Abstract
Background
Identification of epidermal growth factor receptor (EGFR) mutation types is crucial before tyrosine kinase inhibitors (TKIs) treatment. Radiomics is a new strategy to noninvasively predict the genetic status of cancer. In this study, we aimed to develop a predictive model based on 18F-fluorodeoxyglucose positron emission tomography-computed tomography (18F-FDG PET/CT) radiomic features to identify the specific EGFR mutation subtypes.
Methods
We retrospectively studied 18F-FDG PET/CT images of 148 patients with isolated lung lesions, which were scanned in two hospitals with different CT scan setting (slice thickness: 3 and 5 mm, respectively). The tumor regions were manually segmented on PET/CT images, and 1,570 radiomic features (1,470 from CT and 100 from PET) were extracted from the tumor regions. Seven hundred and ninety-four radiomic features insensitive to different CT settings were first selected using the Mann white U test, and collinear features were further removed from them by recursively calculating the variation inflation factor. Then, multiple supervised machine learning models were applied to identify prognostic radiomic features through: (I) a multi-variate random forest to select features of high importance in discriminating different EGFR mutation status; (II) a logistic regression model to select features of the highest predictive value of the EGFR subtypes. The EGFR mutation predicting model was constructed from prognostic radiomic features using the popular Xgboost machine-learning algorithm and validated using 3-fold cross-validation. The performance of predicting model was analyzed using the receiver operating characteristic curve (ROC) and measured with the area under the curve (AUC).
Results
Two sets of prognostic radiomic features were found for specific EGFR mutation subtypes: 5 radiomic features for EGFR exon 19 deletions, and 5 radiomic features for EGFR exon 21 L858R missense. The corresponding radiomic predictors achieved the prediction accuracies of 0.77 and 0.92 in terms of AUC, respectively. Combing these two predictors, the overall model for predicting EGFR mutation positivity was also constructed, and the AUC was 0.87.
Conclusions
In our study, we established predictive models based on radiomic analysis of 18F-FDG PET/CT images. And it achieved a satisfying prediction power in the identification of EGFR mutation status as well as the certain EGFR mutation subtypes in lung cancer.
Keywords: Lung adenocarcinoma, EGFR mutation subtypes, radiomic features, 18F-FDG PET/CT, prediction
Introduction
Lung cancer is the leading cause of cancer-related death worldwide, with an estimated mortality of 142,670 in 2019, and lung adenocarcinoma is the most common histologic type (1,2). Nowadays, to solve the problem of drug resistance to chemotherapy and improve therapeutic effect, molecularly targeted therapy has evolved quickly (3). As an effective therapeutic target, the epidermal growth factor receptor (EGFR) has been well studied in recent decades. Several researches have revealed that, compared to the tumors with EGFR wild type (WT) and other mutation types, a higher response rate of tyrosine kinase inhibitors (TKIs) and longer progression-free survival (PFS) were achieved for cases bearing EGFR mutation-positive tumors (4,5). Therefore, accurate identification of EGFR mutation status is the standard procedure to screen potential patients for EGFR-targeted therapies.
There are more than 200 distinct types of EGFR mutations for non-small cell lung cancer (NSCLC), of which exon 19 deletion (E19 del) and exon 21 L858R missense (E21 mis) are the most common ones and account for approximately 90% (6). Moreover, exon 19 and 21 mutations were sensitive to TKIs, like gefitinib or erlotinib. Apart from these two subtypes, patients with other EGFR mutations may have an unsatisfied response to EGFR TKIs. Interestingly, a previous study showed that tumors with E19 del mutation and E21 mis mutation exhibited different characteristics (7). Some researches have also demonstrated that compared with E21 mis mutation, patients with E19 del mutation harbored longer PFS when receiving EGFR TKIs treatments (8-10). Therefore, accurate evaluation of EGFR mutation subtypes will be increasingly essential to personalize therapy.
At present, the identification of EGFR mutation status is mainly based on the genetic testing of tumor specimens by biopsy. However, in clinic practice, tumor heterogeneity and inadequacy of tumor tissues obtained from biopsies are the barriers to accurately detect the EGFR mutation type (11,12). Analysis of circulating cell-free tumor DNA (ctDNA) is another method to assess the EGFR mutation status (11). Unfortunately, studies have shown that ctDNA testing has a relatively high false-negative rate and is also very costly (13,14). Therefore, reliable methods that can use clinical information to noninvasively detect EGFR mutation subtypes are urgently needed.
Some studies have investigated the relationship between computed tomography (CT) features including air bronchogram, pleural retraction, lesion size and EGFR mutation status in NSCLC (15,16). However, the conclusions of different researches were controversial, which may due to the semiqualitative assessments and limited morphological information from CT. Different from traditional CT analysis, radiomics is a technology for quantitative mapping of medical images and has shown great potential in the prediction of clinical outcome and genomic features (17). From the extraction, analysis and modeling of image features in radiomic analysis, innumerable quantitative features can be extracted by high-throughput computing of medical images. In the past, due to extensive usage of CT in clinical practice, most radiomic researches are based on CT images, and many published articles have built prediction models to assess EGFR mutations status in lung adenocarcinoma using CT data (18-20). Mei et al. showed that area under the curve (AUC) of the combination with clinical information and CT radiomic features to predict EGFR mutations was 0.664 (21). However, CT can only offer the morphological information of the tumor, which may make it difficult to improve the prediction accuracy.
Noninvasive 18F-fluorodeoxyglucose positron emission tomography-computed tomography (18F-FDG PET/CT) is increasingly used in NSCLC patients for staging and therapy evaluation. Compared with traditional CT imaging, 18F-FDG PET/CT reflects the glucose metabolism of tumors. More importantly, the decrease of the tumoral 18F-FDG uptakes demonstrates the efficacy of EGFR inhibitors in clinical settings (22-24), implying that there is a relationship between glucose metabolism and EGFR pathways. In recent years, many articles have analyzed the relationship between some quantitative parameters of 18F-FDG PET/CT and EGFR mutations in NSCLC (22,25). Compared with the clinically used parameters like maximum standardized uptake value (SUVmax), metabolic tumor volume and total lesion glycolysis, texture analysis of 18F-FDG PET/CT images could offer more information about tumoral spatial information and tumor heterogeneity. Therefore, 18F-FDG PET/CT texture analysis is supposed to be related to the tumor microenvironment and tumor phenotype. Several studies have reported that the predictive models derived from 18F-FDG PET/CT features can identify mutative EGFR from WT (26-28). And the accuracy of different predictive models ranged from 60–83%, indicating that 18F-FDG PET/CT radiomics may serve as a noninvasive tool to predict EGFR mutations.
To our knowledge, probably due to the limited data, only a few studies have attempted to predict EGFR mutation subtypes using CT radiomic features (21). Unfortunately, no predictive models based on 18F-FDG PET/CT radiomics have been used to identify the EGFR mutation subtypes (E19 del mutation or E21 mis mutation). The establishment of a prediction model to identify specific EGFR mutation subtypes is of great importance for personalized therapy. Herein, we retrospectively collected 18F-FDG PET/CT data with different scan settings and extracted texture features of 18F-FDG PET/CT images in patients with lung adenocarcinoma. This study aims to develop a predictive model to noninvasively identify E19 del mutation or E21 mis mutation.
Methods
Patients
The study was approved by the Ethics Committee of Fudan University Shanghai Cancer Center (No. 1909207-14-1910) and the data were analyzed anonymously. The requirement of written informed consent was waived. We collected 18F-FDG PET/CT scan data of 178 patients with lung adenocarcinoma between January 2016 and December 2017 from Renji Hospital, School of Medicine, Shanghai Jiao Tong University and Fudan University Shanghai Cancer Center. The inclusion criteria were as follows: (I) pathologically confirmed lung adenocarcinoma; (II) tumors without EGFR mutation, with E19 del mutation or E21 mis mutation; (III) the resected specimens were used for EGFR mutation test; (IV) available clinical characteristics including sex, age, tumor size, and TNM staging; (V) available 18F-FDG PET/CT scan data before treatment. Thereafter, 30 patients were excluded for the following reasons: (I) undergone preoperative treatment; (II) the tumor margin was too difficult to contour; (III) the clinical information was incomplete. Finally, a total of 148 patients were included in the study. And they were randomly assigned into the training group (n=111) and test group (n=37). Patients’ characteristics were shown in Table 1.
Table 1. Clinical characteristics of enrolled patients.
| Characteristics | Total | Exon 19 mutation | Exon 21 mutation | Wild type | Test group | Training group |
|---|---|---|---|---|---|---|
| Number | 148 | 44 | 31 | 73 | 37 | 111 |
| Sex | ||||||
| Male | 85 | 15 | 11 | 59 | 23 | 62 |
| Female | 63 | 29 | 20 | 14 | 14 | 49 |
| Age | ||||||
| Median | 61.2 | 58.9 | 62.4 | 60.9 | 61.6 | 61.1 |
| Min | 36 | 36 | 43 | 36 | 40 | 36 |
| Max | 84 | 82 | 79 | 84 | 82 | 84 |
| TNM staging | ||||||
| II | 50 | 16 | 13 | 21 | 7 | 43 |
| III | 39 | 10 | 7 | 22 | 7 | 32 |
| IV | 59 | 18 | 11 | 30 | 23 | 36 |
| Tumor size (cm) | ||||||
| Median | 3.2 | 3.5 | 2.8 | 2.7 | 3.3 | 3.2 |
| Min | 0.5 | 0.9 | 3.8 | 0.5 | 0.5 | 0.9 |
| Max | 11.3 | 11.3 | 8.2 | 5 | 6.7 | 11.3 |
EGFR mutation detection
The EGFR mutation analyses were performed by experienced pathologists at the Department of Pathology in the hospitals using surgically resected specimens. EGFR exons 18, 19, 20, and 21 were tested using an amplification refractory mutation system real-time technology with ARMS (AmoyDx EGFR Mutations Detection Kit). And we retrospectively obtained the EGFR status from the medical record system of the hospitals.
Image acquisition
In this study, non-time of flight (TOF) 18F-FDG PET/CT scans were performed using a whole-body PET/CT scanner (Biograph mCT, Siemens Medical Systems, Erlangen, Germany) and a regular PET/CT scanner (Biogragh 16 HR, Siemens Medical Systems, Erlangen, Germany). Before 18F-FDG administration, all the patients received glucose level test and the blood glucose levels should be less than 140 mg/dL. Then, patients fasted for at least 6 h before the injection of 18F-FDG (7.4 MBq/kg) and image acquisition was started 1 hour afterward.
For Siemens Biograph mCT PET/CT scanner, a spiral CT scan with a standardized protocol including 120 kV, 140 mA, and a 3-mm slice thickness was conducted followed by a PET scan. And then, for PET scanning, the acquisition time was 3 minutes per bed position and PET image datasets were reconstructed iteratively with CT data for attenuation correction. While for Siemens biograph 16HR PET/CT scanning, CT scanning was first acquired using a low-dose technique (120 kV, 140 mA, 5 mm slice thickness), and PET scan was obtained immediately after the CT scan (2–3 minutes/bed) with gaussian-filter iterative reconstruction method (iterations 4; subsets 8; image size 168).
Image post-acquisition processing
The acquired PET images were normalized using a factor of each patients’ weight and the dose of radioactive tracer. Afterward, intensity discretization was performed on both PET and CT images. Fix bin width approach was used, with a bin width of 25 and 128 respectively for CT and PET images.
Region-of-interest (ROI) segmentation
ROI on CT images was delineated independently by two radiologists using ITK-SNAP software (Version 3.6, United States). And the radiologists were blinded to the pathologic and EGFR mutation test results. Several days after the segmentation, the ROI was confirmed, especially for the contours adjacent to the mediastinum, chest wall, and blood vessels. Only the primary tumor was marked. The delineation was conducted on axial each slice of CT, and then mathematically correspond to PET images.
Radiomic features extraction
The workflow was shown in Figure 1. After manual segmentation, radiomic features were calculated from tumor ROI automatically. Three types of features were included: (I) shape features; (II) first order statistics; (III) texture features. The feature extraction process was conducted in Python 3.6.2 with package Pyradiomics (29). Radiomic features were calculated independently for PET and CT images.
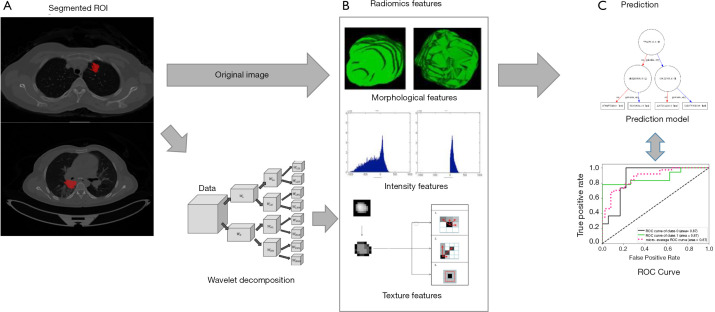
Figure 1.
The workflow of our study. (A) Tumor region of interest (ROI) was segmented by experienced radiologists; (B) radiomic features were extracted from original images and image components after wavelet transformation; (C) prediction of the EGFR mutation.
For CT images, shape features were calculated in both 2D (slice by slice) and 3D (a dicom series as a whole) using original images only. The extraction of other CT features was performed on three different channels: the original images, and the images components derived from two levels of wavelet transformation (wavelet1 and wavelet2). The matrixes and formulas for texture feature calculation were designed according to the definition by the image biomarker standardization initiative (IBSI) (30). In total, 1,470 features were calculated from CT images.
In order to rule out the influence of personal weight and dose, we converted PET image pixels from gray value to SUV value. Considering the low-resolution nature of PET images, radiomic feature extraction of PET was conducted only on the original image with no further transformation or filtering. One hundred PET features were calculated.
Pre-selection: identifying features stable across different image sources
The images we investigated in this study were from two hospitals with different scanners and CT slice thicknesses. To reduce the bias caused by the differences of image sources, it is essential for us to choose features that are relatively consistent between both sources. Before this pre-selection, preprocessing was performed on the calculated features. Firstly, the features concerned with ROI slice numbers were scaled according to the slice thickness. Secondly, the features unrelated to slice numbers were rescaled using the absolute mean to get rid of the scale difference in two image sources.
After preprocessing, we performed the Mann white U test to examine whether these features from different sources can be considered as obeying the same distribution. The features with high confidence of coming from different distributions were eliminated, and the remaining features were preserved for the following feature selection.
Redundant feature elimination and EGFR-related feature selection
Feature selection was performed in the training cohort to remove redundant features and find features most relevant to the targeted EGFR mutation types. Firstly, the variance inflation factor (VIF) (31), a measure for collinearity, was calculated for all features, and feature with the highest VIF was removed. This procedure was performed recursively until the VIF values of all remaining features were below the preset threshold. After the VIF filtering, we compressed the high-dimensional features and removed collinearity. Secondly, a random forest classifier was built to simplify the remaining features. The random forest algorithm is a so-called “ensemble learning” algorithm which combines the result of several weak classifiers to achieve an improved model performance (32), and this algorithm is able to evaluate the relevance of each feature by a feature importance score in the output. Only features with an importance score above a certain threshold were reserved. Thirdly, to further simplify the model and mitigate the overfitting problems, a logistic regression model was introduced to select the optimized feature set for prediction. All thresholds and algorithm model parameters in the procedures above were determined by cross-validation in the training cohort. The operations were conducted in Python 3.6.2 with packages sklearn (33) and stats (34).
Predictive models
The selected optimal features were combined into a radiomic signature. Xgboost, a popular machine learning algorithm (35), was used to build the predictive model. The models for predicting E19 del mutation and E21 mis mutation were constructed separately on the corresponding signature. For each patient, the probability scores for E19 del mutation and E21 mis mutation were computed with the constructed model, based on which we also built an ensemble model to get a general prediction of whether a patient is positive on either E19 del mutation and E21 mis mutation.
For all three models, the model parameters were determined using a 3-fold cross-validation in the training set. Each model’s performance was evaluated on the independent test set using the receiver operating characteristic (ROC) curve, and AUC was calculated to get a quantified performance measurement of each model.
Statistical analysis
Univariate analysis was used to investigate the association of clinical characteristics and selected radiomic features with the targeted EGFR mutations. The Mann-Whitney U test and the chi-square test were performed for continuous variables and categorical variables, respectively. The correlation matrix of selected radiomic features was calculated and illustrated in a heat-map. The AUC of ROC was calculated to evaluate the prediction model performance. The P value of less than 0.05 with a 95% confidence interval was considered statistically significant.
Results
The relevance of clinical characteristics
We finally enrolled 148 patients (61.2±10.4 years, 85 men, 63 women) with primary lung adenocarcinoma who conducted 18FDG PET/CT scan at Renji Hospital and Fudan University Cancer Center. The clinicopathological characteristics of the patients were summarized in Table 1. The percentage of patients with E19 del mutation or E21 mis mutation in the test group and in the training group was 29.7% (11/37), 13.5% (5/37) and 29.7% (33/111), 23.4% (26/111), respectively. And there were no significant differences between the training and test group in terms of age, gender, and stage (P>0.05). Patients with EGFR mutation accounted for 50.6% (75/148), and the percentage of E19 del mutation and E21 mis mutation in EGFR-positive patients was 58.7% (44/75) and 41.3% (31/75), respectively. Of patients with EGFR mutation, 26 patients (26/75, 34.7%) were male and 49 (49/75, 65.3%) patients were female. And there was no significant difference between women and men (P>0.05). The lung cancer stage was determined according to the Eighth Edition Union for International Cancer Control and American Joint Committee on Cancer TNM classification (36). The percentage of patients with stage II, stage III and stage IV was 33.8% (50/148), 26.3% (39/148), and 39.9% (59/148), respectively. Moreover, there were no significant differences in age, sex, tumor size and TNM stage between patients with E19 del mutation and E21 mis mutation. The ANOVA analysis indicated that no associations were found between the different EGFR mutation subtypes and 18F-FDG uptakes.
Feature selection and predictive model
After pre-selection, 718 out of 1,470 CT features and 76 out of 100 PET features were considered robust across the two different image sources. The proportion of feature derivation methods and calculation channels were shown in Tables 2,3.
Table 2. The proportion of feature derivation methods.
| CT feature group | Kept/calculated |
|---|---|
| Shape | 10/14 |
| First order | 218/288 |
| Glcm | 192/352 |
| Glrlm | 48/256 |
| Glszm | 97/256 |
| Gldm | 129/224 |
| Ngtdm | 29/80 |
Table 3. The proportion of feature calculation channels.
| Calculation channels | Percentage of stable features |
|---|---|
| CT feature channels | |
| Original | 63/105 |
| Wavelet components on decomposition level 1 | 308/637 |
| Wavelet components on decomposition level 2 | 347/728 |
| PET feature group | |
| Shape | 14/14, 100% |
| First order | 14/18 |
| Glcm | 16/22 |
| Glrlm | 11/16 |
| Glszm | 11/16 |
| Gldm | 10/14 |
The pre-selection result indicates that general PET features were more robust among different image sources. For CT images, first-order statistics and shape features were generally more stable to different scanner settings compared to the texture features acquired by high-level matrixes. The image channel of feature calculation did not seem to have much influence on the robustness of CT radiomic features.
Feature selection was conducted separately for E19 del mutation and E21 mis mutation. The selected radiomic features of PET/CT were presented in Table 4 and the heatmap of the features was in the Figure S1. Finally, with univariate analysis, we found that five radiomic features were significantly associated with E19 del mutation. The top three features predicting E19 del mutation were CT-wl-fo-Ske, CT-wl-glszm-SZNUN, and CT-wl-glszm-SALGLE (Figure 2). On the other hand, five radiomic features were significantly related with E21 mis mutation (Figure 3). Among them, CT-wl-gldm-LDHGLE, CT-wl-fo-Mean, and CT-orig-fo-Max were the most powerful predictive factors. Thereafter, Xgboost classifiers were built to predict specific EGFR mutation types along with an ensemble model to give a judgment on EGFR general status.
Table 4. Description of selected features in the prediction model.
| Feature name | Description |
|---|---|
| CT_wavelet_HHH_firstorder_Skewness (CT-wl-fo-Ske) | A measure of lack of symmetry |
| CT_wavelet_HLL_gldm_LowGrayLevelEmphasis (CT-wl-gldm-LGLE) | The distribution of the low grey-level runs |
| CT_wavelet-HLL_glszm_SizeZoneNonUniformityNormalized (CT-wl-glszm-SZNUN) | A measure of the variability of size zone volumes throughout the image |
| CT_wavelet2_HHH_glszm_SmallAreaLowGray LevelEmphasis (CT-wl-glszm-SALGLE) | A measure of zone counts where small zone sizes and low grey levels are located |
| PET_original_gldm_DependenceNonUniformity (PET-orig-gldm-DNU) | A measure of the distribution of small dependencies |
| CT_original_firstorder_Maximum (CT-orig-fo-Max) | The maximum gray level intensity in the ROI |
| CT_wavelet_HHH_firstorder_Mean (CT-wl-fo-Mean) | The average gray level intensity in the ROI |
| CT_wavelet_HHH_gldm_LargeDependenceHighGrayLevelEmphasis (CT-wl-gldm-LDHGLE) | A measure of the joint distribution of large dependence with higher gray-level values |
| CT_wavelet2_HHL_firstorder_Median (CT-wl-fo-Median) | The median gray level intensity in the ROI |
| PET_original_glcm_ClusterShade (PET-orig-glcm-CS) | A measure of the skewness and uniformity of the GLCM |
Figure S1.
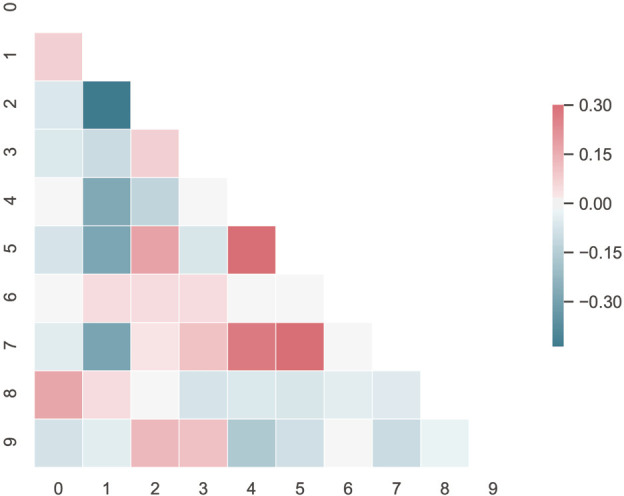
The heatmap of the selected features.
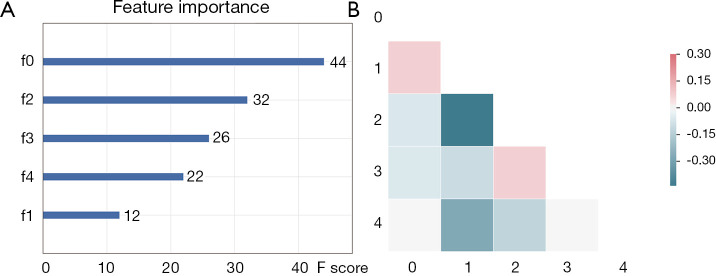
Figure 2.
Selected features in predicting E19 del mutation. (A) Feature importance of selected features; (B) correlation heatmap of selected features. f0, CT-wl-fo-Ske; f2, CT-wl-glszm-SZNUN; f3, CT-wl-glszm-SALGLE; f4, PET-orig-gldm-DNU; f1, CT-wl-gldm-LGLE.
Figure 3.
Selected features in predicting E21 mis mutation. (A) Feature importance of selected features; (B) correlation heatmap of selected features. f7, CT-wl-gldm-LDHGLE; f6, CT-wl-fo-Mean; f5, CT-orig-fo-Max; f8, CT-wl-fo-Median; f9, PET-orig-glcm-CS.
Receiver operating characteristic curve analysis
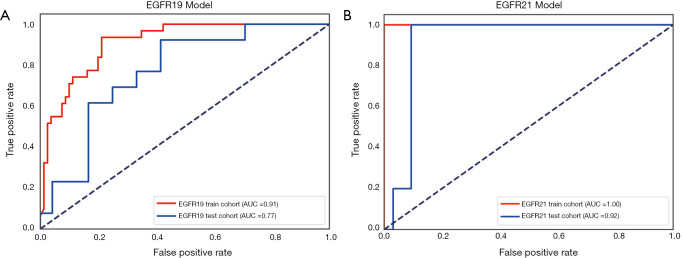
The performance of the predictive model was evaluated with the ROC curve. The ROC curves in the test cohort were shown in Figure 4. Regarding to EGFR mutation, the AUC of the prediction model was 0.93 in the training cohort and 0.87 in the test cohort (Figure 5). For E19 del mutation, the AUC of the predictive model was 0.91 in the training cohort and 0.77 in the test cohort (Figure 6A). Compared with E19 del mutation prediction, the accuracy of the model to identify E21 mis mutation was higher, with a perfect fitting in the training cohort (AUC =1.0) and 0.92 in the test cohort (Figure 6B). The model performance on two different hospital subgroups was also evaluated. As the test cohort is too small to split, we investigated the model in the training cohort. For E19 del mutation, the evaluated AUC of the two patient subgroups were 0.92 and 0.90, respectively (Figure 7). For E21 mis mutation, the model fitted perfectly, so the AUC for each patient subgroup was 1.0. The results suggest that our model shows no preference in any of the patient subgroups.
Figure 4.
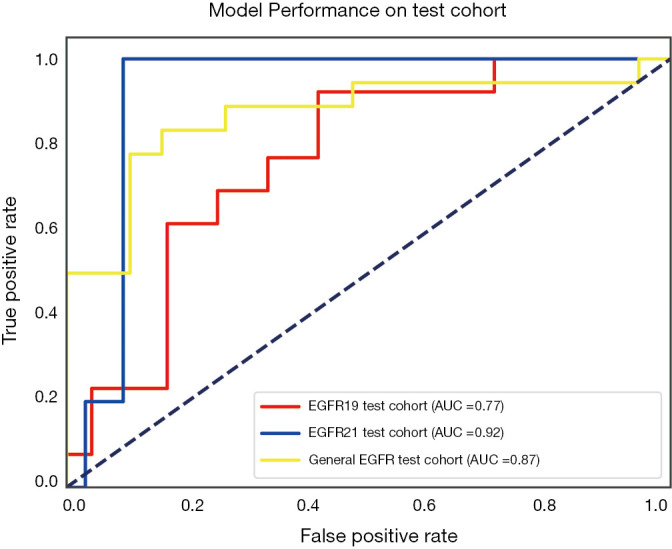
Receiver operating characteristic curve for the predictive model of E19 del mutation and E21 mis mutation in the test cohort.
Figure 5.
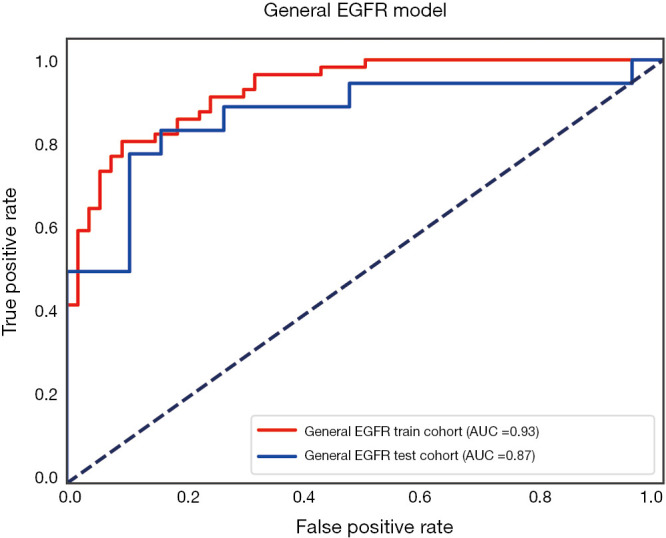
Receiver operating characteristic curve for the EGFR model in the train cohort and test cohort.
Figure 6.
(A) Receiver operating characteristic curve for the predictive model of E19 del mutation; (B) receiver operating characteristic curve for the predictive model of E21 mis mutation.
Figure 7.
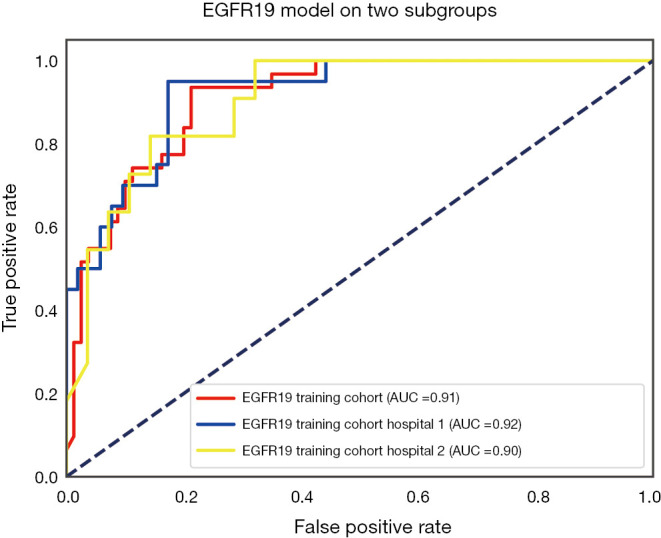
Receiver operating characteristic curve for the predictive model of E19 del mutation on two hospital subgroups.
Discussion
In clinical practice, E19 del mutation and E21 mis mutation are two most common EGFR-mutated subtypes. Increasing evidence has indicated that patients with E19 del mutation may have a longer survival than patients with E21 mis mutation after TKIs treatment (9,37). This may due to the lower plasma concentration of gefitinib in patients with E21 mis mutation (38). Therefore, in order to provide information for individualized therapy and improve prognosis, prediction of the EGFR mutation subtype is crucial. In general, there are two highlights of our study. First, to investigate the robust predictive features, we conducted the radiomic analysis with 18F-FDG PET/CT data acquired from two scanners, which use different acquisition parameters and reconstruction methods. Secondly, to the best of our knowledge, the prediction model for E19 del mutation or E21 mis mutation in lung cancer based on PET/CT radiomic features had not well established.
In our study, we firstly investigated the association between 18F-FDG uptake and EGFR mutation status. The results showed that there was no significant difference between EGFR-positive and -WT patients, and no differences in SUVmax were observed between patients with E19 del mutation and E21 mis mutation. Similarly, Lee et al. (39) and Caicedo et al. (25) have also demonstrated that there were no differences in the SUVmax between the different EGFR mutation subtypes, indicating that SUVmax was not a significant clinical predictor for EGFR mutations. However, the results of previous studies on this topic were contradictory. Some studies have reported that EGFR mutation-positive NSCLCs have relatively lower 18F-FDG uptake compared with WT tumors (40,41), and SUVmax of patients with E21 mis mutation was significantly higher than that of E19 del mutation (42). There are several potential reasons for these conflicting results. On one hand, the TNM stage and histological type of the enrolled patients could significantly affect the results. In comparison, we included only lung adenocarcinoma and patients were mainly with stage II–IV diseases. On the other hand, the small number of patients in our study may explain these discrepant results. As a result, these conflicting results demonstrated that 18F-FDG uptakes may not be a dependable marker for predicting EGFR mutation status.
In recent years, unlike SUVmax, radiomic analysis can reflect the underlying spatial variation and heterogeneity of voxel intensities and tracer uptake within tumors, allowing better tumor characterization. Increasing studies have focused on the relationships of CT features of lung tumor and EGFR mutation status (43,44). The results revealed that radiomic signature could be a better predictor for identifying EGFR mutant than the morphological features that derived from CT images (19). Previous studies have also established CT image-based prediction models to detect EGFR mutation in lung adenocarcinoma (18,29,45). The study by Liu et al. (45) showed that when combined with clinical characteristics (such as sex, pathologic grade, and smoking history), CT-based radiomic features could identify EGFR positive tumors with an AUC value of 0.709. Moreover, the studies of Zhang et al. (46) and Li et al. (47) have validated that a combined radiomic signature with clinical factors exhibited a further improved performance in EGFR mutation differentiation. Compared with these studies, the AUC of our study was a little higher, and we predicted the certain EGFR mutation subtypes (E19 del mutation or E21 mis mutation). However, in our study, there were no significant differences in gender, age and TNM stage between EGFR mutation and WT groups. Thereafter, the above clinical characteristics of patients in our study cannot add the accuracy of the predictive model. It may be attributed to the limited number of patients and select bias.
Furthermore, 18F-FDG PET/CT, as a kind of molecular imaging and harboring higher sensitivity and specificity when compared to CT, has attracted great attention in the field of radiomic analysis. Emerging studies have validated that some PET/CT radiomic features were strongly associated with the EGFR mutation status (27,48). In our study, we found that two PET radiomic features and eight CT features have a relationship with EGFR mutation. Moreover, our study showed that five features (four CT features and one PET feature) and five features (four CT features and one PET features) were associated with the identification of E19 del mutation and E21 mis mutation, respectively. The corresponding CT features mainly described the range, maximum and mean of gray level intensity, as well as the distribution, variability and local homogeneity in the image. The above features have already been certificated in a previous study (21). Moreover, compared with their results, the AUCs of radiomic features to predict E19 del mutation and E21 mis mutation in our study were higher (E19 del mutation: 0.77 vs. 0.65; E21 mis mutation: 0.92 vs. 0.67). This result probably contributed to the combination of PET features in the prediction model. The selected PET features in our research were about tumor homogeneity and symmetry. Yip et al. (27) have also demonstrated the value of 18F-FDG PET radiomic features for predicting EGFR mutation and found that InvDiffmomnor was one of the most predictive features for EGFR mutation status, which was similar to our results.
At present, one of the obstacles hindering the development of radiomic analysis is the lack of available standard clinical imaging data. Therefore, multicenter radiomic analysis is needed in the future. However, it is well known that the accuracy of texture analysis in PET/CT is greatly affected by different scanning protocols, image acquisition parameters, and reconstruction methods. Several researches have verified that the difference in image reconstruction methods has an influence on the predictive efficacy of radiomic features extracted from PET/CT images (49,50). Furthermore, the stability of radiomics features has become a major concern in this field, and there have been numerous publications investigating the influence of slice thicknesses of CT and image reconstruction algorithms of PET on the calculated radiomic features (51,52). Zhao et al. (53) reported that first-order features and shape features were less sensitive to different slice thickness, which were in line with our findings. A previous study also demonstrated that first-order features were more stable than texture features (54). Lasnon et al. (55) showed that image filters, compared with reconstruction methods, had a more evident influence on the features calculated. In our study, we conducted the same filter to PET images from different sources, which may explain why PET features were generally more reproducible than CT features.
In this study, the number of selected features derived from PET images was less than that of a previous study (56). As experimental design has an effect on PET radiomics in predicting somatic mutation status (26,57), we supposed that the reason was that PET/CT data were derived from two scanners with different acquisition parameters and reconstruction methods. Moreover, Kirienko et al. (56) have verified that features from PET images were more likely to be affected by the scanning protocols and reconstruction parameters than CT features. The corresponding results also showed some reliable features including four shape, six statistics, thirteen gray level co-occurrence matrix (GLCM), and six run length matrix (RLM), which were robust to the different experimental settings. Fortunately, the selected features in our study were in consistent with theirs, which supported our results.
There are several limitations in our study. First, as it is a retrospective study, some clinical characteristics like smoking habits were not collected in the study. For further study, we suppose to add more clinical information to improve the prediction efficiency of our model. Second, manual segmentation by radiologists was used in our study, which is time-consuming. However, since the boundary of some lung tumors is rather difficult to draw, manual segmentation is more reliable than the existing automatic method. Definitely, a reliable automatic segmentation method with high accuracy for lung cancer may be developed in the future. Third, the feature extraction of PET and CT were performed separately, which is inherently limited in simultaneously considering both the functional and anatomical information compared with fusing the two modalities together. At last, we should collect more available data to perform an independent test study to confirm our results.
Conclusions
In conclusion, we established predictive models using 18F-FDG PET/CT radiomic features for the identification of E19 del mutation or E21 mis mutation in lung adenocarcinoma, and it obtained a satisfying prediction power. Therefore, 18F-FDG PET/CT radiomic features of lung adenocarcinoma can help to identify EGFR positive tumors, as well as lung adenocarcinoma with E19 del mutation or E21 mis mutation. Moreover, we successfully extracted features with PET/CT data collected from different scan settings, which may be helpful for multicenter radiomic analysis in the future.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was partially supported by the National Natural Science Foundation of China (Nos. 81771861, 81471708, 81830052), 2018 Shanghai Scientific and Technological Innovation Program (No. 18410711200, 19142202100), National Key R&D Program of China (No. 2019YFC1604605) and Shanghai Key Laboratory of Molecular Imaging (18DZ2260400).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Fudan University Shanghai Cancer Center (No. 1909207-14-1910) and the data were analyzed anonymously. The requirement of written informed consent was waived.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr.2020.04.17). The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. 10.1038/nature25183 [DOI] [PubMed] [Google Scholar]
- 3.Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. 10.1016/S0140-6736(16)30958-8 [DOI] [PubMed] [Google Scholar]
- 4.Recondo G, Facchinetti F, Olaussen KA, et al. Making the first move in EGFR-driven or ALK-driven NSCLC: first-generation or next-generation TKI? Nat Rev Clin Oncol 2018;15:694-708. 10.1038/s41571-018-0081-4 [DOI] [PubMed] [Google Scholar]
- 5.da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol 2011;6:49-69. 10.1146/annurev-pathol-011110-130206 [DOI] [PubMed] [Google Scholar]
- 6.Castellanos E, Feld E, Horn L. Driven by Mutations: The Predictive Value of Mutation Subtype in EGFR-Mutated Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:612-23. 10.1016/j.jtho.2016.12.014 [DOI] [PubMed] [Google Scholar]
- 7.Soria JC, Mok TS, Cappuzzo F, et al. EGFR-mutated oncogene-addicted non-small cell lung cancer: current trends and future prospects. Cancer Treat Rev 2012;38:416-30. 10.1016/j.ctrv.2011.10.003 [DOI] [PubMed] [Google Scholar]
- 8.Lim SH, Lee JY, Sun JM, et al. Comparison of clinical outcomes following gefitinib and erlotinib treatment in non-small-cell lung cancer patients harboring an epidermal growth factor receptor mutation in either exon 19 or 21. J Thorac Oncol 2014;9:506-11. 10.1097/JTO.0000000000000095 [DOI] [PubMed] [Google Scholar]
- 9.Sutiman N, Tan SW, Tan EH, et al. EGFR Mutation Subtypes Influence Survival Outcomes following First-Line Gefitinib Therapy in Advanced Asian NSCLC Patients. J Thorac Oncol 2017;12:529-38. 10.1016/j.jtho.2016.11.2225 [DOI] [PubMed] [Google Scholar]
- 10.Choi YW, Jeon SY, Jeong GS, et al. EGFR Exon 19 Deletion is Associated With Favorable Overall Survival After First-line Gefitinib Therapy in Advanced Non-Small Cell Lung Cancer Patients. Am J Clin Oncol 2018;41:385-90. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Chang L, Yang Y, et al. Intratumor heterogeneity comparison among different subtypes of non-small-cell lung cancer through multi-region tissue and matched ctDNA sequencing. Mol Cancer 2019;18:7. 10.1186/s12943-019-0939-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol 2015;16:e342-51. 10.1016/S1470-2045(15)00077-7 [DOI] [PubMed] [Google Scholar]
- 13.Hur JY, Kim HJ, Lee JS, et al. Extracellular vesicle-derived DNA for performing EGFR genotyping of NSCLC patients. Mol Cancer 2018;17:15. 10.1186/s12943-018-0772-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moding EJ, Diehn M, Wakelee HA. Circulating tumor DNA testing in advanced non-small cell lung cancer. Lung Cancer 2018;119:42-7. 10.1016/j.lungcan.2018.02.019 [DOI] [PubMed] [Google Scholar]
- 15.Dai J, Shi J, Soodeen-Lalloo AK, et al. Air bronchogram: A potential indicator of epidermal growth factor receptor mutation in pulmonary subsolid nodules. Lung Cancer 2016;98:22-8. 10.1016/j.lungcan.2016.05.009 [DOI] [PubMed] [Google Scholar]
- 16.Rizzo S, Raimondi S, de Jong EEC, et al. Genomics of non-small cell lung cancer (NSCLC): Association between CT-based imaging features and EGFR and K-RAS mutations in 122 patients-An external validation. Eur J Radiol 2019;110:148-55. 10.1016/j.ejrad.2018.11.032 [DOI] [PubMed] [Google Scholar]
- 17.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14:749-62. 10.1038/nrclinonc.2017.141 [DOI] [PubMed] [Google Scholar]
- 18.Jia TY, Xiong JF, Li XY, et al. Identifying EGFR mutations in lung adenocarcinoma by noninvasive imaging using radiomics features and random forest modeling. Eur Radiol 2019;29:4742-50. 10.1007/s00330-019-06024-y [DOI] [PubMed] [Google Scholar]
- 19.Tu W, Sun G, Fan L, et al. Radiomics signature: A potential and incremental predictor for EGFR mutation status in NSCLC patients, comparison with CT morphology. Lung Cancer 2019;132:28-35. 10.1016/j.lungcan.2019.03.025 [DOI] [PubMed] [Google Scholar]
- 20.Yang X, Dong X, Wang J, et al. Computed Tomography-Based Radiomics Signature: A Potential Indicator of Epidermal Growth Factor Receptor Mutation in Pulmonary Adenocarcinoma Appearing as a Subsolid Nodule. Oncologist 2019;24:e1156-64. 10.1634/theoncologist.2018-0706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mei D, Luo Y, Wang Y, et al. CT texture analysis of lung adenocarcinoma: can Radiomic features be surrogate biomarkers for EGFR mutation statuses. Cancer Imaging 2018;18:52. 10.1186/s40644-018-0184-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benz MR, Herrmann K, Walter F, et al. (18)F-FDG PET/CT for monitoring treatment responses to the epidermal growth factor receptor inhibitor erlotinib. J Nucl Med 2011;52:1684-9. 10.2967/jnumed.111.095257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cook GJ, O'Brien ME, Siddique M, et al. Non-Small Cell Lung Cancer Treated with Erlotinib: Heterogeneity of (18)F-FDG Uptake at PET-Association with Treatment Response and Prognosis. Radiology 2015;276:883-93. 10.1148/radiol.2015141309 [DOI] [PubMed] [Google Scholar]
- 24.De Rosa V, Iommelli F, Monti M, et al. Early (18)F-FDG uptake as a reliable imaging biomarker of T790M-mediated resistance but not MET amplification in non-small cell lung cancer treated with EGFR tyrosine kinase inhibitors. EJNMMI Res 2016;6:74. 10.1186/s13550-016-0229-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caicedo C, Garcia-Velloso MJ, Lozano MD, et al. Role of [(1)(8)F]FDG PET in prediction of KRAS and EGFR mutation status in patients with advanced non-small-cell lung cancer. Eur J Nucl Med Mol Imaging 2014;41:2058-65. 10.1007/s00259-014-2833-4 [DOI] [PubMed] [Google Scholar]
- 26.Yip SSF, Parmar C, Kim J, et al. Impact of experimental design on PET radiomics in predicting somatic mutation status. Eur J Radiol 2017;97:8-15. 10.1016/j.ejrad.2017.10.009 [DOI] [PubMed] [Google Scholar]
- 27.Yip SS, Kim J, Coroller TP, et al. Associations Between Somatic Mutations and Metabolic Imaging Phenotypes in Non-Small Cell Lung Cancer. J Nucl Med 2017;58:569-76. 10.2967/jnumed.116.181826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koyasu S, Nishio M, Isoda H, et al. Usefulness of gradient tree boosting for predicting histological subtype and EGFR mutation status of non-small cell lung cancer on (18)F FDG-PET/CT. Ann Nucl Med 2020;34:49-57. 10.1007/s12149-019-01414-0 [DOI] [PubMed] [Google Scholar]
- 29.van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017;77:e104-7. 10.1158/0008-5472.CAN-17-0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zwanenburg A, Leger S, Vallières M, et al. Image biomarker standardisation initiative. arXiv preprint arXiv:1612.07003.
- 31.O'brien RM. A Caution Regarding Rules of Thumb for Variance Inflation Factors. Qual Quant 2007;41:673-90. 10.1007/s11135-006-9018-6 [DOI] [Google Scholar]
- 32.Breiman L. Random forests. Mach Learn 2001;45:5-32. 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 33.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res 2011;12:2825-30. [Google Scholar]
- 34.Seabold S, Perktold J. “Statsmodels: Econometric and statistical modeling with python.” Proceedings of the 9th Python in Science Conference. 2010. [Google Scholar]
- 35.Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System. In: 22nd SIGKDD Conference on Knowledge Discovery and Data Mining 2016. Available online: https://arxiv.org/abs/1603.02754
- 36.Goldstraw P, Chansky K, Crowley J, et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:39-51. 10.1016/j.jtho.2015.09.009 [DOI] [PubMed] [Google Scholar]
- 37.Lee CK, Davies L, Wu YL, et al. Gefitinib or Erlotinib vs Chemotherapy for EGFR Mutation-Positive Lung Cancer: Individual Patient Data Meta-Analysis of Overall Survival. J Natl Cancer Inst 2017. doi: . 10.1093/jnci/djw279 [DOI] [PubMed] [Google Scholar]
- 38.Okuda Y, Sato K, Sudo K, et al. Low plasma concentration of gefitinib in patients with EGFR exon 21 L858R point mutations shortens progression-free survival. Cancer Chemother Pharmacol 2017;79:1013-20. 10.1007/s00280-017-3285-x [DOI] [PubMed] [Google Scholar]
- 39.Lee SM, Bae SK, Jung SJ, et al. FDG uptake in non-small cell lung cancer is not an independent predictor of EGFR or KRAS mutation status: a retrospective analysis of 206 patients. Clin Nucl Med 2015;40:950-8. 10.1097/RLU.0000000000000975 [DOI] [PubMed] [Google Scholar]
- 40.Cho A, Hur J, Moon YW, et al. Correlation between EGFR gene mutation, cytologic tumor markers, 18F-FDG uptake in non-small cell lung cancer. BMC Cancer 2016;16:224. 10.1186/s12885-016-2251-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lv Z, Fan J, Xu J, et al. Value of (18)F-FDG PET/CT for predicting EGFR mutations and positive ALK expression in patients with non-small cell lung cancer: a retrospective analysis of 849 Chinese patients. Eur J Nucl Med Mol Imaging 2018;45:735-50. 10.1007/s00259-017-3885-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi YJ, Cho BC, Jeong YH, et al. Correlation between (18)f-fluorodeoxyglucose uptake and epidermal growth factor receptor mutations in advanced lung cancer. Nucl Med Mol Imaging 2012;46:169-75. 10.1007/s13139-012-0142-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li M, Zhang L, Tang W, et al. Identification of epidermal growth factor receptor mutations in pulmonary adenocarcinoma using dual-energy spectral computed tomography. Eur Radiol 2019;29:2989-97. 10.1007/s00330-018-5756-9 [DOI] [PubMed] [Google Scholar]
- 44.Ozkan E, West A, Dedelow JA, et al. CT Gray-Level Texture Analysis as a Quantitative Imaging Biomarker of Epidermal Growth Factor Receptor Mutation Status in Adenocarcinoma of the Lung. AJR Am J Roentgenol 2015;205:1016-25. 10.2214/AJR.14.14147 [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, Kim J, Balagurunathan Y, et al. Radiomic Features Are Associated With EGFR Mutation Status in Lung Adenocarcinomas. Clin Lung Cancer 2016;17:441-8.e6. 10.1016/j.cllc.2016.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Zhao X, Zhao Y, et al. Value of pre-therapy (18)F-FDG PET/CT radiomics in predicting EGFR mutation status in patients with non-small cell lung cancer. Eur J Nucl Med Mol Imaging 2020;47:1137-46. 10.1007/s00259-019-04592-1 [DOI] [PubMed] [Google Scholar]
- 47.Li X, Yin G, Zhang Y, et al. Predictive Power of a Radiomic Signature Based on (18)F-FDG PET/CT Images for EGFR Mutational Status in NSCLC. Front Oncol 2019;9:1062. 10.3389/fonc.2019.01062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu L, Yin G, Chen W, et al. Correlation between EGFR mutation status and F(18) -fluorodeoxyglucose positron emission tomography-computed tomography image features in lung adenocarcinoma. Thorac Cancer 2019;10:659-64. 10.1111/1759-7714.12981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ketabi A, Ghafarian P, Mosleh-Shirazi MA, et al. Impact of image reconstruction methods on quantitative accuracy and variability of FDG-PET volumetric and textural measures in solid tumors. Eur Radiol 2019;29:2146-56. 10.1007/s00330-018-5754-y [DOI] [PubMed] [Google Scholar]
- 50.Yan J, Chu-Shern JL, Loi HY, et al. Impact of Image Reconstruction Settings on Texture Features in 18F-FDG PET. J Nucl Med 2015;56:1667-73. 10.2967/jnumed.115.156927 [DOI] [PubMed] [Google Scholar]
- 51.Li Y, Lu L, Xiao M, et al. CT Slice Thickness and Convolution Kernel Affect Performance of a Radiomic Model for Predicting EGFR Status in Non-Small Cell Lung Cancer: A Preliminary Study. Sci Rep 2018;8:17913. 10.1038/s41598-018-36421-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He L, Huang Y, Ma Z, et al. Effects of contrast-enhancement, reconstruction slice thickness and convolution kernel on the diagnostic performance of radiomics signature in solitary pulmonary nodule. Sci Rep 2016;6:34921. 10.1038/srep34921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao B, Tan Y, Tsai WY, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep 2016;6:23428. 10.1038/srep23428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Traverso A, Wee L, Dekker A, et al. Repeatability and Reproducibility of Radiomic Features: A Systematic Review. Int J Radiat Oncol Biol Phys 2018;102:1143-58. 10.1016/j.ijrobp.2018.05.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lasnon C, Majdoub M, Lavigne B, et al. (18)F-FDG PET/CT heterogeneity quantification through textural features in the era of harmonisation programs: a focus on lung cancer. Eur J Nucl Med Mol Imaging 2016;43:2324-35. 10.1007/s00259-016-3441-2 [DOI] [PubMed] [Google Scholar]
- 56.Kirienko M, Cozzi L, Antunovic L, et al. Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. Eur J Nucl Med Mol Imaging 2018;45:207-17. 10.1007/s00259-017-3837-7 [DOI] [PubMed] [Google Scholar]
- 57.Sollini M, Cozzi L, Antunovic L, et al. PET Radiomics in NSCLC: state of the art and a proposal for harmonization of methodology. Sci Rep 2017;7:358. 10.1038/s41598-017-00426-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as