Significance
In photosynthetic organisms, for example, the redox state of various molecules is altered by photosynthetic reactions. Thus, efficient monitoring of the redox state in living cells provides very important information for understanding the activity of the cell. For this purpose, we have designed a redox sensor protein based on the excited-state intramolecular proton transfer (ESIPT) mechanism. The protein having a single excitation peak and a double variable emission peak was named fluorescent protein with redox-dependent change in green/blue (FROG/B). It is just like a green frog that changes its body color according to the environment. This sensor protein allowed us to monitor the real-time redox changes of cyanobacterial cells caused by light/dark transition.
Keywords: biosensor, chloroplast, FRET, redox regulation, thioredoxin
Abstract
The intracellular redox state is one of the key factors regulating various physiological phenomena in the cell. Monitoring this state is therefore important for understanding physiological homeostasis in cells. Various fluorescent sensor proteins have already been developed to monitor intracellular redox state. We also developed fluorescent redox sensor proteins named Oba-Q and Re-Q, the emissions of which are quenched under oxidized and reduced conditions, respectively. Although these sensors were useful to visualize the redox changes in the cell over time, they have the weakness that their emission signals are directly influenced by their in situ expression levels. To overcome this problem, we developed a redox sensor protein with a single excitation peak and dual variable emission peaks. This sensor protein shows green emission under oxidized conditions and blue emission under reduced conditions. We therefore named this sensor FROG/B, fluorescent protein with redox-dependent change in green/blue. By using this sensor, we successfully measured the changes in intracellular redox potentials in cyanobacterial cells quantitatively caused by light/dark transition just by calculating the ratio of emission between green and blue signals.
The Aequorea victoria green fluorescent protein (GFP) has widely been used as a fluorescent indicator for protein localization and gene expression in living cells (1). In addition, many fluorescent sensor proteins have been developed for live-cell imaging by using mutants of GFP (2–4) or other color variants (5, 6).
A number of these fluorescent sensor proteins have been designed based on the Förster resonance energy transfer (FRET) mechanism. They commonly use structural changes of sensor domains caused by detection of the target signal. These changes are then detected as changes in fluorescent intensity ratios of the FRET donor and the FRET acceptor, or decrease in fluorescence life time of the donor protein. These molecular mechanisms were adopted for many sensors, which were categorized as FRET-type sensors, such as Cameleon for calcium ion (7), Ateam for ATP (8), Epac-camps for cAMP (9), GEPII for potassium ion (10), Redoxfluor for redox changes (11), and our latest thioredoxin sensor CROST (12). FRET is hence a major strategy to develop fluorescent sensors. However, these fusion-type sensor proteins have disadvantages. For example, degradation of the sensor proteins in the target cells and a time lag in maturation between FRET donor and acceptor in vivo were observed in the case of ATP sensor. These properties impede accurate measurement (13).
GFP and YFP have two absorption peaks derived from the neutral form and the anionic form of chromophore hydroxyl. Ionization equilibria of these hydroxyl groups are influenced by structural changes of the β-barrel structure of the protein molecule. Many sensors, such as G-GECO for Ca2+ ion (14), pHluorin for pH (15), QUEEN for ATP (13), Hyper for H2O2, SoNar for NADH (16), iNap for NADPH (17), and roGFP (18) and rxYFP (19) for redox changes, have been developed mainly using this property. Therefore, some of these sensors such as Hyper, SoNar, and iNap are sensitive to pH because pH directly affects the ionization state of their chromophore. These sensors except rxYFP are available for ratiometric imaging. However, they are not useful for the real-time monitoring of a nonfixed sample under an optical microscope because two images under different excitation light sources should be collected to obtain ratiometric images. We refer to these sensors as “ionization equilibria type” in this report.
The conformational change of β-barrel structure of the fluorescent protein also influences the quantum yield. In previous studies, we reported the redox-sensitive sensor proteins Oba-Q (20) and Re-Q (21). The fluorescence of these proteins is dramatically quenched under oxidized and reduced conditions. Because these quenching-type sensors do not require chromophore hydroxyl, the basic molecular mechanism of quenching must be applicable to not only GFP or YFP derivatives but also other fluorescent proteins. This implies that various color sensor proteins can be prepared in this way, and some of them must also be stable against pH changes. The disadvantage of this type of sensor is that it is not applicable to ratiometric imaging and cannot be used for quantitative analysis because the fluorescent intensity is directly influenced by the protein expression level.
GEM-GECO is a GFP-based calcium ion sensor (14). The critical difference of this protein from other GECO proteins is that GEM-GECO shows a change of emission color from green to blue caused by the binding of Ca2+ when the molecule is excited at 395 nm (14)). When the chromophore of A. victoria GFP is excited, proton transfer from the chromophore hydroxyl to Glu222 occurs in a hydrogen bond network via the chromophore, a water molecule, Ser205, and Glu222. This mechanism is called excitation state intramolecular proton transfer (ESIPT), and the neutral GFP chromophore is thereby deprotonated, resulting in green fluorescence emission (22, 23). The Thr203 residue can function as part of the ESIPT pathway in GFPS205V mutant. When the chromophore was excited at 395 nm, the double mutant GFPS205V/T203V showed blue but not green emission (24, 25). The emission change of GEM-GECO should be ascribed to the change of ESIPT efficiency according to the structural change of the molecule caused by the calcium ion binding.
deGFP proteins were also reported as ESIPT-based fluorescent sensors (26). deGFP changes the emission color from blue to green according to the increase of pH. Hence, the ESIPT-type sensor is applicable to ratiometric imaging, which can be measured only by a single excitation wavelength. ESIPT-type sensors are therefore more useful to monitor nonfixed samples than ionization equilibrium-type sensors, which require two different excitation wavelengths. In addition, ESIPT-type sensors show two different color emissions from one chromophore. This property confers further advantages that time lag of the chromophore maturation does not affect emission spectrum, and the result must be less influenced by degradation and photobreaching compared with FRET-type sensors. However, despite these advantages, only a few sensor proteins using ESIPT have thus far been reported.
In this study, we developed a redox-sensitive fluorescent protein sensor using the ESIPT mechanism. This sensor protein shows green emission under oxidized conditions and blue emission under reduced conditions. We therefore named this sensor protein FROG/B, fluorescent protein with redox-dependent change in green/blue. By using this sensor protein, we successfully achieved real-time monitoring of the redox changes in cyanobacterial cells.
Results and Discussion
Design of ESIPT-Based Redox-Sensitive Fluorescent Protein Sensor.
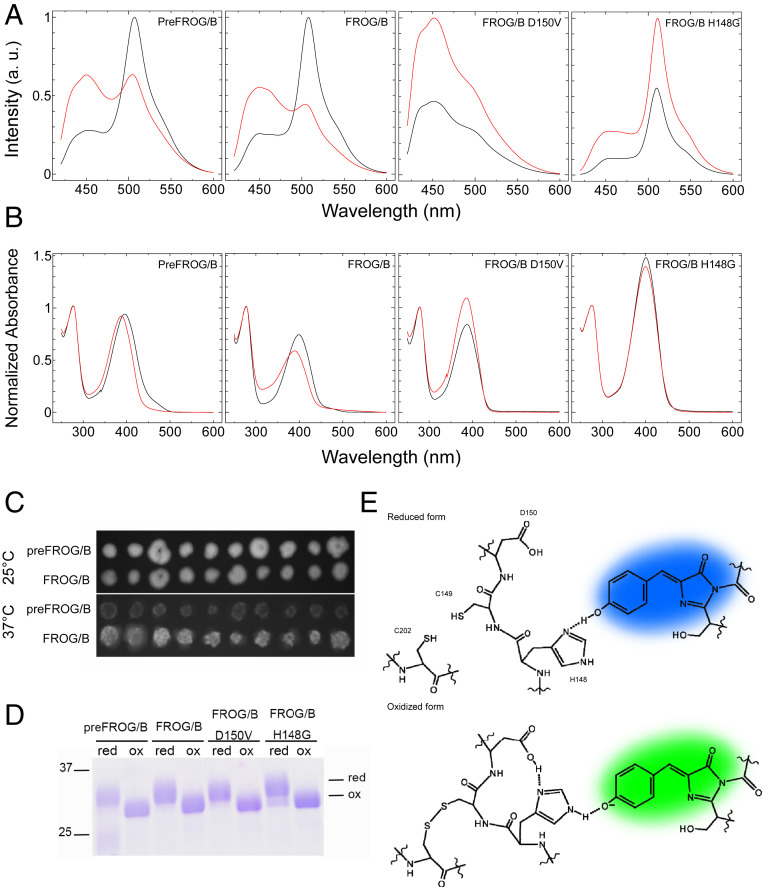
To obtain the ESIPT-based redox sensor, we replaced Thr203 and Ser205 of A. victoria GFP (avGFP) with Val and obtained the mutant GFP (GFPT203V/S205V), which shows an absorption peak at around 400 nm and emits blue fluorescence, as described previously (24). To improve the thermal stability of GFPT203V/S205V, we applied multiple mutations K26R/S30R/Y39N/F64L/N105T/M153T/V163A/I171V/S175G/A206K/H231L according to the mutations used to create Venus (27), Sirius (4), and Superfolder GFP (28). In addition, V224R mutation was introduced to improve the fluorescence quantum yield of blue emission derived from neutral GFP chromophore according to mKalama (29). We then introduced a cysteine pair at positions 149 and 202 of this mutant GFP, which can function as a redox switch, as reported for rxYFP (19). Furthermore, we replaced Val150 with Asp, which can act as a new terminal proton acceptor. This mutant protein, preFROG/B, had an absorption peak at around 400 nm and gave emissions of both green and blue fluorescence according to the change in redox conditions (Fig. 1 A and B). However, preFROG/B showed weak emission in Escherichia coli colonies, which expressed this protein, at 37 °C, perhaps due to the decrease of the thermal stability of the protein molecule by introduction of the V150D mutation (Fig. 1C). The 3D structure of the avGFP (Protein Data Bank ID 1EMB) clearly indicates that Asp150 has the possibility of colliding with Gln69 under reduced conditions. We therefore introduced a mutation, Q69A, into preFROG/B and obtained the desired mutant protein named FROG/B, which emits bright fluorescence in E. coli cells at 37 °C and showed significant emission spectral change (Fig. 1 A–C). Complete reduction and oxidation of preFROG/B and FROG/B by treatments with reduced- and oxidized-form DTT were confirmed by the thiol group modification using 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonate (AMS), followed by nonreducing SDS-polyacrylamide gel electrophoresis (Fig. 1D). Spectroscopic properties of the proteins were shown in Table 1. Molecular absorption coefficients and quantum yields of oxidized and reduced form FROG/B were 1.7 × 104 and 0.14, and 1.4 × 104 and 0.17, respectively. FROG/B showed an absorption peak derived from the neutral GFP chromophore at around 400 nm irrespective of the redox conditions (Fig. 1B). Therefore, redox response of the fluorescent signal from FROG/B was thought to be caused by changes of the ESIPT efficiency but not by changes in the chromophore ionization equilibrium. Asp150 is supposed to be the terminal proton acceptor in the ESIPT pathway of FROG/B. In fact, when Asp150 was returned to Val, the resultant FROG/BD150V showed only blue fluorescence emission irrespective of the redox changes when excited by near-UV light (Fig. 1A and Table 1). His148 should also be critical for switching the ESIPT pathway because the FROG/BH148G mutant showed only green fluorescence emission (Fig. 1A and Table 1). The reason why the FROG/BH148G mutant shows ESIPT through the His148 residue is not yet known. In this mutant, the proton might be transferred from neutral GFP chromophore to Asp150 directly or through a water molecule. In the case of rxYFP, the imidazole side chain of His148 rotates and forms a hydrogen bond with the chromophore phenolic oxygen when the disulfide bond between Cys149 and Cys202 is formed (19). We therefore suspected that His148 may also rotate according to the formation of this disulfide bond and switch the ESIPT pathway in the FROG/B molecule (Fig. 1E). The absorption peaks of PreFROG/B and FROG/B showed a slight red shift under oxidized conditions (Fig. 1B). Because the absorption peaks of reduced-form and oxidized-form FROG/B were close to those of D150V mutant and H148G mutant, respectively, the red shift of the absorption peak may reflect the state change of the chromophore, according to formation of the ESIPT pathway (Fig. 1B and Table 1). Interestingly, the fluorescence intensities of FROG/BD150V and FROG/BH148G decreased under oxidized conditions (Fig. 1A). In a previous study, we already developed a redox-sensitive fluorescent sensor protein, which shows quenching under oxidized conditions; we named this protein Oba-Q (20). It is therefore possible that the fluorescence intensity of the FROG/B mutant was also decreased by conformational change according to the formation of a disulfide bond. The fluorescence intensity of FROG/B should also be influenced by the redox conditions. We might detect emission spectral changes of FROG/B as a result of the changes of both the ESIPT efficiency and the fluorescence intensity.
Fig. 1.
Spectrophotometric properties of FROG/B variants. (A) Emission spectra of the oxidized (black lines) and reduced (red lines) forms of FROG/B and the mutants upon excitation with 400 nm light. (B) Absorption spectra of the oxidized (black lines) and reduced (red lines) forms of FROG/B and the mutants. (C) Fluorescent image of FROG/B expressed in E. coli colonies cultured at 25 and 37 °C. (D) Redox responses of FROG/B and the mutants. Complete oxidation and reduction of FROG/B and the mutants were confirmed by the AMS labeling method followed by nonreducing SDS-polyacrylamide gel electrophoresis. For oxidation, the protein sample was incubated with 20 mM DTTox overnight, and for reduction it was incubated with 10 mM DTTred. (E) Schematic drawings of the expected mechanisms of emission spectral change of reduced and oxidized forms of FROG/B.
Table 1.
Spectroscopic properties of FROG/B and its mutants
| Name | Mutations (comparison with avGFP) | State | Absorption peak (nm) | FG/FB |
| PreFROG/B | K26R/S30R/Y39N/F64L/N105T/D150V/M153T/V163A/I171V/S175G/T203V/S205V/A206K/V224R/H231L | Ox | 395 | 2.1 |
| Red | 387 | 0.65 | ||
| FROG/B | K26R/S30R/Y39N/F64L/Q69A/N105T/D150V/M153/V163A/I171V/S175G/T203V/S205V/A206K/V224R/H231L | Ox | 399 | 2.3 |
| Red | 389 | 0.5 | ||
| PreFROG/BD150V | K26R/S30R/Y39N/F64L/Q69A/N105T/M153T/V163A/I171V/S175G/T203V/S205V/A206K/V224R/H231L | Ox | 386 | 0.33 |
| Red | 386 | 0.30 | ||
| PreFROG/BH148G | K26R/S30R/Y39N/F64L/Q69A/N105T/H148G/D150V/M153/V163A/I171V/S175G/T203V/S205V/A206K/V224R/H231L | Ox | 401 | 3.7 |
| Red | 400 | 2.3 |
Characterization of FROG/B In Vitro.
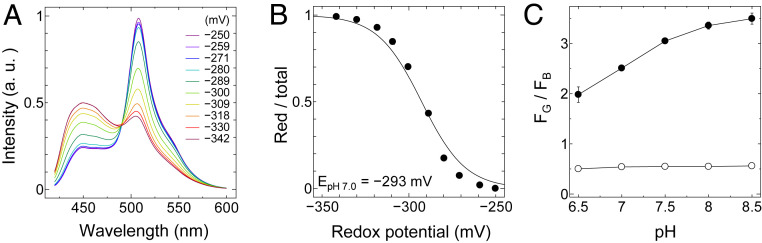
To determine the midpoint redox potential of FROG/B, we measured the emission spectra of the protein in various DTT buffers having different redox potentials. The midpoint redox potential value was then determined from a plot of the fraction of reduced-form protein versus the redox potential, as described by Hanson et al. (18). The midpoint redox potential of FROG/B (Em = −293 mV at pH 7.0) is higher than that in cytoplasm (Em = −320 mV at pH 7.0), indicating that the sensor protein is applicable to monitoring intracellular oxidative stress (Fig. 2 A and B). Although the emission spectra of oxidized-form FROG/B showed remarkable pH dependence (Fig. 2C), the dynamic range of the redox responsive signal was sufficiently large at pH 7 to 8, which fits with the in vivo conditions. The observed pH dependence must be attributed to the pH sensitivity of H148, a key residue of ESIPT pathway of FROG/B but not the ionization of the chromophore itself.
Fig. 2.
Redox potential and pH dependence of FROG/B. (A) Emission spectra of FROG/B in DTT buffers having various redox potentials. The proteins were excited at 400 nm. (B) The reduction levels of FROG/B were quantified as the ratios of the reduced forms to the total proteins and plotted against the calculated redox potentials of DTT buffers (pH 7.0). Data were fitted to the Nernst equation for two electrons exchanged: y = 1/{1 + exp[0.078 (x − Em)]}. (C) The pH dependence of the fluorescence signal ratio (FG/FB) of recombinant FROG/B. FG/FB was calculated from the fluorescence signals from their oxidized form (closed circle) and reduced form (open circle) in 100 mM Tris⋅HCl buffer with various pH levels (pH 6.5 to 8.5). For oxidation, the protein sample was treated with 20 mM DTTox overnight, and for reduction it was incubated with 10 mM DTTred.
Redox Change Imaging in HeLa Cells.
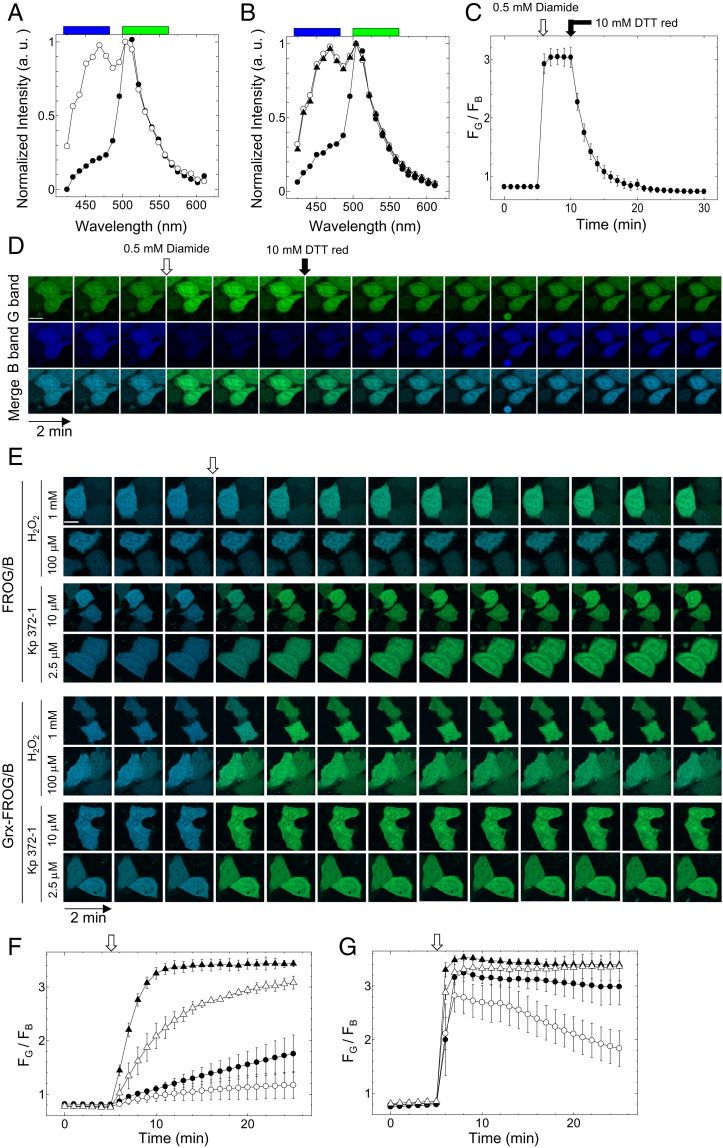
Next, we introduced FROG/B into HeLa cells to examine whether it can detect the redox changes in living cells. The emission spectrum of recombinant FROG/B was measured under a confocal microscope LSM780 (Zeiss) in lambda mode and then normalized at the fluorescent intensity value at 504 nm (Fig. 3A). Because the GaAsP detector equipped in LSM 780 has a lower quantum yield in the shorter-wavelength region, the blue peak of the emission spectrum appeared to show a shift to the longer-wavelength side when compared with the emission spectra obtained from a spectrophotometer (Fig. 1A). For oxidation, FROG/B expressing cells were incubated in the presence of 0.5 mM diamide for 5 min (Fig. 3B, open circle), then 10 mM DTTred was added for reduction, and cells were then incubated for 20 min (Fig. 3B, closed circle). The emission spectra of FROG/B were obtained right after each step (Fig. 3B). The emission spectrum of FROG/B expressed in untreated HeLa cells (Fig. 3B, closed triangle) corresponded to that of the reduced form of the recombinant FROG/B (Fig. 3A, open circle), implying that most of the FROG/B took the reduced form in HeLa cells under normal culture conditions. We also observed the time course of changes in emission intensity ratio (FG/FB) calculated from the intensities at the blue region (FB, 420 to 482 nm) and those at the green region (FG, 500 to 562 nm) when 0.5 mM diamide and 10 mM DTTred were added (Fig. 3 C and D) and could confirm the complete oxidation and reduction of the sensor protein, respectively.
Fig. 3.
Redox response of fluorescence of FROG/B in HeLa cells. HeLa cells expressing FROG/B proteins were observed under a confocal fluorescence microscope. (A) Emission spectra of recombinant FROG/B measured using confocal microscopy for their oxidized (closed circle) and reduced (open circle) forms. Blue and green boxes show the observed wavelengths used for FROG/B as the blue range (B; 420 to 482 nm) and green range (G; 500 to 562 nm). (B) Emission spectra of FROG/B in HeLa cells measured using confocal microscopy. Closed triangle, untreated; closed circle, oxidized by 0.5 mM diamide for 5 min; open circle, rereduced by 10 mM DTTred for 20 min after oxidation. (C and D) The fluorescence signal ratio (FG/FB) changes of FROG/B expressed in HeLa cells; 0.5 mM diamide was added at 5 min, and 10 mM DTT was added at 10 min. (C) FG/FB was plotted against time. The averages of three independent experiments are shown with S. (D). (D) Fluorescence images of the blue range, the green range, and merged images at the indicated time periods are shown. (Scale bar, 20 µm.) (E, F, and G) Imaging of the ROS induction in HeLa cells. Cells expressing FROG/B and Grx-FROG/B were observed under a confocal fluorescence microscope. (E) Typical merged images observed at B range and G range are shown at 2-min intervals. (Scale bar, 20 µm.) FG/FB of FROG/B (F) and Grx-FROG/B (G) before the addition of reagents is shown at time 0 (0 min). At 5 min (indicated as an open arrow), 1 mM H2O2 (closed circle), 0.1 mM H2O2 (open circle), 10 μM Kp372-1 (closed triangle), or 2.5 μM Kp372-1 (open triangle) was added. FG/FB of FROG/B and Grx-FROG/B from images of six cells was averaged at the indicated time.
We then examined whether FROG/B is applicable for observing intracellular redox changes caused by oxidative stress. Redox-sensitive domain of FROG/B was designed based on the previous redox sensor proteins such as roGFP and rxYFP. We therefore predicted that FROG/B can detect redox state of glutathione through glutaredoxin in living cells similar to roGFP and rxYFP (30, 31). Hence, we used “redox states” as a meaning of “glutathione redox states” in this report unless otherwise stated. HeLa cells expressing FROG/B were then incubated in the presence of H2O2 or Kp372-1, and the fluorescence changes were observed under a confocal fluorescent microscope (Fig. 3 E and F). Anticancer reagent Kp372-1 is known to be reduced by NAD(P)H dehydrogenase overexpressed in tumor cells and consequently induces the vigorous generation of reactive oxygen species (ROS) (16). As shown in Fig. 3 E and G, FROG/B expressed in HeLa cells was oxidized slowly in the presence of H2O2 and Kp372-1. Gutscher et al. demonstrated that the fusion of human Grx1 to roGFP2 facilitates redox equilibrium between roGFP and glutathione (30). Accordingly, we prepared the fused protein, Grx-FROG/B, in which human Grx1 and FROG/B were connected by a (GGSGG)6 linker, and observed whether emission ratios of this protein showed a rapid response against the supplemented H2O2 or Kp372-1 (Fig. 3 E and G and SI Appendix, Fig. S1). Consequently, the emission ratio of Grx-FROG/B showed rapid oxidation even in the presence of low concentrations of Kp372-1 or H2O2 (Fig. 3G, open circle and open triangle). We also observed the change in Grx-FROG/B fluorescence signal, which showed the rereduction in the cell upon exposure to the weak oxidative stress conditions (Fig. 3G, open circle). This rereduction might have been caused by stimulation of the glycolytic pathway (21). These results suggest that FROG/B can be useful for monitoring the change in the redox state of Grx as well as roGFP and rxYFP. Hence, Grx-FROG/B is a useful tool for the real-time monitoring of intracellular redox states.
Redox Imaging in Anabaena sp. PCC 7120.
Changes in the intracellular redox status of phototrophs appear to be very drastic, according to the fluctuation of light conditions. We therefore decided to apply this redox sensor protein to monitor intracellular redox changes in cyanobacterial cells. So, the first step was to see if the sensor could be applied to floating cells, such as cyanobacteria.
We then expressed roGFP2 and FROG/B in E. coli cells and observed under a line-scanning confocal microscope. While using roGFP2, two images had to be taken under 488 and 405 nm excitation lights sequentially. Quantitative analysis with roGFP2 in float cells was difficult because the time lag between the two images often changed the position of the cells (red arrows in SI Appendix, Fig. S2A). When using FROG/B, G band image and B band image could be obtained simultaneously using a multichannel detector. Therefore, we were able to measure the intracellular redox state without being affected by cell migration (SI Appendix, Fig. S2A). We also measured redox conditions in E. coli cells in the presence of reduction or oxidation reagents. Western blotting using AMS showed the limited effects of a reductant or oxidant (SI Appendix, Fig. S2B). However, by calculating FG/FB ratio, we were able to monitor the redox state (SI Appendix, Fig. S2C). This indicates that FROG/B is capable of detecting small changes in the redox state of float cells.
Under light conditions, NADPH is mainly produced by oxygen-evolving photosynthesis and is used primarily for carbon dioxide fixation in photosynthetic cells (vegetative cells) of a filamentous, nitrogen-fixing cyanobacterium, Anabaena sp. PCC 7120 (Anabaena 7120). In the absence of available combined nitrogen sources, some vegetative cells separated by 10–20 cells in the filament differentiate into specialized cells named heterocysts, which contain the nitrogenase complex (31); this nitrogenase complex then converts atmospheric nitrogen into ammonium. Heterocysts lack oxygen-evolving photosystem II, and the microoxic environment inside heterocysts is maintained because the nitrogenase complex is easily inactivated by molecular oxygen (31, 32). In heterocysts, NADPH, required for nitrogen-fixation reaction, is mainly produced in the oxidative pentose phosphate pathway using carbohydrates supplied from vegetative cells as substrates (33). The NADPH-generating reaction is catalyzed by glucose 6-phosphate dehydrogenase (G6PDH). Interestingly, this G6PDH activity is redox-regulated by the accessory protein OpcA, and the oxidized form OpcA enhances G6PDH activity. In general, redox-regulated enzymes in phototrophs are up-regulated by reduction, and the G6PDH-OpcA system is an unusual redox-regulated enzyme, although Anabaena 7120 can proceed with nitrogen fixation even under photosynthetic conditions in light. We recently studied the discrepancy of these mechanisms and revealed that the target proteins of thioredoxin, the key protein for redox regulation, are not reduced very well in heterocysts even in light conditions, and therefore, G6PDH is kept active (34).
If this is the case, it would be worth clarifying the redox status in heterocysts and vegetative cells in Anabaena 7120 during light/dark transition. The in vivo glutathione redox balance can be assessed by mass spectrometry or NMR measurements. However, these methods are unable to observe signals from heterocysts and vegetative cells, respectively. When fluorescent sensors were introduced into these cells, the light-dependent redox state changes can be observed under the confocal microscope.
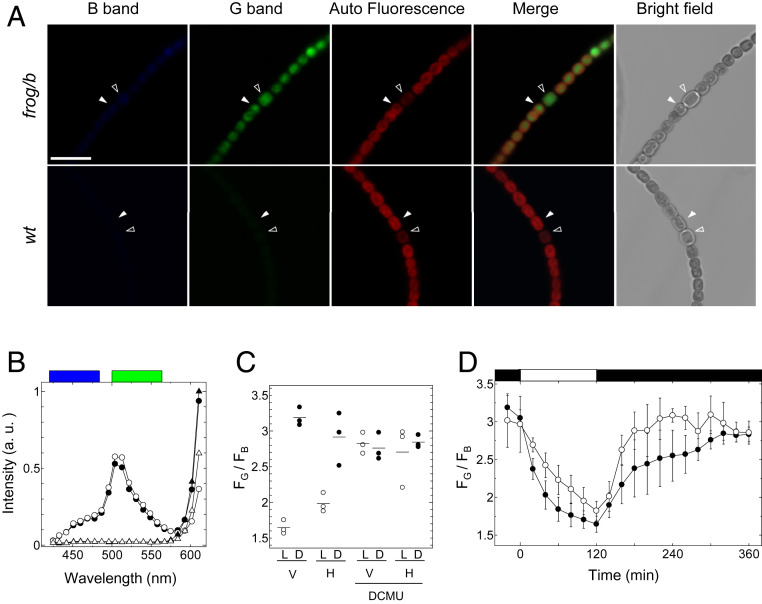
For the redox observation in Anabaena 7120 cells, we used diode 405-nm laser as an excitation light source. To suppress the influence of excitation light to photosynthesis and to avoid the photodamage, we have set the output power of the laser as weak as possible (<0.06%). Because the expression levels of FROG/B were uneven within cells due to the transient expression, we chose some of the Anabaena 7120 cells in which emission signal from the expressed FROG/B was sufficiently bright under a confocal microscope (Fig. 4A). The merged image obtained from B range, G range, and autofluorescence clearly indicates that the fluorescence signal of FROG/B can be observed even in the presence of autofluorescence from light-harvesting pigments. We then successfully obtained the emission spectrum of typical vegetative cells (Fig. 4B, closed circle; obtained from the cell shown in Fig. 4A by a closed arrow) and that of heterocysts (Fig. 4B, open circle; obtained from the cell shown in Fig. 4A by an open arrow). The weak autofluorescence signal of Anabaena 7120 cells did not influence the FROG/B emission spectrum within the measured wavelength range (Fig. 4B, open triangle).
Fig. 4.
Redox responses of fluorescence of FROG/B in Anabaena 7120. (A) Fluorescence images of Anabaena 7120 strain expressing FROG/B (frog/b) transiently and those of wild-type strains (wt). The images were obtained using a diode 405-nm laser for excitation. The images from 420 to 462 nm for the blue range and those from 500 to 562 nm for the green range were observed. Autofluorescence of Anabaena 7120 cells from 651 to 701 nm was also collected, and merged images from these three images were created. Typical heterocyst (open arrow) and vegetative cell (closed arrow) are shown. (Scale bar, 10 μm.) (B) Emission spectra of vegetative cells and heterocysts measured using a confocal microscope in lambda mode. Blue and green boxes show the observed wavelength range used for the blue and green ranges, respectively. Emission spectra of the vegetative cell of frog/b (closed circle; the cells shown in A, closed arrow), the heterocyst of frog/b (open circle; the cells shown in A, open arrow), the vegetative cell of wt (closed triangle; the cells shown in A, closed arrow), and the heterocyst of wt (open triangle; the cells shown in A, open arrow) are shown. (C) FG/FB of the vegetative cells (V) and heterocysts (H) determined under the different conditions. The cyanobacterial cells were incubated under dark conditions (0 μmol photons m−2 s−1, >12 h) or light conditions (50 μmol photons m−2 s−1, 2 h), and the images were obtained. FG/FB values were calculated from these images. Short bars show the averaged values. (D) FG/FB changes of FROG/B under mutual light (50 μmol photons m−2 s−1)/dark conditions. The light period is indicated by the white rectangle above the graph.
Next, we measured the emission spectrum of FROG/B expressed in Anabaena 7120 under the confocal microscope LSM780 (Zeiss) in lambda mode and switched conditions from dark (0 μmol photons m−2 s−1, over 12 h) to light (50 μmol photons m−2 s−1, 2 h) (SI Appendix, Fig. S3). Then, we calculated FG/FB values (Fig. 4C) in the same manner as in the case of HeLa cells, as shown in Fig. 3C. According to the emission spectrum, more than 90% of FROG/B in both vegetative cells and heterocysts appeared to be the oxidized form under dark conditions. This implies that the redox potential in Anabaena 7120 cells was −265 mV or more at 25 °C in the dark, which is calculated from the reduction/oxidation ratio of FROG/B molecule. Unfortunately, the midpoint redox potential of FROG/B (Em = −293 mV at pH 7.0) appeared to be too low to measure the glutathione redox dynamics precisely in Anabaena 7120 cells under light conditions. Nevertheless, 61% of FROG/B in vegetative cells and 46% of that in heterocysts were reduced. Redox potentials were then calculated from these values; −298 mV for vegetative cells and −291 mV for heterocysts at pH 7.0 and 25 °C were obtained. Changes in cellular redox balance revealed by FROG/B signals must reflect electron flow through the electron transport chain in photosynthesis. Indeed, this signal change was clearly inhibited by DCMU (Fig. 4C). We also measured the change in emission spectra over time and calculated emission ratios FG/FB. Under light conditions, the reduction of FROG/B expressed in vegetative cells appeared to take more than 1 h (Fig. 4D, closed circle). In addition, reduction of FROG/B in heterocysts was slower than that in vegetative cells (Fig. 4D, open circle). This difference of reduction kinetics in these cells may reflect the difference of metabolic systems that produce reducing powers. When cells were transferred to dark conditions, FROG/B proteins in both vegetative cells and heterocysts were oxidized. Interestingly, oxidation in heterocysts was faster than that in vegetative cells (Fig. 4D). As shown in Fig. 2C, the fluorescence signal from oxidized FROG/B is affected by pH changes. Since the cytosolic pH of cyanobacteria should increase under light conditions, the FROG/B signal must be partially affected. However, because the majority of FROG/B is reduced under light conditions, which is less sensitive to pH changes, the signal change observed under light conditions is thought to be a change in the reduction level of FROG/B rather than a pH-dependent change.
As mentioned above, we already confirmed that the target proteins of thioredoxin are not reduced very well in heterocysts under light conditions, possibly due to the limited protein expression of thioredoxin m1 and lack of photosystem II (34). Therefore, we suppose that the observed reduction in vegetative cells and heterocysts is due to the glutathione/glutaredoxin system. In the case of Synechocystis sp. PCC 6803 cells, the glutathione/glutaredoxin system plays an important role in physiological phenomena such as arsenate reduction (35). NADPH is also required as the substrate of glutathione reductase to reduce glutathione in Anabaena 7120 cells (36). Although there are no reports on glutaredoxin in Anabaena 7120, whole-genome analysis revealed two glutaredoxin genes, asl3860 and all4873. These two glutaredoxins have the CPYC motif as the catalytic domain, which is similar to human Grx1 (37). In addition, glutathione is used as a reducing power source for antioxidant proteins such as type II Prx, which contains a Grx domain, in this cyanobacterium (38). Therefore, intracellular glutathione oxidation levels may reflect the balance of NADPH production and ROS generation. For further study on the glutathione/glutaredoxin system in Anabaena 7120 cells, our sensor protein appears to be a promising tool.
Advantage of FROG/B.
In this study, we developed an ESIPT-based redox sensor FROG/B, a sensor protein with an absorption peak around 400 nm and an emission spectrum that changes in response to the redox state of the molecule (Fig. 1A). The redox state of FROG/B should be in equilibrium with the Grx-catalyzed glutathione buffer in living cells. Indeed, fusing FROG/B with Grx greatly enhanced the rate of response to changes in the redox state of the cell as well as roGFP (Fig. 3F). The midpoint redox potential of FROG/B (Em = −293 mV at pH 7.0) was lower than roGFP1 (Em = −287 mV at pH 7.0), roGFP2 (Em = −272 mV at pH 7.0) (18), and rxYFP (Em = −262 mV at pH 7.0) (19). Therefore, FROG/B may be more useful for monitoring weak oxidative stress when fused with Grx.
FROG/B uses only one chromophore to detect redox change. This is a great advantage of ESIPT-type sensors because the signal of the sensor is not influenced by the chromophore maturation level. For instance, low-oxygen conditions suppress maturation of the chromophore and extend the maturation time lag of FRET acceptor and FRET donor chromophore. In our case, we could monitor the redox status even inside the heterocyst of Anabaena 7120, which is thought to maintain anaerobic conditions. Although excitation ratiometric type redox sensors such as roGFP also have a single chromophore, this type sensors need to obtain two images using different excitation light sources, and the time lag between first and second imaging is unavoidable (SI Appendix, Fig. S2). This time lag must be excluded to obtain the correct values from the cells, which are not fixed such as cyanobacterium cells in the medium. In the case of FROG/B, we can easily obtain the required images just by separating signals using dichroic mirror or diffraction grating. Thus, this sensor protein must be more useful to the active cells.
Materials and Methods
Protein Expression and Purification.
To obtain FROG/B mutants, genes for avGFP in pET-23a were used as a template, and all point mutations and initial insertions were introduced into the desired genes using PrimeSTAR Mutagenesis Basal Kit (Takara). All FROG/B mutants in pET-23a were expressed in E. coli strain BL21 by allowing cultures to reach an OD600 of between 0.4 and 0.8 at 37 °C, and then the temperature was reduced to 21 °C overnight in the presence of a final concentration of 1 mM IPTG. FROG/B mutants were purified by Ni-NTA affinity chromatography with Ni-NTA agarose (Qiagen) resin. Ni-NTA agarose resin was washed in 50 mM Tris⋅HCl (pH 8.0) containing 20 mM imidazole and eluted with the same buffer containing 250 mM imidazole. Eluates containing FROG/B mutants were applied to a Toyopearl Butyl-650 (Tosoh) column and eluted using 50 mM Tris⋅HCl (pH 8.0) with an ammonium sulfate reverse gradient from 30 to 0%. Peak fractions containing the desired proteins were then collected, dialyzed, and concentrated.
Fluorescence Spectroscopy.
Fluorescence emission spectra were measured using an FP-8500 fluorescence spectrophotometer (Jasco). An excitation wavelength of 400 nm was used to excite FROG/B proteins. Integrated emission fluorescence intensities of FROG/B from 420 to 480 nm (FB) and those from 500 to 560 nm (FG) were calculated from the emission spectra. Absorbance spectra were measured using spectrophotometer V-650 (Jasco). Samples were applied to NAP-5 columns (GE) to remove oxidants or reductants before the measurements.
Measurement of Quantum Yield.
The quantum yields of oxidized and reduced form FROG/B were measured using a Quantaurus-QY (Hamamatsu Photonics). Oxidized and reduced forms of FROG/B proteins were prepared by incubation with 20 mM DTTox and 10 mM DTTred overnight, respectively, and a 400-nm light source was used for excitation.
Expression and Maturation in E. coli.
E. coli strain BL21 transformed with preFROG/B or FROG/B genes in pET-23C (Novagen) was cultured on LB medium plates containing 20 μg/mL kanamycin. Then, 10 colonies each were reproduced using nitrocellulose membrane and cultured on LB medium plates containing 20 μg/mL kanamycin and 1 mM IPTG for 24 h at 25 °C or 37 °C. Next, FROG/B expressed in E. coli colonies was excited using a xenon lamp (LB-LS; Sutter) as an excitation light source through an excitation filter BP360-370 (Olympus) and dichroic mirror DM410 (Olympus). Fluorescent signals were detected using imagEm ×2 (Hamamatsu Photonics) through an emission filter, BA420IF (Olympus).
Determination of the Midpoint Redox Potential of FROG/B.
FROG/B (0.1 μM) was incubated in 100 mM Tris⋅HCl (pH 7.0), 20 mM (for −330 mM to –230 mV), or 8 mM (for −342 mV) DTTox, and various concentrations of DTTred (from 8 μM to 20 mM) overnight at 25 °C. Reduction ratios (R) were calculated using Eq. 1.
| [1] |
F is the ratio of FG to FB for each sample, and Fox and Fred are those at fully oxidized conditions and fully reduced conditions, respectively. The midpoint redox potential values of FROG/B were then calculated by fitting the titration data of R to the Nernst equation according to the methods described in our earlier study (20). A value of −330 mV was used as the midpoint redox potential of DTT at pH 7.0 (39).
Imaging of the Redox Status in HeLa Cells.
FROG/B was expressed in HeLa cells using modified pEGFP-N1 (Addgene) as an expression vector. For Grx1-FROG/B, the expression vector was generated from Grx1-Oba-Qc expression vector, which was prepared in our earlier study (20), by replacing the gene for Oba-Qc with that for FROG/B. Lipofectamine3000 (Life Technologies) was used as a transfection reagent. Cells were then imaged on a confocal microscopy system AxioObserverZ1 with LSM780 (Carl Zeiss). For excitation, a diode 405-nm laser was used. The intensity of the fluorescence image was then digitalized using the image analysis software ImageJ.
Imaging of the Redox Status in E. coli.
E. coli strain BL21 transformed with roGFP2 or FROG/B genes in pET-23a (Novagen) was cultured with OD600 of 0.4 to 0.8 at 37 °C. Then the temperature was lowered to 21 °C, and the culture was continued overnight in the presence of a final concentration of 1 mM IPTG. Cells were then imaged on a confocal microscopy system AxioObserverZ1 with LSM780 (Carl Zeiss). For roGFP excitation, 488 nm Ar laser and 405 diode laser were used. For oxidation of cells, 1 mM diamide was added in the culture medium, and for reduction, 10 mM DTTred was added. As controls for Western blotting, E. coli lysates were treated directly with 1 mM diamide or 10 mM DTTred for 1 h.
Bacterial Strains and Growth Conditions.
Anabaena 7120 and its FROG/B-expressing strain were cultured in BG-11 medium (40) or nitrogen-free medium (BG-110) buffered with 20 mM Hepes-NaOH (pH 7.5). Cultures were grown at 30 °C under continuous light conditions (30 µmol photons m−2 s−1) and bubbled with 1% (vol/vol) CO2.
Construction of Anabaena 7120 Strain Expressing FROG/B.
FROG/B was expressed under control of the petH promoter in Anabaena 7120. The plasmid was constructed by the Hot Fusion method (41). The 316-bp upstream region of petH was used as a petH promoter (42). The promoter region and frog/b were amplified by PCR using the following primer pairs: 5′-tcgtcttcaagaattcaagactcaggactcaggac-3′ and 5′-gcctttcgataccattccttctaccactccgatct-3′, and 5′-atggtatcgaaaggcgaagagttat-3′ and 5′-ttatcaaaaaggatctcacttatagagctcctcca-3′, respectively. The DNA fragments were subsequently cloned into the EcoRI and BamHI restriction sites of the pRL502ΔluxAB vector (43). The resultant plasmid was transferred into Anabaena 7120 by conjugation (44).
Imaging of the Redox Status in Anabaena 7120.
Transformed Anabaena 7120 was cultured in nitrogen-free medium for 72 h before measurements to induce heterocyst differentiation and FROG/B expression. Cells were cultured under dark conditions for 12 h before measurements. Anabaena 7120 cells were then imaged on a confocal microscopy system, AxioObserverZ1 with LSM780 (Carl Zeiss). As an excited light source, a diode 405-nm laser was used. The temperature of samples was kept at 30 °C using a thermostatic chamber during the experiments. The intensity of the fluorescence image was then digitalized using the image analysis software ImageJ.
Data Availability.
All relevant data, associated protocols, and materials are within the manuscript and SI Appendix. If any additional information is needed, it will be available upon request from the corresponding authors.
Supplementary Material
Acknowledgments
We sincerely thank K.-i. Wakabayashi for providing us with some important ideas for this work. We thank the Biomaterials Analysis Division, Tokyo Institute of Technology, for supporting DNA sequencing analysis. This study was supported by Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research (KAKENHI) Grant 16H06556 (to T.H.) and by Dynamic Alliance for Open Innovation Bridging Human, Environment and Materials.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918919117/-/DCSupplemental.
References
- 1.Tsien R. Y., The green fluorescent protein. Annu. Rev. Biochem. 67, 509–544 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Ormö M.et al., Crystal structure of the Aequorea victoria green fluorescent protein. Science 273, 1392–1395 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Heim R., Tsien R. Y., Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 6, 178–182 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Tomosugi W.et al., An ultramarine fluorescent protein with increased photostability and pH insensitivity. Nat. Methods 6, 351–353 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Matz M. V.et al., Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 17, 969–973 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Karasawa S., Araki T., Nagai T., Mizuno H., Miyawaki A., Cyan-emitting and orange-emitting fluorescent proteins as a donor/acceptor pair for fluorescence resonance energy transfer. Biochem. J. 381, 307–312 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyawaki A.et al., Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388, 882–887 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Imamura H.et al., Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc. Natl. Acad. Sci. U.S.A. 106, 15651–15656 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikolaev V. O., Bünemann M., Hein L., Hannawacker A., Lohse M. J., Novel single chain cAMP sensors for receptor-induced signal propagation. J. Biol. Chem. 279, 37215–37218 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Bischof H.et al., Novel genetically encoded fluorescent probes enable real-time detection of potassium in vitro and in vivo. Nat. Commun. 8, 1422 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yano T.et al., A novel fluorescent sensor protein for visualization of redox states in the cytoplasm and in peroxisomes. Mol. Cell. Biol. 30, 3758–3766 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugiura K.et al., The thioredoxin (Trx) redox state sensor protein can visualize Trx activities in the light/dark response in chloroplasts. J. Biol. Chem. 294, 12091–12098 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yaginuma H.et al., Diversity in ATP concentrations in a single bacterial cell population revealed by quantitative single-cell imaging. Sci. Rep. 4, 6522 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y.et al., An expanded palette of genetically encoded Ca2+ indicators. Science 333, 1888–1891 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miesenböck G., De Angelis D. A., Rothman J. E., Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394, 192–195 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Zhao Y.et al., SoNar, a highly responsive NAD+/NADH sensor, allows high-throughput metabolic screening of anti-tumor agents. Cell Metab. 21, 777–789 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tao R.et al., Genetically encoded fluorescent sensors reveal dynamic regulation of NADPH metabolism. Nat. Methods 14, 720–728 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanson G. T.et al., Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 279, 13044–13053 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Ostergaard H., Henriksen A., Hansen F. G., Winther J. R., Shedding light on disulfide bond formation: Engineering a redox switch in green fluorescent protein. EMBO J. 20, 5853–5862 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugiura K.et al., Redox sensor proteins for highly sensitive direct imaging of intracellular redox state. Biochem. Biophys. Res. Commun. 457, 242–248 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Sugiura K., Tanaka H., Kurisu G., Wakabayashi K. I., Hisabori T., Multicolor redox sensor proteins can visualize redox changes in various compartments of the living cell. Biochim. Biophys. Acta Gen. Subj. 1863, 1098–1107 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Chattoraj M., King B. A., Bublitz G. U., Boxer S. G., Ultra-fast excited state dynamics in green fluorescent protein: Multiple states and proton transfer. Proc. Natl. Acad. Sci. U.S.A. 93, 8362–8367 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lossau H.et al., Time-resolved spectroscopy of wild-type and mutant Green Fluorescent Proteins reveals excited state deprotonation consistent with fluorophore-protein interactions. Chem. Phys. 213, 1–16 (1996). [Google Scholar]
- 24.Shu X.et al., An alternative excited-state proton transfer pathway in green fluorescent protein variant S205V. Protein Sci. 16, 2703–2710 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erez Y.et al., Structure and excited-state proton transfer in the GFP S205A mutant. J. Phys. Chem. B 41, 11776–11785 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Hanson G. T.et al., Green fluorescent protein variants as ratiometric dual emission pH sensors. 1. Structural characterization and preliminary application. Biochemistry 41, 15477–15488 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Nagai T.et al., A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Pédelacq J. D., Cabantous S., Tran T., Terwilliger T. C., Waldo G. S., Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Ai H. W., Shaner N. C., Cheng Z., Tsien R. Y., Campbell R. E., Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins. Biochemistry 46, 5904–5910 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Gutscher M.et al., Real-time imaging of the intracellular glutathione redox potential. Nat. Methods 5, 553–559 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Kumar K., Mella-Herrera R. A., Golden J. W., Cyanobacterial heterocysts. Cold Spring Harb. Perspect. Biol. 2, a000315 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicolaisen K., Hahn A., Schleiff E., The cell wall in heterocyst formation by Anabaena sp. PCC 7120. J. Basic Microbiol. 49, 5–24 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Cumino A. C., Marcozzi C., Barreiro R., Salerno G. L., Carbon cycling in Anabaena sp. PCC 7120. Sucrose synthesis in the heterocysts and possible role in nitrogen fixation. Plant Physiol. 143, 1385–1397 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mihara S., Sugiura K., Yoshida K., Hisabori T., Thioredoxin targets are regulated in heterocysts of cyanobacterium Anabaena sp. PCC 7120 in a light-independent manner. J. Exp. Bot. 71, 2018–2027 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.López-Maury L., Sánchez-Riego A. M., Reyes J. C., Florencio F. J., The glutathione/glutaredoxin system is essential for arsenate reduction in Synechocystis sp. strain PCC 6803. J. Bacteriol. 191, 3534–3543 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang F., Hellman U., Sroga G. E., Bergman B., Mannervik B., Cloning, sequencing, and regulation of the glutathione reductase gene from the cyanobacterium Anabaena PCC 7120. J. Biol. Chem. 270, 22882–22889 (1995). [DOI] [PubMed] [Google Scholar]
- 37.Lemaire S. D., Miginiac-Maslow M., The thioredoxin superfamily in Chlamydomonas reinhardtii. Photosynth. Res. 82, 203–220 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Banerjee M., Ballal A., Apte S. K., A novel glutaredoxin domain-containing peroxiredoxin “All1541” protects the N2-fixing cyanobacterium Anabaena PCC 7120 from oxidative stress. Biochem. J. 442, 671–680 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Cleland W. W., Dithiothreitol, a new protective reagent for Sh groups. Biochemistry 3, 480–482 (1964). [DOI] [PubMed] [Google Scholar]
- 40.Rippka R., Deruelles J., Waterbury J. B., Herdman M., Stanier R. Y., Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 111, 1–61 (1979). [Google Scholar]
- 41.Fu C., Donovan W. P., Shikapwashya-Hasser O., Ye X., Cole R. H., Hot fusion: An efficient method to clone multiple DNA fragments as well as inverted repeats without ligase. PLoS One 9, e115318 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valladares A., Muro-Pastor A. M., Fillat M. F., Herrero A., Flores E., Constitutive and nitrogen-regulated promoters of the petH gene encoding ferredoxin:NADP+ reductase in the heterocyst-forming cyanobacterium Anabaena sp. FEBS Lett. 449, 159–164 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Higo A., Isu A., Fukaya Y., Hisabori T., Efficient gene induction and endogenous gene repression systems for the filamentous cyanobacterium Anabaena sp. PCC 7120. Plant Cell Physiol. 57, 387–396 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Elhai J., Wolk C. P., Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167, 747–754 (1988). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data, associated protocols, and materials are within the manuscript and SI Appendix. If any additional information is needed, it will be available upon request from the corresponding authors.