Abstract
Calf diarrhea is one of the considerable infectious diseases in calves, which results in tremendous economic losses globally. To determine the prevalence of Shiga-toxigenic E. coli (STEC) and Enterotoxigenic E. coli (ETEC) incriminated in calf diarrhea, with special reference to Shiga- toxins genes (stx1 and stx2) and enterotoxins genes (lt and sta) that govern their pathogenesis, as well as the virulence genes; eaeA (intimin) and f41(fimbrial adhesion), and the screening of their antibiogram and antimicrobial resistance genes; aadB, sul1, and bla-TEM, a total of 274 fecal samples were collected (April 2018–Feb 2019) from diarrheic calves at different farms in El-Sharqia Governorate, Egypt. The bacteriological examination revealed that the prevalence of E. coli in diarrheic calves was 28.8%. The serotyping of the isolated E. coli revealed 7 serogroups; O26, O128, O111, O125, O45, O119 and O91. Furthermore, the Congo red binding test was carried out, where 89.8% of the examined strains (n = 71) were positive. The antibiogram of the isolated strains was investigated; the majority of E. coli serotypes exhibit multidrug resistance (MDR) to four antimicrobial agents; neomycin, gentamycin, streptomycin, and amikacin. Polymerase chain reaction (PCR) was used to detect the prevalence of the virulence genes; stx1, stx2 lt, sta, f41 and eaeA, as well as the antimicrobial resistance genes; aadB, sul1, and bla-TEM. The prevalence of STEC was 20.2% (n = 16), while the prevalence of ETEC was 30.4% (n = 24). Briefly, the Shiga toxins genes; stx1 and stx2, are the most prevalent virulence genes associated with STEC, which are responsible for the pathogenesis of the disease and helped by the intimin gene (eaeA). In addition, the lt gene is the most prevalent enterotoxin gene accompanied by the ETEC strains, either alone or in combination with sta and/or f41 genes. The majority of pathogenic E. coli incriminated in calf diarrhea possesses the aadB resistance gene, followed by the sul1 gene. Enrofloxacin, florfenicol, amoxicillin-clavulanic acid, and ampicillin-sulbactam, are the most effective antimicrobial agents against the isolated STEC and ETEC strains.
Keywords: ETEC, STEC, diarrhea, calves, virulence genes, antimicrobial resistance genes
1. Introduction
Calf diarrhea is one of the most predominant syndromes in newly born animals all over the world, that are associated with remarkable economic losses, high morbidity and mortality rates [1]. The most prevalent bacterial pathogen which is incriminated in young calf’s diarrhea is Escherichia. coli (E. coli); moreover, the most common viral causes are Rotavirus and Coronavirus [2].
Based on the molecular and pathological criteria, the most common pathotypes of E. coli incriminated in neonatal colibacillosis are; Shiga-toxigenic E. coli (STEC), Enterotoxigenic E. coli (ETEC), and Enterohemorrhagic E. coli (EHEC) [3]. ETEC is a common pathotype associated with infectious diarrhea in calves. The newly born calves exhibit a high affinity to ETEC, which is associated with watery diarrhea. They colonize the small intestine after fimbrial adhesion and predispose to severe watery diarrhea [4]. Two main virulent factors are included; the fimbriae and the enterotoxins [5]. ETEC is skillful in generating 2 major types of enterotoxins; heat-labile (LT) and heat-stable (STa and STb) enterotoxins, in both man and animal [6].
Ruminants are considered the main reservoir of STEC; the severity of infection in ruminants varies depending upon the animal age, immunity, and the gastrointestinal tract conditions. Certain animals undergo the exaggerated shedding of STEC (>104 CFU/g fecal matter), which results in the contamination of the environment and the transmission of the infection [7]. A large number of STEC-outbreaks were reported, due to the ingestion of contaminated vegetables and fruits with the animal feces. STEC infection in humans is mainly associated with hemolytic uremic syndrome and hemorrhagic colitis. STEC has major public health importance, since it is incriminated in causing several food-borne outbreaks [8].
STEC are associated with dysentery in young calves. They produce two various potent types of Shiga-toxins; Stx1 and Stx2, and certain types of STEC have the ability to produce the intimin. The E. coli strains, which possess the eaeA gene and do not produce stx1 and stx2 genes, were defined as Enteropathogenic E. coli [9,10]. The bacteriophages play a major role in the transmission of stx genes. The stx-phages are sharing an identical sequence that analogous to lambdoid-phages. The presence of stx genes in the phage lysis-portion illustrates the link between the production of Shiga-toxins and the release of phage during the lytic growth [3,10].
The antimicrobial resistance is usually associated with pathogenic E. coli that could be attributed to the widespread improper use of antibiotics. The multidrug resistance (MDR) is common in E. coli and primarily associated with several genes like; bla-TEM, blaCTX (β-lactamase genes), sul1 (sulfonamide resistance gene), and aadB (aminoglycoside resistance gene) [11,12].
The current study was performed to investigate the prevalence of Shiga-toxigenic and Enterotoxigenic E. coli incriminated in calf diarrhea, with particular reference to Shiga-toxins genes (stx1 and stx2) and enterotoxins genes (lt and sta) that govern their pathogenesis, as well as the virulence genes; eaeA and f41. In addition, the screening of their antibiogram and antimicrobial resistance genes; aadB (aminoglycosides-resistance gene), sul1 (sulfonamides-resistance gene), and bla-TEM (Extended β-lactamase gene) is conducted, in order to select the antibiotics of choice.
2. Results
2.1. Prevalence and Phenotypic Identification of E. coli
The bacteriological examination of 274 fecal swabs obtained from diarrheic calves revealed that the overall prevalence of E. coli was 28.8% (n = 79), based on the microscopical examination, colonial characters on MacConkey’s agar and eosin-methylene-blue agar, as well as biochemical tests. Concerning the age of the examined calves, the results revealed that 15 diarrheic calves were infected with E. coli in the first 2 months of age (33.3%), 41 calves at 2–4 months old (28.5%), and 23 calves at 4–6 months old (27.1%) (Table 1).
Table 1.
Prevalence of pathogenic E. coli in diarrheic calves at different ages.
| Ages (Months) | Number of Samples | No. of E. coli | Prevalence (%) |
|---|---|---|---|
| 1–2 days | 45 | 15 | 33.3 |
| 2–4 | 144 | 41 | 28.5 |
| 4–6 | 85 | 23 | 27.1 |
| Total | 274 | 79 | 28.8 |
p value = 0.74 (Not significant; p > 0.05).
2.2. Serotyping of E. coli Isolates
In the present study, the serological identification of the retrieved isolates revealed that a total of 64 (81.01%) of the isolated strains were typable with O antisera, while 15 isolates (18.99%) were untypable. The most common serogroup was O128 (16.5%), followed by O111 (13.9%) and O26 (11.4%), O125 (11.4%), O91 (10.1%), O45 (8.9%), and O119 (8.9%). The frequency of E. coli serogroups was illustrated in Table 2. There is no statistically significant difference in the prevalence of E. coli serogroups (p > 0.05).
Table 2.
Frequency of E. coli serogroups in diarrheic calves.
| Serotypes | Number (%) | |
|---|---|---|
| O 128 | 13 (16.5) | |
| O 111 | 11(13.9) | |
| O 125 | 9 (11.4) | |
| O 26 | 9 (11.4) | |
| O 91 | 8 (10.1) | |
| O 45 | 7 (8.9) | |
| O 119 | 7 (8.9) | |
| Total | Typable | 64 (81.01) |
| Untypable | 15 (18.99) | |
p value = 0.73 (Not significant; p > 0.05).
2.3. Congo Red (CR) Binding Test
The Congo red test revealed that 89.8% of the tested E. coli strains (n = 71) were positive, including the members of serotypes; O26, O111, O125, O128, O45, O119 and 15 untypable strains, while the strains of the O91 serogroup were CR negative (n = 8).
2.4. The Antimicrobial Resistance Profiles and the Antimicrobial Resistance Genes of the Isolated E. coli Strains
The antimicrobial susceptibility testing of the isolated E. coli strains (Table 3) showed a remarkable resistance to neomycin (96.2%), gentamycin and streptomycin (95%), and amikacin (93.7%). Furthermore, substantial sensitivity was recorded to enrofloxacin (84.9%), florfenicol (82.4%), and both amoxicillin/clavulanic acid and ampicillin/sulbactam (78.5%, each). The statistical analysis proved that the resistance of the tested strains against various antimicrobial agents was significantly different (p < 0.0001). Regarding the occurrence of the multidrug-resistance and the distribution of the antimicrobial resistance genes among the isolated strains, we noticed that 41.8% of the tested E. coli strains (n = 33) exhibited multidrug resistance to neomycin, gentamicin, streptomycin, and amikacin and harbored the aadB resistance gene. Moreover, 27.8% of the examined strains (n = 22) showed multidrug-resistance to neomycin, gentamicin, streptomycin, amikacin, and trimethoprim/sulfamethoxazole, and harbored both aadB and sul1 resistance genes. In addition, 21.5% of the tested strains (n = 17) exhibited a multidrug-resistance to neomycin, gentamicin, streptomycin, amikacin, amoxicillin/clavulanic acid, and ampicillin/sulbactam, and harbored both aadB and blaTEM resistance genes (Table 4).
Table 3.
In Vitro susceptibility pattern of the isolated E. coli strains (n = 79) against different antimicrobial agents.
| Antibiotics | Sensitive | Intermediate | Resistant | |||
|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | |
| Amoxicillin/Clavulanic acid | 62 | 78.5 | 0 | 0 | 17 | 21.5 |
| Ampicillin/Sulbactam | 62 | 78.5 | 0 | 0 | 17 | 21.5 |
| Enrofloxacin | 67 | 84.9 | 7 | 8.8 | 5 | 6.3 |
| Trimethoprim/Sulfamethoxazole | 30 | 38 | 20 | 25.3 | 29 | 36.7 |
| Gentamycin | 0 | 0 | 4 | 5 | 75 | 95 |
| Neomycin | 0 | 0 | 3 | 3.8 | 76 | 96.2 |
| Florfenicol | 65 | 82.4 | 10 | 12.6 | 4 | 5 |
| Streptomycin | 0 | 0 | 4 | 5 | 75 | 95 |
| Amikacin | 0 | 0 | 5 | 6.3 | 74 | 93.7 |
|
Chi-Square value
p value |
442.4663 p < 0.0001 |
56.4924 p < 0.0001 |
468.007 p < 0.0001 |
|||
Table 4.
The distribution of the multidrug resistance patterns and the antimicrobial resistance genes among the tested E. coli strains (n = 79).
| No of Isolates | % of Isolates | The Multidrug Resistance Patterns | The Antimicrobial Resistance Genes |
|---|---|---|---|
| 33 | 41.8 | Neomycin, gentamicin, streptomycin, and amikacin | aadB |
| 22 | 27.8 | Neomycin, gentamicin, streptomycin, amikacin, and trimethoprim/sulfamethoxazole | aadB, sul1 |
| 17 | 21.5 | Neomycin, gentamicin, streptomycin, amikacin, amoxicillin/clavulanic acid, and ampicillin/sulbactam | aadB, blaTEM |
| 3 | 3.8 | Neomycin, gentamicin, streptomycin, trimethoprim/sulfamethoxazole, enrofloxacin, and florfenicol | sul1 |
| 2 | 2.5 | Amikacin, trimethoprim/sulfamethoxazole, and enrofloxacin | sul1 |
| 1 | 1.3 | Neomycin, trimethoprim/sulfamethoxazole, and florfenicol | sul1 |
| 1 | 1.3 | Trimethoprim/sulfamethoxazole | sul1 |
Concerning the distribution of the antimicrobial resistance genes; the aadB gene is the most predominant antimicrobial resistance gene associated with the retrieved E. coli strains, either alone or in combination with the sul1 gene (27.8%), or in combination with the blaTEM gene (21.5%). Moreover, the sul1 gene was determined alone in 8.8% of the examined strains. Statistically, there is a significant difference in the distribution of the antimicrobial resistance genes among the examined E. coli strains (p < 0.05) (Table 5 and Table 6, and Figure 1).
Table 5.
The distribution of antimicrobial resistance genes in E. coli strains isolated from diarrheic calves (n = 79).
| Antimicrobial Resistance Genes | E. coli | |
|---|---|---|
| No | % | |
| sul1, aadB | 22 | 27.8 |
| aadB | 18 | 22.7 |
| aadB, blaTEM | 17 | 21.5 |
| sul1 | 7 | 8.8 |
| aadB | 15 (untypable strains) | 19 |
p value = 0.04 (Significant; p < 0.05).
Table 6.
The distribution of virulence and antimicrobial resistance genes among the isolated E. coli serotypes.
| Target Genes | O 128 n = 13 |
O 111 n = 11 |
O 125 n = 9 |
O 26 n = 9 |
O 91 n = 8 |
O 45 n = 7 |
O 119 n = 7 |
Untypable n = 15 |
Total | |
|---|---|---|---|---|---|---|---|---|---|---|
| Virulence genes | stx1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 4 |
| stx2 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 3 | |
| stx1 + stx2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | |
| stx1 + stx2 + eaeA | 2 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 7 | |
| lt, f41 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 0 | 10 | |
| sta | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 0 | 7 | |
| lt, sta | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 4 | |
| lt, f41, sta | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 3 | |
| Total | 9 | 4 | 5 | 9 | 5 | 3 | 5 | 0 | 40 | |
| Antibiotic resistance genes | sul1, aadB | 6 | 4 | 4 | 4 | 1 | 1 | 2 | 0 | 22 |
| aadB | 3 | 3 | 2 | 3 | 2 | 2 | 3 | 15 | 33 | |
| aadB, blaTEM | 3 | 4 | 2 | 1 | 3 | 3 | 1 | 0 | 17 | |
| sul1 | 1 | 0 | 1 | 1 | 2 | 1 | 1 | 0 | 7 | |
| Total | 13 | 11 | 9 | 9 | 8 | 7 | 7 | 15 | 79 | |
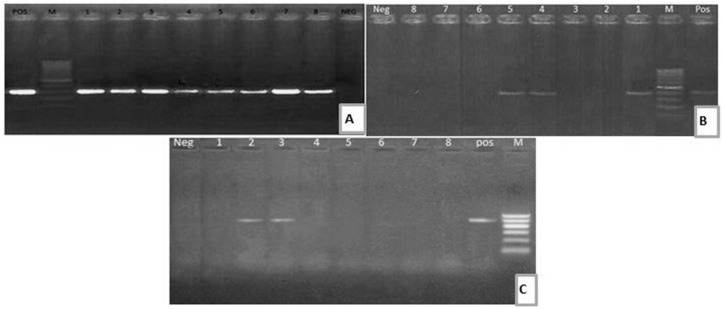
Figure 1.
(A) Electrophoretic pattern of aadB (319 bp): M: 100–1000 bp DNA ladder; POS: Positive control; Neg: Negative control; Lanes: 1–8: positive E. coli strains. (B) Electrophoretic pattern of sul1 gene (433 bp): M: 100–1000 bp DNA ladder, POS: Positive control; Neg: Negative control; Lanes: 1, 4, 5: positive E. coli strains; Lanes: 2, 3, 6, 7, 8: negative E. coli strains. (C) Electrophoretic pattern of blaTEM gene (516 bp): M: 100–600 bp DNA ladder; POS: Positive control; Neg: Negative control; Lanes: 2–3: positive E. coli strains; Lanes: 1, 4, 5, 6, 7, 8: negative E. coli strains.
2.5. The Distribution of Virulence Genes Among the Isolated E. coli Strains
Regarding the distribution of the virulence genes among the examined E. coli strains, the PCR revealed two pathotypes (40/79, 50.6%); Shiga-toxigenic E. coli (STEC) (16/79, 20.2%) and Enterotoxigenic E. coli (24/79, 30.4%). Regarding the STEC; the stx1 gene is the most predominant Shiga-toxin gene, either found alone (n = 4) or in combination with stx2 and eaeA genes (n = 7), or in combination with the stx2 gene (n = 2), while the stx2 gene was detected alone in three E. coli strains. Concerning the ETEC; the lt gene was the most prevalent enterotoxin gene, either found in combination with the f41 gene (n = 10), or in combination with sta gene (n = 4), or in combination with sta and f41 genes (n = 3). In addition, the sta gene was detected alone in seven E. coli strains (Table 6 and Table 7, and Figure 2 and Figure 3). There is no statistically significant difference in the distribution of virulence genes among the isolated E. coli strains (p > 0.05).
Table 7.
The prevalence of the virulence genes in E. coli strains isolated from diarrheic calves (n = 79).
| Pathogenic E. coli | Virulence Genes | E. coli | p-Value | |
|---|---|---|---|---|
| No | % | |||
| Shiga-toxigenic E. coli (STEC) | stx1 | 4 | 5 | 0.19 NS |
| stx2 | 3 | 3.7 | ||
| stx1, stx2 | 2 | 2.5 | ||
| stx1, stx2, eaeA | 7 | 8.9 | ||
| Sub-total | 16 | 20.2 | ||
| Enterotoxigenic E. coli (ETEC) | lt, f41 | 10 | 12.7 | 0.09 NS |
| sta | 7 | 8.9 | ||
| lt, sta | 4 | 5 | ||
| lt, f41, sta | 3 | 3.7 | ||
| Sub-total | 24 | 30.4 | ||
| Total | 40 | 50.6 | ||
NS = Not significant.
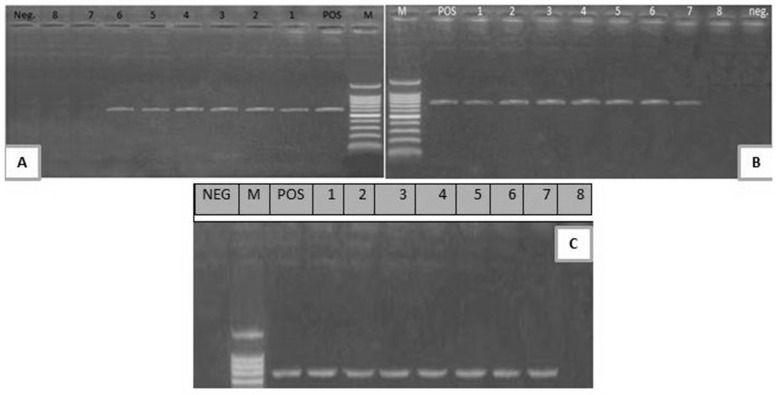
Figure 2.
(A) Electrophoretic pattern of stx1 gene (614 bp); M: 100–1000 bp DNA ladder; POS: Positive control; Neg: Negative control; Lanes 1–6: positive E. coli strains; Lanes 7, 8: negative E. coli strains. (B) Electrophoretic pattern of stx2 gene (779 bp); M: 100–1000 bp DNA ladder, POS: Positive control; Neg: Negative control; Lanes 1–7: positive E. coli strains; Lane 8: negative E. coli strain. (C) Electrophoretic pattern of eaeA gene (248 bp): M: 100–600 bp DNA ladder; POS: Positive control; NEG: Negative control; 1–7: positive E. coli strains; Lane 8: negative E. coli strain.
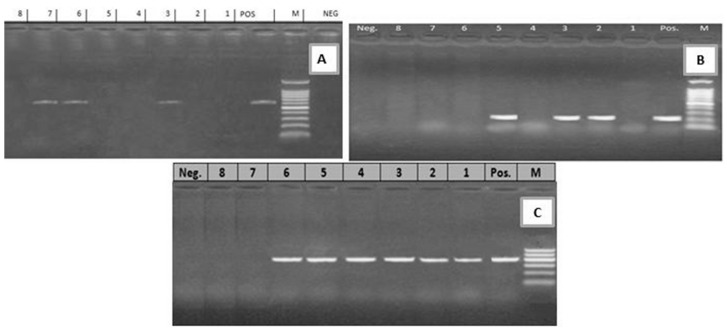
Figure 3.
(A) Electrophoretic pattern of lt gene (605 bp): M: 100–1000 bp DNA ladder; POS: Positive control; Neg: Negative control; Lanes: 3, 6, 7: positive E. coli strains; Lanes 1, 2, 4, 5, 8: negative E. coli strains. (B) Electrophoretic pattern of sta gene (219 bp): M: 100–1000 bp DNA ladder, POS: Positive control; Neg: Negative control; Lanes: 2, 3, 5: positive E. coli strains; Lanes 1, 4, 6, 7, 8: negative E. coli strains. (C) Electrophoretic pattern of f41 gene (380 bp): M: 100–600 bp DNA ladder, POS: Positive control; Neg: Negative control; Lanes: 1–6: positive E. coli strains; Lanes 7, 8: negative E. coli strains.
3. Discussion
This study was aimed at determining the prevalence of STEC and ETEC incriminated in calf diarrhea, with special reference to the Shiga-toxins genes (stx1 and stx2) and enterotoxins genes (lt and sta) that govern their pathogenesis, as well as the virulence genes; eaeA and f41, and the screening of their antimicrobial resistance profiles and antimicrobial resistance genes; aadB, sul1, and bla-TEM. The overall prevalence of E. coli was 28.8%, which is lower than what was reported in diarrheic calves in Egypt (63.6%) [13], Ethiopia (36.8%) [14], Argentina (30.1%) [15] and India (85.04%) [16]; however, a lower prevalence was reported in other previous studies in Korea (22%) [17] and Switzerland (5.5%) [18]. Differences in the prevalence of E. coli may be due to the differences in geography, management practices, floor type, health conditions, and the calf’s age [2,13,15]. Further, the high rate of E. coli isolation in the present study could be attributed to many reasons, such as the mixing of different age groups, poor environmental and hygienic conditions, or the poor quantity and/or quality of colostrum. In addition, the colostrum maternal antibodies cannot neutralize the high dose of pathogenic E. coli infection [14].
Regarding the age of the examined calves, the prevalence of E. coli was 33.3%, 28.5%, and 27.1% in the first 2 months old, 2–4 months old, and 4–6 months old aged calves, respectively. There is no statistically significant difference in the prevalence of E. coli among the different ages (p = 0.74). A previous study reported that the prevalence of E. coli was high in the young calves, and then decreased as the age increased [14].
The serological identification of the retrieved isolates revealed that a total of 64 (81.01%) strains were typable, while 15 isolates (18.99%) were untypable. The most common serogroup was O128 (16.5%), followed by O111 (13.9%) and O26 (11.4%), O125 (11.4%), O91 (10.1%), O45 (8.9%), and O119 (8.9%). The identified serogroups in the current study have a different variety than those previously recorded [13,19,20,21], which are often associated with sick children with diarrhea [19,22].
In the current study, 89.8% of the E. coli strains were CR positive, including serogroups O26, O111, O125, O128, O45, O119 and 15 untypable strains; a finding endorsed by a previous study [23] stated that 95% of E. coli isolates are pathogenic by CR binding, which indicates the virulence of these strains. In other studies, it was reported that 61.9–90% of E. coli isolates were CR positive [24,25].
In the present study, the isolated strains exhibited a remarkable resistance against four antimicrobial agents, including; neomycin (96.2%), gentamycin and streptomycin (95%), and amikacin (93.7%). A previous study from Egypt in diarrheic buffalo-calves farms reported that the most prevalent phenotypic resistance patterns were; ampicillin (71.4%) and amoxicillin (64.3%), as well as trimethoprim/sulfamethoxazole (50%) and gentamicin (42.8%) [26]. A previous study in Iran reported that E. coli strains retrieved from diarrheic calves showed maximum resistance to penicillin, streptomycin, tetracycline, lincomycin and sulfamethoxazole [27]. The resistance pattern observed in our study and the previous other studies indicated that the emergence of multidrug-resistant species has become an increasing problem worldwide, due to the overuse of antimicrobial drugs in both animal and human medicine [28,29]. The increased pattern of multidrug resistance could be attributed to the accumulation of genes encoding for the antibiotic resistance, either on the bacterial chromosome or plasmid [12].
Concerning the occurrence of the multidrug-resistance patterns and the distribution of the antimicrobial resistance genes among the isolated E. coli strains, 41.8% of the tested strains showed multidrug resistance to the aminoglycosides antibiotics; neomycin, gentamicin, streptomycin, and amikacin (harbored aadB gene), while 27.8% exhibited multidrug resistance to the aminoglycosides antibiotics and trimethoprim/sulfamethoxazole (harbored both aadB and sul1 genes). Although clavulanic acid and sulbactam are known as effective β-lactamase inhibitors, 21.5% of the tested strains showed multidrug resistance to amoxicillin/clavulanic acid, ampicillin/sulbactam, and the aminoglycosides antibiotics (harbored both aadB and blaTEM genes). In addition, the most predominant antimicrobial resistance gene was aadB gene, which occurred in 72/79 of the examined strains. These findings are endorsed by a previous study, which reported that an aminoglycoside-resistance determinant (aadB) gene was prevalent among the pathogenic E. coli strains originated from diarrheic calves. Such type of resistance could be attributed to the presence of aadB gene, as well as the widespread improper use of aminoglycosides for the treatment of calf diarrhea in the past years [30]. Genes encoding resistance to aminoglycosides, sulfonamides, and ampicillins were aadB, sul1, and blaTEM genes, respectively [31]. A previous study revealed that the most usually detected β- lactamase gene is blaTEM, occurring in 66/74 of the tested strains; the resistance to the antimicrobial agents could be due to the presence of variable gene variants in the resistant isolates [32].
In the present study, the prevalence of the Enterotoxigenic E. coli was 30.4%, while the prevalence of Shiga-toxigenic E. coli was 20.2%. Concerning the isolated STEC strains, the stx1 gene is the most predominant Shiga-toxin gene, either found alone or in combination with stx2 and eaeA genes, or in combination with the stx2 gene. A previous study from diarrheic calves in India reported that the profile of virulence genes of the STEC isolates was found in diverse combinations, and the combination of hlyA and eaeA genes was most the predominant [16]. Shiga toxins usually inactivate the host cell-ribosomes with subsequent inhibition of protein-biosynthesis. The occurrence of the stx1, eaeA, and stx2 genes together constitute an epidemiological significance, as previous studies reported that the combination of these genes could increase the ability of E. coli to cause severe human illness [16,33].
Regarding the isolated ETEC strains, the lt gene was the most prevalent enterotoxin gene, either found in combination with the f41 gene, or in combination with the sta gene, or in combination with both sta and f41 genes. LT (heat-labile toxin) usually stimulates the adenylate-cyclase enzymatic system, while STa (stable-toxin) activates the guanylate-cyclase system, resulting in severe watery diarrhea. The f41 gene is usually associated with the occurrence of diarrhea in calves and has a higher prevalence in diarrheic calves rather than healthy ones, which warrants a great role of this gene in ETEC pathogenesis [34].
In Argentina, the most prevalent virulence genes of pathogenic E. coli isolated from dairy calves were k99, f41 and f5 [15]. In Brazil, the prevalence of the combination of sta and lt genes was 3.9% in E. coli isolated from diarrheic calves [35]. In China, 15.5% of the ETEC which originated from healthy calves were carried lt and sta genes [36]. Furthermore, in Italy, the sta gene was absent in E. coli that isolated from diarrheic buffalo calves [37]. The diversity in the prevalence of Shiga-toxins genes, enterotoxins genes, and other virulence-related genes in the present study and the other studies may be attributed to the geographical origin of samples, the sample size, the handling of collected samples, the number of examined strains, the type of the examined virulence genes, and the role of the examined virulence genes in the pathogenesis of the disease.
In conclusion, E. coli continues to be one of the major causes of calf diarrhea resulting in severe economic losses. Both Shiga-toxigenic and Enterotoxigenic E. coli are the most prevalent pathotypes incriminated in the disease occurrence. The Shiga toxins genes; stx1 and stx2, are the most prevalent virulence genes associated with STEC, which are responsible for the pathogenesis of the disease and helped by the intimin gene (eaeA). Furthermore, the lt gene is the most prevalent enterotoxin gene which accompanies the ETEC strains isolated from calf diarrhea, either alone or in combination with the sta and or the f41 genes. The majority of the isolated STEC and ETEC harbored aadB antibiotic resistance gene and exhibited a multidrug-resistance pattern to neomycin, gentamycin, streptomycin, and amikacin. Moreover, Enrofloxacin, florfenicol, amoxicillin-clavulanic acid, and ampicillin-sulbactam are the most effective antimicrobial agents against the isolated STEC and ETEC strains. The incessant implementation of the antibiogram profile is needed to determine the most effective antibiotic, due to the high prevalence of the multidrug-resistant E. coli strains.
4. Materials and Methods
4.1. Sampling
A total of 274 fecal specimens were aseptically gathered using sterile rectal swabs from April 2018–Feb 2019, from diarrheic calves aged between 1 day and 6 months, from two private farms (124 calves from farm I and 150 from farm II) at El-Sharqia governorate, Egypt. The collected specimens were rapidly transferred to the bacteriological lab, Faculty of Veterinary Medicine, Suez Canal University, for further analysis. The handling of animals was carried out by well-trained scientists, according to the instructions of the Animal Ethics Review Committee of Suez Canal University (SCU-362-27/12/2016).
4.2. Isolation and Identification of E. coli
The collected specimens were inoculated in nutrient broth and then incubated for 24 h at 37 °C. Loopful from the culture suspension was streaked on MacConkey’s agar and Eosin methylene blue agar (Oxoid, UK). The recovered typical colonies (pink colonies on MacConkey’s agar and metallic green sheen colonies on EMB) were completely identified morphologically and biochemically, as described by Quinn et al. [38].
4.3. Serotyping of Isolated E. coli
The obtained E. coli isolates were serogrouped by the detection of O antigens using the slide agglutination test, according to the method previously described by Edwards and Ewing [39].
4.4. Congo Red Test
The Congo red (CR) binding test has been used to detect the invasive E. coli, using trypticase agar supplemented with 0.03% CR dye (Oxoid, UK). The colonies were streaked on Congo red agar and incubated at 37 °C for 24 hrs, then plates were kept at room temperature for 48 hrs. The demonstration of the invasive strains (red colonies) was observed and recorded according to the methods previously described by Panigrahy and Yushen [24].
4.5. Antimicrobial Susceptibility Testing
The isolated E. coli strains were tested against nine antimicrobial agents; amoxicillin/clavulanic acid (20/10 mcg), ampicillin/sulbactam (10/10 mcg), amikacin (10 mcg), neomycin (30 mcg), enrofloxacin (5 mcg), florfenicol (30 mcg), streptomycin (10 mcg), trimethoprim/sulfamethoxazole (25 mcg) and gentamicin (30 mcg) (Oxoid, UK), using the disc diffusion method. The diameter of the inhibition zone was measured in millimeters and expressed as sensitive, intermediate, and resistant, as described by CLSI [40].
4.6. PCR Based Detection of Virulence Genes and Antmicrobial Resistance Genes
Bacterial DNA of purified bacterial cells was extracted using the QIAamp DNA Mini Kit (Invitrogen, USA). Recovered DNA templates were quantified using a Nanodrop (Nanodrop 1000, Thermo Scientific, Loughborough, UK), adjusted to 100 ng μL−1. To assess the virulence genes (stx1, stx2 lt, sta, f41 and eaeA), as well as the antimicrobial resistance genes (aadB, sul1, and bla-TEM) in the obtained E. coli strains, PCR was performed using specific sets of primers (Metabion, Germany) (Table 8). The PCR reaction (25 μL) consists of 12.5 μL Go Taq® Green Master Mix 2X (Promega, Wisconsin, USA), 1 μL (20 pmol) of each primer, 5 μL DNA extract, and PCR grade water up to 25 μL. The cycling conditions are listed in Table 8. Negative control (no DNA template) and positive control reference strains (previously isolated and kindly provided by A.H.R I, Dokki, Egypt) were used in the PCR assay. Amplified fragments were screened by 1.5% agarose gel electrophoresis (Applichem GmbH, Darmstadt, Germany) for 45 min at 100 V in 1× TAE, visualized using 15 µL of DNA gel stain (Sigma-Aldrich, St. Louis, MI, USA) and photographed under UV transilluminator. A 100 bp ladder (Fermentas, Thermo Scientific, Darmstadt, Germany) was used.
Table 8.
Primers sequences and recycling conditions of PCR assays used for the detection of virulence genes and antimicrobial resistance genes.
| Target Gene | Oligonucleotide Sequence (5′-3′) | Product Size (bp) | Denat. | Cycling (35) | Final E |
References | ||
|---|---|---|---|---|---|---|---|---|
| D | A | E | ||||||
| eaeA | F: GTGGCGAATACTGGCGAGACT R: CCCCATTCTTTTTCACCGTCG |
248 | 95 °C 5 min |
95 °C 30 s |
60 °C 30 s |
72 °C 30 s |
72 °C 10 min |
[41] |
| stx1 | F: ACACTGGATGATCTCAGTGG R: CTGAATCCCCCTCCATTATG |
614 | 95 °C 5 min |
95 °C 3 min |
59 °C 45 s |
72 °C 90 s |
72 °C 10 min |
[42] |
| stx2 | F: CCATGACAACGGACAGCAGTT R: CCTGTCAACTGAGCAGCACTTTG |
779 | 95 °C 5 min |
95 °C 3 min |
59 °C 45 s |
72 °C 90 s |
72 °C 10 min |
[42] |
| sta | F: GAAACAACATGACGGGAGGT R: GCACAGGCAGGATTACAACA |
229 | 94 °C 5 min |
94 °C 30 s |
57 °C 45 s |
72 °C 45 s |
72 °C 10 min |
[43] |
| lt | F: GGTTTCTGCGTTAGGTGGAA R: GGGACTTCGACCTGAAATGT |
605 | 94 °C 5 min |
94 °C 30 s |
57 °C 45 s |
72 °C 45 s |
72 °C 10 min |
[43] |
| f41 | F: GCATCAGCGGCAGTATCT R: GTCCCTAGCTCAGTATTATCACCT |
380 | 94 °C 5 min |
94 °C 30 s |
50 °C 45 s |
72 °C 90 s |
72 °C 10 min |
[44] |
| aadB | F: GAGCGAAATCTGCCGCTCTGG R: CTGTTACAACGGACTGGCCGC |
319 | 94 °C 5 min |
94 °C 30 s |
55 °C 45 s |
72 °C 45 s |
72 °C 10 min |
[45] |
| blaTEM | F: ATCAGCAATAAACCAGC R: CCCCGAAGAAC GTTTTC |
516 | 94 °C 5 min |
94 °C 30 s |
55 °C 45 s |
72 °C 45 s |
72 °C 10 min |
[46] |
| sul1 | F: CGGCGTGGGCTACCTGAACG R: GCCGATCGCGTGAAGTTCCG |
433 | 94 °C 5 min |
94 °C 30 s |
55 °C 45 s |
72 °C 45 s |
72 °C 10 min |
[47] |
D, Denaturation; A, Annealing; E, Extension.
4.7. Statistical Analysis
The Chi-square was performed to analyze the data, to test the null hypothesis of different treatments using the statistical analysis software (SAS® software version 9.4, SAS Institute, Cary, NC, USA). The significance level was (p < 0.05).
Acknowledgments
The authors are grateful to the Researchers Supporting Project number (RSP-2020/53), King Saud University, Riyadh, Saudi Arabia.
Author Contributions
A.M.A. and E.M.R. Designed the study; A.M.A., E.M.R., H.E.M., A.W.E.-K. carried out the Methodology; A.M.A, S.A.A.Y., and W.N.H. Drafted the manuscript; A.M.A., M.M.E., H.E.M., A.W.E.-K., S.A.A.Y., M.O.I.G. and W.N.H Data Analysis, Data Accuracy, Validation, Investigation and Statistical Analysis; A.M.A. Writing, Review and Editing. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Key Contribution
The Shiga toxins genes; stx1 and stx2, are the most prevalent virulence genes associated with Shiga-toxigenic E. coli (STEC), which are responsible for the pathogenesis of the disease and helped by the intimin gene (eaeA). Furthermore, the heat-labile enterotoxin (lt) gene is the most prevalent enterotoxin gene and accompanies the Enterotoxigenic E. coli (ETEC) strains isolated from calf diarrhea, either alone or in combination with the sta and/or the f41 genes.
References
- 1.Tajik J., Nazifi S., Naghib S.M., Ghasrodashti A.R. Comparison of electrocardiographic parameters and serum electrolytes and microelements between single infection of Rotavirus and Coronavirus and concurrent infection of Cryptosporidium parvum with Rotavirus and Coronavirus in diarrheic dairy calves. Comp. Haematol. Int. 2010;21:241–244. doi: 10.1007/s00580-010-1084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho Y.-I., Yoon K.-J. An overview of calf diarrhea-infectious etiology, diagnosis, and intervention. J. Vet. Sci. 2014;15:1–17. doi: 10.4142/jvs.2014.15.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aref N.-E.M., Abdel-Raheem A.-R.A., Kamaly H.F., Hussien S.Z. Clinical and sero-molecular characterization of Escherichia coli with an emphasis on hybrid strain in healthy and diarrheic neonatal calves in Egypt. Open Vet. J. 2018;8:351–359. doi: 10.4314/ovj.v8i4.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foster D.M., Smith G.W. Pathophysiology of diarrhea in calves. Vet. Clin. N. Am. Food Anim. Pract. 2009;25:13–36. doi: 10.1016/j.cvfa.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eid H.M., Algammal A.M., Elfeil W.K., Youssef F.M., Harb S.M., Abd-Allah E.M. Prevalence, molecular typing, and antimicrobial resistance of bacterial pathogens isolated from ducks. Vet. World. 2019;12:677–683. doi: 10.14202/vetworld.2019.677-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olaogun S., Jeremiah O., Jubril A., Adewuyi O. Calf Diarrhea: Epidemiological Prevalence and Bacterial Load in Oyo and Ogun States, Nigeria. Alex. J. Vet. Sci. 2016;51:90. doi: 10.5455/ajvs.232816. [DOI] [Google Scholar]
- 7.Ferens W.A., Hovde C.J. Escherichia coli O157:H7: Animal reservoir and sources of human infection. Foodborne Pathog. Dis. 2011;8:465–487. doi: 10.1089/fpd.2010.0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen V.D., Bennett S.D., Mungai E., Gieraltowski L., Hise K., Gould L.H. Increase in multistate foodborne disease outbreaks—United States, 1973–2010. Foodborne Pathog. Dis. 2015;12:867–872. doi: 10.1089/fpd.2014.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Algammal A.M., Eid H.I., Nasef S.A., Elfeil W., Mansour G.H. Genetic variation among avian pathogenic E. coli Strains isolated from broiler chickens. Asian J. Anim. Vet. Adv. 2016;11:350–356. doi: 10.3923/ajava.2016.350.356. [DOI] [Google Scholar]
- 10.Hashish E.A., El Damaty H.M., Tartor Y.H., Abdelaal A.M. Epidemiological study of diarrheagenic Escherichia coli virulence genes in newborn calves. Pak. Vet. J. 2016;36:54–58. [Google Scholar]
- 11.Algammal A.M., Mahmoud E.E., Fatma M.Y., Shefaa A.S., Mahmoud M.E., Gaber E.B., Wael N.H., Madeha O.I.G. Prevalence, the antibiogram and the frequency of virulence genes of the most predominant bacterial pathogens incriminated in calf pneumonia. AMB Express. 2020;10:1–8. doi: 10.1186/s13568-020-01037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto S., Iwabuchi E., Hasegawa M., Esaki H., Muramatsu M., Hirayama N., Hirai K. Prevalence and molecular epidemiological characterization of antimicrobial-resistant Escherichia coli Isolates from Japanese Black Beef Cattle. J. Food Prot. 2013;76:394–404. doi: 10.4315/0362-028X.JFP-12-273. [DOI] [PubMed] [Google Scholar]
- 13.Enany M., Algammal A.M., Nasef S.A., Abo-Eillil S.A.M., Bin-Jumah M.N., Taha A.E., Allam A. The occurrence of the multidrug resistance (MDR) and the prevalence of virulence genes and QACs resistance genes in E. coli isolated from environmental and avian sources. AMB Express. 2019;9:1–9. doi: 10.1186/s13568-019-0920-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osman K.M., Mustafa A.M., ElHariri M., Abdelhamed G.S. The distribution of Escherichia coli serovars, virulence genes, gene association and combinations and virulence genes encoding serotypes in pathogenic E. coli recovered from diarrhoeic calves, sheep and goat. Transbound Emerg. Dis. 2012;60:69–78. doi: 10.1111/j.1865-1682.2012.01319.x. [DOI] [PubMed] [Google Scholar]
- 15.Gebregiorgis A., Tessema T.S. Characterization of Escherichia coli isolated from calf diarrhea in and around Kombolcha, South Wollo, Amhara Region, Ethiopia. Trop. Anim. Health Prod. 2015;48:273–281. doi: 10.1007/s11250-015-0946-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Picco N.Y., Alustiza F.E., Bellingeri R.V., Grosso M.C., Motta C.E., Larriestra A.J., Vissio C., Tiranti K.I., Terzolo H.R., Moreira A.R., et al. Molecular screening of pathogenic Escherichia coli strains isolated from dairy neonatal calves in Cordoba province, Argentina. Rev. Argent. Microbiol. 2015;47:95–102. doi: 10.1016/j.ram.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivani M., Reddy Y.N., Subramanyam K.V., Reddy M.R., Rao T.S. Prevalence and antimicrobial resistance pattern of Shiga toxigenic Escherichia coli in diarrheic buffalo calves. Vet. World. 2017;10:774–778. doi: 10.14202/vetworld.2017.774-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S.-H., Kim H.-Y., Choi E.W., Kim D. Causative agents and epidemiology of diarrhea in Korean native calves. J. Vet. Sci. 2019;20 doi: 10.4142/jvs.2019.20.e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uhde F.L., Kaufmann T., Sager H., Albini S., Zanoni R., Schelling E., Meylan M. Prevalence of four enteropathogens in the faeces of young diarrhoeic dairy calves in Switzerland. Vet. Rec. 2008;163:362–366. doi: 10.1136/vr.163.12.362. [DOI] [PubMed] [Google Scholar]
- 20.Vu-Khac H., Cornick N.A. Prevalence and genetic profiles of Shiga toxin-producing Escherichia coli strains isolated from buffaloes, cattle, and goats in central Vietnam. Vet. Microbiol. 2008;126:356–363. doi: 10.1016/j.vetmic.2007.07.023. [DOI] [PubMed] [Google Scholar]
- 21.Vagh A., Jani R. Prevalence and comparative studies of some major serotype of E. coli from cattle and buffalo calf scour. Vet. World. 2010;3:458. [Google Scholar]
- 22.Wani S., Hussain I., Beg S., Rather M., Kabli Z., Mir M., Nishikawa Y. Diarrhoeagenic Escherichia coli and Salmonella in calves and lambs in Kashmir: Absence, prevalence and antibiogram. Rev. Sci. Tech. 2013;32:833–840. doi: 10.20506/rst.32.2.2213. [DOI] [PubMed] [Google Scholar]
- 23.Käppeli U., Hächler H., Giezendanner N., Beutin L., Stephan R. Human infections with non-O157 Shiga toxin–producing Escherichia coli, Switzerland. Emerg. Infect. Dis. 2011;17:180–185. doi: 10.3201/eid1702.100909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osman K.M., Hessain A.M., Abo-Shama U.H., Girh Z.M., Kabli S.A., Hemeg H.A., Moussa I.M. An alternative approach for evaluating the phenotypic virulence factors of pathogenic Escherichia coli. Saudi J. Boil. Sci. 2018;25:195–197. doi: 10.1016/j.sjbs.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panigrahy B., Yushen L. Differentiation of pathogenic and nonpathogenic Escherichia coli isolated from poultry. Avian Dis. 1990;34:941. doi: 10.2307/1591387. [DOI] [PubMed] [Google Scholar]
- 26.Mousa M., Akeila M., Khaliel S., Abdellrazeq G. Virulence factors of Escherichia coli isolated from diarrheic sheep and goats. J. Vet. Sci. 2010;30:137–147. [Google Scholar]
- 27.Hakim A., Omara S., Syame S., Fouad E.A. Serotyping, antibiotic susceptibility, and virulence genes screening of Escherichia coli isolates obtained from diarrheic buffalo calves in Egyptian farms. Vet. World. 2017;10:769–773. doi: 10.14202/vetworld.2017.769-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shahrani M., Dehkordi F.S., Momtaz H. Characterization of Escherichia coli virulence genes, pathotypes and antibiotic resistance properties in diarrheic calves in Iran. Biol. Res. 2014;47:28. doi: 10.1186/0717-6287-47-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen M.L. Changing patterns of infectious disease. Nature. 2000;406:762–767. doi: 10.1038/35021206. [DOI] [PubMed] [Google Scholar]
- 30.Szmolka A., Nagy B. Multidrug resistant commensal Escherichia coli in animals and its impact for public health. Front. Microbiol. 2013;4:258. doi: 10.3389/fmicb.2013.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du X., Shen Z., Wu B., Xia S., Shen J. Characterization of class 1 integrons-mediated antibiotic resistance among calf pathogenic Escherichia coli. FEMS Microbiol. Lett. 2005;245:295–298. doi: 10.1016/j.femsle.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen T.D., Vo T.T., Vu-Khac H. Virulence factors in Escherichia coli isolated from calves with diarrhea in Vietnam. J. Vet. Sci. 2011;12:159–164. doi: 10.4142/jvs.2011.12.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jørgensen R.L., Friis-Møller A., Fjeldsøe-Nielsen H., Schønning K., Nielsen J.B. Prevalence and molecular characterization of clinical isolates of Escherichia coli expressing an AmpC phenotype. J. Antimicrob. Chemother. 2010;65:460–464. doi: 10.1093/jac/dkp484. [DOI] [PubMed] [Google Scholar]
- 34.Blanco M., Blanco J., Mora A., Dahbi G., Alonso M.P., Gonzalez E.A., Bernardez M.I., Blanco J. Serotypes, virulence genes, and intimin types of Shiga toxin (Verotoxin)-producing Escherichia coli Isolates from cattle in Spain and identification of a new intimin variant gene (eae-ξ) J. Clin. Microbiol. 2004;42:645–651. doi: 10.1128/JCM.42.2.645-651.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolenda R., Burdukiewicz M., Schierack P. A systematic review and meta-analysis of the epidemiology of pathogenic Escherichia coli of calves and the role of calves as reservoirs for human pathogenic E. coli. Front. Microbiol. 2015;5:23–35. doi: 10.3389/fcimb.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huasai S., Chen A., Wang C.-J., Li Y., Tongrige B. Occurrence and characteristics of virulence genes of Escherichia coli strains isolated from healthy dairy cows in Inner Mongolia, China. Braz. J. Microbiol. 2012;43:528–534. doi: 10.1590/S1517-83822012000200013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borriello G., Lucibelli M., De Carlo E., Auriemma C., Cozza D., Ascione G., Scognamiglio F., Iovane G., Galiero G. Characterization of enterotoxigenic E. coli (ETEC), Shiga-toxin producing E. coli (STEC) and necrotoxigenic E. coli (NTEC) isolated from diarrhoeic Mediterranean water buffalo calves (Bubalus bubalis) Res. Vet. Sci. 2012;93:18–22. doi: 10.1016/j.rvsc.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quinn P.J., Markey B.K., Leonard F.C., Hartigan P., Fanning S., FitzPatrick E. Veterinary Microbiology and Microbial Disease. John Wiley & Sons; Hoboken, NJ, USA: 2011. [Google Scholar]
- 39.Edwards P.R., Ewing W.H. Identification of Enterobacteriaceae. Elsevier Science; Amsterdam, The Netherlands: 1962. [Google Scholar]
- 40.Watts J.L., Shryock T.R., Apley M., Bade D.J., Brown S.D., Gray J.T., Heine H., Hunter R.P., Mevius D.J., Papich M.G., et al. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals: Approved Standard. National Committee for Clinical Laboratory Standards; Dublin, OH, USA: 2008. [Google Scholar]
- 41.Bisi-Johnson M., Obi C.L., Vasaikar S., Baba K.A., Hattori T. Molecular basis of virulence in clinical isolates of Escherichia coli and Salmonella species from a tertiary hospital in the Eastern Cape, South Africa. Gut Pathog. 2011;3:9. doi: 10.1186/1757-4749-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhanashree B., Mallya P.S. Detection of shiga-toxigenic Escherichia coli (STEC) in diarrhoeagenic stool & meat samples in Mangalore, India. Indian J. Med. Res. 2008;128:271. [PubMed] [Google Scholar]
- 43.Lee J.H., Hur J., Stein B. Occurrence and characteristics of enterohemorrhagic Escherichia coli O26 and O111 in calves associated with diarrhea. Vet. J. 2008;176:205–209. doi: 10.1016/j.tvjl.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 44.Franck S.M., Bosworth B.T., Moon H.W. Multiplex PCR for enterotoxigenic, attaching and effacing, and Shiga toxin-producing Escherichia coli strains from calves. J. Clin. Microbiol. 1998;36:1795–1797. doi: 10.1128/JCM.36.6.1795-1797.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frana T.S., Carlson S.A., Griffith R.W. Relative distribution and conservation of genes encoding aminoglycoside-modifying enzymes in Salmonella enterica serotype Typhimurium phage type DT104. Appl. Environ. Microbiol. 2001;67:445–448. doi: 10.1128/AEM.67.1.445-448.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colom K., Pérez J., Alonso R., Fernã¡ndez-Aranguiz A., Lariño E., Cisterna R., Pérez J., Cisterna R. Simple and reliable multiplex PCR assay for detection of blaTEM, blaSHV and blaOXA-1 genes in Enterobacteriaceae. FEMS Microbiol. Lett. 2003;223:147–151. doi: 10.1016/S0378-1097(03)00306-9. [DOI] [PubMed] [Google Scholar]
- 47.Ibekwe A.M., Murinda S.E., Graves A.K. Genetic diversity and antimicrobial resistance of Escherichia coli from human and animal sources uncovers multiple resistances from human sources. PLoS ONE. 2011;6:e20819. doi: 10.1371/journal.pone.0020819. [DOI] [PMC free article] [PubMed] [Google Scholar]