INTRODUCTION
The field of Biochemical Engineering is vast. From its historical origins in the microbial production of antibiotics in the 1940’s, today’s Biochemical Engineer may contribute to advances in a wide range of technical areas including biomaterials, synthetic biology, tissue engineering, pharmaceutical production, food science, and bioenergy, among others. The industrial biotechnology sector, traditionally the province of biochemical engineering, is estimated at >$100 billion per year in the US with over 10% growth rate (Carlson, 2016). There are many grand challenges that will require solutions that involve biotechnology such as: energy, water, waste, carbon utilization, food, healthcare etc. The opportunities for biotechnology to positively impact life on earth have never been higher.
The recent Biochemical and Molecular Engineering XXI conference held in Mont Tremblant, Quebec, focused on “The Next Generation of Biochemical and Molecular Engineering: The role of emerging technologies in tomorrow’s products and processes” (July 2019). At this conference, a panel of biochemical engineers was convened to discuss grand challenges for the field. The composition of the panel was designed to cover a range of research areas, feature speakers with variable years of experience in the field, and include academic and industrial practitioners. The panel contributed eighteen topical areas (2 per panelist) for consideration in advance of the meeting, and conference attendees voted to select nine of these (1 per panelist) for further discussion. To aid in voting, short descriptions were provided for each topic through a polling app recommended by Engineering Conferences International (ECI). Attendees could also offer comments that could be read and endorsed by other attendees. The selected topics therefore represented the consensus view of the attendees of the most significant option of each pair. For each selection, perspectives were offered by the panel and broadly discussed by the attendees in a robust moderated dialogue. The goal was to capture and cross-fertilize ideas of the different conference sessions that might contribute to emerging research areas or grand challenges.
This Perspective article synthesizes these grand challenge topical areas to five broad thematic areas (Table 1) where concentrated efforts and focus by the field are needed, recognizing that many opportunities across the discipline exist. Perhaps the most consistent theme was the need to move beyond traditional products (therapeutic proteins) and model organisms/cells (Chinese Hamster Ovary (CHO), Escherichia coli, Saccharomyces cerevisiae). Many grand challenges in environmental and food sustainability, personalized health, and others, emerged that could be solved by biochemical engineers skilled in the techniques and methodologies of modern biotechnology. In order to do so, the field must develop new tools, funding, and drivers to expand into these new areas. The prevailing sentiment was that we must push past the traditional limits of biochemical synthesis, with the paradigm of one cell type producing one product. Broad challenges, for example, within this specific thematic area include: developing rules for hybrid biochemical/chemical conversion bioprocesses; predictive control of metabolic pathway spatial assembly; and the use of alternative biomanufacturing paradigms for enhancing biological conversion processes, such as microbial consortia, designed co-cultures, or cell-free systems. Other thematic areas include: bioprocess development for individualized medicine, forward-engineering for cellular control and predictable cell behaviors, which includes data-driven machine learning approaches for accelerating design, and engineering to understand & exploit new biology.
Table 1:
Thematic and topical areas considered for this perspective.
| THEMATIC AREAS | Novel Products and Non-traditional organisms | Pushing past the limits of biochemical synthesis | Bioprocess development for individualized medicine | Forward engineering for cellular and biomolecular control | Engineering to understand & exploit new biology |
|---|---|---|---|---|---|
| Topical Area (Green - Selected; Blue - Unselected) | Non-model organism development | Combining chemical catalysis with biochemical conversion | Bioprocess development for individualized medicine | Integration of mechanistic based models with data driven approaches for protein- and cell-based engineering | The biology and biotechnology of extracellular vesicles |
| Valorization of waste streams | Dynamic spatial assembly of metabolons | Integrating biotherapeutic products and medical devices | Transforming cellular control and predictable cell behaviors through synthetic biology | Building and exploiting interface between electronics and biology | |
| Biochemical engineering opportunities in food & beverage production | Consortia & Co-cultures – New modality for synthesis | Gene Therapy: The next leap in Biopharma Technology | Genetically encoded biosensors | ||
| Point of care cell-free production modalities | Intgrating computational and experimental protein design | ||||
| Chassis development for plant medicinal pathways | Melding heterogeneous biological systems data into a decision framework |
The topical areas listed below are by no means a comprehensive portrait of all current activities by biochemical engineers, nor is this the only current technical roadmap (e.g. https://roadmap.ebrc.org/). Rather, this Perspective is meant to synthesize one possible vision on where investment in research areas is needed for biotechnology to continue contributing to some of the world’s grand challenges.
Thematic areas
Novel products and non-traditional organisms
Much of our view of biology and what is possible in biotechnology is shaped by what we learn in a small collection of well-characterized model cells like E. coli, S. cerevisiae, and CHO cells. Most educational resources are based on the discoveries made in these systems, and thus our view of life is often viewed in the context of these cells. Therefore, the fields of metabolic engineering and synthetic biology frequently turn to this short list of model cells as “chassis” for technology development. This has led to fantastic accomplishments, with undoubtedly great new advances on the horizon.
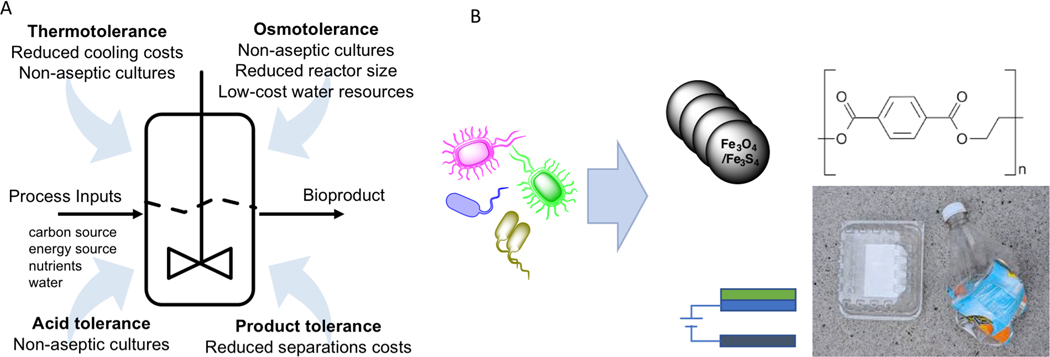
By contrast, investigation of non-model organisms, development of genetic tools in non-model organisms, and development of non-model organisms for use as chassis has been more limited. There are many important reasons why we need to expand applied research activities with non-model cells and organisms (Figure 1).
Figure 1.
Novel traits in non-conventional microbial hosts can be exploited to create a new generation of biochemical processes. (A) Many non-conventional fungi and bacteria exhibit high tolerance to various environmental stresses that can occur during bioprocessing. Matching stress tolerant traits with critical bioprocessing challenges can save process costs and enable new designs that enhance product titer, rate, and yield. (B) Non-conventional hosts can be exploited for non-conventional processes like formation of magnet nanoparticles, bioelectro-synthesis, and valorization of plastic waste streams.
Alternative cells provide new opportunities for metabolic engineering and synthetic biology. Non-model cells may serve as superior “chassis” organisms as they can thrive in extreme environments and are already evolved for optimized performance of various capabilities. Non-model cells can provide different capabilities like stress-tolerant phenotypes and enhanced catabolic breadth (described in a recent review(Thorwall, Schwartz, Chartron, & Wheeldon, 2020)). Thus, alternative chassis may prove to be more suitable for future applications, , including the use of cell-free systems(Silverman, Karim, & Jewett, 2019).
Non-model organisms are already involved in a wide variety of well-established and scaled bioprocesses like wastewater treatment, metal mining, nitrogen fixation, and food production. Further investigation into the organisms found in existing bioprocesses will lead to new understandings of critical mechanisms, metabolic capacities, microbial competition, and mechanisms for robustness of cell-cell communication networks.
Advances in biotechnology often arise from advances in basic biology, and important insights have been gained from non-model cells. Classic examples include restriction endonucleases and polymerases from thermotolerant extremophiles (Frock & Kelly, 2012). A more recent example is the discovery and engineering of a poly-ethylene terephthalate (PET) plastic degrading pathway found in a bacterium isolated from a bottle recycling facility(Yoshida et al., 2016). It is likely that new genomes and metagenomic sequence information from unculturable microbes and viruses in extreme and unusual environments can enable discovery of new biological capabilities and inspire new biochemical technologies.
A critical future goal of the biochemical and molecular engineering community will be the investigation, development, and engineering of non-model cells, components, and processes. This requires advances in computational tools for pathway prediction and large-scale systems biology data analysis to enable forward engineering. Such advances and research focus would especially benefit biotechnologies on the horizon such as biological/computer interfaces, waste recycling, and extra-terrestrial exploration (see a partial list in Table 2). As synthetic biology further expands into new organisms and microbial ecosystems it will be critical to replicate and even expand the biosafety strategies that have been used in the development of the classic model cells. There has already been interest in introducing biocontainment features into future generations of engineered cells (J. W. Lee, Chan, Slomovic, & Collins, 2018). In the sections that follow, we consider the near-future biotechnologies of sustainable protein production, and biological valorization of waste streams.
Table 2.
Selected non-conventional microbial hosts and cell-free systems for next generation bioprocessing
| Bacteria | Desirable phenotype |
|---|---|
| Halomonas campaniensis | Thermo-, osmo-, and alkaline tolerance |
| Clostridium thermocellum | Thermotolerance; lignocellulosic biomass breakdown |
| Clostridium spec. | Use of CO / CO2 as sole carbon sources |
| Methanotrophs | Use of gaseous alkanes as sole carbon sources |
| Pseudomonas putida | Solvent tolerant; catabolism of aromatics |
| Acidothiobacillus ferrooxidans | Acid tolerant; extracellular electron transfer |
| Shewanella oneidensis | Extracellular electron transfer |
| Yeast and Fungi | |
| Kluyveromyces marxianus | Acid and thermotolerance; rapid growth |
| Issatchenkia orientalis | Acid and thermotolerance |
| Yarrowia lipolytica | Lipid catabolism |
| Pichia pastoris | Heterologous protein expression |
| Neocallimastigomycota | Lignocellulosic biomass breakdown |
| Cell-free systems | |
| Platforms | High-yielding, cost-effective, scalable bacterial systems for probing cellular function and biomanufacturing (E. coli, Vibrio natriegens, Streptomyces sp., clostridia, CHO, yeast, pichia pastoris, plants ) |
Valorization of waste streams
Streams from municipal, agricultural, food, and plastic waste materials constitute a burden for communities, industries, nations, climate change and the environment more broadly. Increasingly, such streams are also viewed as an opportunity for utilizing the enormous quantities of chemical energy stored within them(Tuck, Pérez, Horváth, Sheldon, & Poliakoff, 2012). Many of these streams will be eventually converted to the greenhouse gases methane and CO2 (e.g., in solid-waste disposal facilities or anaerobic wastewater treatment facilities) with very low, or zero, capture efficiency.
Generation of methane (biogas) from waste streams involves semi-solid or liquid-stream methanogenic anaerobic digestion, largely based on the development of natural microbial consortia. Such processes are slow, not very effective, and thus not widely adopted. Challenges of producing fuels and chemicals from diverse feedstocks include the necessity of expensive biomass hydrolysis for effective fermentation, the loss of significant electrons generated from substrate catabolism to H2, and extensive CO2 loss due to decarboxylation of pyruvate to produce acetyl-CoA, the key starting intermediate for the production of most chemicals and fuels.
The ability to simultaneously use biomass substrates and gaseous substrates (renewable H2 or syngas from various sources, such as from gasification of municipal or agricultural wastes) is of major technological significance as it would result in exceptional levels of substrate-carbon and electron utilization thus leading to high product yields. There are opportunities for combining biological and non-biological (e.g. catalytic/electrocatalytic) processes to achieve this goal. Technologies for utilizing both solid/semisolid and gaseous waste streams are therefore of major interest and should be the target of additional research investment.
In certain respects, valorization of plastic waste is an easier problem because waste is concentrated through commercial recycling operations with reasonable batch-batch consistency. Biological conversion and upgrading of polyester and polyurethane waste plastic streams is particularly attractive because (i.) ester and urethane bonds are accessible by enzymes; (ii.) plastic waste is much cheaper on a per mass basis than most existing carbohydrate feedstocks; (iii.) biological conversion routes are compatible with typical contaminants in plastic waste streams; and (iv.) monomers have similar reducing equivalents with current feedstocks. For example, the PET monomer ethylene terephthalate (C4H8O4) has the same degree of reduction as glucose. Specific biochemical engineering challenges include developing enzymes that can efficiently deconstruct plastics to constituent monomers, and designing non-model organisms that can catabolize plastic monomers while also withstanding the necessary processing conditions. Additional challenges include a distributed “supply chain” and heterogeneity of contaminants in the waste streams. This will be a fertile ground for bioprocess engineers, protein engineers, synthetic biologists, and metabolic engineers.
The pressing environmental implications, and the need to move forward the concept of circular economy make it imperative that new thinking, new players and new investments are necessary to enable high-end and efficient processes to solve a problem of enormous global importance.
Biochemical engineering opportunities in food and beverage production
Biochemical engineering has a long and storied history of supplying innovation for the food and beverage industry, including large-scale cultivation of microorganisms for nutrition. This development of such ‘single cell protein’ was winding down as a research area before several authors on this perspective were born (Solomons & Litchfield, 1983). However, a resurgence of this topical area is led by commercialization of plant-based and cell-based meat products palatable to the end consumer.
As an example, the most publicized ingredient in the Impossible burger is genetically modified Pichia pastoris protein-rich extract containing a legume heme protein; when formulated in the burger this ingredient adds reddish color and flavor. This unapologetic use of genetically modified microorganisms opens the door for biochemical innovation in food products. Engineering microbial proteins that are more nutritious and yet still mimic the mouth feel of meat, or that can taste like sugar(Ming & Hellekant, 1994), or designing microbes with distinct flavor profiles tailored by metabolic engineering(Denby et al., 2018) are examples of innovations needed on the cellular engineering side. While large-scale fermentation processes for food and beverages exist, scale-up and bioprocess challenges for microbe-based protein are daunting: supplanting even 1% of US daily protein consumption by single cell protein would require 750 metric tons of cells per day. More efficient cell harvesting and dewatering unit operations, programmed cell lysis, bioreactor design, and use of alternative feedstocks will be necessary before widespread deployment occurs.
The same rationale is valid for application as single-cell protein in present-day aquaculture. While aquaculture is the most-efficient and fastest growing protein generator for human consumption, one of its most relevant feedstocks is fishmeal which is limited in supply due to overfishing and therefore significantly compromises future sustainability of the aquaculture industry. Single-cell protein tailored to the specific needs of farmed fish and crustacean species might offer a solution.
Cultivated meat, by contrast, involves the in vitro production of cells present in meat used for human consumption. The cells used to produce cultivated meat include cell types present in meat such as skeletal myocytes and adipocytes from the mammalian, avian, and piscine cell lines of any meat-harvested species(E. A. Specht, Welch, Clayton, & Lagally, 2018). Recently, the National Academies of Science, Engineering, and Medicine noted the high growth potential of cultivated meat and identified it as an emerging biotechnology area(National Academies of Sciences & Medicine, 2017). Efforts to achieve commercialization within the decade will require considerable attention to scale-up and large-scale manufacturing(M. J. Post, 2012). These practices include cell line selection and development, scaffolding, bioreactor design, cell culture medium optimization, and management of supply chain and distribution. One of the dominant barriers for cultivated meat to reach competitive prices with conventional meat is the cost of cell-culture media(National Academies of Sciences & Medicine, 2017). Traditionally, cell-culture media incorporated serum to promote cell growth, via the action of growth factors and other often non-defined components. Although serum-free and animal-origin-free media are able to support cell survival, proliferation, and differentiation(M. Post & van der Weele, 2014), a drastic cost reduction of both the basal medium and the growth factors would be required for economic viability at scale(L. Specht, 2020). Efforts directed towards drastically reducing the amount of growth factors needed, or the production of these factors in recombinant organisms, or the development of cheap protein mimotopes of these growth factors could offer a way out of this challenge. Metabolic modeling also offers an attractive avenue for benchmarking different ways of formulating a growth medium using either defined ingredients-only or supplemented with cell extracts (i.e., yeast or microalgae(Sathasivam, Radhakrishnan, Hashem, & Abd_Allah, 2019)).
Pushing past the limits of biochemical synthesis
Combining chemical catalysis with biochemical conversion
Whenever the production of a new complex molecule is required from a given precursor there exists significant creative tension between chemists and biochemical engineers. Chemistry offers advantages in throughput, toxic intermediate tolerance, freedom to operate at high temperatures and the ability to leverage an existing chemical processing infrastructure. In contrast, biology allows for simpler processes, self-regulated pathways, making chemical changes in specific locations even for highly functionalized molecules. The recent review article by G-M Lin and colleagues highlights many of the new advances and remaining challenges (Lin, Warden-Rothman, & Voigt, 2019). It is worth noting that continuous progress over the last few decades toward expanding the utility of enzymes, including advances in protein engineering, artificial enzyme development, and high-throughput screening have opened new opportunities for chemoenzymatic synthesis in both aqueous and nonaqueous media.
While there are famous examples where both chemical catalysis and biochemistry were brought to bear (Anbarasan et al., 2012; Karp et al., 2017; Paddon et al., 2013), generally the two modes of production are deployed in isolation of one another. A number of retrosynthetic algorithms are available(Campodonico, Andrews, Asenjo, Palsson, & Feist, 2014; Henry, Broadbelt, & Hatzimanikatis, 2010; Kumar, Wang, Ng, & Maranas, 2018) for identifying a sequence of steps to a product using both existing and novel enzymatic steps. At the same time rapid progress has been made for chemical synthesis using rules-based pathway design (Klucznik et al., 2018). What is lacking is an integrated workflow for making decisions as to what steps will be carried out through biochemical conversions and which steps will be left to chemical catalysis (Wheeldon, Christopher, & Blanch, 2017).
How can we harness both chemistry and biology to produce previously unobtainable molecules? One potential new direction is the use of cell-free systems to create hybrid molecule products composed of elements derived from both chemical and biological synthesis strategies in the absence of viability constraints(Swartz, 2012). In another direction, repurposing the translation apparatus (including the ribosome and the associated factors needed for polymerization) to make sequence defined polymers comprised of novel monomers could lead to new classes of materials of defined atomic sequence, exact monodisperse length, and programmed stereochemistry. For example, synthesis of polyamides (outside of polypeptides) or aramid polymers could open new opportunities at the intersection of materials science and synthetic biology (Ad et al., 2019; J. Lee et al., 2019).
Dynamic spatial assembly of metabolons and metabolic pathways
The design and assembly of so-called metabolons (structural-metabolic cellular complexes) and organelles mimics one of nature’s strategies for maximizing productivity and carbon flux through biochemical pathways, and is a rich area of research for biochemical and biomolecular engineers. Metabolons or metabolosomes are multienzyme complexes that allow the direct passage of a product from one enzymatic reaction to a consecutive enzyme in a metabolic pathway, which in some cases may benefit from substrate channeling (e.g., when a side reaction competes for an intermediate in the bulk or an inhibitor is present that interferes with a reaction step (Wheeldon et al., 2016)). Coordinated assembly and disassembly of these metabolons is an important factor in optimizing production of the desired metabolites. Natural organelle engineering has been effective in clustering key groups of enzymes—in peroxisomes and carboxysomes—and biochemical pathways believed to capitalize to at least some degree on enzyme localization and/or sequestration include tryptophan synthesis, the citric acid cycle, glycolysis, and purine synthesis.
The engineering concepts and physicochemical processes underlying the function of metabolons represent a scaled-down version of classical reaction engineering, and biochemical engineers have already made important contributions in modeling the behavior of systems ranging from 1D scaffolds to 3D microcompartments on multiple scales. Substrate channeling(Wheeldon et al., 2016), enzyme clustering(Castellana et al., 2014), and bacterial microcompartments (Jakobson, Tullman-Ercek, Slininger, & Mangan, 2017) have been the subjects of excellent modeling work, and these studies have provided important mechanistic insights and identified design criteria under which biochemical pathways will benefit from proximity and encapsulation effects. However, there are relatively few direct comparisons between such models and experimental systems, in part because well-characterized, precisely controlled experimental systems remain difficult to come by. Developing better techniques and methods to effect and control the assembly of scaffolded and compartmentalized systems both in vitro and in vivo is an exciting opportunity at the frontier of biomolecular engineering and related fields.
Many questions and challenges surrounding synthetic metabolons and organelles remain to be addressed, and several that emerge from the literature (Castellana et al., 2014; Jakobson et al., 2017; Kerfeld, Aussignargues, Zarzycki, Cai, & Sutter, 2018; Wheeldon et al., 2016) include the following:
Controlling transport of substrates and products across the compartment shell/membrane
Predicting the membrane permeability of a given small molecule metabolite
Precisely controlling the number and location of encapsulated proteins
Harnessing experimental methods to analyze the physical configuration and molecular organization of the metabolon
Quantifying the kinetic effects of enzyme clustering and compartmentalization
The new fundamental knowledge of how nature optimizes the productivity of biochemical pathways, together with the opportunities that such knowledge will afford for optimally engineering new pathways of practical interest, combine to make this area very fertile terrain for biochemical engineers.
Microbial consortia & Co-cultures
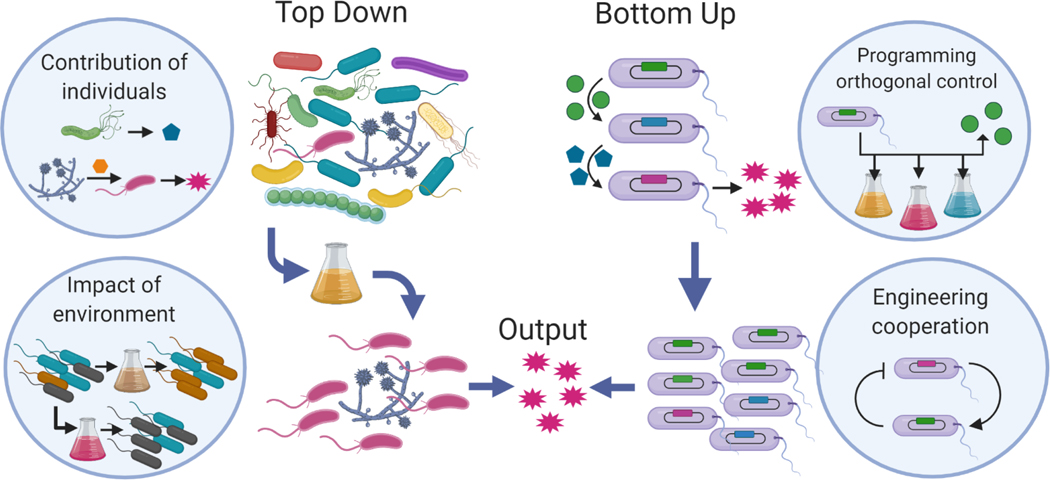
Many challenges in industrial biotechnology can be tackled by organizing microorganisms as “directed” consortia or even more well-defined “microconsortia”, such as synthetic co-cultures. These systems can be engineered using a more traditional top-down approach wherein microbe rich feedstocks are interrogated, prodded, and selected for specific purposes (Figure 2, (Gilmore et al., 2019)). Genomics-based methodologies and modeling are now being developed for the functional identification of the most useful consortia(Zuñiga et al., 2019), where the molecular bases for their intended functions are revealed and maintained. Importantly, complex initial sources, such as from anaerobic environments, can be accommodated(Solomon et al., 2016). Then, by using methodologies that reveal useful components for synthesis(Haitjema et al., 2017), “tuned” consortia might then be placed into processing environments for production. In this way, biomass feedstocks, particularly those that might otherwise be agricultural or municipal wastes, can be turned into useful, high value products. A major challenge to address for these applications is to maintain the consortia, or specifically, the precise composition of microbes (e.g., bacteria, fungi, protozoans) that is needed to carry out the specific function, particularly if the processing conditions require extended time periods in industrial (non-native) environments where population instability is well-known.
Figure 2.
Microbial consortia or “microconsortia” can be designed using top-down or bottom-up approaches. In top down approaches, consortia exhibiting desired properties are obtained from natural environments and tuned or directed for the desired function or output. This approach would benefit from a better understanding of the contribution of individuals in the original consortia and environment, as well as a better understanding of how the environment affects the consortia composition and function. An alternate approach to designing mini-consortia uses bottom-up strategies. Here, individual strains or species are engineered to perform specific functions that are part of a larger task. In this approach, tools or strategies to guarantee the behavior of the individual strains despite changing or unknown environmental conditions are needed. Further, methods to engineer communication and feedback between strains could allow for maintenance of the consortia composition and function over time.
In these situations, a bottom-up approach may be more advantageous (Figure 2). In this scenario, co-cultures or other “mini consortia” can be assembled of sets of engineered cells forming highly functional cell systems that are programmed to execute specific tasks(Bittihn, Din, Tsimring, & Hasty, 2018; Jones et al., 2016; Lindemann et al., 2016; Shong, Diaz, & Collins, 2012). Additional design space is available for such systems relative to a monoculture engineered to perform the same task; each cell or strain can be optimally designed for executing a particular part of an overall task. In turn, the distribution of engineered cell subpopulations provides additional flexibility in the overall process design. For example, a hypothetical production process may be distributed among three cell types: one that employs raw materials and makes an intermediate, a second strain may also use a raw material, but also uses the intermediate synthesized by the first population to make a second intermediate, and the third strain might finish the overall process. The relative numbers of the three strains can then be a control variable that is manipulated to ensure efficient production overall.
In both top-down and bottom-up situations, methodologies to coordinate subpopulation dynamics will be needed. These might involve external process inputs such as the addition of an inducer, an adjustment in oxygen or pH, or perhaps even process vessels that allow for fluid segregation or differential mixing. Conversely, in another novel approach, subpopulation dynamics could be created by rewiring native molecular communication systems like quorum sensing to autonomously control composition(Stephens, Pozo, Tsao, Hauk, & Bentley, 2019).
Specifically, new methodologies that recognize and interrogate the interplay between the external microenvironment and cell physiology will yield new insight on how to control cell behavior, particularly cell behavior that changes due to context. A cell’s response, for example, to a molecular cue might be completely different depending on the redox potential in its microenvironment or on the identity of the neighboring cells. For example, in the human microbiome environmental factors, e.g. chemicals, diets, etc. are known to impact the genotype-phenotype relationship and the development of diseases(Go, Nguyen, Harris, & Paul Lee, 2005). Thus far, they have been studied mostly for their involvement in metabolism (Sadler et al., 2018; Srivastava & Chan, 2008), signaling (and regulatory mechanism) (Yang & Chan, 2009), and even biophysical interactions(Cho et al., 2019). However, it is becoming increasingly apparent that diets and environmental factors alter the microbiome (Lewis et al., 2015) as well as the epigenetic landscape (Cowley Jr et al., 2012; Herceg, 2007) via DNA methylation patterns or histone tails to modulate the activity of genes and drive the development of disease. To investigate these new mechanisms, novel computational tools are needed to (i.) decipher the microbiome and microbial communities (Kim, Koh, & Rho, 2015) and how they impact the environment (diet)-gene-phenotype, and (ii.) integrate data from the genetic, epigenetic, transcriptional, post-transcriptional, and metabolic levels and their interaction with the microbiota in the development of diseases. The differences between anaerobic, micro-aerobic and aerobic physiologies are well known, but are these conditions purposely manipulated to guide behavior? How are signal molecules perceived at the molecular level and how can we design consortia or guide microbiomes to adapt to and utilize cues to assemble valuable behaviors, synthesize valuable compounds, or degrade xenobiotics or other problematic compounds, or even guide human health? With additional tools that enable predictive biology and that exploit external inputs, we might better control systems that are comprised of microbiomes or consortia in a variety of places, not just in human locales, but in the rhizosphere and fresh or saltwater environments. Efforts in these areas are ripe for the talents of biochemical engineers who want to build on their strengths to address challenging problems that are sure to have a great impact on human health and our society.
Bioprocess development for individualized medicine
Individualized medicine heralded a breakthrough when the FDA approved Kymriah(Dolgin, 2017), the first CAR T cell immunotherapy and the first gene therapy in the United States. Following closely on the heels of cell-based gene therapies, directly administered viral vector-based gene therapy Luxturna for the treatment of a monogenic inherited vision loss disorder was approved by the FDA in 2018 (Food & Administration, 2017). Currently in 2020, there are 17 FDA-approved cell and gene therapies (https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/approved-cellular-and-gene-therapy-products), with further growth in this sector expected in the next decades. Cell and viral vector cell production for personalized medicine constitutes new challenges and opportunities for bioprocess engineers. In conventional bioprocessing, biomolecules are typically produced in stirred tanks that can be scaled up to meet demand. In the case of personalized medicine, particularly for autologous cell products, the challenge becomes scaling out production because each patient requires their own bioreactor. In many ongoing clinical trials, cell production is also decentralized and labor intensive: clinical teams at hospitals handle in-hospital cell manufacturing, often using batch cultures with little monitoring of cell culture variables such as cell density, pH, partial pressure of oxygen, and nutrient consumption rates. These process variables, when monitored, are often done off-line using sporadic culture sampling. Manual handling of cell therapy products using functionally open cell culture systems such as T-flasks remains commonplace. More automated systems are available, such as those utilized for autologous adoptive immunotherapies (Harrison, Ruck, Medcalf, & Rafiq, 2017; Iyer, Bowles, Kim, & Dulgar-Tulloch, 2018), but even these have limited on-line monitoring and feed-back control over cell culture parameters. Automation and regulatory requirements to minimize risks of contamination as well as product variability create a strong drive towards the use of closed cell culture systems such as cell culture bags. As most pre-clinical studies are conducted in polystyrene vessels, the transition to bag-based cultures can lead to changes in cell-surface interactions and other culture parameters such as gas exchange(Fekete, Béland, Campbell, Clark, & Hoesli, 2018). There is a strong need to use scale-down culture systems during preclinical development which better reflect manufacturing methods and culture vessels at clinical scale.
For allogeneic cell products, scale-up can be performed and can rely on bioreactor designs that approach more conventional biomanufacturing. However, the challenge of on-line monitoring of a cell-based product remains. Moreover, many allogeneic cell therapy products such as mesenchymal stem cells or induced pluripotent stem cell-derived products are anchorage-dependent cells. Scale-up thus often relies on increasing the surface area for cell adhesion, for example using microcarriers, hollow fiber bioreactors or stacked vessels – increasing the complexity of automated handling.
Viral vector production – whether for transduction of cells ex vivo or in vivo – at clinical scales with high reproducibility also remains challenging(McCarron, Donnelley, McIntyre, & Parsons, 2016). Many research-scale viral vector production system utilize anchorage-dependent cells which require hollow fiber bioreactor or microcarrier systems which are much more complex to scale up. With cell lines adapted to suspension culture such as human embryonic kidney cells, process intensification is an area of focus. Productivity does not only require high yields of viral particles, but also of properly assembled viral particles that maintain their functional capacity to transduce and express transgenes in target cells. In-line or rapid off-line monitoring of viral particles would significantly accelerate upstream process optimization. Finally, novel downstream purification methods that are scalable and that can resolve functional from non-functional viral particles are needed.
Although there have been significant advances in adapting culture systems to challenging cell therapy products over recent years, some of the practical questions that need to be addressed are:
Can we formulate a list of overarching cell culture parameter ranges required for cell and therapy products in adherent versus suspension culture?
What biomaterial approaches or genetic engineering methods may we employ to control the homogeneity of the desired cell populations?
How can we make current culture systems more flexible and adaptable by end-users (including clinical centers) to facilitate manufacturing of several cell therapy products with a single system?
What in-line methods could we employ to better assess and control cell and gene product quality?
Cellular therapy is set to revolutionize the treatment of cancer and conditions where small molecules and other biologics have not led to a cure to date. The growing list of approved cell therapy products (https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/approved-cellular-and-gene-therapy-products) not only for people suffering from blood disorders and cancers, but also for cartilage, retinal and other tissue defects portends a new era in the treatment of degenerative disease. Groundbreaking clinical trials are testing the safety and efficacy of embryonic stem cell-derived products transplanted in various encapsulation devices to treat type 1 diabetes(Moeun et al., 2019). Addressing the bioprocessing challenges listed above is critical in assuring the safety, efficacy and accessibility of these life-saving products.
Forward engineering for cellular and biomolecular control
Integration of mechanistic based models with data driven approaches for protein- and cell-based engineering
Since the advent of the biochemical engineering discipline mechanistic models based on kinetics and thermodynamic constraints have guided experiments. We now have a torrent of high quality data from myriad omics technologies, deep mutational experiments of protein and RNA-encoding sequences(Kowalsky et al., 2015), and facile high-throughput strain development in many organisms spurred in part by the CRISPR revolution (Schwartz, Hussain, Blenner, & Wheeldon, 2016). To what extent could these new large datasets, with potential for more modern machine learning approaches, enhance current modeling techniques? Compared with current models, what kind of biological knowledge could we gain by using machine learning?
An illustrative example comes from protein science. The protein folding problem is typically formulated as predicting an accurate atomic structure of a protein given its sequence of amino acids. In 2018, the winners of the blind prediction CASP challenge were a group of Alphabet engineers without specific training in this area. The team, dubbed AlphaFold, outperformed all other scientific groups in the world and really advanced the field by about 2–3 years(AlQuraishi, 2019). Importantly, they used the mechanistic insight that positions that are close in distance tend to co-evolve together. This insight is not new and has been developed in the literature over the past two decades(Morcos et al., 2011). They were successful in large part because the existing datasets of tens of millions of accurate protein sequences and over a hundred thousand protein structures were vast, centralized, and curated. They used deep learning to learn a differentiable potential between co-evolving residues that is specific for each protein.
This example is particularly instructive because it tells us a few things about how our community should approach this opportunity. First, we want good data and heaps of it, no matter the source. Methodological advances should be encouraged for collecting large amounts of phenotypic and genotypic data on engineered strains and activities and biophysical properties of proteins. Similarly, strong efforts to centralize already existing literature datasets should be supported, perhaps as a community effort. As an example, the protein engineering field now does this with ProtaBank(Wang et al., 2018). Second, the AlphaFold team improved on existing mechanistic insights into how co-evolution of residues predicts distance in the folded polypeptide chain using their deep learning approach. They also used an ensemble model with existing structure-based prediction using physically realistic potentials in the macromolecular modeling software package Rosetta. The field should embrace ensemble models and related techniques may be applied to nail down the thermodynamic driving forces for resolving kinetics of intracellular fluxes(Gopalakrishnan, Dash, & Maranas, 2019) or better use of evolutionary and/or co-evolutionary networks and other mechanistic insights to engineer stability in enzymes(Ritter & Hackel, 2019). Here is where deep learning may be particularly useful in identifying very strong mechanistic bases for why outcomes look the way they do, given a range of potential inputs. Third, the AlphaFold team originally looked at much more complicated machine learning models using features that do not have such mechanistic insight, which they discarded because of the strength of the simpler and more powerful co-evolutionary analysis. Simpler features grounded in physico-chemical or evolutionary mechanisms will ultimately be more useful, more likely to lead to biological insights that can be exploited, as most of what we do is grounded with strong constraints set by physical chemistry.
Finally, we should be realistic about the data we have and can generate. Existing linear and non-linear regression based models work well in a variety of contexts. For example, one of us (T.A.W., unpublished) has found in protein engineering that linear regression seems to work fairly well for prediction of protein activity, consistent with reports from more limited datasets(Fox et al., 2007). These simpler regression models also have the advantage of being more interpretable.
For cell engineering specifically, there are clear recommendations for efficiently exploring the vast genetic space to achieve actionable and or valuable cell engineering outcomes:
Mine existing data sets: Many large, unbiased genetic characterization studies have been conducted to date on model organisms and have been published. We need to leverage what has already been done to find patterns. This requires us to aggregate and organize the datasets across multiple studies and leverage searchable databases. It also requires a higher level of engagements/knowledge sharing from industry. Here community efforts to centralize such datasets, as mentioned above, should be strongly supported and encouraged. As an example, some studies have comprehensively tested the genetic landscape for host organisms (e.g. genetic transcription engineering). Can we retroactively review these studies and outcomes to learn what worked and perhaps why? Can we leverage those findings to understand how to effectively truncate a genetic search space without losing quality/positive outcomes?
In silico tools: Meticulous experimental studies are time and resource consuming. Search space is more efficiently managed using good in silico models. We need to continue to enhance metabolic models, and pressure test the quality of models that are developed using diverse metabolic pathways (i.e. not just central carbon metabolism). Going forward, there should be more emphasis on comprehensive, complex metabolic functions (glycosylation, lipids, polyphenols, etc.), which complements the complex products the field is now interested in producing using cellular hosts.
Understand what is host/cell line specific vs. biologically universal: A lot of excellent studies are published on one cell line/host to understand or fix specific biology. We need to understand when these findings can be leveraged for a different cell line/product, and when we can avoid repeating cellular optimization/engineering efforts. To build this understanding, we should consider vertical organism testing, i.e. progressing an optimization with a specific outcome in mind first through a single celled, prokaryotic organism, then through a single celled eukaryotic organism, and finally through a multi-celled eukaryotic cellular host.
Beware of model protein products! Proof of concept work on simple proteins may not translate to complex targets. It could mask/mislead/not scale to the desired, applications and products. We should incorporate this consideration into study designs to ensure the best quality information is captured.
Transforming cellular control and predictable cell behaviors through synthetic biology
A major issue in biomanufacturing and bioprocessing is heterogeneity and lack of control in cell behavior manifesting in alterations of process parameters and product quality. We need to understand and control the sources and mechanisms of heterogeneity to achieve better process control, reproducibility and reliability. One way to address this challenge is to build orthogonal, tunable tools that operate on time scales faster than the process being controlled in order to make cells more readily manipulated and directed towards the generation of desirable products.
Engineering cellular systems with predictable behavior requires diversification of tools to achieve control at the molecular level. Current tools to control cell behavior are mainly based on transcriptional regulators and have been successfully evolved through a variety of protein engineering methods. There is a pressing need currently to identify new tools and new methods for identifying appropriate dynamic control elements to use in larger systems. Protein-mediated regulation typically operates over faster time scales than transcriptional and translational control and may be coupled directly to endogenous pathways and without the need for genomic integration(Budihardjo, Oliver, Lutter, Luo, & Wang, 1999), enabling dynamic control. Repurposed CRISPR-Cas molecules have also been explored(Xu & Qi, 2019). Despite successful methods for exogenous control over CRISPR system, methods for internal controls remain a challenge. Efficient tools for tuning CRISPR activity, such as the recently discovered anti-CRISPRs, are needed for the future development of synthetic CRISPR-mediated circuits(Nakamura et al., 2019). Finally, naturally occurring epigenetic programs underlying cellular differentiation and development provide new opportunities for the design of control systems based on molecular writers and readers of chromatin signatures(Park, Patel, Keung, & Khalil, 2019).
Larger control systems can be assembled as more control elements are developed. Yet, there are many open operational questions for how cellular pathways detect and process input signals. First, the quantitative and dynamic input features that are perceived by natural and synthetic control systems are not always fully characterized for systems. It has become increasingly apparent that input dynamics rather than absolute values play significant roles in shaping the ultimate cellular outcome. Second, the system design needs to be carefully determined: extrapolating the design rules of classic microbial two-component systems to predict more complex signaling networks has proved to be a non-trivial endeavor, requiring tuning of control elements guided by deterministic and stochastic modelling carefully deployed to predict system behavior.
Predicting pathway behavior has proven to require quantitative modeling to develop an accurate understanding of even relatively simple systems (Ha & Ferrell, 2016). Ligand-controlled responses such as growth factor pathways, for instance, can respond to input concentrations with a diverse range of sensitives, pointing to the critical need to build operational models of cellular systems based on quantitative descriptions of the input-output properties of each signaling pathway.
Critical recommendations related to progress in the development of cellular control systems include:
Experiments should focus on single cell analyses to avoid confounding effects of population heterogeneity. Because cellular behaviors are often unsynchronized, it is also important to explore the dynamic response of single cells to avoid artifacts from static single cell or population measurements. Additionally, where possible, researchers should capitalize on gene-editing technology to reduce population heterogeneity.
Studies of cellular control systems should rely on reconstitution of minimal versions of circuits and gene networks; isolation of minimal version of cellular pathways from natural inputs and outputs enables studying signal processing capabilities systematically and generating predictive models that recapitulate the governing features of different control networks.
As larger scale genetic circuit engineering remains challenging, it is important to leverage the predictive power of mathematical modeling and integrate models and experiments to explore the behavior of complex cellular systems across parameter regimes.
Engineering to understand & exploit new biology
Building and exploiting interface between electronics and biology
Semiconductor technologies have transformed our abilities to access, store, process, and communicate information by enabling increasingly smaller, cheaper, more powerful, interconnected, and easier-to-use electronic devices. Synthetic biology will enable the extension of these modalities to interface with electronics – by rewiring and programming cellular processes that manipulate chemical information at the molecular scale, using redox as a vector of information transfer(Liu et al., 2017) - in ways that facilitate information exchange with electrodes (Tschirhart et al., 2017; VanArsdale et al., 2019). There has already been remarkable progress in spanning biological and electronic communications for an important subset of problems involving the ionic electrical modality, including advances in neuro-prosthetics and in understanding and mapping brain function. Molecularly based information transfer and notably, redox enabled communication is widespread in biology: it is used by the immune system for inflammation and wound healing; it underpins communication within the gut, and potentially between the gut and brain; and it enables communication in the biosphere (e.g., cells in the plant roots can detect/respond to activities in its rhizosphere), to name a few. To enable redox-based communication, future opportunities lie in the fabrication of ‘smart’ materials interfaces that integrate biological recognition and computation while facilitating information transfer to and from the devices at length and time scales that are often viewed as discordant.
There is tremendous potential for the development of devices that seamlessly transfer information to and within biology. To provide just one example, wearable devices such as smart watches that provide actual chemical information in addition to what is currently available (i.e., moisture, temperature, and cardiovascular function) will radically transform our everyday lives. New efforts in electron transfer, redox biology, materials and surface characterization and assembly, will be needed in addition to traditional expertise in mass and momentum transfer, reaction kinetics, and thermodynamics, to create effective systems for information transfer into and out of the biological system.
The biology and biotechnology of extracellular vesicles
Extracellular vesicles (EVs) are membrane vesicles that carry RNAs, proteins, lipids and sometimes DNA from their parent cells(Kao & Papoutsakis, 2019). EV generation takes place under cellular activation or stress. Cells use EVs to communicate with other cells by delivering signals through their content and surface proteins. Besides mammalian cells, outer membrane vesicles (OMVs) (Anand & Chaudhuri, 2016), derived from Gram-negative bacteria, are involved in stress response, promoting survival, pathogenesis, and interaction between bacteria in a community. Gram-positive bacteria generate a large number of EVs, as well, but their role in intercellular communication remains largely unexplored. Over the last few years, EVs have emerged as important mediators of intercellular communication regulating an ever-expanding range of biological processes, both on normo- and patho-physiology. The former includes enhancing and accelerating native developmental programs in immunology, vascular repair and angiogenesis, while the latter include carcinogenesis and cancer metastasis, neurodegenerative disorders, and infectious and cardiovascular diseases.
Based on their currently known biology, EVs are suitable for a broad range of applications, from minimally invasive diagnostic applications to therapeutic interventions, including cell therapies and macromolecular drug delivery. In addition, there are two new emerging EV subfields. One is the role of microbial EVs in microbial consortia activities, including those of the microbiomes, and in the plant-to-microbe interactions. The other is based on the metabolic activities of EVs independently of the parent cells. The latter can be the basis for designing and employing efficient cell-free systems for advanced biocatalysis including combinatorial biosynthesis, but distinct from the current technologies that are based on in vitro transcription and translation.
Both EV cargo and membranes can be independently engineered and used for various applications (Kao & Papoutsakis, 2019). In order to pursue such applications involving EVs, better EV characterization, as well as better understanding of the mechanisms of cell targeting(Jiang, Kao, & Papoutsakis, 2017) and methods for EV biomanufacturing are needed. This is a relatively new field, especially regarding microbial EVs, but there is great potential in a broad spectrum of applications, thus making EV-funding investments a worthy cause.
Perspective
Biochemical engineers are involved in solving many of the world’s greatest challenges. This perspective synthesizes where research investment should be strengthened to enhance the impact by the discipline. For each thematic area there are clear recommendations moving forward.
First, further and more sustained investment and research is needed in developing efficient ways to build new genetic tools in non-model organisms. Novel products requiring non-model organisms or cell-free systems should be particularly supported. Additionally, novel technologies enabling microbial process scale-up and downstream processing are strongly desired.
Second, developing truly sustainable bioprocesses requires circumventions of current limitations on cellular biochemical synthesis. High on the list are methods or workflows to determine how to split a process between biochemical conversion and chemical catalysis. Cell-free systems creating hybrid chemical/biological synthesis is one approach to remove cellular constraints; continued development of such systems should be supported. There are a number of fundamental questions on metabolons that can be addressed with careful experimentation. Finally, control mechanisms should be discovered and engineered for tailoring precise, stable, compositions of microbial consortia for various bioprocesses.
Third, several aspects of bioprocess development for individualized medicine need to be studied, including determining cell culture parameter ranges for adherent vs. suspension cultures, improving the homogeneity of the cell populations, continued innovation for increasing flexibility and adaptability of cell culture systems, and developing better in-line methods for assessing and controlling product quality while assuring accessibility to these life-saving therapies.
A fourth thematic area in forward engineering for cellular engineering and biomolecular control already commands significant research support, which should continue, but with several clear recommendations. The current published deluge of high quality phenotypic and genotypic data on engineered strains and proteins should be centralized, perhaps as a community effort. Machine learning approaches to analyze, evaluate, and predict properties should be undergirded by evolutionary and/or physical chemistry principles. Researchers should be wary about using model cell lines and protein products to extract out biologically universal principles. Experiments with engineered networks should focus on single cell analyses as well as engineered homogeneous cell populations with robust mathematical modeling to guide understanding of the phenotypic parameter space.
The fifth and final thematic area involves engineering to understand and exploit new biology. Here new topical areas in merging electronics and biology and exploitation of extracellular vesicles were discussed, along with the attendant challenges inherent in these new fields.
Progress on these thematic areas is necessary for solving grand challenges in environmental & energy sustainability, and the next generation of safe, effective medicines.
Acknowledgments
The authors wish to acknowledge NSF CBET Award #1929518 to C.C. used to support the conference, academic speakers, and students. The authors also thank Steven Peretti and Amine Kamen for helpful discussions. Research reported in this publication was supported by the National Institute Of Allergy And Infectious Diseases of the National Institutes of Health under Award Number R01AI141452 to T.A.W; U.S. Army Research Laboratory and the U.S. Army Research Office MURI Award #W911NF1410263 to S.B. and I.W.; NSF CBET Award # 1802992 to C.C. C.H. was supported by a Canada Research Chair in Cellular Therapy Bioprocess Engineering and is a member of ThéCell (The Quebec Network for Cell, Tissue and Gene Therapy), the Quebec Center for Advanced Materials, PROTEO (The Quebec Network for Research on Protein Function), the McGill Regenerative Medicine network, the Montreal Diabetes Research Center and the Bioencapsulation Research Group; and DOE Center for Bioenergy Innovation Award # DE-AC05-000R22725 to C.M. M.C.J. was supported by ARO Award #W911NF-16-1-0372 & W911NF-19-1-0298, DOE Grant Award #DE-SC0018249, NSF Award #MCB-1716766. MCJ also gratefully acknowledges the David and Lucile Packard Foundation and the Camille Dreyfus Teacher-Scholar Program. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing interests
The authors declare no competing interests.
REFERENCES
- Ad O, Hoffman KS, Cairns AG, Featherston AL, Miller SJ, Söll D, & Schepartz A (2019). Translation of diverse aramid-and 1, 3-dicarbonyl-peptides by wild type ribosomes in vitro. ACS central science, 5(7), 1289–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AlQuraishi M (2019). AlphaFold at CASP13. Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand D, & Chaudhuri A (2016). Bacterial outer membrane vesicles: New insights and applications. Molecular membrane biology, 33(6–8), 125–137. [DOI] [PubMed] [Google Scholar]
- Anbarasan P, Baer ZC, Sreekumar S, Gross E, Binder JB, Blanch HW, . . . Toste FD (2012). Integration of chemical catalysis with extractive fermentation to produce fuels. nature, 491(7423), 235–239. [DOI] [PubMed] [Google Scholar]
- Bittihn P, Din MO, Tsimring LS, & Hasty J (2018). Rational engineering of synthetic microbial systems: from single cells to consortia. Current opinion in microbiology, 45, 92–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budihardjo I, Oliver H, Lutter M, Luo X, & Wang X (1999). Biochemical pathways of caspase activation during apoptosis. Annual review of cell and developmental biology, 15(1), 269–290. [DOI] [PubMed] [Google Scholar]
- Campodonico MA, Andrews BA, Asenjo JA, Palsson BO, & Feist AM (2014). Generation of an atlas for commodity chemical production in Escherichia coli and a novel pathway prediction algorithm, GEM-Path. Metabolic engineering, 25, 140–158. [DOI] [PubMed] [Google Scholar]
- Carlson R (2016). Estimating the biotech sector’s contribution to the US economy. Nature biotechnology, 34(3), 247–255. [DOI] [PubMed] [Google Scholar]
- Castellana M, Wilson MZ, Xu Y, Joshi P, Cristea IM, Rabinowitz JD, . . . Wingreen NS (2014). Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nature biotechnology, 32(10), 1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Stanzione F, Oak A, Kim GH, Yerneni S, Qi L, . . . Chan C (2019). Intrinsic structural features of the human IRE1α transmembrane domain sense membrane lipid saturation. Cell reports, 27(1), 307–320. e305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley AW Jr, Nadeau JH, Baccarelli A, Berecek K, Fornage M, Gibbons GH, . . . O’Connor DT (2012). Report of the national heart, lung, and blood institute working group on epigenetics and hypertension. Hypertension, 59(5), 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denby CM, Li RA, Vu VT, Costello Z, Lin W, Chan LJG, . . . Petzold CJ (2018). Industrial brewing yeast engineered for the production of primary flavor determinants in hopped beer. Nature communications, 9(1), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgin E (2017). Epic $12 billion deal and FDA’s approval raise CAR-T to new heights. In: Nature Publishing Group. [DOI] [PubMed] [Google Scholar]
- Fekete N, Béland AV, Campbell K, Clark SL, & Hoesli CA (2018). Bags versus flasks: a comparison of cell culture systems for the production of dendritic cell–based immunotherapies. Transfusion, 58(7), 1800–1813. [DOI] [PubMed] [Google Scholar]
- Food U, & Administration D (2017). FDA approves novel gene therapy to treat patients with a rare form of inherited vision loss. US Dep Heal Hum Serv. [Google Scholar]
- Fox RJ, Davis SC, Mundorff EC, Newman LM, Gavrilovic V, Ma SK, . . . Muley S (2007). Improving catalytic function by ProSAR-driven enzyme evolution. Nature biotechnology, 25(3), 338. [DOI] [PubMed] [Google Scholar]
- Frock AD, & Kelly RM (2012). Extreme thermophiles: moving beyond single-enzyme biocatalysis. Current opinion in chemical engineering, 1(4), 363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore SP, Lankiewicz TS, Wilken SE, Brown JL, Sexton JA, Henske JK, . . . O’Malley MA (2019). Top-down enrichment guides in formation of synthetic microbial consortia for biomass degradation. ACS synthetic biology, 8(9), 2174–2185. [DOI] [PubMed] [Google Scholar]
- Go VLW, Nguyen CT, Harris DM, & Paul Lee W-N (2005). Nutrient-gene interaction: metabolic genotype-phenotype relationship. The Journal of nutrition, 135(12), 3016S–3020S. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan S, Dash S, & Maranas C (2019). K-FIT: An accelerated kinetic parameterization algorithm using steady-state fluxomic data. bioRxiv, 612994. [DOI] [PubMed] [Google Scholar]
- Ha S, & Ferrell J (2016). Thresholds and ultrasensitivity from negative cooperativity. science, 352(6288), 990–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haitjema CH, Gilmore SP, Henske JK, Solomon KV, De Groot R, Kuo A, . . . Zhao Z (2017). A parts list for fungal cellulosomes revealed by comparative genomics. Nature microbiology, 2(8), 17087. [DOI] [PubMed] [Google Scholar]
- Harrison RP, Ruck S, Medcalf N, & Rafiq QA (2017). Decentralized manufacturing of cell and gene therapies: Overcoming challenges and identifying opportunities. Cytotherapy, 19(10), 1140–1151. [DOI] [PubMed] [Google Scholar]
- Henry CS, Broadbelt LJ, & Hatzimanikatis V (2010). Discovery and analysis of novel metabolic pathways for the biosynthesis of industrial chemicals: 3‐hydroxypropanoate. Biotechnology and bioengineering, 106(3), 462–473. [DOI] [PubMed] [Google Scholar]
- Herceg Z (2007). Epigenetics and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis, 22(2), 91–103. [DOI] [PubMed] [Google Scholar]
- Iyer RK, Bowles PA, Kim H, & Dulgar-Tulloch A (2018). Industrializing autologous adoptive immunotherapies: manufacturing advances and challenges. Frontiers in medicine, 5, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobson CM, Tullman-Ercek D, Slininger MF, & Mangan NM (2017). A systems-level model reveals that 1, 2-Propanediol utilization microcompartments enhance pathway flux through intermediate sequestration. PLoS computational biology, 13(5), e1005525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Kao C-Y, & Papoutsakis ET (2017). How do megakaryocytic microparticles target and deliver cargo to alter the fate of hematopoietic stem cells? Journal of Controlled Release, 247, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JA, Vernacchio VR, Sinkoe AL, Collins SM, Ibrahim MH, Lachance DM, . . . Koffas MA (2016). Experimental and computational optimization of an Escherichia coli co-culture for the efficient production of flavonoids. Metabolic engineering, 35, 55–63. [DOI] [PubMed] [Google Scholar]
- Kao C-Y, & Papoutsakis ET (2019). Extracellular vesicles: exosomes, microparticles, their parts, and their targets to enable their biomanufacturing and clinical applications. Current opinion in biotechnology, 60, 89–98. [DOI] [PubMed] [Google Scholar]
- Karp EM, Eaton TR, i Nogué VS, Vorotnikov V, Biddy MJ, Tan EC, . . . Manker LP (2017). Renewable acrylonitrile production. science, 358(6368), 1307–1310. [DOI] [PubMed] [Google Scholar]
- Kerfeld CA, Aussignargues C, Zarzycki J, Cai F, & Sutter M (2018). Bacterial microcompartments. Nature Reviews Microbiology, 16(5), 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Koh I, & Rho M (2015). Deciphering the human microbiome using next-generation sequencing data and bioinformatics approaches. Methods, 79, 52–59. [DOI] [PubMed] [Google Scholar]
- Klucznik T, Mikulak-Klucznik B, McCormack MP, Lima H, Szymkuć S, Bhowmick M, . . . Gajewska EP (2018). Efficient syntheses of diverse, medicinally relevant targets planned by computer and executed in the laboratory. Chem, 4(3), 522–532. [Google Scholar]
- Kowalsky CA, Klesmith JR, Stapleton JA, Kelly V, Reichkitzer N, & Whitehead TA (2015). High-resolution sequence-function mapping of full-length proteins. PLoS One, 10(3), e0118193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Wang L, Ng CY, & Maranas CD (2018). Pathway design using de novo steps through uncharted biochemical spaces. Nature communications, 9(1), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Schwieter KE, Watkins AM, Yu H, Schwarz KJ, Lim J, . . . Ellington AD (2019). Expanding the limits of the second genetic code with ribozymes. Nature communications, 10(1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Chan CT, Slomovic S, & Collins JJ (2018). Next-generation biocontainment systems for engineered organisms. Nature chemical biology, 14(6), 530–537. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Chen EZ, Baldassano RN, Otley AR, Griffiths AM, Lee D, . . . Hoffmann C (2015). Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell host & microbe, 18(4), 489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G-M, Warden-Rothman R, & Voigt CA (2019). Retrosynthetic design of metabolic pathways to chemicals not found in nature. Current Opinion in Systems Biology. [Google Scholar]
- Lindemann SR, Bernstein HC, Song H-S, Fredrickson JK, Fields MW, Shou W, . . . Beliaev AS (2016). Engineering microbial consortia for controllable outputs. The ISME journal, 10(9), 2077–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Tsao CY, Kim E, Tschirhart T, Terrell JL, Bentley WE, & Payne GF (2017). Using a Redox Modality to Connect Synthetic Biology to Electronics: Hydrogel‐Based Chemo‐Electro Signal Transduction for Molecular Communication. Advanced healthcare materials, 6(1), 1600908. [DOI] [PubMed] [Google Scholar]
- McCarron A, Donnelley M, McIntyre C, & Parsons D (2016). Challenges of up-scaling lentivirus production and processing. Journal of biotechnology, 240, 23–30. [DOI] [PubMed] [Google Scholar]
- Ming D, & Hellekant G (1994). Brazzein, a new high‐potency thermostable sweet protein from Pentadiplandra brazzeana B. FEBS letters, 355(1), 106–108. [DOI] [PubMed] [Google Scholar]
- Moeun BN, Da Ling S, Gasparrini M, Rutman AK, Negi S, Paraskevas S, & Hoesli CA (2019). Islet Encapsulation: A Long-Term Treatment for Type 1 Diabetes. [Google Scholar]
- Morcos F, Pagnani A, Lunt B, Bertolino A, Marks DS, Sander C, . . . Weigt M (2011). Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proceedings of the National Academy of Sciences, 108(49), E1293–E1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Srinivasan P, Chavez M, Carter MA, Dominguez AA, La Russa M, . . . Zhao D (2019). Anti-CRISPR-mediated control of gene editing and synthetic circuits in eukaryotic cells. Nature communications, 10(1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Academies of Sciences, E., & Medicine. (2017). Preparing for future products of biotechnology: National Academies Press. [PubMed] [Google Scholar]
- Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, . . . Eng D (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. nature, 496(7446), 528–532. [DOI] [PubMed] [Google Scholar]
- Park M, Patel N, Keung AJ, & Khalil AS (2019). Engineering epigenetic regulation using synthetic read-write modules. Cell, 176(1–2), 227–238. e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post M, & van der Weele C (2014). Principles of tissue engineering for food In Principles of tissue engineering (pp. 1647–1662): Elsevier. [Google Scholar]
- Post MJ (2012). Cultured meat from stem cells: Challenges and prospects. Meat science, 92(3), 297–301. [DOI] [PubMed] [Google Scholar]
- Ritter SC, & Hackel BJ (2019). Validation and Stabilization of a Prophage Lysin of Clostridium perfringens by Using Yeast Surface Display and Coevolutionary Models. Appl. Environ. Microbiol, 85(10), e00054–00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler NC, Webb-Robertson B-JM, Clauss TR, Pounds JG, Corley R, & Wright AT (2018). High-fat diets alter the modulatory effects of xenobiotics on cytochrome P450 activities. Chemical research in toxicology, 31(5), 308–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathasivam R, Radhakrishnan R, Hashem A, & Abd_Allah EF (2019). Microalgae metabolites: A rich source for food and medicine. Saudi journal of biological sciences, 26(4), 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz CM, Hussain MS, Blenner M, & Wheeldon I (2016). Synthetic RNA polymerase III promoters facilitate high-efficiency CRISPR–Cas9-mediated genome editing in Yarrowia lipolytica. ACS synthetic biology, 5(4), 356–359. [DOI] [PubMed] [Google Scholar]
- Shong J, Diaz MRJ, & Collins CH (2012). Towards synthetic microbial consortia for bioprocessing. Current opinion in biotechnology, 23(5), 798–802. [DOI] [PubMed] [Google Scholar]
- Silverman AD, Karim AS, & Jewett MC (2019). Cell-free gene expression: an expanded repertoire of applications. Nature Reviews Genetics, 1–20. [DOI] [PubMed] [Google Scholar]
- Solomon KV, Haitjema CH, Henske JK, Gilmore SP, Borges-Rivera D, Lipzen A, . . . Theodorou MK (2016). Early-branching gut fungi possess a large, comprehensive array of biomass-degrading enzymes. science, 351(6278), 1192–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomons G, & Litchfield JH (1983). Single cell protein. Critical Reviews in Biotechnology, 1(1), 21–58. [Google Scholar]
- Specht EA, Welch DR, Clayton EMR, & Lagally CD (2018). Opportunities for applying biomedical production and manufacturing methods to the development of the clean meat industry. Biochemical Engineering Journal, 132, 161–168. [Google Scholar]
- Specht L (2020). An analysis of culture medium costs and production volumes for cultivated meat. [Google Scholar]
- Srivastava S, & Chan C (2008). Application of metabolic flux analysis to identify the mechanisms of free fatty acid toxicity to human hepatoma cell line. Biotechnology and bioengineering, 99(2), 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens K, Pozo M, Tsao C-Y, Hauk P, & Bentley WE (2019). Bacterial co-culture with cell signaling translator and growth controller modules for autonomously regulated culture composition. Nature communications, 10(1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz JR (2012). Transforming biochemical engineering with cell‐free biology. AIChE Journal, 58(1), 5–13. [Google Scholar]
- Thorwall S, Schwartz C, Chartron JW, & Wheeldon I (2020). Stress-tolerant non-conventional microbes enable next-generation chemical biosynthesis. Nature chemical biology, 16(2), 113–121. [DOI] [PubMed] [Google Scholar]
- Tschirhart T, Kim E, McKay R, Ueda H, Wu H-C, Pottash AE, . . . Payne GF (2017). Electronic control of gene expression and cell behaviour in Escherichia coli through redox signalling. Nature communications, 8, 14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck CO, Pérez E, Horváth IT, Sheldon RA, & Poliakoff M (2012). Valorization of biomass: deriving more value from waste. science, 337(6095), 695–699. [DOI] [PubMed] [Google Scholar]
- VanArsdale E, Tsao C. y., Liu Y, Chen C. y., Payne GF, & Bentley WE (2019). Redox-Based Synthetic Biology Enables Electrochemical Detection of the Herbicides Dicamba and Roundup via Rewired Escherichia coli. ACS sensors, 4(5), 1180–1184. [DOI] [PubMed] [Google Scholar]
- Wang CY, Chang PM, Ary ML, Allen BD, Chica RA, Mayo SL, & Olafson BD (2018). ProtaBank: A repository for protein design and engineering data. Protein Science, 27(6), 1113–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeldon I, Christopher P, & Blanch H (2017). Integration of heterogeneous and biochemical catalysis for production of fuels and chemicals from biomass. Current opinion in biotechnology, 45, 127–135. [DOI] [PubMed] [Google Scholar]
- Wheeldon I, Minteer SD, Banta S, Barton SC, Atanassov P, & Sigman M (2016). Substrate channelling as an approach to cascade reactions. Nature chemistry, 8(4), 299. [DOI] [PubMed] [Google Scholar]
- Xu X, & Qi LS (2019). A CRISPR–dCas toolbox for genetic engineering and synthetic biology. Journal of molecular biology, 431(1), 34–47. [DOI] [PubMed] [Google Scholar]
- Yang X, & Chan C (2009). Repression of PKR mediates palmitate-induced apoptosis in HepG2 cells through regulation of Bcl-2. Cell research, 19(4), 469–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Hiraga K, Takehana T, Taniguchi I, Yamaji H, Maeda Y, . . . Oda K (2016). A bacterium that degrades and assimilates poly (ethylene terephthalate). science, 351(6278), 1196–1199. [DOI] [PubMed] [Google Scholar]
- Zuñiga C, Li C-T, Yu G, Al-Bassam MM, Li T, Jiang L, . . . Zengler K (2019). Environmental stimuli drive a transition from cooperation to competition in synthetic phototrophic communities. Nature microbiology, 4(12), 2184–2191. [DOI] [PubMed] [Google Scholar]