Significance
Bromine is an essential trace element, required for the formation of sulfilimine cross-links in collagen IV, a basement membrane protein. Sulfilimine cross-link formation depends on the production of hypobromous acid by peroxidasin. Once the cross-links are formed, bromide is released, but we suspected that hypobromous acid might brominate bystander molecules in the basement membrane. NanoSIMS imaging of mouse kidneys revealed peroxidasin-dependent bromine enrichment in the basement membrane, and proteomics studies uncovered peroxidasin-dependent bromination of a specific tyrosine in collagen IV. Our studies show that bromine, an essential element, is largely restricted to basement membranes in mammalian tissues. Our studies also show that peroxidasin brominates bystander tyrosines in addition to promoting the formation of collagen IV sulfilimine cross-links.
Keywords: collagen IV, sulfilimine cross-links, NanoSIMS imaging, bromine, bromotyrosine
Abstract
Bromine and peroxidasin (an extracellular peroxidase) are essential for generating sulfilimine cross-links between a methionine and a hydroxylysine within collagen IV, a basement membrane protein. The sulfilimine cross-links increase the structural integrity of basement membranes. The formation of sulfilimine cross-links depends on the ability of peroxidasin to use bromide and hydrogen peroxide substrates to produce hypobromous acid (HOBr). Once a sulfilimine cross-link is created, bromide is released into the extracellular space and becomes available for reutilization. Whether the HOBr generated by peroxidasin is used very selectively for creating sulfilimine cross-links or whether it also causes oxidative damage to bystander molecules (e.g., generating bromotyrosine residues in basement membrane proteins) is unclear. To examine this issue, we used nanoscale secondary ion mass spectrometry (NanoSIMS) imaging to define the distribution of bromine in mammalian tissues. We observed striking enrichment of bromine (79Br, 81Br) in basement membranes of normal human and mouse kidneys. In peroxidasin knockout mice, bromine enrichment of basement membranes of kidneys was reduced by ∼85%. Proteomic studies revealed bromination of tyrosine-1485 in the NC1 domain of α2 collagen IV from kidneys of wild-type mice; the same tyrosine was brominated in collagen IV from human kidney. Bromination of tyrosine-1485 was reduced by >90% in kidneys of peroxidasin knockout mice. Thus, in addition to promoting sulfilimine cross-links in collagen IV, peroxidasin can also brominate a bystander tyrosine. Also, the fact that bromine enrichment is largely confined to basement membranes implies that peroxidasin activity is largely restricted to basement membranes in mammalian tissues.
Six years ago, bromine was shown to be an essential element, required for the formation of sulfilimine cross-links in collagen IV (1). Collagen IV is a trimeric molecule generated from six different collagen IV isomers (α1–α6) and is a crucial component for basement membranes underlying epithelial cells, endothelial cells, and muscle cells (2). There are three types of collagen IV trimers: α12–α2, α3–α4–α5, and α52–α6. Sulfilimine cross-links link two collagen IV trimers, forming a hexamer structure. The sulfilimine cross-linking reaction is mediated by peroxidasin, an extracellular matrix (ECM) peroxidase that uses two substrates, hydrogen peroxide (H2O2) and bromide, to generate hypobromous acid (HOBr) (3). It has been proposed that HOBr forms a bromosulfonium intermediate in the side chain of methionine-1570 (numbering throughout this paper refers to COL4A2) located within the carboxyl-terminal noncollagenous domain (NC1 domain) of collagen IV. The bromosulfonium intermediate then promotes the formation of a sulfilimine cross-link between the methionine and a spatially adjacent hydroxylysine residue (lysine-1689) in a partner collagen IV trimer (1, 4). While bromide is required for the formation of the sulfilimine cross-links, bromide is simply an intermediate in the cross-linking reaction. H2O2 is consumed during the cross-linking reaction, but, following cross-linking, the bromide is released into the extracellular space (1).
The peroxidasin- and bromide-catalyzed sulfilimine cross-links buttress the structural integrity of the basement membrane (5) and are crucial in all metazoans, including in Drosophila melanogaster, Caenorhabditis elegans, and mammals (2, 4, 6). In Drosophila, an absence of sulfilimine cross-links, due either to peroxidasin deficiency or a deficiency in bromine, causes disorganization of basement membranes of muscle cells, leading to gut rupture and larval death (1, 7). In humans and mice, a deficiency of peroxidasin causes severe developmental abnormalities in the anterior chamber of the eye by disrupting the integrity of basement membranes lining lens epithelial cells (8–10).
The production of hypohalous acids by peroxidases is not unique to peroxidasin. Myeloperoxidase in polymorphonuclear leukocytes produces hypochlorous acid, which is important for destroying microorganisms within leukocyte phagolysosomes (11, 12). Eosinophil peroxidase produces HOBr (13), which assists in controlling parasitic infections (14–16). Hypohalous acids are highly reactive and known to oxidize amino acids, carbohydrates, and nucleic acids in microorganisms, explaining their utility in fighting infections, but they also react with bystander molecules of the host (7, 17). For example, the release of eosinophil peroxidase into the lung parenchyma results in HOBr, which in turn forms bromotyrosine residues (18, 19). Similarly, the myeloperoxidase in atherosclerotic plaques results in the formation of chlorotyrosine residues in bystander proteins (20, 21).
The fact that myeloperoxidase and eosinophil peroxidase can cause oxidative damage to bystander host proteins naturally raises questions about peroxidasin biology. The first is whether the HOBr produced by peroxidasin is entirely selective for promoting sulfilimine cross-links in collagen IV or whether it also covalently modifies bystander molecules within the ECM (e.g., forms bromotyrosines in basement membrane proteins). Several groups have raised the possibility that peroxidasin activity could damage bystander proteins (7, 22, 23), but there has been little experimental evidence to support this idea. Second, if peroxidasin were to covalently modify bystander proteins, would those modifications be confined to the basement membrane, or would they be more widespread within the underlying stromal/interstitial ECM between cells? Ig domains within peroxidasin are thought to facilitate indirect interactions between peroxidasin and collagen IV within the basement membrane (6). However, at this point, it is unclear whether peroxidasin activity is confined to the basement membrane or is ubiquitous within the ECM. Third, if bromination of ECM proteins occurs in mammalian tissues, would the extent of bromination be reduced by a deficiency of peroxidasin?
In the current study, our goal was to address these issues, taking advantage of a combination of mass spectrometry-based imaging (nanoscale secondary ion mass spectrometry [NanoSIMS] imaging) of mammalian tissues and mass spectrometry-based proteomic studies of basement membrane proteins. We focused our efforts on the kidney because basement membrane composition, structure, and function are highly relevant to chronic kidney disease. Mutations in three collagen IV chain genes (COL4A3, COL4A4, and COL4A5) cause Alport syndrome, a glomerulopathy associated with deafness (24–26). These mutations interfere with the formation of the α3–α4–α5 collagen IV trimer, which is generated by podocytes and is critical for the function of the glomerulus. Heterozygous mutations in COL4A3 and COL4A4 cause a related disease, thin basement membrane nephropathy (TBMN) (27). Autoantibodies against the glomerular basement membrane (GBM) collagen IV cause Goodpasture’s disease (anti-GBM disease) (28). In patients with diabetic kidney disease, GBMs are thickened and dysfunctional (29).
Results
Distribution of Bromine in Mouse Tissues.
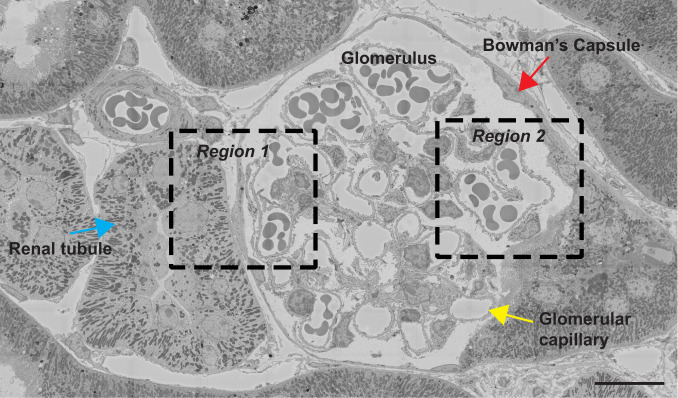
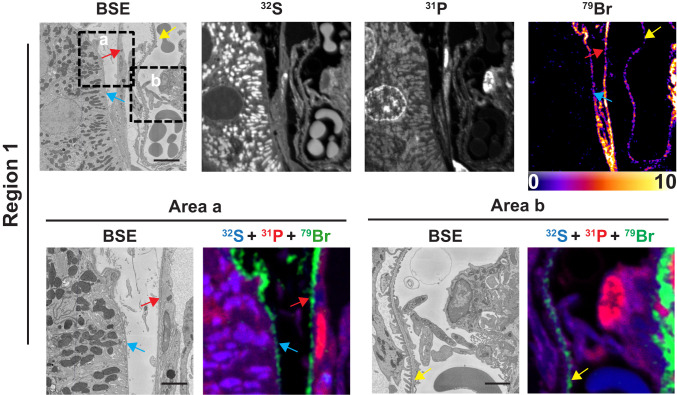
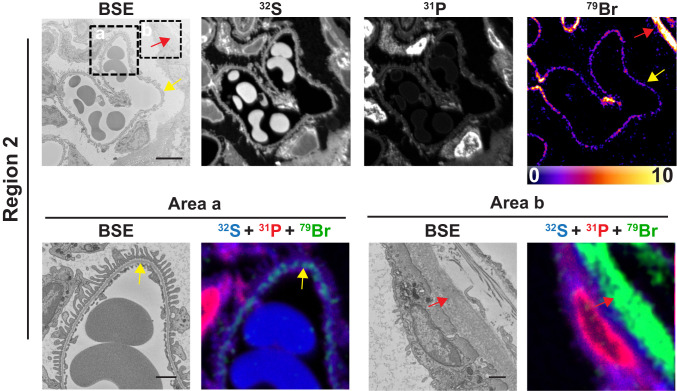
Bromine is an essential element in metazoans (1), but its tissue distribution has never been defined. We examined 500-nm-thick sections of resin-embedded mouse kidney by both backscattered electron (BSE) imaging (providing high-resolution morphology of cells and tissues) and NanoSIMS analyses (generating high-resolution images based on isotopic content). Fig. 1 shows a BSE image of a glomerulus and adjacent renal tubules from a wild-type mouse. Higher magnification electron micrographs of two boxed regions (region 1 and region 2) are shown in the Upper panels of Figs. 2 and 3, respectively, along with 32S, 31P, and 79Br NanoSIMS images. The 32S and 31P images are useful for cellular and tissue morphology, and the 31P images are useful for identifying cell nuclei. NanoSIMS images of region 1 (Fig. 2, Upper) revealed striking 79Br enrichment along the edge of a renal tubular epithelial cell, along the edge of a parietal cell in Bowman’s capsule, and along a glomerular capillary. NanoSIMS images of region 2 (Fig. 3, Upper) revealed marked 79Br enrichment along a glomerular capillary and along a parietal cell in Bowman’s capsule.
Fig. 1.
Tiled BSE image of a glomerulus and surrounding renal tubules from a kidney of a wild-type mouse. Boxed regions were imaged by NanoSIMS (see Figs. 2 and 3). Red arrow, Bowman’s capsule; blue arrow, renal tubule; yellow arrow, glomerular capillary. (Scale bar: 15 μm.)
Fig. 2.
Bromine is enriched in basement membranes of mouse kidney, as judged by BSE imaging and NanoSIMS analyses. Region 1 of a renal glomerulus (see Fig. 1) was mapped by BSE imaging and NanoSIMS. 31P and 32S images were useful for defining tissue morphology; the 79Br image depicts bromine distribution. (Lower) Close-up images of boxed regions (area a and area b) in the Upper. BSE images and composite 31P (red), 32S (blue), and 79Br (green) NanoSIMS images of area a and area b reveal bromine enrichment in the GBM (yellow arrows), the renal tubule basement membrane (blue arrows), and the basement membrane of Bowman’s capsule (red arrows). (Scale bars: Upper, 5 μm; Lower, 2 μm.)
Fig. 3.
Bromine is enriched in basement membranes of mouse kidney, as judged by BSE imaging and NanoSIMS analyses. Region 2 of a renal glomerulus (see Fig. 1) was mapped by BSE imaging and NanoSIMS. 31P and 32S images were useful for defining tissue morphology; the 79Br image depicts bromine distribution within the kidney. (Lower) Within each region, close-up images of boxed regions (area a, area b) in the Upper. BSE images and composite 31P (red), 32S (blue), and 79Br (green) NanoSIMS images of area a and area b reveal bromine enrichment in the GBM (yellow arrows) and the basement membrane of Bowman’s capsule (red arrows). (Scale bars: Upper, 5 μm; Lower, 1 μm.)
Higher magnification electron micrographs of the boxed regions in the Upper panels of Figs. 2 and 3 (area a and area b) are shown in the Lower panels, along with composite 31P, 32S, and 79Br NanoSIMS images. The higher magnification images show that 79Br is confined to basement membranes, including renal tubule basement membranes (TBMs), Bowman’s capsule basement membranes (BCBMs), and the GBM between podocytes and endothelial cells of glomerular capillaries.
Additional correlative BSE and NanoSIMS images of mouse glomeruli are shown in SI Appendix, Figs. S1 and S2, again revealing bromine enrichment in the TBMs, BCBMs, and GBMs. NanoSIMS images for 79Br (50.7% natural abundance) and 81Br (49.3% natural abundance) were virtually identical (SI Appendix, Fig. S1). Quantification of the signals revealed that levels of 79Br and 81Br, relative to 13C, were greater in basement membranes than inside cells (SI Appendix, Figs. S1 and S2). Levels of 79Br secondary ions were ∼2.6% greater than levels of 81Br, consistent with the natural abundance of the two isotopes. There was little or no 127I enrichment in basement membranes, relative to cells (SI Appendix, Fig. S2). We also observed bromine enrichment in the basement membranes lining capillary endothelial cells of the lung (SI Appendix, Fig. S3).
Bromine Localization in Normal and Diseased Human Glomeruli.
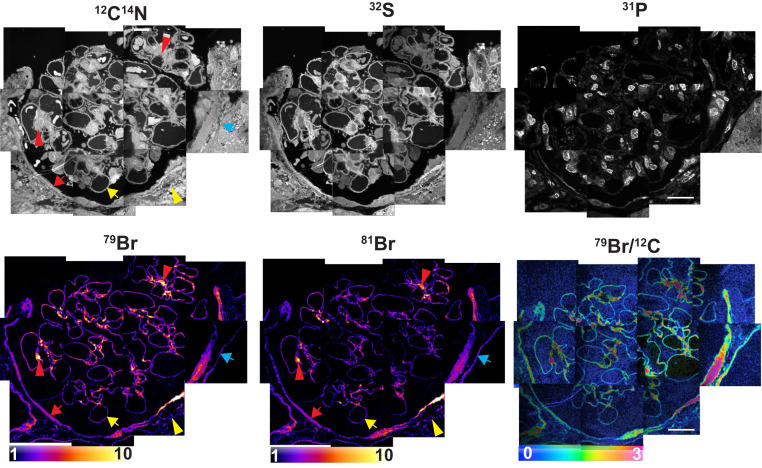
Kidney biopsies (obtained for diagnostic purposes) are routinely processed for light and electron microscopy. We obtained archived resin-embedded kidney biopsy specimens and cut 500-nm sections for NanoSIMS analyses. We examined two biopsy specimens from allograft kidneys deemed free of histopathologic abnormalities (Figs. 4 and 5A). Mirroring our findings in mouse kidneys, NanoSIMS analyses of normal human kidneys revealed striking 79Br and 81Br enrichment in TBMs, BCBMs, and GBMs (Figs. 4 and 5A). We also observed intense bromine enrichment in the mesangial matrix within the glomerulus. We found lower, but detectable, bromine enrichment within the ECM between renal tubules. We did not observe 127I enrichment in basement membranes of normal human kidney (SI Appendix, Fig. S4).
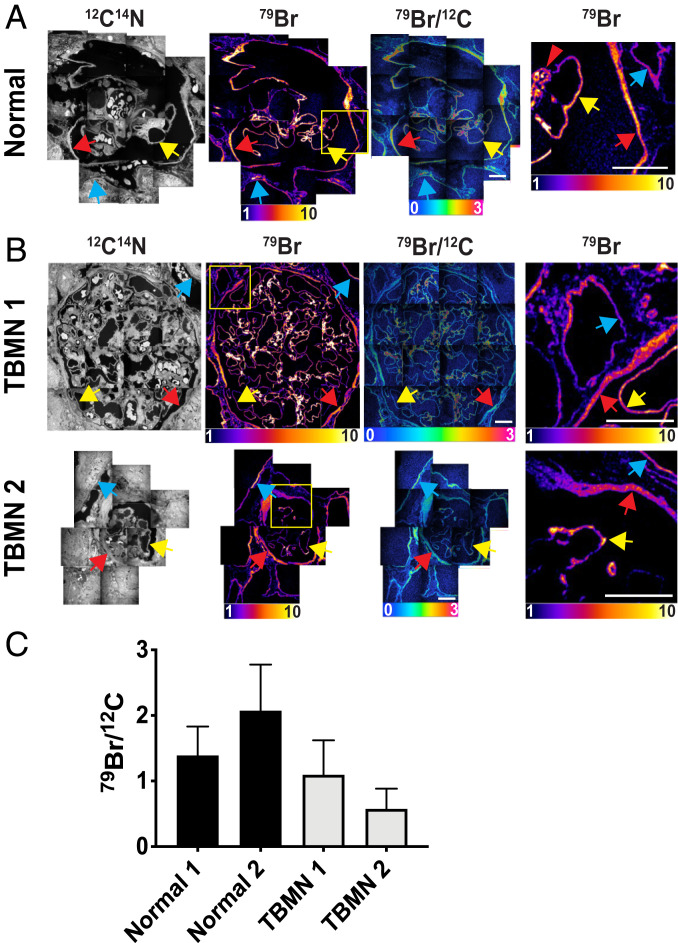
Fig. 4.
Bromine enrichment in basement membranes of human kidney. A normal human kidney specimen was processed for NanoSIMS imaging. 12C14N, 32S, and 31P NanoSIMS images were useful for visualizing morphology. 79Br, 81Br, and 79Br/12C images revealed bromine enrichment in the basement membrane of glomerular capillaries (yellow arrows), the basement membrane of renal tubules (blue arrows), and the basement membrane of Bowman’s capsule (red arrows). There were several areas of intense bromine enrichment in the mesangial matrix of the glomerulus (red arrowheads). We also observed low amounts of bromine enrichment in the ECM outside of Bowman’s capsule (yellow arrowheads). Quantification of the 79Br/12C ratios in glomerular capillaries is reported in Fig. 5. 79Br/12C ratios were multiplied by 10,000. (Scale bars: 20 μm.)
Fig. 5.
Bromine enrichment of basement membranes from a normal human kidney (A) and two specimens from patients with TBMN (B). 12C14N NanoSIMS images were useful for visualizing morphology; 79Br and 79Br/12C images were created to show bromine distribution and bromine enrichment in the GBM (yellow arrows), the renal tubule basement membrane (blue arrows), the basement membrane of Bowman’s capsule (red arrows), and the mesangial matrix of the glomerulus (red arrowheads). The 79Br images in the right-hand column are higher magnification images of the boxed regions of the images in the second column. 79Br/12C ratios are multiplied by 10,000. (Scale bars: 20 μm.) (C) Quantification of 79Br/12C ratios (mean ± SD) in glomerular capillary basement membranes for two normal human kidneys. NanoSIMS images for Normal 1 are shown in Fig. 4; images for Normal 2 are shown in A. Images for two TBMN specimens (TBMN 1, TBMN 2) are shown in B. The bar graph depicts the 79Br/12C ratio in 222 ROIs in glomerular capillaries of Normal 1; 57 ROIs for Normal 2; 442 ROIs for TBMN 1; and 28 ROIs for TBMN 2.
We performed NanoSIMS analyses (Fig. 5B) on kidney specimens from two patients with TBMN, which had been diagnosed based on both clinical findings and transmission electron microscopy measurements of GBM thickness (30). Many patients with TBMN have heterozygous mutations in the genes for α3 collagen IV (COL4A3) or α4 collagen IV (COL4A4) (30, 31), but genetic studies of TBMN are not routine and had not been performed in the two cases that we examined. 79Br enrichment in the GBMs was lower in the two TBMN specimens than in the two normal specimens that we had examined (Fig. 5C); however, drawing firm conclusions about bromine enrichment in TBMN GBMs would require analyzing many more cases. In an Alport syndrome kidney specimen, we observed localized regions of bromine enrichment in the mesangial matrix, but bromine enrichment in the GBM was lower than in the two normal kidney specimens (SI Appendix, Fig. S5A). Again, however, we would not draw conclusions about bromine enrichment from a single case of Alport syndrome. We observed high levels of bromine enrichment in the mesangial matrix in a glomerulus from a patient with anti-GBM disease (SI Appendix, Fig. S5B).
Because the HOBr from eosinophil peroxidase is known to brominate proteins, we performed NanoSIMS analyses on a kidney specimen from a patient with biopsy-proven allergic-type acute interstitial nephritis with active injury and prominent eosinophilic infiltrates (SI Appendix, Fig. S6). We observed bromine enrichment in basement membranes; however, bromine enrichment in the interstitial ECM between tubules (where inflammatory infiltrates were present) was not greater than in the ECM of normal kidney specimens. This result was surprising, but it is conceivable that turnover of brominated proteins in the ECM is quite rapid or that the acute nature of the disease did not allow sufficient time for widespread bromination of the interstitial ECM. Drawing firm conclusions about protein bromination of the interstitial ECM in eosinophilic nephritis would require examining additional biopsy specimens, including specimens obtained at different time points during the course of the disease.
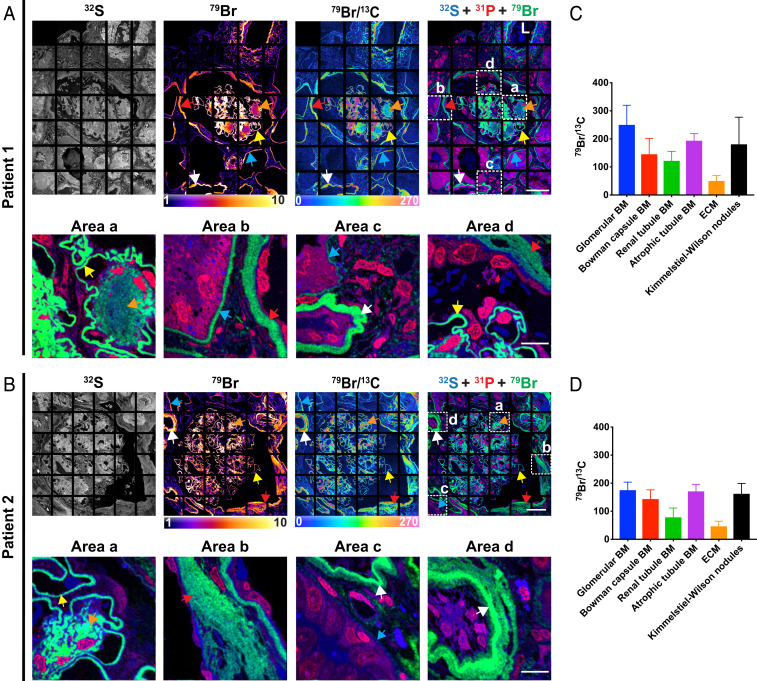
Because thickening of the GBM is a hallmark of diabetic nephropathy, we performed NanoSIMS imaging on kidney biopsy specimens from three patients with diabetic nephropathy (Fig. 6 and SI Appendix, Figs. S7, S8, and S9). As expected, the thickness of the GBMs, as judged by NanoSIMS images, was higher in the three diabetic nephropathy specimens (mean thickness of 967 nm, 29 capillaries measured) than in the two normal kidney specimens shown in Figs. 4 and 5A (mean thickness of 593 nm, 24 capillaries measured). Bromine enrichment in GBMs, normalized to carbon signals (12C or 13C), was ∼37% higher in the diabetic nephropathy specimens (Fig. 6 and SI Appendix, Fig. S9) than in the normal kidney specimens (Figs. 4 and 5A). In the diabetic nephropathy specimens, we also observed bromine enrichment in the TBMs (Fig. 6). In the diabetic nephropathy specimen shown in Fig. 6 and SI Appendix, Fig. S9, bromine enrichment in GBMs was approximately twofold higher than in TBMs. Bromine enrichment was also observed in Kimmelstiel–Wilson nodules (Fig. 6). Bromine enrichment was also high in the basement membranes of atrophic renal tubules (Fig. 6 and SI Appendix, Fig. S8). In the specimen shown in Fig. 6B, the thick multilayered TBMs (typically found in atrophic tubules) was highly enriched in bromine.
Fig. 6.
NanoSIMS images reveal marked bromine enrichment in the GBMs of kidney biopsy specimens from two patients with diabetic nephropathy. (A and B) 32S, 79Br, 79Br/13C, and composite 31P (red), 32S (blue), and 79Br (green) NanoSIMS images of the kidney biopsies from two patients with diabetic nephropathy. Individual 13C, 12C14N, 31P, 32S, and 79Br NanoSIMS images of the specimens in A and B are shown, respectively, in SI Appendix, Figs. S7 and S8. The Lower within A and B represent close-up images of boxed regions (area a, area b, area c, and area d) in the Upper. 79Br, 79Br/13C, and 31P (red), 32S (blue), and 79Br (green) composite NanoSIMS images were generated to show bromine in the GBM (yellow arrows), the renal tubule basement membrane (BM) (blue arrows), the BM of Bowman’s capsule (red arrows), the BM of atrophic renal tubules (white arrows), and Kimmelstiel–Wilson nodules (orange arrows). (C) Quantification of 79Br/13C ratio for ROIs (mean ± SD) in patient 1 specimen. Glomerular BM, n = 144 ROIs; Bowman capsule BM, n = 67 ROIs; renal tubule BM, n = 191 ROIs; atrophic renal tubule BM, n = 17 ROIs; ECM between renal tubules (n = 105 ROIs); Kimmelstiel–Wilson nodules, n = 86 ROIs. (D) Quantification of 79Br/13C ratio for ROIs (mean ± SD) in patient 2. Glomerular BM, n = 196 ROIs; Bowman capsule BM, n = 105 ROIs; renal tubule BM, n = 12 ROIs; atrophic renal tubule BM, n = 54 ROIs; ECM, n = 33 ROIs; and Kimmelstiel–Wilson nodules, n = 191 ROIs. 79Br/13C ratios were multiplied by 10,000. L, artery lumen. (Scale bars: Upper, 45 μm; Lower, 10 μm.)
Bromine Enrichment of Basement Membranes Is Peroxidasin-Dependent.
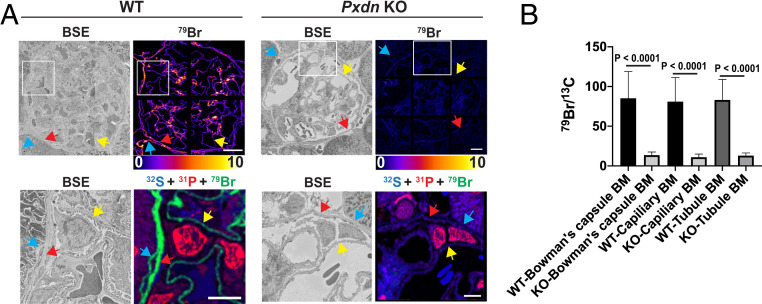
We used NanoSIMS analyses to assess 79Br enrichment and distribution in kidneys of wild-type and in peroxidasin knockout mice (Pxdn−/−; homozygous for a Pxdn allele harboring a gene-trap insertion in intron 8) (Materials and Methods) (5). We observed no evidence of renal pathology in Pxdn−/− mice by BSE imaging (SI Appendix, Fig. S10). However, we observed markedly reduced amounts of 79Br in GBMs from Pxdn−/− mice (Fig. 7A and SI Appendix, Fig. S11). Quantitative analyses of 79Br– secondary ions revealed that bromine enrichment in basement membranes was reduced by ∼85% in Pxdn−/− mice (Fig. 7B); the extent of the reduction was similar in the TBMs, BCBMs, and GBMs (Fig. 7). The existence of some basement membrane bromination in the Pxdn−/− mice, combined with earlier observation of some sulfilimine bond formation in the same mice (32), raised our suspicions that the Pxdn gene-trap allele was leaky (with splicing around the gene-trap insertion). Indeed, quantitative RT-PCR studies of an amplicon spanning exons 8 and 9 sequences (across the insertional mutation) revealed evidence for the expression of normal Pxdn transcripts in gene-trap Pxdn−/− mice at a level of ∼6.7% of those found in wild-type mice.
Fig. 7.
Reduced amounts of bromine in BMs of kidneys from peroxidasin knockout (Pxdn KO) mice, as judged by NanoSIMS analyses. (A) Correlative BSE imaging and 79Br NanoSIMS imaging of glomeruli in wild-type (WT) and Pxdn KO mice. (Lower) Higher magnification images of the boxed regions outlined in Upper. BSE images and composite 31P (red), 32S (blue), and 79Br (green) NanoSIMS images of the same regions show bromine enrichment in the GBM (yellow arrows), the renal tubule BM (blue arrows), and the BM of Bowman’s capsule (red arrows) of WT mice. Bromine enrichment was markedly reduced in the basement membranes of Pxdn KO mouse kidneys. (Scale bars, 10 μm.) (B) Quantification of bromine enrichment in basement membranes (mean ± SD) in kidneys of WT and Pxdn KO mice. 79Br signals, normalized to 13C signals, in wild-type Bowman’s capsule basement membranes (WT-Bowman’s capsule BM; n = 123 ROIs), Pxdn KO Bowman’s capsule basement membranes (KO-Bowman’s capsule BM; n = 88 ROIs), wild-type capillary basement membranes (WT-Capillary BM; n = 699 ROIs), Pxdn KO capillary basement membranes (KO-Capillary BM; n = 271 ROIs), wild-type kidney tubule basement membrane (WT-tubule BM; n = 102 ROIs), and Pxdn KO kidney tubule basement membrane (KO-tubule BM; n = 40 ROIs). 79Br/13C ratios were multiplied by 10,000.
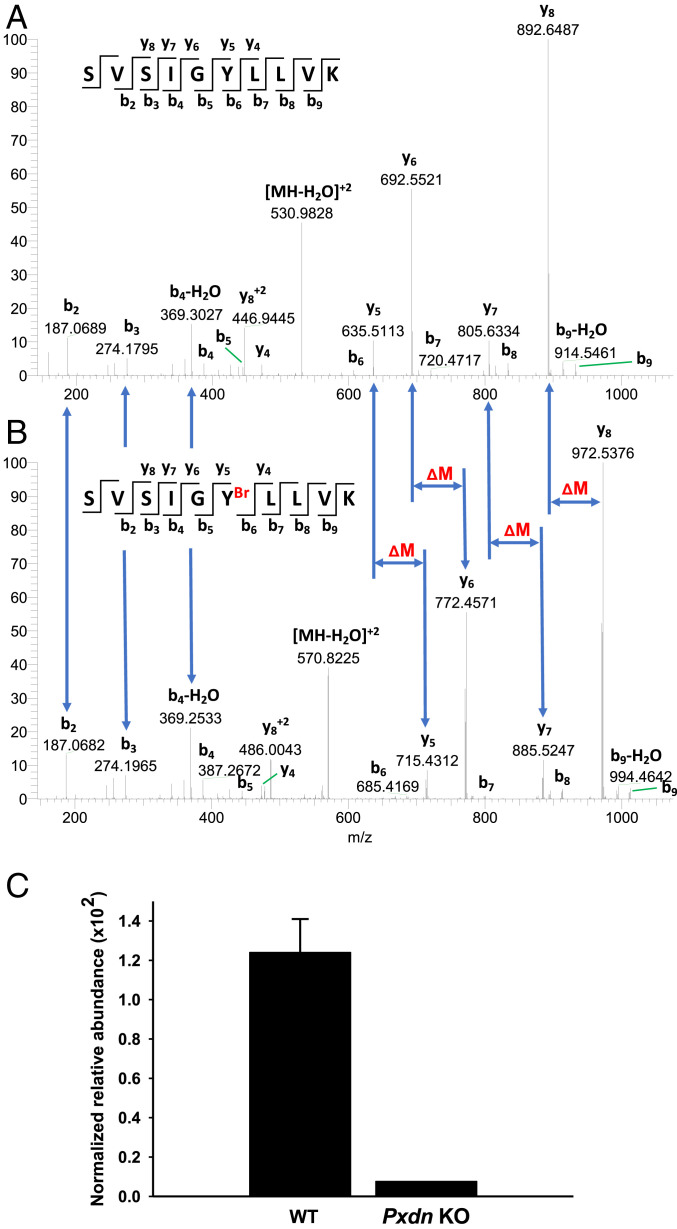
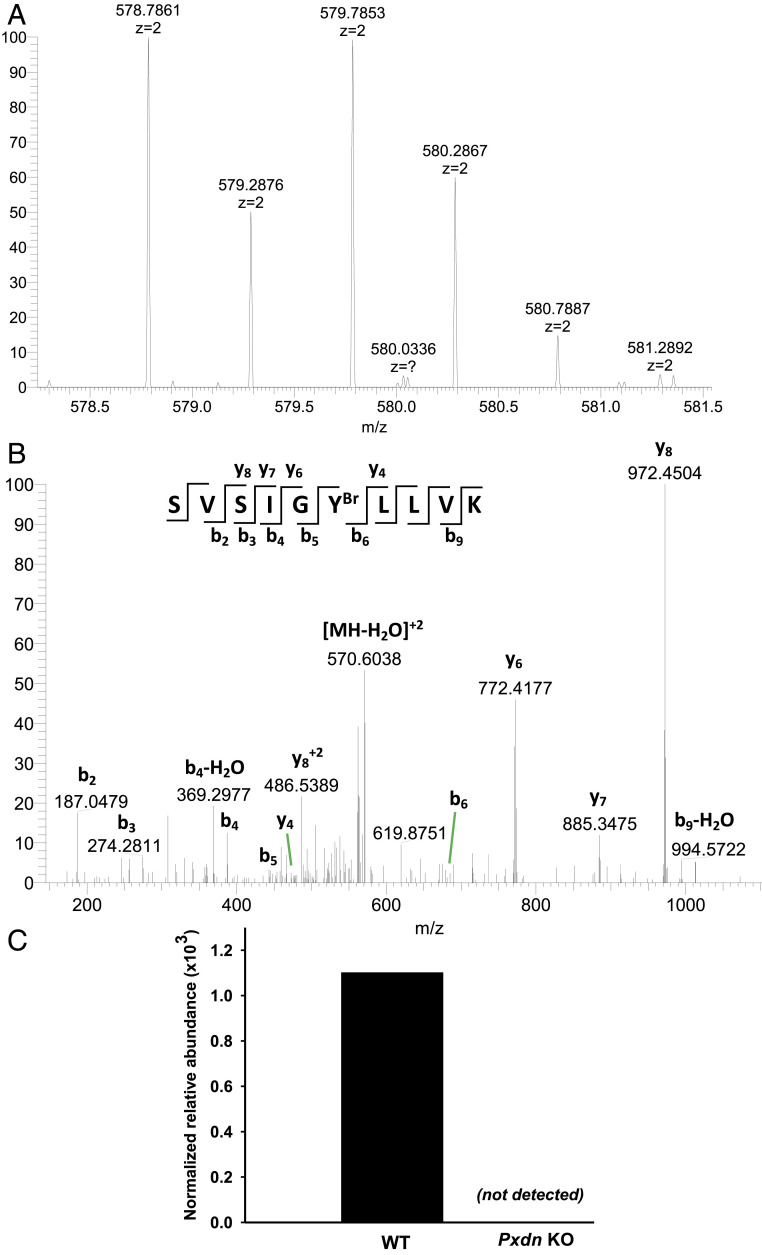
Because hypobromous acid is known to brominate tyrosine residues in proteins (33), we suspected that the reduced bromine enrichment in the basement membranes of Pxdn−/− mice would be mirrored by reduced amounts of bromotyrosine in basement membrane proteins. We identified, by liquid chromatography–tandem mass spectrometry (LC-MS/MS), a tyrosine bromination site in the carboxyl-terminal noncollagenous 1 (NC1) domain of the α2 collagen IV chain (COL4A2). Analysis of tryptic peptides revealed a bromine modification of COL4A2-Tyr1485 (Fig. 8 and SI Appendix, Table S1). The very same tyrosine was brominated in human kidney COL4A2 (SI Appendix, Table S1 and Fig. S12). The bromination of COL4A2-Tyr1485 was peroxidasin-dependent; bromination of that residue was reduced by ∼93% in the NC1 domain of α2 collagen IV in kidneys of Pxdn−/− mice (Fig. 8C) (5). Bromination of Tyr1485 was undetectable in the NC1 domains isolated from the ECM of Pxdn-null mouse PFHR9 cells (32) (where peroxidasin expression was eliminated by CRISPR/CAS9 genome editing and sulfilimine bonds are not formed) (Fig. 9)
Fig. 8.
Bromination site in the α2NC1 domain of collagen IV from mouse kidney and its dependence on peroxidasin expression. NC1 domains of collagen IV were isolated from WT and Pxdn−/− mice and analyzed by LC-MS/MS as described in Materials and Methods. (A and B) Evidence for bromination of Tyr1485 in a tryptic peptide of the α2NC1 domain of mouse collagen IV [α2(IV)NC1]. Shown are MS2 spectra of unmodified (A) and brominated (B) peptide from the α2NC1 domain, including peak assignments. The mass shifts (ΔM) in y-ion series starting from y5 and indicating the sequence position of brominated tyrosine residue are also shown. Masses of 79Br and 81Br isotopes calculated from the data in A and B were 78.9107 ± 0.0271 and 80.9086 ± 0.0143, respectively. The standard isotopic masses for 79Br and 81Br are 78.9183 and 80.9163, respectively. (Insets) The sequence of unmodified (A) and brominated (B) precursor peptide and the MS2 fragmentation pattern producing specific y- and b-ions. The y axis in A and B is relative abundance. (C) Quantitation of the same brominated peptide in α2(IV)NC1 preparations from WT and Pxdn−/− mice. Normalized relative abundance represents modified peptide/(modified peptide + unmodified peptide). The WT data represent mean ± SD for three independent kidney NC1 preparations; the Pxdn−/− mouse data are from a preparation generated from the pooled kidneys of five Pxdn−/− mice.
Fig. 9.
Bromination site in the α2NC1 domain of collagen IV from the ECM deposited by cultured PFHR9 cells and its dependence on peroxidasin expression. NC1 domains of collagen IV were isolated from ECM deposited by WT and Pxdn-deficient PFHR9 cells (Pxdn KO) and analyzed by LC-MS/MS as described in Materials and Methods. (A) Isotopic envelope of brominated precursor peptide, revealing a characteristic bromine pattern deriving from the presence of 79Br and 81Br isotopes (mass accuracy is 3.3 parts per million [ppm]). (B) The MS2 spectra of the brominated peptide, including peak assignments. (Inset) The sequence of brominated precursor peptide and the MS2 fragmentation pattern producing specific y- and b-ions. (C) Quantification of tyrosine bromination in the tryptic peptide from α2(IV)NC1 preparations derived from the matrix of WT PFHR9 cells and Pxdn-deficient PFHR9 cells. Normalized relative abundance represents modified peptide/(modified peptide + unmodified peptide). The WT and Pxdn KO data represent single preparations, each deriving from the ECM collected from four 150-mm cell culture dishes.
Discussion
The formation of sulfilimine cross-links between the NC1 domains of collagen IV trimers depends on the capacity of peroxidasin to use H2O2 and bromine substrates to produce HOBr, a potent oxidizing agent (1, 7, 13). Sulfilimine cross-links are crucial for the integrity of both collagen IV and the basement membrane (1, 3, 5). A longstanding puzzle, however, has been whether the HOBr released by peroxidasin acts selectively to promote collagen IV sulfilimine cross-links or whether it also results in oxidative damage to bystander molecules in the ECM (7, 22, 23): for example, by brominating tyrosine residues within proteins. After sulfilimine cross-links have been formed, bromide is presumably released into the ECM and is available for recycling in the same cross-linking reaction. However, we hypothesized that we might find evidence for bromination of bystander ECM proteins by using NanoSIMS imaging (34–37) to visualize bromine distribution within tissues. In the current studies, we discovered, by NanoSIMS analyses, that the basement membranes in wild-type mouse kidneys and normal human kidneys are markedly enriched in bromine. We also observed bromine enrichment of basement membranes in kidney specimens from patients with TBMN, Alport syndrome, anti-GBM disease, and diabetes mellitus. We discovered that bromine enrichment of basement membranes is peroxidasin-dependent. In kidneys of Pxdn−/− mice, bromine enrichment of basement membranes was reduced by 85%. When we purified collagen IV NC1 domains from mouse and human kidney and examined tryptic peptides by mass spectrometry, we observed bromination of a tyrosine within the NC1 domain of COL4A2 (Tyr1485 in mouse and Tyr1490 in human). Bromination of Tyr1485 Pxdn−/− kidneys was 93% lower than in wild-type kidneys. In NC1 domains purified from a peroxidasin-null cell line, bromination of Tyr1485 was undetectable. Thus, peroxidasin is responsible for the bromination of basement membranes (as judged by NanoSIMS analyses) and for bromination of a bystander tyrosine residue within the NC1 domain of α2 collagen IV (as judged by LC-MS/MS studies). Of note, bromination of tyrosine residues presumably represents only a subset of peroxidasin-mediated oxidation reactions. HOBr does not always generate bromine adducts; for example, it rapidly reacts with methionine and cysteine residues to generate methionine sulfoxide and cysteine sulfenic acid, respectively (38).
In the current studies, we examined Pxdn−/− mice (5) that had a gene-trap insertion in intron 8 (Materials and Methods). These “gene-trap” Pxdn−/− mice, like peroxidasin-deficient humans (8) and Pxdn−/− mice harboring a premature stop codon within the peroxidase domain (9), exhibited severe dysgenesis of the anterior segment of the eye. Despite the severity of the eye pathology in the gene-trap Pxdn−/− mice, we suspected (even before embarking on our studies) that the gene-trap allele might yield trace amounts of peroxidasin expression. This view is based upon the fact that collagen IV cross-links were easily detectable (about 30% of normal) in gene-trap Pxdn−/− mice (5) but were virtually absent in Pxdn−/− mice harboring a 2-base pair (bp) deletion near the beginning of the peroxidasin domain coding sequences (39). Our current studies heightened our suspicions that the gene-trap Pxdn allele might be “leaky.” By NanoSIMS analyses, bromine enrichment in GBMs of the gene-trap Pxdn−/− mice was only reduced by ∼85% (rather than being eliminated), and, by mass spectrometry, bromination of Tyr1485 of COL4A2 from Pxdn−/− kidneys was reduced by 93% (rather than being eliminated). We suspect that bromine enrichment of the basement membrane would be completely eliminated in knockout mice harboring a bona fide Pxdn-null allele, given that bromination of Tyr1485 was undetectable in a peroxidasin-null cell line.
In Drosophila, peroxidasin is located in the basement membrane of muscle cells lining the gut (1, 3). In mammals, peroxidasin is assumed to be located in the ECM (23), but the localization of peroxidasin within the ECM has never been precisely defined, owing to an absence of useful antibody reagents. The fact that bromine enrichment in our NanoSIMS images of mice was largely restricted to basement membranes implies that peroxidasin activity is largely confined to the basement membrane. However, given that we observed small amounts of bromine in the interstitial spaces between renal tubules, it is possible that small amounts of peroxidasin are located outside of basement membranes. Of note, we observed peroxidasin-mediated bromine enrichment in basement membranes in Bowman’s capsule (which contain α1α2α1 and α5α6α5 collagen IV networks) (40) and in the basement membranes between podocytes and endothelial cells of glomerular capillaries (which contain separate α1α2α1 and α3α4α5 collagen IV networks) (41, 42). Despite differences in sulfilimine bond formation in wild-type and Pxdn−/− mice (32) and the extent of bromination of kidney basement membranes, we observed no histologic differences by electron microscopy or NanoSIMS imaging. The only overt histopathologic abnormality resulting from peroxidasin deficiency is dysgenesis of the anterior chamber of the eye (9).
The fact that we were able to detect, by LC-MS/MS, peroxidasin-dependent bromination of Tyr1485 in the NC1 domain of mouse kidney α2 collagen IV indicates that the HOBr from peroxidasin modifies bystander tyrosine residues in proteins in addition to promoting sulfilimine cross-links. However, it is noteworthy that the bromination of the bystander tyrosine residues was not extensive. In our proteomic studies, only ∼1.5% of α2 chains of collagen IV in mouse kidney contained brominated Tyr1485, and we never observed a doubly brominated tyrosine at that residue. Also, there are 58 tyrosine residues in the NC1 domains of α1α2α1 collagen IV hexamers, and only two of them (the Tyr1485 residues in the α2 chains) are brominated. Peroxidasin mutagenesis studies suggested that there are indirect interactions between peroxidasin Ig domains and collagen IV (6). We speculate that any such interactions are quite specific, such that the catalytic domain of peroxidasin—from which HOBr is released—is positioned in close proximity to Tyr1485 of the α2 (IV) chain and close to the methionine and hydroxylysine residues involved in sulfilimine cross-links. However, because Tyr1485 and the sulfilimine cross-links are located on opposite sides of α2 NC1 domains, peroxidasin positioning could be complicated and involve conformational changes.
Does bromination of Tyr1485 in the α2 collagen IV NC1 domain fully account for the 79Br and 81Br enrichment in mammalian basement membranes? Our data cannot answer this question in a definitive manner, simply because the NanoSIMS instrument merely records the release of 79Br– and 81Br– secondary ions. However, we are skeptical that the bromination of Tyr1485 is fully responsible for bromine enrichment of basement membranes. We suspect that the HOBr released by peroxidasin brominates other domains within collagen IV as well as other basement membrane proteins (perhaps including peroxidasin itself). If so, defining the location of additional bromotyrosine residues could yield valuable insights into the positioning of peroxidasin’s catalytic site within the basement membrane as well as the spatial relationships between different basement membrane proteins.
Our NanoSIMS analyses of diabetic nephropathy specimens were intriguing. The mechanism for GBM thickening in diabetes is not fully understood, but both increased synthesis and reduced turnover of basement membrane proteins have been proposed (43, 44). In our studies, we observed a high degree of bromine enrichment in the glomerular capillaries in diabetic nephropathy specimens. While this phenomenon could be due to increased expression of collagen IV and/or other basement membrane proteins, we are attracted to the idea that it is caused, at least in part, by reduced turnover of basement membrane proteins. Collagen IV as well as other basement membrane proteins (e.g., laminin, nidogen) are long-lived proteins (45), but a slower turnover rate would presumably increase tyrosine bromination by prolonging exposure to the HOBr produced by peroxidasin. It is possible, perhaps even likely, that the thickening of basement membranes in diabetes, along with peroxidasin-mediated bromination of basement membrane proteins, underlies higher bromotyrosine levels in the urine of patients with diabetes mellitus (46). In future studies, it should be possible to pursue that possibility in mouse models of diabetes, including in mice that are homozygous for Pxdn-null alleles.
Materials and Methods
Mouse Strains and Cells.
Wild-type mice (strain C57BL/6) were purchased from The Jackson Laboratory. Peroxidasin-deficient mice (Pxdn−/−; strain C57BL/6) were generated by Bhave et al. (5). These Pxdn−/− mice were generated with a targeting vector from the Knockout Mouse Project (KOMP) Repository (Project ID CSD 80013; https://www.komp.org/) and contained a gene-trap cassette in intron 8. Additional details regarding Pxdn mouse models and experimental methods for quantifying Pxdn transcripts in mouse tissues are described in SI Appendix, Materials and Methods. Pxdn-null PFHR9 cells were generated and characterized by Colon and Bhave (32).
Sample Preparation of Mouse Tissues for Electron Microscopy and NanoSIMS Analyses.
Mouse tissues were fixed at 4 °C overnight in a solution containing 4% paraformaldehyde, 2.5% glutaraldehyde, and 2.1% sucrose in 0.1 M sodium cacodylate. On the following day, tissues were rinsed five times for 3 min with 0.1 M sodium cacodylate and then incubated in a 0.1-M sodium cacodylate solution containing 2% osmium tetroxide and 1.5% potassium ferricyanide for 1 h at 4 °C. The samples were then rinsed five times for 3 min with distilled water before being incubated with 1% thiocarbohydrazide for 20 min at room temperature. Next, samples were rinsed five times for 3 min with distilled water and then incubated with 2% osmium tetroxide for 30 min at room temperature. The samples were then rinsed five times for 3 min each with distilled water and incubated with 2% uranyl acetate at 4 °C overnight. On the following day, the samples were rinsed with distilled water and then dehydrated by incubating the samples in progressively increasing amounts of ethanol (30, 50, 70, 85, 95, and 100%) for 10 min each followed by two additional 10-min incubations with 100% ethanol. Next, the samples were infiltrated with Embed812 resin (Electron Microscopy Sciences) by incubating the samples in 33% resin (diluted in anhydrous acetone) for 2 h, 66% resin overnight, and 100% resin for 4 h. Samples were then embedded in fresh resin with polypropylene molds (Electron Microscopy Sciences) and polymerized in a vacuum oven at 65 °C for 48 h.
Preparation of Human Kidney Biopsies for NanoSIMS Analyses.
Archived resin blocks from renal biopsies were obtained from the Translational Pathology Core Laboratory (TPCL), a College of American Pathologists/Clinical Laboratory Improvement Acts (CAP/CLIA)-certified research facility that was established as an institutional resource by the David Geffen School of Medicine at the University of California, Los Angeles (UCLA). The TPCL is administered by UCLA’s Department of Pathology and Laboratory Medicine. Permission to utilize archived kidney biopsy material for this collaborative study was requested by J.E.Z., a UCLA renal pathologist and coauthor on this paper. J.E.Z.’s request was reviewed and approved by UCLA’s Human Use Institutional Review Board (IRB) (no. 19-002243). No other coauthor was aware of patient information. Kidney biopsies from patients with biopsy-proven TBMN, Alport syndrome, anti-GBM disease, allergic-type acute interstitial nephritis, and diabetic nephropathy were identified; the biopsy diagnoses were confirmed based on a review of original biopsy material and patient information by J.E.Z. Kidney donor transplant biopsy specimens with no significant pathologic changes were used for the “normal” controls.
At the time of original biopsy, fresh tissue was placed in a 0.1-M sodium cacodylate buffer, pH 7.4, containing 2.5% glutaraldehyde and 2.0% paraformaldehyde for a minimum of 2 h at room temperature and then washed with 0.1 M sodium cacodylate. Tissue was fixed in 1% osmium tetroxide in 0.1 M sodium cacodylate for 1.5 to 2 h and again washed in the 0.1 M sodium cacodylate buffer. Next, the tissue was dehydrated through an acetone gradient and infiltrated with epon resin. Epon-embedded tissue was warmed in a 65 to 85 °C oven for 40 min and embedded into predried molds. Polymerization was allowed to occur at 65 to 85 °C overnight.
Scanning Electron Microscopy.
Five hundred-nanometer sections of resin-embedded mouse tissues were prepared with a Leica UC6 ultramicrotome and mounted onto silicon wafers. BSE images were obtained with an FEI Verios scanning electron microscope (Thermo Fisher) with a 1-kV beam with the current at 200 pA.
NanoSIMS Analyses.
Five hundred-nanometer sections of resin-embedded tissues were cut with a Leica UC6 ultramicrotome and mounted onto silicon wafers. Sections were then coated with 5 nm of gold and analyzed with either NanoSIMS 50 or NanoSIMS 50L instruments (CAMECA). Samples were bombarded with a focused 16-KeV 133Cs+ beam, and both secondary electrons (SEs) and secondary ions (12C– or 13C–,12C14N– or 13C14N–, 31P–, 32S–, 79Br– and/or 81Br–, 127I–) were collected. In some experiments, correlative BSE imaging and NanoSIMS analyses were performed on the same tissue section. NanoSIMS instrument settings are described in SI Appendix, Materials and Methods.
Images were processed and prepared with the OpenMIMS plugin in ImageJ. 79Br, 81Br, and 127I images were processed with a Gaussian Blur filter in ImageJ with a radius of 1 to reduce noise. For image quantification, regions of interest (ROIs) were drawn manually based on the morphological features in NanoSIMS and BSE images. 79Br, 81Br, and 127I in ROIs, normalized to 13C or 12C, were extracted using the OpenMIMS plugin and processed by GraphPad Prism 7.0. Comparisons between bromine enrichment measurements normalized to 12C or 13C (natural abundance, 1.1%) were calculated by 79Br/12C = (79Br/13C)/89.9.
Isolation of Collagen IV NC1 Domains from Mouse Kidneys and the ECM Deposited by Cultured Cells.
Collagen IV NC1 domains were isolated from kidney tissue as described (47) with minor modifications. Kidneys were homogenized in 50 mM Tris⋅HCl buffer, pH 7.4, supplemented with 150 mM NaCl, 20 mM ethylenediaminetetraacetic acid, 25 mM ε-aminocaproic acid, 5 mM benzamidine, 5 mM N-ethylmaleimide, 1 mM phenylmethylsulfonylfluoride (PMSF), and 1 μg/mL each of three protease inhibitors (leupeptin, pepstatin A, aprotinin), followed by stirring for 1 h at 4 °C and centrifugation at 8,000 × g for 15 min. This procedure was repeated three times with fresh buffer, and the pellets were resuspended in ice-cold water with 0.05% sodium azide and 1 mM PMSF. The suspension was stirred for 1 h at 4 °C, followed by centrifugation at 8,000 × g for 15 min. The pellets were then resuspended in 1 M NaCl and incubated with 200 Kunitz U/mL of DNase, followed by stirring for 2 h at room temperature and centrifugation at 8,000 × g for 15 min. The pellet was resuspended in ice-cold water, gradually adding sodium deoxycholate to a final concentration of 1% and stirring at room temperature for 2 h. Pellets were digested with collagenase (20 U/mL) for 24 h at 37 °C with stirring (48). The digested material was dialyzed against 50 mM Tris⋅HCl, pH 7.5, and then loaded onto a diethylaminoethyl (DEAE) Sepharose column. A pass-through fraction from the DEAE column was collected and purified with a Superdex 200 10/300 GL gel-filtration column (GE Healthcare) (49).
Isolation of collagen IV NC1 domains from the ECM deposited by cultured PFHR9 cells was performed as described (50). Briefly, confluent parental PFHR9 cells or Pxdn-deficient PFHR9 cells (32) were maintained in Dulbecco’s modified Eagle’s medium for 7 d in the presence of 50 μg/mL ascorbic acid with daily medium changes. Cells were then washed in phosphate-buffered saline and scraped into 1% (wt/vol) sodium deoxycholate lysis buffer. The lysates were sonicated to shear genomic DNA, and the insoluble material was collected by centrifugation at 20,000 × g, washed twice in 1 M NaCl and 10 mM Tris⋅HCl, and then resuspended in 50 mM Tris⋅HCl, pH 7.4. The matrix material was digested with bacterial collagenase (20 U/mL; Worthington Biochemical Corporation) in a buffer containing 5 mM CaCl2, 5 mM benzamidine, 25 mM ε-aminocaproic acid, and 0.4 mM PMSF. Digests were cleared by centrifugation at 15,000 × g for 5 min, dialyzed against 50 mM Tris⋅HCl (pH 7.5), and purified on a DEAE Sepharose column, followed by a Superdex 200 10/300 GL gel-filtration column (49). Purified NC1 collagen IV hexamers were concentrated with Amicon Ultra Centrifugal Filter Units (10,000 molecular weight cutoff).
LC-MS/MS Analyses of Bromination Sites.
Bromination sites in the collagen IV NC1 domains from kidneys or from the ECM deposited by cultured cells were analyzed by LC-MS/MS in the Proteomics Core of the Vanderbilt Mass Spectrometry Research Center (51). Sample preparations and MS2 spectra generation and analyses are described in SI Appendix, Materials and Methods.
Institutional Approvals.
Animal protocols were approved by institutional Animal Research Committees (UCLA, Vanderbilt). Mice were fed a chow diet and housed in AAALAC (Association for Assessment and Accreditation of Laboratory Animal Care International)-approved facilities with a 12-h light–dark cycle. Archived samples of human kidney biopsies were obtained with approval from UCLA’s Institutional Review Board (IRB no. 19-002243).
Statistics.
Statistical analyses were performed with GraphPad Prism 7.0 software. Differences were assessed using a Student’s t test with Welch’s correction.
Data Availability.
Data are included in the figures of this paper.
Supplementary Material
Acknowledgments
This work was supported by National Heart, Lung, and Blood Institute Grants HL090553, HL087228, and HL125335 (to S.G.Y.); an Australian Research Council Discovery Early Career Researcher Award (to H.J.); NIH Grant R01 DK116964 and a Burroughs Welcome Fund Career Award for Medical Scientists (13030995) (to G.B.); and NIH Grants R01 DK065138 (to B.G.H. and P.A.V.) and R24 DK103067 (to B.G.H., in part). We thank Dr. Vadim Pedchenko (Division of Nephrology, Vanderbilt University Medical Center) for providing samples of NC1 domains from human renal cortical shavings, Dr. Kristie Lindsey Rose (Mass Spectrometry Research Center, Vanderbilt University) for suggestions on experimental design and data analysis, and Dr. Maximiliano Gutierrez (The Francis Crick Institute) for helpful discussions. We thank the Centre for Microscopy, Characterisation & Analysis at the University of Western Australia, which is funded by the University and both the State and Commonwealth Governments.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007749117/-/DCSupplemental.
References
- 1.McCall A. S.et al., Bromine is an essential trace element for assembly of collagen IV scaffolds in tissue development and architecture. Cell 157, 1380–1392 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fidler A. L.et al., Collagen IV and basement membrane at the evolutionary dawn of metazoan tissues. eLife 6, e24176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhave G.et al., Peroxidasin forms sulfilimine chemical bonds using hypohalous acids in tissue genesis. Nat. Chem. Biol. 8, 784–790 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fidler A. L.et al.; Aspirnauts , A unique covalent bond in basement membrane is a primordial innovation for tissue evolution. Proc. Natl. Acad. Sci. U.S.A. 111, 331–336 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhave G., Colon S., Ferrell N., The sulfilimine cross-link of collagen IV contributes to kidney tubular basement membrane stiffness. Am. J. Physiol. Renal Physiol. 313, F596–F602 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ero-Tolliver I. A., Hudson B. G., Bhave G., The ancient immunoglobulin domains of peroxidasin are required to form sulfilimine cross-links in collagen IV. J. Biol. Chem. 290, 21741–21748 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colon S., Page-McCaw P., Bhave G., Role of hypohalous acids in basement membrane homeostasis. Antioxid. Redox Signal. 27, 839–854 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi A.et al., Novel mutations in PXDN cause microphthalmia and anterior segment dysgenesis. Eur. J. Hum. Genet. 23, 337–341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan X.et al., Peroxidasin is essential for eye development in the mouse. Hum. Mol. Genet. 23, 5597–5614 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan K.et al., Homozygous mutations in PXDN cause congenital cataract, corneal opacity, and developmental glaucoma. Am. J. Hum. Genet. 89, 464–473 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klebanoff S. J., Kettle A. J., Rosen H., Winterbourn C. C., Nauseef W. M., Myeloperoxidase: A front-line defender against phagocytosed microorganisms. J. Leukoc. Biol. 93, 185–198 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen N. E., Karle H., Andersen V., Malmquist J., Hoff G. E., Neutrophilic granulocytes in acute bacterial infection. Sequential studies on lysozyme, myeloperoxidase and lactoferrin. Clin. Exp. Immunol. 26, 463–468 (1976). [PMC free article] [PubMed] [Google Scholar]
- 13.Weiss S. J., Test S. T., Eckmann C. M., Roos D., Regiani S., Brominating oxidants generated by human eosinophils. Science 234, 200–203 (1986). [DOI] [PubMed] [Google Scholar]
- 14.Nogueira N. M., Klebanoff S. J., Cohn Z. A., T. cruzi: Sensitization to macrophage killing by eosinophil peroxidase. J. Immunol. 128, 1705–1708 (1982). [PubMed] [Google Scholar]
- 15.Buys J., Wever R., van Stigt R., Ruitenberg E. J., The killing of newborn larvae of Trichinella spiralis by eosinophil peroxidase in vitro. Eur. J. Immunol. 11, 843–845 (1981). [DOI] [PubMed] [Google Scholar]
- 16.Specht S.et al., Lack of eosinophil peroxidase or major basic protein impairs defense against murine filarial infection. Infect. Immun. 74, 5236–5243 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colon S.et al., Peroxidasin and eosinophil peroxidase, but not myeloperoxidase, contribute to renal fibrosis in the murine unilateral ureteral obstruction model. Am. J. Physiol. Renal Physiol. 316, F360–F371 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aldridge R. E.et al., Eosinophil peroxidase produces hypobromous acid in the airways of stable asthmatics. Free Radic. Biol. Med. 33, 847–856 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Wu W.et al., Eosinophils generate brominating oxidants in allergen-induced asthma. J. Clin. Invest. 105, 1455–1463 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohiuddin I.et al., Nitrotyrosine and chlorotyrosine: Clinical significance and biological functions in the vascular system. J. Surg. Res. 133, 143–149 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Hazen S. L., Crowley J. R., Mueller D. M., Heinecke J. W., Mass spectrometric quantification of 3-chlorotyrosine in human tissues with attomole sensitivity: A sensitive and specific marker for myeloperoxidase-catalyzed chlorination at sites of inflammation. Free Radic. Biol. Med. 23, 909–916 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Bathish B., Turner R., Paumann-Page M., Kettle A. J., Winterbourn C. C., Characterisation of peroxidasin activity in isolated extracellular matrix and direct detection of hypobromous acid formation. Arch. Biochem. Biophys. 646, 120–127 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Péterfi Z.et al., Peroxidasin is secreted and incorporated into the extracellular matrix of myofibroblasts and fibrotic kidney. Am. J. Pathol. 175, 725–735 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kruegel J., Rubel D., Gross O., Alport syndrome–Insights from basic and clinical research. Nat. Rev. Nephrol. 9, 170–178 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Funk S. D., Lin M. H., Miner J. H., Alport syndrome and Pierson syndrome: Diseases of the glomerular basement membrane. Matrix Biol. 71–72, 250–261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hudson B. G., Tryggvason K., Sundaramoorthy M., Neilson E. G., Alport’s syndrome, Goodpasture’s syndrome, and type IV collagen. N. Engl. J. Med. 348, 2543–2556 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Rana K.et al., The genetics of thin basement membrane nephropathy. Semin. Nephrol. 25, 163–170 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Pedchenko V.et al., Molecular architecture of the Goodpasture autoantigen in anti-GBM nephritis. N. Engl. J. Med. 363, 343–354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papadopoulou-Marketou N., Chrousos G. P., Kanaka-Gantenbein C., Diabetic nephropathy in type 1 diabetes: A review of early natural history, pathogenesis, and diagnosis. Diabetes Metab. Res. Rev. 33, e2841 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Haas M., Alport syndrome and thin glomerular basement membrane nephropathy: A practical approach to diagnosis. Arch. Pathol. Lab. Med. 133, 224–232 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Plevová P., Gut J., Janda J., Familial hematuria: A review. Medicina (Kaunas) 53, 1–10 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Colon S., Bhave G., Proprotein convertase processing enhances peroxidasin activity to reinforce collagen IV. J. Biol. Chem. 291, 24009–24016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu W., Chen Y., d’Avignon A., Hazen S. L., 3-Bromotyrosine and 3,5-dibromotyrosine are major products of protein oxidation by eosinophil peroxidase: Potential markers for eosinophil-dependent tissue injury in vivo. Biochemistry 38, 3538–3548 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Jiang H.et al., High-resolution imaging of dietary lipids in cells and tissues by NanoSIMS analysis. J. Lipid Res. 55, 2156–2166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang H.et al., Stable isotope imaging of biological samples with high resolution secondary ion mass spectrometry and complementary techniques. Methods 68, 317–324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He C.et al., NanoSIMS analysis of intravascular lipolysis and lipid movement across capillaries and into cardiomyocytes. Cell Metab. 27, 1055–1066.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He C.et al., High-resolution imaging and quantification of plasma membrane cholesterol by NanoSIMS. Proc. Natl. Acad. Sci. U.S.A. 114, 2000–2005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pattison D. I., Davies M. J., Kinetic analysis of the reactions of hypobromous acid with protein components: Implications for cellular damage and use of 3-bromotyrosine as a marker of oxidative stress. Biochemistry 43, 4799–4809 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Sirokmány G.et al., Peroxidasin-mediated crosslinking of collagen IV is independent of NADPH oxidases. Redox Biol. 16, 314–321 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ninomiya Y.et al., Differential expression of two basement membrane collagen genes, COL4A6 and COL4A5, demonstrated by immunofluorescence staining using peptide-specific monoclonal antibodies. J. Cell Biol. 130, 1219–1229 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heidet L., Cai Y., Guicharnaud L., Antignac C., Gubler M. C., Glomerular expression of type IV collagen chains in normal and X-linked Alport syndrome kidneys. Am. J. Pathol. 156, 1901–1910 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suleiman H.et al., Nanoscale protein architecture of the kidney glomerular basement membrane. eLife 2, e01149 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kriz W.et al., Accumulation of worn-out GBM material substantially contributes to mesangial matrix expansion in diabetic nephropathy. Am. J. Physiol. Renal Physiol. 312, F1101–F1111 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Ziyadeh F. N., Renal tubular basement membrane and collagen type IV in diabetes mellitus. Kidney Int. 43, 114–120 (1993). [DOI] [PubMed] [Google Scholar]
- 45.Liu P., Xie X., Jin J., Isotopic nitrogen-15 labeling of mice identified long-lived proteins of the renal basement membranes. Sci. Rep. 10, 5317 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kato Y.et al., Quantification of modified tyrosines in healthy and diabetic human urine using liquid chromatography/tandem mass spectrometry. J. Clin. Biochem. Nutr. 44, 67–78 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fox J. W., Butkowski R. J., Hudson B. G., Detergent-prepared glomerular basement membrane is composed of a heterogeneous group of polypeptides. J. Biol. Chem. 256, 9313–9315 (1981). [PubMed] [Google Scholar]
- 48.Wieslander J., Kataja M., Hudson B. G., Characterization of the human Goodpasture antigen. Clin. Exp. Immunol. 69, 332–340 (1987). [PMC free article] [PubMed] [Google Scholar]
- 49.Kahsai T. Z.et al., Seminiferous tubule basement membrane. Composition and organization of type IV collagen chains, and the linkage of alpha3(IV) and alpha5(IV) chains. J. Biol. Chem. 272, 17023–17032 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Boudko S. P., Danylevych N., Hudson B. G., Pedchenko V. K., Basement membrane collagen IV: Isolation of functional domains. Methods Cell Biol. 143, 171–185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Madu H.et al., Pyridoxamine protects proteins from damage by hypohalous acids in vitro and in vivo. Free Radic. Biol. Med. 89, 83–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are included in the figures of this paper.