Abstract
Background
Urinary bladder cancer (UBC) is one of few cancers with an established gene-environment interaction (GxE) with smoking. However, it is unknown whether the interaction with tobacco use is present non-muscle invasive bladder cancer (NMIBC) and characteristics of prognostic relevance. We aimed to investigate if smoking status and/or smoking intensity interact with the effect of discovered variants on key NMIBC characteristics of tumor grade, stage, size, and patient age within the Bladder Cancer Prognosis Programme (BCPP) cohort.
Methods
Analyzed sample consisted of 546 NMIBC patients with valid smoking data from the BCPP. In a previous genome-wide association study (GWAS), we have identified 61 single nucleotide polymorphisms (SNPs) potentially associated with the NMIBC characteristics of tumor stage, grade, size, and patient age. In the current analysis, we have tested these SNPs for GxE with smoking.
Results
Out of 61 SNPs, 10 have showed suggestion (statistical significance level of P<0.05) for GxE with NMIBC tumor size rs35225990, rs188958632, rs180910528, rs74603364, rs187040828, rs144383242, rs117587674, rs113705641, rs2937268, and chromosome 14:38247577. All SNPs were located across loci of 1p31.3, 3p26.1, 6q14.1, 14q21.1, and 13q14.13. In addition, two of the tested polymorphisms were suggestive for interaction with smoking intensity (chromosome 14:38247577 and rs2937268).
Conclusions
Our study suggests interaction between genetic variance and smoking behavior for increased NMIBC tumor size at the time of diagnosis. Further replication is required to validate these findings.
Keywords: Bladder cancer, gene-environment interaction (GxE), smoking, SNP, tumor size
Introduction
Current evidence suggests that the effect of urinary bladder cancer (UBC) related single nucleotide polymorphisms (SNPs) may depend on smoking exposure (1-4). However, it remains unknown if gene-environment interactions (GxE) can be differential across UBC subtypes and characteristics.
To date, studies on gene-tobacco interactions (GxT) for UBC have only considered the overall risk of UBC as an outcome. These investigations have resulted in overlapping findings, collectively reporting multiple significant loci (2-4). Some of the most commonly reported genes (NAT2, UGT1A6) are regulatory of phase II detoxification, a pathway for metabolizing tobacco-related carcinogens (i.e., aromatic amines) (2,3,5). As polymorphisms in NAT2 and UGT1A6 alter smoking-related carcinogen clearance, their effect naturally becomes limited to smokers. Concurrently, the effect is reduced (2) or almost absent for never smokers (5). Importantly, the level of smoking exposure has shown to be an important aspect in GxT, revealing an important pattern of certain genotypes interacting with smoking in a dose-dependent manner (2,4-7). Some genotypes, namely GSTM1-null status (3), appear more important for non-smokers, although significant effects are present regardless of tobacco use. Expectedly, the effect of these genotypes on UBC risk does not vary within smoking intensity categories (6).
UBC is one of few cancers with an established GxE (4), yet previous research has not distinguished between muscle invasive bladder cancer (MIBC) and non-muscle invasive bladder cancer (NMIBC) groupings. Given our continually-evolving knowledge of the pathogenesis of MIBC and NMIBC (8), not stratifying for these categories in genetic analyses could result in overlooking important biological mechanisms. Furthermore, it is now known that genetic loci for UBC risk are not relevant for UBC prognosis (9), which continues to highlight the complexity of genetic associations. Finally, little evidence is present for genetic associations with specific characteristics such as tumor size, grade, stage or patient age (2,5). However, these characteristics are especially influential for NMIBC outcomes (10), and are more specific entities than the more broadly-defined prognostic outcomes such as recurrence or progression.
We have previously carried out a genome-wide association study (GWAS) on NMIBC tumor and patient characteristics in the Bladder Cancer Prognosis Programme (BCPP) cohort, potentially identifying novel genetic associations (11). In the current study, we have investigated if smoking status and/or smoking intensity interact with the effect of discovered variants on key NMIBC characteristics of tumor grade, stage, size and patient age within the BCPP cohort.
Methods
Participants and genotyping
BCPP is a prospective cohort of 1,544 participants (12). Baseline clinical tumor characteristics (stage, grade, size) were collected from medical records, while demographic and smoking data were retrieved from records of semi-structured interviews conducted at baseline. Largest tumor size was estimated visually during cystoscopy. Blood samples of 888 participants were genotyped on the Illumina Infinium OmniExpress-24 BeadChip array (previously known as The HumanOmniExxpress-24 BeadChip) at deCODE genetics (Reykjavik, Iceland) (13).
Quality control (QC)
QC for the initial GWAS was carried out in PLINK v1.90 (released 17th November 2016) (14,15) and is described in more detail elsewhere (11). In brief, genotype samples yielding inconclusive sex calls, excessive missingness, low or increased heterozygosity rate were excluded from the analysis. Related individuals and participants presenting as population outliers [identified in the principal component analysis (PCA)] were additionally excluded from the study. Genetic markers deviating from the Hardy-Weinberg equilibrium (HWE), those with high missingness rate and low minor allele frequency (MAF) were excluded. All QC steps resulted in a pre-imputed dataset consisting of 653 individuals and 597,764 SNPs.
Imputation
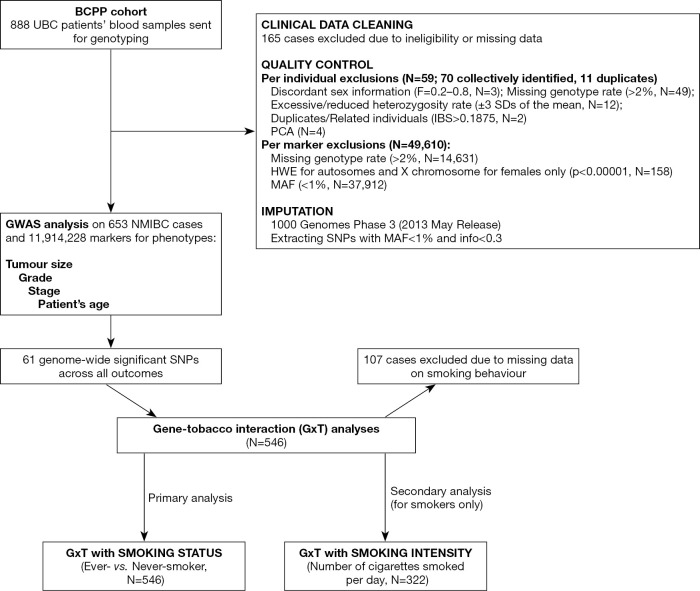
A two-step imputation was performed with Eagle v2.3.2 (16) for haplotype phasing and IMPUTE2 (17) for genotype imputation, using 1000 Genomes Phase 3 (released 2nd May 2013) (18) as a reference panel in the genome build 19 (GRCh37/hg19). Post-imputation QC consisted of deleting markers having info score of <0.3 and MAFs of <1%, resulting in a dataset containing 11,914,228 markers that were used for GWAS analysis with tumor and patient baseline characteristics. The exact thresholds applied and number of exclusions per step are outlined in detail in Figure 1.
Figure 1.
Flowchart for data analysis on GxT in the BCPP cohort. GxT, gene-tobacco interaction; BCPP, Bladder Cancer Prognosis Programme; HWE, Hardy-Weinberg equilibrium; SD, standard deviation; IBS, identity by state; PCA, principal component analysis; MAF, minor allele frequency; NMIBC, non-muscle invasive bladder cancer; PCA, principal component analysis; SD, standard deviation; SNP, single nucleotide polymorphism.
Outcomes and exposure variables
As studies on genetic associations with NMIBC baseline characteristics are scarce, we have used results of a previously-reported GWAS in the BCPP cohort (11). Hence, a total of 61 discovered variants were used to test for GxT. The outcomes were as follows: tumor size (centimeters), stage (T1 vs. Ta/Tis), grade (G3/G2 vs. G1), and patients’ age [years as a continuous outcome and a binary variable with sample mean as a cut-off value (</≥69.9 years, representing sample mean age)]. Our analyses were restricted to NMIBC cases only (corresponding to recorded stage at the time of diagnosis of Ta, T1, or Tis) who also had valid records on tobacco use, resulting in 546 patients.
Smoking exposure was modelled as two variables: smoking status (ever vs. never smokers) and smoking intensity (number of cigarettes smoked daily for smokers only). GxT with smoking status was considered as a primary analysis (including 546 subjects), while analysis on smoking intensity was considered secondary (consisting of 322 smokers with valid data on number of cigarettes smoked per day).
Statistical analysis
Differences in baseline sample characteristics were calculated by applying chi-square distribution test for categorical variables (age, tumor size, stage, grade) and t-test for continuous variables (age, tumor size, smoking intensity).
To test the interaction terms that account for the uncertainty of imputed genotypes, QUICKTEST software was used (19). All analyses were adjusted for gender and first five genetic principal components to increase estimate precision.
A total set of 61 SNPs that were discovered in the previous GWAS analysis of the BCPP cohort (11), were tested. GxT term (depicted as SNP × smoking or SNP × smoking intensity) was modelled for each discovered association. An interaction term was deemed to be significant if P value <0.05.
Functional annotation and network analysis
All identified significant SNPs were annotated to an overlapping or closest gene using a web-based SNPnexus tool (20,21), with Ensembl (22) (version 74) as a functional annotation system. Gene-Tissue Expression (GTEx) database (23) was queried for all significant SNPs to identify expression quantitative loci (eQTL).
Results
BCPP patient characteristics are presented in Table 1. Out of the total sample of 546 participants, 72 were never-smokers, whilst 474 had a history of tobacco use. Participant gender was predominantly male (chi-square statistic =9.97, P=0.002). No other statistically significant differences in baseline characteristics between smokers and never-smokers were observed.
Table 1. Descriptive characteristics of the BCPP sample for GxE with smoking analysis.
| Characteristics | Never smokers (N=72) | Ever smokers (N=474) | P value* |
|---|---|---|---|
| Tumor size (cm), mean (SD) (N=533) | 2.69 (2.14) | 2.48 (1.86) | 0.4 |
| Gender (N=546) | 0.002 | ||
| Males (%) | 46 (10.8) | 381 (89.2) | |
| Females (%) | 26 (21.8) | 93 (78.2) | |
| Age (years), mean (SD) (N=546) | 72.2 (10.9) | 69.7 (10.2) | 0.07 |
| Grade (N=539) | 0.7 | ||
| G3 (%) | 49 (13.6) | 312 (86.4) | |
| G2 and G1 (%) | 22 (12.4) | 156 (87.6) | |
| Tumor stage (N=546) | 0.6 | ||
| Ta and Tis (%) | 47 (12.6) | 326 (87.4) | |
| T1 (%) | 25 (14.5) | 148 (85.5) | |
| Cigarettes smoked/day, mean (SD) (N=322) | N/A | 15.8 (12.9) | N/A |
*, P value was calculated with chi-square test for categorical variables and t-test for continuous outcomes. BCPP, Bladder Cancer Prognosis Programme; GxE, gene-environment interaction.
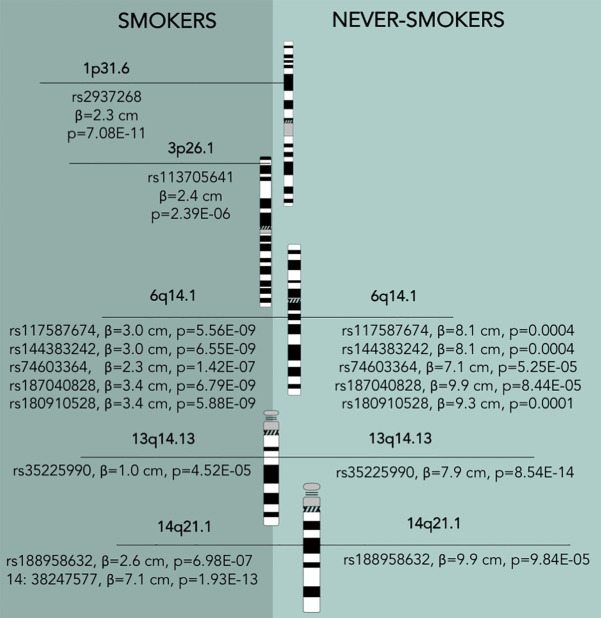
Out of 61 SNPs tested for a GxT in the BCPP cohort, 10 have reached P<0.05, all associated with tumor size (Table 2, Figure 2). Five of these SNPs were located in 6q14.1, two in 14q21.1; while the rest were mapped to 1p31.3, 3p26.1, and 13q14.13.
Table 2. SNPs attaining statistical significance for interaction with smoking status in the BCPP cohort.
| Phenotype | rsID | BP | Locus | REF allele | EFF allele | MAF | β (cm) (SNP) | P (SNP) | β (cm) (never smokers) | P (never smokers) | β (cm) (smokers) | P (smokers) | P (SNP × smoking status)a | P (SNP × smoking intensity)b |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tumor size (cm) | Not yet assigned | 38247577 | 14q21.1 | CTGG | C | 0.01 | 8.1 | 9.41E–14 | –1.6 | 1 | 7.1 | 1.93E–13 | 0.0008 | 0.07 |
| rs35225990 | 46117489 | 13q14.13 | C | T | 0.07 | 1.5 | 5.17E–09 | 7.9 | 8.54E–14 | 1.0 | 4.52E–05 | 0.005 | 0.37 | |
| rs188958632 | 38266174 | 14q21.1 | G | A | 0.01 | 2.8 | 1.02E–06 | 9.9 | 9.84E–05 | 2.6 | 6.98E–07 | 0.008 | 0.29 | |
| rs180910528 | 79821806 | 6q14.1 | A | C | 0.01 | 3.9 | 2.64E–11 | 9.9 | 8.44E–05 | 3.4 | 5.88E–09 | 0.02 | 0.33 | |
| rs74603364 | 79509518 | 6q14.1 | C | T | 0.02 | 2.5 | 1.88E–08 | 7.1 | 5.25E–05 | 2.3 | 1.42E–07 | 0.02 | 0.20 | |
| rs187040828 | 79802426 | 6q14.1 | T | C | 0.01 | 3.9 | 3.58E–11 | 9.3 | 0.0001 | 3.4 | 6.79E–09 | 0.02 | 0.33 | |
| rs144383242 | 79489625 | 6q14.1 | G | T | 0.01 | 3.5 | 5.74E–11 | 8.1 | 0.0004 | 3.0 | 6.55E–09 | 0.03 | 0.24 | |
| rs117587674 | 79432536 | 6q14.1 | G | A | 0.01 | 3.5 | 4.67E–11 | 8.1 | 0.0004 | 3.0 | 5.56E–09 | 0.03 | 0.24 | |
| rs113705641 | 5375733 | 3p26.1 | A | G | 0.02 | 9.5 | 2.68E–27 | –3.1 | 1 | 2.4 | 2.39E–06 | 0.03 | 0.95 | |
| rs2937268 | 66553607 | 1p31.3 | C | T | 0.04 | 2.6 | 3.28E–11 | 1.4 | 0.29 | 2.3 | 7.08E–11 | 0.04 | 0.06 |
a, Interaction with smoking status (never or ever smokers); b, interaction with smoking intensity (number of cigarettes smoked per day). Restricted to smokers only. SNP, single nucleotide polymorphism; BCPP, Bladder Cancer Prognosis Programme; rsID, SNP ID; BP, base pair; REF, reference; EFF, effect; MAF, minor allele frequency; cm, centimeters.
Figure 2.
Group-specific patterns of significant loci for gene-environment interaction for NMIBC tumour size in the BCPP cohort. Idiogram credit: David Adler, Available online: http://www.pathology.washington.edu/research/cytopages/idiograms/human/. NMIBC, non-muscle invasive bladder cancer; BCPP, Bladder Cancer Prognosis Programme.
The most significant interaction with smoking status was observed for a SNP in 14q21.1 locus [base pair (BP): 14:38247577, P=0.0008], that maps to the TTC6 gene (Table 3). Another SNP in the same locus has also yielded statistical significance for interaction (rs188958632, P=0.008, TTC6), but the two variants show differential results for smokers and non-smokers (Figure 2). SNP in 14:38247577 has an effect on tumor size only among smokers (β=7.1 cm, P=1.93E-13). However, rs188958632 is significant among both tobacco use groups, with a larger effect size for never-smokers [β (never-smokers) =9.9 cm, P=9.84E–05; β (ever-smokers) =2.6 cm, P=6.98E–07] (Table 2, Figure 2).
Table 3. Functional annotation of the variants that reached statistical significance for GxE with smoking status.
| Phenotype | rsID | BP | Locus | REF allele | EFF allele | Overlapped gene | Nearest upstream gene | Distance (BP) | Nearest downstream gene | Distance (BP) |
|---|---|---|---|---|---|---|---|---|---|---|
| Tumor size (cm) | Not yet assigned | 38247577 | 14q21.1 | CTGG | C | TTC6 (intronic) | – | – | – | – |
| rs35225990 | 46117489 | 13q14.13 | C | T | FAM194B (intronic) | – | – | – | – | |
| rs188958632 | 38266174 | 14q21.1 | G | A | TTC6 (intronic) | – | – | – | – | |
| rs180910528 | 79821806 | 6q14.1 | A | C | – | PHIP (protein coding) | 33853 | HMGN3 (protein coding) | 89,156 | |
| rs74603364 | 79509518 | 6q14.1 | C | T | – | RP3–390M24.1 (lincRNA) | 193765 | RP11–173D14.3 (lincRNA) | 9,914 | |
| rs187040828 | 79802426 | 6q14.1 | T | C | – | PHIP (protein coding) | 14473 | HMGN3 (protein coding) | 108,536 | |
| rs144383242 | 79489625 | 6q14.1 | G | T | – | RP3–390M24.1 (lincRNA) | 173872 | RP11–173D14.3 (lincRNA) | 29,807 | |
| rs117587674 | 79432536 | 6q14.1 | G | A | – | RP3–390M24.1 (lincRNA) | 116783 | RP11–173D14.3 (lincRNA) | 86,896 | |
| rs113705641 | 5375733 | 3p26.1 | A | G | – | AC026202.5 (lincRNA) | 77287 | AC027119.1 | 628,796 | |
| rs2937268 | 66553607 | 1p31.3 | C | T | PDE4B (intronic) | – | – | – | – |
GxE, gene-environment interaction; BP, base pair; rsID, SNP ID; SNP, single nucleotide polymorphism; REF, reference; EFF, effect.
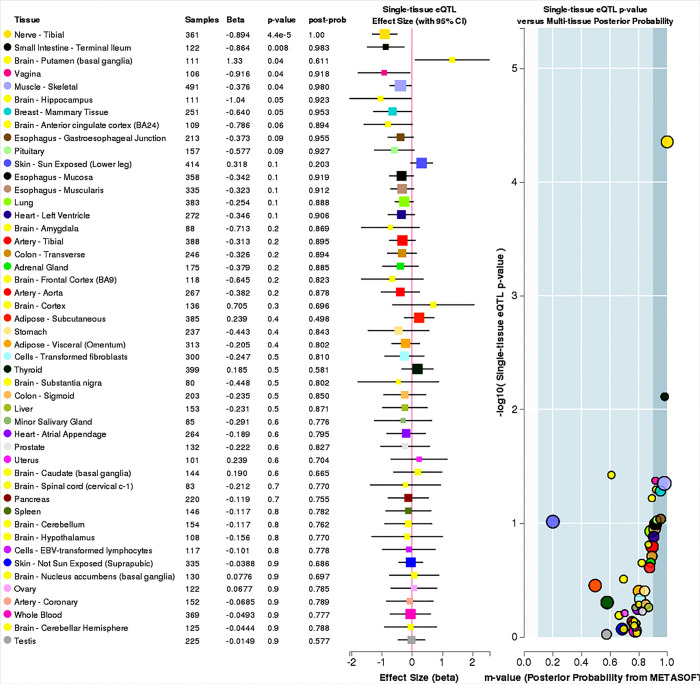
Five variants on 6q14.1 were statistically significantly associated with NMIBC tumor size among both strata of smoking behavior; however, β estimates for never-smokers were universally higher, more than doubling the effect sizes for ever-smokers (Table 2, Figure 2). All five SNPs (rs180910528, rs74603364, rs187040828, rs144383242, and rs117587674) have mapped to intergenic regions (Table 3). Interestingly, two of these SNPs (rs144383242 and rs117587674) are recorded in the GTEx database as having an effect on the expression of HMGN3-AS1 in the tibial nerve tissue (Figures S1,S2, available online) (23). None of the 6q14.1-located SNPs have shown significant interaction between tumor size and smoking intensity in our sample.
Figure S1.
eQTL effect of rs144383242 (6q14.1) across multiple tissues (source: GTEx). eQTL, expression quantitative loci; GxE, gene-environment interaction.
Figure S2.
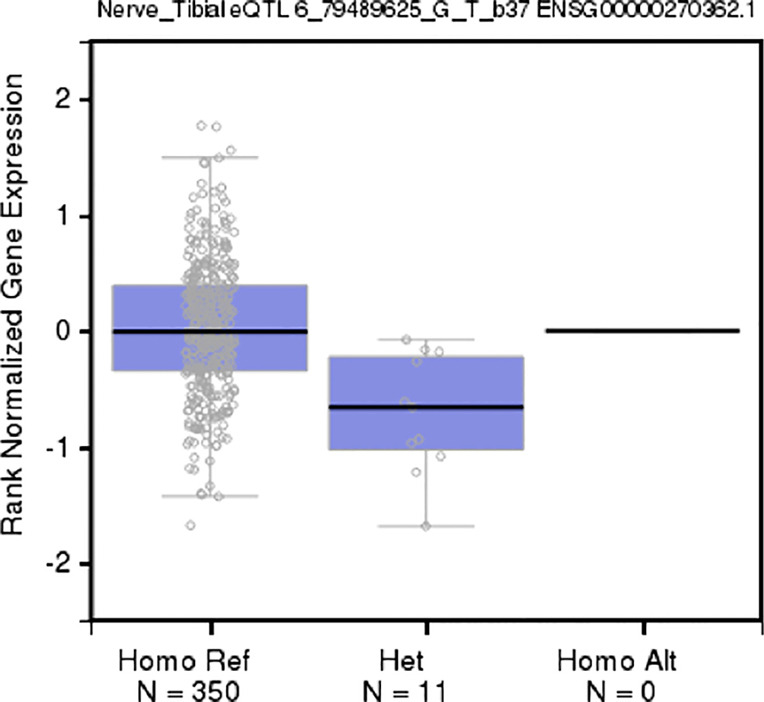
Boxplot of an eQTL effect of rs144383242 (6q14.1) in nerve tissue (source: GTEx). eQTL, expression quantitative loci; GxE, gene-environment interaction.
Rs113705641 on 3p26.1 (intergenic region) has shown to only be statistically significantly associated with tumor size among ever-smokers (β=2.4 cm, P=2.39E–06), and no interaction with smoking intensity. Similarly, rs2937268 on 1p31.3, mapped to the PDE4B gene, has not only reached significance among ever-smokers (β=2.3 cm, P=7.08E–11), but has also shown suggestive interaction with smoking intensity (P=0.06) among tobacco users.
It is notable that variants associated with a stronger effect among never-smokers, had higher estimates overall (ranging from 7.1 to 9.9 cm). In contrast, rs2937268 (1p31.3, PDE4B) and rs113705641 (3p26.1) variants, shown to be significant only for smokers, have been observed to carry a more modest increase in tumor size. The exception would be the 14:38247577 variant in 14q21.1 that was not significant among never-smokers, but resulted in a very large tumor size increase of 7.1 cm in smokers (Table 2, Figure 2).
Discussion
We hereby report findings for GxE with smoking status for SNPs previously discovered in a GWAS on NMIBC baseline characteristics (11). The study provides indication for ten variants interacting with smoking status for tumor size at the time of NMIBC diagnosis, and two of those additionally having suggestive interactions with smoking intensity.
Previous studies on GxT for UBC risk show repeating patterns that highlight potential biological mechanisms (2,4,24). Smoker-specific UBC risk genes are associated with pathways of metabolite detoxification (24), offering a plausible explanation on why the effect is more penetrant among smokers. Alternatively, genes having more importance among never smokers are more often enriched in those regulating cell cycle and DNA integrity (24). Additionally, results across multiple GxT studies on UBC demonstrate that variants significant for never-smokers carry a large effect (i.e., multiplicative interaction), while SNPs that are more important for smokers usually exhibit a milder effect (additive interaction) (5).
Among smokers, the strongest evidence for GxT in our sample was observed for variants rs113705641 (3p26.1) and rs2937268 (1p31.3, PDE4B).
Rs113705641 maps to a long non-coding RNA molecule, and reports on its function are lacking, therefore we are unable to postulate on a potential biological mechanism of interaction.
On the other hand, phosphodiesterase 4B (PDE4B) enzymes are well described to regulate cyclic adenosine 3’,5’-monophosphate (cAMP) concentration in cells by breakdown to non-active molecules (25). The PDE4 family is cAMP-specific, and four existing isoforms of PDE4B account for most cAMP-degradation in a cell (25).
The beneficial effect of PDE inhibitors is well-known and they are commonly used for a variety of disorders: inflammatory conditions [namely chronic obstructive pulmonary disease (COPD) (26,27), asthma (26), and psoriasis (26,27)], overactive bladder (28,29), and various cancer types (30-35). Importantly, studies suggest tumor PDE4 expression can serve as a prognostic marker in colorectal cancer (35). As cAMP signaling is pleiotropic, the mechanisms behind the observed benefit have garnered more than one explanation. For example, elevated cAMP levels suppress cell invasion and migration by disrupting the microtubule cytoskeleton in bladder cancer cells (36), increase phosphatidylinositol 3-kinase (PI3K/AKT)-dependent apoptosis in B-cell malignancies (37), and prohibit proliferation via protein kinase (PKA) and cAMP response element binding (CREB) protein pathways in ovarian cancer (38).
Interestingly, tobacco exposure also increases cAMP signaling in epithelial cells (39), specifically by polycyclic aromatic hydrocarbons (PAHs, found in tobacco smoke) binding to aryl hydrocarbon receptors (AhR) (39). Stimulation of cAMP results in increased levels of amphiregulin (AR), which is hypothesized to eventually form a self-sustainable loop of survival for neoplastic cells (39-41).
In our analysis, cAMP seems to be the overlapping component between PDE4B and smoking, which results in a counter-effect of cAMP regulation (i.e., overexpressed PDE4B decreases cAMP, while smoking increases cAMP). Thus, we hypothesize that smoking interacts with PDE4B mutation status and results in a smaller tumor size increase than the PDE4B mutation itself. This pattern would explain the difference between a main effect exhibited by the SNP in PDE4B (resulting in 2.6 cm increase in tumor size) and a surprising reduced estimate of a joint effect of the SNP and smoking exposure (2.1 cm increase in tumor size).
Smoking, when considered as an independent factor, is the main external risk factor for developing UBC (42). Our described interaction is conditional on the overexpression of PDE4B enzyme; thus, it should be viewed not as a case for the benefits of smoking, but one describing a potential cAMP-related pathway of bladder cancer pathogenesis.
An additional variant in 14q21.1 (BP: 38247577) reached statistical significance among smokers only. However, a SNP in the same locus (rs188958632) showed contradicting results and had a significant effect regardless of smoking exposure. We suspect that the SNP on 14:38247577 is more likely to be a random result due to a lower imputation accuracy (info =0.59) than rs188958632 (info =0.95) (remaining SNPs have an info score of ≥0.90, with the exception for rs113705641, with info =0.62).
In contrast, variants on 13q14.13 and 6q14.1 have reached significant values for main effects among all participants. However, the clinical importance might be higher for never smokers as they showed substantially higher effect sizes than for smokers. The mapped FAM194B (ERICH6B) gene (13q14.13) is recorded in the GTEx database as excessively expressed in testis tissue (23), but other reports on gene function or phenotype associations are scarce, thus precluding a discussion on its potential biological pathway. SNPs in 6q14.1 were mapped to intergenic regions, with rs180910528 and rs187040828 located between protein coding genes of PHIP and HMGN3. Interestingly, four of the total five SNPs in 6q14 region (rs180910528, rs187040828, rs144383242, and rs117587674) are recorded in the GTEx database as having a cis-regulatory effect on an RNA gene of HMGN3-AS1 (23). Antisense RNAs bind to messenger RNA molecules, preventing them from being translated into protein (43). Thus, lower expression of HMGN3-AS1 may cause higher levels of proteins coded by HMGN3 gene. HMGN is a family of nucleosome-binding proteins that alter chromatin structure and regulate essential cell functions, such as differentiation and development (44).
Our study has several limitations. Current analysis was carried out on a relatively small sample of individuals, and false-positives are naturally not to be excluded. The associations between genetic variation and NMIBC tumor size should be interpreted with caution, as they lack validation (11). Besides tobacco, other variables, such as occupational-related exposures with aromatic amines might have independent GxE effects and were not adjusted for in our analyses (45). Larger samples with detailed clinical data are a next step to exploring and validating reported GxEs with smoking for NMIBC. Until robust evidence exists for including genetic variance into NMIBC prognostication tools, our study is complementary to other proof-of-principle reports (2).
In summary, our study suggests smoking might alter the outcome between genetic variance and NMIBC tumor characteristics. These results may provide more evidence on the complexity of the joint influence of genetic background and external exposures on bladder cancer.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-19-523). RTB has contributed to advisory boards for Olympus Medical Systems and Janssen. The other authors have no conflicts of interest to declare.
References
- 1.Selinski S. The post GWAS era: strategies to identify gene-gene and gene-environment interactions in urinary bladder cancer. EXCLI J 2014;13:1198-203. [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia-Closas M, Rothman N, Figueroa JD, et al. Common genetic polymorphisms modify the effect of smoking on absolute risk of bladder cancer. Cancer Res 2013;73:2211-20. 10.1158/0008-5472.CAN-12-2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothman N, Garcia-Closas M, Chatterjee N, et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat Genet 2010;42:978-84. 10.1038/ng.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Figueroa JD, Han SS, Garcia-Closas M, et al. Genome-wide interaction study of smoking and bladder cancer risk. Carcinogenesis 2014;35:1737-44. 10.1093/carcin/bgu064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.García-Closas M, Malats N, Silverman D, et al. NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet 2005;366:649-59. 10.1016/S0140-6736(05)67137-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore LE, Baris DR, Figueroa JD, et al. GSTM1 null and NAT2 slow acetylation genotypes, smoking intensity and bladder cancer risk: results from the New England bladder cancer study and NAT2 meta-analysis. Carcinogenesis 2011;32:182-9. 10.1093/carcin/bgq223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lubin JH, Kogevinas M, Silverman D, et al. Evidence for an intensity-dependent interaction of NAT2 acetylation genotype and cigarette smoking in the Spanish Bladder Cancer Study. Int J Epidemiol 2007;36:236-41. 10.1093/ije/dym043 [DOI] [PubMed] [Google Scholar]
- 8.Czerniak B, Dinney C, McConkey D. Origins of bladder cancer. Annu Rev Pathol 2016;11:149-74. 10.1146/annurev-pathol-012513-104703 [DOI] [PubMed] [Google Scholar]
- 9.Grotenhuis AJ, Dudek AM, Verhaegh GW, et al. Independent replication of published germline polymorphisms associated with urinary bladder cancer prognosis and treatment response. Bladder Cancer 2016;2:77-89. 10.3233/BLC-150027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babjuk M, Böhle A, Burger M, et al. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder: update 2016. Eur Urol 2017;71:447-61. 10.1016/j.eururo.2016.05.041 [DOI] [PubMed] [Google Scholar]
- 11.Lipunova N, Wesselius A, Cheng KK, et al. Genome-wide association study for tumour stage, grade, size, and age at diagnosis of non-muscle-invasive bladder cancer. Eur Urol Oncol 2019;2:381-9. 10.1016/j.euo.2018.08.020 [DOI] [PubMed] [Google Scholar]
- 12.Zeegers MP, Bryan RT, Langford C, et al. The West Midlands Bladder Cancer Prognosis Programme: rationale and design. BJU Int 2010;105:784-8. 10.1111/j.1464-410X.2009.08849.x [DOI] [PubMed] [Google Scholar]
- 13.Hakonarson H, Gulcher JR, Stefansson K. deCODE genetics, Inc. Pharmacogenomics 2003;4:209-15. 10.1517/phgs.4.2.209.22627 [DOI] [PubMed] [Google Scholar]
- 14.Chang CC, Chow CC, Tellier LC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015;4:7. 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559-75. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loh PR, Danecek P, Palamara PF, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet 2016;48:1443-8. 10.1038/ng.3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009;5:e1000529. 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke L, Fairley S, Zheng-Bradley X, et al. The international Genome sample resource (IGSR): A worldwide collection of genome variation incorporating the 1000 Genomes Project data. Nucleic Acids Res 2017;45:D854-9. 10.1093/nar/gkw829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson T, Kutalik Z. QUICKTEST user guide v0.94. 2008. Available online: http://toby.freeshell.org/software/quicktest.shtml
- 20.Dayem Ullah AZ, Lemoine NR, Chelala C. SNPnexus: a web server for functional annotation of novel and publicly known genetic variants (2012 update). Nucleic Acids Res 2012;40:W65-70. 10.1093/nar/gks364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dayem Ullah AZ, Lemoine NR, Chelala C. A practical guide for the functional annotation of genetic variations using SNPnexus. Brief Bioinform 2013;14:437-47. 10.1093/bib/bbt004 [DOI] [PubMed] [Google Scholar]
- 22.Aken BL, Ayling S, Barrell D, et al. The Ensembl gene annotation system. Database (Oxford) 2016;2016:baw093. [DOI] [PMC free article] [PubMed]
- 23.GTEx Consortium . The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580-5. 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwender H, Selinski S, Blaszkewicz M, et al. Distinct SNP combinations confer susceptibility to urinary bladder cancer in smokers and non-smokers. PLoS One 2012;7:e51880. 10.1371/journal.pone.0051880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conti M, Richter W, Mehats C, et al. Cyclic AMP-specific PDE4 phosphodiesterases as critical components of cyclic AMP signaling. J Biol Chem 2003;278:5493-6. 10.1074/jbc.R200029200 [DOI] [PubMed] [Google Scholar]
- 26.Page CP, Spina D. Phosphodiesterase inhibitors in the treatment of inflammatory diseases. Handb Exp Pharmacol 2011;(204):391-414. 10.1007/978-3-642-17969-3_17 [DOI] [PubMed] [Google Scholar]
- 27.Abdel-Magid AF. Selective Inhibitors of Phosphodiesterase 4B (PDE-4B) May Provide a Better Treatment for CNS, Metabolic, Autoimmune, and Inflammatory Diseases. ACS Med Chem Lett 2017;8:1132-3. 10.1021/acsmedchemlett.7b00425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahnama'i MS, Ückert S, Hohnen R, et al. The role of phosphodiesterases in bladder pathophysiology. Nat Rev Urol 2013;10:414-24. 10.1038/nrurol.2013.101 [DOI] [PubMed] [Google Scholar]
- 29.Agis-Torres Á, Recio P, López-Oliva ME, et al. Phosphodiesterase type 4 inhibition enhances nitric oxide- and hydrogen sulfide-mediated bladder neck inhibitory neurotransmission. Sci Rep 2018;8:4711. 10.1038/s41598-018-22934-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong H, Claffey KP, Brocke S, et al. Inhibition of breast cancer cell migration by activation of cAMP signaling. Breast Cancer Res Treat 2015;152:17-28. 10.1007/s10549-015-3445-9 [DOI] [PubMed] [Google Scholar]
- 31.Powers GL, Hammer KD, Domenech M, et al. Phosphodiesterase 4D inhibitors limit prostate cancer growth potential. Mol Cancer Res 2015;13:149-60. 10.1158/1541-7786.MCR-14-0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Domvri K, Zarogoulidis K, Zogas N, et al. Potential synergistic effect of phosphodiesterase inhibitors with chemotherapy in lung cancer. J Cancer 2017;8:3648-56. 10.7150/jca.21783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooney JD, Aguiar RC. Phosphodiesterase 4 inhibitors have wide-ranging activity in B-cell malignancies. Blood 2016;128:2886-90. 10.1182/blood-2016-09-737676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suhasini AN, Wang L, Holder KN, et al. A phosphodiesterase 4B-dependent interplay between tumor cells and the microenvironment regulates angiogenesis in B-cell lymphoma. Leukemia 2016;30:617-26. 10.1038/leu.2015.302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsunoda T, Ota T, Fujimoto T, et al. Inhibition of phosphodiesterase-4 (PDE4) activity triggers luminal apoptosis and AKT dephosphorylation in a 3-D colonic-crypt model. Mol Cancer 2012;11:46. 10.1186/1476-4598-11-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ou Y, Zheng X, Gao Y, et al. Activation of cyclic AMP/PKA pathway inhibits bladder cancer cell invasion by targeting MAP4-dependent microtubule dynamics. Urol Oncol 2014;32:47.e21-8. 10.1016/j.urolonc.2013.06.017 [DOI] [PubMed] [Google Scholar]
- 37.Smith PG, Wang F, Wilkinson KN, et al. The phosphodiesterase PDE4B limits cAMP-associated PI3K/AKT-dependent apoptosis in diffuse large B-cell lymphoma. Blood 2005;105:308-16. 10.1182/blood-2004-01-0240 [DOI] [PubMed] [Google Scholar]
- 38.Gong S, Chen Y, Meng F, et al. Roflumilast restores cAMP/PKA/CREB signaling axis for FtMt-mediated tumor inhibition of ovarian cancer. Oncotarget 2017;8:112341-53. 10.18632/oncotarget.22866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du B, Altorki NK, Kopelovich L, et al. Tobacco smoke stimulates the transcription of amphiregulin in human oral epithelial cells: evidence of a cyclic AMP-responsive element binding protein-dependent mechanism. Cancer Res 2005;65:5982-8. 10.1158/0008-5472.CAN-05-0628 [DOI] [PubMed] [Google Scholar]
- 40.Zuo WL, Yang J, Gomi K, et al. EGF-Amphiregulin Interplay in Airway Stem/Progenitor Cells Links the Pathogenesis of Smoking-Induced Lesions in the Human Airway Epithelium. Stem Cells 2017;35:824-37. 10.1002/stem.2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bryan RT, Hussain SA, James ND, et al. Molecular pathways in bladder cancer: part 1. BJU Int 2005;95:485-90. 10.1111/j.1464-410X.2005.05325.x [DOI] [PubMed] [Google Scholar]
- 42.Burger M, Catto JW, Dalbagni G, et al. Epidemiology and risk factors of urothelial bladder cancer. Eur Urol 2013;63:234-41. 10.1016/j.eururo.2012.07.033 [DOI] [PubMed] [Google Scholar]
- 43.Park H, Yoon Y, Suk S, et al. Effects of different target sites on antisense RNA-mediated regulation of gene expression. BMB Rep 2014;47:619-24. 10.5483/BMBRep.2014.47.11.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furusawa T, Cherukuri S. Developmental function of HMGN proteins. Biochim Biophys Acta 2010;1799:69-73. [DOI] [PMC free article] [PubMed]
- 45.Murta-Nascimento C, Schmitz-Dräger BJ, Zeegers MP, et al. Epidemiology of urinary bladder cancer: from tumor development to patient's death. World J Urol 2007;25:285-95. 10.1007/s00345-007-0168-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as