Abstract
Allogeneic hematopoietic stem cell transplantation (HSCT) and xenotransplantation are accompanied by viral reactivations and virus-associated complications resulting from immune deficiency. Here, in a Mauritian cynomolgus macaque model of fully MHC-matched allogeneic HSCT, we report reactivations of cynomolgus polyomavirus, lymphocryptovirus, and cytomegalovirus, macaque viruses analogous to HSCT-associated human counterparts BK virus, Epstein-Barr virus, and human Cytomegalovirus. Viral replication in recipient macaques resulted in characteristic disease manifestations observed in HSCT patients, such as polyomavirus-associated hemorrhagic cystitis and tubulointerstitial nephritis or lymphocryptovirus-associated post-transplant lymphoproliferative disorder. However, in most cases, the reconstituted immune system, alone or in combination with short-term pharmacological intervention, exerted control over viral replication, suggesting engraftment of functional donor-derived immunity. Indeed, the donor-derived reconstituted immune systems of two long-term engrafted HSCT recipient macaques responded to live attenuated yellow fever 17D vaccine (YFV 17D) indistinguishably from untransplanted controls, mounting 17D-targeted neutralizing antibody responses and clearing YFV 17D within 14 days. Together, these data demonstrate that this macaque model of allogeneic HSCT recapitulates clinical situations of opportunistic viral infections in transplant patients, and provides a pre-clinical model to test novel prophylactic and therapeutic modalities.
Keywords: Hematopoietic stem cell transplantation, opportunistic infections, transplant complications, nonhuman primates, Mauritian cynomolgus macaques, lymphocryptovirs, cytomegalovirus, polyomavirus, yellow fever virus
INTRODUCTION
Infectious disease complications are a major cause of morbidity and mortality after allogeneic hematopoietic stem cell transplantation (HSCT) and xenotransplantation {Haustein:2008ws, Boneva:2001gc, 1,2. BK Polyomavirus (BKV), Epstein-Barr virus (EBV), and human Cytomegalovirus (HCMV) are ubiquitous, persistent, opportunistic viruses and major causative agents of disease in allogeneic HSCT patients3-9. In the immunodeficient setting of allogeneic HSCT, lack of effective antiviral immunity can result in viral reactivation and serious virus-associated complications, including BKV-associated hemorrhagic cystitis and nephropathy, EBV-associated post-transplant lymphoproliferative disorder (EBV-PTLD), and HCMV-associated pneumonia and colitis10-16. Limited antivirals and therapies exist to address these viral complications in allogeneic HSCT patients and are accompanied with significant toxicitiy12. Thus, new treatments are needed to reduce opportunistic infection-mediated patient morbidity and mortality. The development of such novel antiviral therapeutics would be greatly accelerated by an animal model that recapitulates human viral reactivations in the setting of allogeneic HSCT.
We recently developed a nonhuman primate model of allogeneic HSCT using Mauritian cynomolgus macaques17, and sought to determine the competence of reconstituted immune systems post-transplant. To this end, we first examined the extent of viral reactivation and disease emergence in transplanted recipient macaques, including both SIV-naïve macaques and SIV-infected macaques maintained on antiretroviral therapy with fully suppressed SIV viremia. Macaques are ubiquitously infected with polyomaviruses, lymphocryptoviruses, and cytomegaloviruses, viruses closely related to human viruses BK, EBV, and HCMV, respectively18. Due to the close evolutionary and physiological relationship between humans and macaques and between human and macaque viruses19,20, we hypothesized that allogeneic HSCT recipient Mauritian cynomolgus macaques would experience reactivations of macaque viruses analogous to human viruses known to reactivate and cause disease in human allogeneic HSCT recipients. Second, to directly test the competence of post-transplant reconstituted immune systems, we examined the response to live attenuated yellow fever vaccine 17D (YFV 17D) in two allogeneic HSCT recipient macaques vaccinated 26-29 months post-transplant and possessing fully donor-derived immune systems. Our results demonstrate (1) reactivation of human-analogous, clinically relevant opportunistic viruses in Mauritian cynomolgus macaques undergoing allogeneic HSCT, and (2) the ability of HSCT-reconstituted macaque immune systems to eventually control opportunistic viral replication and respond to and clear a live attenuated virus vaccine. Thus, this pre-clinical animal model can be used in the development of novel antivirals and therapeutic regimens for allogeneic HSCT-associated opportunistic infections. Furthermore, because viral reactivation and disease are major adverse events in preclinical macaque models of xenotransplantation during immunosuppression, the results presented here will benefit these translational animal studies by improving diagnosis and treatment of opportunistic viral infections.
MATERIALS AND METHODS
Animals
Mauritian cynomolgus macaques (Macaca fascicularis), 4–10 years of age, were housed at the Oregon National Primate Research Center (ONPRC) under the approval of the Oregon Health and Science University (OHSU) West Campus Institutional Animal Care and Use Committee (IACUC). All macaques in this study were managed according to the ONPRC animal care program, which is fully accredited by AAALAC International and is based on the laws, regulations, and guidelines set forth by the United States Department of Agriculture (e.g., the Animal Welfare Act and its regulations, and the Animal Care Policy Manual), Institute for Laboratory Animal Research (e.g., Guide for the Care and Use of Laboratory Animals, 8th edition), and the Public Health Service Policy on Humane Care and Use of Laboratory Animals. The nutritional plan utilized by the ONPRC is based on National Research Council recommendations and supplemented with a variety of fruits, vegetables, and other edible objects as part of the environmental enrichment program established by the Behavioral Management Unit. Paired/grouped animals exhibiting incompatible behaviors were reported to the Behavioral Management staff and managed accordingly. All efforts were made to minimize suffering through the use of minimally invasive procedures, anesthetics, and analgesics when appropriate. During the peri-transplant period, animals had indwelling subclavian catheters and were placed on a tether catheter protection system, which allowed for full range of motion as well as blood collection and drug delivery without sedation17. Animals were acclimated to the tether catheter protection system -- approximately 30-40 days prior to transplantation, prior to catheterization, animals were placed in the jacket and on tether. Where indicated, animals were painlessly euthanized with sodium pentobarbital and euthanasia was assured by exsanguination and bilateral pneumothorax, consistent with the recommendations of the American Veterinary Medical Guidelines on Euthanasia (2013).
Pre-transplant conditioning, peri-transplant care, and immunosuppression.
HSCT recipient Mauritian cynomolgus macaques were transplanted with leukapheresis product from mobilized, MHC-matched unrelated donors as previously described17. This study includes 11 HSCT recipient macaques, including 4 SIV-naïve macaques and 7 SIVmac239-infected treated with once daily injectable combination antiretroviral therapy consisting of 5.1 mg/kg tenofovir disoproxil fumarate, 40 mg/kg emtricitabine, and 2.5 mg/kg dolutegravir. In all SIVmac239-infected macaques, SIV viremia was fully suppressed throughout the study described here. Detailed transplant regimens for each HSCT recipient macaque studied are shown in Figure S4. Nine of eleven studied HSCT recipient Mauritian cynomolgus macaques (MCM) underwent reduced intensity conditioning with a previously described regimen consisting of CD3-immunotoxin (acquired through the NHP Reagent Resource, Boston, MA), busulfan, and total body irradiation (TBI, delivered at 10-25 cGy/min using an Elektra Synergy system) 17. Two of eleven studied HSCT recipient MCM underwent reduced intensity conditioning with a slightly altered regimen of CD3-immunotoxin and two doses of busulfan, without TBI. All eleven studied recipient MCM received anti-GVHD prophylaxis consisting of daily tacrolimus given as an intramuscular (IM) injection SID, with dose adjusted to maintain a tacrolimus trough level of 5-15 ng/ml in whole blood (measured by immunoassay, Abbott Architect i2000). Prior to tacrolimus discontinuation, tacrolimus dose was tapered by 20% each day until dose fell below 0.002 mg/kg, at which point administration was discontinued. Ten of eleven studied HSCT recipients also received additional anti-GVHD prophylaxis of post-transplant cyclophosphamide and belatacept. On days of cyclophosphamide treatment, animals received IV Normosol at approximately 15-20 ml/kg over 6-8 hours as well as bladder protectant MESNA (40 mg/kg IV) to prevent sterile hemorrhagic cystitis. One of eleven studied HSCT recipients (ID 35132) received an altered anti-GVHD prophylaxis regimen consisting of tacrolimus and abatacept, without post-transplant cyclophosphamide. Peri-transplant care for all recipients consisted of polymyxin B, neomycin sulfate, cefazolin, and trimethoprim-sulfamethoxazole (30 mg/kg PO, BID from day of cefazolin discontinuation to approximately day +60). Enrofloxacin was administered (10 mg/kg IV, SID) when absolute neutrophil count was less than 1,000/ml of whole blood and continued until absolute neutrophil count exceeded 1,000/ml of whole blood for two consecutive complete blood counts. Some macaques received ondansetron (0.1-0.2 mg/kg PO, SID or BID) and dronabinol (2.5 mg PO, SID to BID) to treat nausea and anorexia. Four of eleven HSCT recipients were treated with prednisone (up to 4 mg/kg/day PO) or methylprednisolone sodium succinate (up to 5 mg/kg/day) to treat suspected or confirmed GVHD. Two macaques received rhesus recombinant anti-CD8α depleting monoclonal antibody (M-T807R1, acquired through the NHP Reagent Resource). Four macaques received rhesus recombinant anti-CD20 depleting monoclonal antibody (CD20-2B8R1, acquired through the NHP Reagent Resource). Serum chemistry and complete blood counts were performed 2-3 times weekly or as clinically indicated. Leukoreduced whole blood or packed red blood cells were transfused when hemoglobin was less than 6 g/dl or hematocrit was less than 20%. Platelet-rich plasma was transfused when platelet counts were less than 20,000 cells/μl blood or if signs of coagulopathy were observed on physical examination. All blood product transfusions were matched according to the ABO blood group antigen system. Animals were treated with intravenous fluids and nephrotoxic drug doses were adjusted as needed when azotemia was detected. Animals received appropriate supportive veterinary care under the direction of an ONPRC veterinarian. Any animal suspected of being in discomfort received analgesics.
Virus detection and monitoring
To detect and measure viral DNA and/or RNA, plasma, urine, or cell culture supernatant was concentrated by centrifugation at >20,000 rcf at 4 °C for 1 h, after which supernatant was removed to leave 200 μl volume. Viral nucleic acid was extracted from plasma using QIAamp MinElute Virus Spin Kit (Qiagen, Venlo, Netherlands) according to manufacturer’s instructions. Polyomavirus DNA was initially detected by polymerase chain reaction (PCR), which was carried out in 25 μl reactions by combining 8.5 μl extracted viral nucleic acid, 12.5 μl Phusion Hot Start Flex 2X mastermix (New England Biolabs, Inc., Ipswich, MA), and 2 μl of each 5 μM primer (forward: 5′-TAGGTGCCAACCTATGGAAC-3′; reverse: 5′-GGAAAGTCTTTAGGGTCTTCTACC-3′) with the following cycling conditions: 98°C 02:00/40 cycles: 98°C 00:15, 52°C 00:20, 72°C 01:00/72°C 3:00. Primers were designed based on previously published sequences for cynomolgus polyomavirus21. Reactions were run on a 2% agarose gel alongside TrackIt 100 bp DNA ladder (Invitrogen, Carlsbad, CA). For sequencing, the 182-bp bands corresponding to polyomavirus large T antigen amplicons were excised, purified using the Nucleospin Gel and PCR cleanup (Macherey-Nagel, Bethlehem, PA), Sanger sequenced using the same primers, and analyzed in Geneious (Biomatters Ltd., Auckland, New Zealand). Quantitative PCR probes for polyomavirus were designed based on the resulting sequences (Table S4). Quantitative PCR primers and probes targeting CyLCV, CyCMV, and YFV 17D were designed based on previously published sequences [Genbank accession numbers KP676001.1 (CyLCV), KX689265 (CyCMV), U17066.1 (YFV 17D), Table S4]. For YFV 17D viral load, cDNA was reverse-transcribed from 10 μl extracted viral nucleic acid prior to quantitative PCR using the RevertAid first strand cDNA synthesis kit (Thermo Scientific, Waltham, MA) as per manufacturer’s instructions, after which 30 μl water was added to dilute the cDNA. Quantitative PCR targeting was carried out in 20 μl reactions by combining 5 μl of extracted viral nucleic acid (polyomavirus, CyLCV, CyCMV) or cDNA (YFV17D), 10 μl TaqMan Fast Advanced Master Mix (Life Technologies, Carlsbad, CA), 1.5 μl of each 6 μM primer, and 2 μl of 2.5 μM probe. Sample reactions were run in duplicate in MicroAmp Fast Optical 96-well reaction plates (Applied Biosystems, Foster City, CA) alongside no template control reactions and standard reactions containing ten-fold serial dilutions of standard (1 copy to 107 copies per reaction, in duplicate). Plasmids (pCEP4 or pUC57) containing each target region served as standards. Plates were run on a StepOnePlus Real-Time PCR system (Applied Biosystems) with the following conditions: 95 °C 00:20, followed by 40 cycles of 95 °C 00:01, 60 °C 00:20. Quantities in each sample reaction were calculated according to the standard curve. For polyomavirus, CyLCV, and CyCMV viral loads, quantities were adjusted for the proportion of viral nucleic acid added to the reaction and initial sample volume to obtain genome copies per milliliter plasma, urine, or cell cultures supernatant. For YFV 17D plasma viral loads, quantities were adjusted for the proportion of viral nucleic acid added to the cDNA synthesis, proportion of cDNA added to the quantitative PCR reaction, and the initial plasma volume to obtain RNA copies/ml plasma. Theoretical limits of detection (LODs) are based on the ability of quantitative PCR assays to reliably detect standard reactions containing 10 copies. For CD20+ cell-associated CyLCV assays, DNA was extracted from CD20+ cells sorted from ACK-treated blood or from tissue mononuclear cell suspensions (see “Flow cytometric analysis and cell sorting” section) using Qiagen DNeasy kit (Qiagen) and eluted in 50 μl water. Quantitative PCR reactions were modified to include primers and probe targeting the cynomolgus macaque albumin gene (Genbank accession number U17066.1, Figure S3): 5 μl of extracted cell-associated DNA, 10 μl TaqMan Fast Advanced Master Mix (Life Technologies), 0.25 μl of each 36 μM primer (CyLCV-F, CyLCV-R, albumin-F, albumin-R, and 2 μl of each 2.5 μM probe (CyLCV and albumin). Cell number input for each quantitative PCR reaction was calculated by dividing the albumin quantity by 2. Cell-associated viral load (copies per 1×106 cells) was calculated by dividing the CyLCV quantity by cell number input, then multiplying by 1×106.
CyCMV treatment
When CyCMV plasma viral loads exceeded 1,000 copies/ml, treatment with valganciclovir (10 mg/kg PO) was initiated SID. If plasma viral loads were not decreased by the subsequent timepoint (3-4 days later), valganciclovir treatment was escalated to BID and continued until plasma viral loads fell below 1,000 copies/ml. If plasma viral loads increased above 15,000 copies/ml despite BID valganciclovir treatment, cidofovir was administered weekly (5 mg/kg IV) with SID valganciclovir until viremia remained below 15,000 copies/ml for one week, at which point cidofovir was discontinued and BID valganciclovir was administered until complete resolution of viremia. Nine HSCT recipient MCM experiencing CMV viremia of >1,000 copies/ml plasma were treated with valganciclovir. Two HSCT recipient MCM experiencing CMV viremia of >15,000 copies/ml were treated with valganciclovir and cidofovir, which was administered with kidney protectant probenecid (500 mg PO BID on the day of cidofovir treatment).
CyCMV immunity
CMV-specific T cell responses were assessed by interferon-γ enzyme-linked immunospot assay (IFN-γ ELISPOT) as previously described17 using Monkey IFN-γ ELISPOT Plus ALP kit (Mabtech, Cincinnati, OH), following manufacturer’s instructions. ELISPOT assays were performed in duplicate, stimulating 100,000 peripheral blood mononuclear cells per well with rhesus CMV viral lysate and pools of 15-mer peptides (11 amino-acid overlap, 7–13 peptides per pool) spanning the RhCMV pp65a, pp65b, IE-1, and IE-2 proteins (Genbank accession #AY186194). Negative control wells were incubated without antigen and served as background controls; positive control wells were incubated with Concanavalin A.
Flow cytometric analysis and cell sorting
Immune subset frequencies and T cell activation in HSCT recipients were monitored longitudinally by whole blood staining as previously described17. EDTA-treated whole blood (50-100μl) was washed twice with PBS and stained for CD45, CD3, CD20, CD4, CD8, CD14, CD28 and CD95, and viability (Live/dead Yellow Fixable, Invitrogen) at room temperature for 30 minutes. After staining, whole blood was resuspended in 1ml FACS Lysing Solution (BD Biosciences) to lyse red blood cells and fix remaining cells, incubated at room temperature for 8 minutes, washed three times with FACS buffer, and collected on a BD LSRII instrument. All flow cytometric analysis was performed using Flow Jo (Tree Star, Inc., Ashland, OR). CD20+ frequency was defined as the percentage of CD20+CD3− cells within the live CD45+ singlet population. CD8+ T cell frequency was defined as the percentage of CD3+CD8+CD4− cells within the live CD45+ singlet population. Absolute cell counts were calculated by multiplying whole blood staining frequencies by the white blood cell count, as determined by complete blood count performed on an additional aliquot of EDTA-treated whole blood. To assess levels of CD8+ T-cell activation in YFV 17D-vaccinated macaques, EDTA-treated whole blood (50-100μl) was washed twice with PBS and stained with two panels: (1) CD45, CD3, CD4, CD8, HLA-DR, CD38, and viability (Live/dead Yellow Fixable, Invitrogen), (2) CD45, CD3, CD4, CD8 at room temperature for 30 minutes. After staining, whole blood was resuspended in 1ml FACS Lysing Solution (BD Biosciences) to lyse red blood cells and fix remaining cells, incubated at room temperature for 8 minutes, and washed three times with FACS buffer. Tubes stained with panel 2 were stained intracellularly as follows: Pellets were resuspended in 500 μl FACS perm (FACS Lysing solution with 0.05% Tween-20), incubated for 10 minutes, washed three times with FACS buffer, and stained for Ki67 and Bcl2 for 45 minutes. After staining, cells were washed once with FACS buffer. Stained samples were collected on a BD LSRII instrument and analyzed using Flow Jo (Tree Star, Inc.). HLA-DR+CD38+ CD8+ T-cell frequency was defined as the percentage of HLA-DR+, CD38+, CD3+, CD8+, CD4− cells within the live CD45+ singlet population. Ki67+Bcl2− CD8+ T-cell frequency was defined as the percentage of Ki67+, Bcl2-, CD3+, CD8+, CD4− cells within the live CD45+ singlet population. Absolute counts of HLA-DR+CD38+ CD8+ T-cells and Ki67+Bcl2− CD8+ T-cells were calculated by multiplying whole blood staining frequencies by the white blood cell count, as determined by complete blood count performed on an additional aliquot of EDTA-treated whole blood. To assess levels of CD20+ cell-associated LCV, whole blood cells or tissue mononuclear cell suspensions were sorted on a BD FACS Aria Fusion instrument. Whole blood was treated with ACK Lysing Solution (Fisher Scientific, Hampton, NH) to lyse red blood cells, incubated at 5 minutes at room temperature, and washed twice with PBS. Tissues were processed into mononuclear cell preparations, as previously described22. ACK-treated whole blood or tissue mononuclear cells were stained for CD45, CD3, CD20, and viability (Live/dead near-IR, Invitrogen) for 30 minutes at room temperature and subsequently washed with PBS prior to sorting. Cells were kept at 4°C until sorting. CD20+ cells were defined and sorted as live, CD45-hi, SSC-lo, CD3−, CD20+ events. For each sorted population, 12,000-200,000 cells (>95% pure) were collected and used for DNA extraction. The following monoclonal antibodies were used in these studies: a) from BD Biosciences, D058-1283 (CD45; APC), SP34-2 (CD3; Alexa700, PacBlu), 2H7 (CD20; APC-H7, FITC), L200 (CD4; PerCP-Cy5.5, Alexa700), RPA-T8 (CD8; PacBlu), SK1 (CD8, TruRed, APC-Cy7), 28.2 (CD28; PE), DX2 (CD95; FITC, PE-Cy7), G46-6 (HLA-DR; FITC), B56 (Ki67, Per-CP-Cy5.5), Bcl-2/100 (Bcl2, PE-CF594), b) from Beckman Coulter, RMO52 (CD14; ECD), and (c) from NHP Reagent Resource, OKT10 (CD38; PE).
Histology
Immunohistochemistry was performed on the intelliPATH FLX® automated slide stainer (Biocare Medical, Pacheco, CA). Deparaffinized slides were pretreated with 3% H2O2/diH2O for 10 minutes, and then antigen was retrieved using microwave heated citrate buffer (pH 6.0). Immunohistochemistry for SV40 was performed using the ABC-Immunoperoxidase procedure as follows. On the intelliPATH FLX®, slides were then incubated with avidin (Biocare Medical), biotin (Biocare Medical), and protein block (Dako; Agilent, Santa Clara, CA) for 10 minutes each, respectively. Anti-SV40 T Antigen (Ab-2) Mouse monoclonal antibody (clone PAb416, Sigma-Aldrich/Millipore Sigma, Burlington, MA) was diluted to a working 1:12,500 dilution with Dako Antibody Diluent with background reducing components (Dako) and incubated for 2 hours at room temperature. Following this, slides were incubated for 30 minutes at room temperature with Biotinylated Horse Anti-Mouse IgG (Vector Laboratories; Maravai LifeSciences, Burlingame, CA). Peroxidase step was performed using Vectastain ABC HRP standard kit (Vector Laboratories). Finally, the reaction was visualized by 3,3′-diaminobenzidine (DAB) and counterstained with hematoxylin. Multiplex immunohistochemistry for EBNA2/CD20 was performed using the multiplex micro-polymer HRP and AP detection systems. After the pre-treatment and antigen retrieval steps described above, slides were incubated with protein block (Dako) for 10 minutes on the intelliPATH FLX®. Mouse Monoclonal Anti-EBNA2 antibody (EBNA2) (clone PE2, Abcam Inc., Cambridge, MA) was diluted to a working 1:1000 dilution with Dako Antibody Diluent with background reducing components (Dako) and applied for 30 minutes at room temperature. Following this, Mach 2 Mouse HRP-polymer (Biocare Medical) was applied for 30 minutes. EBNA2 was then visualized by Betazoid DAB (Biocare Medical) for 5 minutes. Next, slides were eluted using a denaturing solution (diluted 1 part A with 2 parts B) (Biocare Medical) for 5 minutes. Monoclonal Mouse Anti-Human CD20cy (clone L26, Dako) was diluted to a working 1:125 dilution with Dako Antibody Diluent with background reducing components (Dako) and incubated for 2 hours at room temperature. Slides were then treated with Mach 2 Mouse AP-polymer (Biocare Medical) for 30 minutes. CD20 was visualized via intelliPATH™ Warp Red™ for 7 minutes (Biocare Medical) and counterstained with hematoxylin.
Yellow fever 17D vaccination and neutralizing antibody detection
Macaques were vaccinated with 1×105 plaque-forming units of yellow fever strain 17D subcutaneously. The preparation of yellow fever strain 17D (substrain 17DD) used in this study was obtained from David Watkins (University of Miami) and used previously in NHP studies23,24. To detect neutralizing antibodies in serum of macaques, a YFV foci reduction neutralization assay was performed. Briefly, YFV-17D at 1.2×103 FFU/ml was pre-incubated with serial dilutions of heat inactivated macaque serum at 37°C for 2 hours in Dulbecco modified Eagle medium with 5% fetal bovine serum (FBS), penicillin, and streptomycin. Virus-serum mixtures containing 30-40 FFU were added in to individual wells of 96-well tissue culture plates containing Vero cell monolayers at ∼90% confluence (prepared by seeding 1.5×104 cells in 100 μl/well one day prior to assay setup) and incubated at 37°C for 1 hour. The wells were overlaid with 200 μl of 1% methylcellulose in DMEM containing 2% FBS. After 3 days of incubation at 37°C, the methylcellulose overlay was removed by gently pipetting and the wells were washed two times with 200 μl of PBS. The wells were fixed by incubating with 200 μl of 1% formaldehyde in PBS for 10 minutes at room temperature followed by washing with 200 μl of PBS. The cells were permeabilized by washing with 200 μl of Permwash consisting of 0.1% saponin, 0.1% FBS and 0.1% sodium azide in PBS. The cells were stained intracellularly for 2 hours with 50 μl/well of biotinylated YFV-specific mouse monoclonal antibody (clone 3A8.B6) prepared in Permwash. After washing with 200 μl of Permwash twice, the wells were incubated for 1–2 hours with horseradish peroxidase (HRP)-conjugated streptavidin (Invitrogen) at 0.31 μg/ml. The wells were washed with 200 μl of Perm wash buffer two times and the foci were visualized by adding 50 μl of ImmPACT VIP Peroxidase Substrate (Vector Laboratories) according to manufacturer’s instructions. The plates were dried and foci were counted by visual inspection under a stereomicroscope after drying the plates for 2-4 hrs. The 50% focus reduction neutralization titer (NT50) was calculated using the reciprocal of the serum dilution that reduced the number of foci of infected cells by at least 50%.
RNA-sequencing and gene-wise differential expression analyses
Per sample, RNA was extracted from 1 million PBMCs using the Qiagen RNAeasy kit (Qiagen, Venlo, Netherlands) according to the manufacturer’s protocol. An Agilent Bioanalyzer 2100 was used to evaluate RNA quality and RIN score (Agilent, Santa Clara, CA). RNA-seq library construction was performed by MedGenome (MedGenome, Inc., Foster City, CA), using the TruSeq v2 mRNA kit (Illumina, San Diego, CA), followed by sequencing on an Illumina HiSeq (Illumina, San Diego, CA). An average of 52,133,947 single end 100bp reads were obtained per sample (range: 39,990,202 to 60,708,782). The files were imported into ONPRC’s DISCVR-Seq25, LabKey Server26 based system, which managed raw data and most analyses. Trimmomatic27 was used to remove any remaining Illumina adapters and trim the 3’ read ends. Reads were aligned to the rhesus macaque reference genome (rheMac8, NCBI assembly GCF_000772875.2) using the STAR aligner in two-pass mode with default parameters28. Ensembl gene annotations were used (release 87). The per-sample gene count tables generated by STAR were merged to form a single gene count matrix. We performed differential expression (DE) analyses using three experimental contrasts: A) Controls post-vaccination (day 3 and 14 post-HSCT) vs. controls at day 0, B) HSCT macaques post-infection vs. controls at day 0 and C) HSCT macaques post-infection vs. HSCT macaques at day 0. These analyses were performed directly with the raw gene counts using R version 3.5.129,30. We performed DE analysis using two commonly used methods Limma31 and DESeq232, and combined the significant genes from each. These methods rely on different mathematical theories and assumptions, and we expect each method to capture different projections of the true biology. The Limma package fits multiple gene-wise linear models using weighted least squares, while DESeq2 fits multiple gene-wise negative binomial generalized linear models. This combined approach was done for two main reasons: first, in this pilot study a significance threshold was not estimated a priori and second, there was a small number of animals in each group. For each DE algorithm, we selected a reasonable p-value (<0.001) threshold heuristically to achieve none-zero gene sets and then combined across all of the contrasts. The union of the two methods ensures the capture of the majority of possible DE genes. To remove false-positive genes, we derived the final set by intersecting our day 3 and day 14 genes which yielded the 72-gene signature. External gene names as well as descriptions pertaining to these genes were identified using the useEnsembl() function of biomaRT package33.
RESULTS
Polyomavirus is commonly shed by HSCT recipient macaques post-transplant and can be associated with hemorrhagic cystitis and nephritis
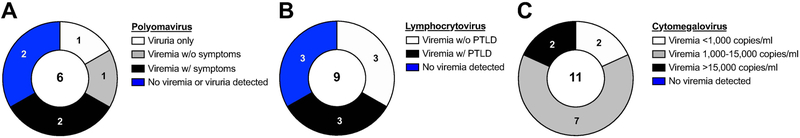
To assess the frequency of polyomavirus shedding in HSCT recipient macaques and identify macaques at risk of polyomavirus-associated complications post-HSCT, we monitored urine and plasma of HSCT recipient macaques for polyomavirus large T antigen DNA by PCR and quantitative PCR using primers previously reported to amplify cynomolgus polyomavirus (CPV) DNA in immunosuppressed cynomolgus macaques21. Overall, polyomavirus shedding was detected in 67% of analyzed recipients post-HSCT (4/6 macaques, Figure 1A, Table S1), in line with previously reported frequencies of BKV shedding in HSCT patients11,34-38. Cases ranged in severity from asymptomatic viruria only to viremia accompanied by hemorrhagic cystitis and nephritis (Figure 1A, 2).
Figure 1. Frequency of viral reactivations in HSCT recipient Mauritian cynomolgus macaques.
Pie charts summarize findings among HSCT recipient macaques analyzed for A, Polyomavirus, B, Lymphocryptovirus, and C, Cytomegalovirus reactivation. Numbers on each slice correspond to the number of macaques belonging to each group as described in the corresponding legend. Numbers in the middle of each chart represent the number of recipients analyzed. Viremia and viruria were defined as >60 copies/ml plasma or urine, the limit of detection for quantitative PCR assays. Polyomavirus symptoms include hematuria, hemorrhagic cystitis, and/or kidney dysfunction, defined as a serum blood urea nitrogen and/or creatinine levels exceeding the normal range of 10 to 21 and 0.6 to 1.3 milligrams per deciliter, respectively. Post-transplant lymphoproliferative disorder (PTLD) was diagnosed by tissue histology.
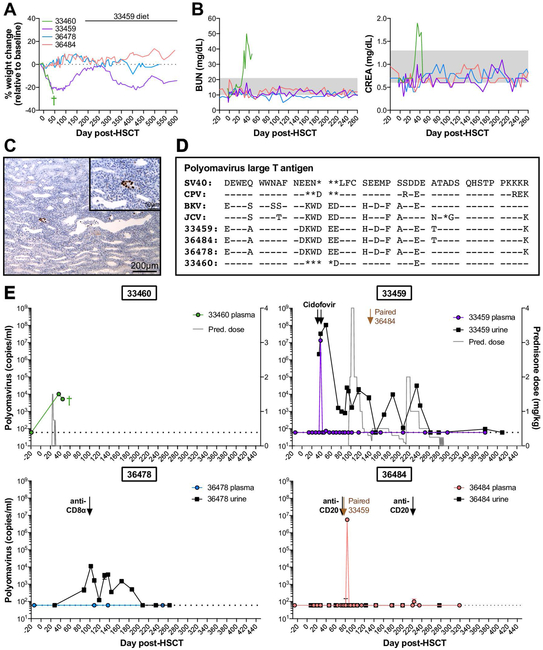
Figure 2. Polyomavirus reactivation varies among HSCT recipient macaques and can result in hemorrhagic cystitis and nephropathy.
A, Weight changes in four HSCT recipient macaques with detectable polyomavirus. B, Longitudinal blood urea nitrogen (BUN) and creatinine (CREA) in serum of HSCT recipient macaques shown in part A. Gray boxes indicated the normal ranges. C, SV40 T antigen immunohistochemistry of 33460 kidney at time of euthanasia. Main (10X magnification): Multifocally, nuclei within infected epithelial cells are immunoreactive to SV40 T antigen. Inset (40X magnification): Infected renal tubular cells exhibit nuclear positivity. D, Amino acid sequence alignment of large T antigens from human polyomaviruses (BK, JC), previously identified macaque polyomaviruses (SV40, CPV), and polyomaviruses from HSCT recipient macaques shown in part A. Sequences shown correspond to amino acids 89 to 131 of SV40 large T antigen. Genbank accession numbers for SV40, CPV, BK virus Dunlop strain, and JC virus are provided in Materials & Methods section. Dashes indicate identical amino acids; asterisks indicate deletions. E, Longitudinal Polyomavirus viral loads in plasma (colored circles) and urine (black squares) of HSCT recipient macaques shown in part A, measured by quantitative PCR. Undetectable viral loads are graphed as 60 DNA copies/ml, the limit of detection for the assay, indicated by horizontal dotted lines. Based on sequence variation among detected Polyomavirus amplicons (see part E), a distinct probe sequence was required for quantitative PCR of 33460, described in Materials & Methods section. Arrows indicate intravenous cidofovir for treatment of CMV reactivation. Gray trace on 33460 and 33459 graphs indicates daily doses of oral prednisone (pred.). Labeled black arrows indicate treatment with depleting antibodies targeted CD20 and CD8α. Labeled brown arrows indicate timepoint 33459 and 36484 were paired. Green cross indicates timepoint of 33460 euthanasia.
In the first case, HSCT recipient macaque 33460 presented with gross hematuria, anorexia, and sustained weight loss around 30 days post-HSCT (Figure 2A). Hematuria was confirmed microscopically and by urine dipstick, and serum chemistry showed elevated levels of blood urea nitrogen (BUN) and creatinine (CREA) indicative of renal dysfunction (Figure 2B). Despite multiple rounds of intravenous fluid diuresis, symptoms persisted, and the macaque became lethargic and was euthanized due to weight loss (>20%, Figure 2A) and persistent cystitis. Gross and histological analysis of necropsy tissues demonstrated active tubulointerstitial nephritis and hemorrhagic cystitis (Figure S1). In addition, intranuclear inclusions indicative of viral infection were observed in renal tubule cells of both kidneys. While the macaque had experienced CyCMV reactivation, CyCMV plasma viremia was minimal and resolved with valganciclovir treatment by time of euthanasia, and tissue sections did not stain positive for CMV antigens via immunohistochemistry (data not shown). Instead, renal epithelial cells in 33460 kidney sections stained positive for SV40 T antigen by immunohistochemistry (Figure 2C), suggesting ongoing polyomavirus replication. In contrast, kidney sections of a non-transplanted control cynomolgus macaque did not show any intranuclear inclusions or SV40 T antigen reactivity (Figure S1). We detected polyomavirus large T antigen DNA in 33460’s plasma post-HSCT, but not in plasma from prior to transplant (Figure S2). These results were confirmed by quantitative PCR, which detected moderate levels of polyomavirus in 33460 plasma at 37 and 45 days post-HSCT, but not in pre-HSCT plasma (Figure 2E).
Polyomavirus shedding was detected in three additional macaques post-transplant: 33459, 36478, and 36484 (Figure 1A, 2E, Figure S2). In each case, no polyomavirus viremia was detected prior to HSCT. Macaque 33459 presented with mild clinical symptoms, including weight loss and low-grade, intermittent hematuria between 14 and 50 days post-HSCT. Two rounds of intravenous fluid diuresis were administered as supportive care around 30 days post-HSCT. Unlike the case of macaque 33460, weight loss and hematuria in macaque 33459 resolved, and no other clinical symptoms of polyomavirus reactivation were observed throughout the monitoring period (Figure 2A, B). Polyomavirus reactivation in HSCT recipient macaques 36478 and 36484 was not accompanied by weight loss or other clinical symptoms of polyomavirus reactivation (Figure 2A, B). With the exception of 33460, all HSCT recipient macaques eventually controlled polyomavirus replication to undetectable levels (<60 copies per milliliter plasma and urine, Table S1), suggesting effective antiviral immunity was eventually restored.
In the process of designing probes for polyomavirus quantitative PCR assays, we sequenced large T antigen PCR amplicons from macaques 33460, 33459, 36478 and 36484. Interestingly, sequencing revealed differences among the polyomavirus large T antigens detected in HSCT recipient cynomolgus macaques as well as differences between these sequences and the previously described CPV large T antigen (Figure 2D). While large T antigen sequences in 33459 and 36484 were 100% identical at the nucleotide level, this sequence differed from those found in macaques 36478 and 33460. In macaques 33459/36484 and 36478, the amplified large T antigen regions were very similar to each other (95% nucleotide identity, 98% amino acid identity), and were most similar to BKV Dunlop strain (84% and 87% amino acid identity, respectively). However, this region in macaque 33460 was divergent from macaques 33459/36484 and 36478 (70% and 74% nucleotide identity, and 69% and 71% amino acid identity, respectively). In addition, 33460’s sequence matched more closely with macaque polyomaviruses SV40 and CPV (93% and 90% amino acid identity, respectively). Thus, polyomavirus sequences vary among individual cynomolgus macaques, a factor which must be considered in viral nucleic acid detection assays.
Development of post-transplant lymphoproliferative disorder is associated with cynomolgus lymphocryptovirus reactivation
In order to examine the incidence of LCV reactivation among HSCT recipient cynomolgus macaques in this study, we developed a quantitative PCR assay to detect cynomolgus lymphocryptovirus (CyLCV) by adapting a previously reported quantitative PCR assay for the rhesus lymphocryptovirus EBER-1 coding region39,40. We detected cynomolgus LCV (CyLCV) plasma viremia in 67% (6/9) of studied HSCT recipient cynomolgus macaques post-HSCT (Figure 1B, Table S1), which is in line with EBV reactivation rates reported in T cell-depleted HSCT patients41.
Due to the ability of lymphocryptoviruses to latently infect and transform B-cells, immunosuppressed transplant patients are at high-risk for developing EBV-associated post-transplant lymphoproliferative disorder (PTLD), a spectrum of disorders ranging from B-cell hyperplasia to monoclonal B-cell lymphoma42. In this study, 50% (3 of 6) of cynomolgus macaques positive for CyLCV plasma viremia developed PLTD, as diagnosed by tissue histology (Figure 1B, 3, Table S1). A representative case of LCV-PTLD is shown in Figure 3. Approximately 45 days post-HSCT, recipient macaque 35132 presented with anorexia, axillary and inguinal lymphoadenopathy, and splenomegaly. Abdominal ultrasound at 53 days post-HSCT showed enlarged, nodular spleen and enlarged, irregular mesenteric lymph nodes. In addition, complete blood count revealed thrombocytopenia, anemia, and 11% lymphoblastic cells. The macaque was euthanized due to suspicion of PTLD. Lymphoma consistent with monomorphic PTLD was found in the spleen, liver, kidney, adrenal glands, pancreaticosplenic, and several periaortic lymph nodes (kidney shown in Figure 3A). Some tissues contained both overtly neoplastic lymphocytes forming masses and infiltrates more consistent with polymorphic PTLD, with mixed populations of lymphoid cells. By immunohistochemistry, the majority of infiltrating neoplastic cells in kidney stained positive for B-cell marker CD20 and lymphocryptovirus Epstein Barr Nuclear Antigen-2 (EBNA-2) (Figure 3B), demonstrating lymphocryptovirus-associated B-cell lineage lymphoproliferative disease and consistent with the vast majority of PTLD cases in transplant patients and previously described cases of PTLD in transplanted cynomolgus macaques43-45. In contrast, kidney sections from a non-transplanted control cynomolgus macaque did not show infiltrating neoplastic cells and did not stain positive for CD20 or EBNA-2 (Figure S3).
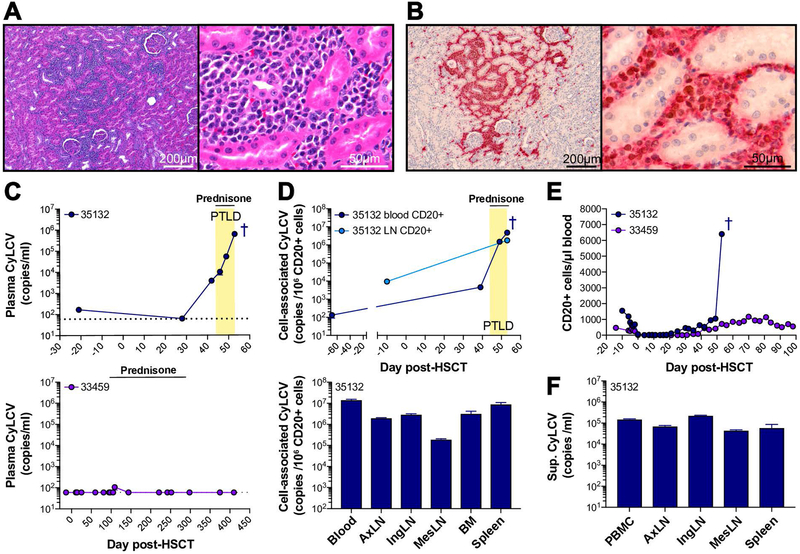
Figure 3. Post-transplant lymphoproliferative disorder is associated with lymphocryptovirus reactivation.
A, Hematoxylin and eosin stain of HSCT recipient macaque 35132 kidney. Left (10X magnification): Sheets of neoplastic lymphocytes infiltrate and expand the renal interstitium. Right (63X magnification): Infiltrating neoplastic lymphocytes have scant amounts of basophilic cytoplasm and round to oval nuclei with clumped chromatin and large, often multiple, eosinophilic nuclei. There is mild anisokaryosis. B, Immunohistochemistry of HSCT recipient macaque 35132 kidney for CD20 (red) and lymphocryptovirus EBNA-2 (brown) at 10X (left) and 40X (right) magnification. The majority of infiltrating neoplastic lymphocytes are both immunoreactive for CD20 (red membranous and cytoplasmic staining) and EBNA-2 (brown nuclear staining), indicating B-cell lineage lymphocytes infected with LCV. C, Longitudinal CyLCV plasma viral loads in 35132 (top) compared to 33459 (bottom), an HSCT recipient macaque who did not develop PTLD, as measured by quantitative PCR. Undetectable plasma viral loads are graphed as 60 DNA copies/ml, the limit of detection (LOD) for the assay, indicated by horizontal dotted lines. Yellow box indicates period where clinical symptoms consistent with PTLD were observed, including lymphadenopathy and splenomegaly. Horizontal solid lines indicate period of oral prednisone treatment. Dark blue cross indicates timepoint of 35132 euthanasia. D, CyLCV CD20+ cell-associated viral loads in 35132, shown longitudinally in blood and lymph node (top) and in tissues at time of euthanasia (bottom). LOD for cell-associated viral loads varied among assays due to variable cell number input. Of note, blood CD20+ cell-associated LCV was undetectable 5 days pre-HSCT and 25 days post-HSCT (LODs 7,324 and 4,048 DNA copies per 1 × 106 CD20+ cells, respectively). E, Longitudinal blood CD20+ cell counts in HSCT recipients shown in part C. F, CyLCV viral loads in supernatants from cultures of single cell suspensions isolated from necropsy tissues.
Quantitative PCR showed detectable CyLCV plasma viremia in macaque 35132 at 42 days post-HSCT, which continued to increase until time of euthanasia (Figure 3C). A low level of CyLCV DNA was found in pre-HSCT plasma, raising the possibility of ongoing lytic viral replication in this macaque prior to transplantation. In contrast to macaque 35132, HSCT recipient macaque 33459 did not exhibit any clinical signs of PTLD and had only one small blip of detectable CyLCV viremia (<200 copies per milliliter plasma) throughout the monitoring period. Next, we measured cell-associated CyLCV in isolated CD20+ B-cells from 35132’s blood and tissues. Levels of cell-associated CyLCV in blood and lymph node CD20+ cells increased after transplant and until time of euthanasia, and high levels of CD20+ cell-associated CyLCV were detected in blood, lymph nodes, bone marrow, and spleen at time of euthanasia (Figure 3D). Consistent with B-cell-origin lymphoproliferative disease, longitudinal absolute CD20+ cell counts in blood of macaque 35132 revealed a large increase in blood B-cells at time of euthanasia, in contrast to the slow B-cell reconstitution that typically occurs post-HSCT and exemplified by macaque 33459 (Figure 3E). To further demonstrate the existence of LCV-transformed cells in macaque 35132, we cultured single cell suspensions from necropsy tissues in R10 medium without any additional cytokines or growth factors. After over two months of culture with passage and media replacement once or twice weekly, all cultures resembled B-lymphoblastoid cell lines and clarified supernatants were positive for CyLCV DNA (Figure 3F). Cultures continued to grow and secrete CyLCV indefinitely, over 20 months. Together, these data demonstrate the development of CyLCV-associated PTLD in a cynomolgus macaque undergoing HSCT.
Cytomegalovirus reactivates early post-HSCT, but is subsequently controlled
To assess cynomolgus CMV (CyCMV) reactivation in HSCT recipient cynomolgus macaques, we developed a quantitative PCR for CyCMV UL121 DNA based on the sequence of CyCMV isolates from Mauritian cynomolgus macaques46. CyCMV plasma viremia was detected in 100% of analyzed recipient macaques within 30 days post-transplant (Figure 1C, 4A, Table S1). Representative macaques are shown in Figure 4A. Plasma viral load in 2 of 11 macaques remained below 1,000 copies per milliliter (copies/ml) and dropped below detectable levels within 30 days of transplant (Figure 4A, 36478). As standard preemptive therapy, treatment with HCMV antiviral valganciclovir was initiated when CyCMV plasma viral load exceeded 1,000 copies/ml (9/11 macaques) and continued until viremia was no longer detectable. Valganciclovir administration alone led to sustained decreases in CyCMV plasma viral load in 7 of 9 treated macaques. CyCMV plasma viral loads fell below detectable levels within 50 days of treatment in six treated macaques (Figure 4A, 36484); one macaque was euthanized prior to complete CyCMV control due development of LCV-PTLD (Figure 4A, 35132). In 2 of 9 treated macaques, valganciclovir alone was unable to control CyCMV replication -- CyCMV plasma viral loads increased above 15,000 copies/ml despite twice daily valganciclovir administration (Figure 4A, 33459). This was associated with lower absolute counts of blood CD8+ T cells early post-HSCT in macaque 33459 compared to macaques that did not require cidofovir treatment to control viremia (Figure S5). These macaques received weekly infusions of intravenous cidofovir until viremia remained below 15,000 copies/ml for one week. Combination valganciclovir and cidofovir treatment effectively controlled CyCMV plasma viremia to undetectable levels in both macaques. After treatment discontinuation, all recipients controlled CyCMV plasma viremia to low levels (<500 copies/ml) and eventually achieved complete control of viremia within 30 days of treatment interruption and for the remainder of the monitoring period. No symptoms of CMV disease were noted in any HSCT recipient macaque throughout the monitoring period.
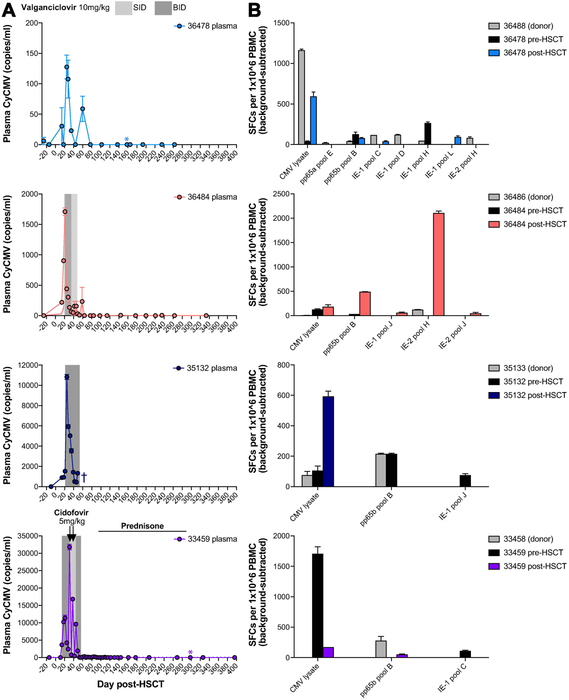
Figure 4. Cytomegalovirus consistently reactivates early post-transplant, but is subsequently controlled.
A, Longitudinal cynomolgus Cytomegalovirus (CyCMV) plasma viral loads in four representative HSCT recipient macaques, measured by quantitative PCR (limit of detection 60 DNA copies/ml). Gray boxes indicate periods of oral valganciclovir treatment. Arrows indicate intravenous cidofovir treatment. Colored asterisks indicate timepoints at which post-HSCT PBMC was tested for CMV-specific T cell responses, shown in part B. Dark blue cross indicates timepoint of 35132 euthanasia. SID = once daily, BID = twice daily. B, CMV-specific T cell responses measured by interferon-gamma ELISPOT of peripheral blood mononuclear cells (PBMC) stimulated with CMV lysate or pools of 15-mer peptides spanning IE-1, IE-2, and pp65 (11 amino acid overlap). Graphs show results for HSCT recipient macaques shown part A, pre-HSCT (black) and post-HSCT (colored), and their HSCT donors (gray). SFCs = spot-forming units.
The observed control of CyCMV viremia after reactivation suggested successful reconstitution of effective anti-CMV immunity post-HSCT. Virus-specific T-cells are critical for controlling CMV replication and disease in humans and macaques47-52. Thus, we used interferon-gamma ELISPOT to screen for T cell responses against CMV lysate and pools of 15-mer peptides spanning CMV antigens IE-1, IE-2, and pp65 (Figure 4B). Indeed, we readily detected CMV-specific T cell responses in PBMC of recipient macaques post-HSCT as well as PBMC of recipients pre-HSCT and donor macaques. In some cases, we observed CMV peptide-specific T cell responses in recipients post-HSCT that were not detectable prior to HSCT or in the HSCT donor, raising the possibility that new CMV-specific T cell responses were primed after transplant.
HSCT recipient macaques respond to live attenuated yellow fever virus 17D vaccination indistinguishably from non-transplanted controls
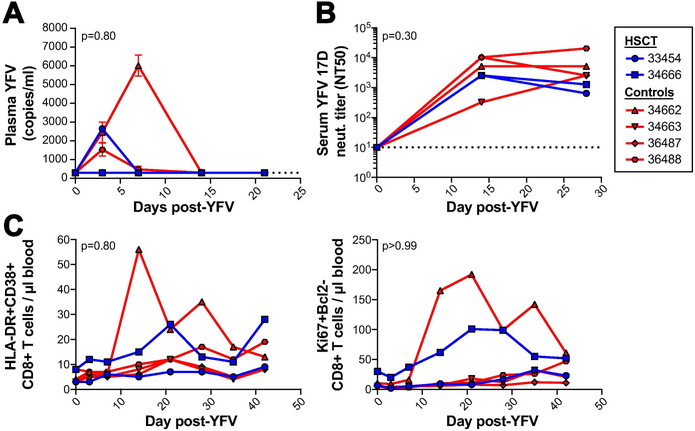
The observed reactivation and subsequent control of polyomavirus, CyLCV, and CyCMV in HSCT recipient cynomolgus macaques suggested post-transplant reconstitution of effective antiviral immunity. However, as not all macaques possessed a fully donor-derived immune system at the timepoints studied above (data not shown), we could not rule out the possibility that post-transplant antiviral immunity had expanded from surviving recipient-derived immune responses. To directly test the ability of HSCT recipient macaque immune systems to prime new immune responses, we used live attenuated yellow fever vaccine 17D (YFV 17D). In healthy humans and rhesus macaques, YFV 17D vaccination results in minimal, short-term viremia and induces vaccine-specific neutralizing antibody and T cell responses23,24,53-57. Here, we compared the response to YFV 17D vaccination in six Mauritian cynomolgus macaques: two HSCT recipient macaques (862 and 806 days post-HSCT at time of vaccination) and four non-transplanted control macaques. Of note, the immune systems of both HSCT recipient macaques were 100% donor-derived prior to vaccination17. First, we assessed the ability of the macaques to control YFV 17D replication. Quantitative reverse transcription PCR detected low levels of YFV 17D RNA in plasma of 1 of 2 HSCT macaques and 3 of 4 control macaques within 7 days of vaccination (Figure 5A). By 14 days post-vaccination, YFV 17D RNA was no longer detectable in the plasma of any macaque, suggesting all macaques had cleared YFV 17D virus. Indeed, all macaques mounted a YFV 17D-targeted neutralizing antibody response by 14 days post-vaccination, and we found similar titers of neutralizing antibodies in the serum of all six macaques at 14 and 28 days post-vaccination (Figure 5B). In addition, we did not observe any differences in CD8+ T cell activation in blood between HSCT and control groups, as measured by HLA-DR+CD38+ and Ki67+Bcl2− CD8+ T-cells (Figure 5C).
Figure 5. Long-term engrafted HSCT recipient macaques respond to live-attenuated yellow fever 17D vaccination similarly to untransplanted macaques.
A, Longitudinal yellow fever (YFV) plasma viral loads in two long-term engrafted HSCT recipient macaques (blue) compared to four control untransplanted Mauritian cynomolgus macaques (red) after live-attenuated yellow fever 17D vaccination. Undetectable plasma viral loads graphed as 300 RNA copies/ml, the limit of detection (LOD) for the assay, indicated by horizontal dotted line. B, Longitudinal serum YFV 17D neutralizing antibody titers. NT50 = highest fold serum dilution where YFV 17D plaque number was reduced by at least 50%. The lowest dilution of serum tested was 10-fold, indicated by the horizontal dotted line. C, Longitudinal counts of activated CD8+ T-cells in blood post-vaccination, as measured by HLA-DR and CD38 (left) and Ki67 and Bcl2 (right). No statistically significant difference in plasma viral loads or CD8+ T cell activation were observed between the HSCT and control groups, analyzed by comparing areas under the curve (AUC) using the Mann-Whitney test (p-value displayed on graphs). No statistically significant difference in neutralizing antibody titers were observed between the HSCT and control groups at any timepoint, analyzed using repeated measures ANOVA (overall group factor p-value displayed on graph, Sidak-adjusted p-values for timepoints 0, 14, and 28 are >0.99, 0.76, and 0.37, respectively).
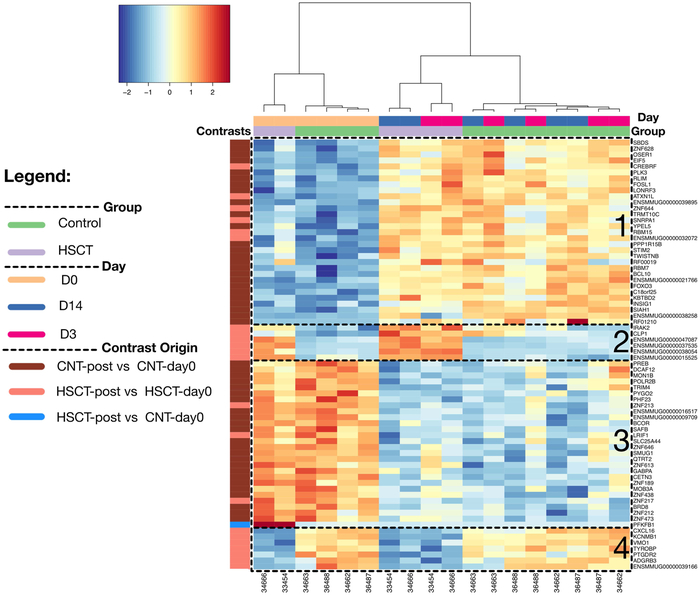
Previous studies have reported transcriptomic changes following vaccination with YFV 17D in humans and rhesus macaques55,58. To determine whether a gene expression signature was present in our animals following vaccination, and to determine if this signature differed between HSCT and control macaques, we performed bulk RNA-seq on peripheral blood mononuclear cells (PBMC) from vaccinated macaques at days 0, 3 and 14 post-vaccination. These data revealed a total of 655 differentially expressed gene (DEG) candidates across the experimental contrasts in this study (Table S2); the intersect of these DE gene sets highlights a signature of 72 differentially expressed genes (DEGs) (Figure 6, Table S3) with two distinct patterns across the subjects. The first pattern contained the majority of DEGs (59 of 72) and separated the pre- and post-vaccination samples (Figure 6, clusters 1 and 3). For all but one of these genes, expression was consistently increased (n=30) or decreased (n=28) after vaccination in both HSCT and control groups (Figure 6, clusters 1 and 3). Expression of one gene, 6-phosphofructo-2-kinase (PFKFB1), an activator of the glycolysis pathway, was elevated in the HSCT group compared to the control group prior to vaccination, but decreased to similar levels to controls after vaccination. PFKFB1 expression in the control group did not change after vaccination. The second, minor pattern contained genes (13 of 72) that were significantly different between HSCT and controls prior to vaccination (Figure 6, clusters 2 and 4). Gene expression was increased (n=6) or decreased (n=7) in the HSCT macaques relative to the controls, but did not consistently change after vaccination in either group. Thus, while HSCT recipient macaques have some baseline gene expression differences from non-transplanted controls, changes in gene expression induced by YFV 17D vaccination in control macaques also occurred in HSCT recipient macaques, suggesting the vaccine response in HSCT recipient macaques was not impaired. Together, these data demonstrate priming of effective donor-derived antiviral immunity in HSCT recipient macaques, who by all measures responded to YFV 17D vaccination indistinguishably from non-transplanted controls.
Figure 6: HSCT and control macaques experience similar changes in gene expression after YFV 17D vaccination.
A heatmap of differentially expressed genes (rows) and samples (columns) demonstrating the identified 72-gene transcriptomic signature on the rlog-transformed expression matrix; pair-wise hierarchical clustering with Ward.D2 using Euclidean distances. There are 2 experimental groups (HSCT vs. controls), collected at 3 timepoints: day 0, 3, and 14, which are labeled according to the colors in the legend. The genes are obtained by identifying the overlap of 3 contrasts pertaining to the experimental design: A) Control macaques post-vaccination (Days 3 and 14) versus pre-vaccination (Day 0), B) HSCT macaques post-vaccination versus control macaques pre-vaccination (day 0), and C) HSCT macaques post-vaccination (Days 3 and 14) versus pre-vaccination (Day 0). Genes grouped into four clusters based on their expression pattern across groups.
DISCUSSION
In this study of viral reactivations in Mauritian cynomolgus macaques undergoing allogeneic HSCT, we focused on macaque analogues of BKV, EBV, and HCMV, which are among the most common viral etiologies of infectious disease complications in allogeneic HSCT patients59. These viral pathogens are also significant obstacles in solid organ xenotransplantation18,60. Multiple polyomaviruses have been reported to ubiquitously infect macaques, including cynomolgus polyomavirus (CPV) and simian virus 40 (SV40) 21,61. Lymphocryptoviruses (LCVs) analogous to EBV ubiquitously infect many species of non-human primates, with virtually all macaques harboring LCV by 2 years of age62,63. Species-specific CMVs analogous to HCMV have been isolated from many nonhuman primate species, and virtually all individuals are infected within the first year of life46,64-66. We observed frequent reactivations of polyomavirus, CyLCV, and CyCMV in transplanted cynomolgus macaques, which occasionally resulted in clinical manifestations identical to those observed in human patients, including polyomavirus-associated hemorrhagic cystitis and nephropathy and lymphocryptovirus-associated PTLD. To our knowledge, this is the first report of polyomavirus and lymphocryptovirus reactivation and disease in a macaque model of HSCT.
Similar to allogeneic HSCT in the clinic59, viral reactivation and disease in this study typically occurred early post-transplant, and in most cases, was eventually brought under control, presumably due to reconstitution of effective antiviral immunity. In support of this, we found CMV-specific T cell immunity after resolution of post-transplant CMV viremia in all studied allogeneic HSCT recipient cynomolgus macaques. Associations of antiviral T-cells with viral control post-transplant have been reported for CMV, BKV, EBV, and adenovirus67-77, and numerous studies have demonstrated safety and efficacy of allogeneic virus-specific T cell infusion in treating viral reactivations with minimal or no exacerbation of GVHD78-88. Due to the simplified MHC genetics of Mauritian cynomolgus macaques89-91, this model provides an opportunity to conduct detailed investigations of T cell-based therapies for viral complications of allogeneic HSCT.
Finally, in further support of reconstitution of functional immunity in our macaque model of allogeneic HSCT, we show safe and effective vaccination of HSCT recipient macaques with live attenuated virus vaccine YFV 17D, in line with previous reports of YFV 17D vaccination in allogeneic HSCT patients92-94. Further, whole transcriptome RNA-seq of YFV 17D-vaccinated HSCT recipient and non-transplanted control macaques showed identical changes in gene expression after immunization. Previous studies of gene expression changes after YFV 17D vaccination in rhesus macaques were conducted with microarray technology and reported significant changes in 39 genes following YFV-17D vaccination58, and our DEG set overlapped with 7 of these: IFIT3, MARKS, HERC6, HERC5, CMPK2, RSAD2, EIF2AK2, LILRA2. This limited overlap is likely due to the very distinct technologies of microarray versus whole transcriptome RNA-seq where discrepancies are not uncommon95. Regardless, the detection of consistent transcriptional changes between groups of macaques in this study suggests that the response to YFV 17D vaccination in HSCT recipient macaques is not impaired.
Despite vast improvements in managing infectious disease complications of allogeneic HSCT, many challenges still exist. Between 2015 and 2016, approximately 11 to 24% of deaths after allogeneic HSCT were due to infection1,2. While implementation of viral surveillance methods has improved early diagnosis and intervention for virus-associated complications, most measurements have poor predictive value for disease. Namely, patients with high viral loads do not always progress to disease, and disease can occasionally manifest without high viral loads in blood37,41,96-102. In addition, there are major challenges in treating viral complications, including lack of effective treatments, drug toxicity, and simultaneous management of GVHD, which is treated with intensified immunosuppression that can exacerbate viral reactivation and disease103-105. Similar challenges exist in solid organ and xenotransplantation, where intensified immunosuppression is often required to prevent rejection18,60. Overall, the findings in this study demonstrate that this Mauritian cynomolgus macaque model recapitulates viral disease and immunity observed in human transplant patients. Therefore, this macaque model of allogeneic HSCT can be used to explore novel strategies for early diagnosis, prevention, and treatment of viral complications after allogeneic and xenogeneic transplantation.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to acknowledge the animals that contributed to this study and express our respect and gratitude for their invaluable role in this research. We thank Leslie Kean for helpful discussions on CMV treatment, David Watkins for providing yellow fever vaccine 17D, Amgen, Inc. for providing Neupogen, Bristol-Myers Squibb for providing Belatacept, Roger Wiseman and David O’Connor for assistance with MCM MHC typing, and Margaret Terry, Wendy Price, and Erinn Stefanich for technical assistance with histology and immunohistochemistry. This work was supported by National Institutes of Health grants R21 AI112433 and R01 AI129703 awarded to J.B.S., AI079898 awarded to M.K.S., and P51 OD011092 awarded to the Oregon National Primate Research Center. The ONPRC Integrated Pathology Core provided support services for the research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
ABBREVIATIONS
- HSCT
hematopoietic stem cell transplantation
- MHC
major histocompatibility complex
- LCV
lymphocryptovirus
- CyLCV
cynomolgus lymphocryptovirus
- CMV
cytomegalovirus
- CyCMV
cynomolgus cytomegalovirus
- HCMV
human cytomegalovirus
- BKV
BK virus
- EBV
Epstein-Barr virus
- PTLD
post-transplant lymphoproliferative disorder
- SIV
simian immunodeficiency virus
- YFV 17D
yellow fever virus strain 17D vaccine
- GVHD
graft-versus-host disease
- PBMC
peripheral blood mononuclear cells
- BKV-HC
BK virus-associated hemorrhagic cystitis
- BKVN
BK virus-associated nephropathy
- PCR
polymerase chain reaction
- CPV
cynomolgus polyomavirus
- SV40
simian virus 40
- HLA
human leukocyte antigen
- ATG
antithymocyte globulin
- EBNA-2
Epstein-Barr nuclear antigen 2
- DEG
differently expressed gene
- SID
once daily
- BID
twice daily
- IV
intravenous
- PO
per os
- IM
intramuscular
- SC
subcutaneous
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors of this manuscript have no conflicts of interest to disclose as described by the Xenotransplantation.
REFERENCES
- 1.D’Souza A, Frethman C. Current Uses and Outcomes of Hematopoietic Cell Transplantation (HCT): CIBMTR Summary Slides, 2018. https://www.cibmtr.org.
- 2.Gratwohl A, Brand R, Frassoni F, et al. Cause of death after allogeneic haematopoietic stem cell transplantation (HSCT) in early leukaemias: an EBMT analysis of lethal infectious complications and changes over calendar time. Bone Marrow Transplant. 2005;36(9):757–769. doi: 10.1038/sj.bmt.1705140. [DOI] [PubMed] [Google Scholar]
- 3.Tomblyn M, Chiller T, Einsele H, et al. Guidelines for preventing infectious complications among hematopoietic cell transplantation recipients: a global perspective. Biol Blood Marrow Transplant. 2009;15(10):1143–1238. doi: 10.1016/j.bbmt.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirsch HH, Steiger J. Polyomavirus BK. Lancet Infect Dis. 2003;3(10):611–623. doi: 10.1016/S1473-3099(03)00770-9. [DOI] [PubMed] [Google Scholar]
- 5.Knowles WA. Discovery and epidemiology of the human polyomaviruses BK virus (BKV) and JC virus (JCV). Adv Exp Med Biol. 2006;577(Chapter 2):19–45. doi: 10.1007/0-387-32957-9_2. [DOI] [PubMed] [Google Scholar]
- 6.Tzellos S, Farrell PJ. Epstein-barr virus sequence variation-biology and disease. Pathogens. 2012;1(2):156–174. doi: 10.3390/pathogens1020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smatti MK, Al-Sadeq DW, Ali NH, Pintus G, Abou-Saleh H, Nasrallah GK. Epstein-Barr Virus Epidemiology, Serology, and Genetic Variability of LMP-1 Oncogene Among Healthy Population: An Update. Front Oncol. 2018;8:211. doi: 10.3389/fonc.2018.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cannon MJ, Schmid DS, Hyde TB. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev Med Virol. 2010;20(4):202–213. doi: 10.1002/rmv.655. [DOI] [PubMed] [Google Scholar]
- 9.Adland E, Klenerman P, Goulder P, Matthews PC. Ongoing burden of disease and mortality from HIV/CMV coinfection in Africa in the antiretroviral therapy era. Front Microbiol. 2015;6:1016. doi: 10.3389/fmicb.2015.01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilis L, Morisset S, Billaud G, et al. High burden of BK virus-associated hemorrhagic cystitis in patients undergoing allogeneic hematopoietic stem cell transplantation. 2014;49(5):664–670. doi: 10.1038/bmt.2013.235. [DOI] [PubMed] [Google Scholar]
- 11.Abudayyeh A, Hamdi A, Lin H, et al. Symptomatic BK Virus Infection Is Associated With Kidney Function Decline and Poor Overall Survival in Allogeneic Hematopoietic Stem Cell Recipients. Am J Transplant. 2016;16(5):1492–1502. doi: 10.1111/ajt.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin R, Liu Q. Diagnosis and treatment of viral diseases in recipients of allogeneic hematopoietic stem cell transplantation. J Hematol Oncol. 2013;6(1):94. doi: 10.1186/1756-8722-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Styczynski J, Einsele H, Gil L, Ljungman P. Outcome of treatment of Epstein-Barr virus-related post-transplant lymphoproliferative disorder in hematopoietic stem cell recipients: a comprehensive review of reported cases. Transpl Infect Dis. 2009;11(5):383–392. doi: 10.1111/j.1399-3062.2009.00411.x. [DOI] [PubMed] [Google Scholar]
- 14.Bitan M Epstein-Barr virus infection after stem cell transplantation: new concepts are needed both for the donor and the recipient. Clin Infect Dis. 2006;43(7):896–897. doi: 10.1086/507032. [DOI] [PubMed] [Google Scholar]
- 15.Ljungman P, Hakki M, Boeckh M. Cytomegalovirus in hematopoietic stem cell transplant recipients. Hematol Oncol Clin North Am. 2011;25(1):151–169. doi: 10.1016/j.hoc.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Camargo JF, Komanduri KV. Emerging concepts in cytomegalovirus infection following hematopoietic stem cell transplantation. Hematol Oncol Stem Cell Ther. 2017;10(4):233–238. doi: 10.1016/j.hemonc.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Burwitz BJ, Wu HL, Abdulhaqq S, et al. Allogeneic stem cell transplantation in fully MHC-matched Mauritian cynomolgus macaques recapitulates diverse human clinical outcomes. Nature Communications. 2017;8(1):1418. doi: 10.1038/s41467-017-01631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haustein SV, Kolterman AJ, Sundblad JJ, Fechner JH, Knechtle SJ. Nonhuman primate infections after organ transplantation. ILAR J. 2008;49(2):209–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phillips KA, Bales KL, Capitanio JP, et al. Why primate models matter. Am J Primatol. 2014;76(9):801–827. doi: 10.1002/ajp.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estes JD, Wong SW, Brenchley JM. Nonhuman primate models of human viral infections. Nat Rev Immunol. 2018;18(6):390–404. doi: 10.1038/s41577-018-0005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Gorder MA, Pelle Della P, Henson JW, Sachs DH, Cosimi AB, Colvin RB. Cynomolgus polyoma virus infection: a new member of the polyoma virus family causes interstitial nephritis, ureteritis, and enteritis in immunosuppressed cynomolgus monkeys. Am J Pathol. 1999;154(4):1273–1284. doi: 10.1016/S0002-9440(10)65379-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burwitz BJ, Reed JS, Hammond KB, et al. Technical advance: liposomal alendronate depletes monocytes and macrophages in the nonhuman primate model of human disease. J Leukoc Biol. 2014;96(3):491–501. doi: 10.1189/jlb.5TA0713-373R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonaldo MC, Martins MA, Rudersdorf R, et al. Recombinant Yellow Fever Vaccine Virus 17D Expressing Simian Immunodeficiency Virus SIVmac239 Gag Induces SIV-Specific CD8+ T-Cell Responses in Rhesus Macaques. Journal of Virology. 2010;84(7):3699–3706. doi: 10.1128/JVI.02255-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mudd PA, Piaskowski SM, Neves PCC, et al. The live-attenuated yellow fever vaccine 17D induces broad and potent T cell responses against several viral proteins in Indian rhesus macaques—implications for recombinant vaccine design. Immunogenetics. 2010;62(9):593–600. doi: 10.1007/s00251-010-0461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DISCVR-Seq. BimberLab/DISCVRSeq. https://github.com/BimberLab/DISCVRSeq.
- 26.Nelson EK, Piehler B, Eckels J, et al. LabKey Server: an open source platform for scientific data integration, analysis and collaboration. BMC Bioinformatics. 2011;12(1):71. doi: 10.1186/1471-2105-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Team RC. A Language and Environment for Statistical Computing. https://www.r-project.org/.
- 30.Xie Y knitr: A General-Purpose Package for Dynamic Report Generation in R. https://yihui.name/knitr/. [Google Scholar]
- 31.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47–e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4(8):1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arthur RR, Shah KV, Baust SJ, Santos GW, Saral R. Association of BK viruria with hemorrhagic cystitis in recipients of bone marrow transplants. N Engl J Med. 1986;315(4):230–234. doi: 10.1056/NEJM198607243150405. [DOI] [PubMed] [Google Scholar]
- 35.Arthur RR, Shah KV, Charache P, Saral R. BK and JC virus infections in recipients of bone marrow transplants. J Infect Dis. 1988;158(3):563–569. [DOI] [PubMed] [Google Scholar]
- 36.Bedi A, Miller CB, Hanson JL, et al. Association of BK virus with failure of prophylaxis against hemorrhagic cystitis following bone marrow transplantation. J Clin Oncol. 1995;13(5):1103–1109. doi: 10.1200/JCO.1995.13.5.1103. [DOI] [PubMed] [Google Scholar]
- 37.O’Donnell PH, Swanson K, Josephson MA, et al. BK virus infection is associated with hematuria and renal impairment in recipients of allogeneic hematopoetic stem cell transplants. Biol Blood Marrow Transplant. 2009;15(9):1038–1048.e1. doi: 10.1016/j.bbmt.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knowles WA, Pillay D, Johnson MA, Hand JF, Brown DW. Prevalence of long-term BK and JC excretion in HIV-infected adults and lack of correlation with serological markers. Journal of Medical Virology. 1999;59(4):474–479. [PubMed] [Google Scholar]
- 39.Rivailler P, Carville A, Kaur A, et al. Experimental rhesus lymphocryptovirus infection in immunosuppressed macaques: an animal model for Epstein-Barr virus pathogenesis in the immunosuppressed host. Blood. 2004;104(5):1482–1489. doi: 10.1182/blood-2004-01-0342. [DOI] [PubMed] [Google Scholar]
- 40.Kamperschroer C, Gosink MM, Kumpf SW, O’Donnell LM, Tartaro KR. The genomic sequence of lymphocryptovirus from cynomolgus macaque. Virology. 2016;488:28–36. doi: 10.1016/j.virol.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 41.van Esser JW, van der Holt B, Meijer E, et al. Epstein-Barr virus (EBV) reactivation is a frequent event after allogeneic stem cell transplantation (SCT) and quantitatively predicts EBV-lymphoproliferative disease following T-cell--depleted SCT. Blood. 2001;98(4):972–978. [DOI] [PubMed] [Google Scholar]
- 42.Snow AL, Martinez OM. Epstein-Barr virus: evasive maneuvers in the development of PTLD. Am J Transplant. 2007;7(2):271–277. doi: 10.1111/j.1600-6143.2006.01650.x. [DOI] [PubMed] [Google Scholar]
- 43.Rasche L, Kapp M, Einsele H, Mielke S. EBV-induced post transplant lymphoproliferative disorders: a persisting challenge in allogeneic hematopoetic SCT. Bone Marrow Transplant. 2013;49(2):163–167. doi: 10.1038/bmt.2013.96. [DOI] [PubMed] [Google Scholar]
- 44.McInnes EF, Jarrett RF, Langford G, et al. Posttransplant lymphoproliferative disorder associated with primate gamma-herpesvirus in cynomolgus monkeys used in pig-to-primate renal xenotransplantation and primate renal allotransplantation. Transplantation. 2002;73(1):44–52. [DOI] [PubMed] [Google Scholar]
- 45.Schmidtko J, Wang R, Wu C-L, et al. Posttransplant lymphoproliferative disorder associated with an Epstein-Barr-related virus in cynomolgus monkeys. Transplantation. 2002;73(9):1431–1439. [DOI] [PubMed] [Google Scholar]
- 46.Burwitz BJ, Malouli D, Bimber BN, et al. Cross-Species Rhesus Cytomegalovirus Infection of Cynomolgus Macaques. Murphy EA, ed. PLoS Pathog. 2016;12(11):e1006014. doi: 10.1371/journal.ppat.1006014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li CR, Greenberg PD, Gilbert MJ, Goodrich JM, Riddell SR. Recovery of HLA-restricted cytomegalovirus (CMV)-specific T-cell responses after allogeneic bone marrow transplant: correlation with CMV disease and effect of ganciclovir prophylaxis. Blood. 1994;83(7):1971–1979. [PubMed] [Google Scholar]
- 48.Quinnan GV, Kirmani N, Rook AH, et al. Cytotoxic t cells in cytomegalovirus infection: HLA-restricted T-lymphocyte and non-T-lymphocyte cytotoxic responses correlate with recovery from cytomegalovirus infection in bone-marrow-transplant recipients. N Engl J Med. 1982;307(1):7–13. doi: 10.1056/NEJM198207013070102. [DOI] [PubMed] [Google Scholar]
- 49.Reusser P, Riddell SR, Meyers JD, Greenberg PD. Cytotoxic T-lymphocyte response to cytomegalovirus after human allogeneic bone marrow transplantation: pattern of recovery and correlation with cytomegalovirus infection and disease. Blood. 1991;78(5):1373–1380. [PubMed] [Google Scholar]
- 50.Kaur A, Hale CL, Noren B, Kassis N, Simon MA, Johnson RP. Decreased Frequency of Cytomegalovirus (CMV)-Specific CD4+ T Lymphocytes in Simian Immunodeficiency Virus-Infected Rhesus Macaques: Inverse Relationship with CMV Viremia. Journal of Virology. 2002;76(8):3646–3658. doi: 10.1128/JVI.76.8.3646-3658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaur A, Kassis N, Hale CL, et al. Direct relationship between suppression of virus-specific immunity and emergence of cytomegalovirus disease in simian AIDS. Journal of Virology. 2003;77(10):5749–5758. doi: 10.1128/JVI.77.10.5749-5758.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Einsele H, Roosnek E, Rufer N, et al. Infusion of cytomegalovirus (CMV)-specific T cells for the treatment of CMV infection not responding to antiviral chemotherapy. Blood. 2002;99(11):3916–3922. [DOI] [PubMed] [Google Scholar]
- 53.Poland JD, Calisher CH, Monath TP, Downs WG, Murphy K. Persistence of neutralizing antibody 30-35 years after immunization with 17D yellow fever vaccine. Bull World Health Organ. 1981;59(6):895–900. [PMC free article] [PubMed] [Google Scholar]
- 54.Akondy RS, Monson ND, Miller JD, et al. The yellow fever virus vaccine induces a broad and polyfunctional human memory CD8+ T cell response. J Immunol. 2009;183(12):7919–7930. doi: 10.4049/jimmunol.0803903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Querec TD, Akondy RS, Lee EK, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2008;10(1):116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller JD, van der Most RG, Akondy RS, et al. Human Effector and Memory CD8+ T Cell Responses to Smallpox and Yellow Fever Vaccines. Immunity. 2008;28(5):710–722. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 57.Slifka MK, Leung DYM, Hammarlund E, et al. Transcutaneous yellow fever vaccination of subjects with or without atopic dermatitis. J Allergy Clin Immunol. 2014;133(2):439–447. doi: 10.1016/j.jaci.2013.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Engelmann F, Josset L, Girke T, et al. Pathophysiologic and Transcriptomic Analyses of Viscerotropic Yellow Fever in a Rhesus Macaque Model. Geisbert T, ed. PLoS Negl Trop Dis. 2014;8(11):e3295–16. doi: 10.1371/journal.pntd.0003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ogonek J, Kralj Juric M, Ghimire S, et al. Immune Reconstitution after Allogeneic Hematopoietic Stem Cell Transplantation. Front Immunol. 2016;7(19):507. doi: 10.3389/fimmu.2016.00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boneva RS, Folks TM, Chapman LE. Infectious disease issues in xenotransplantation. Clinical Microbiology Reviews. 2001;14(1):1–14. doi: 10.1128/CMR.14.1.1-14.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bofill-Mas S, Albiñana-Giménez N, Pipkin PA, Minor PD, Girones R. Isolation of SV40 from the environment of a colony of cynomolgus monkeys naturally infected with the virus. Virology. 2004;330(1):1–7. doi: 10.1016/j.virol.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 62.Carville A, Mansfield KG. Comparative pathobiology of macaque lymphocryptoviruses. Comp Med. 2008;58(1):57–67. [PMC free article] [PubMed] [Google Scholar]
- 63.Fujimoto K, Honjo S. Presence of antibody to Cyno-EBV in domestically bred cynomolgus monkeys (Macaca fascicularis). J Med Primatol. 1991;20(1):42–45. [PubMed] [Google Scholar]
- 64.Vogel P, Weigler BJ, Kerr H, Hendrickx AG, Barry PA. Seroepidemiologic studies of cytomegalovirus infection in a breeding population of rhesus macaques. Lab Anim Sci. 1994;44(1):25–30. [PubMed] [Google Scholar]
- 65.Jones-Engel L, Engel GA, Heidrich J, et al. Temple monkeys and health implications of commensalism, Kathmandu, Nepal. Emerging Infect Dis. 2006;12(6):900–906. doi: 10.3201/eid1206.060030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arvin A, Campadelli-Fiume G, Mocarski E, et al. Primate betaherpesviruses. 2007. [PubMed]
- 67.Lilleri D, Gerna G, Fornara C, Lozza L, Maccario R, Locatelli F. Prospective simultaneous quantification of human cytomegalovirus-specific CD4+ and CD8+ T-cell reconstitution in young recipients of allogeneic hematopoietic stem cell transplants. Blood. 2006;108(4):1406–1412. doi: 10.1182/blood-2005-11-012864. [DOI] [PubMed] [Google Scholar]
- 68.Lilleri D, Fornara C, Chiesa A, Caldera D, Alessandrino EP, Gerna G. Human cytomegalovirus-specific CD4+ and CD8+ T-cell reconstitution in adult allogeneic hematopoietic stem cell transplant recipients and immune control of viral infection. Haematologica. 2008;93(2):248–256. doi: 10.3324/haematol.11912. [DOI] [PubMed] [Google Scholar]
- 69.Gratama JW, Brooimans RA, van der Holt B, et al. Monitoring cytomegalovirus IE-1 and pp65-specific CD4+ and CD8+ T-cell responses after allogeneic stem cell transplantation may identify patients at risk for recurrent CMV reactivations. Cytometry B Clin Cytom. 2008;74(4):211–220. doi: 10.1002/cyto.b.20420. [DOI] [PubMed] [Google Scholar]
- 70.Tormo N, Solano C, Benet I, et al. Reconstitution of CMV pp65 and IE-1-specific IFN-γ CD8(+) and CD4(+) T-cell responses affording protection from CMV DNAemia following allogeneic hematopoietic SCT. Bone Marrow Transplant. 2011;46(11):1437–1443. doi: 10.1038/bmt.2010.330. [DOI] [PubMed] [Google Scholar]
- 71.Boeckh M, Leisenring W, Riddell SR, et al. Late cytomegalovirus disease and mortality in recipients of allogeneic hematopoietic stem cell transplants: importance of viral load and T-cell immunity. Blood. 2003;101(2):407–414. doi: 10.1182/blood-2002-03-0993. [DOI] [PubMed] [Google Scholar]
- 72.Krause H, Hebart H, Jahn G, Müller CA, Einsele H. Screening for CMV-specific T cell proliferation to identify patients at risk of developing late onset CMV disease. Bone Marrow Transplant. 1997;19(11):1111–1116. doi: 10.1038/sj.bmt.1700801. [DOI] [PubMed] [Google Scholar]
- 73.Schachtner T, Müller K, Stein M, et al. BK virus-specific immunity kinetics: a predictor of recovery from polyomavirus BK-associated nephropathy. Am J Transplant. 2011;11(11):2443–2452. doi: 10.1111/j.1600-6143.2011.03693.x. [DOI] [PubMed] [Google Scholar]
- 74.Chakera A, Bennett S, Lawrence S, et al. Antigen-specific T cell responses to BK polyomavirus antigens identify functional anti-viral immunity and may help to guide immunosuppression following renal transplantation. Clin Exp Immunol. 2011;165(3):401–409. doi: 10.1111/j.1365-2249.2011.04429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zandvliet ML, Falkenburg JHF, van Liempt E, et al. Combined CD8+ and CD4+ adenovirus hexon-specific T cells associated with viral clearance after stem cell transplantation as treatment for adenovirus infection. Haematologica. 2010;95(11):1943–1951. doi: 10.3324/haematol.2010.022947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Annels NE, Kalpoe JS, Bredius RGM, et al. Management of Epstein-Barr virus (EBV) reactivation after allogeneic stem cell transplantation by simultaneous analysis of EBV DNA load and EBV-specific T cell reconstitution. Clin Infect Dis. 2006;42(12):1743–1748. doi: 10.1086/503838. [DOI] [PubMed] [Google Scholar]
- 77.Trydzenskaya H, Sattler A, Müller K, et al. Novel approach for improved assessment of phenotypic and functional characteristics of BKV-specific T-cell immunity. Transplantation. 2011;92(11):1269–1277. doi: 10.1097/TP.0b013e318234e0e5. [DOI] [PubMed] [Google Scholar]
- 78.Walter EA, Greenberg PD, Gilbert MJ, et al. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T-cell clones from the donor. N Engl J Med. 1995;333(16):1038–1044. doi: 10.1056/NEJM199510193331603. [DOI] [PubMed] [Google Scholar]
- 79.Schmitt A, Tonn T, Busch DH, et al. Adoptive transfer and selective reconstitution of streptamer-selected cytomegalovirus-specific CD8+ T cells leads to virus clearance in patients after allogeneic peripheral blood stem cell transplantation. Transfusion. 2011;51(3):591–599. doi: 10.1111/j.1537-2995.2010.02940.x. [DOI] [PubMed] [Google Scholar]
- 80.Feuchtinger T, Opherk K, Bethge WA, et al. Adoptive transfer of pp65-specific T cells for the treatment of chemorefractory cytomegalovirus disease or reactivation after haploidentical and matched unrelated stem cell transplantation. Blood. 2010;116(20):4360–4367. doi: 10.1182/blood-2010-01-262089. [DOI] [PubMed] [Google Scholar]
- 81.Feuchtinger T, Matthes-Martin S, Richard C, et al. Safe adoptive transfer of virus-specific T-cell immunity for the treatment of systemic adenovirus infection after allogeneic stem cell transplantation. Br J Haematol. 2006;134(1):64–76. doi: 10.1111/j.1365-2141.2006.06108.x. [DOI] [PubMed] [Google Scholar]
- 82.Icheva V, Kayser S, Wolff D, et al. Adoptive transfer of epstein-barr virus (EBV) nuclear antigen 1-specific t cells as treatment for EBV reactivation and lymphoproliferative disorders after allogeneic stem-cell transplantation. J Clin Oncol. 2013;31(1):39–48. doi: 10.1200/JCO.2011.39.8495. [DOI] [PubMed] [Google Scholar]
- 83.Moosmann A, Bigalke I, Tischer J, et al. Effective and long-term control of EBV PTLD after transfer of peptide-selected T cells. Blood. 2010;115(14):2960–2970. doi: 10.1182/blood-2009-08-236356. [DOI] [PubMed] [Google Scholar]
- 84.Bollard CM, Heslop HE. T cells for viral infections after allogeneic hematopoietic stem cell transplant. Blood. 2016;127(26):3331–3340. doi: 10.1182/blood-2016-01-628982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Heslop HE, Slobod KS, Pule MA, et al. Long-term outcome of EBV-specific T-cell infusions to prevent or treat EBV-related lymphoproliferative disease in transplant recipients. Blood. 2010;115(5):925–935. doi: 10.1182/blood-2009-08-239186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Doubrovina E, Oflaz-Sozmen B, Prockop SE, et al. Adoptive immunotherapy with unselected or EBV-specific T cells for biopsy-proven EBV+ lymphomas after allogeneic hematopoietic cell transplantation. Blood. 2012;119(11):2644–2656. doi: 10.1182/blood-2011-08-371971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Leen AM, Bollard CM, Mendizabal AM, et al. Multicenter study of banked third-party virus-specific T cells to treat severe viral infections after hematopoietic stem cell transplantation. Blood. 2013;121(26):5113–5123. doi: 10.1182/blood-2013-02-486324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tzannou I, Papadopoulou A, Naik S, et al. Off-the-Shelf Virus-Specific T Cells to Treat BK Virus, Human Herpesvirus 6, Cytomegalovirus, Epstein-Barr Virus, and Adenovirus Infections After Allogeneic Hematopoietic Stem-Cell Transplantation. J Clin Oncol. 2017;35(31):3547–3557. doi: 10.1200/JCO.2017.73.0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Budde ML, Wiseman RW, Karl JA, Hanczaruk B, Simen BB, O’Connor DH. Characterization of Mauritian cynomolgus macaque major histocompatibility complex class I haplotypes by high-resolution pyrosequencing. Immunogenetics. 2010;62(11-12):773–780. doi: 10.1007/s00251-010-0481-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.O’Connor SL, Blasky AJ, Pendley CJ, et al. Comprehensive characterization of MHC class II haplotypes in Mauritian cynomolgus macaques. Immunogenetics. 2007;59(6):449–462. doi: 10.1007/s00251-007-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Greene JM, Burwitz BJ, Blasky AJ, et al. Allogeneic lymphocytes persist and traffic in feral MHC-matched mauritian cynomolgus macaques. Unutmaz D, ed. PLoS One. 2008;3(6):e2384. doi: 10.1371/journal.pone.0002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sicre de Fontbrune F, Arnaud C, Cheminant M, et al. Immunogenicity and Safety of Yellow Fever Vaccine in Allogeneic Hematopoietic Stem Cell Transplant Recipients After Withdrawal of Immunosuppressive Therapy. J Infect Dis. 2018;217(3):494–497. doi: 10.1093/infdis/jix564. [DOI] [PubMed] [Google Scholar]
- 93.Gowda R, Cartwright K, Bremner JAG, Green ST. Yellow fever vaccine: a successful vaccination of an immunocompromised patient. Eur J Haematol. 2004;72(4):299–301. doi: 10.1111/j.1600-0609.2004.00218.x. [DOI] [PubMed] [Google Scholar]
- 94.Yax JA, Farnon EC, Cary Engleberg N. Successful immunization of an allogeneic bone marrow transplant recipient with live, attenuated yellow Fever vaccine. J Travel Med. 2009;16(5):365–367. doi: 10.1111/j.1708-8305.2009.00336.x. [DOI] [PubMed] [Google Scholar]
- 95.Zhao S, Fung-Leung W-P, Bittner A, Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. Zhang S-D, ed. PLoS One. 2014;9(1):e78644. doi: 10.1371/journal.pone.0078644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gulley ML, Tang W. Laboratory assays for Epstein-Barr virus-related disease. J Mol Diagn. 2008;10(4):279–292. doi: 10.2353/jmoldx.2008.080023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gulley ML, Tang W. Using Epstein-Barr Viral Load Assays To Diagnose, Monitor, and Prevent Posttransplant Lymphoproliferative Disorder. Clinical Microbiology Reviews. 2010;23(2):350–366. doi: 10.1128/CMR.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Frías C, Lauzurica R, Bayés B, Ausina V. Prospective follow-up of Epstein-Barr virus load in adult kidney transplant recipients by semiquantitative polymerase chain reaction in blood and saliva samples. Eur J Clin Microbiol Infect Dis. 2001;20(12):892–895. [DOI] [PubMed] [Google Scholar]
- 99.Savoie A, Perpête C, Carpentier L, Joncas J, Alfieri C. Direct correlation between the load of Epstein-Barr virus-infected lymphocytes in the peripheral blood of pediatric transplant patients and risk of lymphoproliferative disease. Blood. 1994;83(9):2715–2722. [PubMed] [Google Scholar]
- 100.Gärtner BC, Schäfer H, Marggraff K, et al. Evaluation of use of Epstein-Barr viral load in patients after allogeneic stem cell transplantation to diagnose and monitor posttransplant lymphoproliferative disease. J Clin Microbiol. 2002;40(2):351–358. doi: 10.1128/JCM.40.2.351-358.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wagner H-J, Cheng YC, Huls MH, et al. Prompt versus preemptive intervention for EBV lymphoproliferative disease. Blood. 2004;103(10):3979–3981. doi: 10.1182/blood-2003-12-4287. [DOI] [PubMed] [Google Scholar]
- 102.Green ML, Leisenring W, Stachel D, et al. Efficacy of a viral load-based, risk-adapted, preemptive treatment strategy for prevention of cytomegalovirus disease after hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2012;18(11):1687–1699. doi: 10.1016/j.bbmt.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ortiz A, Justo P, Sanz A, et al. Tubular cell apoptosis and cidofovir-induced acute renal failure. Antivir Ther. 2005;10(1):185–190. [PubMed] [Google Scholar]
- 104.Leather HL. Drug interactions in the hematopoietic stem cell transplant (HSCT) recipient: what every transplanter needs to know. Bone Marrow Transplant. 2004;33(2):137–152. doi: 10.1038/sj.bmt.1704316. [DOI] [PubMed] [Google Scholar]
- 105.Gyurkocza B, Sandmaier BM. Conditioning regimens for hematopoietic cell transplantation: one size does not fit all. Blood. 2014;124(3):344–353. doi: 10.1182/blood-2014-02-514778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.