Abstract
The CAGI-5 PCM1 challenge aimed to predict the effect of 38 transgenic human missense mutations in the Pericentriolar Material 1 (PCM1) protein implicated in schizophrenia. Participants were provided with 16 benign variants (negative controls), 10 hypomorphic, and 12 loss of function variants. Six groups participated and were asked to predict the probability of effect and standard deviation associated to each mutation. Here, we present the challenge assessment. Prediction performance were evaluated using different measures to conclude in a final ranking which highlights the strengths and weaknesses of each group. The results show a great variety of predictions where some methods performed significantly better than others. Benign variants played an important role as negative controls, highlighting predictors biased to identify disease phenotypes. The best predictor, Bromberg lab used a neural-network based method able to discriminate between neutral and non-neutral single nucleotide polymorphisms. The CAGI-5 PCM1 challenge allowed us to evaluate the state of the art techniques for interpreting the effect of novel variants for a difficult target protein.
Keywords: Critical assessment, community challenge, missense mutations, effect prediction, variant interpretation, bioinformatics tools
INTRODUCTION
Next generation sequence techniques produce new gene and genome sequences every day, providing lots of genetic information that is still unanalyzed (Niroula & Vihinen, 2016). Furthermore, genetic analysis is performed more frequently to study human diseases and consequently thousands of variants of unknown significance (VUS) appear. The scientific community has been making a big effort in developing computational tools that allow a better interpretation of VUS and genomic information. However, there is still plenty of work which has to be done to improve the current state of art. Critical Assessment of Genome Interpretation (CAGI) experiment has been running since 2010 with the aim to assess the state of art of computational methods which try to predict the phenotypic impact of genomic variations.
Here, we present the assessment of the CAGI-5 Pericentriolar Material 1 (PCM1) challenge. Predictors were asked to predict the pathogenicity of 38 transgenic human missense mutations in the PCM1 gene. The PCM1 gene maps to the human chromosome 8p22. The protein encoded by this gene is localized on centriolar satellites and has an important role in the radial organization of microtubules and the recruitment of proteins to the centrosome (Dammermann & Merdes, 2002; Villumsen et al., 2013). PCM1 is recruited to the centrosome to form a complex with the Bardet-Biedl syndrome 4 (BBS4) and Disrupted in Schizophrenia-1 (DISC1) proteins (Ansley et al., 2003; Guo et al., 2006). Suppression of one of these proteins could lead to neuronal migration defects (Kamiya et al., 2008). PCM1 is a large protein of 2,024 amino acids without known crystal structures. Database annotations in UniProt (The UniProt Consortium, 2017) show several coiled coil regions, while MobiDB (Piovesan et al., 2018) predicts regions of intrinsic disorder accounting for about 40% of the sequence. Linkage analysis has shown that the PCM1 gene has a role in susceptibility to schizophrenia in humans and is associated with orbitofrontal gray matter volumetric deficits (Gurling et al., 2006). Indeed, a candidate pathogenic mutation on this gene has been reported in an affected family (Kamiya et al., 2008). The effects of PCM1 haploinsufficiency have been studied on model animals, whereas affected mice show a significant reduction in brain volume and behavioral alterations (Zoubovsky et al., 2015). In addition to being risk factors for schizophrenia, several studies have also implicated some PCM1 component in genetic susceptibility to cancers and other mental diseases (Kamiya et al., 2008; Zoubovsky et al., 2015).
Ventricular enlargement is one of the most consistent abnormal structural brain findings in schizophrenia. A set of 38 transgenic human PCM1 missense mutations implicated in schizophrenia were assayed in a zebrafish model to determine their impact on the posterior ventricle area. The CAGI challenge aims to predict whether variants implicated in schizophrenia impact zebrafish brain development determining a reduction in the ventricular area of the brain. In particular, in addition to classifying benign variants, predictors have to distinguish between loss of function and hypomorphic variants. This challenge presents new difficulties for current state of the art predictors using different strategies to predict variant effects, while the variability of results suggests that we are far from a general pathogenicity predictor, some groups have promising results in this challenge.
MATERIALS AND METHODS
Experimental data
The Katsanis lab assessed 38 PCM1 missense mutations in a zebrafish model. The native zebrafish embryo PCM1 protein was suppressed by injecting morpholino (MO) antisense oligonucleotides to inhibit translation of mRNA of the PCM1 gene. MOs are stable molecules consisting of a large, non ribose morpholine backbone with four DNA bases pairing stably with mRNA at either the translation start site (to disrupt protein synthesis) or at intron-exon boundaries (to disrupt mRNA splicing) (Summerton & Weller, 1997). Morpholinos have been shown to bind and block translation of mRNA in vitro, in tissue culture cells, and, in vivo (Davis, Frangakis, & Katsanis, 2014). Embryos deficient in PCM1 function show an absence of brain ventricle formation.
For each mutation, the Katsanis lab injected a group of embryos with MO and the mRNA of the human gene carrying the mutation (MO+VAR). Brain ventricle formation of the group of (MO+VAR) animals was compared to brain ventricle formation measured in a group of animals with MO alone and a group with MO+WT. The ventricle space is filled with a fluorescent dye and imaged by brightfield and fluorescence microscopy to access the effect on mutations on ventricle size (Gutzman & Sive, 2009; Niederriter et al., 2013). Each image was processed with an automated image processing tool to quantify the ventricle structure volume (Mikut et al., 2013; Näslund & Johnsson, 2016). P-values for statistically significantly different brain ventricle volumes between pairs of conditions (Lowery, De Rienzo, Gutzman, & Sive, 2009) were obtained using Student’s t-test with a confidence level of 95%. The functional effect of each variant was then assigned as follows. When the p-value for (MO+VAR) is not significantly different from MO (p-value > 0.05), but significantly different from MO+WT (p-value < 0.05), the variant is pathogenic or loss of function. If the p-value (MO+VAR) is significantly different from MO, but not from MO+WT, the variant is benign. When the p-value for (MO+VAR) is significantly different from MO, and also significantly different from MO+WT, the variant is hypomorphic or partial loss of function.
The experiment was performed in duplicate, blind to injection and the experimental data provided by the Katsanis lab is shown in Table 1. The dataset is composed of 16 benign variants (negative controls), 10 hypomorphic, and 12 loss of function variants. In percentages, 42% of the variants are benign and 58% have some functional effect (~32% loss of function and ~26% hypomorphic).
Table 1:
PCM1 experimental data. Variant nomenclature refers to PCM1 mRNA (Refseq transcripts: NM_001315507.1, NM_006197.3). Each variant is associated with the corresponding p-values in the two evaluated experimental conditions (MO and MO+WT) and the resulting functional effect. Loss of function and hypermorphic variantes were evaluated together as a single category.
| Nucleotide variant | Protein variant | p-value from MO | p-value from MO+WT | Functional effect (class) | Functional effect (description) |
|---|---|---|---|---|---|
| NM_001315507.1:c.17G>A | p.(Gly6Asp) | 0.067 | 0.0001 | 2 | loss of function |
| NM_001315507.1:c.69G>C | p.(Glu23Asp) | 0.0004 | 0.0007 | 1 | hypomorph |
| NM_001315507.1:c.229A>G | p.(Thr77Ala) | 0.57 | 0.0001 | 2 | loss of function |
| NM_001315507.1:c.436A>G | p.(Met146Val) | 0.0001 | 0.13 | 0 | benign |
| NM_001315507.1:c.467C>T | p.(Ala156Val) | 0.0001 | 0.0099 | 1 | hypomorph |
| NM_001315507.1:c.599T>C | p.(Met200Thr) | 0.28 | 0.0001 | 2 | loss of function |
| NM_001315507.1:c.600G>A | p.(Met200Ile) | 0.0022 | 0.0049 | 1 | hypomorph |
| NM_001315507.1:c.641A>G | p.(Asp214Gly) | 0.0005 | 0.0013 | 1 | hypomorph |
| NM_001315507.1:c.742G>C | p.(Glu248Gln) | 0.53 | 0.0001 | 2 | loss of function |
| NM_001315507.1:c.931G>C | p.(Glu311Gln) | 0.0012 | 0.0036 | 1 | hypomorph |
| NM_001315507.1:c.1106A>G | p.(Glu369Gly) | 0.059 | 0.0001 | 2 | loss of function |
| NM_001315507.1:c.1168C>T | p.(Pro390Ser) | 0.38 | 0.0001 | 2 | loss of function |
| NM_001315507.1:c.1414C>G | p.(Leu472Val) | 0.039 | 0.0003 | 1 | hypomorph |
| NM_001315507.1:c.1445G>T | p.(Gly482Val) | 0.0002 | 0.0012 | 1 | hypomorph |
| NM_001315507.1:c.1627G>A | p.(Glu543Lys) | 0.0001 | 0.64 | 0 | benign |
| NM_001315507.1:c.1721A>G | p.(Asp574Gly) | 0.0044 | 0.0021 | 1 | hypomorph |
| NM_001315507.1:c.1811G>T | p.(Arg604Leu) | 0.0001 | 0.55 | 0 | benign |
| NM_001315507.1:c.1870G>A | p.(Glu624Lys) | 0.0001 | 0.58 | 0 | benign |
| NM_001315507.1:c.1977C>G | p.(Ile659Met) | 0.0001 | 0.62 | 0 | benign |
| NM_001315507.1:c.2410A>C | p.(Ser804Arg) | 0.0001 | 0.69 | 0 | benign |
| NM_001315507.1:c.2498G>C | p.(Arg833Thr) | 0.0001 | 0.71 | 0 | benign |
| NM_001315507.1:c.2626T>C | p.(Cys876Arg) | 0.0033 | 0.59 | 0 | benign |
| NM_006197.3:c.2674G>A | p.(Gly892Arg) | 0.16 | 0.0007 | 2 | loss of function |
| NM_001315507.1:c.2750A>G | p.(Glu917Gly) | 0.19 | 0.0001 | 2 | loss of function |
| NM_001315507.1:c.2862G>C | p.(Lys954Asn) | 0.0001 | 0.92 | 0 | benign |
| NM_001315507.1:c.3374A>G | p.(Asn1125Ser) | 0.0001 | 0.11 | 0 | benign |
| NM_001315507.1:c.3823A>G | p.(Lys1275Glu) | 0.0001 | 0.32 | 0 | benign |
| NM_001315507.1:c.4055A>T | p.(His1352Leu) | 0.012 | 0.0045 | 1 | hypomorph |
| NM_001315507.1:c.4082G>A | p.(Cys1361Tyr) | 0.0003 | 0.61 | 0 | benign |
| NM_001315507.1:c.4469C>G | p.(Ala1490Gly) | 0.0001 | 0.55 | 0 | benign |
| NM_001315507.1:c.4603G>A | p.(Glu1535Lys) | 0.0001 | 0.59 | 0 | benign |
| NM_001315507.1:c.4658C>G | p.(Ala1553Gly) | 0.0015 | 0.0034 | 1 | hypomorph |
| NM_006197.3:c.4667G>A | p.(Gly1556Asp) | 0.36 | 0.0001 | 2 | loss of function |
| NM_006197.3:c.5583A>C | p.(Lys1861Asn) | 0.0001 | 0.13 | 0 | benign |
| NM_006197.3:c.5625T>G | p.(Asn1875Lys) | 0.087 | 0.0001 | 2 | loss of function |
| NM_006197.3:c.5720G>A | p.(Arg1907His) | 0.0001 | 0.12 | 0 | benign |
| NM_006197.3:c.5738C>T | p.(Pro1913Leu) | 0.75 | 0.0027 | 2 | loss of function |
| NM_006197.3:c.5935G>T | p.(Ala1979Ser) | 0.72 | 0.0027 | 2 | loss of function |
Dataset and classifications
The challenge presents 38 transgenic human PCM1 missense mutations implicated in schizophrenia (Experimental data URL: https://genomeinterpretation.org/content/PCM1). These variants were assayed in a zebrafish model to determine their impact on the posterior ventricle area as previously explained. Each variant codes for a single amino acid substitution, showing no insertions or deletions. The variant number used in this work refers to PCM1 mRNA (GenBank identifier: NM_001315507). Participants were asked to predict the probability (p-value) of the effect of the variants on zebrafish brain development. These p-values were predicted considering the two different case scenarios: the probability that the variant (MO+VAR) is significantly different from MO and the probability that the variant is significantly different from MO+WT. In addition, predictors were also allowed to specify the standard deviation (SD) which defines the confidence of each prediction. Large SD means low confidence, while small SD means that the predictor is confident about the submitted prediction. According to the predicted probabilities and their interpretation, the participants had to inform the functional effect of the variant which could be: pathogenic, hypomorphic or benign. Six out of seven submissions reported for all the variants the p-values, SD and functional effect.
Performance assessment
The performance evaluation of bioinformatics tools aiming to predict VUS is a non-trivial problem, as the assessment should be more than a discrimination between good and bad predictions. In this challenge participants were requested to predict the p-values associated to each variant under two different conditions. According to the data provider results, the functional effects of each variant could be: benign, pathogenic (loss of function) and hypomorphic (partial loss of function). Even though one of the challenge was to predict the p-values relative to the changes from MO and MO+WT, it was a very difficult task to begin with. After analyzing the correlation between experimental and predicted p-values in the two experimental conditions, we found that Pearson correlation coefficients range between −0.29 and 0.23 for different submissions, showing that there is no relationship between experimental and predicted p-values. Predicted p-values were therefore not taken into account to perform the assessment and consequently the use of global evaluation metrics as ROC or precision-recall curves was not possible. This is why we only used the predicted variant effect informed by the authors to address the final ranking.
To assess further the prediction reliability in a medical setting, a binary classification was used based on the variant predicted effects. The three variant effects mentioned above were reorganized as a binary classification, benign and pathogenic (loss of function and hypomorphic were considered together). A set of measures were implemented in order to perform a thorough assessment and to obtain a better description about predictor performance (Vihinen, 2012). The aim was to produce a global overview of the strengths and weaknesses of each method. For each submission we calculate five different scores to assess the quality of the binary prediction: Balanced Accuracy (BACC), Matthews Correlation Coefficient (MCC), F1 score (F1), True Positive Rate (TPR) and True Negative Rate (TNR). All measures are defined in more detail in the Suppl. Material. The final ranking of predictor performances was the average of the individual rankings produced by each measure. To assess the statistical significance of each performance index, we generated 10,000 random predictions and used these data to estimate an empirical continuous score probability distribution (s). The p-value is then calculated by defining the proportion of random predictions scoring > s.
The R scripts used to perform the assessment are publicly available from the GitHub repository at URL: https://github.com/BioComputingUP/CAGI-PCM1-assessment.
Groups description
This challenge received 7 submissions from 6 different groups which were assessed blindly. Only one group (Bromberg lab) contributed with two submissions. Group 3 submitted an empty template and method description and consequently was not considered in the assessment. After completing the assessment, all groups provided their name and affiliations. Table 2 lists the participating groups, ID, name, and method used. Group 1 (Casadio lab) based their predictions on the Disease Index matrix (Casadio, Vassura, Tiwari, Fariselli, & Luigi Martelli, 2011), which measures how protein stability is affected by mutations. Group 2 (Lichtarge lab) uses their Evolutionary Action approach (Katsonis & Lichtarge, 2014) to relate the variant effect with the evolutionary fitness effect. Group 4 (Bromberg lab) performed the predictions for their first submission using SNAP (Bromberg & Rost, 2007; Bromberg, Yachdav, & Rost, 2008), a neural network-based method for the prediction of the functional effects of non-synonymous SNPs. In their second submission, predictions were depending on fuNTRp (Miller et al., submitted), a Random Forest-based method to classify protein positions based on the expected range of possible mutational impacts per position (Neutral positions = no or weak effects; Rheostat positions = range of effects, i.e. functional tuning; Toggle positions = mostly strong effects). Group 5 (Carter lab) analyzed each variant with VEST (Carter, Douville, Stenson, Cooper, & Karchin, 2013), assigning to each mutation a score indicating confidence in a functional mutation. Group 6 (BioFolD unit) used the SNPs&GO (Emidio Capriotti et al., 2013) and PhD-SNP (E. Capriotti, Calabrese, & Casadio, 2006) methods. A more detailed description of the methods used by each group can be found in the Suppl. Material.
Table 2:
Predictions overview. Each submission is associated to the predictor group and a summary of the features used for the prediction.
| Submission ID | Group ID | Prediction features |
|---|---|---|
| Submission 1.1 | Group 1 (Casadio lab) | Protein stability |
| Submission 2.1 | Group 2 (Lichtarge lab) | Evolutionary action |
| Submission 3.1 | Group 3 | No predictions made |
| Submission 4.1 | Group 4 (Bromberg lab) | Conservation, annotation |
| Submission 4.2 | Group 4 (Bromberg lab) | Conservation, annotation |
| Submission 5.1 | Group 5 (Carter lab) | Annotation |
| Submission 6.1 | Group 6 (BioFolD unit) | Metaprediction |
RESULTS
Participation and similarity between predictions
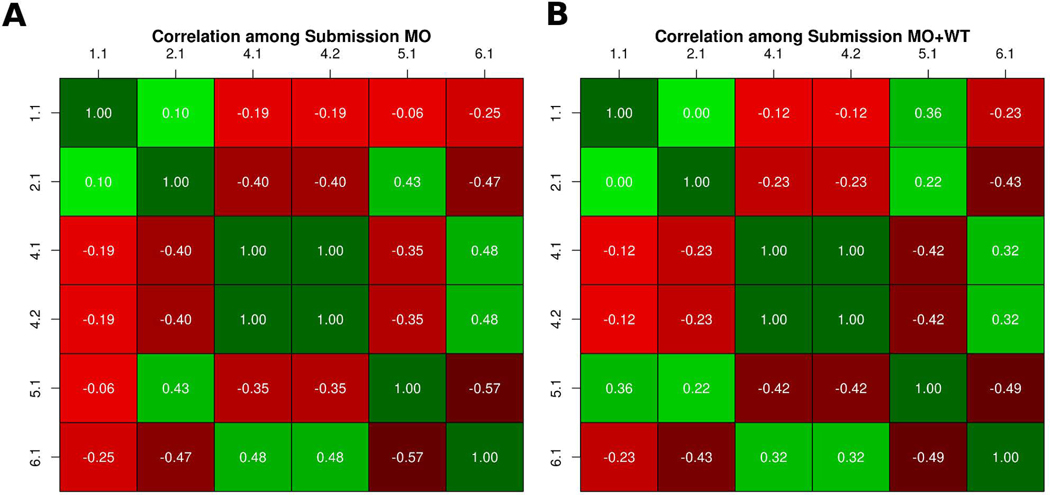
In the PCM1 CAGI-5 challenge, participants were requested to predict the probability of the effect caused by 38 variants on zebrafish brain development. Essentially, the predicted probability allowed to infer three kinds of functional effects associated to each variant: benign, hypomorphic (partial loss of function), and loss of function. We performed a correlation analysis between submissions to address the similarity. Then we divided the predictions in two subsets: variants predicted as loss of function and predicted as hypomorphic. Figure 1 shows the two predictions submitted by Bromberg lab obtained the same probability values for each variant. Both predictions used SNAP (Bromberg & Rost, 2007; Bromberg et al., 2008) to predict the p-values but differed in the way the variant is classified. Their submission 2 used fuNTRp (Miller et al., submitted), a tool based on random forest that predicts position types (i.e. expected range of variant effects per position). Another observation from this analysis is that most groups predicted very different p-values, highlighting difficult of this challenge. We can also observe some weak positive and negative correlations between groups. On one hand we have a weak positive correlation between groups 4 and 6, possibly because predicted p-values are quite similar in some variants. Groups 2 and 5 also show a positive weak correlation possibly because predicted p-values in both groups are close to zero. On the other hand, we have some weak negative correlations between groups which have predicted opposite probability values for some variants, such as groups 2 and 5 versus groups 4 and 6.
Figure 1:
Similarity between predicted p-values. A-B) Each cell shows the Pearson correlation coefficient between two submissions, with a color scale ranging from green (+1, perfect correlation) to red (0, no correlation) and black (−1, perfect anti-correlation).
Assessment criteria and performance evaluation
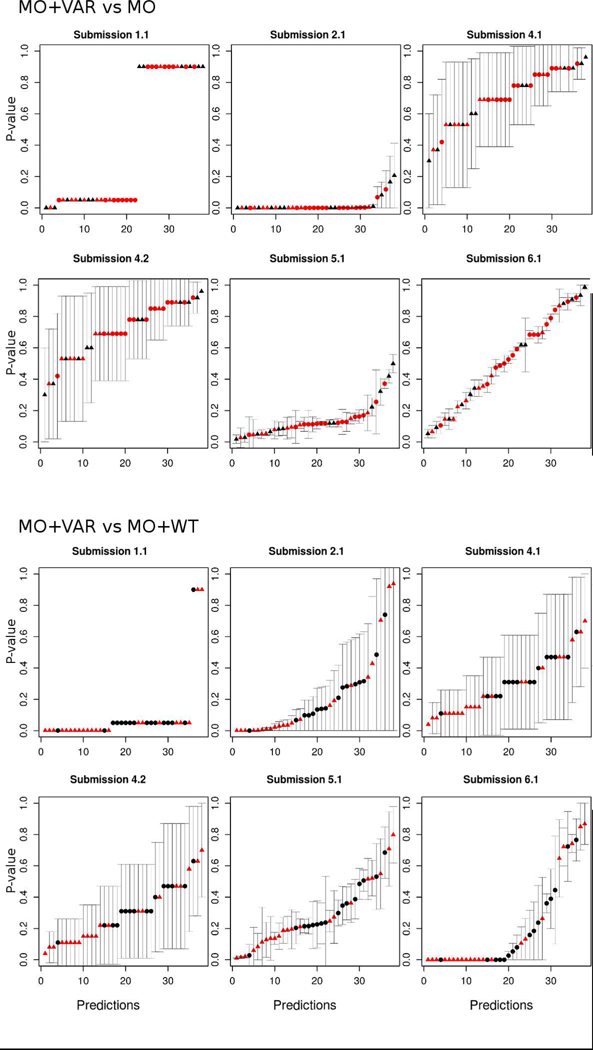
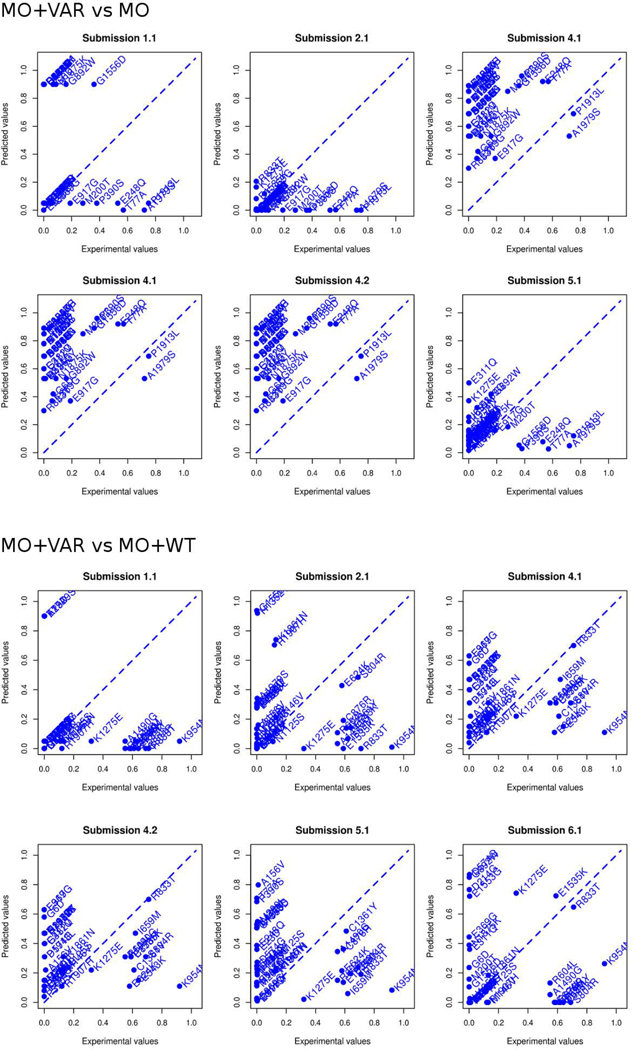
The evaluation criteria used to assess a CAGI challenge directly influence perceptions gained from the test. In order to highlight predictor performance and their practical relevance, we performed the evaluation only considering the predicted functional effect of each variant provided by the participants. As most submissions reported the predicted p-values, we tried first to perform the assessment as an inherently continuous prediction challenge. After some exploratory analysis, we concluded that predicted p-values among all submissions did not agree at all with the experimental p-values and also with the interpretation of the p-values to infer the functional classes (Figure 2 and Figure 3). For this reason, we decided to perform the assessment using only the predicted functional class of each mutation.
Figure 2:
Predicted p-values with their corresponding standard deviation for each experimental condition by group. The x-axis is from 1 to 38 and represents the predicted p-values for a particular position (sequentially ordered by the position on the sequence). The y-axis is the value of the predicted p-value.
Dot shapes represent the variant effect, with triangles for pathogenic and circles for benign. The color indicates the experimental p-value, red for p-value < 0.05 and black for p-value ≥ 0.05.
Figure 3:
Predicted vs. experimental p-values for all submissions. The predicted value (y-axis) is plotted against the experimental value (x-axis) for all variants (in the two experimental conditions) in each of the 6 submissions.
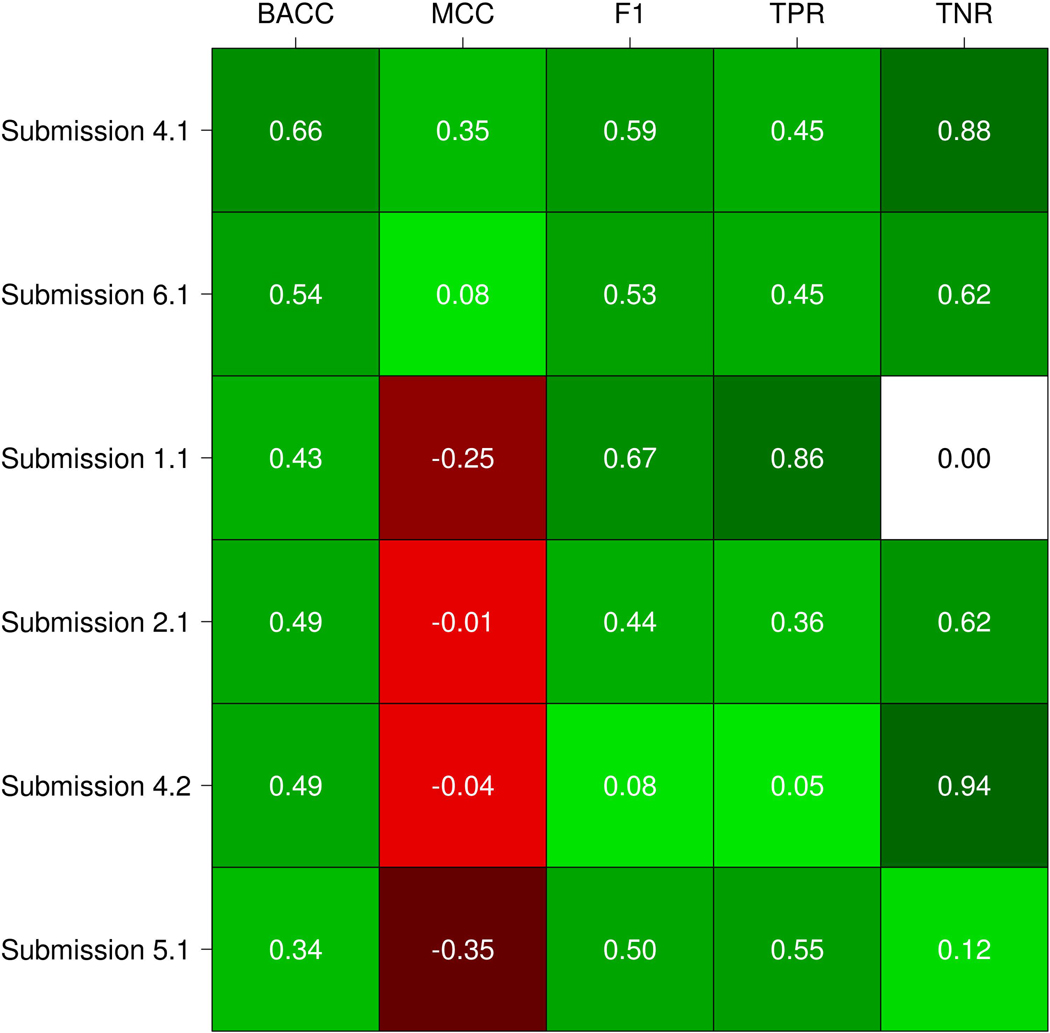
The performance was evaluated using five standard measures as described above. Our assessment shows that the six submissions achieved in general a poor performance. This is highlighted by the MCC values (Figure 4), where most of the submissions have values close or below zero. As the average among all submissions is −0.06, this means that the correlation between the experimental and predicted variant functional effect is no better than random predictions in most of the cases. The highest MCC value is 0.35 and was reached by submission 4.1 (Bromberg lab). This submission correctly predicted 10 out of 22 pathogenic variants and 14 out of 16 benign variants (Table 3). Then, submission 5.1 (Carter lab) obtained the lowest MCC value (−0.35), correctly predicting 12 disease mutations but only 2 benigns (Table 3).
Figure 4:
Submissions performance evaluation. Each cell represents the value of a measure for a specific submission. The color scale ranges from dark green (+1, perfect performance) to red (−1, perfect anticorrelation just for MCC). White means zero in terms of performance.
Table 3:
Confusion matrices for all submissions. Disease category contains hypomorph and loss of function variants.
| Submission 1.1 | Submission 2.1 | Submission 4.1 | ||||||
| Obs. Disease | Obs. Benign | Obs. Disease | Obs. Benign | Obs. Disease | Obs. Benign | |||
| Pred. Disease | 19 | 16 | Pred. Disease | 8 | 6 | Pred. Disease | 10 | 2 |
| Pred. Benign | 3 | 0 | Pred. Benign | 14 | 10 | Pred. Benign | 12 | 14 |
| Submission 4.2 | Submission 5.1 | Submission 6.1 | ||||||
| Obs. Disease | Obs. Benign | Obs. Disease | Obs. Benign | Obs. Disease | Obs. Benign | |||
| Pred. Disease | 1 | 1 | Pred. Disease | 12 | 14 | Pred. Disease | 10 | 6 |
| Pred. Benign | 21 | 15 | Pred. Benign | 10 | 2 | Pred. Benign | 12 | 10 |
For BACC, we can observe that submissions 4.1 (Bromberg lab) and 6.1 (BioFolD), performed better than other methods, also considering their MCC values (Figure 4). Since a method could be biased to predict the more frequent class, BACC is a good way to calculate the accuracy evaluating if the predictor takes advantage or not of an imbalanced test set. Consequently, F1 shows values higher than 0.50 for three out seven submissions. F1 measure considers the precision and recall of the test, submission 1.1 (Casadio lab) obtained the highest F1 value of 0.67, followed by submissions 4.1 and 6.1 with 0.59 and 0.53 respectively. However, if we observe the TNR and confusion matrix of submission 1.1 (see Table 3), this predictor presents a biased confusion matrix and was not able to identify any benign variants.
To perform a global assessment of each predictor performance we need to take into account all performance measures together instead of just comparing them separately. We decided to observe the ranking achieved for each submission on each considered measure. Moreover, this allows non-expert users to better understand the results of the assessment. The Bromberg lab (Submission 4.1) achieved the best overall performance comparing with all other predictors, ranking first in BACC and MCC measures, second in F1 and TNR and sharing the third place in TNR (see Table 4). BioFolD (submission 6.1) ranked second in overall performance, second in BACC and MCC, and third in the other measures. The Casadio lab achieved the best rank in F1 and TNR measures and ranking third in overall performance. However, their prediction was biased toward diseases phenotypes, with no benign variant correctly predicted (Table 3). Something similar but opposite happened with the Bromberg lab (Submission 4.2), where the prediction was biased towards benign variants and only one disease variant predicted correctly (Table 3). In addition, we can observe that MCC values for the two submissions mentioned above are negative (i.e. negatively correlated) and almost zero (i.e. close to random). Observing the confusion matrices, we can conclude that most submissions produced unbalanced predictions biased towards the prediction of disease phenotypes.
Table 4:
Submissions ranking. Individual and overall rankings among all submissions based on the performance measures considered. Each cell contains the ranking of a submission for a specific performance measure and in brackets the performance value. The overall final ranking is obtained by the average rank achieved for each submission considering all the performance measures.
| Submission ID | BACC | MCC | F1 | TPR | TNR | Avg. Ranking | Final rank |
|---|---|---|---|---|---|---|---|
| Submission 4.1 | 1 (0.67) | 1 (0.35) | 2 (0.59) | 3.5 (0.46) | 2 (0.88) | 1.9 | 1 |
| Submission 6.1 | 2 (0.54) | 2 (0.08) | 3 (0.53) | 3.5 (0.46) | 3.5 (0.63) | 2.8 | 2 |
| Submission 1.1 | 5 (0.43) | 5 (−0.25) | 1 (0.67) | 1 (0.86) | 6 (0) | 3.6 | 3 |
| Submission 2.1 | 3 (0.49) | 3 (−0.01) | 5 (0.44) | 5 (0.36) | 3.5 (0.63) | 3.9 | 4 |
| Submission 4.2 | 4 (0.49) | 4 (−0.04) | 6 (0.08) | 6 (0.05) | 1 (0.94) | 4.2 | 5 |
| Submission 5.1 | 6 (0.34) | 6 (−0.35) | 4 (0.5) | 2 (0.55) | 5 (0.13) | 4.6 | 6 |
Considering the poor performance of most predictors, we only calculated the statistical significance of submission 4.1 (Bromberg lab) for the BACC, MCC and F1 measures. A bootstrap with 10,000 replicas was used to test whether the performance of submission 4.1 could be achieved by chance. We can conclude that it performs better than random (p-value < 0.05) for MCC and BACC measures (see Suppl. Figure S1). The only exception is F1, denoting unbalanced predicted classes from the real data.
Another interesting aspect of this challenge is to see how each group correctly predicted the real disease effect, loss of function and hypomorph. In Suppl. Table S1 we can see the contingency matrices split into three categories. Most of the groups had difficulties identifying the correct disease class. Submission 4.1 correctly predicted 4 hypomorph variants and no loss of function one. Submission 6.1 correctly identified one loss of function and one hypomorph variant. On the other hand, submission 1.1, which was biased to predict disease variants, correctly predicted 6 hypomorph and 4 loss of function.
Difficult variants
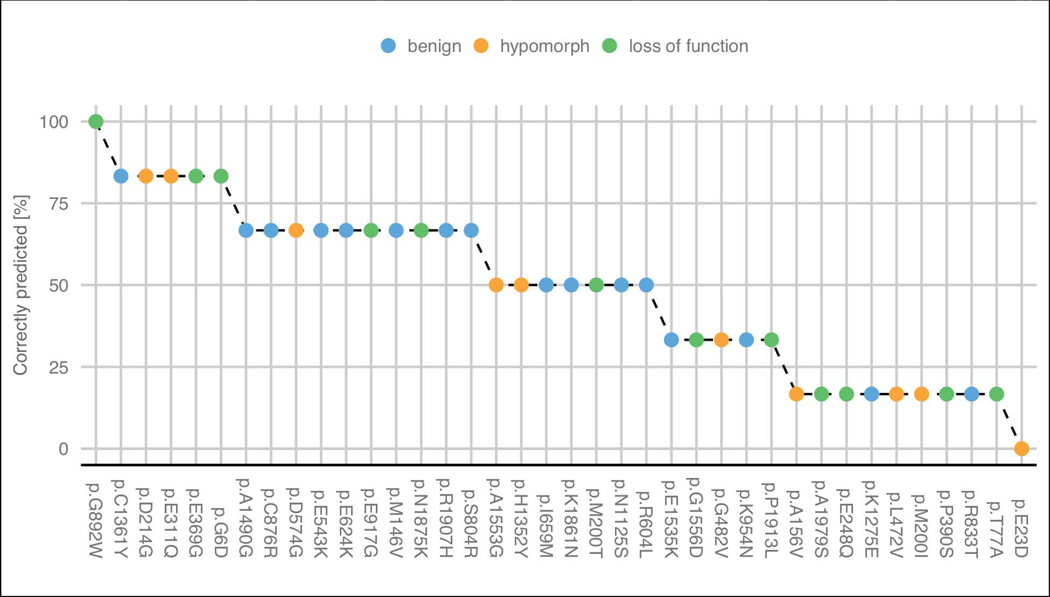
Looking at the predicted functional effects for each variant, we can see that some were particularly complex to be predicted (Figure 5). The functional effect (benign and pathogenic) was well predicted for 41% of the proposed variants by more than half of the predictors. Due to the limited structural characterization of PCM1 is difficult to analyze the structural properties of each residue. We tried to explore further some properties of PCM1 using FELLS (Piovesan, Walsh, Minervini, & Tosatto, 2017). Disease variant p.G892W was correctly predicted by all submissions and that position presents high propensity to be coil and disordered. On the other hand, disease variant p.E23D was not identified by any predictor and shows high propensity to be disordered.
Figure 5:
Percentage of groups which correctly predicted the effect of each variant. Hypomorph and loss of function variants were considered as disease in group predictions. The variants are colored by their experimental effect.
There are 15 variants where most groups failed to correctly predict their effect (<50% correctly predicted): 4 benign, 5 hypomorphs (disease) and 6 loss-of-function (disease). Interestingly, submission 1.1 (Casadio lab) predicted correctly five of these 15 disease mutations. However, this predictor was biased towards pathogenic variants and not able to identify any benign. The PCM1 challenge highlights how some variants are really hard targets for most of the methods.
DISCUSSION
The determination of novel variant effects is a key challenge of great value for clinicians. Due to the diversity and complexity of the biological systems, a variant could impact at different levels such as protein function, subcellular localization, metabolic pathways, among others (Hamp & Rost, 2012). The best predictor should be able to discriminate between pathogenic and benign variants. Here, we presented the assessment of the CAGI-5 PCM1 challenge. This challenge is based on the prediction of the probability of missense variant effects, in analogy to the CAGI-3 p16 challenge (Carraro et al., 2017). While the p16 challenge was testing the ability to predict cell proliferation rate, the PCM1 challenge is focused on predicting the probable variant effect on zebrafish brain development. PCM1 is a component of centriolar satellites occurring around centrosomes in vertebrate cells (Dammermann & Merdes, 2002; Kubo & Tsukita, 2003). It also interacts with BBS4 and DISC1 (Kamiya et al., 2008; Miyoshi et al., 2004) and has an important role in centrosome formation, which is needed for proper neurodevelopment (Ayala, Shu, & Tsai, 2007; Gupta, Tsai, & Wynshaw-Boris, 2002; Mochida & Walsh, 2004; Solecki, Govek, Tomoda, & Hatten, 2006; Tsai & Gleeson, 2005). The Katsanis lab provided experimental data for 38 missense mutations in PCM1 in a zebrafish model. The experimental effect determined by the data providers is unambiguous and resulted of brain brain ventricle volumes between MO and MO+WT. This kind of comparison studies have been performed in the past and the specificity/sensitivity metrics have been reported to be high (Zaghloul et al., 2010).
Submissions were compared with experimental data to evaluate their prediction performance. Using a set of performance measures highlighting strengths and weaknesses of each predictor similar to previous CAGI assessments (Carraro et al., 2017).
From a technical point of view, the groups used different approaches to predict p-values and variant effect, ranging from machine learning to position-specific scoring matrices. The assessment suggests that most state-of-art predictors participating in this challenge were not sufficient to perform reliable variant effect predictions. The absence of structural information and high disorder content make this protein challenging, especially for predictors based on structural information. The MCC values reached by different submissions are subpar, close to random prediction. MCC is one of the best measures to handle unbalanced data, since some predictions were biased to identify disease or benign phenotypes (Boughorbel, Jarray, & El-Anbari, 2017). The best MCC and BACC values were reached by submission 4.1 (Bromberg lab), showing also the best overall ranking. They correctly predicted 10 out of 22 disease variants and 14 out of 16 benign variants (Table 3). However, if we look at the disease class considering loss of function and hyphomorph, submission 4.1 correctly predicted only 4 hypomorph variants. Showing again the difficulty in p-values interpretation (Suppl. Table S1). Anyway, these results suggest that SNAP (Bromberg & Rost, 2007; Bromberg et al., 2008), a neural network-based method, may be a useful method to screen big datasets for pathogenic variants in a similar context.
Interestingly, group 1 (Casadio Lab) obtained a promising TPR of 0.86 and predicted 19 out 22 disease variants but they could not identify any benign variants. Nevertheless, they identified the highest number of loss of function variants (Suppl. Table S1). Conversely, group 4 submission 2 reached a high TNR score and predicted 15 out 16 benign variants but identified only one disease variant. Group 6 (BioFolD unit) well predicted 10 out 16 benign variants and 10 out 22 disease, scoring second considering the overall rank and MCC value. We should emphasise here that data imbalance frequently occurs in biomedical applications and the use of inadequate performance metrics could lead to misinterpretation of predictors performance (Boughorbel et al., 2017).
This CAGI-5 PCM1 challenge evidences that there is still plenty of work to improve the pathogenicity prediction of VUS. Despite the generally low performance of predictors, some identified a good number of disease and benign variants. However, we still have to improve our prediction methods if we want a generic pathogenicity predictor. We expect that the CAGI challenges which help motivate research, improving the current methods and generating new ideas.
Supplementary Material
ACKNOWLEDGMENTS
The CAGI experiment coordination is supported by NIH U41 HG007346 and the CAGI conference by NIH R13 HG006650. This work was partially supported by the Italian Ministry of Health grants GR-2011-02347754 and GR-2011-02346845 to E.L and S.C.E.T., respectively; by the Fondazione Istituto di Ricerca Pediatrica - Città della Speranza, Grant 18-04 to E.L. PK and OL were supported by the National Institutes of Health (GM079656 and GM066099); E.C. was supported by an FFABR grant from the Ministry of Education, Universities and Research (MIUR). A.M.M. is funded by the research program “MSCA Seal of Excellence @UniPD”. Y.B., Y.W., and M.M. were supported by the NIH U01 GM115486 grant.
REFERENCES
- Ansley SJ, Badano JL, Blacque OE, Hill J, Hoskins BE, Leitch CC, … Katsanis N (2003). Basal body dysfunction is a likely cause of pleiotropic Bardet–Biedl syndrome. Nature, 425, 628. [DOI] [PubMed] [Google Scholar]
- Ayala R, Shu T, & Tsai L-H (2007). Trekking across the brain: the journey of neuronal migration. Cell, 128(1), 29–43. [DOI] [PubMed] [Google Scholar]
- Boughorbel S, Jarray F, & El-Anbari M (2017). Optimal classifier for imbalanced data using Matthews Correlation Coefficient metric. PloS One, 12(6), e0177678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y, & Rost B (2007). SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Research, 35(11), 3823–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y, Yachdav G, & Rost B (2008). SNAP predicts effect of mutations on protein function. Bioinformatics, 24(20), 2397–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E, Calabrese R, & Casadio R (2006). Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics, 22(22), 2729–2734. [DOI] [PubMed] [Google Scholar]
- Capriotti E, Calabrese R, Fariselli P, Martelli PL, Altman RB, & Casadio R (2013). WS-SNPs&GO: a web server for predicting the deleterious effect of human protein variants using functional annotation. BMC Genomics, 14 Suppl 3, S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro M, Minervini G, Giollo M, Bromberg Y, Capriotti E, Casadio R, … Tosatto SCE (2017). Performance of in silico tools for the evaluation of p16INK4a (CDKN2A) variants in CAGI. Human Mutation, 38(9), 1042–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter H, Douville C, Stenson PD, Cooper DN, & Karchin R (2013). Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genomics, 14 Suppl 3, S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadio R, Vassura M, Tiwari S, Fariselli P, & Luigi Martelli P (2011). Correlating disease-related mutations to their effect on protein stability: a large-scale analysis of the human proteome. Human Mutation, 32(10), 1161–1170. [DOI] [PubMed] [Google Scholar]
- Dammermann A, & Merdes A (2002). Assembly of centrosomal proteins and microtubule organization depends on PCM-1. The Journal of Cell Biology, 159(2), 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis EE, Frangakis S, & Katsanis N (2014). Interpreting human genetic variation with in vivo zebrafish assays. Biochimica et Biophysica Acta, 1842(10), 1960–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Yang Z, Song W, Chen Q, Wang F, Zhang Q, & Zhu X (2006). Nudel contributes to microtubule anchoring at the mother centriole and is involved in both dynein-dependent and -independent centrosomal protein assembly. Molecular Biology of the Cell, 17(2), 680–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Tsai L-H, & Wynshaw-Boris A (2002). Life is a journey: a genetic look at neocortical development. Nature Reviews. Genetics, 3(5), 342–355. [DOI] [PubMed] [Google Scholar]
- Gurling HMD, Critchley H, Datta SR, McQuillin A, Blaveri E, Thirumalai S, … Dolan RJ (2006). Genetic association and brain morphology studies and the chromosome 8p22 pericentriolar material 1 (PCM1) gene in susceptibility to schizophrenia. Archives of General Psychiatry, 63(8), 844–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzman JH, & Sive H (2009). Zebrafish brain ventricle injection. Journal of Visualized Experiments: JoVE, (26). 10.3791/1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamp T, & Rost B (2012). Alternative protein-protein interfaces are frequent exceptions. PLoS Computational Biology, 8(8), e1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya A, Tan PL, Kubo K-I, Engelhard C, Ishizuka K, Kubo A, … Sawa A (2008). Recruitment of PCM1 to the centrosome by the cooperative action of DISC1 and BBS4: a candidate for psychiatric illnesses. Archives of General Psychiatry, 65(9), 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsonis P, & Lichtarge O (2014). A formal perturbation equation between genotype and phenotype determines the Evolutionary Action of protein-coding variations on fitness. Genome Research, 24(12), 2050–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, & Tsukita S (2003). Non-membranous granular organelle consisting of PCM-1: subcellular distribution and cell-cycle-dependent assembly/disassembly. Journal of Cell Science, 116(Pt 5), 919–928. [DOI] [PubMed] [Google Scholar]
- Lowery LA, De Rienzo G, Gutzman JH, & Sive H (2009). Characterization and classification of zebrafish brain morphology mutants. Anatomical Record, 292(1), 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikut R, Dickmeis T, Driever W, Geurts P, Hamprecht FA, Kausler BX, … Peyriéras N (2013). Automated processing of zebrafish imaging data: a survey. Zebrafish, 10(3), 401–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Asanuma M, Miyazaki I, Diaz-Corrales FJ, Katayama T, Tohyama M, & Ogawa N (2004). DISC1 localizes to the centrosome by binding to kendrin. Biochemical and Biophysical Research Communications, 317(4), 1195–1199. [DOI] [PubMed] [Google Scholar]
- Mochida GH, & Walsh CA (2004). Genetic basis of developmental malformations of the cerebral cortex. Archives of Neurology, 61(5), 637–640. [DOI] [PubMed] [Google Scholar]
- Näslund J, & Johnsson JI (2016). Environmental enrichment for fish in captive environments: effects of physical structures and substrates. Fish and Fisheries, 17(1), 1–30. [Google Scholar]
- Niederriter AR, Davis EE, Golzio C, Oh EC, Tsai I-C, & Katsanis N (2013). In vivo modeling of the morbid human genome using Danio rerio. Journal of Visualized Experiments: JoVE, (78), e50338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niroula A, & Vihinen M (2016). Variation Interpretation Predictors: Principles, Types, Performance, and Choice. Human Mutation, 37(6), 579–597. [DOI] [PubMed] [Google Scholar]
- Piovesan D, Tabaro F, Paladin L, Necci M, Micetic I, Camilloni C, … Tosatto SCE (2018). MobiDB 3.0: more annotations for intrinsic disorder, conformational diversity and interactions in proteins. Nucleic Acids Research, 46(D1), D471–D476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piovesan D, Walsh I, Minervini G, & Tosatto SCE (2017). FELLS: fast estimator of latent local structure. Bioinformatics, 33(12), 1889–1891. [DOI] [PubMed] [Google Scholar]
- Solecki DJ, Govek E-E, Tomoda T, & Hatten ME (2006). Neuronal polarity in CNS development. Genes & Development, 20(19), 2639–2647. [DOI] [PubMed] [Google Scholar]
- Summerton J, & Weller D (1997). Morpholino antisense oligomers: design, preparation, and properties. Antisense & Nucleic Acid Drug Development, 7(3), 187–195. [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium. (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Research, 45(D1), D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai L-H, & Gleeson JG (2005). Nucleokinesis in neuronal migration. Neuron, 46(3), 383–388. [DOI] [PubMed] [Google Scholar]
- Villumsen BH, Danielsen JR, Povlsen L, Sylvestersen KB, Merdes A, Beli P, … Bekker-Jensen S (2013). A new cellular stress response that triggers centriolar satellite reorganization and ciliogenesis. The EMBO Journal, 32(23), 3029–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaghloul NA, Liu Y, Gerdes JM, Gascue C, Oh EC, Leitch CC, … Katsanis N (2010). Functional analyses of variants reveal a significant role for dominant negative and common alleles in oligogenic Bardet-Biedl syndrome. Proceedings of the National Academy of Sciences of the United States of America, 107(23), 10602–10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoubovsky S, Oh EC, Cash-Padgett T, Johnson AW, Hou Z, Mori S, … Jaaro-Peled H (2015). Neuroanatomical and behavioral deficits in mice haploinsufficient for Pericentriolar material 1 (Pcm1). Neuroscience Research, 98, 45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.