Abstract
Background
Expression of circulating serum microRNAs has not been studied in a cohort of patients with papillary renal cell carcinoma (pRCC) so far. We hypothesized that miRNA deregulation in malignant tissue is reflected in serum and could be used for non-invasive diagnosis of pRCC as well as differentiation between type 1 and type 2 pRCC.
Methods
We selected 11 differentially regulated miRNAs from the Cancer Genome Atlas (TCGA) pRCC data set as potential serum validation candidates. Serum miRNA expression was determined by qRT-PCR in a total of 34 pRCC type 1, 33 pRCC type 2 and 33 control subjects of three german high-volume medical centers.
Results
Heatmap and principal component analysis showed that miRNA expression did not cluster the samples into distinct sample groups and that miRNA levels did not significantly discriminate healthy individuals from patients with pRCC, nor between patients with type 1 and type 2 pRCC. However, miR-21-5p levels were significantly increased in patients with advanced pRCC (>pT3, and/or pN+ and/or pM+) in comparison to localized pRCC. Moreover, adding the expression of miR-210-3p, which was significantly down-regulated in localized pRCC sera in comparison to healthy sera, additionally increased diagnostic accuracy in our study cohort.
Conclusions
In our multicenter cohort, we were not able to identify a single miRNA serum marker for pRCC including its subclasses. However, our study revealed that miR-21-5p levels were elevated in advanced disease (with added diagnostic accuracy via addition of miR-210-3p expression), proposing these two miRs as potential biomarkers in pRCC.
Keywords: miRNA, papillary renal cell carcinoma (pRCC), multicenter
Introduction
Accounting for about 3% of adult malignancies, renal cell carcinoma (RCC) is the most common neoplasm of the adult kidney (1). Additionally, it bears a comparably high mortality rate of up to 40%, which stresses the clinical importance of this entity (2). Papillary renal cell carcinoma (pRCC) accounts for 15–20% of all RCCs and represents a heterogeneous disease regarding disease progression, therapy response and patient outcome (3-5). To more accurately stratify aggressive disease from indolent tumours, Delahunt and Eble proposed a morphological subtyping of pRCCs in 1997 into two subgroups: first, there is type 1 pRCC, which is often found multifocal, holding small cuboid cells arranged into layers and a predominantly indolent clinical course; on the contrary, there is type 2 pRCC, with voluminous eosinophilic cells in pseudostratification patterns and a more aggressive clinical behaviour (6). This subclassification pattern and its prognostic value was confirmed in various clinical studies (7).
Unfortunately, besides morphological characterization, there have been no investigations whether serum biomarkers might be useful to detect and/or distinguish pRCC and its subtypes from other malign entities of the kidney. MiRNAs are predestined for this particular task: their expression profiles not only allow distinguishing malignant and non-malignant tissue, but also different tumor entities (8). MiRNAs are circulating in a cell-free form in blood (9-11), very likely in exosomes which protect them against RNase degradation (9,11). MiRNA-signatures in blood are similar in men and women, as well as individuals of different age (10). Furthermore, miRNA expression levels are comparable both in plasma and serum, and are not disturbed by repeated freeze/thaw cycles as well as by prolonged storage at room temperature (11).
So far, circulating miRNAs have not been investigated in patients with pRCC regarding their potential diagnostic (compared to healthy controls) or discriminative (regarding the two subtypes) implications. Several groups analysed miRNA-expression in pRCC tissue specimen with conflicting results (12-14). We therefore designed our study to identify and validate potential miRNA serum-candidates in a multicenter setting.
Methods
Ethics statement
This study was approved by the local human research ethics committees of the three participating medical centres (Charité Berlin: No. EA1/134/12, University of Bonn: No. 036/08, University of Würzburg: No. 136/08) and was conducted according to the standards set by the declaration of Helsinki; all patients provided written informed consent before being enrolled.
Patients and serum samples
We collected preoperative serum samples of n=67 patients suffering from pRCC, divided into n=34 pRCC type 1 and n=33 pRCC type 2 cases who consecutively underwent surgery at the participating centers between 2008–2014. The clinical and pathological patient characteristics are shown in Table 1. Sera of healthy subjects (n=33) not suffering from urological or other malignancies were used as controls.
Table 1. Clinical characteristics of the patient cohort.
| Characteristics | pRCC type 1 (n=34) | pRCC type 2 (n=33) | Both (n=67) | P value |
|---|---|---|---|---|
| Patient age (years) | 63.6 [35–78] | 67.4 [46–81] | 65 [35–81] | 0.06 |
| Sex | ||||
| Male | 30 (88.3%) | 28 (84.9%) | 58 | |
| Female | 4 (11.7%) | 5 (15.1%) | 9 | 0.12 |
| Tumor stage | ||||
| pT1 | 27 (79.4%) | 23 (69.8%) | 50 | |
| pT2 | 4 (11.7%) | 5 (15.1%) | 9 | |
| pT3 | 3 (8.9%) | 5 (15.1%) | 8 | fw ANOVA |
| pT4 | 0 (0%) | 0 (0%) | 0 | 0.132 |
| Nodal stage | ||||
| pN0 | 32 (94.1%) | 24 (72.7%) | 56 | |
| pN1 | 0 (0%) | 5 (15.1%) | 5 | <0.01 |
| pNX | 2 (5.9%) | 4 (12.2%) | 6 | |
| Metastasis | ||||
| pM0 | 33 (97.1%) | 32 (94.1%) | 65 | |
| pM1 | 1 (2.9%) | 1 (5.9%) | 2 | 0.1 |
| WHO grading | ||||
| G1 | 3 (8.9%) | 1 (5.9%) | 4 | |
| G2 | 29 (85.2%) | 23 (64%) | 52 | |
| G3 | 2 (5.9%) | 8 (24.2%) | 10 | fw ANOVA |
| G4 | 0 (0%) | 1 (5.9%) | 1 | 0.141 |
fw ANOVA, forward analysis of variance.
RNA extraction, qRT-PCR and normalization
To centralize sample processing, RNA extraction and qRT-PCR for microRNAs let-7b, miR-10a-3p, miR-10b-5p, miR-21-5p, miR-126-3p, miR-127-3p, miR-142-3p, miR-155-5p, -199a-3p, miR-210-3p and miR-425-5p were executed by Exiqon RNA services (http://www.exiqon.com). Samples were spiked with UniSp2, UniSp4, and UniSp to exclude technical errors during RNA isolation and cDNA synthesis, haemolysis was controlled by measuring the optical density of the samples at 414 nm (cut-off value 0.3) in a monochromator and by calculating the ΔCt of miR-23a-3p (unaffected by haemolysis) and miR-451a-5p (expressed in erythrocytes). The geometric mean of miR-23a-3p, miR-191-5p and miR-103a-3p was used for normalization.
Statistics and data analysis
Statistical analysis was performed using R 3.10 (URL: http://www.r-project.org), figures were drawn either with R or Graphpad Prism 7.
Pearson’s chi-square test was used to realize intergroup comparison. If two means of normally distributed data had to be compared, two-sided unpaired student’s t-test was used. More than two group means were differentiated by analysis of variance (ANOVA) with post-hoc testing (Tukey’s test) if significant differences occurred. Principal component analysis (PCA), hierarchical clustering, heat maps and correlation matrices were realized with the packages corrplot, Amelia, ROCR, My.stepwise, ggplot2, limma. lattice, and gplots. Significant associations were set as P≤0.05 (*).
Results
Candidate miRNA selection and quality control
Potential miRNAs were identified based on their elevated expression in pRCC, their elevated expression in either type 1 or type 2 pRCC and high overall expression levels on the basis of previously published results (12-14). To allow the identification of a robust and sensitive serum marker, we further selected only miRNAs that were already present in serum in detectable levels in healthy controls (11,15). An overview of the experimental setup is shown in Figure 1A.
Figure 1.
Candidate miRNA selection and quality control. (A) Description of the multicenter study design. (B) Haemolysis control for miR-23 (not expressed in erythrocytes), miR-451 (highly expressed in erythrocytes) and present haemoglobin (Hb) in the samples. (C) Normalized miR-expression in relation to the participating center.
Of the selected miRNAs, miR-210-3p, miR-21-5p were previously described as serum markers (13,14), whereas selection of let-7b-3p, miR-10a-5p, miR-10b-5p, miR-126-3p, miR-127-3p, miR-142-3p, miR-155-5p, miR-199a-3p, and miR-425-5p was solely based on elevated tissue expression. The miRNAs let-7b, miR-10a-5p, miR-10b-5p, miR-126-3p, miR-127-3p, and miR-199a-3p, and miR-425-5p were deregulated in pRCC type 1 vs. pRCC type 2 (12).
To ensure quality of the samples, we tested for haemolysis by comparing the expression values of miR-23, which is not expressed in erythrocytes and thus not influenced by haemolysis and miR-451, which is highly expressed in erythrocytes. We further correlated expression of all miRNAs with the serum haemolysis values and found that beside miR-451, miR-425-5p, and miR-210-3p were correlated with haemolysis in our samples (Figure 1B).
Comparability validation: normalized relative miR-expression in relation to their center of origin
To validate comparability between the different samples in regard to the participating centres, we assessed the relative, normalized expression of all tested miRNAs and compared it in regard to the participating center. We found that one of the tested miRNAs was significantly differently expressed between the participating centres: miR-425-5p was significantly lower expressed in the Charité cohort in comparison to the other centers, and was therefore excluded from further analysis (Figure 1C).
Diagnostic power of miRNA expression in control vs. pRCC serum samples
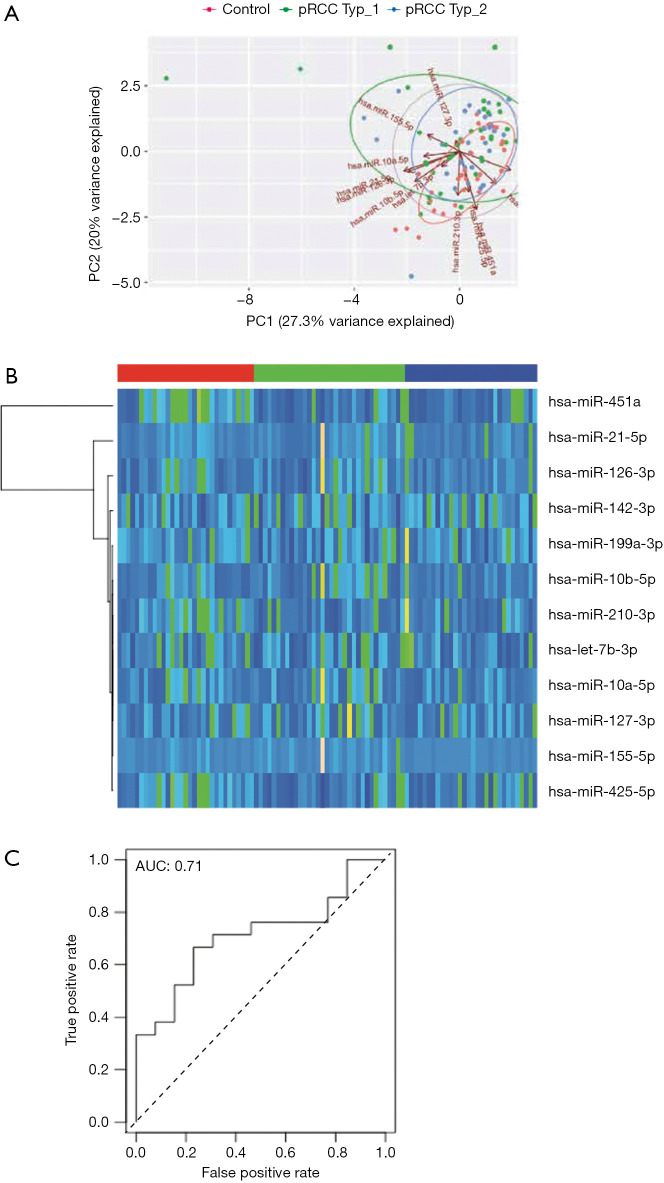
We performed PCA of normalized miRNAs to identify relevant patients’ subgroups. However, miRNA expression was not able to cluster serum samples and the first two components were only able to explain 50% of the observed variance (Figure 2A). Likewise, supervised hierarchical clustering clearly shows that miRNA expression did not differentiate tumor samples from healthy controls (Figure 2B). The hierarchical clustering also indicated that no single miRNA was clearly differentially expressed between subtype 1 and subtype 2 sera (compare samples marked in red and blue), which was confirmed by direct comparison of each miRNAs in the pRCC samples and healthy controls (Figure S1).
Figure 2.
Diagnostic power of miRNA expression in control vs. pRCC serum samples. (A) Principal component analysis (PCA) of normalized miR-expression in relation to control sera (red), pRCC type 1 (green) and type 2 sera (blue). (B) Hierarchical supervised clustering of normalized miR-expression in relation to control sera (red, left), pRCC type 1 (green, middle) and type 2 sera (blue, right). (C) Logistic regression analysis of a model comprising all normalized miR expressions in regard to diagnostic power to detect pRCC.
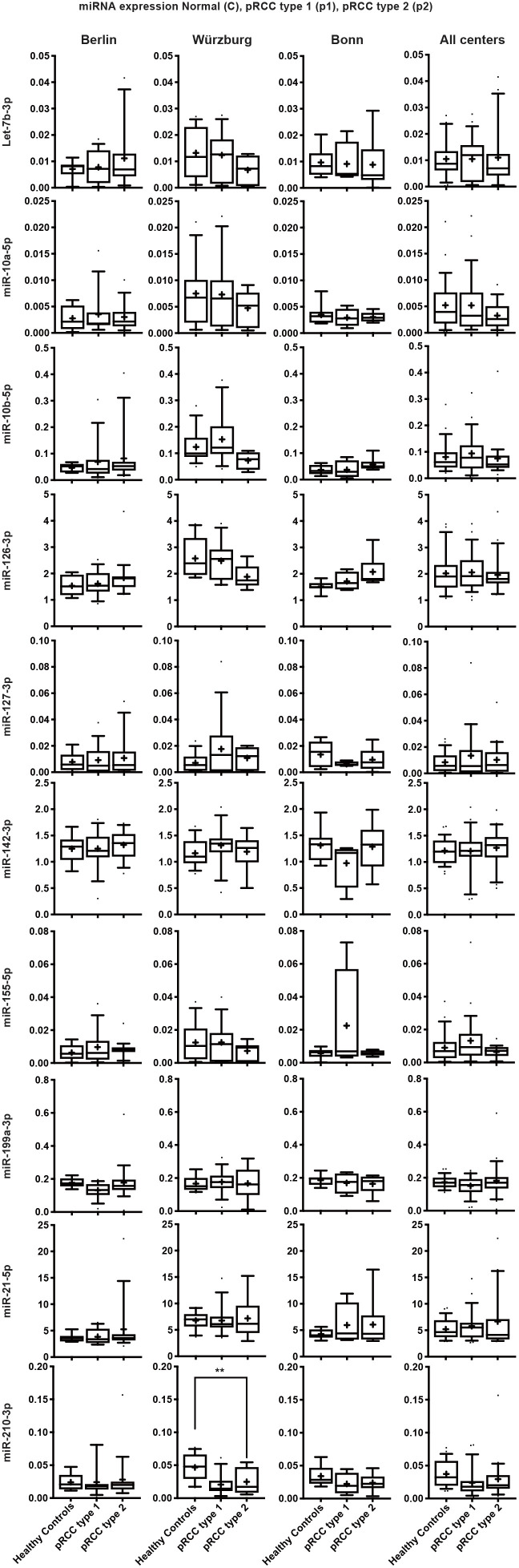
Figure S1.
Normalized expression of all tested miRs in healthy control sera, pRCC type 1 and type 2 subtypes, divided into the respective participating centers (left plots) and subsequent cross-cohort comparison (right plots).
In line with the PCA and hierarchical clustering, logistic regression analysis did not identify a single independent diagnostic marker and a model with all miRNA only resulted in an AUC of 0.71 in ROC analysis (Figure 2C).
Detailed expression data with regard to centres and subclasses are presented in Figure S1.
Association of miRNA expression and clinicopathological parameters in pRCC sera
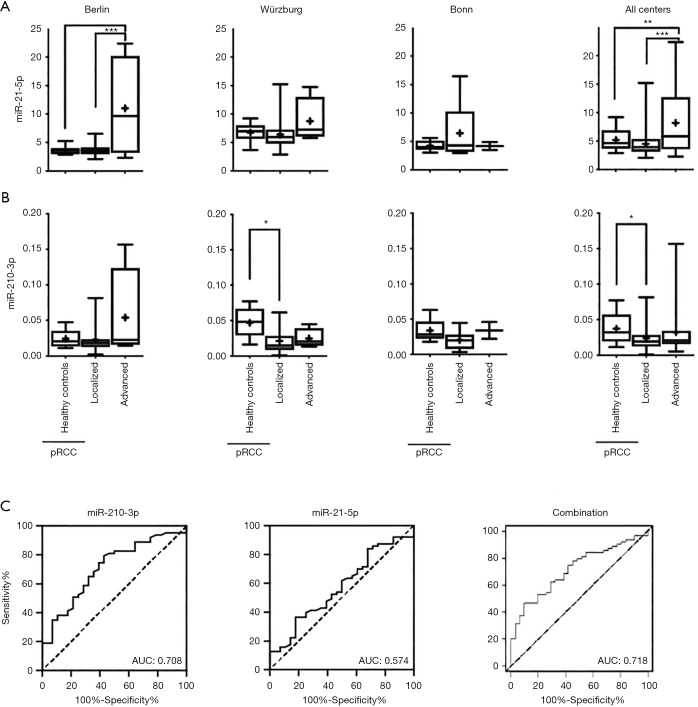
To explore prognostic liabilities of the assessed miRNA expression profiles, we correlated the expression of all examined serum miRNAs with clinicopathological parameters, grouping patients into a localized (pT1-2 and/or pN0 and/or pM0) or advanced (defined as pT>3 and/or pN+ and/or pM+) disease group. miR-21-5p was significantly higher expressed in advanced samples in comparison to non-advanced samples and control sera in the Berlin cohort as well as in the cross cohort comparison (Figure 3) and showed a borderline significance in the Wurzburg cohort. As the Bonn cohort only contained two advanced disease sample, we did not consider this cohort separately. While several other miRNAs appeared differentially expressed in advanced samples in one cohort all of the results were not confirmed when combining the data of all cohorts. For example: while miR-10b-5p appeared highly expressed in advanced samples in the Charité and Würzburg cohorts it failed to show a statistically significant result in the cross cohort comparison (see Figure S2 for an overview of all miRNA expression patterns grouped into the single centres and combined).
Figure 3.
Association of miRNA expression and clinicopathological parameters in pRCC sera. (A) Normalized miR-21-5p and (B) miR-210-3p expression in healthy control sera, localized and advanced pRCC disease, divided into the respective participating centers (left plots) and subsequent cross-cohort comparison (right plots). (C) AUC curves regarding the diagnostic accuracy of normalized miR-210-3p (left plot), miR-21-5p (middle plot) expression and a combination of both (right plot) regarding localized and advanced pRCC. *, P<0.05; **, P<0.01; ***, P<0.001.
Figure S2.
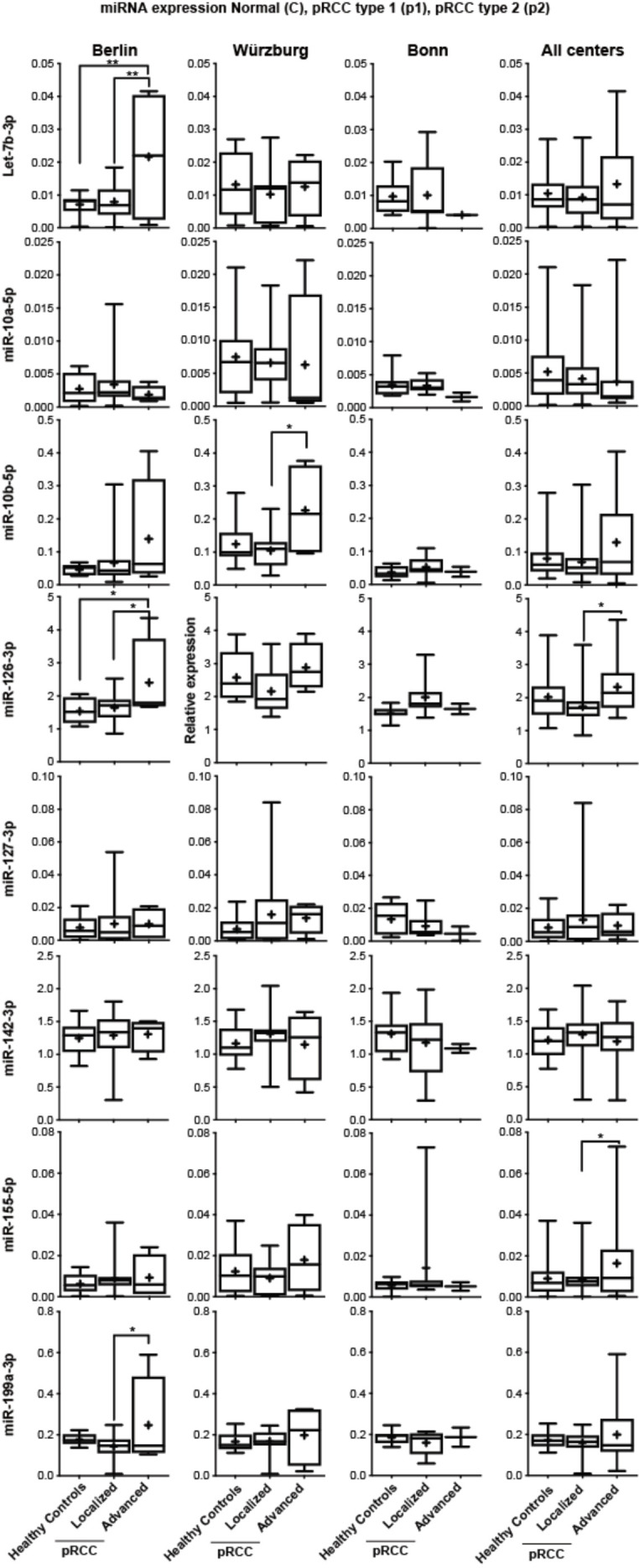
Normalized expression of all tested miRs in healthy control sera, localized and advanced pRCC disease, divided into the respective participating centers (left plots) and subsequent cross-cohort comparison (right plots).
miR-210-3p, which was not significantly regulated in pRCC in comparison to healthy control did show a statistically significantly lower expression in patients with localized tumors in comparison to healthy controls, while expression in patients with advanced tumors were comparable to healthy controls. ROC curve analysis resulted in an AUC of 0.708 for miR-210-3p, while miR-21-5p did not result in significantly better AUC (0.574). A combined model including both miR-210-3p and miR-21-5p slightly improved AUC in comparison to miR-210-3p alone (AUC: 0.718).
Discussion
The potential of circulating miRNAs as diagnostic and/or prognostic biomarker has been investigated in RCC in the past years. However, these studies focussed on the most common subtype clear cell RCC (16,17). The analysis of serum/plasma miRNAs in pRCC patients was limited to few patients so far and these were usually part of a larger cohort comprising of patients with all histological subtypes (18-20). As a result, there is no reliable information regarding the relevance of circulating miRNAs in these patients. We thus studied serum miRNA expression in a multi-centre cohort including 67 pRCC patients and 33 healthy individuals.
We chose 11 miRNAs for analysis in serum samples based on their potential role as diagnostic (13,14) or pRCC type 1 vs. type 2 discriminative tissue markers (12) in tissue. Somewhat disappointingly we did not observe a significant different expression of any serum miRNA in patients with pRCC and healthy controls. However, miR-21-5p levels were significantly increased in patients with advanced pRCC. In general, miR-21 is one of the most frequently upregulated miRNAs in solid malignancies (21) and considered to be a typical “onco-miR” as most of its targets are tumour suppressors (22). Notably, miR-21 serum levels were earlier correlated with advanced clinical stage in ccRCC patients (23). Tusong et al. further reported that serum miR-21 levels decreased following nephrectomy in patients with ccRCC (24). We thus assume that serum miR-21 expression could be helpful to identify RCC patients with advanced disease.
Our study has several strengthens: the multi-centre designs allowed us to analyse the expression of serum miRNAs in a relevant number (n=67) of pRCC patients. In addition, the multi-centre setting stands out from most of earlier studies reported in patients with RCC so far. Finally, the quality control by analysing haemolysis-associated miRNAs (miR-23a-3p and miR-451a-5p) as well as the analysis of three reference miRNAs (miR-23a-3p, miR-191-5p and miR-103a-3p) for normalization was superior to other studies in the field of RCC (16,17).
However, there are also limitations to this study. We did not perform global screening of miRNA expression in pRCC tissue in comparison to healthy controls. This resulted in lack of identifying a sensitive miRNA-based marker for pRCC detection and subtype differentiation. Our selection process was based on the hypothesis that upregulation of miRNA expression in pRCC tissue should be reflected by higher miRNAs serum levels. miRNAs can either be actively secreted by the miRNA-expressing cells in in vesicles or bound to Ago proteins or they can be passively released from apoptotic and necrotic cells (17). Passive release of miRNAs should result in serum miRNA levels reflecting the observed deregulation of miRNAs in tissue. Active release on the other hand will result in a different distribution of miRNAs in serum and tissue. Our findings report apparent discrepancies in tissue and serum, making a global screening in serum mandatory for the identification of potential non-invasive miRNA markers.
Our data also raise the question on whether serum miRNA serum levels are sensitive enough to be used as diagnostic markers. While we observed reduced miR-210-3p serum levels in localized and elevated serum miR-21 levels only in advanced cancers, the discriminative power of both miRNA biomarkers even in combination was moderate. We also observed an increased but not statistically significant elevation for a number of other miRNAs in advanced disease, raising the possibility that only few miRNAs will be released in sufficient levels to the blood stream to be suitable for cancer diagnosis. We will add however that we only studied a limited number of miRNAs. It might well be that we will identify miRNAs, whose levels are elevated in patients with early stage disease by global screening approaches in future studies. Further, we suggest the validation of our findings for miR-21-5p and miR-210-5p in a larger cohort.
Conclusions
The analysis of serum miRNAs did not provide diagnostic information in patients with pRCC and was not able to differentiate type 1 and type 2 subtypes. Serum miR-21-5p expression was increased in patients with advanced pRCC, whereas miR-210-3p expression was down-regulated in localized pRCC in our multicenter approach. We thus propose that these two miRs – after validation in larger patient cohorts—could be useful as prognostic biomarkers in this subtype of a clinically relevant disease.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This publication was funded by a twinning research grant of the Berlin Institute of Health.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the local human research ethics committees of the three participating medical centres (Charité Berlin: No. EA1/134/12, University of Bonn: No. 036/08, University of Würzburg: No. 136/08). All patients provided written informed consent prior to being enrolled in the study.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau.2020.03.18). The authors have no conflicts of interest to declare.
References
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Znaor A, Lortet-Tieulent J, Laversanne M, et al. International variations and trends in renal cell carcinoma incidence and mortality. Eur Urol 2015;67:519-30. 10.1016/j.eururo.2014.10.002 [DOI] [PubMed] [Google Scholar]
- 3.Saleeb RM, Brimo F, Farag M, et al. Toward Biological Subtyping of Papillary Renal Cell Carcinoma With Clinical Implications Through Histologic, Immunohistochemical, and Molecular Analysis. Am J Surg Pathol 2017;41:1618-29. 10.1097/PAS.0000000000000962 [DOI] [PubMed] [Google Scholar]
- 4.Saleeb RM, Plant P, Tawedrous E, et al. Integrated Phenotypic/Genotypic Analysis of Papillary Renal Cell Carcinoma Subtypes: Identification of Prognostic Markers, Cancer-related Pathways, and Implications for Therapy. Eur Urol Focus 2018;4:740-8. 10.1016/j.euf.2016.09.002 [DOI] [PubMed] [Google Scholar]
- 5.Courthod G, Tucci M, Di Maio M, et al. Papillary renal cell carcinoma: A review of the current therapeutic landscape. Crit Rev Oncol Hematol 2015;96:100-12. 10.1016/j.critrevonc.2015.05.008 [DOI] [PubMed] [Google Scholar]
- 6.Delahunt B, Eble JN. Papillary renal cell carcinoma: a clinicopathologic and immunohistochemical study of 105 tumors. Mod Pathol 1997;10:537-44. [PubMed] [Google Scholar]
- 7.Cheville JC, Lohse CM, Zincke H, et al. Comparisons of outcome and prognostic features among histologic subtypes of renal cell carcinoma. Am J Surg Pathol 2003;27:612-24. 10.1097/00000478-200305000-00005 [DOI] [PubMed] [Google Scholar]
- 8.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257-61. 10.1073/pnas.0510565103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chim SS, Shing TK, Hung EC, et al. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem 2008;54:482-90. 10.1373/clinchem.2007.097972 [DOI] [PubMed] [Google Scholar]
- 10.Hunter MP, Ismail N, Zhang X, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One 2008;3:e3694. 10.1371/journal.pone.0003694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513-8. 10.1073/pnas.0804549105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research N , Linehan WM, Spellman PT, et al. Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. N Engl J Med 2016;374:135-45. 10.1056/NEJMoa1505917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wach S, Nolte E, Theil A, et al. MicroRNA profiles classify papillary renal cell carcinoma subtypes. Br J Cancer 2013;109:714-22. 10.1038/bjc.2013.313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Youssef YM, White NM, Grigull J, et al. Accurate molecular classification of kidney cancer subtypes using microRNA signature. Eur Urol 2011;59:721-30. 10.1016/j.eururo.2011.01.004 [DOI] [PubMed] [Google Scholar]
- 15.Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem 2010;56:1733-41. 10.1373/clinchem.2010.147405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellinger J, Gevensleben H, Muller SC, et al. The emerging role of non-coding circulating RNA as a biomarker in renal cell carcinoma. Expert Rev Mol Diagn 2016;16:1059-65. 10.1080/14737159.2016.1239531 [DOI] [PubMed] [Google Scholar]
- 17.Fendler A, Stephan C, Yousef GM, et al. The translational potential of microRNAs as biofluid markers of urological tumours. Nat Rev Urol 2016;13:734-52. 10.1038/nrurol.2016.193 [DOI] [PubMed] [Google Scholar]
- 18.Fedorko M, Stanik M, Iliev R, et al. Combination of MiR-378 and MiR-210 Serum Levels Enables Sensitive Detection of Renal Cell Carcinoma. Int J Mol Sci 2015;16:23382-9. 10.3390/ijms161023382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redova M, Poprach A, Besse A, et al. MiR-210 expression in tumor tissue and in vitro effects of its silencing in renal cell carcinoma. Tumour Biol 2013;34:481-91. 10.1007/s13277-012-0573-2 [DOI] [PubMed] [Google Scholar]
- 20.Wulfken LM, Moritz R, Ohlmann C, et al. MicroRNAs in renal cell carcinoma: diagnostic implications of serum miR-1233 levels. PLoS One 2011;6:e25787. 10.1371/journal.pone.0025787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfeffer SR, Yang CH, Pfeffer LM. The Role of miR-21 in Cancer. Drug Dev Res 2015;76:270-7. 10.1002/ddr.21257 [DOI] [PubMed] [Google Scholar]
- 22.Javanmardi S, Aghamaali MR, Abolmaali SS, et al. miR-21, An Oncogenic Target miRNA for Cancer Therapy: Molecular Mechanisms and Recent Advancements in Chemo and Radio-resistance. Curr Gene Ther 2017;16:375-89. 10.2174/1566523217666170102105119 [DOI] [PubMed] [Google Scholar]
- 23.Cheng T, Wang L, Li Y, et al. Differential microRNA expression in renal cell carcinoma. Oncol Lett 2013;6:769-76. 10.3892/ol.2013.1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tusong H, Maolakuerban N, Guan J, et al. Functional analysis of serum microRNAs miR-21 and miR-106a in renal cell carcinoma. Cancer Biomark 2017;18:79-85. 10.3233/CBM-160676 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as