Abstract
Background
The pathogenesis underlying the alterations of orbital architecture in Graves’ orbitopathy (GO) is not yet fully understood. The present study aimed to investigate the association of DNA methylation in peripheral blood and orbital volumetry in Chinese patients with GO.
Methods
A total of 35 GO subjects (70 orbits) were subjected to CT scan. The total cross-sectional area of the extraocular muscles (orbital muscles, OM), total orbit area (TOA), and the exophthalmometry were measured and OM/TOA ratio was calculated. Targeted bisulfite sequencing was performed on seven candidate genes.
Results
No significant correlation was established between the DNA methylation levels of these genes and exophthalmometry. The MBP methylation level was found to be correlated with OM/TOA ratio (P < 0.05). Multiple linear regression analysis on parameters including age, sex, TRAb, duration of GO, and DNA methylation levels of seven genes with OM/TOA ratio confirmed that MBP and OM/TOA ratio had a significant correlation (P < 0.05). The partial least squares analysis showed that the top three genes with the highest loadings were MBP, BOLL, and BECN1 and that OM/TOA ratio affected the DNA methylation block than exophthalmometry.
Conclusions
This study provided preliminary evidence that MBP is a potential gene associated with OM enlargement in GO patients according to the combination of DNA methylation sequencing and orbital CT measurement.
Keywords: graves’ orbitopathy, extraocular muscles, orbit, DNA methylation
Introduction
Graves’ orbitopathy (GO) or thyroid-associated ophthalmopathy (TAO) is an autoimmune inflammatory disease exerting severe functional and psychosocial influence. Approximately 30% of the patients with Graves’ disease (GD) develop inflammation of the orbital tissues, which is known as GO (1). It occurs in patients with hyperthyroidism, hypothyroidism, or normal thyroid function. However, the pathogenesis underlying the disease has not yet been clarified. Several factors, including genetic susceptibility, and multiple environmental factors, such as gender, age, and smoking, have been demonstrated to be involved in the development of GO (2). Our previous study identified several genomic loci that had significant differences in DNA methylation (DNAm) patterns associated with GO (3, 4). BOLL, MBP, and CDK5 are potential genes associated with GO, according to the genome-scale screening of DNAm and the single methylation sequencing validation using enlarged samples (5). These findings provided in-depth insights into this disease.
GO has various clinical manifestations that might result in severe damage to orbital architecture and vision. The enlargement of orbital fat and extraocular muscles (ocular muscles, OM) is the hallmark of GO, as is evident from surgical pathology and MRI or CT imaging (6, 7). Orbit CT scan facilitates the visualization of the fat, muscle, and the cortical bone/calcium (8). It also provides objective and accurate data for the measurement of the orbital architecture of GO patients. Accumulating evidence postulated two subtypes of GO: one with predominant extraocular muscle enlargement and the other one with predominant retrobulbar connective and fat tissue involvement (9, 10). Some studies have shown that OM enlargement is associated with impaired motility, impaired vision, more proptosis, older age, and high TSH binding inhibitory immunoglobulin (TBII) values and that fat volume increase is related to proptosis and less diplopia (10). Another study also showed that a long duration of GO is associated with a high orbital fat volume (FV), while extraocular muscle volume (MV) is related to the severity of GO (11). However, the pathogenesis underlying the altered orbital architecture is yet to be clarified.
Although the understanding of the pathogenesis of GO has improved (12, 13, 14), the treatments are not accurate because they do not precisely target the pathogenic mechanisms of the disease (15). DNAm, the most studied epigenetic mechanism, has been associated with stable alterations of gene expression and implicated in the pathogenesis of several autoimmune diseases (16). Moreover, DNAm is a chemical modification process mediated by DNA methyltransferases (DNMTs). The base sequence of the DNA after methylation is not changed, but the expression of the gene is affected. Conversely, epigenetic modifications are reversible and could be the targets for drug intervention. Although some potential genes associated with GO based on the DNAm have been identified (5), further studies are essential to elucidate the precise roles in this disease. Hence, the present study aimed to investigate the association between DNAm and orbital architecture (proptosis and OM enlargement) of GO patients. The findings might help to further explore the pathogenesis of this disease and provide novel treatment and prevention strategies.
Materials and methods
Study participants
The present study was performed at the Department of Endocrinology, Beijing Tongren Hospital, Capital Medical University, Beijing, China. A total of 35 GO subjects (70 orbits) were enrolled between June 2016 and March 2017. The cohort consisted of 17 females and 18 males, aged 22–71 years (average, 47.9 ± 10.6). The diagnosis of GO was established according to the European Group on Graves’ Orbitopathy (EUGOGO) Guidelines (17). These enrolled GO subjects had not received any treatment for ocular discomfort, and their medical history was within 1 year. Among them, 28 subjects were hyperthyroid, 4 were euthyroid, and 3 were hypothyroid. All subjects with hyperthyroidism received only antithyroid drugs (Thyrozol, Merck Company) and underwent CT scan of the orbits, which excluded other orbital diseases, such as tumors or extraocular myositis. The characteristics of the subjects, including age, gender, weight, height, and ethnicity, were recorded. To evaluate the thyroid function and examine the thyroid autoantibody, peripheral blood was collected from the subjects after an overnight fast. The levels of serum T3, T4, FT3, FT4, and thyroid stimulating hormone (TSH) were assayed using ADVIA Centaur XP Immunoassay System (Siemens Diagnostics) and that of thyroperoxidase antibody (TPOAb), antithyroglobulin antibody (TgAb), thyrotropin receptor antibody (TRAb) were evaluated by electrochemiluminescence immunoassay using Cobas e601 (Roche Diagnostics). The reference ranges, provided by the manufacturer were as follows: 3.5–6.5 pmol/L for FT3, 11.5–22.7 pmol/L for FT4, 0.92–2.79 nmol/L for T3, 57.9–140 nmol/L for T4, 0.55–4.78 µIU/mL for TSH, 0–40 IU/mL for TPOAb, 0–115 IU/mL for TgAb, and 0–1.75 IU/L for TRAb.
This study was approved by the Ethics Committee of Beijing Tongren Hospital, Capital Medical University (Registration No. TRECKY2016-003). Written informed consent was obtained from all the patients.
Measurement of the cross-sectional area of OM, the total orbit area (TOA), and exophthalmometry
All subjects were examined using a Philips Brilliance 64 CT scanner. The CT of the orbital soft tissue was set at tube voltage 120 kV, 250 mAs/layer, scanning resolution high, collimation width 64 × 0.625 mm, pitch 0.639, acquisition and reconstruction matrix 512 × 512, layer thickness 0.67 mm, layer spacing 0.33 mm, reconstruction Filtered back projection (FBP), field of view (FOV) 180 mm × 180 mm, reconstruction algorithm standard (B), edge enhancement factor 0, window width 350 HU, and window bit 40 HU.
The patient lay in a supine position on the CT scan bed, while the head was kept still, and the scan baseline was parallel to the auditory basal line. The scan path followed from the upper edge of the orbit to the lower edge of the orbit. In order to prevent uneven OM contraction, the patient was required to maintain the first eye position and keep the eyes closed during the scan. The image was reconstructed using the extended image brilliance workspace (EBW). The cross-sectional plane reconstruction baseline was parallel to the intraorbital segment of the optic nerve, with a layer thickness of 2 mm and a layer spacing of 2 mm. The coronal plane was reconstructed on each side of the orbit. The baseline of the reconstructed unilateral orbital coronal plane was perpendicular to the transverse plane of the intraorbital segment of the optic nerve and the sagittal plane of the intraorbital segment of the optic nerve, with a layer thickness of 1 mm and a layer spacing of 1 mm.
On the coronal plane, 2 mm plane was found behind the ball with the following parameters: the back of the eyeball as the tangent line, the layer thickness 1 mm, and the pitch 1 mm. This section provided a clear OM and TOA. The cross-sectional area of the orbit and each OM was measured by AutoCAD software three times to obtain an average value. Because the superior rectus and the upper levator muscles were indistinguishable in the majority of the inner orbital segments, the method of measurement was complex. Finally, the total cross-sectional area of the OM and the ratio of orbital muscles/total orbit area (OM/TOA) was calculated (Fig. 1A) (7). According to the standard, the center of the lens and the intraorbital segment of the optic nerve were displayed on the same side, and when the outer edge of the orbit was at the lowest point, the exophthalmometry was measured on the image workstation (Fig. 1B). The largest value among OM/TOA ratio and exophthalmometry for both orbits was selected.
Figure 1.
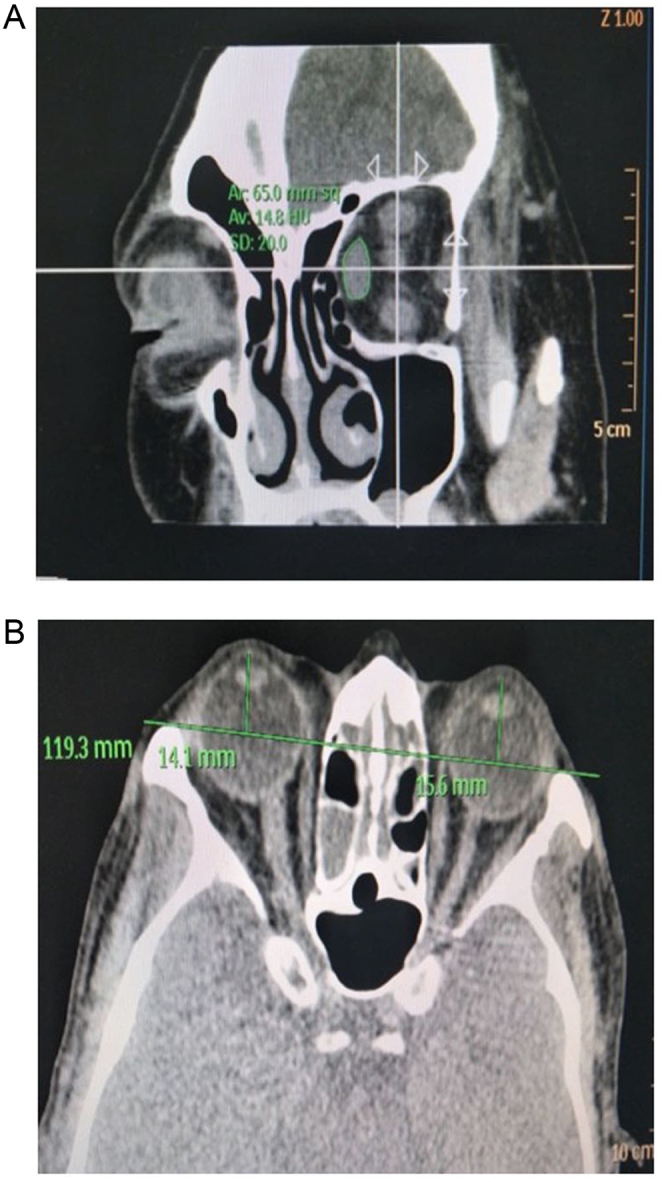
Coronal and axial CT images of a representative patient with GO. (A) Measurement of the cross-sectional area of the OM and the orbit. On the coronal plane of CT scan of the orbit, with the back of the eyeball as the tangent line, layer thickness 1 mm, and pitch 1 mm, 2-mm plane was found behind the ball. Superior, lateral, inferior, and medial extraocular muscle areas were observed. The cross-sectional area of the orbit and each extraocular muscle was measured by AutoCAD software, and each extraocular muscle was measured three times to obtain an average value. (B) Measurement of exophthalmometry. Exophthalmometry was measured according to the standard that the center of the lens and the intraorbital segment of the optic nerve are displayed on the same side and the outer edge of the orbit was at the lowest point.
Identification of candidate genes
In a previous study, a genome-scale screening of DNA methylation was performed on the peripheral blood sample from six patients with GO and six controls and 841 differentially methylated regions (DMRs) were extracted (3). Subsequently, the low methylated genes (LMGs) and the high methylated genes (HMGs) were used to construct the gene-regulated networks. Accordingly, the top ten genes with outstanding topological properties were selected as candidate genes. Next, a large sample consisting of 48 patients with GO and 24 normal controls was utilized to validate the association among these extracted candidate genes. A targeted bisulfite sequencing assay showed that the loci at ANGEL1, BECN1, BOLL, CDK5, IL17RE, LYAR, and MBP differed significantly in DMRs between GO patients and controls (5).
Targeted bisulfite sequencing assay
Briefly, the DNAm level was analyzed by MethylTarget (Genesky Biotechnologies Inc., Shanghai, China), a next-generation sequencing (NGS)-based multiple-target CpG methylation analysis method. Specifically, the genomic regions of interest were analyzed and transformed into bisulfite-converted sequences using geneCpG software. The primers for the PCR were designed from the bisulfite-converted DNA using the Methylation Primer software (18). Genomic DNA was subjected to sodium bisulfite treatment using the EZ DNA Methylation-Gold Kit (Zymo Research). Multiplex PCR was performed with optimized primers. Libraries from different samples were quantified and pooled together, followed by sequencing on the Illumina MiSeq platform. Filtered reads were mapped to the genome using BLAST. After recalibrating the reads with USEARCH, methylation and haplotype were analyzed using the Perl script, as described previously (5).
Statistical analysis
The normally distributed measurement data were expressed as mean ± s.d. The two groups were compared using a t-test. The non-normally distributed data were expressed by the median and interquartile range (IQR). The comparison between the two groups was conducted using the Mann–Whitney U-test. The association analysis between DNA methylation levels of the identified candidate genes and OM/TOA ratio or exophthalmometry was performed by Pearson’s or Spearman’s correlation test. Multiple linear regression analysis was used to analyze the association of clinical data, DNA methylation levels of genes, and OM/TOA ratio. The partial least squares analysis was performed to establish the correlation between DNA methylation block and CT index block. K-means clustering method was utilized to cluster the samples based on DNA methylation levels of the seven candidate genes. K-means clustering is one of the unsupervised learning algorithms, in which grouping is based on a set of numerical features of the objects. It is optimal when the data points are centered around the specific cluster and each data point belongs to the closest cluster. Generally, the selection of ‘K’ value is based on the experience or multiple experimental results. Owing to the small sample size in this study, we set K = 2 to ensure that sufficient number of samples were allocated to each cluster for downstream further analysis.
All statistical analyses were conducted using the SPSS version 11.5 software package (SPSS Inc.) for Windows and mixOmics package of R software (http://www.r-project.org). A two-sided P-value of < 0.05 was considered statistically significant.
Results
Clinical data
The characteristics of 35 GO subjects are summarized in Table 1, including age, gender, clinical activity score (CAS), thyroid function, and thyroid autoantibody. In these subjects (70 eyes), the areas of the section of the superior, inferior, lateral, medial OM, the area of orbits, and values of exophthalmometry were measured. Every OM/TOA ratio was calculated. The subjects’ values for each OM area, TOA, OM/TOA, and exophthalmometry are shown in Table 2.
Table 1.
Clinical characteristics of GO subjects.
| Sex (M/F) | Age (year) | Height (cm) | Weight (kg) | GO Duration (months) | CAS | FT3 (pmol/L) | FT4 (pmol/L) | T3 (pmol/L) | T4 (pmol/L) | TSH (uIU/mL) | TPOAb (IU/mL) | TGAb (IU/mL) | TRAb (IU/L) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case (35) | 18/17 | 47.9±10.6 | 167.9±7.8 | 68.6±10.0 | 6 (4, 12) | 3 (2, 3) | 5.32 (4.67, 6.62) | 16.6 (13.8, 18.7) | 1.87 (1.61, 2.36) | 109 (94, 137) | 0.42 (0.03, 2.67) | 12.1 (7.5, 208) | 16.3 (10.9, 226) | 4.18 (1.22, 11.2) |
Values are expressed as n, means ± s.d., median (25th percentile, 75th percentile).
CAS, clinical activity score; TGAb, antithyroglobulin antibody; TPOAb, thyroperoxidase antibody; TRAb, thyrotrophin receptor antibody; TSH, thyroid stimulating hormone.
Table 2.
Extraocular muscle area, orbital area and exophthalmometry measurement in GO subjects.
| SR (mm2) | IR (mm2) | MR (mm2) | LR (mm2) | Left eye | Right eye | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TOA (mm2) | OM/TOA | Exophthalmometry (mm) | SR (mm2) | IR (mm2) | MR (mm2) | LR (mm2) | TOA (mm2) | OM/TOA | Exophthalmometry (mm) | |||||
| Mean | 53.6 | 41.1 | 41.7 | 30.0 | 911.3 | 0.19 | 19.7 | 49.5 | 40.1 | 41.7 | 31.8 | 887.5 | 0.19 | 19.4 |
| s.d. | 23.8 | 17.2 | 17.6 | 7.5 | 141.3 | 0.06 | 3.1 | 18.5 | 17.4 | 22.4 | 8.4 | 164.3 | 0.06 | 3.4 |
IR, inferior rectus; LR, lateral rectus; MR, medial rectus; OM, orbital muscles; SR, superior rectus; TOA, total orbit area.
DNA methylation levels at the identified candidate loci
In our previous study, a genome-scale screening of DNA methylation in GO subjects was performed, followed by the construction of LMG- and HMG-regulated networks and the top LMGs and HMGs were extracted based on the topological properties. Next, the extracted candidate genes were validated in a large population. The DNA methylation levels of the genes identified in the GO subjects in this study were tested (Supplementary Table 1), see section on supplementary materials given at the end of this article).
Correlation analysis between DNA methylation levels of candidate genes and CT index
The correlation analysis was performed to find the potential association between the DNA methylation levels of seven candidate genes and exophthalmometry or OM/TOA ratio. The results showed that there was no significant correlation between the DNA methylation levels of these genes and exophthalmometry. However, the MBP methylation level was found to be correlated with OM/TOA ratio (P = 0.039) (Fig. 2).
Figure 2.
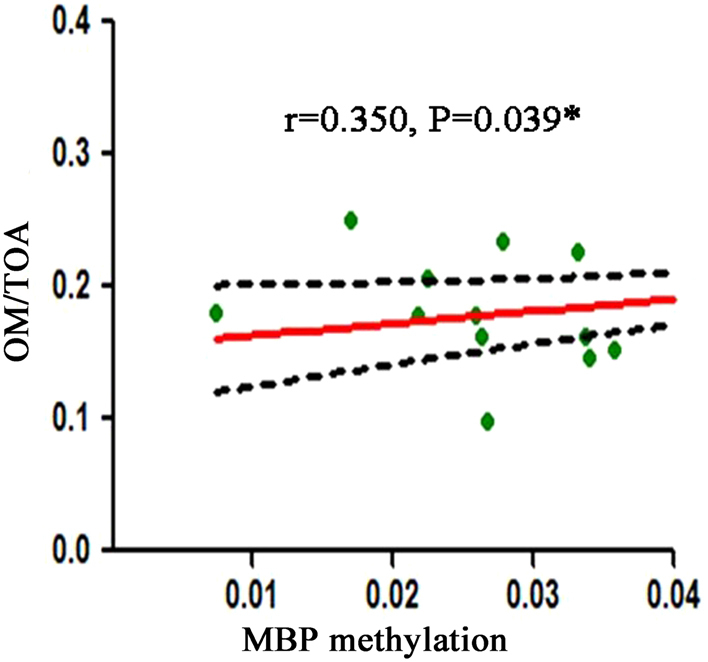
Correlation between MBP DNA methylation level and OM/TOA ratio. OM/TOA, orbital muscles/total orbit area.
Multiple linear regression analysis of clinical characteristics, DNA methylation levels of candidate genes, and OM/TOA ratio
Next, we performed multiple linear regression analysis including age, sex, duration of GO, TRAb level, and DNA methylation levels of seven genes with OM/TOA ratio and confirmed a significant correlation only between MBP and OM/TOA ratio (P = 0.035) (Table 3). This phenomenon indicated that OM/TOA ratio increases by 1.293 units if the DNA methylation level of MBP increased by 1 unit.
Table 3.
Multiple linear regression analysis of clinical characteristics, DNA methylation levels of candidate genes and OM/TOA ratio.
| Variables | B | s.e. | Beta | t | P |
|---|---|---|---|---|---|
| (constant) | 0.131 | 0.100 | 1.311 | 0.205 | |
| Sex | −0.020 | 0.022 | −0.177 | −0.877 | 0.392 |
| Age | 0.000 | 0.001 | −0.093 | −0.496 | 0.626 |
| Duration | −0.003 | 0.002 | −0.475 | −2.030 | 0.057 |
| TRAb | 0.000 | 0.001 | 0.044 | 0.192 | 0.850 |
| BECN1 | 3.485 | 8.318 | 0.093 | 0.419 | 0.680 |
| BOLL | 1.859 | 2.197 | 0.220 | 0.846 | 0.408 |
| MBP | 1.293 | 0.571 | 0.496 | 2.264 | 0.035a |
| ANGEL1 | 0.141 | 2.030 | 0.016 | 0.070 | 0.945 |
| CDK5 | 1.309 | 1.131 | 0.295 | 1.158 | 0.261 |
| IL17RE | 0.735 | 1.880 | 0.075 | 0.391 | 0.700 |
| LYAR | 0.403 | 2.120 | 0.044 | 0.190 | 0.851 |
B, unstandardized coefficients; Beta, standardized coefficients; s.e., standard error of unstandardized coefficients; TRAb, thyrotrophin receptor antibody.
aP < 0.05.
Partial least squares analysis
The partial least squares analysis was performed to find the association between the gene block and the CT index block (Fig. 3A). The loadings of the gene markers indicated the association strength to the first principal component (named as comp 1) of the CT index block. The X-axis indicates the loadings, and the Y-axis indicates the name of the gene markers. A gene with a large absolute value of the loadings suggested a strong association with the CT index block. Genes with positive loading indicated a positive association with the CT index block, whereas genes in the opposite direction indicated a negative association with the CT index block. Similarly, loadings of the CT index indicated the association strength to the first principal component (names as comp 1) of the gene block (seven gene markers). The X-axis indicates the loadings, and Y-axis indicates the name of the CT index. A CT index with a high absolute value of the loadings indicated a strong correlation between this index and the gene block. OM/TOA with positive loading indicated a positive association with gene block, whereas exophthalmometry in the opposite direction indicated a negative association with the gene block. These results revealed that the top three genes with the highest loadings were MBP, BOLL, and BECN1, indicating that these genes exert significant effects on the CT index block. Similarly, the OM/TOA ratio had greater loading than exophthalmometry, indicating that it exerted a marked effect on the gene block. In addition, for the two principal components (comp 1 and comp 2) extracted by partial least squares analysis, MBP had the highest variable importance in the projection (VIP) value, hinting toward its significant effect on CT indicators (Fig. 3B).
Figure 3.
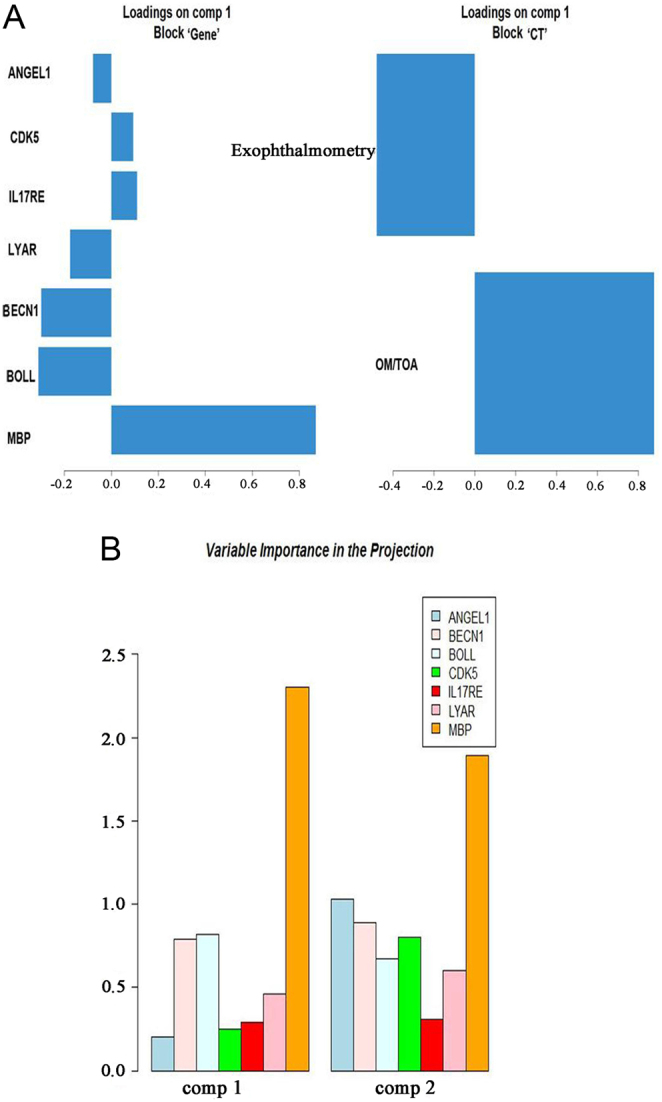
Association between DNA methylation levels of seven candidate genes and CT indicators based on partial least squares analysis. (A) The loadings of levels of seven candidate genes and CT indicators. (B) The VIP values of seven genes in two principal components. OM/TOA, orbital muscles/total orbit area; VIP, variable importance in the projection.
K-means clustering based on DNA methylation levels of candidate genes
The K-means clustering method was applied to assign the samples into two clusters (cluster 1 and cluster 2) based on the DNA methylation levels of the seven genes (Fig. 4). We compared the clinical data, CT indicators, and DNA methylation levels between the two clusters and found that the subjects in cluster 1 had significantly higher MBP methylation and lower TRAb levels (P < 0.001 and P = 0.025). Furthermore, OM/TOA ratio was higher (0.21 ± 0.06 vs 0.18 ± 0.04) and exophthalmometry value (19.7 ± 2.9 vs 21.1 ± 3.6) was lower in cluster 1 than cluster 2, albeit not significantly (P = 0.132 and 0.221, respectively) (Table 4). This result indicated that MBP plays a major role in gene block consisting of seven candidate genes with the highest loading and VIP value in partial least squares analysis, as well as the correlation between MBP methylation level and OM/TOA ratio in the correlation analysis. Higher TRAb level in cluster 2 showed that it might be correlated with a high level of exophthalmometry (mainly FV), but not OM/TOA ratio (MV), as compared to cluster 1. TRAb stimulates orbital and periorbital tissues and is also crucial for the development of GO (19). Nonetheless, the correlation between TRAb and inflammation of different retrobulbar tissue needs to be further explored.
Figure 4.
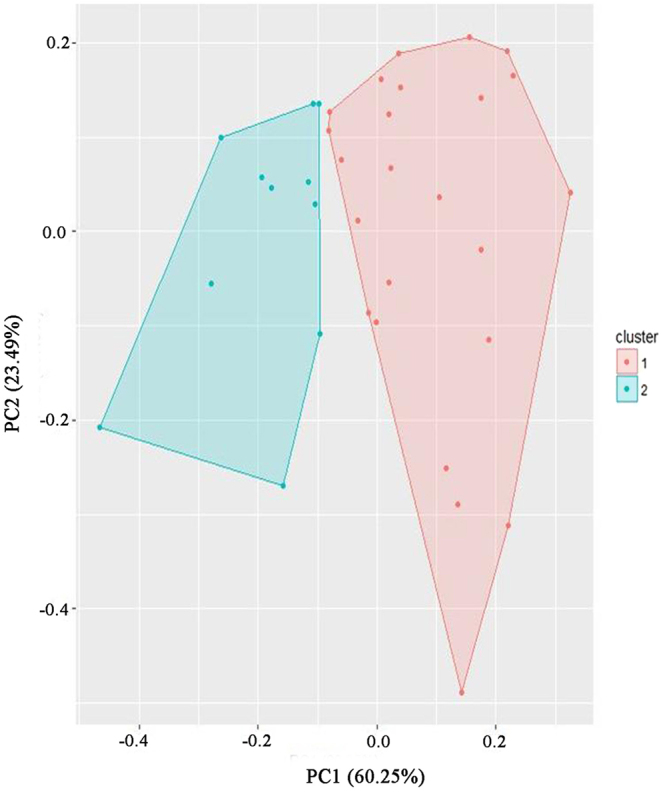
K-means clustering to cluster the samples into two clusters based on the DNA methylation levels of seven candidate genes. Candidate genes: ANGEL1, BECN1, BOLL, CDK5, IL17RE, LYAR, and MBP.
Table 4.
Comparison of clinical characteristics and DNA methylation levels of candidate genes between cluster 1 and cluster 2 by K-means clustering.
| Variables | Cluster 1 (n = 24) | Cluster 2 (n = 11) | P |
|---|---|---|---|
| Sex | 14/10 | 5/6 | 0.716 |
| Age (year) | 46.6 ± 11.9 | 50.5 ± 7.2 | 0.337 |
| Duration (month) | 6 (6, 9) | 8 (6, 12) | 0.188 |
| CAS | 3 (2, 3) | 3 (2, 3) | 0.969 |
| TPOAb (IU/mL) | 15.2 (7.5, 248) | 10.4 (7.6, 123) | 0.479 |
| TGAb (IU/mL) | 18.0 (10.2, 153) | 14.3 (11.2, 267) | 0.774 |
| TRAb (IU/L) | 2.81 (0.78, 8.20) | 9.91 (3.03, 40.0) | 0.025a |
| Exophthalmometry (mm) | 19.7 ± 2.9 | 21.1 ± 3.6 | 0.221 |
| OM/TOA ratio | 0.21 ± 0.06 | 0.18 ± 0.04 | 0.132 |
| DNA methylation level (×102) | |||
| BECN1 | 0.63 ± 0.11 | 0.73 ± 0.19 | 0.054 |
| BOLL | 1.15 ± 0.49 | 1.57 ± 0.90 | 0.173 |
| MBP | 5.91 ± 1.64 | 2.53 ± 0.82 | <0.001a |
| ANGEL1 | 0.64 ± 0.52 | 0.81 ± 0.79 | 0.465 |
| CDK5 | 0.90 ± 1.04 | 1.69 ± 1.68 | 0.097 |
| IL17RE | 1.92 ± 0.52 | 2.15 ± 0.65 | 0.265 |
| LYAR | 0.84 ± 0.53 | 1.07 ± 0.71 | 0.302 |
Values are expressed as n, means (s.d.), and median (interquartile range).
CAS, Clinical Activity Score; OM/TOA, orbital muscles/total orbit area ratio; TGAb, antithyroglobulin antibody; TPOAb, thyroperoxidase antibody; TRAb, thyrotrophin receptor antibody; TSH, Thyroid Stimulating Hormone .
aP < 0.05.
Discussion
The clinical features of GO vary from mild grittiness of the eyes to disfiguring proptosis, severe diplopia, and even loss of vision (20, 21). Some patients with GO have enlarged muscles (edema and inflammation), some have increased orbital fat, and others have both enlarged muscles and increased orbital fat (8, 22). However, the pathogenesis underlying the alterations of orbital architecture is not yet understood. In this study, the MBP methylation level was found to be correlated with OM/TOA ratio. Also, a significant correlation was identified between the MBP methylation level and OM/TOA ratio by multiple linear regression analyses. MBP, BOLL, and BECN1 were the top three genes that affected the CT index.
A large number of molecular immunology studies have elucidated the inflammation of retrobulbar tissue; however, less attention has been paid to the differential involvement of OM and orbital fat (11). The former is mainly associated with a high extraocular MV, while the latter is associated with a high FV, as displayed by the CT image. A previous study showed that patients with increased MV had more impaired ductions than those without increased MV (10). In patients with dysthyroid optic neuropathy (DON), the most severe manifestation of GO, apical muscle enlargement, might be more important than orbital fat enlargement (8). Other studies also showed that extraocular MV was related to the severity of GO (7, 11). Moreover, a high orbital FV was associated with a prolonged duration of GO in CT images, deeming it as a late phenomenon (11). In conclusion, the measurement of orbital soft tissue volumes in patients with GO is helpful in understanding the etiology and pathogenesis (8) and in estimating the clinical severity and activity of the disease.
Hitherto, there is no consensus on how to measure extraocular MV and FV. In some studies, the largest portion of the cross-section of the eye muscle from serial sections of the coronal image of CT was selected for estimation of the muscle area (23). In another study, 3D image analysis measured the volume and density of the intraorbital and extraorbital FV and MV (24). However, because of the time required and the complexity, these measurements are suitable only for research purposes (25). In the present study, commercial software was used with a segmentation technique to calculate OM and TOA, chosen at 2 mm backward at the globe pole tangent on a coronal CT scan of the orbit (7). It is a simple and relatively accurate method to determine the cross-section areas of the MV and orbit and could be applied in clinical practice. The results showed that the methylation level of MBP was correlated with OM/TOA ratio, but no significant correlation was established between the DNA methylation levels of the genes and the exophthalmometry value. The OM/TOA ratio determines the MV, while exophthalmometry determines both MV and FV. This might lead to a discrepancy in the correlation between DNA methylation for OM/TOA and proptosis.
Although the MBP gene has not yet been reported in GO pathogenesis, it was validated as the significant DMR between GO and controls (5) and its methylation level was found to be correlated with OM/TOA ratio in this study. Intriguingly, MBP is highly expressed in the CNS, and MBP-related transcripts are also present in the bone marrow and the immune system. The isoforms of the gene may be involved in signaling pathways in T-cells and neural cells. Some studies have shown that myelin basic protein (MBP) induces inflammation in rats’ eyes after s.c. injection. LEW rats injected with MBP developed experimental autoimmune encephalomyelitis (EAE) and associated anterior uveitis (AU), which was mediated by CD4 (+) T cells. After reinjection with MBP, the rats developed AU without EAE (26). Th1 cells produced the major inflammatory mediators in the rat model: IL-2 and IFN-γ (27). The MBP-induced models of T-cell-mediated autoimmune CNS disease exhibited a number of symptoms similar to multiple sclerosis (MS) involving the brain and eye (28). Although the specific mechanisms underlying this phenomenon are not yet elucidated, studies have suggested that minor epitopes of MBP are involved by the expansion of the sensitized T cells (27, 29). Therefore, further studies are needed to elucidate the precise roles of MBP in the inflammation of retrobulbar tissue in GO patients.
Nevertheless, the study has some limitations. First, the small sample size might be insufficient for conclusions regarding the methylation status in GO subjects. The correlation coefficient between the MBP methylation level and OM/TOA displays a weak association, which could become obvious if a certain number of samples was utilized. Although the power is close to 80% according to the current sample size, correlation coefficient, and significance level, a large sample size is needed to validate this finding. Second, we could not validate the significant difference of OM/TOA ratio in two of the clusters by K-means clustering based on the DNA methylation levels of the candidate genes. Thus, large cohorts of GO subjects need to be enrolled for genetic and orbital imaging analyses to confirm the current findings. Finally, although increased methylation is known to decrease the gene expression levels in some diseases, those of the candidate genes need to be analyzed further.
In summary, this study provided preliminary evidence that MBP might be a potential gene associated with OM enlargement in GO patients, as assessed by the combination of DNA methylation sequencing and orbital CT measurement. This study was the first clinical investigation of the association between MBP gene and the phenotype of GO. However, further studies are needed to elucidate the mechanism underlying this disease.
Supplementary Material
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Acknowledgements
The authors thank all the participants and staff involved in the study.
Funding
This work was supported by the Beijing Municipal Hospital Research and Development Program (PX2016063), the Expert Promotion Program of Beijing Health Systems (2015-3-017) to Zhong Xin, and Thyroid Research Program of Young and Middle-aged Physicians by China Health Promotion Foundation (2019) to Ya-Fen Hu.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Author contribution statement
Z X and L H contributed to develop research methodology, analysis and discussion, and wrote the draft of the manuscript. Y F H, X T, Y L Y, T T S, Y F L and Z Y Y coordinated to collect and analyze research data. Y F H and Z X contributed to the discussion. Z X contributed to the design of the study, research data analysis and discussion, and wrote the manuscript and coordinated submission. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was conducted with the approval from the Ethics Committee of Beijing Tongren Hospital, Capital Medical University. Informed written consents were obtained from all individual participants included in this study.
References
- 1.Piantanida E, Tanda ML, Lai A, Sassi L, Bartalena L. Prevalence and natural history of Graves’ orbitopathy in the XXI century. Journal of Endocrinological Investigation 2013. 36 444–449. ( 10.3275/8937) [DOI] [PubMed] [Google Scholar]
- 2.Leo M, Menconi F, Rocchi R, Latrofa F, Sisti E, Profilo MA, Mazzi B, Albano E, Nardi M, Vitti P, et al Role of the underlying thyroid disease on the phenotype of Graves’ orbitopathy in a tertiary referral center. Thyroid 2015. 25 347–351. ( 10.1089/thy.2014.0475) [DOI] [PubMed] [Google Scholar]
- 3.Xin Z, Hua L, Shi TT, Tuo X, Yang FY, Li Y, Cao X, Yang JK. A genome-wide DNA methylation analysis in peripheral blood from patients identifies risk loci associated with Graves’ orbitopathy. Journal of Endocrinological Investigation 2018. 41 719–727. ( 10.1007/s40618-017-0796-6) [DOI] [PubMed] [Google Scholar]
- 4.Xin Z, Hua L, Yang YL, Shi TT, Liu W, Tuo X, Li Y, Cao X, Yang FY. A pathway analysis based on genome-wide DNA methylation of chinese patients with Graves’ orbitopathy. BioMed Research International 2019. 2019 9565794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi TT, Hua L, Xin Z, Li Y, Liu W, Yang YL. Identifying and validating genes with DNA methylation data in the context of biological network for Chinese patients with Graves’ orbitopathy. International Journal of Endocrinology 2019. 2019 6212681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kahaly GJ. Imaging in thyroid-associated orbitopathy European Journal of Endocrinology 2001. 145 107–118. ( 10.1530/eje.0.1450107) [DOI] [PubMed] [Google Scholar]
- 7.Le Moli R, Pluchino A, Muscia V, Regalbuto C, Luciani B, Squatrito S, Vigneri R. Graves’ orbitopathy: extraocular muscle/total orbit area ratio is positively related to the clinical activity score. European Journal of Ophthalmology 2012. 22 301–308. ( 10.5301/ejo.5000018) [DOI] [PubMed] [Google Scholar]
- 8.Al-Bakri M, Rasmussen AK, Thomsen C, Toft PB. Orbital volumetry in Graves’ orbitopathy: muscle and fat involvement in relation to dysthyroid optic neuropathy. ISRN Ophthalmology 2014. 2014 435276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagy EV, Toth J, Kaldi I, Damjanovich J, Mezosi E, Lenkey A, Toth L, Szabo J, Karanyi Z, Leovey A. Graves’ ophthalmopathy: eye muscle involvement in patients with diplopia. European Journal of Endocrinology 2000. 142 591–597. ( 10.1530/eje.0.1420591) [DOI] [PubMed] [Google Scholar]
- 10.Regensburg NI, Wiersinga WM, Berendschot TT, Potgieser P, Mourits MP. Do subtypes of Graves’ orbitopathy exist? Ophthalmology 2011. 118 191–196. ( 10.1016/j.ophtha.2010.04.004) [DOI] [PubMed] [Google Scholar]
- 11.Potgieser PW, Wiersinga WM, Regensburg NI, Mourits MP. Some studies on the natural history of Graves’ orbitopathy: increase in orbital fat is a rather late phenomenon. European Journal of Endocrinology 2015. 173 149–153. ( 10.1530/EJE-14-1140) [DOI] [PubMed] [Google Scholar]
- 12.Smith TJ, Hegedus L, Douglas RS. Role of insulin-like growth factor-1 (IGF-1) pathway in the pathogenesis of Graves’ orbitopathy. Best Practice and Research Clinical Endocrinology and Metabolism 2012. 26 291–302. ( 10.1016/j.beem.2011.10.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiersinga WM. Autoimmunity in Graves’ ophthalmopathy: the result of an unfortunate marriage between TSH receptors and IGF-1 receptors? Journal of Clinical Endocrinology and Metabolism 2011. 96 2386–2394. ( 10.1210/jc.2011-0307) [DOI] [PubMed] [Google Scholar]
- 14.Yin X, Latif R, Bahn R, Davies TF. Genetic profiling in Graves’ disease: further evidence for lack of a distinct genetic contribution to Graves’ ophthalmopathy. Thyroid 2012. 22 730–736. ( 10.1089/thy.2012.0007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartalena L. Graves’ orbitopathy: imperfect treatments for a rare disease. European Thyroid Journal 2013. 2 259–269. ( 10.1159/000356042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arakawa Y, Watanabe M, Inoue N, Sarumaru M, Hidaka Y, Iwatani Y. Association of polymorphisms in DNMT1, DNMT3A, DNMT3B, MTHFR and MTRR genes with global DNA methylation levels and prognosis of autoimmune thyroid disease. Clinical and Experimental Immunology 2012. 170 194–201. ( 10.1111/j.1365-2249.2012.04646.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartalena L, Baldeschi L, Boboridis K, Eckstein A, Kahaly GJ, Marcocci C, Perros P, Salvi M, Wiersinga WM. The 2016. European Thyroid Association; /European Group on Graves’ Orbitopathy guidelines for the management of Graves’ orbitopathy. European Thyroid Journal 2016. 5 9–26. ( 10.1159/000443828) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Tang D, Shen P, Xu H, Qiu H, Wu T, Gao X. Analysis of DNA methylation in chondrocytes in rats with knee osteoarthritis. BMC Musculoskeletal Disorders 2017. 18 377 ( 10.1186/s12891-017-1739-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seo S, Sanchez Robledo M. Usefulness of TSH receptor antibodies as biomarkers for Graves’ ophthalmopathy: a systematic review. Journal of Endocrinological Investigation 2018. 41 1457–1468. ( 10.1007/s40618-018-0945-6) [DOI] [PubMed] [Google Scholar]
- 20.Bartalena L, Pinchera A, Marcocci C. Management of Graves’ ophthalmopathy: reality and perspectives. Endocrine Reviews 2000. 21 168–199. ( 10.1210/edrv.21.2.0393) [DOI] [PubMed] [Google Scholar]
- 21.Kim HC, Yoon SW, Lew H. Usefulness of the ratio of orbital fat to total orbit area in mild-to-moderate thyroid-associated ophthalmopathy. British Journal of Radiology 2015. 88 20150164 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKeag D, Lane C, Lazarus JH, Baldeschi L, Boboridis K, Dickinson AJ, Hullo AI, Kahaly G, Krassas G, Marcocci C, et al Clinical features of dysthyroid optic neuropathy: a European Group on Graves’ Orbitopathy (EUGOGO) survey. British Journal of Ophthalmology 2007. 91 455–458. ( 10.1136/bjo.2006.094607) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang TC, Huang KM, Hsiao YL, Tzeng SS, Kao SC. Relationships of orbital computed tomographic findings and activity scores to the prognosis of corticosteroid therapy in patients with Graves’ ophthalmopathy. Acta Ophthalmologica Scandinavica 1997. 75 301–304. ( 10.1111/j.1600-0420.1997.tb00779.x) [DOI] [PubMed] [Google Scholar]
- 24.Byun JS, Moon NJ, Lee JK. Quantitative analysis of orbital soft tissues on computed tomography to assess the activity of thyroid-associated orbitopathy. Graefes Archive for Clinical and Experimental Ophthalmology 2017. 255 413–420. ( 10.1007/s00417-016-3538-0) [DOI] [PubMed] [Google Scholar]
- 25.Bijlsma WR, Mourits MP. Radiologic measurement of extraocular muscle volumes in patients with Graves’ orbitopathy: a review and guideline. Orbit 2006. 25 83–91. ( 10.1080/01676830600675319) [DOI] [PubMed] [Google Scholar]
- 26.Manczak M, Jiang S, Orzechowska B, Adamus G. Crucial role of CCL3/MIP-1alpha in the recurrence of autoimmune anterior uveitis induced with myelin basic protein in Lewis rats. Journal of Autoimmunity 2002. 18 259–270. ( 10.1006/jaut.2002.0591) [DOI] [PubMed] [Google Scholar]
- 27.Gasparin F, Takahashi BS, Scolari MR, Gasparin F, Pedral LS, Damico FM. Experimental models of autoimmune inflammatory ocular diseases. Arquivos Brasileiros de Oftalmologia 2012. 75 143–147. ( 10.1590/S0004-27492012000200016) [DOI] [PubMed] [Google Scholar]
- 28.Adamus G, Chan CC. Experimental autoimmune uveitides: multiple antigens, diverse diseases. International Reviews of Immunology 2002. 21 209–229. ( 10.1080/08830180212068) [DOI] [PubMed] [Google Scholar]
- 29.Adamus G, Sugden B, Arendt A, Hargrave PA. Importance of cryptic myelin basic protein epitopes in the pathogenicity of acute and recurrent anterior uveitis associated with EAE. Journal of Neuroimmunology 2001. 113 212–219. ( 10.1016/S0165-5728(00)00439-2) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.



 This work is licensed under a
This work is licensed under a