Abstract
Background
To investigate whether radiomic features from (18F)-fluorodeoxyglucose positron emission tomography/computed tomography [(18F)-FDG PET/CT] can predict epidermal growth factor receptor (EGFR) mutation status and prognosis in patients with lung adenocarcinoma.
Methods
One hundred and seventy-four consecutive patients with lung adenocarcinoma underwent (18F)-FDG PET/CT and EGFR gene testing were retrospectively analyzed. Radiomic features combined with clinicopathological factors to construct a random forest (RF) model to identify EGFR mutation status. The mutant/wild-type model was trained on a training group (n=139) and validated in an independent validation group (n=35). The second RF classifier predicting the 19/21 mutation site was also built and evaluated in an EGFR mutation subset (training group, n=80; validation group, n=25). Radiomic score and 5 clinicopathological factors were integrated into a multivariate Cox proportional hazard (CPH) model for predicting overall survival (OS). AUC (the area under the receiver characteristic curve) and C-index were calculated to evaluate the model’s performance.
Results
Of 174 patients, 109 (62.6%) harbored EGFR mutations, 21L858R was the most common mutation type [55.9% (61/109)]. The mutant/wild-type model was identified in the training (AUC, 0.77) and validation (AUC, 0.71) groups. The 19/21 mutation site model had an AUC of 0.82 and 0.73 in the training and validation groups, respectively. The C-index of the CPH model was 0.757. The survival time between targeted therapy and chemotherapy for patients with EGFR mutations was significantly different (P=0.03).
Conclusions
Radiomic features based on (18F)-FDG PET/CT combined with clinicopathological factors could reflect genetic differences and predict EGFR mutation type and prognosis.
Keywords: Lung adenocarcinoma, radiomics, positron emission tomography, epidermal growth factor receptor (EGFR), prognosis
Introduction
Lung cancer is one of the most common and malignant tumors. The proportion of non-small cell lung cancer (NSCLC) is approximately 85% in patients with lung cancer, and nearly 80% of patients are in the advanced stage when they are diagnosed. The main histological type of NSCLC is adenocarcinoma and has a poor prognosis (1-3). According to the recommendations of the National Comprehensive Cancer Network guidelines for the diagnosis and treatment of NSCLC (4), platinum-containing, two-drug chemotherapy is the standard treatment for most patients with advanced lung adenocarcinoma without gene mutation(s) (5). However, for some patients with sensitive gene mutations, the benefits of chemotherapy are limited. With more extensive research investigating the nature of tumors using molecular genetic and biology techniques, molecular typing based on tumor gene characteristics has brought new hope for the treatment of NSCLC. Research investigating the genes driving lung adenocarcinoma has led to significant breakthroughs in recent years and has ushered the treatment of NSCLC into a new era of personalized targeted therapy.
Epidermal growth factor receptor (EGFR)—a tyrosine receptor that can be used as a therapeutic target to inhibit its overexpression in tumor cells—has become an important predictor of treatment efficacy. Research has shown that the effect of tyrosine kinase inhibitors (TKIs) in patients with EGFR mutation was significantly better than that in patients with wild-type (6,7), demonstrating benefits in patients with mutations on disease-free survival and overall survival (OS) (8,9). Owing to the high mutation load of lung cancer, EGFR mutations can occur in exons 18 to 21. The most common sensitive mutations include exon 19 deletions (19DEL) and exon 21L858R, which account for 90% of EGFR mutations. The sensitivity and efficacy of the treatment response to TKIs largely depends on mutant genotypes; therefore, the identification of EGFR mutation types is essential for individualized molecular targeted therapy (10-14). Several clinical studies have shown that 19DEL mutations are more sensitive to TKIs than 21L858R mutations, resulting in longer survival times (15). However, tissue biopsy samples required for the detection of EGFR mutation status is not always available, especially in patients with low Karnofsky Performance Scale scores. Although liquid biopsy is convenient, rapid, and inexpensive, its sensitivity and stability are not optimal (16). Therefore, accurate and non-invasive approaches to predict EGFR mutation status are desired for clinical use.
18Fluorine-fluoro-D-glucose positron emission tomography/computed tomography ((18F)-FDG PET/CT), as an emerging imaging technique, can not only depict anatomical structure, but also reflect the biological characteristics of tumors. However, the potential of PET/CT to predict mutation status of EGFR remains controversial (17-20). Radiomics, which has emerged as a promising technique to identify genetic phenotypes in several types of tumors, is inspired by the concept of extracting underlying imaging features from medical images for quantitative imaging analysis (21). Recently, several studies focusing on radiomics for the prediction of EGFR status have been published (22-24). However, to our knowledge, very few have investigated the ability of PET/CT imaging-based radiomic features to predict EGFR mutation status and prognosis (25).
Therefore, the purpose of the present study was to investigate whether radiomic features from (18F)F-FDG PET/CT can predict EGFR mutation status, subtype, and clinical outcome in patients with lung adenocarcinoma.
Methods
Patients
The Institutional Review Board of Jinling Hospital, Medical School of Nanjing University (Nanjing, China) approved this study (No. 2016NZKYKS-004-01). Given the retrospective nature of the investigation and the use of anonymized patient data, requirements for informed consent were waived. Data were collected from 174 consecutive patients who were diagnosed with lung adenocarcinoma at Jinling Hospital, Medical School of Nanjing University between July 2009 and August 2016. The inclusion criteria were as follows: PET/CT examination within 1 month before surgery or biopsy; no anti-tumor treatment before PET/CT examination; with surgical (n=41) or biopsy (n=133) specimens confirmed by pathology; and available EGFR mutation detection results. Patients with poor image quality or lung metastatic tumor(s) were excluded. Clinicopathological data were obtained from medical records. Follow-up was from July 2009 to January 2019. OS in this study was defined as the period from the date of PET/CT examination to the date of telephone follow-up or the date of patient death. Among those with EGFR mutations, 65 patients underwent targeted therapy and 33 underwent chemotherapy. Twenty-nine patients with 19DEL and 36 with 21L858R mutations underwent targeted therapy. Thirty-three patients with EGFR mutations and 51 with wild-type EGFR underwent chemotherapy.
PET/CT acquisition, analysis, and parameter extraction
All patients underwent (18F)-FDG PET/CT before any treatment for the purposes of tumor staging and target volume delineation, which was performed using a PET/CT system (Biogragh 16, Siemens, Erlangen, Germany) 75 min after the injection of (18F)-FDG (EBCO’s TR19 medical cyclotron, Canada). Each patient fasted for at least 6 h before the test, and blood glucose levels were controlled to <6.7 mmol/L. The intravenous injection dose of (18F)-FDG was 3.7–6.6 MBq/kg. After the injection, the patient rested quietly for 1 h, consumed approximately 500–1,000 mL of water, and then voided the bladder before scanning. The scan ranged from the skull base to the upper femur. The CT scan (scan parameters: slice thickness, 5 mm; pitch 0.75; tube voltage, 120 kV; tube current, 140 mAs, tube rotation speed, 0.8 s/rotation) was performed before intravenous injection, followed by a PET emission scan of 3 min per bed position. An iterative algorithm was used for image reconstructions to obtain transverse, sagittal, and coronal fusion images from CT, PET, and PET/CT.
(18F)-FDG PET/CT analysis
A volume of interest (VOI) was drawn semi-automatically around the tumor by one observer with 5 years’ experience in nuclear medicine who was blinded to patient outcomes using a Radiomics prototype (Radiomics, Frontier, Siemens). Segmentation was semi-automatically produced by drawing a line along the boundary of the tumor and manually adjusted by the observer in a three-dimensional domain on the Radiomics prototype. PET/CT metabolic parameters were based on the high FDG metabolic area of the lesion using the MS viewer software and by manually delineating the region of interest (ROI) to measure the metabolic tumor volume (MTV), maximal standard uptake value (SUVmax) and mean standard uptake value (SUVmean), and total lesion glycolysis (TLG) was calculated using the following equation: TLG = SUVmean × MTV.
EGFR gene detection
EGFR genetic mutations were tested from the affected tumor tissue sample obtained by surgical resection or biopsy. The tissue samples were fixed in formalin, dehydrated, embedded in paraffin, and serially sectioned for hematoxylin and eosin staining. The amplification refractory mutation system polymerase chain reaction method was used to detect mutation sites in four exons (exons 18–21) in the coding region of the EGFR gene, the results of which were acquired according to the interpretation principle provided by the reference test kit (ACCB Biotech Ltd, Beijing, China). If any exon mutation was detected, the tumor was identified as an EGFR mutant, otherwise, the tumor was identified as EGFR wild type.
Feature extraction
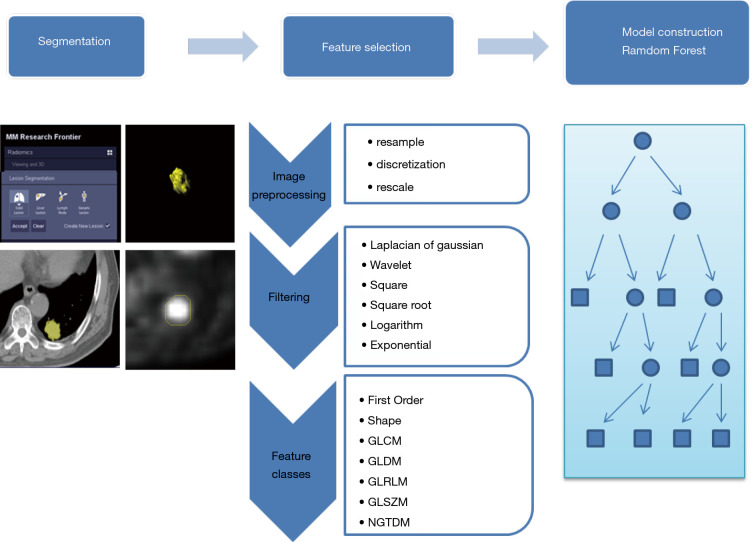
The features used for model development included both clinical and radiomic features. Clinical features included sex, age, smoking history, family history, TNM staging, and carcinoembryonic antigen level. In total, 1,672 CT-derived radiomic features and another 1,672 PET-derived radiomic features were extracted by the prototype (Radiomics, Frontier, Siemens, Figure 1). The extracted feature categories were 18 first-order statistics, 16 shape and size, and 74 texture features. In addition, these features were filtered using 6 different filters, and wavelet analysis were performed to extract more dimensional information. Finally, 11 features were ranking with high importance in the model (Table 1).
Figure 1.
The process of the radiomics method included the following steps. The radiomics workflow in the flowchart showed three basic modules. The lesions were segmented in the first step on Siemens radiomics prototype semiautomatically. In the second step, 3,344 features including first order features, shape related features and texture features were extracted in the software after imaging pre-processing. The random forest (RF) algorithm was applied in the next step to construct the model in the training group, then validate in the testing group
Table 1. Feature importance in RF model.
| Feature name | Importance |
|---|---|
| CT_log-sigma-4-5-mm-3D_glszm_SmallAreaHighGrayLevelEmphasis | 0.1209 |
| CT_wavelet-HLH_glcm_Idn | 0.1207 |
| PET_wavelet-LHH_glszm_LowGrayLevelZoneEmphasis | 0.1124 |
| PET_wavelet-HHH_gldm_GrayLevelVariance | 0.1060 |
| PET_log-sigma-0-5-mm-3D_firstorder_Maximum | 0.0940 |
| PET_wavelet-HLH_firstorder_Range | 0.0876 |
| CT_wavelet-LLH_firstorder_Uniformity | 0.0838 |
| CT_logarithm_firstorder_RootMeanSquared | 0.0793 |
| CT_log-sigma-0-5-mm-3D_glrlm_LongRunEmphasis | 0.0791 |
| PET_log-sigma-0-5-mm-3D_gldm_LowGrayLevelEmphasis | 0.0715 |
| CT_wavelet-HLH_glszm_SmallAreaHighGrayLevelEmphasis | 0.0447 |
RF, random forest.
Model construction
Two models were constructed based on clinical features, PET metabolic features, CT-derived radiomic features and PET-derived radiomics features using the random forest (RF) method to predict EGFR mutant status and the 19DEL/21L858R mutation site. In the first mutant/wild-type model, patients with the mutant status 19DEL or 21L858R were defined as positive and others as negative, while in the second model, the 19DEL/21L858R mutation site model focused on patients with mutant status, from which patients with the 19DEL were labelled as positive and those with 21L858R as negative. The RF algorithm has a comparably low tendency to overfit and is well suited for data sets with a large number of heterogeneous predictors and cluster-correlated observations, thus, it was adopted for a machine learning-based predictive model. The created RF consisted of 100 trees, and split quality was measured according to Gini impurity. Prior probability for each class was set equal, and the number of features per node was set according to the square root of the total number of features. The generalization capacity of the machine learning-based model was inherently evaluated using 10-fold cross-validation.
Model testing
The created RF model was evaluated on an additional independent cohort study, which was randomly selected with potentially unseen test cases. Model performance was assessed according to receiver operating characteristic (ROC) curve, area under the ROC curve (AUC), sensitivity, and specificity.
Clinical outcome prediction
For the purpose of predicting OS, the least absolute shrinkage and selection operator (LASSO) Cox regression model was used to select the most useful prognostic features in the entire dataset. A radiomic score (R-score) was computed for each patient through a linear combination of the selected features weighted according to their respective coefficient. The clinicopathological factors combined with R-scores were then applied in a multivariate Cox regression model for the prediction of OS. The C-index was calculated to evaluate the performance of the model. The prognosis of targeted therapy and chemotherapy in patients with different EGFR mutation status was compared.
Statistical analysis
Statistical analysis was used to select significant clinicopathological factors and to validate the performance of the models. SPSS version 25.0 (IBM Corporation, Armonk, NY, USA) was used for analysis and the radiomics models were built using the authors’ in-house software programmed with the Python Scikit-learn package (Python version 3.7, Scikit-learn version 0.21, http://scikit-learn.org/).Quantitative data subject to normal distribution are expressed as mean ± standard deviation. An independent sample t test was used for comparisons between the two groups. Qualitative data are expressed as number of cases and percentage [n (%)]. The chi-squared test was used for comparisons between the two groups. When the chi-squared test was not met, the Fisher’s exact test was used. Significant clinicopathological factors (P<0.05) were selected and combined with radiomic features for model establishment. The model performance was assessed using ROC curve, AUC, sensitivity, and specificity. Univariate and multivariate analyses with Cox proportional hazards (CPHs) regression determined the predictors of OS. Variables that achieved statistical significance in the univariate analysis were considered in the multivariate model. The Kaplan-Meier method was used and compared by a two-sided log-rank tests; a two-sided P<0.05 was considered to be statistically significant.
Results
Patient information
Among the 174 patients [93 males (53.45%), 81 females (46.65%)] with lung adenocarcinoma, 109 (62.64%) harbored mutations and 65 (37.36%) were wild type; 44 had the 19DEL and 61 had the 21L858R mutation. The mean ages of the mutant and wild-type patients were 60.24±8.97 and 64.21±12.08 years, respectively, and the difference was statistically significant (P=0.023). Compared with the EGFR wild type, EGFR mutations were more common in women 61 (75.31%) and non-smokers 82 (71.30%) (Table 2). Among clinical features, sex and smoking history demonstrated a significant association with EGFR mutation status. These two clinical features were, therefore, added to the construction of the models.
Table 2. Association between clinical characteristics and the EGFR status in lung adenocarcinoma.
| Characteristics | EGFR wild-type | EGFR mutant-type | Total | P value |
|---|---|---|---|---|
| Age, years | 64.21±12.08 [23–85] | 60.24±8.97 [37–81] | 61.72±10.39 [23–85] | 0.023 |
| Sex, n (%) | <0.001 | |||
| Male | 45 (48.39) | 48 (51.61) | 93 (53.45) | |
| Female | 20 (24.69) | 61 (75.31) | 81 (46.65) | |
| Family history, n (%) | 0.620 | |||
| No | 62 (37.80) | 102 (62.20) | 164 (94.35) | |
| Yes | 3 (30.00) | 7 (70.00) | 10 (5.75) | |
| Smoking status, n (%) | 0.001 | |||
| No | 33 (28.70) | 82 (71.30) | 115 (66.09) | |
| Yes | 32 (54.24) | 27 (45.76) | 59 (33.91) | |
| Lymph node metastasis, n (%) | 0.102 | |||
| No | 23 (46.94) | 26 (53.06) | 49 (28.16) | |
| Yes | 42 (33.60) | 83 (66.40) | 125 (71.84) | |
| Distant metastasis, n (%) | 0.076 | |||
| No | 27 (46.55) | 31 (53.45) | 58 (33.33) | |
| Yes | 38 (32.76) | 78 (67.24) | 116 (66.67) | |
| Stage, n (%) | 0.102 | |||
| I/II | 13 (52.00) | 12 (48.00) | 25 (14.37) | |
| III/IV | 52 (34.90) | 97 (65.10) | 149 (85.63) | |
| CEA, n (%) | 0.136 | |||
| <5.05 | 35 (43.21) | 46 (56.79) | 81 (46.55) | |
| ≥5.05 | 30 (32.26) | 63 (67.74) | 93 (53.45) | |
| Size, n (%) | 0.748 | |||
| <3 cm | 27 (36.00) | 48 (64.00) | 75 (43.10) | |
| ≥3 cm | 38 (38.38) | 61 (61.62) | 99 (56.90) | |
| Site, n (%) | 0.049 | |||
| Left upper lobe | 16 (34.78) | 30 (65.22) | 46 (26.44) | |
| Left lower lobe | 19 (61.29) | 12 (38.71) | 31 (17.82) | |
| Right upper lobe | 18 (32.14) | 38 (67.86) | 56 (32.18) | |
| Right middle lobe | 2 (22.22) | 7 (77.78) | 9 (5.17) | |
| Right lower lobe | 10 (31.25) | 22 (68.75) | 32 (18.39) | |
EGFR, epidermal growth factor receptor; CEA, carcinoembryonic antigen.
Model performance
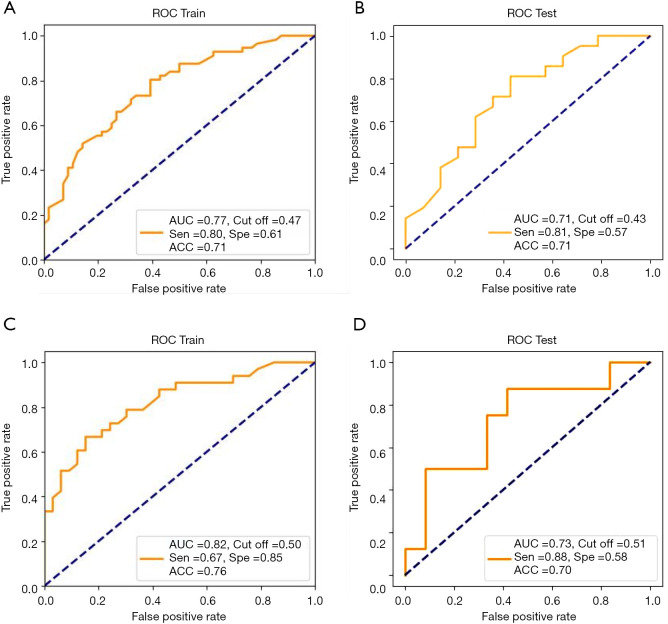
A total of 3,344 (including CT-derived and PET-derived) radiomic features combined with two clinical features (sex and smoking history) and four PET/CT metabolic parameters (MTV, SUVmax, SUVmean and TLG) were used to build the models. The mutant/wild model was trained on a set of 139 patients and validated on an independent test group of 35 patients. After 10-fold cross-validation, the AUC was 0.77, with a sensitivity of 0.80 and a specificity of 0.61 in the training group, while the AUC in the test group was 0.71, with a sensitivity 0.81 and a specificity 0.57 (Figure 2A,B). The second model for the prediction of 19/21 mutation site was trained on a subset of 80 patients with EGFR mutations and tested in an independent subset of 25 patients. The performance of the second model demonstrated an AUC of 0.82, a sensitivity of 0.67 and a specificity of 0.85 in the training cohort, and an AUC of 0.73, a sensitivity of 0.88 and specificity of 0.58 in the test cohort (Figure 2C,D).
Figure 2.
The predictive performance of models. (A) AUC of the first model to predict EGFR mutant/wild status in the training group is 0.77, sensitivity 0.80, specificity 0.61; (B) AUC of the first model in the independent validation group is 0.71, sensitivity 0.81, specificity 0.57; (C) AUC of the second model to predict EGFR mutation site (19/21) in the training group, sensitivity 0.67, specificity 0.85; (D) AUC of the second model in the validation group, sensitivity 0.88, specificity 0.58. EGFR, epidermal growth factor receptor.
Predictors of survival
The median survival time for the 174 patients was 34.00 months [interquartile range (IQR), 18.00–45.00 months]. In total, 104 patients died during follow-up, 7 were lost to follow-up, and 63 were alive at the end of follow-up. The 1- and 5-year OS rates were 89.66% and 13.22%, respectively. R-score, family history, smoking status, lymph node metastasis, mutation status, TLG, MTV, and stage were assessed as survival predictors using multivariate Cox regression. Results revealed that R-score (OR 2.71; 95% CI, 1.90–3.91), family history (OR 0.98; 95% CI, 0.39–2.44), smoking status (OR 1.21; 95% CI, 0.73–2.01), lymph node metastasis (OR 2.47; 95% CI, 1.28–4.78), mutation status (OR 0.51; 95% CI, 0.35–0.76), MTV (OR 1.72; 95% CI, 1.09–2.73), and SUVmax (OR 1.17; 95% CI, 1.07–1.28) were independent predictors of OS. R-scores combined with Cox proportional hazard (CPH) demonstrated a good predictive survival performance, with a C-index of 0.757.
Among those with EGFR mutations, 65 patients underwent targeted therapy and 33 underwent chemotherapy. The mean survival time was 40.72 months and the median survival time of targeted therapy was 38.00 months (IQR, 29.00–46.50 months). For chemotherapy, the mean survival time was 30.55 months and the median survival time was 31.00 months (IQR, 13.00–42.50 months). The prognosis for targeted therapy was better than for chemotherapy. There was a significant difference in survival time between targeted therapy and chemotherapy for patients with EGFR mutations (P=0.03). Survival curves for both treatments are presented in Figure 3.
Figure 3.

Survival curve of targeted therapy and chemotherapy in patients with EGFR mutation. Kaplan-Meier survival analysis of the patients with EGFR mutation in the targeted therapy and chemotherapy groups. The patients with EGFR mutation were stratified into targeted therapy and chemotherapy groups based on their treatment method (P=0.03, log-rank test). EGFR, epidermal growth factor receptor.
Twenty-nine patients with 19DEL and 36 with 21L858R mutations underwent targeted therapy. The mean and median survival times for 19DEL were 42.69 months and 40.00 months (IQR, 29.50–52.00 months), respectively. The mean and median survival times for 21L858R were 39.11 and 37.00 months (IQR, 26.00–45.00 months), respectively. There was no significant difference in survival time between those with 19DEL and 21L858R mutations for targeted therapy (P=0.40). Survival curves for both mutation sites are presented in Figure 4.
Figure 4.
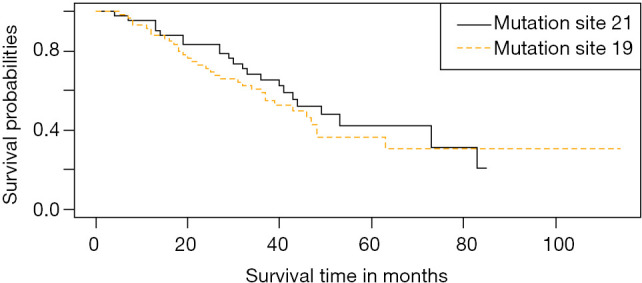
Survival curve of targeted therapy in patients with 19DEL and 21L858R mutation. Kaplan-Meier survival analysis of the patients with 19DEL and 21L858R mutation in the targeted therapy. The patients were stratified into 19DEL and 21L858R mutation groups based on their mutation sites (P=0.40, log-rank test).
Thirty- three patients with EGFR mutations and 51 with wild-type EGFR underwent chemotherapy. The mean and median survival times for EGFR mutations were 45.36 and 42.00 months (IQR, 36.00–50.00 months), respectively. The mean and median survival times for EGFR wild-type were 53.91 and 59.00 months (IQR, 34.00–67.00 months), respectively. There was no significant difference in survival time between those with EGFR mutations and EGFR wild type after chemotherapy (P=0.30). Survival curves for both mutations are presented in Figure 5.
Figure 5.
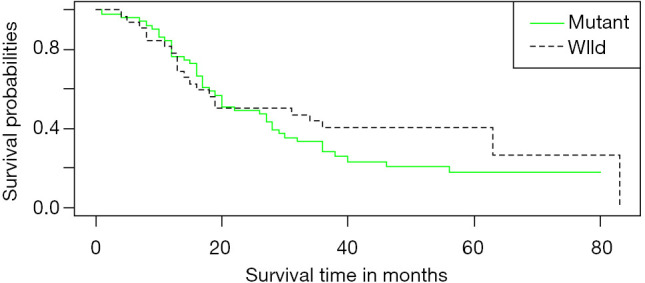
Survival curve of chemotherapy in patients with EGFR mutation and EGFR wild. Kaplan-Meier survival analysis of the patients with EGFR mutation and EGFR wild in the chemotherapy (P=0.30, log-rank test). EGFR, epidermal growth factor receptor.
Discussion
Radiomics has recently emerged as a new method of quantitative image analysis by extracting large numbers of high-dimensional image features. Compared with tissue biopsy, radiomics is non-invasive, rapid, low-cost, and easy to perform (26,27). Moreover, radiomics can provide complementary information for lesions with intratumor heterogeneity. With advances in precision medicine, treatment planning can be improved by adding tumor phenotype information acquired from medical imaging in the future (28,29). Therefore, the purpose of our study was to investigate whether radiomics features from (18F)-FDG PET/CT in patients with lung adenocarcinoma combined with clinical data can predict EGFR mutation status and the EGFR mutation site in exons 19DEL and 21L858R. The third model was then developed using the combination of R-scores and 5 clinicopathological factors to build the multivariate CPH model to predict OS, aiming to provide a reference for individualized molecular targeted therapy.
Although several studies have attempted to build a CT imaging-based model to predict EGFR mutation status in patients with lung adenocarcinoma, few have investigated the performance of the models based on PET/CT imaging features. Lv et al. (25) performed multivariate logistic regression analysis on PET/CT metabolic parameters and clinical data to predict EGFR mutation status. They reported an AUC of 0.557 when using metabolic parameters alone, but an AUC of 0.697 after combination with clinical data. However, there were no radiomic features analyzed in the study, moreover, their results were not tested in an independent group and accuracy may have been reduced if new samples were different from those in the training group. Our study revealed an AUC of 0.77 in the training group and 0.71 in further validation in the test group to predict EGFR mutation status. This was slightly more favorable than in previous studies, and may contribute to combining radiomic features of PET and CT imaging, and implementing a cross-validation approach and our RF classifier, which has been proposed in several previous studies to achieve better model performance (30,31). Moreover, the clinical features of sex and smoking history were selected to construct the models, which was consistent with previous studies (32,33). Combining clinical features may improve model performance, and has been confirmed in several studies (34,35).
In addition, our study built a second model to predict mutations between 19DEL and 21L858R sites using an RF algorithm. Patients with different mutation sites exhibit different response rates to EGFR-TKI therapy and, thus, outcomes are influenced. However, to the best of our knowledge, very few investigations have focused on the identification of the EGFR mutation site. Mei et al. (36) investigated whether radiomic features can be surrogate biomarkers for EGFR mutation status between the 19DEL and 21L858R sites using logistic regression. The analysis yielded AUCs for combining clinical and radiomic features to predict 19DEL mutation, 21L858R mutation and EGFR mutation of 0.655, 0.675, and 0.664, respectively. However, independent testing was not applied to evaluate the performance of the model, and no PET/CT imaging-related studies have, to our knowledge, not been performed. Our second radiomics model to predict mutation at the 19DEL and 21L858R sites achieved better predictive performance, with an AUC of 0.73 (sensitivity, 0.88; specificity, 0.58; accuracy, 0.70) for validation in the test group. However, the diagnostic power of the radiomics approach for mutation site prediction is still not sufficiently reliable for clinical use and, therefore, further study is needed.
Our study also developed a CPH model by combining R-scores with five clinicopathological factors for prediction of OS. R-scores combined with CPH were found to provide good prediction of survival, with a C-index of 0.757. EGFR mutation status and MTV were independent predictors of OS, the results of mutation status analysis revealed an OR of 0.51, which suggests that EGFR mutation status is closely related to prognosis. As a parameter that reflects the metabolic load of the whole body of the tumor, MTV can stratify patients more effectively than other metabolic parameters in order to conduct accurate prognosis evaluation. Our study revealed as OR for MTV of 1.72, indicating that MTV is also an important prognostic factor for patients with NSCLC. This is consistent with previous studies (37,38).
Our results demonstrated that radiomic features not only had the potential ability to predict EGFR mutation status and mutation site, but also indicated the prognosis of lung adenocarcinoma. In addition, the survival analysis revealed that the prognosis of targeted therapy was better than chemotherapy in patients with EGFR mutation(s), which was consistent with previous reference reports (9,39-41). The mean survival time of targeted therapy for 19DEL and 21L858R mutations was revealed to be not significantly different, however, the 19DEL survival rate was greater than that for 21L858R, referring to the long-term survival time (after 80 months). Although this can be explained by the small sample size, the higher survival rate of 19DEL after 80 months could still imply better prognosis for 19DEL than 21L858R, which agrees with the study by Yang et al. (42). The mean survival time after chemotherapy for EGFR mutant and EGFR wild-type demonstrated no statistically significant difference. This indicated that patient survival time was primarily related to the treatment method, and had little to do with EGFR mutation status.
Regarding strengths of our study, our findings not only predicted EGFR mutation status, but also predicted mutation at the 19DEL and 21L858R sites, which has rarely been reported. Furthermore, the effects of different treatment methods on OS of patients with EGFR mutations were further analyzed, which could provide a reference for individualized molecular targeted therapy and provide a good foundation for clinical application(s) of radiomics. Second, all cases in the present study underwent imaging using the same PET/CT instrument and a standardized protocol, which avoided the heterogeneity of image acquisition from different scans and reconstruction parameters, thus leading to more stable and reliable results. Moreover, semi-automatic segmentation tools were implemented in our radiomics research prototype; therefore, individual differences in manual drawing were minimized. Third, repeated cross-validation was used for training to reduce biased estimation, and testing was performed in an independent group to evaluate the performance of our model. As such, our models are more robust and reliable. However, our study also had several limitations, the first of which were its retrospective design and the possibility of some selection bias in patient selection. Secondly, the sample size was small, and patients with mutations in exons 18 and 20 were not included because of small populations. Thirdly, our study with no external validation, which may led to generalization capacity of model was not so well. In future studies, we will expand the sample size to investigate other genetic mutations and conduct multicenter study
Conclusions
Radiomic features from (18F)-FDG PET/CT in patients with lung adenocarcinoma combined with clinical factors demonstrated moderate classification performance in predicting EGFR mutate status and prognosis. Therefore, it may provide reference for personalized molecular targeted therapy.
Supplementary
The article’s supplementary files as
Acknowledgments
We would like to thank all the personnel from the Department of Nuclear Medicine and Department of Medical Imaging, Jinling Hospital, Medical school of Nanjing University for their collective efforts on the image post-processing, clinical management, and follow-up. Additionally, we would like to thank Dr. Shaofeng Duan for his guidance in our project. The authors also thank Editage (www.editage.jp) for English language editing.
Funding: This work was supported by the National Key Research and Development Program of China (2017YFC0113400 for LJZ). And Natural Science Foundation of Jiangsu Province (BK2011665).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The Institutional Review Board of Jinling Hospital, Medical School of Nanjing University approved this retrospective study (No. 2016NZKYKS-004-01) and waived requirements for informed consent from the patients.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-19-592). The authors have no conflicts of interest to declare.
References
- 1.Reck M, Popat S, Reinmuth N, et al. Metastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2014;25:iii27-39. 10.1093/annonc/mdu199 [DOI] [PubMed] [Google Scholar]
- 2.Parums DV. Current status of targeted therapy in non-small cell lung cancer. Drugs Today (Barc) 2014;50:503-5. 10.1358/dot.2014.50.07.2185913 [DOI] [PubMed] [Google Scholar]
- 3.Fan TWM, Lane AN, Higashi RM, et al. Metabolic profiling identifies lung tumor responsiveness to erlotinib. Exp Mol Pathol 2009;87:83-6. 10.1016/j.yexmp.2009.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ettinger DS, Aisner DL, Wood DE, et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 5.2018. J Natl Compr Canc Netw 2018;16:807-21. 10.6004/jnccn.2018.0062 [DOI] [PubMed] [Google Scholar]
- 5.Novello S, Barlesi F, Califano R, et al. Metastatic non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2016;27:v1-27. 10.1093/annonc/mdw326 [DOI] [PubMed] [Google Scholar]
- 6.Miller VA, Riely GJ, Zakowski MF, et al. Molecular characteristics of bronchioloalveolar carcinoma and adenocarcinoma, bronchioloalveolar carcinoma subtype, predict response to erlotinib. J Clin Oncol 2008;26:1472-8. 10.1200/JCO.2007.13.0062 [DOI] [PubMed] [Google Scholar]
- 7.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. 10.1056/NEJMoa0810699 [DOI] [PubMed] [Google Scholar]
- 8.Han JY, Park K, Kim SW, et al. First-SIGNAL: first-line single-agent iressa versus gemcitabine and cisplatin trial in never-smokers with adenocarcinoma of the lung. J Clin Oncol 2012;30:1122-8. 10.1200/JCO.2011.36.8456 [DOI] [PubMed] [Google Scholar]
- 9.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. 10.1016/S1470-2045(11)70393-X [DOI] [PubMed] [Google Scholar]
- 10.Yang JC, Sequist LV, Zhou C, et al. Effect of dose adjustment on the safety and efficacy of afatinib for EGFR mutation-positive lung adenocarcinoma: post hoc analyses of the randomized LUX-Lung 3 and 6 trials. Ann Oncol 2016;27:2103-10. 10.1093/annonc/mdw322 [DOI] [PubMed] [Google Scholar]
- 11.Zhao FN, Zhao YQ, Han LZ, et al. Clinicoradiological features associated with epidermal growth factor receptor exon 19 and 21 mutation in lung adenocarcinoma. Clin Radiol 2019;74:80.e7-80.e17. 10.1016/j.crad.2018.10.001 [DOI] [PubMed] [Google Scholar]
- 12.Bell DW, Gore I, Okimoto RA, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet 2005;37:1315-6. 10.1038/ng1671 [DOI] [PubMed] [Google Scholar]
- 13.Sharma SV, Bell DW, Settleman J. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169-81. 10.1038/nrc2088 [DOI] [PubMed] [Google Scholar]
- 14.Zhu JQ, Zhong WZ, Zhang GC, et al. Better survival with EGFR exon 19 than exon 21 mutations in gefitinib-treated non-small cell lung cancer patients is due to differential inhibition of downstream signals. Cancer Lett 2008;265:307-17. 10.1016/j.canlet.2008.02.064 [DOI] [PubMed] [Google Scholar]
- 15.Choi YJ, Rho JK, Jeon BS, et al. Combined inhibition of IGFR enhances the effects of gefitinib in H1650: a lung cancer cell line with EGFR mutation and primary resistance to EGFR-TK inhibitors. Cancer Chemother Pharmacol 2010;66:381-8. 10.1007/s00280-009-1174-7 [DOI] [PubMed] [Google Scholar]
- 16.Goldman JW, Noor ZS, Remon J, et al. Are liquid biopsies a surrogate for tissue EGFR testing? Ann Oncol 2018;29:i38-i46. 10.1093/annonc/mdx706 [DOI] [PubMed] [Google Scholar]
- 17.Putora PM, Szentesi K, Glatzer M, et al. SUVmax and tumour location in PET-CT predict oncogene status in lung cancer. Oncol Res Treat 2016;39:681-6. 10.1159/000450622 [DOI] [PubMed] [Google Scholar]
- 18.Desseroit MC, Visvikis D, Tixier F, et al. Development of a nomogram combining clinical staging with (18)F-FDG PET/CT image features in non-small-cell lung cancer stage I-III. Eur J Nucl Med Mol Imaging 2016;43:1477-85. 10.1007/s00259-016-3325-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang D, Zhang M, Gao X. Prognostic value of baseline 18F-FDG PET/CT functional parameters in patients with advanced lung adenocarcinoma stratified by EGFR mutation status. PLoS One 2016;11:e0158307. 10.1371/journal.pone.0158307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao Z, Ren G, Liu H, et al. PET of EGFR expression with an 18F-labeled affibody molecule. J Nucl Med 2012;53:1110-8. 10.2967/jnumed.111.100842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho A, Hur J, Moon YW, et al. Correlation between EGFR gene mutation, cytologic tumor markers, 18F-FDG uptake in non-small cell lung cancer. BMC Cancer 2016;16:224. 10.1186/s12885-016-2251-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dou TH, Coroller TP, van Griethuysen JJM, et al. Peritumoral radiomics features predict distant metastasis in locally advanced NSCLC. PLoS One 2018;13:e0206108. 10.1371/journal.pone.0206108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee G, Lee HY, Park H, et al. Radiomics and its emerging role in lung cancer research, imaging biomarkers and clinical management: state of the art. Eur J Radiol 2017;86:297-307. 10.1016/j.ejrad.2016.09.005 [DOI] [PubMed] [Google Scholar]
- 24.Nair VS, Gevaert O, Davidzon G, et al. NF-κB protein expression associates with (18)F-FDG PET tumor uptake in non-small cell lung cancer: a radiogenomics validation study to understand tumor metabolism. Lung Cancer 2014;83:189-96. 10.1016/j.lungcan.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lv Z, Fan J, Xu J, et al. Value of F-FDG PET/CT for predicting EGFR mutations and positive ALK expression in patients with non-small cell lung cancer: a retrospective analysis of 849 Chinese patients. Eur J Nucl Med Mol Imaging 2018;45:735-50. 10.1007/s00259-017-3885-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar V, Gu Y, Basu S, et al. Radiomics: the process and the challenges. Magn Reson Imaging 2012;30:1234-48. 10.1016/j.mri.2012.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gillies RJ, Kinahan PE. Radiomics: images are more than pictures, they are data. Radiology 2016;278:563-77. 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14:749-62. 10.1038/nrclinonc.2017.141 [DOI] [PubMed] [Google Scholar]
- 29.Keek SA, Leijenaar RT, Jochems A. A review on radiomics and the future of theranostics for patient selection in precision medicine. Br J Radiol 2018;91:20170926. 10.1259/bjr.20170926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang X, Dong X, Wang J, et al. Computed tomography-based radiomics signature: a potential indicator of epidermal growth factor receptor mutation in pulmonary adenocarcinoma appearing as a subsolid nodule. Oncologist 2019;24:e1156-e1164. 10.1634/theoncologist.2018-0706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia TY, Xiong JF, Li XY, et al. Identifying EGFR mutations in lung adenocarcinoma by noninvasive imaging using radiomics features and random forest modeling. Eur Radiol 2019;29:4742-50. 10.1007/s00330-019-06024-y [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Kim J, Balagurunathan Y, et al. Radiomic features are associated with EGFR mutation status in lung adenocarcinomas. Clin Lung Cancer 2016;17:441-448.e6. 10.1016/j.cllc.2016.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou JY, Zheng J, Yu ZF, et al. Comparative analysis of clinicoradiologic characteristics of lung adenocarcinomas with ALK rearrangements or EGFR mutations. Eur Radiol 2015;25:1257-66. 10.1007/s00330-014-3516-z [DOI] [PubMed] [Google Scholar]
- 34.Zhang L, Chen B, Liu X, et al. Quantitative biomarkers for prediction of epidermal growth factor receptor mutation in non-small cell lung cancer. Transl Oncol 2018;11:94-101. 10.1016/j.tranon.2017.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee EY, Khong PL, Lee VH, et al. Metabolic phenotype of stage IV lung adenocarcinoma: relationship with epidermal growth factor receptor mutation. Clin Nucl Med 2015;40:e190-e195. 10.1097/RLU.0000000000000684 [DOI] [PubMed] [Google Scholar]
- 36.Mei D, Luo Y, Wang Y. CT texture analysis of lung adenocarcinoma: can Radiomic features be surrogate biomarkers for EGFR mutation statuses. Cancer Imaging 2018;18:52. 10.1186/s40644-018-0184-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao S, Penney BC, Zhang H, et al. Prognostic value of the quantitative metabolic volumetric measurement on 18F-FDG PET/CT in stage IV nonsurgical small-cell lung cancer. Acad Radiol 2012;19:69-77. 10.1016/j.acra.2011.08.020 [DOI] [PubMed] [Google Scholar]
- 38.Yoo SW, Kim J, Chong A, et al. Metabolic tumor volume measured by F-18 FDG PET/CT can further stratify the prognosis of patients with stage IV non-small cell lung cancer. Nucl Med Mol Imaging 2012;46:286-93. 10.1007/s13139-012-0165-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu YL, Zhou C, Hu CP, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol 2014;15:213-22. 10.1016/S1470-2045(13)70604-1 [DOI] [PubMed] [Google Scholar]
- 40.Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013;31:3327-34. 10.1200/JCO.2012.44.2806 [DOI] [PubMed] [Google Scholar]
- 41.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. 10.1016/S1470-2045(11)70184-X [DOI] [PubMed] [Google Scholar]
- 42.Yang JC, Wu YL, Schuler M, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol 2015;16:141-51. 10.1016/S1470-2045(14)71173-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as