Abstract
Group A Streptococcus (GAS) displays cell-surface proteins that resemble human collagen. We find that a fluorophore-labeled collagen mimetic peptide (CMP) labels GAS cells but not Escherichia coli or Bacillus subtilis cells, which lack such proteins. The CMP likely engages in a heterotrimeric helix with endogenous collagen, as the nonnatural d enantiomer of the CMP does not label GAS cells. To identify a molecular target, we used reverse genetics to “knock-in” the GAS genes that encode two proteins with collagen-like domains, Scl1 and Scl2, into B. subtilis. The fluorescent CMP labels the cells of these B. subtilis strains. Moreover, these strains bind tightly to a surface of mammalian collagen. These data are consistent with streptococcal collagen forming triple helices with damaged collagen in a wound bed, and thus have implications for microbial virulence.
Graphical abstract

For decades, collagen was thought to be a protein found only in animals. Recently, however, collagen has been discovered on the surface of several microbes, including Streptococcus spp., Bacillus anthracis, Clostridium difficile, Legionella pneumococci, and Burkholderia spp.1–3 The most thoroughly characterized of these microbial collagens are those from group A Streptococcus pyogenes (GAS).
GAS is a Gram-positive bacterium that is responsible for a wide range of ailments, ranging from mild infections such as pharyngitis (i.e., “strep throat”) and impetigo (which is a localized skin infection) to serious infections such as rheumatic fever, scarlet fever, and necrotizing fasciitis (“flesh-eating disease”).4,5 GAS infections can manifest in a variety of host environments. Accordingly, much research has been done to reveal molecular interactions between bacterium and host.6–9 Of particular interest in this context are bacterial cell-surface proteins known as adhesins, which are involved in adherence and colonization during infection.9
Pathogenic bacteria like GAS often display variety and redundancy in their adhesins.5,7,10 The “streptococcal collagen-like” adhesins Scl1 and Scl2 of GAS, which were discovered by Björck,11 Musser,12,13 and their coworkers, are no exception. The Scl1 and Scl2 proteins are anchored on the bacterial surface via a membrane-binding domain. Immediately adjacent to that domain is a collagen-like sequence that contains canonical Xaa-Yaa-Gly repeats. Finally, on the N terminus is a variable domain. The collagen-like sequence forms a triple helix that holds the variable domain in a trimeric complex, leading the proteins to adopt a lollypop-like shape (Figure 1).14–16 These Scl proteins have been implicated in binding to a wide variety of factors on the surface of host cells, including fibronectin, laminin, integrins α2β1 and α11β1, thrombin-activatable fibrinolysis inhibitor, LDL, ApoB, and Factor H.3,17–25 In addition to binding to host factors, Scl proteins can play a role in biofilm formation, neutrophil evasion, and protection against host defense peptides.3,26,27 Notably, GAS in which Scl1 is knocked out shows diminished virulence in a mouse infection model.12
Figure 1.
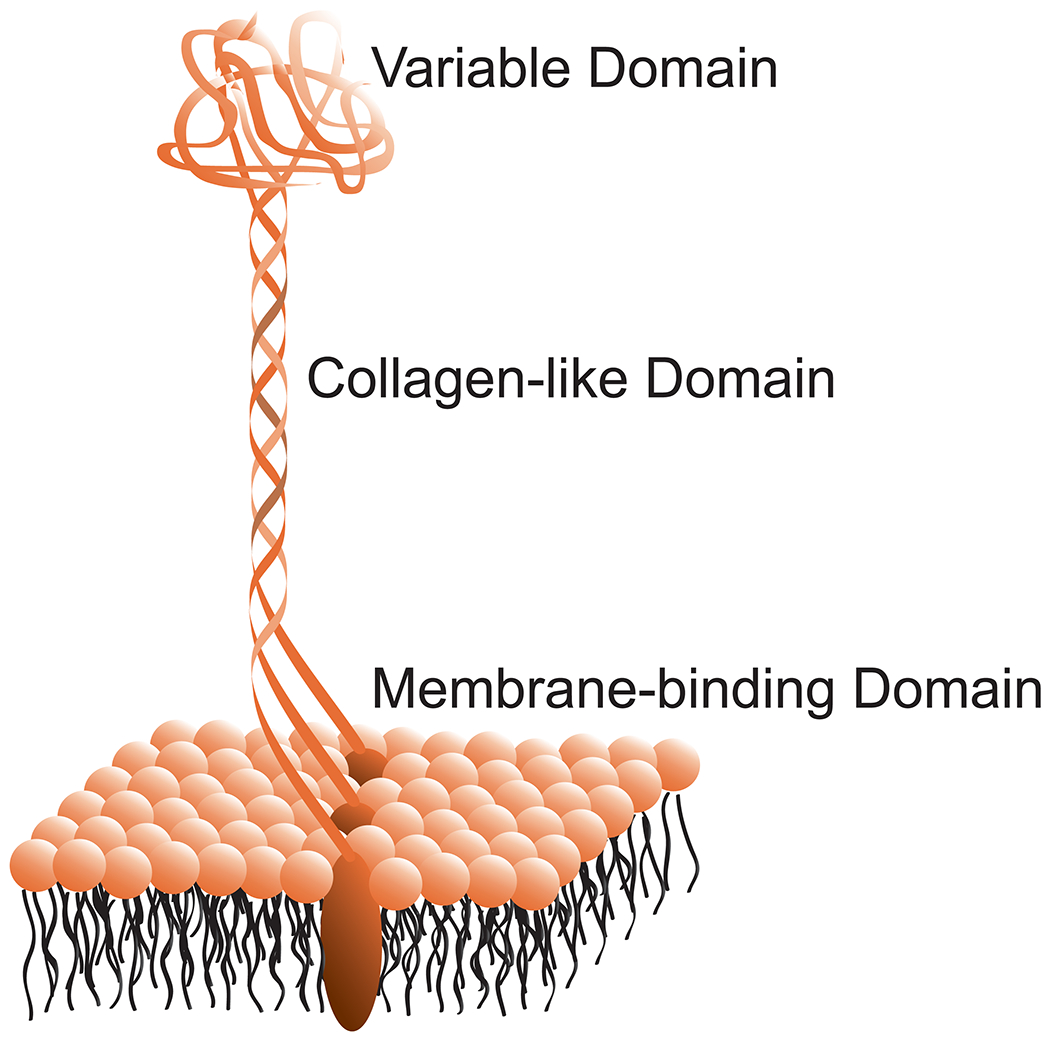
Anatomy of Scl proteins on the GAS cell surface.
Known interactions between Scl proteins and host cells occur through the variable domain.3 The collagen-like domain of Scl proteins has been thought to act merely as a strut that orients the variable domain outward from the cell surface.3,14,18,28 We reasoned that the collagen-like sequences of Scl1 and Scl2 could also mediate interactions with the extracellular matrix (ECM) of a host.
Collagen is the major component of the ECM and the most abundant protein in the human body, comprising a third of all protein by dry weight and ¾ of the protein in skin.29,30 Skin is the first barrier of protection against bacteria. When skin becomes damaged, a human is more vulnerable to infection. In wounds, collagen loses its triple-helical structure, and collagen mimetic peptides (CMPs) can adhere to the damaged collagen.31–40 Hence, we surmised that GAS could likewise use its endogenous collagen-like domains to bind to damaged collagen.
The thermostability of mammalian collagen is enhanced greatly by preorganization endowed by its prevalent proline and 4-hydroxyproline residues.29,30 In contrast, the collagen domains of Scl1 and Scl2 contain few proline residues and no hydroxylated proline. Their thermostability derives instead from relatively weak noncovalent interactions.41,42 Strikingly, the collagen domains of the Scl proteins have a Tm value near 37 °C (where Tm refers to the temperature at the midpoint of the thermal transition between native and denatured states).14,42 Thus, at human body temperature, half of these domains might be single strands that could adhere to damaged collagen.
To test our hypothesis, we used a fluorophore-labeled CMP as a probe for collagen strands on the surface of live GAS cells. To identify the target for cell-surface binding, we expressed the genes that encode S. pyogenes Scl1 and Scl2 in Bacillus subtilis, which is also a Gram-positive bacterium but lacks a collagen-like protein and has numerous divergencies in its cell envelope.43,44 We then probed the surface of these transgenic microbes, as well as their ability to adhere to mammalian collagen. Our findings suggest new strategies for detecting and antagonizing GAS infections.
RESULTS AND DISCUSSION
Scl1 and Scl2 Bind to Collagen Mimetic Peptides.
We synthesized a CMP conjugated to a rhodamine fluorophore. This conjugate, Red-lCMP, had the sequence Ac-Lys(Rhodamine Red™)-(Ser-Gly)3-(l-Pro-l-Pro-Gly)7. We chose this sequence because a (l-Pro-l-Pro-Gly)7 peptide does not form a stable homotrimeric helix (Tm = 6–7 °C),45 but does anneal to damaged collagen.31,33,35 We used this conjugate to probe the surface of GAS cells. Specifically, we grew bacterial cells for 24 h and incubated them with Red-lCMP for 1 h at 37 °C. Then, we washed the cells and visualized using confocal microscopy. Red fluorescence would be indicative of a binding interaction between CMP and the cell surface. To show that this interaction is unique to GAS, we also evaluated binding to B. subtilis and Escherichia coli, which is a Gram-negative bacterium that does not produce collagen-like proteins. We found that GAS was the only bacterium that exhibited red fluorescence, indicative of an interaction with Red-lCMP (Figure 2).
Figure 2.

Confocal microscopy images of live GAS, E. coli, and B. subtilis cells incubated for 1 h at 37 °C with Red-lCMP (3 μM), Red-dCMP (3 μM), or left untreated, and washed (3×). Scale bar: 10 μm.
In addition to the bacterial controls, we also employed a CMP control. If adherence to the cell surface is due to triple-helix formation between Red-lCMP and a bacterial protein, then the enantiomeric dCMP should not label the cell. If, however, the labeling is due to electrostatic or hydrophobic interactions, then dCMP should behave the same as lCMP. Accordingly, we synthesized Red-dCMP, which is Ac-Lys(Rhodamine Red™)-(Ser-Gly)3-(d-Pro-d-Pro-Gly)7. We found that GAS cells were not labeled with Red-dCMP (Figure 2). This selectivity for lCMP over dCMP is consistent with an interaction that requires collagen triple-helix formation.
Effect of Temperature.
Our initial conception of the adherence by a GAS cell to damaged collagen took into consideration the Tm value of the collagen-like triple helix of the Scl proteins.14,42 We reasoned that at low temperatures, the Scl proteins would be engaged in a triple helix (Figure 1) and therefore not able to bind to Red-lCMP. Nonetheless, the collagen-like domains of Scl1 (50 Xaa-Yaa-Gly repeats) and Scl2 (38 repeats) are much longer than the CMP (7 repeats), and the integrity of their triple helices might not be complete throughout. To assess that integrity on the surface of live cells, we repeated the Red-lCMP labeling experiment on cells incubated at different temperatures. We observed significant labeling at all temperatures with microscopy (Figure 3). We quantified the binding with cytometry. Again, we found that GAS cells were labeled by Red-lCMP at all temperatures (Figure 4). These data suggest that segments of the collagen-like domains of the Scl adhesins are not triple-helical on live cells.
Figure 3.

Confocal microscopy images of live GAS cells incubated with Red-lCMP (3 μM), incubated for 1 h at 5, 25, 37, or 45 °C, and washed (3×). Scale bar: 10 μm.
Figure 4.

Flow cytometry of live GAS and B. subtilis cells incubated with Red-lCMP or Red-dCMP, or left untreated. Conditions are as described in Figures 2 and 3. Labeling is represented as the percentage of cell population within the gates for Rhodamine Red™ and SYTO 9 fluorescence.
Scl1 and Scl2 B. subtilis Knock-ins.
GAS has variety of adhesins that enable binding to a host.17–25 Some of these adhesins can bind to collagen.46–50 Knowing that Red-lCMP binds to GAS cells (Figures 2–4), we next sought to demonstrate that the binding is due to collagen-like proteins Scl1 and Scl2 rather than to another factor. Our strategy was to isolate these two adhesins and see if the CMP-labeling of GAS cells was recapitulated inAnother background. Having tested B. subtilis and seen no labeling (Figures 2 and 4), we created inducible B. subtilis knock-ins of Scl1 or Scl2.51 We evaluated the production of Scl1 and Scl2 in B. subtilis by assessing mRNA production with qPCR. Upon induction, we observed a 3.7-fold increase in the expression of Scl1 and a 4.0-fold increase in the expression of Scl2 (Figure 5).
Figure 5.

Change in expression of Scl1 and Scl2 upon induction of their encoded genes in B. subtilis as evaluated by qPCR. Values are relative to the uninduced strain.
Upon repeating the CMP-labeling experiment with the knock-in B. subtilis cells, we expected to see cells fluoresce red if the interaction is indeed due to the heterologous collagen-like proteins. By detecting labeling with confocal microscopy (Figure 6) and flow cytometry (Figure 7), we did indeed observe labeling by Red-lCMP, and more so upon induction. Thus, Scl1 and Scl2 confer the phenotype in B. subtilis cells that was observed in GAS cells. This concurrence is strong evidence that these collagen-like proteins are responsible for the interaction between GAS and a CMP. Again, the near-absence of binding by the enantiomeric Red-dCMP is indicative of binding by Red-lCMP being due to triple-helix formation.
Figure 6.

Confocal microscopy images of live B. subtilis Scl1 and Scl2 knock-in cells exposed to Red-lCMP or Red-dCMP. Cells were induced (+) or not induced (−) to express a gene that encodes an Scl protein. Scale bar: 10 μm.
Figure 7.

Flow cytometry of live B. subtilis Scl1 and Sc12 knock-in cells exposed to Red-lCMP or Red-dCMP, or left untreated. Cells were induced (+) or not induced (−) to express a gene that encodes an Scl protein. Labeling is represented as the percentage of cells within the gate for both Rhodamine Red™ and SYTO 9 fluorescence.
Scl1 and Scl2 Bind to a Collagen Surface.
Although we have shown that the collagen-like proteins on the surface of GAS can interact with a single-stranded CMP, our data do not report on how these proteins interact with a collagen surface, like that of human skin. To evaluate adhesion to a collagen surface we employed an assay using a crystal violet stain.52,53 This assay was performed by growing cells on a surface of mammalian collagen. The surface was then washed, leaving only adhered bacteria. Then, the surface was submerged in an aqueous solution of crystal violet (0.1% w/v). Excess crystal violet was washed away, leaving behind crystal violet that had been internalized by bacterial cells. The dye was extracted from the cells with ethanol, allowing for a colorimetric readout of the number of cells that were retained on the surface.
We found that the Scl1 and Scl2 knock-ins showed significantly greater adherence to the collagen-coated surface than did wild-type B. subtilis (Figure 8). In addition, significant binding occurred only in wells coated with collagen. Apparently, the enhanced bacterial adherence is due to an interaction between the displayed collagen-like proteins and the collagen surface. As seen with CMP-labeling (Figures 6 and 7), there is an increase in bacterial adherence to the collagen surface upon induction. These data support Scl proteins as being responsible for the binding of bacterial cells to a collagen surface.
Figure 8.

Adherence of wild-type B. subtilis and B. subtilis Scl1 and Scl2 knock-in cells to uncoated and collagen-coated wells, as determined by the absorbance of extracted crystal violet at 620 nm. Knock-in cells were induced (+; diamonds) or not induced (−; circles) to express a gene that encodes an Scl protein. Data are the mean ± SD; *, P < 0.05.
CONCLUSION
The collagen-like domains of Scl1 and Scl2 on GAS cells can both bind to synthetic collagen-mimetic peptides and adhere to mammalian collagen. These attributes could enhance microbial virulence as well as serve as the basis of new means to detect streptococcal cells and antagonize their interaction with host organisms. This deployment of collagen-mimetic peptides could also be efficacious with the many other microbes1–3 that display collagen on their cell surface.
MATERIALS AND METHODS
General.
Amino acid derivatives and HOBt were from Chem-Impex International (Wood Dale, IL). Fmoc-Gly-2-chlorotrityl resin was from EMD Millipore (La Jolla, CA). Rhodamine Red™-X, succinimidyl ester, 5-isomer was from Thermo Fischer Scientific (Waltham, MA). Nuclei lysis solution and protein precipitation solution were from Promega (Fitchburg, WI). miRCURY RNA isolation kit was from Qiagen (Hilden, Germany). All other reagents were from Sigma–Aldrich (St. Louis, MO) and were used without further purification.
DMF was dried with a Glass Contour system from Pure Process Technology (Nashua, NH) and passed through an associated isocyanate “scrubbing” column to remove any amines. Water was purified with an Arium Pro system from Sartorius (Göttingen, Germany).
PCR was performed with a 1:30 min extension time (annealing temperature 58 °C, 30 cycles).
All procedures were performed in air at ambient temperature (~22 °C) and pressure (1.0 atm) unless indicated otherwise.
Instrumentation.
Solid-phase peptide synthesis was performed at the University of Wisconsin–Madison Biotechnology Center with a Prelude peptide synthesizer from Protein Technologies (Tucson, AZ). Synthetic peptides were purified by HPLC with a Prominence instrument from Shimadzu (Kyoto, Japan) equipped with a VarioPrep 250/21 C18 column from Macherey–Nagel (Düren, Germany). Molecular mass was determined by matrix-assisted laser desorption/ionization–time-of-flight (MALDI–TOF) mass spectrometry on an α-cyano-4-hydroxycinnamic acid matrix with a Voyager DE-Pro instrument from Thermo Fischer Scientific at the Biophysics Instrumentation Facility at the University of Wisconsin–Madison. Purity analyses were performed with an Acquity UPLC® H-Class system from Waters (Waltham, MA) that was equipped with an Acquity photodiode array detector, Acquity quaternary solvent manager, Acquity sample manager with a flow-through needle, Acquity UPLC® BEH C18 column (2.1 × 50 mm, 1.7-μm particle size), and Empower 3 software. Cells were imaged with a Eclipse Ti inverted confocal microscope from Nikon (Melville, NY) at the Biochemistry Optical Core of the University of Wisconsin−Madison. Flow cytometry was performed with an LSR Fortessa flow cytometer from BD Biosciences (San Jose, CA) at the Carbone Cancer Center at the University Wisconsin–Madison. qPCR was performed with a QuantStudio 7 instrument from Thermo Fischer Scientific. Absorbance was measured with an Infinite M1000 plate reader from Tecan (Männedorf, Switzerland).
Peptide Synthesis.
Red-lCMP, which is Ac-Lys(Rhodamine Red™)-(Ser-Gly)3-(l-Pro-l-Pro-Gly)7, was synthesized as described previously.35 Red-dCMP, which is Ac-Lys(Rhodamine Red™)-(Ser-Gly)3-(d-Pro-d-Pro-Gly)7, was synthesized as follows.
Ac-Lys-(Ser-Gly)3-(d-Pro-d-Pro-Gly)7.
Using the Fmoc-d-Pro-d-Pro-Gly-OH tripeptide synthesized in solution without chromatography54 and an Fmoc-d-Pro-OH monomer, Ac-Lys-(Ser-Gly)3-(d-Pro-d-Pro-Gly)7 was synthesized by two additions of monomer followed by six segment-condensations of tripeptide on preloaded Fmoc-Gly-2-chlorotrityl resin (0.19 mmol/g). Fmoc-deprotection was achieved by treatment with piperidine (20% v/v) in DMF. The tripeptide or amino acid monomer (4 equiv) was converted to an active ester by using HATU and NMM. Each residue was double-coupled between Fmoc-deprotections. The peptide was cleaved from the resin with 96.5:2.5:1.0 TFA/H2O/TIPSH (5 mL), precipitated from diethyl ether at −80 °C, and isolated by centrifugation. The peptide was purified by preparative HPLC using a gradient of 10–50% v/v B over 50 min (A: H2O containing 0.1% v/v TFA; B: acetonitrile containing 0.1% v/v TFA). MALDI (m/z): [M + H]+ calcd, 2380.6; found, 2380.0. A 0.05-mmol scale synthesis afforded 18.2 mg (15%) of Ac-Lys-(Ser-Gly)3-(d-Pro-d-Pro-Gly)7 after purification.
Ac-Lys(Rhodamine Red™)-(Ser-Gly)3-(d-Pro-d-Pro-Gly)7.
Ac-Lys-(Ser-Gly)3-(d-Pro-d-Pro-Gly)7 (3.4 mg, 1.43 μmol) was dissolved in 2.0 mL of DMSO. Rhodamine Red™-X, succinimidyl ester, 5-isomer (200 μL, 1.30 μmol) was added as a 5.0 mg/mL solution in DMSO. DIEA (200 μL, 1.14 mmol) was added dropwise. The reaction mixture was allowed to stir for 8 h. The solution was then diluted with 7 mL of H2O, frozen, and lyophilized. Red-dCMP was purified by preparative HPLC using a gradient of 65–95% v/v B over 55 min (A: H2O containing 0.1% v/v TFA; B: methanol containing 0.1% v/v TFA) to yield 0.5 mg (11%) of Red-dCMP after purification. MALDI (m/z): [M + Na]+ calcd, 3056.42; found, 3056.36.
The purity of Red-lCMP and Red-dCMP was assessed with UPLC as being >95% (Figure S1).
Cell Growth.
Cultures of group A S. pyogenes (SF 370) were grown for 24 h in Brain Heart Infusion broth at 37 °C. Cultures of E. coli (RP437) were grown for 16 h in Luria–Bertani (LB) medium at 37 °C on a platform shaker. Cultures of B. subtilis (Ol 1085) were grown overnight in LB at 37 °C on a platform shaker. B. subtilis knock-in cultures were grown in LB medium. Knock-in cultures were induced after 4 h by addition ITPG to a final concentration of 1.0 mM. All experiments with knock-in cultures were done after 8 h of growth.
Cell Labeling for Microscopy and Cytometry.
Aliquots (1.0 mL) of bacterial culture (vide supra) were treated with either Red-lCMP or Red-dCMP to a final concentration of 3.25 μM. Mixtures were allowed to incubate at 37 °C for 1 h. Cultures were pelleted, and the supernatant was removed and resuspended in water. This procedure was repeated three times to wash the cultures. Finally, cells were resuspended in 1.0 mL of water.
For microscopy, cells were spotted on a thin layer of LB agarose on a microscope slide. Images were merged from brightfield and fluorescence (560 nm laser with a 595/50 filter) microscopy. Three biological replicates of each experiment were performed, and recorded images were representative.
For cytometry, cells were washed, resuspended, and exposed to 1.0 μL of a solution of SYTO 9 (5 mM) in DMSO. Cells (50,000) were counted by using 488 nm and 561 nm lasers with 530/30 nm and 586/15 nm filters with a flow cytometer.
Creation of B. subtilis Knock-ins of Scl1 and Scl2.
To achieve heterologous expression, the coding sequence for Scl1 (SPy1983) and Scl2 (SPy1054) from the M1 serotype of GAS were integrated at the amyE locus in the B. subtilis chromosome (with translation of the mRNA being ensured by the addition of the tufA ribosome-binding site) and controlled expression with the IPTG-inducible Physpank promoter. The presence of the genes was verified by PCR, and chromosomal integration was confirmed by plating on starch agar. Integration of the construct leads to the disruption of the gene coding for amylase and hence to the loss of the ability to degrade starch.
To isolate genomic DNA, cells from 1.0 mL of a turbid culture were harvested by centrifugation (2 min at maximum rpm). Cells were suspended in 480 μL of 50 mM EDTA (pH 8.0). Lysozyme (60 μL of a 10 mg/mL solution) was added, and the resulting solution was mixed gently. The solution was then incubated for 30 min at 37 °C, and 600 μL of nuclei lysis solution (Promega A7943) was added. The resulting mixture was incubated for 2–3 min at room temperature, 200 μL of protein precipitation solution (Promega A7951) was added, and the mixture was vortexed for 20 s. The mixture was incubated on ice for 5 min and then clarified by centrifugation for 3 min at maximum rpm. A 900-μL aliquot of the supernatant was transferred to a new tube containing 600 μL of isopropyl alcohol, and the tube was inverted gently. The resulting precipitate was collected by centrifugation for 2 min at maximum rpm. The pellet was washed with 500 μL of aqueous EtOH (70% v/v) and collected by centrifugation for 1 min at maximum rpm. The dry pellet was suspended in 100 μL of H2O, and the resulting solution was incubated for 30 min at 37 °C. The isolated genomic DNA was stored at −20 °C.
To generate an IPTG-inducible construct of Scl1 (i.e., plasmid pFK1), the spy1983 gene was PCR-amplified from genomic DNA by using primers 5′-TCAGCATGCTTAGTTGTTTTCTTGACGTTTTGC-3′ and 5′-TCAAAGCTTTAAGGAGGATTTTAGAATGTTGAC ATCAAAGCACCATAATC-3′, and digested with HindIII and SphI. The ensuing fragment was ligated into the HindIII/SphI sites of plasmid pDP111, which contains the Physpank promoter, the lacI gene encoding the LacI repressor, and a kanamycin-resistance cassette between the arms of the amyE gene. The tufA ribosome-binding site was added to the spy1983 gene within the forward primer to increase its expression, and the resulting construct was transformed into DK1042 B. subtilis cells55 to generate strain DK4458 (amyE::Physpank-RBSimpscl1 kan).
To generate an IPTG-inducible construct of Scl2 (i.e., plasmid pFK3), the spy1054 gene was PCR-amplified from genomic DNA by using primers 5′-TCAGCATGCTTAGTTGTTTTCTTGACGTTTTGC-3′ and 5′-TCAAAGCTTTAAGGAGGATTTTAGATTGCTGAC CTTTGGAGGTGC-3′, and digested with HindIII and SphI. The ensuing fragment was ligated into the HindIII/SphI sites of pDP111, which contains the Physpank promoter, the lacI gene encoding the LacI repressor, and a kanamycin-resistance cassette between the arms of the amyE gene. The tufA ribosome-binding site was added to the spy1054 gene within the forward primer to increase its expression, and the construct was transformed in to DK1042 B. subtilis cells55 to generate strain DK4460 (amyE::Physpank-RBSimpscl2 kan).
qPCR Analysis of B. subtilis Knock-ins.
Prior to qPCR, RNA was isolated from cultures using miRCURY RNA isolation kit and complementary DNA was prepared from the RNA by reverse transcription using an Applied Biosystems High-Capacity cDNA Reverse Transcription kit. Samples were prepared in duplicate and added to Quanta PerfeCTa SYBR Green Fastmix Low Rox master mix. These samples were amplified by using an Applied Biosystems QuantStudio 7 kit. Relative gene expression was calculated with the 2−ΔΔCt method,56 normalized against RspJ, RspE, and yoxA.
qPCR primers were designed by using the program Primer357–59 and are listed in Table S1. cDNA and primers were tested against each other and analyzed by gel electrophoresis on a gel to ensure purity and integrity.
Bacterial Adherence Assays.
Cultures (2.0 mL) of the B. subtilis knock-ins and wild-type Ol 1085 cells were grown in the wells of a 24-well microplate, which were coated with rat-tail collagen (product A1142802 from Thermo Fisher Scientific). Gene expression was induced after 4 h of growth. All cultures were removed from the wells, which were washed three times with water. An aqueous solution (1.0 mL) of crystal violet (0.1% w/v) was added to the wells. After 3 min, the crystal violet solution was removed, and the wells washed three times with water. Ethanol (500 μL) was added to the wells, and an aliquot (200 μL) from each well was transferred to a 96-well plate and the absorbance at 620 nm was recorded with a plate reader. Data were analyzed with Prism software from GraphPad (San Diego, CA). Significance was assessed by applying an unpaired Student’s t test to paired comparators.
Supplementary Material
ACKNOWLEDGMENT
We are grateful to Professor Douglas B. Weibel for advice and contributive discussions. We thank Emily R. Garnett for help with qPCR analysis and Drs. Rachael T. Sheridan, Elle K. Grevstad, and Darrell R. McCaslin for facility support. This work was supported by Grants R56 AR044276 and R35 GM131783 (NIH). AJE was supported by Chemistry–Biology Interface (CBI) Training Grant T32 GM008505 (NIH). The UWCCC Flow Cytometry Laboratory was supported by Grant P30 CA014520 (NIH).
Footnotes
The authors declare no competing financial interests.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.9b00351.
REFERENCES
- (1).Rasmussen M; Jacobsson M; Björck L Genome-based identification and analysis of collagen related structural motifs in bacterial and viral proteins. J. Biol. Chem 2003, 27, 32313–32316. [DOI] [PubMed] [Google Scholar]
- (2).Shubin N Your Inner Fish: A Journey into the 3.5-Billion-Year History of the Human Body. Random House: New York, NY, 2008; pp. 116–138. [Google Scholar]
- (3).Lukomski S; Bachert BA; Squeglia F; Berisio R Collagen-like proteins of pathogenic streptococci. Mol. Microbiol 2017, 103, 919–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Cunningham MW Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev 2000, 13, 470–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Walker MJ; Barnett TC; McArthur JD; Cole JN; Gillen CM; Henningham A; Sriprakash KS; Sanderson-Smith ML; Nizet V Disease manifestations and pathogenic mechanisms of group A Streptococcus. Clin. Microbiol. Rev 2014, 27, 264–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Finlay BB; Cossart P Exploitation of mammalian host cell functions by bacterial pathogens. Science 1997, 276, 718–725. [DOI] [PubMed] [Google Scholar]
- (7).Pizarro-Cerda J; Cossart P Bacterial adhesion and entry into host cells. Cell 2006, 124, 715–727. [DOI] [PubMed] [Google Scholar]
- (8).Ribet D; Cossart P How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect. 2015, 17, 173–183. [DOI] [PubMed] [Google Scholar]
- (9).Dickey SW; Cheung GYC; Otto M Different drugs for bad bugs: Antivirulence strategies in the age of antibiotic resistance. Nat. Rev. Drug Discov 2017, 16, 457–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kline KA; Falker S; Dahlberg S; Normark S; Henriques-Normark B Bacterial adhesins in host–microbe interactions. Cell Host Microbe 2009, 5, 580–592. [DOI] [PubMed] [Google Scholar]
- (11).Rasmussen M; Edén A; Björck L SclA, a novel collagen-like surface protein of Streptococcus pyogenes. Infect. Immun 2000, 68, 6370–6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Lukomski S; Nakashima K; Abdi I; Cipriano VJ; Ireland RM; Reid SD; Adams GG; Musser JM Identification and characterization of the Scl gene encoding a gsilroup A Streptococcus extracellular protein virulence factor with similarity to human collagen. Infect. Immun 2000, 68, 6542–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Lukomski S; Nakashima K; Abdi I; Cipriano VJ; Shelvin BJ; Graviss EA; Musser JM Identification and characterization of a second extracellular collagen-like protein made by group A Streptococcus: Control of production at the level of translation. Infect. Immun 2001, 69, 1729–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Xu Y; Keene DR; Bujnicki JM; Hook M; Lukomski S Streptococcal Scl1 and Scl2 proteins form collagen-like triple helices. J. Biol. Chem 2002, 277, 27312–27318. [DOI] [PubMed] [Google Scholar]
- (15).Yu Z; An B; Ramshaw JA; Brodsky B Bacterial collagen-like proteins that form triple-helical structures. J. Struct. Biol 2014, 186, 451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Squeglia F; Bachert B; De Simone A; Lukomski S; Berisio R The crystal structure of the streptococcal collagen-like protein 2 globular domain from invasive M3-type group A Streptococcus shows significant similarity to immunomodulatory HIV protein gp41. J. Biol. Chem 2014, 289, 5122–5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Humtsoe JO; Kim JK; Xu Y; Keene DR; Hook M; Lukomski S; Wary KK A streptococcal collagen-like protein interacts with the α2β1 integrin and induces intracellular signaling. J. Biol. Chem 2005, 280, 13848–13857. [DOI] [PubMed] [Google Scholar]
- (18).Han R; Caswell CC; Lukomska E; Keene DR; Pawlowski M; Bujnicki JM; Kim JK; Lukomski S Binding of the low-density lipoprotein by streptococcal collagen-like protein Scl1 of Streptococcus pyogenes. Mol. Microbiol 2006, 61, 351–367. [DOI] [PubMed] [Google Scholar]
- (19).Caswell CC; Lukomska E; Seo NS; Hook M; Lukomski S Scl1-dependent internalization of group A Streptococcus via direct interactions with the α2β1 integrin enhances pathogen survival and re-emergence. Mol. Microbiol 2007, 64, 1319–1331. [DOI] [PubMed] [Google Scholar]
- (20).Pahlman LI; Marx PF; Morgelin M; Lukomski S; Meijers JC; Herwald H Thrombin-activatable fibrinolysis inhibitor binds to Streptococcus pyogenes by interacting with collagen-like proteins A and B. J. Biol. Chem 2007, 282, 24873–24881. [DOI] [PubMed] [Google Scholar]
- (21).Caswell CC; Han R; Hovis KM; Ciborowski P; Keene DR; Marconi RT; Lukomski S The Scl1 protein of M6-type group A Streptococcus binds the human complement regulatory protein, Factor H, and inhibits the alternative pathway of complement. Mol. Microbiol 2008, 67, 584–596. [DOI] [PubMed] [Google Scholar]
- (22).Caswell CC; Barczyk M; Keene DR; Lukomska E; Gullberg DE; Lukomski S Identification of the first prokaryotic collagen sequence motif that mediates binding to human collagen receptors, integrins α2β1 and α11β1. J. Biol. Chem 2008, 283, 36168–36175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Caswell CC; Oliver-Kozup H; Han R; Lukomska E; Lukomski S Scl1, the multifunctional adhesin of group A Streptococcus, selectively binds cellular fibronectin and laminin, and mediates pathogen internalization by human cells. FEMS Microbiol. Lett 2010, 303, 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Chen S-M; Tsai Y-S; Wu C-M; Liao S-K; Wu L-C; Chang C-S; Liu Y-H; Tsai P-J Streptococcal collagen-like surface protein 1 promotes adhesion to the respiratory epithelial cell. BMC Microbiol. 2010, 10, 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Reuter M; Caswell CC; Lukomski S; Zipfel PF Binding of the human complement regulators CFHR1 and Factor H by streptococcal collagen-like protein 1 (Scl1) via their conserved C termini allows control of the complement cascade at multiple levels. J. Biol. Chem 2010, 285, 38473–38485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Döhrmann S; Anik S; Olson J; Anderson EL; Etesami N; No H; Snipper J; Nizet V; Okumura CYM Role for streptococcal collagen-like protein 1 in M1T1 group A Streptococcus resistance to neutrophil extracellular traps. Infect. Immun 2014, 82, 4011–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).LaRock CN; Nizet V Cationic antimicrobial peptide resistance mechanisms of streptococcal pathogens. Biochim. Biophys. Acta 2015, 1848, 3047–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).In some serotypes, the collagen sequence contains a binding motif for collagen-binding integrins on the host cell surface (see: refs 17, 19, 22, and 24).
- (29).Shoulders MD; Raines RT Collagen structure and stability. Annu. Rev. Biochem 2009, 78, 929–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Bella J Collagen structure: New tricks from a very old dog. Biochem. J 2016, 473, 1001–1025. [DOI] [PubMed] [Google Scholar]
- (31).Chattopadhyay S; Murphy CJ; McAnulty JF; Raines RT Peptides that anneal to natural collagen in vitro and ex vivo. Org. Biomol. Chem 2012, 10, 5892–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Li Y; Yu SM Targeting and mimicking collagens via triple helical peptide assembly. Curr. Opin. Chem. Biol 2013, 17, 968–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Chattopadhyay S; Guthrie KM; Teixeira L; Murphy CJ; Dubielzig RR; McAnulty JF; Raines RT Anchoring a cytoactive factor in a wound bed promotes healing. J. Tissue Eng. Regen. Med 2014, 10, 1012–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Bennink LL; Li Y; Kim B; Shin IJ; San BH; Zangari M; Yoon D; Yu SM Visualizing collagen proteolysis by peptide hybridization: From 3D cell culture to in vivo imaging. Biomaterials 2018, 183, 67–76. [DOI] [PubMed] [Google Scholar]
- (35).Ellison AJ; Raines RT A pendant peptide endows a sunscreen with water-resistance. Org. Biomol. Chem 2018, 16, 7139–7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Li Y; Yu SM In situ detection of cegraded and denatured collagen via triple helical hybridization: New tool in histopathology. Methods Mol. Biol 2019, 1944, 135–144. [DOI] [PubMed] [Google Scholar]
- (37).Dones JM; Tanrikulu IC; Chacko JV; Schroeder AB; Hoang TT; Gibson ALF; Eliceiri KW; Raines RT Optimization of interstrand interactions enables burn detection with a collagen-mimetic peptide. Org. Biomol. Chem 2019, 17, 9906–9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Tanrikulu IC; Westler WM; Ellison AJ; Markley JL; Raines RT Templated collagen “double helices” maintain their structure. J. Am. Chem. Soc 2020, 142, 1137–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Song JY; Pineaultz KM; Dones JM; Raines RT; Wellik DM Hox genes maintain critical roles in the adult skeleton. Proc. Natl. Acad. Sci. USA 2020, 117, 7296–7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Ellison AJ; Tanrikulu IC; Dones JM; Raines RT Cyclic peptide mimetic of damaged collagen. Biomacromolecules 2020, 21, 1539–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Mohs A; Silva T; Yoshida T; Amin R; Lukomski S; Inouye M; Brodsky B Mechanism of stabilization of a bacterial collagen triple helix in the absence of hydroxyproline. J. Biol. Chem 2007, 282, 29757–29765. [DOI] [PubMed] [Google Scholar]
- (42).Xu C; Yu Z; Inouye M; Brodsky B; Mirochnitchenko O Expanding the family of collagen proteins: Recombinant bacterial collagens of varying composition form triple-helices of similar stability. Biomacromolecules 2010, 11, 348–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Silhavy TJ; Kahne D; Walker S The bacterial cell envelope. Cold Spring Harb. Perspect. Biol 2010, 2, a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Dufresne K; Paradis-Bleau C Biology and assembly of the bacterial envelope. Adv. Exp. Med. Biol 2015, 883, 41–76. [DOI] [PubMed] [Google Scholar]
- (45).Shaw BR; Schurr JM The association reaction of collagen model polypeptides (Pro-Pro-Gly)n. Biopolymers 1975, 14, 1951–1985. [Google Scholar]
- (46).Dinkla K; Rohde M; Jansen WTM; Carapetis JR; Chhatwal GS; Talay SR Streptococcus pyogenes recruits collagen via surface-bound fibronectin: A novel colonization and immune evasion mechanism. Mol. Microbiol 2003, 47, 861–869. [DOI] [PubMed] [Google Scholar]
- (47).Dinkla K; Rohde M; Jansen WT; Kaplan EL; Chhatwal GS; Talay SR Rheumatic fever-associated Streptococcus pyogenes isolates aggregate collagen. J. Clin. Invest 2003, 111, 1905–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Kreikemeyer B; Nakata M; Oehmcke S; Gschwendtner C; Normann J; Podbielski A Streptococcus pyogenes collagen type I-binding Cpa surface protein. Expression profile, binding characteristics, biological functions, and potential clinical impact. J. Biol. Chem 2005, 280, 33228–33239. [DOI] [PubMed] [Google Scholar]
- (49).Bober M; Morgelin M; Olin AI; von Pawel-Rammingen U; Collin M The membrane bound LRR lipoprotein Slr, and the cell wall-anchored M1 protein from Streptococcus pyogenes both interact with type I collagen. PLoS One 2011, 6, e20345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Reissmann S; Gillen CM; Fulde M; Bergmann R; Nerlich A; Rajkumari R; Brahmadathan KN; Chhatwal GS; Nitsche-Schmitz DP Region specific and worldwide distribution of collagen-binding M proteins with PARF motifs among human pathogenic streptococcal isolates. PLoS One 2012, 7, e30122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Notably S pyogenes is a BSL-2 organism, whereas B. subtilis is a BSL-1 organism. [Google Scholar]
- (52).Gram HC Über die isolierte Färbung der Schizomyceten in Schnitt- un Trockenpräparaten. Fortschr. Med 1884, 2, 185–189. [Google Scholar]
- (53).Bohinc K; Dražić G; Fink R; Oder M; Jevšnik M; Nipič D; Godič-Torkar K; Raspor P Available surface dictates microbial adhesion capacity. Int. J. Adhes. Adhes 2014, 50, 265–272. [Google Scholar]
- (54).Ellison AJ; VanVeller B; Raines RT Convenient synthesis of collagen-related tripeptides for segment condensation. Peptide Sci. 2015, 104, 674–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Konkol MA; Blair KM; Kearns DB Plasmid-encoded ComI inhibits competence in the ancestral 3610 strain of Bacillus subtilis. J. Bacteriol 2013, 195, 4085–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Livak KJ; Schmittgen TD Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT. Methods 2001, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- (57).Koressaar T; Remm M Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [DOI] [PubMed] [Google Scholar]
- (58).Untergasser A; Cutcutache I; Koressaar T; Ye J; Faircloth BC; Remm M; Rozen SG Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Koressaar T; Lepamets M; Kaplinski L; Raime K; Andreson R; Remm M Primer3_masker: Integrating masking of template sequence with primer design software. Bioinformatics 2018, 34, 1937–1938. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


