22.2% in PT, LNM and TPV, respectively) (Figure 5A,B), of which the deletions had similar size (the median size was 6,452 bp, 6,191 bp in optical mapping and WGS) (Figure 5C, Figure S3). The median size of non-overlapping SVs in optical mapping was distinct from the non-overlapping ones detected by WGS (8,875 bp, 143 bp in optical mapping and WGS respectively) (Figure 5C, Figure S3). Specifically, Optical mapping is more capable of detecting large SVs (>5,000 bp) (Figure 5D). Generally, WGS can detect SVs at a high resolution of base but has many limitations: it depends on a short-read sequencing technique, needs a reference genome, and challenges of computational and bioinformatics algorithms exist. In contrast, optical mapping detects large and complex SVs using high molecular weight (HMW) DNA which are longer, ranging from 0.1 to 2Mb. The results suggested that the combination of WGS and optical mapping used for detecting SVs allows to a more comprehensive understanding of structural variants among tumor cells within different sites and demonstrated optical mapping is more sensitive for detection of large SVs.
Figure 5.
Comparison of structural variants detected by WGS and optical mapping. (A) The number of structural variants detected by whole-genome sequencing and optical mapping. (B) The number of different types of structural variants detected by whole-genome sequencing and optical mapping in TPV. (C) Size distribution of deletions in TPV. (D) The number of large structural variants (>5,000 bp) detected by whole-genome sequencing and optical mapping in TPV. TPV, tumor thrombus in pulmonary vein.
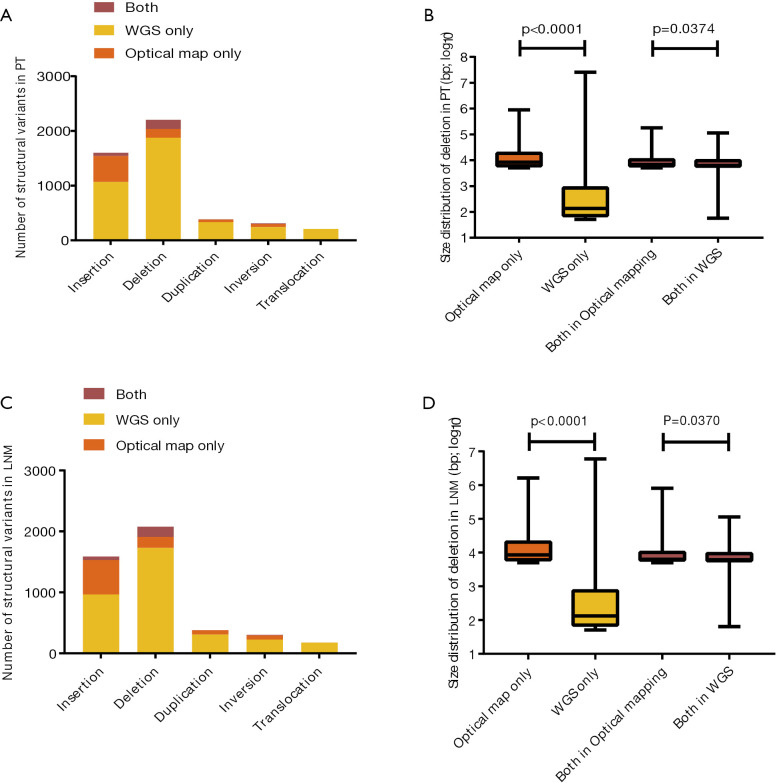
Figure S3.
The number of different types of structural variants detected by whole-genome sequencing and optical mapping in PT (A) and LNM (C), of which size distribution of deletions in PT (B) and LNM (D). PT, primary tumor; LNM, lymph node metastases.
ITGH in the form of SVs
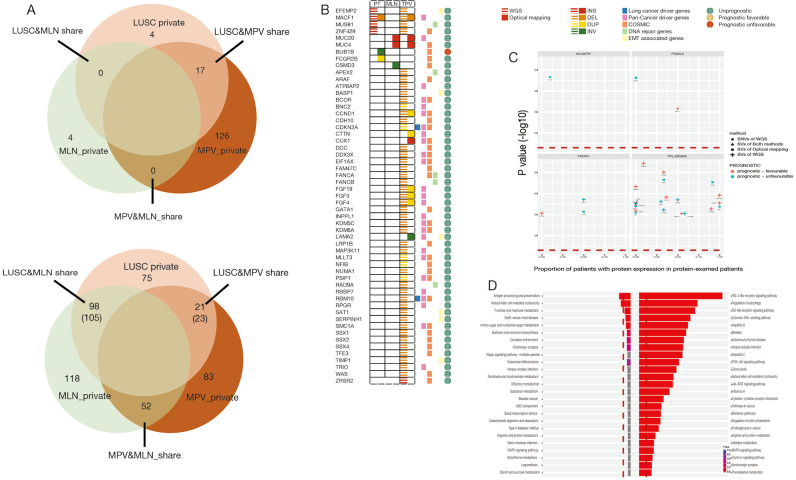
We did an comparison among PT, LNM and TPV based on SVs detected by WGS and SVs detected by optical mapping, identifying a greater amount of private SVs in TPV (126 from WGS, 83 from optical mapping) than in either PT (4 from WGS, 75 from optical mapping) or LNM (4 from WGS, 118 from optical mapping) (Figure 6A), consistent with the results of SNVs and indels analysis. There was no overlap between private SVs identified by WGS and private SVs identified by optical mapping in each of tumors except TPV (7 private SVs from optical mapping overlapped with 6 private SVs from WGS). Smaller number of SVs in TPV (17 from WGS, 23 from optical mapping) overlapped with SVs of PT than those in LNM (105 from optical mapping). Specifically, 52 SVs from optical mapping undetected in PT were shared between LNM and TPV.
Figure 6.
Intratumoral genetic heterogeneity in form of structural variants. (A) Overlap of structural variants detected by whole-genome sequencing (upper) and optical mapping (lower) among PT, LNM and TPV. (B) Genes associated with tumorigenesis and progression affected by structural variants detected by whole-genome sequencing and optical mapping in PT, LNM and TPV. (C) Genes associated with prognosis of lung cancer affected by structural variants detected by whole-genome sequencing and optical mapping. (Red dotted line represents P value >0.05) (D) KEGG enrichment of genes only affected by metastases-specific structural variants. (Red dotted line represents adjusted P value >0.05). PT, primary tumor; LNM, lymph node metastases; TPV, tumor thrombus in pulmonary vein.
We further explored whether these SVs overlap with genes previously associated with tumorigenesis and progression (Figure 6B). Several private SVs of TPV detected by either WGS or optical mapping were associated with DNA repair genes including APEX2, FANCA, FANCB and RAD9A suggesting that mutations in DNA repair genes may play a role in progression of metastatic lung cancer by generating chromosomal instability. We also identified several EMT associated genes including BASP1, LAMA2, SAT1, SERPINH1 and TIMP1 were affected by SVs only detected in TPV. Completely different with TPV, only CSMD3, a frequently mutated gene in LUSC (47,48) was affected by private SVs of LNM. Loss of CSMD3 was reported to be associated with the proliferation of airway epithelial cells (47) and mutations in CSMD3 is associated with a better prognosis in patients with LUSC (48). Compared with the gene expression and survival data in The Human Protein Atlas (HPA) (39-41), we also identified 21 other genes affected by SVs previously unrecognized as tumor associated genes, of which expression was significantly associated with the prognosis of lung cancer patients (Figure 6C).
Furthermore, to comprehensively understand the functional consequence of genomic alterations only found in tumor cells in metastatic sites, we performed a KEGG enrichment analysis based on genes only affected by SNVs, indels and SVs in metastases (Figure 6D). Specifically, genes involved in the PI3K-Akt pathway which has an important role in tumorigenesis and progression (49), were significantly affected by variants in TPV.
Discussion
SNVs and CNVs detected by next-generation sequencing in multiregional tumors has improved our understanding of ITGH (8-10,46,50), while studies focusing on the analysis of ITGH in the form of SVs among tumor cells in primary and different metastatic sites are limited. Previous studies detected SVs through WGS (51,52). WGS, relying on sequencing by synthesis, is based on short reads. The DNA molecules are fragmented to countless reads and amplified by polymerase chain reaction (PCR), to meet the requirement of the high-throughput. And then we detect the SVs based on the read-pair or SR. That is, WGS detects the SVs on the basis of incomplete structure of DNA, which may miss some SVs in specific locations of chromosome or those with large size (53). In contrast, the integrity of DNA molecular is crucial for optical mapping to detect the SVs, with specific site labeled HMW DNA and nano-channel imaging system, optical mapping could de novo identify SVs without the bias of PCR amplification. Therefore, optical mapping and WGS could complement mutually.
To our knowledge, our study is the first study applying WGS and optical mapping to multiregional samples of a LUSC patient, aiming to compressively investigate the intratumoral heterogeneity within one patient. We do observe a significant difference in the variants burden between primary tumor and metastases and between metastases in different sites. Like SNVs and indels, SVs play an indispensable role in heterogeneity. Combination of WGS and optical mapping allows us to gain a more comprehensive understanding of structural variants, especially large SVs. Compared with the analysis of SVs detected by WGS, optical mapping were more informative in identifying private SVs for ITGH.
Variants shared between primary tumor and metastases indicate that mutations in primary tumor subclones with metastatic potential accumulated before metastasizing. Among them, mutations shared between TPV and PT which affect genes associated with tumorigenesis and progression, may enable tumor cells in the primary site to metastasize and live in hemato-microenvironment. Tumor cells harbor mutations identified both in PT and TPV may have more capability to metastasize and settle down in lymph node.
Meanwhile, private variants detected in different groups of tumors suggest genetic mutations occurred both before and after metastasis. Mutations unique to LNM or TPV indicate an interaction between tumor cells and microenvironment in metastatic sites. Private variants in TPV, especially those affected genes associated with DNA repair and epithelial-mesenchymal transition (EMT), are much more frequently identified than in PT or LNM. This suggests that tumor cells in hemato-microenvironment bear a higher degree of chromosomal instability and has more potential to act as a metastases relay station between primary tumor and metastases of distant organs, previously observed by Ferronika et al. (54).
It should be noted that the major limitation of our study is that analysis only based on one individual. The main reason is that most LUSC patients received surgery are at early stage and non-metastatic. In clinical practice, metastatic lymph node and tumor thrombus collected from the same patient in this study is rare to obtain by surgical resection. And biopsy sampling of multiple metastatic regions has not been widely accepted due to the potential risks for the prognosis of patients (55). Additionally, previous studies confirmed that analysis in a small number of cases even in one patient could reveal ITGH (6,10,15).
Notwithstanding its limitation, our results do demonstrate the ability of optical mapping in detection of large SVs to make up the deficiency of WGS and reveal that SVs are as crucial in describing ITGH as SNVs and indels.
Supplementary
The article’s supplementary files as
Acknowledgments
We thank the patient to provide the samples for this study; Litao Han and Ben Ma for advice to manuscript. We also thank Lili Tan for excellent technical assistance; Hainan Cheng for bioinformatics analysis.
Funding: This work was supported by Ministry of Science and Technology of the People’s Republic of China (2017YFA0505500; 2016YFA0501800), Science and Technology Commission of Shanghai Municipality (19XD1401300).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Fudan University Shanghai Cancer Center Institutional Review Board (No. 090977-1) and written informed consent was obtained from all patients.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-19-401). The authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607-16. 10.1038/ng.3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meng F, Zhang L, Ren Y, et al. The genomic alterations of lung adenocarcinoma and lung squamous cell carcinoma can explain the differences of their overall survival rates. J Cell Physiol 2019;234:10918-25. 10.1002/jcp.27917 [DOI] [PubMed] [Google Scholar]
- 4.Langer CJ, Obasaju C, Bunn P, et al. Incremental Innovation and Progress in Advanced Squamous Cell Lung Cancer: Current Status and Future Impact of Treatment. J Thorac Oncol 2016;11:2066-81. 10.1016/j.jtho.2016.08.138 [DOI] [PubMed] [Google Scholar]
- 5.Gandara DR, Hammerman PS, Sos ML, et al. Squamous cell lung cancer: from tumor genomics to cancer therapeutics. Clin Cancer Res 2015;21:2236-43. 10.1158/1078-0432.CCR-14-3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer cell 2015;27:15-26. 10.1016/j.ccell.2014.12.001 [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, Fujimoto J, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 2014;346:256-9. 10.1126/science.1256930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma P, Fu Y, Cai MC, et al. Simultaneous evolutionary expansion and constraint of genomic heterogeneity in multifocal lung cancer. Nat Commun 2017;8:823. 10.1038/s41467-017-00963-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Zhang J, Li L, et al. Genomic heterogeneity of multiple synchronous lung cancer. Nat Commun 2016;7:13200. 10.1038/ncomms13200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leong TL, Gayevskiy V, Steinfort DP, et al. Deep multi-region whole-genome sequencing reveals heterogeneity and gene-by-environment interactions in treatment-naive, metastatic lung cancer. Oncogene 2019;38:1661-75. 10.1038/s41388-018-0536-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vignot S, Frampton GM, Soria JC, et al. Next-generation sequencing reveals high concordance of recurrent somatic alterations between primary tumor and metastases from patients with non-small-cell lung cancer. J Clin Oncol 2013;31:2167-72. 10.1200/JCO.2012.47.7737 [DOI] [PubMed] [Google Scholar]
- 12.Tubio JMC. Somatic structural variation and cancer. Brief Funct Genomics 2015;14:339-51. 10.1093/bfgp/elv016 [DOI] [PubMed] [Google Scholar]
- 13.Inaki K, Liu ET. Structural mutations in cancer: mechanistic and functional insights. Trends Genet 2012;28:550-9. 10.1016/j.tig.2012.07.002 [DOI] [PubMed] [Google Scholar]
- 14.Dixon JR, Xu J, Dileep V, et al. Integrative detection and analysis of structural variation in cancer genomes. Nat Genet 2018;50:1388-98. 10.1038/s41588-018-0195-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaratlerdsiri W, Chan EKF, Petersen DC, et al. Next generation mapping reveals novel large genomic rearrangements in prostate cancer. Oncotarget 2017;8:23588-602. 10.18632/oncotarget.15802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589-95. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078-9. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213-9. 10.1038/nbt.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.NCBI Resource Coordinators . Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2014;42:D7-17. 10.1093/nar/gkt1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012;491:56-65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Schulz-Trieglaff O, Shaw R, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 2016;32:1220-2. 10.1093/bioinformatics/btv710 [DOI] [PubMed] [Google Scholar]
- 24.English AC, Salerno WJ, Hampton OA, et al. Assessing structural variation in a personal genome-towards a human reference diploid genome. BMC Genomics 2015;16:286. 10.1186/s12864-015-1479-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research Network . Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. 10.1038/nature11404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47-53. 10.1038/nature14664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Research Network . Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285-91. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sondka Z, Bamford S, Cole CG, et al. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer 2018;18:696-705. 10.1038/s41568-018-0060-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The Gene Ontology Consortium .Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res 2017;45:D331-8. 10.1093/nar/gkw1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maupin KA, Sinha A, Eugster E, et al. Glycogene expression alterations associated with pancreatic cancer epithelial-mesenchymal transition in complementary model systems. PLoS One 2010;5:e13002. 10.1371/journal.pone.0013002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menge T, Zhao Y, Zhao J, et al. Mesenchymal stem cells regulate blood-brain barrier integrity through TIMP3 release after traumatic brain injury. Sci Transl Med 2012;4:161ra150. 10.1126/scitranslmed.3004660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chu IM, Michalowski AM, Hoenerhoff M, et al. GATA3 inhibits lysyl oxidase-mediated metastases of human basal triple-negative breast cancer cells. Oncogene 2012;31:2017-27. 10.1038/onc.2011.382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liberzon A, Birger C, Thorvaldsdottir H, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417-25. 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liberzon A, Subramanian A, Pinchback R, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27:1739-40. 10.1093/bioinformatics/btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiozaki A, Bai XH, Shen-Tu G, et al. Claudin 1 mediates TNFalpha-induced gene expression and cell migration in human lung carcinoma cells. PLoS One 2012;7:e38049. 10.1371/journal.pone.0038049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tam WL, Lu H, Buikhuisen J, et al. Protein kinase C alpha is a central signaling node and therapeutic target for breast cancer stem cells. Cancer cell 2013;24:347-64. 10.1016/j.ccr.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pontén F, Jirstrom K, Uhlen M. The Human Protein Atlas--a tool for pathology. J Pathol 2008;216:387-93. 10.1002/path.2440 [DOI] [PubMed] [Google Scholar]
- 40.Uhlén M, Bjorling E, Agaton C, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics 2005;4:1920-32. 10.1074/mcp.M500279-MCP200 [DOI] [PubMed] [Google Scholar]
- 41.Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science 2017;357. doi: . 10.1126/science.aan2507 [DOI] [PubMed] [Google Scholar]
- 42.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gehring JS, Fischer B, Lawrence M, et al. SomaticSignatures: inferring mutational signatures from single-nucleotide variants. Bioinformatics 2015;31:3673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ginestet CJJotRSS . ggplot2: Elegant Graphics for Data Analysis by H. Wickham. 2011;174:245-6. [Google Scholar]
- 45.Cancer Genome Atlas Research Network . Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. 10.1038/nature11404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu K, Zhang X, Li F, et al. Frequent alterations in cytoskeleton remodelling genes in primary and metastatic lung adenocarcinomas. Nat Commun 2015;6:10131. 10.1038/ncomms10131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu P, Morrison C, Wang L, et al. Identification of somatic mutations in non-small cell lung carcinomas using whole-exome sequencing. Carcinogenesis 2012;33:1270-6. 10.1093/carcin/bgs148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.La Fleur L, Falk-Sorqvist E, Smeds P, et al. Mutation patterns in a population-based non-small cell lung cancer cohort and prognostic impact of concomitant mutations in KRAS and TP53 or STK11. Lung cancer 2019;130:50-8. 10.1016/j.lungcan.2019.01.003 [DOI] [PubMed] [Google Scholar]
- 49.Fruman DA, Chiu H, Hopkins BD, et al. The PI3K Pathway in Human Disease. Cell 2017;170:605-35. 10.1016/j.cell.2017.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan Q, Cui J, Huang J, et al. Genomic Alteration During Metastasis of Lung Adenocarcinoma. Cell Physiol Biochem 2016;38:469-86. 10.1159/000438644 [DOI] [PubMed] [Google Scholar]
- 51.Quigley DA, Dang HX, Zhao SG, et al. Genomic Hallmarks and Structural Variation in Metastatic Prostate Cancer. Cell 2018;174:758-769.e9. 10.1016/j.cell.2018.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murphy SJ, Aubry MC, Harris FR, et al. Identification of independent primary tumors and intrapulmonary metastases using DNA rearrangements in non-small-cell lung cancer. J Clin Oncol 2014;32:4050-8. 10.1200/JCO.2014.56.7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ewing A, Semple C. Breaking point: the genesis and impact of structural variation in tumours. F1000Res 2018;7. doi: . 10.12688/f1000research.16079.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferronika P, Hof J, Kats-Ugurlu G, et al. Comprehensive Profiling of Primary and Metastatic ccRCC Reveals a High Homology of the Metastases to a Subregion of the Primary Tumour. Cancers (Basel) 2019;11. doi: . 10.3390/cancers11060812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 2018;15:81-94. 10.1038/nrclinonc.2017.166 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as