Significance
Anterior–posterior axis patterning is a fascinating but poorly understood developmental process. Single-cell sequencing was utilized to interrogate patterning abnormalities in a mesoderm-specific Hedgehog (Hh) pathway mutant, which revealed selective anterior mesoderm defects. We found that Hh signaling was required for fibroblast growth factor (FGF) pathway activity in nascent mesoderm during gastrulation. Mouse genetics, cell biology, and genomic experiments indicate that FGF signaling acts downstream of Hh signaling in nascent mesoderm for anterior mesoderm morphogenesis. This work demonstrates the utility of applying single-cell technologies to resolve complex developmental phenotypes. Here, it revealed a role for Hh signaling from the node in organizing anterior mesoderm morphogenesis, independent of its role in left–right determination, by promoting FGF signaling in gastrulating mesoderm.
Keywords: FGF signaling, mesoderm, embryonic node, Hedgehog signaling, single-cell sequencing
Abstract
The mechanisms used by embryos to pattern tissues across their axes has fascinated developmental biologists since the founding of embryology. Here, using single-cell technology, we interrogate complex patterning defects and define a Hedgehog (Hh)–fibroblast growth factor (FGF) signaling axis required for anterior mesoderm lineage development during gastrulation. Single-cell transcriptome analysis of Hh-deficient mesoderm revealed selective deficits in anterior mesoderm populations, culminating in defects to anterior embryonic structures, including the pharyngeal arches, heart, and anterior somites. Transcriptional profiling of Hh-deficient mesoderm during gastrulation revealed disruptions to both transcriptional patterning of the mesoderm and FGF signaling for mesoderm migration. Mesoderm-specific Fgf4/Fgf8 double-mutants recapitulated anterior mesoderm defects and Hh-dependent GLI transcription factors modulated enhancers at FGF gene loci. Cellular migration defects during gastrulation induced by Hh pathway antagonism were mitigated by the addition of FGF4 protein. These findings implicate a multicomponent signaling hierarchy activated by Hh ligands from the embryonic node and executed by FGF signals in nascent mesoderm to control anterior mesoderm patterning.
One of the first challenges of metazoan development is the generation and distribution of mesoderm across the anterior–posterior (A-P) axis during gastrulation (1–4). Presumptive mesoderm migrates through the primitive streak and is patterned during migration toward the anterior embryonic pole (1, 3–7). A-P patterning of the mesoderm is determined by secreted signals from the node and primitive streak. These signals include members of the nodal, fibroblast growth factor (FGF), and Wnt pathways, all of which are required for the patterning of the anterior mesoderm (6, 8). While the requirement for these pathways in gastrulation has been historically defined by loss-of-function assays, the multicomponent cross-talk between signaling axes that dictates A-P patterns remain largely unknown.
The Hedgehog (Hh) signaling pathway is required for the morphogenesis of organs derived from all three germ layers in most metazoans (9, 10). Hh signaling was first described in a classic forward genetic screen for genes that determine A-P segment polarity during early Drosophila melanogaster development (11). Hh pathway activation in mammals is initiated by the binding of Hh ligands (SHH, IHH, or DHH) to PTCH1, relieving its inhibition on Smoothened (SMO). Activated SMO then promotes nuclear translocation of full-length GLI2/3 transcription factors (TFs) that drive Hh target gene transcription (12, 13). Conversely, the absence of Hh signaling triggers proteolysis and truncation of GLI2/3 proteins to repressive isoforms GLI2R/3R, repressing Hh target gene transcription (13, 14). Hh signaling participates in the patterning of a diverse array of structures including the Drosophila wing disk (15), cnidarian pharyngeal musculature (16), tetrapod forelimb (17), vertebrate central nervous system (18), and heart (19–21), although it has not been implicated in patterning the early A-P axis in vertebrates.
Removal of all Hh signaling through germline deletion of Smo in mice revealed an essential role for the Hh pathway in early mammalian development (22). Smo−/− embryos exhibit cardiac and somitic defects, absence of the anterior dorsal aorta, and failure to establish left/right (L-R) axis asymmetry (22, 23). Furthermore, treatment of zebrafish embryos with a small-molecule Smo antagonist, cyclopamine, during early but not late gastrulation resulted in cardiac hypoplasia, suggesting that Hh signaling is essential during early embryogenesis for proper development of mesoderm-derived organs (24). While Shh is the only Hh ligand necessary for L-R axis patterning in mammals (25), compound Shh−/−;Ihh−/− mutants produce phenotypes indistinguishable from Smo−/− mutants (22). This suggests an important but poorly understood role for combined Shh and Ihh signaling during early embryogenesis, independent of L-R patterning.
We investigated the role for Hh signaling in mesoderm development during gastrulation. Using single-cell RNA sequencing (scRNA-seq) to profile WT and Hh-mutant mesoderm at the end of gastrulation, we observed a deficiency in the population of anterior mesoderm lineages in Hh-mutant embryos. Mesoderm-intrinsic and germline Hh pathway mutants both demonstrated defects selective to anterior mesoderm-derived structures, including the head, heart, pharyngeal arches, and anterior somites, which suggests a novel requirement for Hh signaling in anterior mesoderm development. Through genetic-inducible fate mapping and single-cell transcriptome analysis, we observed that onset of Hh-reception from the node coincided with the generation of Mesp1+ nascent mesoderm. Transcriptional profiling of Hh-deficient mesoderm during gastrulation revealed widespread down-regulation of FGF signaling pathway genes, required for patterning at the primitive streak and the generation of specific anterior mesoderm lineages. Mesoderm-specific Fgf4/Fgf8 double-mutants recapitulated anterior mesoderm defects and Hh-dependent GLI transcription factors modulated enhancer activity proximal to the shared Fgf3, Fgf4, and Fgf15 locus. Small-molecule Hh pathway inhibition in chicken embryos caused dose-dependent mesoderm migration defects, which could be mitigated by the addition of exogenous FGF4 protein. These findings resolve a Hh to FGF node to primitive streak midline signaling axis required for the migration and patterning of the anterior mesoderm during gastrulation.
Results
Hh Signaling Is First Active in the Organizing Centers of Embryonic Axis Determination.
We investigated the role of early Hh pathway activity on mesoderm development. We confirmed the earliest tissues to receive Hh signaling by evaluating β-galactosidase (β-gal) expression from the Ptch1LacZ reporter allele (26). No evidence of Ptch1 reporter activity was observed prior to embryonic day (E) 7.0 (SI Appendix, Fig. S1). β-gal becomes apparent in the node at E7.25, consistent with previous reports (SI Appendix, Fig. S1) (27). At E8.25, Ptch1 reporter activity expands to the notochord (SI Appendix, Fig. S1) and shortly thereafter appears throughout the neural floorplate, somites, dorsal aorta, and the second heart field (SHF) at E8.5 (SI Appendix, Fig. S1). These observations confirm that Hh signaling is first active in the node, which is essential for axis determination in mesoderm lineages, and later becomes active in several differentiated mesodermal lineages.
Based on the importance of signals sent from the node for mesoderm development, we assessed the necessity of Hh signaling in this process by driving the Cre-dependent coexpression of a dominant-negative transcriptional repressor of the Hh pathway, Gli3R, with a Venus fluorophore in the nascent mesoderm using Mesp1Cre (28, 29). We observed severe head and heart tube defects in Mesp1Cre/+;R26Gli3R-IRES-Venus/+ (Mesp1Cre-Gli3R) (30) mutants, which phenocopied defects characteristic of Smo−/− mutants (Fig. 1A) (22). The observation of defects beyond L-R patterning in both Smo−/− and Mesp1Cre-Gli3R mutant embryos suggest an essential role for Hh signaling in early mesoderm development.
Fig. 1.
scRNA-seq identifies Hh-dependent selective defects to anterior mesoderm progenitor populations. (A) Whole-mount left lateral views of control and Hh-mutant embryos comparing defects observed between Smo−/− mutants (Upper Right) and Mesp1Cre/+;R26Gli3R-IRES-Venus/+ (Mesp1Cre-Gli3R) mutants (Lower Right). (Scale bars, 200 μm.) (B) Experimental schema for collecting mutant Mesp1Cre-Gli3R and control Mesp1Cre/+;R26tdTomato/+ (Mesp1Cre-tdTomato) Mesp1Cre-labeled cells by FACS for scRNA-seq. (C, Left) UMAP plots for all Mesp1Cre-tdTomato and Mesp1Cre-Gli3R cells used in this study including both extraembryonic (gray and light blue) and embryonic (dark blue) mesoderm lineages. (Right) UMAP for embryonic mesoderm color-coded by cell lineage. (D) UMAP for embryonic mesoderm color-coded for either Mesp1Cre-tdTomato (blue) or Mesp1Cre-Gli3R (orange) genotypes. (E) Represents the proportion of Mesp1Cre-tdTomato (blue) and Mesp1Cre-Gli3R (orange) cells in each annotated lineage from D, where the number linked to each column represents absolute cell counts. Mut, mutant. (F) Heatmap of marker gene expression for aSHF (red), pSHF (green), and cardiomyocytes (blue) where each column represents a single cell. (G) Expression of aSHF and pSHF markers Isl1 and Wnt2, respectively, superimposed on UMAP plots of C. (H) Proportion of Mesp1Cre-tdTomato (blue) and Mesp1Cre-Gli3R (orange) cells across aSHF, pSHF, and cardiomyocyte (CM) populations. (I) Genetic fate map for aSHF-specific Cre (Mef2cAHF-Cre) with a Cre-dependent lacZ reporter (R26R) in Smo+/+ and Smo−/− backgrounds. Images represent the posterior margin of the aSHF at low and high power in the Upper and Lower, respectively. The arrow points to dorsal mesenchyme marker by Mef2cAHFCre, which is absent in the Smo−/− background. (Scale bars, 200 µm.) At, atrium; HT, heart tube; LV, left ventricle.
Drop-Seq Reveals Selective Anterior Mesoderm Deficits in Hh Pathway Mutants.
To examine the consequence of disrupting mesoderm-intrinsic Hh signaling, we performed scRNA-seq by Drop sequencing (Drop-seq) (31) on cells isolated by FACS from mutant Mesp1Cre-Gli3R and control Mesp1Cre/+;R26tdTomato/+ (Mesp1Cre-tdTomato) (32) embryos at the onset of organogenesis (E8.25) (33, 34) (Fig. 1B). We jointly processed and analyzed 9,843 Mesp1Cre-tdTomato and 10,663 Mesp1Cre-Gli3R cells from two biological replicates. We observed similar gene detection across them (SI Appendix, Fig. S2). Using unsupervised clustering to identify distinct cell populations (35, 36), we assigned cells to either embryonic mesoderm lineages (44%; 8,937 of 20,506) or hepatic stellate cells and extraembryonic tissues involved in early blood development (56%, 11,569 of 20,506) (Fig. 1C and SI Appendix, Fig. S3) (37). We focused on the embryonic mesoderm and identified all expected Mesp1Cre-derived lineages in both mutant and control embryos using a combination of canonical marker gene detection and comparison with extant scRNA-seq datasets (Fig. 1C and SI Appendix, Figs. S4–S6) (29, 38–40). When displayed on a uniform manifold approximation and projection (UMAP) plot, the proximity between clusters roughly recapitulated developmental relationships before and after batch-correction by sample integration (35) (Fig. 1C and SI Appendix, Fig. S7). Specifically, we observed that the majority of embryonic mesoderm cells contributed to mesenchyme (4,619, 51.7%) (dark blue in Fig. 1C) followed by cardiac (1,109, 12.4%) (orange in Fig. 1C), lateral plate (887, 9.92%) (gray-blue in Fig. 1C), allantoic (734, 8.21%) (pale orange in Fig. 1C), somitic (641, 7.17%) (light-green in Fig. 1C), pharyngeal (433, 4.85%) (dark-green in Fig. 1C), cranial (311, 3.48%) (red in Fig. 1C), and intermediate (203, 2.27%) (salmon in Fig. 1C) mesoderm lineages (Fig. 1C and SI Appendix, Figs. S4 and S5).
Expression of Gli3R altered the distribution of Mesp1Cre-derived cells (Fig. 1 D and E). We observed a selective deficiency in the proportional contribution of cells to anterior mesoderm lineages, including cranial, pharyngeal, and somitic mesoderm (Fig. 1E and SI Appendix, Fig. S5). Cranial mesoderm, marked by Otx2 expression (41), is the anterior-most mesoderm lineage and demonstrated the most pronounced deficiency, demonstrating a more than fourfold reduction in lineage contribution from mutant Mesp1Cre-Gli3R cells compared to Mesp1Cre-tdTomato controls (1.33% vs. 6.06% of cells). Somitic mesoderm, which is marked by Meox1 and Tcf15 (42, 43), was reduced in mutants nearly threefold (3.82% vs. 11.2%). Pharyngeal mesoderm, marked by Col2a1 and Sox9 (44, 45), demonstrated approximately twofold reduction within mutants (3.43% vs. 6.53%). Therefore, a deficiency in Mesp1Cre-derived cell contributions was observed within multiple independent anterior mesoderm lineages following expression of the Hh-pathway transcriptional repressor Gli3R.
Although the heart, which is derived from the anterior mesoderm, is severely malformed in Mesp1Cre-Gli3R embryos, the relative number of cells in the cardiac lineage was not reduced (Fig. 1E). Cardiac progenitors include cells in the first heart field (FHF) and SHF. The SHF is divided anatomically into anterior (aSHF) and posterior (pSHF) domains (46). We hypothesized that the aSHF may be preferentially affected in Mesp1Cre-Gli3R embryos. We analyzed the cardiac lineage from the Drop-seq in isolation and observed that it was comprised of three distinct subclusters: Functional cardiomyocytes from the FHF marked by Tnnt2 and Myh6 (47, 48); the aSHF, defined by Fgf8, Fgf10, and Isl1 expression (Fig. 1 F and G) (49); and the pSHF, defined by Wnt2, Sfrp1, and Tbx5 expression (Fig. 1 F and G) (50, 51). Within the SHF populations in Mesp1Cre-Gli3R embryos, fewer cells contributed to the aSHF (57.3%) compared to controls (73.9%) (Fig. 1H). We complemented these analyses by examining whether Hh mutants exhibited decreased aSHF cellularity. We performed a genetic fate-mapping study of the aSHF progenitor pool in Hh mutants utilizing Mef2cAHF-Cre (44) and Cre-dependent reporter (R26R) (45) alleles and observed β-gal expression in Smo+/+ and Smo−/− mutant embryos. We observed a substantial reduction of β-gal in the aSHF of Mef2cAHF-Cre;R26Rc/+;Smo−/− mutants compared to Mef2cAHF-Cre;R26Rc/+;Smo+/+ controls at E9.5, when the aSHF is normally well established (Fig. 1I). These data indicate that reduced Hh signaling caused defective population of the anterior cardiac lineage in Mesp1Cre-Gli3R embryos.
Hh Signaling Is Selectively Required for Anterior Mesoderm Development.
Drop-seq analysis indicated that anterior mesoderm lineages were reduced in Mesp1Cre-Gli3R mutants during organogenesis. We hypothesized that this disruption would culminate in specific phenotypic defects later in development. Anterior embryonic mesoderm lineages arise during gastrulation as undifferentiated epiblast cells migrate through a transient structure known as the primitive streak (1, 3–5). The earliest cells to contribute to the embryonic mesoderm enter the streak during early to midstreak stage and migrate furthest toward the anterior embryonic pole (4, 5), where they subsequently differentiate into specific lineages within the Mesp1 domain including, from anterior to posterior, cranial, pharyngeal, cardiac, and somitic mesoderm (Fig. 2A). We directly analyzed the development of anterior mesoderm-derived structures in both mesoderm-intrinsic and germline Hh pathway mutants. Mesoderm-intrinsic Hh mutants, including Mesp1Cre/+;Smof/− (29, 52) and Mesp1Cre-Gli3R embryos, exhibited cardiac and pharyngeal arch hypoplasia compared to Mesp1Cre/+;Smof/+ controls (Fig. 2 B–D). The anterior-most somites in mesoderm-intrinsic Hh mutants were positioned normally but exhibited severe morphological defects and failed to compact compared to more posterior somites (Fig. 2 F and G). In contrast, anterior and posterior somites were similar in Mesp1Cre/+;Smof/+ controls (Fig. 2E). The anterior-most somites of germline Smo−/− mutants fail to form and are replaced by loosely packed mesenchyme, while posterior somites are relatively well-formed (Fig. 2M). Smo−/− embryos also demonstrated the most severe anterior defects, including agenesis of the first pharyngeal arches and cardiac defects (Fig. 2J) (22). This indicates that anterior mesoderm defects intrinsic to Mesp1Cre-Gli3R mutants are attributable to early Hh pathway disruption, rather than a result of possible anterior mesoderm-restriction of Mesp1Cre activity. Surprisingly, removal of Shh failed to phenocopy the severe cardiac chamber or somite defects observed in Smo−/− mutants (Fig. 2I). Instead, Shh mutants only exhibited previously described pharyngeal arch hypoplasia (53) and relatively mild cardiac defects with respect to WT controls (Fig. 2 H, I, K, and L) (53). These results demonstrate that reduced Hh signaling by mutations to Smo, but not Shh, disrupts the development of anterior mesoderm.
Fig. 2.
Hh signaling is required for anterior mesoderm morphogenesis. (A) Diagrammatic representation of the developmental ontogeny for anterior mesoderm lineages with estimated scale bars for each embryonic stage. (B–G) Phenotypic abnormalities in conditional Hh mutants, Mesp1Cre/+Smof/− (C and F) and Mesp1Cre/+; R26Gli3R-IRES-Venus/+ (Mesp1Cre-Gli3R) (D and G) with their respective control, Mesp1Cre/+; Smof/+ (B and E) at E9.5. (H–M) Germline Hh pathway mutants, Shh−/− (I and L) and Smo−/− (J and M) and their respective Wt control (H and K) at E9.5. (B–D and H–J) Frontal whole-mount views of embryos (Left) with corresponding sagittal histology sections of the pharyngeal arch (Upper Right) and heart (Lower Right). Black bars link the whole-mount views of the pharyngeal arch and heart to their corresponding histology section. (E–G and K–M) Low-power sagittal histology views of both anterior and posterior somites (Left). High-power views of anterior and posterior somites are represented in the Upper Right and Lower Right, respectively. Somite 1 is indicated by the asterisk (*) in each, Upper Right. (Scale bars, 200 µm.) A, anterior; At, atrium; D, dorsal; IFT, inflow tract; LV, left ventricle; OFT, outflow tract; P, posterior; PA, pharyngeal arch; V, ventral.
Embryonic Mesoderm Lineages Receive Functional Hh Signals during Anterior Mesoderm Formation.
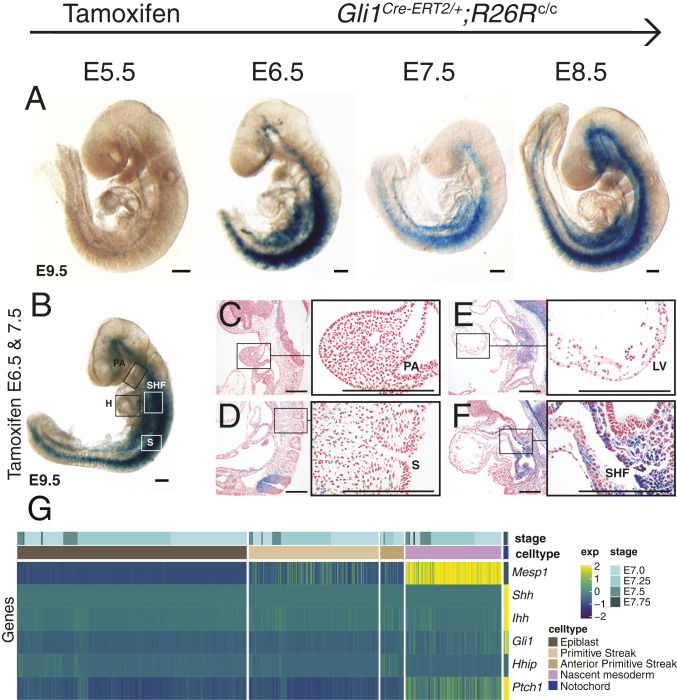
We interrogated which lineages received direct Hh signaling during anterior mesoderm development using genetic-inducible fate mapping (GIFM). We marked Hh-receiving cells and their descendants by GIFM using a tamoxifen (TM) -inducible Cre at the Gli1 locus (Gli1CreERT2) (54). Pregnant females were administered a single dose of TM on days ranging from E5.5 to E8.5 and Gli1CreERT2/+;R26Rc/c embryos were analyzed at E9.5 (Fig. 3A). We observed a near-absence of marked cells when TM was administered 1 d preceding gastrulation at E5.5 (Fig. 3A). Administration of TM during early gastrulation at either early (E6.5) or late (E7.5) stages of primary gastrulation resulted in robust β-gal expression throughout the neural tube and lateral mesenchyme, with notably few positive clones appearing in the pharyngeal mesoderm, somite bodies, or heart (Fig. 3A). TM administered after primary gastrulation at E8.5 demonstrated roughly the same staining pattern, albeit with more stained cells and the inclusion of some paraxial mesoderm (Fig. 3A). Administration of TM twice, at both E6.5 and E7.5, did not increase the number of marked lineages, although we did observe a higher proportion of stained cells within previously marked lineages (Fig. 3B). Serially sectioned embryos confirmed staining throughout mesenchymal tissues with relatively minor contributions to anterior mesoderm structures, such as the first pharyngeal arch (Fig. 3C), anterior somite bodies (Fig. 3D), or heart (Fig. 3E). For example, while the heart was sparsely labeled (Fig. 3E and SI Appendix, Table S1), SHF cells were robustly labeled, consistent with previous studies (Fig. 3F and SI Appendix, Table S1) (21, 55, 56).
Fig. 3.
Nascent embryonic mesoderm receives active Hh signaling during gastrulation. (A) Genetic-inducible fate maps for Hh-receiving cells in Gli1CreERT2/+;R26Rc/c embryos from whole-mount left lateral views at E9.5. Embryos were harvested from pregnant dams given a single TM dose at the times indicated at the top. (B–F) Whole-mount (B) and sagittal sections (C–F) from embryos harvested from pregnant dams dosed with TM at both E6.5 and E7.5. (C–F) Sagittal sections of the pharyngeal arch (C), anterior somites (D), left ventricle (E), and SHF (F) at low (Left) and high (Right) magnification; black boxes in the low-magnification field correspond to the area of high magnification. (G) A heatmap using reanalyzed data from, a scRNA-seq dataset spanning gastrulation (40). Cells are clustered by defined lineages spanning nascent mesoderm development through developmental stages E7.0 to E7.75. Each column represents a single cell and each row represents the denoised expression values of individual genes within the Hh signaling pathway in addition to Mesp1. (Scale bars, 200 um.) H, heart; S, somite.
We attempted to identify a population of Hh-receiving cells in the nascent mesoderm. We reanalyzed a publicly available scRNA-seq dataset spanning gastrulation in WT embryos for Hh signaling-dependent gene expression (40) and focused our analysis on embryonic stages coinciding with the generation of the anterior mesoderm (E7.0 to E7.75) (7). We plotted denoised expression values for genes associated with Hh signaling against cell populations across the epiblast and nascent mesoderm differentiation trajectory (Fig. 3G). Although no Hh targets were expressed in the epiblast or primitive streak, the nascent mesoderm demonstrated some cells with coexpression of Mesp1 with Hh targets Ptch1 and Gli1, but with much lower coverage than the known Hh expression lineages of the node and notochord (Fig. 3G). Together, the single-cell analysis and GIFM indicate that a subset of nascent mesoderm cells receives Hh signaling from the node throughout anterior mesoderm development. However, the GIFM indicated that this reception was sparse within anterior mesoderm progenitors, suggesting the role of additional Hh-dependent pathways in a cell nonautonomous mechanism for anterior mesoderm development (Fig. 3 B–G).
Transcriptional Profiling Identified Hh-Dependent TFs and Signaling Pathways Necessary for Mesoderm Morphogenesis.
To identify Hh-dependent pathways in the embryonic mesoderm, we performed bulk RNA-seq on mesoderm from Mesp1Cre/+;R26Gli3R-IRES-Venus/+ (Mesp1Cre-Gli3R) mutants during late gastrulation at E7.5. We utilized a Cre-dependent dual-color system to separate Mesp1Cre-marked red fluorophore-expressing WT Mesp1Cre/+;R26tdTomato (Mesp1Cre-Tomato) cells, from Mesp1Cre-marked yellow fluorophore-expressing mutant (Mesp1Cre-Gli3R) cells from litter-matched RNA-seq samples using FACS (n = 3) (Fig. 4A). We observed consistent differential expression between biological replicates and identified 190 dysregulated genes by mRNA-seq (false-discovery rate [FDR] ≤ 0.10) (Fig. 4B). We observed widespread down-regulation of classic targets for Hh signaling, such as Gli1, Ptch1, and Hhip (SI Appendix, Fig. S8). Many of the down-regulated genes are known to be critical for primitive streak function and anterior mesoderm morphogenesis, including Wnt3a, Msgn1, Cripto (Tdgf1), Fgf4, Fgf8, and Smad3 (57–61). Additionally, classic factors for both A-P patterning and midline development, Brachyury (T) and Hnf3β (FoxA2), were also down-regulated (62–64). These results demonstrate that Hh signaling is required for the normal expression of many genes required for anterior mesoderm development, further supporting a role for Hh signaling in organizing this process.
Fig. 4.
Disruption of Hh signaling causes major disruptions in genetic pathways for mesoderm development. (A) Breeding strategy to produce litter-matched controls of Hh mutant Mesp1Cre/+;R26Gli3R-IRES-Venus/+ (Mesp1Cre-Gli3R) and WT Mesp1Cre/+;R26tdTomato (Mesp1Cre-tdTomato) mesoderm by FACS-sorting Mesp1Cre-marked cells by yellow and red channels, respectively. (B) Heat maps are labeled with row z-score–normalized heatmap for dysregulated genes (FDR ≤ 0.10), where each column represents individual embryos. (C) A volcano plot for dysregulated genes in Hh mutants. (D) GO analysis for genes down-regulated genes from B and C. (E) Heatmap for all dysregulated genes under the Mesoderm Development (GO:007498) term (FDR ≤ 0.30).
Gene ontology (GO) analysis for down-regulated genes (FDR < 0.10) (Fig. 4D) corroborated the assessment that mesoderm morphogenesis was perturbed, as nearly all top terms were associated with early mesoderm development, including the term “Mesoderm Development” (GO:0007498) (Fig. 4 D and E). The majority of significantly dysregulated genes (FDR ≤ 0.30) classified under Mesoderm Development were down-regulated, which suggested widespread dysfunction to primitive streak function and mesoderm morphogenesis. GO analysis for Hh-dependent TFs identified somitogenesis and paraxial mesoderm as the most highly represented terms (Fig. 5A). Genes disrupted within the GO term for somitogenesis (GO:0001756; FDR ≤ 0.3) revealed down-regulation of TFs that either play an isolated role in somite development—including, Hes7 (relative transcript expression [RTE] = 0.081, FDR = 1.86E-2), Tcf15 (RTE = 0.715, FDR = 2.85E-1), Dll3 (RTE = 0.316, FDR = 7.22E-8), and Ripply2 (RTE = 0.101, FDR = 5.62 E-13) (65—60)—or factors that also play a dual role in somite development and gastrulation, including Msgn1 (RTE = 0.249, FDR = 3.00E-2), Tbx6 (RTE = 0.342, FDR = 2.69E-1), Mesp2 (RTE = 0.357, FDR = E8.25E-2), FoxA2 (RTE = 0.019, FDR = 7.60E-3), T (RTE = 0.254, FDR = 6.42E-3), and Smad3 (RTE = 0.608, FDR = 9.61E-2) (57, 62–64) (Fig. 5B). Using RNA in situ hybridization, we analyzed the spatial expression patterns for key genes responsible for shared paraxial and mesoderm development (Tbx6, Msgn1, and Mesp2) along with Dll3, a gene responsible primarily for somitogenesis. The dual-function transcription factors Tbx6, Msgn1, and Mesp2 lost large portions of their lateral expression domains, while the somite-specific Dll3 gene only appeared to lose a small portion of right-sided expression distal to the streak (Fig. 5C) and may suggest the loss of a general mesoderm transcriptional program.
Fig. 5.
Targeted pathway analysis reveals widespread dysfunction of nascent mesoderm and FGF pathways. (A) GO analysis performed on down-regulated genes classified as TFs in E7.5 Mesp1Cre-Gli3R embryos. (B) Heatmap generated from dysregulated genes (FDR ≤ 0.30) overlapping with top GO term, Somitogenesis (GO:0001756). (C) In situ hybridization for the somitogenesis genes Msgn1, Tbx6, Mesp2, and Dll1 in control (Mesp1+/+R26Gli3R/+) and Mut (Mesp1Cre/+;R26Gli3R/+) embryos at E7.5. (D) GO analysis for down-regulated genes classified as signaling molecules. (E) Heatmap for dysregulated genes (FDR ≤ 0.30) for the top GO term, FGF Receptor Signaling Pathway (GO:0008543. (F) In situ hybridization for FGF ligands Fgf4, Fgf8, Fgf15, and Fgf3 in control (Mesp1+/+R26Gli3R/+) and Mut (Mesp1Cre/+;R26Gli3R/+) embryos at E7.5. (Scale bars, 200 um.) A, anterior; L, left; P posterior.
The Hh Pathway Is Upstream of FGF Signaling for Anterior Mesoderm Morphogenesis.
We identified FGF signaling as a Hh-dependent signaling pathway in developing mesoderm that may mediate a cell nonautonomous requirement for Hh signaling in anterior mesoderm development. We performed GO analysis for down-regulated genes classified as either signal ligands or ligand receptors in the Fantom consortium database (70) (Fig. 5D). The FGF pathway comprised the top candidate using this approach and differential expression of genes in the GO category “FGF Receptor signaling pathway” (GO:0008543) revealed a striking pattern of down-regulation among all included FGF ligands (FDR ≤ 0.30) (Fig. 5E). The expression of multiple FGF ligand genes were highly down-regulated in the Hh-deficient mesoderm, including Fgf3 (RTE = 0.254, FDR = 3E-16), Fgf4 (RTE = 0.095, FDR = 1.51E-3), Fgf8 (RTE = 0.406, FDR = 4.14E-3), and Fgf15 (RTE = 0.389, FDR = 6.74 E-11) (Fig. 5E). Furthermore, the expression domains of Fgf4 and Fgf8 were qualitatively diminished, specifically loss of their anterior-most expression domains, by in situ hybridization (Fig. 5F).
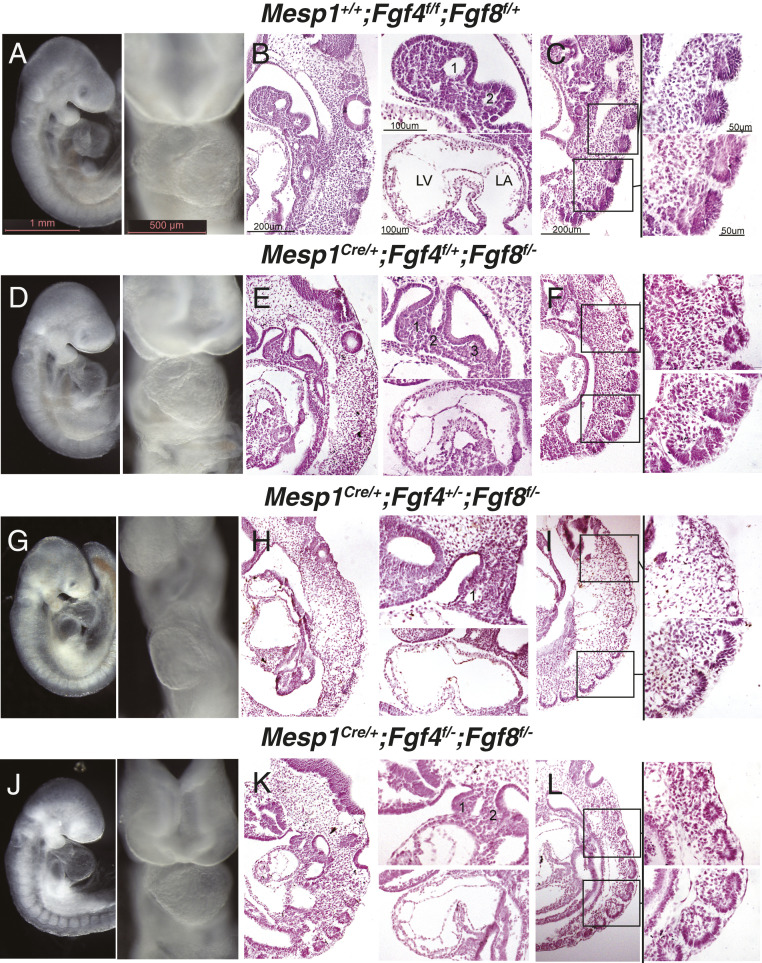
We hypothesized that Fgf4 and Fgf8 may act downstream of Hh signaling are required within the Mesp1 domain for anterior mesoderm development. To test this, we generated multiple allelic combinations for mesoderm-specific Fgf4/Fgf8 loss-of-function, including Mesp1Cre/+;Fgf4f/+;Fgf8f/− (Fig. 6 D–F), Mesp1Cre/+;Fgf4+/−;Fgf8f/− (Fig. 6 G–I), and Mesp1Cre/+;Fgf4f/−;Fgf8f/− embryos (Fig. 6 J–L). In all three mutant genotypes, we observed anterior-selective defects of head, heart, and pharyngeal mesoderm, compared to Mesp1+/+;Fgf4f/f;Fgf8f/+ control embryos (Fig. 6 A–C). The defects to anterior structures included reduced head structures (Fig. 6 D, G, and J), hypoplastic pharyngeal and cardiac mesoderm (Fig. 6 E, H, and K), and anterior somites that were more severely malformed than the posterior somites (Fig. 6 F, I, and L). Taken together, this constellation of defects resembled the anterior-specific defects observed in Hh mutant embryos, albeit with less severity (Figs. 2 and 6).
Fig. 6.
Fgf4 and Fgf8 are required in the Mesp1Cre domain for normal morphogenesis of anterior mesoderm-derived structures. (A, D, G, and J) Right-sided and ventral whole-mount views of representative E9.5 embryos reveal gross defects in the head, pharyngeal arches, hearts, and somites of Mesp1Cre-conditional FGF pathway mutants: (D) Mesp1Cre/+; Fgf4f/+; Fgf8f/−, (G) Mesp1Cre/+;Fgf4+/−;Fgf8f/−, and (J) Mesp1Cre/+;Fgf4f/−;Fgf8f/− compared to A Mesp1+/+;Fgf4f/f; Fgf8+/f control embryos. (B–L) Sagittal H&E sections of embryos shown in whole-mount views. (B, E, H, and K) Low-magnification views with head at top, dorsal to right, and higher-magnification images of pharyngeal arches and left side of hearts, bisecting the atrioventricular canal; pharyngeal arches are numbered. (C, F, I, and L) Low-magnification images of sagittal sections depicting somites (Left); high-magnification views of anterior and posterior somites (boxed) are presented in the adjacent Upper and Lower Right, respectively. Scale bars shown in A–C also apply to the complementary panels of the mutant embryos (D–L). LA, left atrium; LV, left ventricle.
We next assessed the developmental compartment in which FGF and Hh pathways intersect during gastrulation. We reanalyzed a publicly available gastrulation scRNA-seq dataset (40) to assess the expression of FGF ligands Fgf3, Fgf4, Fgf8, and Fgf15 and FGF-dependent genes Dusp6 and Spry4 in addition to Hh pathway components (Fig. 7A and SI Appendix, Fig. S9). Using denoised expression values, FGF ligand expression was largely excluded from the epiblast or primitive streak population with the exception of Fgf8, which was expressed prior to mesoderm specification in the primitive streak. However, the onset of Fgf3, Fgf4, and Fgf15 expression was concomitant with Hh-dependent gene expression (Ptch1) in Mesp1+ nascent mesoderm, as observed in clustered individual cells (Fig. 7A and SI Appendix, Fig. S9) and when aggregated by tissue type (SI Appendix, Fig. S10). In conjunction with the early mesoderm profiling of Mesp1Cre-Gli3R mutants, this pattern of gene expression indicated that Hh signaling was upstream of Fgf3, Fgf4, and Fgf15, which reside in the same topologically associating domain (71, 72).
Fig. 7.
Hh signaling directly regulates Fgf ligand transcript levels within nascent mesoderm. (A) Heatmap displaying denoised expression values for Mesp1, FGF, and Hh pathway genes in single cells from a reanalyzed scRNA-seq dataset spanning anterior mesoderm development (40). (B) IGV Genome browser snapshot of the Fgf3/4/15 locus with peak calls from GLI1 ChIP-seq (30) and SHF ATAC-seq (73) datasets; selected enhancers are highlighted in yellow. (C) Shows relative GLI1-dependant luciferase activity for multiple DNA sequences where “Control” represents a scrambled insertion, “Wt” represents the endogenous enhancer sequence from B and “Mut” represents the ‘WT’ enhancer sequence with poly(T) replacement of GLI-binding sites. Three of four assayed WT enhancers demonstrated significant activation over their paired Mut enhancer (Bonferroni-corrected α = 0.0125).
We hypothesized that Hh signaling directly regulated the expression of Fgf3, Fgf4, and Fgf15. To identify candidate enhancers at the Fgf3/Fgf4/Fgf15 locus, we interrogated GLI3 chromatin immunoprecipitation sequencing (ChIP-seq) and ATAC-seq chromatin accessibility data from publicly available embryonic mesoderm datasets (30, 73). Regions with accessible chromatin and GLI-bound sequences with at least one GLI-binding site within 100 kb upstream of the Fgf4 promoter were selected for analysis (Fig. 7B). Four candidate enhancers were identified based on these criteria and were examined by luciferase assays. These elements were examined for GLI-dependent transcriptional regulation by coexpression with GLI1, an obligate transcriptional activator for Hh pathway-dependent genes, in HEK293T cells. Three of four enhancers showed significant activation by GLI1 compared to paired mutant enhancers, in which endogenous GLI binding sites were replaced by a size-equivalent poly(T) nucleotide sequence (average luciferase activation for WT vs. mutant enhancers: Enhancer 1: 40.3 vs. 1.99, SEM = 0.029, P = 5.19E-8; enhancer 2: 1.04 vs. 0.792, SEM = 0.073, P = 0.038; enhancer 3: 2.26 vs. 1.16, SEM = 0.031, P = 1.37E-4; enhancer 4: 4.71 vs. 0.768, SEM = 0.014, P = 3.97E-7; Bonferroni-corrected α = 0.0125) (Fig. 7C). These findings suggested that FGF ligand expression may be directly controlled by GLI transcription factors.
Cell Migration Defects Caused by Hh Pathway Antagonism Are Rescued by FGF4.
We hypothesized that Hh signaling drives FGF pathway activity for anterior mesoderm migration. Studies in early mammalian development have shown that the FGF pathway directs the migration of nascent mesoderm toward the anterior embryonic pole (59, 74). To assess whether mesoderm cells exhibited migratory defects in Hh mutant mouse embryos, we labeled mesoderm cells using Mesp1Cre in both Smof/− and Smof/+ backgrounds (Fig. 8 A and B). Mesp1Cre/+;Smof/−;R26Rc/+ embryos demonstrated a profound deficiency in Mesp1Cre-labled anterior mesoderm (Fig. 8A) compared to MespCre/+;Smof/+;R26Rc/+ controls (Fig. 8B), which could reflect a requirement for Hh signaling in anterior mesoderm migration.
Fig. 8.
Hh signaling is required for anterior cell migration during gastrulation upstream of the FGF pathway. (A and B) Genetic fate maps for Mesp1Cre in Smof/+ (A) and Smof/− (B) genetic backgrounds using a R26R LacZ reporter allele. (C) Placement of two lipophilic dyes at the posterior portion of a HH3 chicken embryo (dorsal view). (D–F) The trajectory of postingression mesoderm cells in vehicle control-treated chicken embryos (D), where cells positionally mapped (E) and binned (F) across the A-P axis. (G and H) Addition of the Smo antagonist cyclopamine at 25 µM (G) and 50 µM (H). (I) Quantification of inhibitory effect of cyclopamine between 0 µM and 100 µM (***P < 0.001; n.s., not significant). (G–I) Embryos treated with 25 µM of cyclopamine had heparin beads coated with FGF4 placed toward the anterior-right pole of the embryo with BSA-coated heparin beads placed on the anterior-left portion of the embryo to serve as a contralateral control (J). After 16 h of gastrulation, 7 of 11 embryos demonstrated a marked rescue of specific directional migration on the FGF4-treated side (K), which was quantified to significantly differ from the control P = 0.0057 (L). (Scale bars: A and B, 200 μm; D–K, 1 mm.)
We examined whether impaired Hh signaling disrupts mesoderm migration, utilizing a chicken embryonic model of gastrulation. We measured the migratory movements of newly generated mesoderm and endoderm by labeling cells in the chick primitive streak with DiI and DiO at Hamburger–Hamilton (75) stage 3 (HH3). Following 16 h of incubation, we generated a position map of all labeled cells and scored the spread of DiI/DiO along the embryonic A-P axis (Fig. 8C). Embryos treated with DMSO vehicle demonstrated classic anterior-lateral movement of labeled cells consistent with well-established fate and migration maps of the nascent mesoderm (Fig. 8 D–F) (59, 76). We added the Hh pathway antagonist cyclopamine (77) to HH3 chicken embryos, prior to the onset of endogenous Hh signaling in the streak, and observed a dose-dependent reduction of nascent mesendoderm migration (Fig. 8 G–I). Embryos treated with 25 µM cyclopamine demonstrated a significant reduction in DiI/DiO cell distribution across the A-P axis regardless of the initial position of the labels within the primitive streak (P < 0.001) (Fig. 8G). At concentrations of 50 µM cyclopamine or higher migratory defects were compounded by widespread phenotypic disruptions, including greatly reduced embryo size (Fig. 8H).
We attempted to rescue the anterior-specific mesoderm migration effects caused by abrogated Hh signaling using exogenous FGF ligand. To test whether FGF-directed cell migration was downstream of the Hh pathway, we placed beads coated with BSA or FGF4 protein on opposite sides of the anterior midline in 25-µM cyclopamine-treated embryos (Fig. 8J). We determined the effect of FGF4 on migration by monitoring the anterior excursion of DiO or DiI dyes from the primitive streak over a 16-h period of gastrulation. Importantly, labeled cells ipsilateral to the FGF4-coated beads showed a statistically significant increase in A-P spread when compared with cells migrating through the contralateral side of the primitive streak toward BSA-coated control beads (n = 11; P = 0.0057) (Fig. 8 K and L). Thus, the migratory defects resulting from cyclopamine treatment can be mitigated by FGF4. Together, these experiments provide evidence that Hh signaling is required upstream of FGF signaling for the development of anterior mesoderm structures by promoting morphogenetic movements through the primitive streak during gastrulation.
Discussion
Patterning the embryonic axis is one of the earliest and most important morphogenetic events during development. Despite its importance, attempts to study this process in mammals have been fundamentally limited, perhaps because of the complexity of the phenotypes resulting from early embryonic axis disruption. The application of single-cell sequencing can unravel complex patterning defects by independently interrogating distinct cell types in a single assay. Here, we apply single-cell sequencing to interrogate complex patterning defects caused by removal of Hh signaling from the early mesoderm by resolving heterogenous cell admixtures. Leveraging this resource, we uncovered a role for Hh signaling in the development of anterior mesoderm lineages during gastrulation. We next identified a critical FGF pathway for gastrulation downstream of Hh signaling and demonstrate that exogenous FGF replacement can rescue migration defects caused by blocking Hh signaling during gastrulation. These observations indicate that Hh signals originating from the embryonic node are essential for the induction of FGF signals in the nascent mesoderm that, in turn, drive the allocation of mesoderm to developing anterior tissues including the head, heart, pharynx, and anterior somites.
In this study, we implicate FGF signaling as a major Hh-dependent pathway for anterior mesoderm migration. We also demonstrated that deletion of Fgf4/8 within the Mesp1Cre domain mimics the anterior-selective defects seen in embryos with decreased Hh signaling in the Mesp1Cre domain (Figs. 2 and 6). Multiple FGF ligands were down-regulated in Mesp1Cre-Gli3R embryos and several of them of them share genomic loci proximal to GLI-dependent enhancers (Fig. 7 B and C). Finally, we show that Hh-dependent migratory defects can be rescued by direct addition of FGF4 ligand (Fig. 8 K and L). Together, these data suggest that the Hh signaling controls anterior mesoderm development through an FGF pathway for mesoderm migration. This work is consistent with prior investigations into the role of FGF signaling in early embryonic development. For example, Fgf8 and Fgfr1 germline mutants exhibit defects to both cell migratory behavior during gastrulation and anterior tissue development (60, 74). Although our work does not preclude a role for Hh signaling upstream of FGF-directed mitogenic activity, our proposed mechanism is in line with both the described role for the FGF pathway in embryonic tissue migration (59, 60, 74) and the observation that migratory defects during gastrulation culminate selective defects to anterior mesendoderm-derived tissues (78).
The compendium of gene networks downstream of Hh signaling required for anterior mesoderm development is complex. It is notable that Mesp1Cre-conditional knockout of Fgf4/8 ligands produces milder phenotypes than Mesp1Cre-conditional knockout of Smo or overexpression of Gli3R (Figs. 2 and 6). This observation indicates that other FGF ligands or independent Hh-dependent pathways are functionally required in addition to Fgf4 and Fgf8 in anterior mesoderm development. Consistently, we observed that several other FGF ligands are expressed in the nascent mesoderm in a Hh-dependent manner, including Fgf3 and Fgf15, which may share GLI-dependent enhancers at the Fgf3, Fgf4, Fgf15 locus (Figs. 5 and 7). Furthermore, we identified candidate Hh-dependent genes for anterior mesoderm development that are outside the FGF pathway, including, Tdgf1, Smad3, FoxA2, and T, all of which are down-regulated in Mesp1Cre-Gli3R mutants at E7.5, and have been previously implicated in A-P mesoderm patterning and axis formation (Fig. 4 C and E) (61–64, 79). Future studies focused on genetic interactions between the Hh pathway and these genes will yield insight into the gene regulatory networks required for anterior mesoderm development.
Perhaps the most intriguing inference of this work involves the link between signals originating from the node and anterior mesoderm morphogenesis. The importance of Hh signaling in embryonic axis patterning beyond L-R patterning was first demonstrated by compound mutations to Shh and Ihh, which have overlapping expression in the node (22). However, the role of the node itself in embryonic axis formation has been controversial. The first transplantation study of the mammalian node demonstrated its potential to induce ectopic neuroectoderm and somite formation in the epiblast of late-gastrulation embryos (80). However, much of the later debate about the node’s potential as an anterior organizer centered on the ability to ectopically induce anterior neuroectoderm in similar transplantation experiments (81–83). Although the node does not appear to be required for endogenous neurectoderm formation and patterning (84), these prior studies reached the limited conclusion that the node’s role as an organizer is principally in L-R determination. Attempts to evaluate the effect of physical node ablation did not directly assay the effect on embryonic A-P patterning and were performed after the onset of Hh pathway activity (85). However, absolute loss of node formation through genetic ablation Foxa2 (HNF-3β) caused profound defects to anterior tissues and truncation of anterior structures beyond the otic vesicle (62, 63, 84). Although deletion of Foxa2 may also result in node-independent defects, this work is consistent with a role for the embryonic node in A-P patterning. Our work suggests that combined Ihh/Shh expression in the node produces anterior organizing signals through downstream control of the FGF pathway in nascent mesoderm.
In this study we demonstrate that Hh signaling in the early embryo serves a previously uncharacterized role in the development of specific anterior mesoderm lineages. We note that cardiac, cranial, pharyngeal, and anterior somitic mesoderm migrate through the primitive streak at approximately the same time as Hh pathway activity arises within the embryonic node (7). We provide a mechanistic link between Hh signaling from the node and anterior mesoderm development by establishing the Hh-dependence of FGF pathway activity for cell migration during gastrulation (Fig. 9A). Perturbations to early Hh signaling diminish the expression of migratory FGF signals within the nascent mesoderm, leading to A-P patterning defects through dysfunctional mesoderm migration (Fig. 9B). This study demonstrates a functional link between Hh signaling from the embryonic node and FGF signaling in the nascent mesoderm for the formation of anterior embryonic structures. Furthermore, we provide evidence that signals from the mammalian node can influence embryonic A-P axis patterning in addition to its role in L-R determination. These findings have the potential to describe a general mechanism by which the node and primitive streak coordinate proper A-P patterning during gastrulation.
Fig. 9.
Midline Hh signaling from the node drives an FGF pathway for anterior mesoderm morphogenesis. (A and B) A diagrammatic representation for the role of Hh signaling in anterior mesoderm patterning. (A) In WT embryos, Hh signaling is active early in embryonic node which is required for the full activation of a midline FGF pathway activity in the nascent mesoderm to drives mesoderm migration. (B) In Hh pathway mutants, attenuation of FGF pathway activity disrupts the migration and patterning of anterior mesoderm lineages. Ant, anterior; Post, posterior.
Materials and Methods
Mouse Lines.
Gli1CreERT2 (54) mice were obtained from the Joyner laboratory (Sloan Kettering Institute, New York, NY). Rosa26Gli3R-IRES-Venus (30), Rosa26Lacz (45), Rosa26tdTomato (32), PtchLZ (26), Shh− (86), Smo− (22), and Smof (52) lines were obtained from the Jackson Laboratory. Mef2cAHF-Cre (44) and Mesp1Cre (29) lines were reported previously. The above mouse lines were maintained on a mixed genetic background later outcrossing to CD-1 lines for multiple generations and housed at the University of Chicago (Institutional Animal Care and Use Committee protocol #71737). Fgf4− and Fgf8− mice were generated by germline Cre recombination of Fgf4f (87) and Fgf8f (88) mice, respectively. Fgf4/8 mouse lines were maintained on a mixed C57BL/6J-SV129 background and housed at the Weis Center for Research (Institutional Animal Care and Use Committee protocol #203). Mice were genotyped by PCR according to specifications from the Jackson laboratory or previous publications. See SI Appendix for full details.
TM Administration and X-Gal Staining.
TM-induced activation of Gli1CreERT2 was accomplished by intraperitoneal injection of a 2 g:1 g TM (MP Biomedicals):progesterone (Sigma) mixture in corn oil (Sigma). X-gal staining of β-gal-expressing embryos was performed as previously described (21); see SI Appendix for full details.
Histology.
Embryos were fixed in 4% paraformaldehyde, embedded in paraffin wax, and sectioned to 5-μM thickness. For normal histology, tissue was counter-stained with H&E. For histology sections of X-gal–stained embryos, tissue was counter-stained with Nuclear Fast Red. Histologic sections for Figs. 1–3 were processed by the University of Chicago Human Tissue Resource Center. Histologic sections for Fig. 6 were processed by the A.M.M. laboratory at the Weis Center for Research.
Transcriptional Profiling of Early Embryos.
Embryos used for bulk transcriptome profiling were harvested at E7.5 in 1× ice-cold PBS, pooled with their littermates, dissociated with TrypLE (Fisher) reagent for 5 min at 37 °C while shaking at 1,400 rpm in a Fisher Thermomixer, followed by inactivation by addition of 10% FBS in 1× PBS. Cells were spun down at 800 × g for 5 min, resuspended in 10% FBS with a Near-IR dead dye (Life Technologies), and strained through a 40-µm mesh prior to FACS. Next, 1,000 live cells were sorted directly into cell lysis buffer from either the tdTomato or YFP channel for biological replicates of control (Mesp1Cre/+;R26tdTomato/+) and mutant (Mesp1Cre/+;R26Gli3R-IRES-Venus/+) cells, respectively. cDNA libraries were generated using SMARTer Ultra Low RNA Kit for Illumina sequencing (Clontech) and sequencing libraries were constructed using Nextera XT DNA Library Preparation Kit (Illumina). Quality control was performed both after cDNA synthesis and library preparation using a Bioanalyzer (Agilent). RNA-seq Libraries were sequenced on the HiSeq2500 in the University of Chicago Functional Genomics Facility.
Bulk RNA-Seq Data Analysis.
FASTQ files were aligned to the mm9 Mus musculus genome using TopHat2 (89) running standard parameters. FeatureCounts in the SubRead (90) package was used to generate read counts from the aligned bam files and subsequently analyzed using edgeR (91) for differential expression. Significant differentially expressed genes between mutant Mesp1Cre/+;R26Gli3R-IRES-Venus/+ (n = 3) and control Mesp1Cre/+;R26tdTomato/+ (n = 3) embryos were identified using a Benjamini–Hochberg (92) -corrected statistical thresholds of FDR ≤ 0.10. Differentially expressed genes were also used to identify associated GO terms using Panther classification system (93). See SI Appendix for full details.
In Situ Hybridization.
Mouse in situ hybridization was performed as previously described (21), with a reduction in Proteinase K digestion time to 1 min for early embryos. See SI Appendix for full details.
Drop-Seq.
Embryos were harvested at E8.25 in ice-cold 1× PBS. Between 7 and 13 litters were pooled for each biological replicate. Embryo dissociation and was performed as described for E7.5 embryos, except with a 10-min TrypLE digest. Mesp1Cre-marked cells were isolated by FACS and collected in cold 1× PBS; 0.01% BSA solution. Cell processing on the Drop-seq platform and subsequent data analysis was performed as previously described (94). See SI Appendix for full details.
Luciferase Assays.
pCIG expression vectors for Gli1 were previously described (19). Putative GLI-dependent enhancers were cloned into the pGL4.23 vectors (Promega). Expression and reporter vectors were transfected into HEK293T cells using FuGENE (Promega). Luciferase activity was measured relative to cells transfected with empty pCIG vectors. Cells were cultured for 48 h after transfection and then were lysed and assayed using the Dual-Luciferase Reporter Assay System (Promega). All technical replicates were performed in triplicate. See SI Appendix for full details.
Migration Assay in Chicken Embryo.
Chicken embryos were isolated at HH2/3 and cultured ventral-side up on albumin agar plates according to the technique described in Chapman et al. (95). DiI and DiO (ThermoFisher Scientific) were microinjected through the ventral endoderm proximal to the primitive streak using a Femotjet pressure injector and Injectman micromanipulator (Eppendorf), as described in Bressan et al. (76). Following 16 h of incubation, embryos were photographed on a Lecia M165 FC fluorescent stereo microscope using a Hamamatsu Orca-flash 4.0 camera. A-P spread was quantified using ImageJ software (v2.0.0). Briefly, the center-of-mass for all fluorescent particles present within the embryonic area pellucida was calculated and this position was normalized to the posterior extent of the embryonic midline. For Hh inhibition studies, cyclopamine (Sigma-Aldrich) was dissolved in 45% cyclodextrin (Sigma-Aldrich) Pannet/Compton’s saline at concentrations, described in the text. Next, 500 μL of cyclopamine solution was then added to each embryo. For FGF rescue experiments, Heparin-agarose beads (Sigma-Aldrich) were soaked for 1 h at room temperature in either 1 mg/mL BSA or FGF4 (R&D Systems). Beads were then implanted between the epiblast and hypoblast of HH2/3 embryos as depicted in Fig. 6 just prior to treatment with 25 μM cyclopamine.
Data Deposition.
Bulk and single-cell RNA-sequencing data have been deposited in the Gene Expression Omnibus (accession nos. GSE147868 for RNA-seq for E7.5 Hh-deficient mesoderm and GSE149335 for the Drop-seq experiment for E8.25 Hh-deficient mesoderm).
Supplementary Material
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Both bulk and single-cell RNA-seq data sets have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE147868 and GSE149335).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914167117/-/DCSupplemental.
References
- 1.Tam P. P., Behringer R. R., Mouse gastrulation: The formation of a mammalian body plan. Mech. Dev. 68, 3–25 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Martinez V., Schoenwolf G. C., Primitive-streak origin of the cardiovascular system in avian embryos. Dev. Biol. 159, 706–719 (1993). [DOI] [PubMed] [Google Scholar]
- 3.Lawson K. A., Meneses J. J., Pedersen R. A., Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development 113, 891–911 (1991). [DOI] [PubMed] [Google Scholar]
- 4.Tam P. P., Parameswaran M., Kinder S. J., Weinberger R. P., The allocation of epiblast cells to the embryonic heart and other mesodermal lineages: The role of ingression and tissue movement during gastrulation. Development 124, 1631–1642 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Parameswaran M., Tam P. P., Regionalisation of cell fate and morphogenetic movement of the mesoderm during mouse gastrulation. Dev. Genet. 17, 16–28 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Tam P. P., Loebel D. A., Gene function in mouse embryogenesis: Get set for gastrulation. Nat. Rev. Genet. 8, 368–381 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Kinder S. J., et al., The orderly allocation of mesodermal cells to the extraembryonic structures and the anteroposterior axis during gastrulation of the mouse embryo. Development 126, 4691–4701 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Arnold S. J., Robertson E. J., Making a commitment: Cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 10, 91–103 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Irimia M., et al., Comparative genomics of the Hedgehog loci in chordates and the origins of Shh regulatory novelties. Sci. Rep. 2, 433 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ingham P. W., Placzek M., Orchestrating ontogenesis: Variations on a theme by sonic hedgehog. Nat. Rev. Genet. 7, 841–850 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Nüsslein-Volhard C., Wieschaus E., Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801 (1980). [DOI] [PubMed] [Google Scholar]
- 12.Briscoe J., Thérond P. P., The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14, 416–429 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Robbins D. J., Fei D. L., Riobo N. A., The Hedgehog signal transduction network. Sci. Signal. 5, re6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falkenstein K. N., Vokes S. A., Transcriptional regulation of graded Hedgehog signaling. Semin. Cell Dev. Biol. 33, 73–80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabata T., Kornberg T. B., Hedgehog is a signaling protein with a key role in patterning Drosophila imaginal discs. Cell 76, 89–102 (1994). [DOI] [PubMed] [Google Scholar]
- 16.Walton K. D., Warner J., Hertzler P. H., McClay D. R., Hedgehog signaling patterns mesoderm in the sea urchin. Dev. Biol. 331, 26–37 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tickle C., Towers M., Sonic hedgehog signaling in limb development. Front. Cell Dev. Biol. 5, 14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balaskas N., et al., Gene regulatory logic for reading the Sonic Hedgehog signaling gradient in the vertebrate neural tube. Cell 148, 273–284 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann A. D., et al., Foxf genes integrate tbx5 and hedgehog pathways in the second heart field for cardiac septation. PLoS Genet. 10, e1004604 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie L., et al., Tbx5-hedgehog molecular networks are essential in the second heart field for atrial septation. Dev. Cell 23, 280–291 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann A. D., Peterson M. A., Friedland-Little J. M., Anderson S. A., Moskowitz I. P., Sonic hedgehog is required in pulmonary endoderm for atrial septation. Development 136, 1761–1770 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X. M., Ramalho-Santos M., McMahon A. P., Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R symmetry by the mouse node. Cell 106, 781–792 (2001). [PubMed] [Google Scholar]
- 23.Vokes S. A., et al., Hedgehog signaling is essential for endothelial tube formation during vasculogenesis. Development 131, 4371–4380 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Thomas N. A., Koudijs M., van Eeden F. J., Joyner A. L., Yelon D., Hedgehog signaling plays a cell-autonomous role in maximizing cardiac developmental potential. Development 135, 3789–3799 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsukui T., et al., Multiple left-right asymmetry defects in Shh(-/-) mutant mice unveil a convergence of the shh and retinoic acid pathways in the control of Lefty-1. Proc. Natl. Acad. Sci. U.S.A. 96, 11376–11381 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodrich L. V., Milenković L., Higgins K. M., Scott M. P., Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277, 1109–1113 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Daane J. M., Downs K. M., Hedgehog signaling in the posterior region of the mouse gastrula suggests manifold roles in the fetal-umbilical connection and posterior morphogenesis. Dev. Dyn. 240, 2175–2193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saga Y., Kitajima S., Miyagawa-Tomita S., Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc. Med. 10, 345–352 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Saga Y., et al., MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development 126, 3437–3447 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Vokes S. A., Ji H., Wong W. H., McMahon A. P., A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 22, 2651–2663 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macosko E. Z., et al., Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madisen L., et al., A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaufman M. H., Bard J. B., The Anatomical Basis of Mouse Development (Gulf Professional Publishing, 1999). [Google Scholar]
- 34.Ibarra-Soria X., et al., Defining murine organogenesis at single-cell resolution reveals a role for the leukotriene pathway in regulating blood progenitor formation. Nat. Cell Biol. 20, 127–134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stuart T., et al., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoder M. C., Inducing definitive hematopoiesis in a dish. Nat. Biotechnol. 32, 539–541 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Devine W. P., Wythe J. D., George M., Koshiba-Takeuchi K., Bruneau B. G., Early patterning and specification of cardiac progenitors in gastrulating mesoderm. eLife 3, e03848 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lescroart F., et al., Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat. Cell Biol. 16, 829–840 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pijuan-Sala B., et al., A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ang S. L., Conlon R. A., Jin O., Rossant J., Positive and negative signals from mesoderm regulate the expression of mouse Otx2 in ectoderm explants. Development 120, 2979–2989 (1994). [DOI] [PubMed] [Google Scholar]
- 42.Reijntjes S., Stricker S., Mankoo B. S., A comparative analysis of Meox1 and Meox2 in the developing somites and limbs of the chick embryo. Int. J. Dev. Biol. 51, 753–759 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Blanar M. A., et al., Meso1, a basic-helix-loop-helix protein involved in mammalian presomitic mesoderm development. Proc. Natl. Acad. Sci. U.S.A. 92, 5870–5874 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verzi M. P., McCulley D. J., De Val S., Dodou E., Black B. L., The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev. Biol. 287, 134–145 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Soriano P., Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71 (1999). [DOI] [PubMed] [Google Scholar]
- 46.Kelly R. G., Buckingham M. E., Moorman A. F., Heart fields and cardiac morphogenesis. Cold Spring Harb. Perspect. Med. 4, a015750 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Q., Reiter R. S., Huang Q. Q., Jin J. P., Lin J. J., Comparative studies on the expression patterns of three troponin T genes during mouse development. Anat. Rec. 263, 72–84 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Ruiz J. C., Conlon F. L., Robertson E. J., Identification of novel protein kinases expressed in the myocardium of the developing mouse heart. Mech. Dev. 48, 153–164 (1994). [DOI] [PubMed] [Google Scholar]
- 49.Watanabe Y., et al., Fibroblast growth factor 10 gene regulation in the second heart field by Tbx1, Nkx2-5, and Islet1 reveals a genetic switch for down-regulation in the myocardium. Proc. Natl. Acad. Sci. U.S.A. 109, 18273–18280 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Francou A., et al., Second heart field cardiac progenitor cells in the early mouse embryo. Biochim. Biophys. Acta 1833, 795–798 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Vincent S. D., Buckingham M. E., How to make a heart: The origin and regulation of cardiac progenitor cells. Curr. Top. Dev. Biol. 90, 1–41 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Long F., Zhang X. M., Karp S., Yang Y., McMahon A. P., Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development 128, 5099–5108 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Yamagishi C., et al., Sonic hedgehog is essential for first pharyngeal arch development. Pediatr. Res. 59, 349–354 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Ahn S., Joyner A. L., Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell 118, 505–516 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Dyer L. A., Kirby M. L., Sonic hedgehog maintains proliferation in secondary heart field progenitors and is required for normal arterial pole formation. Dev. Biol. 330, 305–317 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goddeeris M. M., et al., Intracardiac septation requires hedgehog-dependent cellular contributions from outside the heart. Development 135, 1887–1895 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nowotschin S., Ferrer-Vaquer A., Concepcion D., Papaioannou V. E., Hadjantonakis A. K., Interaction of Wnt3a, Msgn1 and Tbx6 in neural versus paraxial mesoderm lineage commitment and paraxial mesoderm differentiation in the mouse embryo. Dev. Biol. 367, 1–14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boulet A. M., Capecchi M. R., Signaling by FGF4 and FGF8 is required for axial elongation of the mouse embryo. Dev. Biol. 371, 235–245 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang X. S., Dormann D., Munsterberg A. E., Weijer C. J., Cell movement patterns during gastrulation in the chick are controlled by positive and negative chemotaxis mediated by FGF4 and FGF8. Dev. Cell 3, 425–437 (2002). [DOI] [PubMed] [Google Scholar]
- 60.Sun X., Meyers E. N., Lewandoski M., Martin G. R., Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes Dev. 13, 1834–1846 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ding J., et al., Cripto is required for correct orientation of the anterior-posterior axis in the mouse embryo. Nature 395, 702–707 (1998). [DOI] [PubMed] [Google Scholar]
- 62.Ang S. L., Rossant J., HNF-3 beta is essential for node and notochord formation in mouse development. Cell 78, 561–574 (1994). [DOI] [PubMed] [Google Scholar]
- 63.Weinstein D. C., et al., The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell 78, 575–588 (1994). [DOI] [PubMed] [Google Scholar]
- 64.Beddington R. S. P., Rashbass P., Wilson V., Brachyury—A gene affecting mouse gastrulation and early organogenesis. Dev. Suppl., 157–165 (1992). [PubMed] [Google Scholar]
- 65.Sewell W., et al., Cyclical expression of the Notch/Wnt regulator Nrarp requires modulation by Dll3 in somitogenesis. Dev. Biol. 329, 400–409 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rowton M., et al., Regulation of mesenchymal-to-epithelial transition by PARAXIS during somitogenesis. Dev. Dyn. 242, 1332–1344 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Burgess R., Rawls A., Brown D., Bradley A., Olson E. N., Requirement of the paraxis gene for somite formation and musculoskeletal patterning. Nature 384, 570–573 (1996). [DOI] [PubMed] [Google Scholar]
- 68.Chan T., et al., Ripply2 is essential for precise somite formation during mouse early development. FEBS Lett. 581, 2691–2696 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Bessho Y., et al., Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 15, 2642–2647 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramilowski J. A., et al., A draft network of ligand-receptor-mediated multicellular signalling in human. Nat. Commun. 6, 7866 (2015). Correction in: Nat. Commun.7, 10706 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dixon J. R., et al., Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nora E. P., et al., Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rowton M., et al., Hedgehog signaling controls progenitor differentiation timing during heart development. bioRxiv:10.1101/270751 (23 February 2018).
- 74.Ciruna B., Rossant J., FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev. Cell 1, 37–49 (2001). [DOI] [PubMed] [Google Scholar]
- 75.Hamburger V., Hamilton H. L., A series of normal stages in the development of the chick embryo. J. Morphol. 88, 49–92 (1951). [PubMed] [Google Scholar]
- 76.Bressan M., Liu G., Mikawa T., Early mesodermal cues assign avian cardiac pacemaker fate potential in a tertiary heart field. Science 340, 744–748 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen J. K., Taipale J., Cooper M. K., Beachy P. A., Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 16, 2743–2748 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pauli A., et al., Toddler: An embryonic signal that promotes cell movement via Apelin receptors. Science 343, 1248636 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tanaka S., Kunath T., Hadjantonakis A. K., Nagy A., Rossant J., Promotion of trophoblast stem cell proliferation by FGF4. Science 282, 2072–2075 (1998). [DOI] [PubMed] [Google Scholar]
- 80.Beddington R. S., Induction of a second neural axis by the mouse node. Development 120, 613–620 (1994). [DOI] [PubMed] [Google Scholar]
- 81.Kinder S. J., et al., The organizer of the mouse gastrula is composed of a dynamic population of progenitor cells for the axial mesoderm. Development 128, 3623–3634 (2001). [DOI] [PubMed] [Google Scholar]
- 82.Tam P. P., Steiner K. A., Anterior patterning by synergistic activity of the early gastrula organizer and the anterior germ layer tissues of the mouse embryo. Development 126, 5171–5179 (1999). [DOI] [PubMed] [Google Scholar]
- 83.Yang Y. P., Klingensmith J., Roles of organizer factors and BMP antagonism in mammalian forebrain establishment. Dev. Biol. 296, 458–475 (2006). [DOI] [PubMed] [Google Scholar]
- 84.Klingensmith J., Ang S. L., Bachiller D., Rossant J., Neural induction and patterning in the mouse in the absence of the node and its derivatives. Dev. Biol. 216, 535–549 (1999). [DOI] [PubMed] [Google Scholar]
- 85.Davidson B. P., Kinder S. J., Steiner K., Schoenwolf G. C., Tam P. P., Impact of node ablation on the morphogenesis of the body axis and the lateral asymmetry of the mouse embryo during early organogenesis. Dev. Biol. 211, 11–26 (1999). [DOI] [PubMed] [Google Scholar]
- 86.St-Jacques B., et al., Sonic hedgehog signaling is essential for hair development. Curr. Biol. 8, 1058–1068 (1998). [DOI] [PubMed] [Google Scholar]
- 87.Moon A. M., Boulet A. M., Capecchi M. R., Normal limb development in conditional mutants of Fgf4. Development 127, 989–996 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Park E. J., et al., Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development 133, 2419–2433 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim D., et al., TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 91.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Benjamini Y., Hochberg Y., Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995). [Google Scholar]
- 93.Mi H., et al., PANTHER version 11: Expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45, D183–D189 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Selewa A., et al., Systematic comparison of high-throughput single-cell and single-nucleus transcriptomes during cardiomyocyte differentiation. Sci. Rep. 10, 1535 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chapman S. C., Collignon J., Schoenwolf G. C., Lumsden A., Improved method for chick whole-embryo culture using a filter paper carrier. Dev. Dyn. 220, 284–289 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.