Significance
All bacteria inevitably encounter conditions that are not conducive to growth and, in response, they can enter into quiescent phenotypes. The nucleotides guanosine tetraphosphate and pentaphosphate ((p)ppGpp) play a key role in this response by inhibiting DNA replication, GTP biosynthesis, and transcription. (p)ppGpp also mediates down-regulation of protein synthesis by reducing ribosome biogenesis. However, whether (p)ppGpp also regulates active translation, the most energy intensive process in the cell, during quiescence is not known. Here, we show that (p)ppGpp directly inhibits translation initiation when bacteria stop growing rapidly by binding the essential GTPase IF2. The present study identifies translation initiation as a regulatory target during quiescence in bacteria and establishes the mechanistic basis of this regulation.
Keywords: protein synthesis, dormancy, IF2, translational regulation
Abstract
Many bacteria exist in a state of metabolic quiescence where energy consumption must be minimized so as to maximize available resources over a potentially extended period of time. As protein synthesis is the most energy intensive metabolic process in a bacterial cell, it would be an appropriate target for down-regulation during the transition from growth to quiescence. We observe that when Bacillus subtilis exits rapid growth, a subpopulation of cells emerges with very low protein synthetic activity. This phenotypic heterogeneity requires the production of the nucleotides (p)ppGpp, which we show are sufficient to inhibit protein synthesis in vivo. We then show that one of these molecules, ppGpp, inhibits protein synthesis by preventing the allosteric activation of the essential GTPase Initiation Factor 2 (IF2) during translation initiation. Finally, we demonstrate that the observed attenuation of protein synthesis during the entry into quiescence is a consequence of the direct interaction of (p)ppGpp and IF2.
Most microbial life exists in a nonproliferating state of quiescence that enables survival during nutrient limitation and in stressful environments (1, 2). A major challenge facing a quiescent cell is how to minimize energy consumption so as to maximize available resources over a potentially extended period of time. As protein translation is the most energy intensive metabolic process in a cell, accounting for as much as ∼70% of total energy consumption in bacteria (3, 4), it would be an appropriate target for down-regulation during the transition from growth to quiescence. Metabolic conditions that may be present during this transition such as amino acid limitation would reduce protein translation, but whether it is also actively inhibited remains an open question.
Several highly conserved, essential GTPases participate in translation (5) and are appealing regulatory targets for quiescence-dependent attenuation. In the bacterium Bacillus subtilis undergoing sporulation, phosphorylation of EF-Tu inhibits its ability to hydrolyze GTP, an activity essential for its function and, thereby, blocks translation (6). How else could these GTPases be targeted? Binding of a ligand other than GTP to the GTP binding site could affect protein function. For example, in vitro, the hyperphosphorylated nucleotides guanosine tetraphosphate and pentaphosphate can bind translational GTPases (7). Collectively termed (p)ppGpp, these alarmones mediate the stringent response, the mechanism used by bacteria to coordinate a response to cell stresses including nutrient deprivation (8, 9). The primary source of (p)ppGpp in most bacteria is the ribosome-associated RelA synthase that is stimulated by the presence of uncharged transfer RNAs (tRNAs) in the ribosome A site, presumably reflective of amino acid starvation. (p)ppGpp synthesis results in the down-regulation of metabolic processes including transcription, replication, GTP synthesis, and ribosome assembly. These effects are mediated by direct interactions of (p)ppGpp with RNA polymerase in Gram-negative bacteria (10, 11), DNA primase (12), the GTP biosynthetic enzymes HprT and Gmk (13), and several ribosome assembly factors (14), respectively. (p)ppGpp inhibits both transcription of ribosomal RNAs and proteins (15–18) and ribosome assembly, any of which would reduce ribosome number and, thereby, overall protein synthesis. However, other than several early, intriguing observations (19, 20), much less attention has been paid to the large number of fully formed active ribosomes and if a direct (p)ppGpp-dependent mechanism inhibits their activity. Furthermore, although myriad translational- and ribosome-associated GTPases have been shown in vitro to be inhibited by (p)ppGpp, the identification of a verified in vivo target and its role during quiescence related attenuation requires further study.
Consistent with an inhibitory effect on translation, overexpression of a truncated RelA protein that synthesizes (p)ppGpp in the absence of amino acid limitation results in a rapid decrease in [35S]methionine incorporation (21). However, further investigation of a direct effect on translation in vivo has been complicated by several factors. First, (p)ppGpp affects synthesis of ribosomal proteins and RNAs in Escherichia coli at least in part by inhibiting RNA polymerase (22). Second, studies examining the effect of (p)ppGpp produced by RelA typically use compounds that greatly increase the cellular pools of uncharged tRNAs in order to stimulate RelA (20, 23–26). However, as these compounds themselves directly arrest translation, they are not appropriate for investigating (p)ppGpp effects.
The ability of (p)ppGpp to inhibit translation in vitro similarly to nonhydrolysable GTP analogs (27) suggests that (p)ppGpp targets a translational GTPase. Consistently, (p)ppGpp inhibits the GTPase activity of IF2 and EF-Tu (7) as well as several GTPases involved in ribosome assembly (14, 28), by acting as a competitive inhibitor. Furthermore, (p)ppGpp is capable of binding translational GTPases including EF-Tu, EF-G (29), IF2 (30, 31), and the ribosome assembly GTPase ObgE (32) at affinities commensurate with the reported in vivo concentration. Of note, the affinity of IF2 for (p)ppGpp as compared to EF-G (31) and the observation that (p)ppGpp interferes with IF2 function (30) suggests that IF2 may be a principal target in vivo. However, this is not known.
Here, we show that during the transition into a nonproliferative state, B. subtilis exhibits a large reduction in protein synthesis that is dependent on (p)ppGpp. We further show that (p)ppGpp inhibits protein synthesis in vivo and in vitro by targeting translation directly. Next, we identify mutations in IF2 that allow us to demonstrate in vitro that it is a direct target of (p)ppGpp during translation. We then show that binding of ppGpp fails to allosterically stabilize a conformation of IF2 that is typically triggered by binding of GTP and that enables IF2 to stably bind the ribosomal small, or 30S, subunit initiation complex (IC) and catalyze rapid joining of the ribosomal large, or 50S, subunit to the 30S IC. Finally, we demonstrate in vivo that binding of (p)ppGpp to IF2 mediates the observed (p)ppGpp-dependent inhibition of protein synthesis as B. subtilis exits growth.
Results
Protein Synthesis Is Inhibited in a (p)ppGpp-Dependent Manner during Stationary Phase.
B. subtilis grows exponentially in Lysogeny broth (LB) until a stereotypic cell density when growth slows in the transition phase, ultimately culminating in the nonproliferative state of stationary phase (Fig. 1A). We assayed protein synthesis during these different growth phases by measuring incorporation of the puromycin analog O‐propargyl‐puromycin (OPP) that can be visualized and quantified in single cells following addition of a fluorophore using click chemistry (33). Incorporation of OPP results in the accumulation of fluorescently tagged nascent polypeptide chains that directly reflects the rate of translation (33). Although puromycin causes premature termination of protein synthesis resulting from its incorporation into the nascent polypeptide chain, global protein synthesis continues following the addition of OPP under our conditions (SI Appendix, Fig. S1). Consistently, the concentration of OPP used (13 μM) is ∼3× lower than the IC50 of puromycin (34), and OPP is ∼3× less potent than puromycin in inhibiting protein synthesis (33). Other methods for measuring protein synthesis exist, but they rely on growth in the absence of amino acids (specifically methionine). This poses a particular problem for strains which exhibit auxotrophies to these amino acids, such as those lacking (p)ppGpp (35).
Fig. 1.
(p)ppGpp mediates inhibition of protein synthesis upon departure from growth. B. subtilis (JDB 1772) cells were labeled with OPP at different time points following exponential phase. (A) B. subtilis WT and isogenic (p)ppGpp0 (JDB 4294) strains grow equivalently to each other during exponential and transition phases, but (p)ppGpp0 strain lyses upon entry into stationary phase (means ± SDs). (B) Representative pictures of WT (Upper) and (p)ppGpp0 (Lower) strains labeled with OPP at different time points. (C) Distributions of mean-cell fluorescence of WT (gray) and (p)ppGpp0 (blue) cells. Time points in B and C are indicated by black dashed lines in A.
We labeled B. subtilis cultures at a series of time points during growth in LB (Fig. 1A; dashed black lines). As expected, cells exhibited a decrease in OPP incorporation soon after departure from exponential growth (early transition) that continued during late transition (Fig. 1 B, Upper). This trend is apparent in the average cellular fluorescence at these time points as well as in stationary phase (SI Appendix, Fig. S2A, gray bars). However, at the late transition time point, a substantial fraction of cells in the population appeared to lose all fluorescent signal (Fig. 1 B, Upper). This loss of signal indicates an absence of protein synthesis, resulting in a population whose single-cell distribution of protein synthetic activity is roughly bimodal (Fig. 1C; gray). This heterogeneity is consistent with the inhibition of protein synthesis being a direct effect in a subpopulation of cells rather than a consequence of amino acid limitation in the growth medium that would be expected to cause a homogenous effect across the entire population. That is, a subpopulation of cells in the late transition phase culture exhibited a near total inhibition of protein synthesis, whereas a separate subpopulation exhibited relatively greater protein synthesis activity (Fig. 1C, gray).
We speculated that a mechanism known to control growth rate might be important for the shutdown of protein synthesis in postexponential growth. An attractive candidate is (p)ppGpp, a molecule involved in regulating diverse processes that affect cells growing under suboptimal conditions (36). Furthermore, many bacteria synthesize (p)ppGpp when they depart from exponential phase (37, 38). To investigate the possible role of (p)ppGpp, we utilized a strain that lacks (p)ppGpp via genetic deletion of the three known B. subtilis (p)ppGpp synthetases, relA, sasA, and sasB (39, 40). Under our growth conditions, this strain ((p)ppGpp0) grows equivalently to the parent WT strain during exponential phase as well as early and late transition phase. Incorporation of OPP in this strain in exponential phase is indistinguishable from the parent (Fig. 1 B and C), indicating similar protein synthesis activity. In contrast, early in transition phase, some WT cells incorporated substantially less OPP than (p)ppGpp0 cells. This trend continues late in transition phase where a significant number of cells in the (p)ppGpp0 strain (blue) have higher rates of protein synthesis compared to the WT parent (gray). This effect is evident although cells lacking (p)ppGpp do not display a different growth rate up to this point (Fig. 1 B and C and SI Appendix, Fig. S2 A and B). We further confirmed the difference between the WT and (p)ppGpp0 strains using a fluorescently tagged methionine analog (HPG). Although incorporation of this analog is not very efficient in media containing methionine ((p)ppGpp0 strain requires methionine to grow; ref. 35), the (p)ppGpp0 strain incorporated comparatively more HPG during late transition phase, consistent with a higher rate of protein synthesis (SI Appendix, Fig. S2C). Thus, taken together, these data demonstrate that (p)ppGpp is necessary for the observed inhibition of protein synthesis during late transition phase.
(p)ppGpp Is Sufficient to Inhibit Protein Synthesis.
We investigated whether production of (p)ppGpp is sufficient to inhibit protein synthesis by introducing an inducible (p)ppGpp synthetase into the chromosome of B. subtilis. As previously observed (41), induction of sasA and the subsequent elevated cellular (p)ppGpp concentration decreases growth rate, culminating in the cessation of growth (SI Appendix, Fig. S3A). To determine how (p)ppGpp affects protein synthesis, cells were labeled with OPP at 30-min time intervals following inducer (xylose) addition. At the time of addition (T0), there is not a significant difference in protein synthesis between the induced and uninduced cultures (SI Appendix, Fig. S3 B and C). In contrast, at 30 min following inducer addition (T30) and at a later time (T60), protein synthesis is significantly reduced in the induced cultures as compared to the uninduced cultures (Fig. 2 A and B and SI Appendix, Fig. S3 B and C). This result indicates that (p)ppGpp negatively affects protein synthesis, but it does not demonstrate that this effect is a consequence of a direct inhibition of translation.
Fig. 2.
(p)ppGpp inhibits protein translation specifically. The effects of (p)ppGpp on protein translation were tested in vivo using expression of an inducible (p)ppGpp synthase and in vitro using a reconstituted transcription translation coupled reaction. (A) Representative pictures of OPP-labeled induced and uninduced cultures of Pxyl-sasA strain (JDB4295) 30 min after induction (T30). (B) Total fluorescence of OPP-labeled induced and uninduced cultures of Pxyl-sasA strain (means ± SDs). (C) The Pxyl-sasA Pveg-luc strain (JDB4296) was grown in duplicate, and 0.05% xylose was added after 60 min of growth (T0). Growth and luminescence were measured for 90 min post induction of sasA (means ± SDs). (D) Production of CotE-FLAG protein by a PUREexpress reaction (NEB) in the absence or presence of ppGpp was assayed via Western blot with α-FLAG (means ± SDs). n.s., not significant; **P < 0.01; ***P < 0.001.
(p)ppGpp Inhibits Translation Specifically.
We reasoned that measuring both the synthesis of a single protein and the transcription of its gene would allow us to definitively demonstrate that inhibition of protein synthesis occurs by directly targeting translation. We chose the B. subtilis Pveg promoter, which is insensitive to (p)ppGpp (42), and firefly luciferase as a reporter protein. The half-life of luciferase in B. subtilis is only 6 min (43), so measurements of its activity closely reflect the kinetics of its synthesis. We grew strains that contained an inducible copy of sasA (Pxyl-sasA) as well the Pveg-luc reporter and compared luciferase activity in the presence and absence of inducer. While luciferase activity is easily detected during exponential growth, sasA induction is quickly followed by a drastic decrease in luciferase activity per cell as compared to cells in the absence of induction (Fig. 2C). The rapidity of this inhibition is not consistent with an effect on ribosome synthesis and/or assembly, which are likely targets of (p)ppGpp (44). This decrease does not occur at the level of transcription as the amount of luc mRNA on a per cell level is similar before and after induction (SI Appendix, Fig. S4A). Furthermore, this decrease is not dependent on changes in GTP levels, which on their own could inhibit translation, as levels of the GTP-sensitive rrnB P1 promoter have not yet significantly changed, indicating that GTP levels have not dropped significantly at this time point (SI Appendix, Fig. S4A). Thus, (p)ppGpp is sufficient to rapidly reduce synthesis of a particular protein without affecting transcription of its mRNA, and these effects occur before GTP levels have greatly decreased, consistent with a direct effect on translation.
We extended these in vivo observations by using the PURExpress in vitro reconstituted, coupled transcription-translation system (New England Biolabs, NEB) that utilizes a defined mix of purified transcription and E. coli translation components to transcribe and translate a specific mRNA (45). Addition of ppGpp to PURExpress reactions inhibited synthesis of a reporter protein (CotE) in a dose-dependent fashion (Fig. 2D). Concentrations of ppGpp sufficient to significantly inhibit CotE synthesis (∼1 mM) are similar to those of (p)ppGpp observed during stringent response induction in E. coli (46). qRT-PCR analysis demonstrated that ppGpp had no effect on cotE transcription at concentrations where CotE synthesis was significantly impaired (SI Appendix, Fig. S4B), consistent with the known insensitivity of the T7 RNA polymerase used in the PURExpress reaction to ppGpp (47). Thus, ppGpp directly inhibits translation in vitro.
IF2 Is a Target of (p)ppGpp.
Given these observations, we wished to identify the component(s) of the translation machinery that is (are) targeted by ppGpp in vivo. The translational GTPases EF-Tu (29), EF-G (31), and IF2 (30, 48) as well as the ribosome-associated GTPases including Obg (49, 50), RsgA (14, 51), RbgA (14, 28), Era (14), and HflX (14, 51) have all been reported to bind ppGpp. However, since ppGpp inhibits protein synthesis by the PURExpress system (Fig. 2D), which contains only IF2, EF-Tu, and EF-G, inhibition of one or more of these three proteins is likely sufficient to account for the in vivo inhibitory effect of (p)ppGpp on translation.
We first investigated whether IF2 was a target under our conditions because it has been proposed to serve as a “metabolic sensor” of (p)ppGpp (30). We therefore attempted to identify mutations in IF2 that would affect (p)ppGpp binding without disrupting GTP binding sufficiently to impair normal function. E. coli EF-G and IF2 have differential affinity for (p)ppGpp (31) and similar, but not absolutely conserved, G domains. So, if we could identify residue(s) that affect this difference, this information might allow us to construct a B. subtilis IF2 allele less sensitive to (p)ppGpp. We focused on those IF2 residues which display a shift in nuclear magnetic resonance (NMR) spectra upon binding of ppGpp as compared to GDP (Fig. 3B, blue residues) (30) since GDP interacts with multiple residues in the G domain of IF2 (52). We aligned the region containing those residues (i.e., the G1 motif) with the homologous region in EF-G, which has lower affinity for (p)ppGpp than IF2 (Fig. 3A). We noted that one of the blue residues in IF2, Gly-226, is an alanine residue (Ala-18) in EF-G (Fig. 3B, red). In addition, the histidine residue (His-230) in IF2 that is adjacent to the two blue residues is an alanine (Ala-21) in EF-G (Fig. 3B, red). These differences suggested that substituting the IF2 residues with the corresponding residues found in EF-G would affect the ability of IF2 to bind (p)ppGpp. We therefore compared the affinity of WT and mutant IF2 for radiolabeled (p)ppGpp using the DRaCALA filter binding assay and observed that the double mutant IF2 (G226A H230A) bound (p)ppGpp significantly less well than the WT protein (Fig. 3C).
Fig. 3.
IF2 is a target of ppGpp. IF2 was validated in vitro as a direct target of ppGpp using IF2 mutations that reduce ppGpp binding. (A) Affinity of B. subtilis EF-G and IF2 for (p)ppGpp was compared using the differential radial capillary action of a ligand assay (DRaCALA) (53). (means ± SDs). (B) Alignment of G1 domains of B. subtilis IF2 and EF-G. Residues in blue denote those most shifted upon binding of ppGpp versus GDP (30). Residues in red are those that differ in EF-G and IF2 and were used to engineer a mutant IF2 with reduced affinity for ppGpp (G226A H230A). (C) DRaCALA-based comparison of (p)ppGpp affinity for WT and mutant IF2 (means ± SDs). (D) In vitro sensitivity of WT and mutant IF2 was assessed using the PURExpress system (NEB). WT and mutant IF2 were added at equimolar amounts to separate PURExpress reactions in the presence of 1 mM ppGpp, and protein synthesis was monitored by Western blot (means ± SDs). **P < 0.01.
To test the functional consequence of mutating these residues in B. subtilis IF2, we used a PURExpress kit that lacks IF2 (ΔIF2). We first confirmed that the ΔIF2 kit, which contains purified E. coli translation factors, works equivalently whether the added IF2 is derived from E. coli or B. subtilis (SI Appendix, Fig. S5B). When supplied as the sole source of IF2 in this reaction, the double mutant B. subtilis IF2 produced an equivalent amount of protein as WT B. subtilis IF2 (SI Appendix, Fig. S5B), demonstrating that the slight reduction in GTP binding (SI Appendix, Fig. S5A) did not substantially affect IF2 function in translation. However, the double mutant B. subtilis IF2 was significantly less sensitive to ppGpp inhibition than its WT counterpart (Fig. 3D). Taken together, these results indicate that ppGpp binding to IF2 accounts for a substantial portion of the inhibition of translation by ppGpp.
ppGpp Fails to Allosterically Activate IF2 for Rapid Subunit Joining.
Previously, Milon et al. demonstrated that ppGpp inhibits IF2’s ability to catalyze 30S IC assembly and subunit joining (30). To elucidate the structural basis of this inhibition, we wished to determine whether and how ppGpp influences the binding of E. coli IF2 to the E. coli 30S IC and the conformational dynamics of 30S IC-bound IF2. Briefly, IF2 promotes binding of initiator tRNA (fMet-tRNAfMet) to the 30S subunit and uses its domain IV (dIV) to directly contact the N-formyl-methionine and aminoacyl acceptor stem of fMet-tRNAfMet, resulting in the formation of an IF2-tRNA subcomplex on the intersubunit surface of the 30S IC (54). The presence of GTP in the G domain and recognition of fMet-tRNAfMet by dIV “activate” 30S IC-bound IF2 for rapid subunit joining (55). We previously used an IF2-tRNA single-molecule fluorescence resonance energy transfer (smFRET) signal (56) to show that activation of IF2 for rapid subunit joining involves a GTP- and fMet-tRNAfMet-dependent conformational change of IF2 that results in an increase in the affinity of IF2 for the 30S IC and an increase in the rate of subunit joining (57). To understand how ppGpp affected this interaction, we began by comparing the affinities of GTP-bound IF2 (IF2(GTP)) and ppGpp-bound IF2 (IF2(ppGpp)) for the 30S IC. As in our previous studies (56, 57), FRET efficiency (EFRET) versus time trajectories recorded for individual 30S ICs fluctuate between a zero FRET state (IF2-free 30S IC) and a nonzero FRET state (IF2-bound 30S IC) (Fig. 4, Lower Middle). Initial inspection of these trajectories and the corresponding surface contour plots of the postsynchronized time evolution of population FRET reveals that, while IF2(GTP) exhibits relatively long-lived and stable binding events on the 30S IC, IF2(ppGpp) exhibits significantly shorter-lived and unstable binding events (Fig. 4, Lower Middle and Bottom). To quantitatively compare the affinities of IF2(GTP) and IF2(ppGpp) for the 30S IC, we extracted kinetic and thermodynamic parameters from the smFRET data describing the binding of IF2 to the 30S IC. We find that IF2(ppGpp) has a significantly lower affinity for the 30S IC compared to IF2(GTP), with an equilibrium dissociation constant (Kd) that is ∼100-fold higher (SI Appendix, Table S1).
Fig. 4.
ppGpp inhibits IF2 function in catalyzing rapid 50S subunit joining. The binding of IF2 to the 30S IC and the conformation of 30S IC-bound IF2 in the presence of GTP (A) and ppGpp (B) were directly observed by smFRET using an IF2-tRNA smFRET signal. (Top) Cartoon representations of 30S ICs assembled using Cy3 FRET donor fluorophore-labeled fMet-tRNAfMet and Cy5 FRET acceptor fluorophore-labeled IF2(GTP) or IF2(ppGpp). (Upper Middle) Plots of Cy3 (green) and Cy5 (red) fluorescence emission intensity versus time trajectories. (Lower Middle) Plots of the EFRET versus time trajectories corresponding to the plots of Cy3 and Cy5 fluorescence intensity trajectories in Upper Middle. (Bottom) Surface contour plots of the postsynchronized time evolution of population FRET. These plots are generated by superimposing the EFRET versus time trajectories of individual IF2 binding events such that the start of each event is computationally postsynchronized to time = 1 s, thereby allowing visualization of the time evolution of population FRET for the entire population of IF2 binding events. “N” indicates the total number of individual 30S ICs analyzed and “n” indicates the total number of individual IF2 binding events analyzed. A.U., arbitrary units.
We then compared the conformations of IF2(ppGpp) and IF2(GTP) on the 30S IC by plotting histograms of the EFRET values observed under each condition (SI Appendix, Fig. S6). Consistent with our previous studies, we observed a single nonzero peak centered at an <EFRET> of ∼0.74 for 30S IC-bound IF2(GTP), corresponding to a distance between our labeling positions of ∼46.2 Å (assuming a Förster distance, R0, of ∼55 Å; ref. 58) (SI Appendix, Fig. S6A). In contrast, we observed a single nonzero peak centered at a significantly lower <EFRET> of ∼0.53 (P < 0.005) for 30S IC-bound IF2(ppGpp), corresponding to a distance between our labeling positions of ∼53.9 Å, an increase of ∼7.7 Å relative to IF2(GTP) (SI Appendix, Fig. S6B). Notably, the EFRET distribution of 30S IC-bound IF2(ppGpp) closely resembles that of 30S IC-bound IF2(GDP) (57). Thus, the 30S IC-bound IF2(ppGpp) exhibits a conformation that is similar to an IF2 inactive for rapid subunit joining (i.e., IF2(GDP)).
(p)ppGpp Binding to IF2 Mediates Translational Inhibition during Transition Phase.
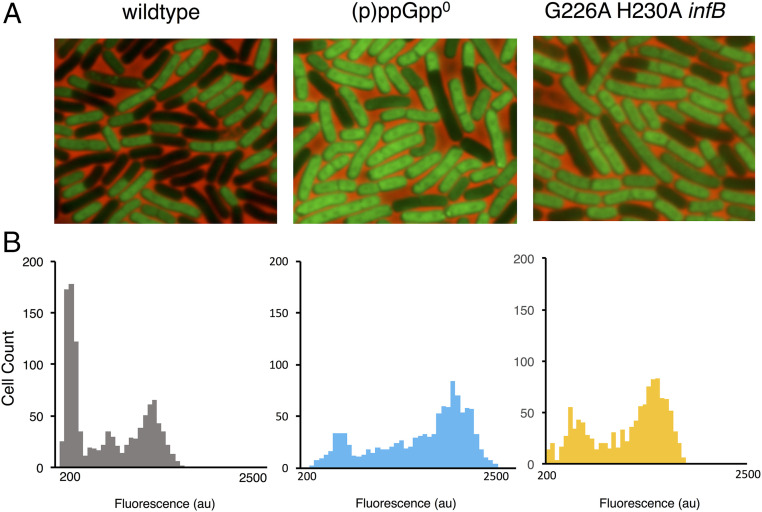
Our identification of an IF2 allele that is less sensitive to (p)ppGpp in vitro enabled us to test our initial hypothesis that (p)ppGpp accumulation reduces protein synthesis in vivo because (p)ppGpp binds IF2 and inhibits its function in translation. We generated a B. subtilis strain expressing, as the sole source of IF2, a mutant protein containing the mutations G226A and H230A that in vitro exhibits reduced ppGpp binding without substantially affecting IF2 function (Fig. 3C and SI Appendix, Fig. S5B). This strain grows equivalently to the WT parent throughout all phases of growth, validating that the mutant IF2 is functional in vivo (SI Appendix, Fig. S7A). We first tested how these mutations affect protein synthesis during late transition phase since in the WT background, protein synthesis is strongly inhibited in a subpopulation of cells during this period (Fig. 1C). Mutations in IF2 that affect its binding to (p)ppGpp appear to significantly attenuate this phenotype (Fig. 5 A and B and SI Appendix, Fig. S7 B and C). This attenuation is similar to that observed in the complete absence of (p)ppGpp (Fig. 5 A and B), consistent with it resulting from a direct interaction of (p)ppGpp with IF2. Thus, during transition phase, (p)ppGpp binding to IF2 is sufficient to substantially inhibit translation and, thereby, reduce protein synthesis.
Fig. 5.
IF2 mediates inhibition of protein synthesis by (p)ppGpp during late transition phase. IF2 was validated as an in vivo target of (p)ppGpp by measuring protein synthesis in a strain expressing an IF2 G226A H230A double mutant. (A) Representative pictures of WT (JDB 1772), (p)ppGpp0 (JDB 4294), and G226A H230A infB (JDB 4297) strains labeled with OPP during late transition phase. (B) Distributions of mean cell fluorescence of WT, (p)ppGpp0, and G226A H230A infB strains during late transition phase (same time as in Fig. 1).
Discussion
Here, we demonstrate that protein synthesis is actively attenuated in B. subtilis during exit from rapid growth in a subpopulation of cells. (p)ppGpp is both necessary and sufficient for this phenomenon and acts, at least in part, by a direct inhibitory effect on the translational GTPase IF2. Thus, the regulatory mechanism of endogenous (p)ppGpp synthesis mediates the active down-regulation of the most energy-consuming process in cells as they enter quiescence.
The role of (p)ppGpp under basal conditions such as those examined in this study has become increasingly appreciated (59). In Synechococcus elongatus, loss of (p)ppGpp leads to increases in the global translation rate per cell under basal conditions (60). (p)ppGpp synthesis is also relevant to physiological situations including survival of pathogenic (61) and commensal bacteria (62). “Persisters” (rare bacterial cells with increased tolerance to antibiotics) are also thought to have relatively higher levels of (p)ppGpp (63). While the basis for this heterogeneous accumulation of (p)ppGpp is not understood, an interesting question raised by our observations is whether the (p)ppGpp-dependent inhibition of protein synthesis affects the increased antibiotic tolerance of these cells.
We show that the sensitivity of IF2 to (p)ppGpp can be altered by mutations that affect binding. (p)ppGpp also binds numerous ribosome-associated GTPases (ObgE, refs. 32, 49, 50; BipA, ref. 64; RbgA, refs. 14, 28; HflX, ref. 14; and Era, ref. 14) and affects their function in vitro. The importance of (p)ppGpp binding to these proteins in vivo remains a critical question. Our demonstration that point mutations in IF2 affect (p)ppGpp binding and subsequent in vitro and in vivo function suggests that introducing similar mutations into the highly conserved G1 motif of other GTPases might be informative. Interestingly, the residues in the IF2 G1 motif that altered (p)ppGpp binding and inhibition (Fig. 3B) are conserved in EF-Tu. This conservation, along with the similarities in binding affinities between E. coli EF-Tu and IF2 (29, 31), suggest that EF-Tu may also be a target for regulation during quiescence. A similar mutagenic strategy was reported recently for the (p)ppGpp-binding protein E. coli PurF glutamine amido-phosphoribosyltransferase (65).
Our studies also reveal the structural basis through which ppGpp inhibits IF2. We find that ppGpp stabilizes a conformation of IF2 that is different from that which is stabilized by GTP, resulting in a reduced affinity for the 30S IC. Consequently, ppGpp may interfere with the formation and/or stabilization of the IF2-tRNA subcomplex on the 30S IC, the formation of which plays a key role in stabilizing the binding and/or positioning of fMet-tRNAfMet on the 30S IC (54, 66). Previously, we demonstrated that this interaction is crucial for the allosteric activation of IF2(GTP), thereby enabling IF2 to catalyze rapid subunit joining (57). Thus, combined with the decreased affinity of IF2(ppGpp) for the 30S IC, our results suggest that the conformation of 30S IC-bound IF2(ppGpp) directly interferes with the ability of IF2 to promote rapid subunit joining to the 30S IC.
Finally, what is the source of the observed heterogeneity of protein synthesis activity in single cells? During exponential growth, rare (∼1%) cells have high levels of sasA expression with concomitant physiological effects including induction of (p)ppGpp-dependent genes and enhanced antibiotic tolerance (67). However, the roughly bimodal character (∼50%) observed in our study suggests the presence of bistability (68) at the population level, a phenomenon often attributed to the presence of positive nonlinear autoregulation or two mutually repressive repressors (69). Intriguingly, SasB, one of the two nonribosomally associated (p)ppGpp synthases in B. subtilis, is subject to positive allosteric regulation (70), which could generate a sharp threshold-like response. Thus, the potential role of SasB in the heterogeneity of protein synthesis under nonexponential growth will be the subject of future investigation.
Methods
See SI Appendix, Methods for detailed descriptions of experimental conditions. Strains, plasmids, and primers used in this study are listed in SI Appendix, Tables S2–S4.
Growth Curves.
All growth curves were performed at 37 °C with continuous shaking, and OD600 measurements were made every 5 min.
OPP/HPG Labeling.
Click-iT Protein Synthesis Assay Kits (Invitrogen) were used to label cells with OPP or HPG following manufacturer’s instructions.
Luminescence Growth Curves.
Luminescence growth curves were performed as above with the addition of 4.7 mM d-luciferin (Goldbio) and using 96-well flat bottom white-sided plates (Greiner Bio-One).
RNA Quantification.
RNA was isolated from cultures grown as above using the Direct-Zol RNA miniprep kit (Zymo Research). cDNAs were generated with the High Capacity cDNA Reverse Transcription kit (Applied Biosystems) and qPCRs were performed using SYBR green.
In Vitro Translation Assays.
Translation assays used the PURExpress system (NEB) following the manufacturer’s protocol and a plasmid encoding a CotE-FLAG fusion protein as template DNA (6). WT and mutant IF2s were purified as previously described (71). Band intensities were analyzed using ImageJ.
DRaCALA Binding Assays.
Radiolabeled (p)ppGpp was generated as described (14). DRaCALA binding assays were carried out essentially as described (14, 53).
smFRET Experiments.
smFRET experiments were carried out essentially as described (57).
Data Availability Statement.
All data for the paper are contained in the main text or SI Appendix. All plasmids and strains used will be made available upon request.
Supplementary Material
Acknowledgments
We thank the members of our laboratories for helpful discussions and comments on the manuscript. S.D. was supported by NIH Grant R01GM114213-03S1 and in part by predoctoral training grant support from the Columbia University Graduate Program in Microbiology, Immunology and Infection (Grant R01 AI106711, Program Directors D. Fidock and L. Symington); J.R. and R.L.G. were supported by NIH Grants R01GM084288 and R01GM119386; and J.D. was supported by NIH Grants R01GM114213 and R21AI135427 and is a Burroughs-Welcome Investigator in the Pathogenesis of Infectious Diseases.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920013117/-/DCSupplemental.
References
- 1.Lennon J. T., Jones S. E., Microbial seed banks: The ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 9, 119–130 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Rittershaus E. S., Baek S. H., Sassetti C. M., The normalcy of dormancy: Common themes in microbial quiescence. Cell Host Microbe 13, 643–651 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tempest D. W., Neijssel O. M., The status of YATP and maintenance energy as biologically interpretable phenomena. Annu. Rev. Microbiol. 38, 459–486 (1984). [DOI] [PubMed] [Google Scholar]
- 4.Szaflarski W., Nierhaus K. H., Question 7: Optimized energy consumption for protein synthesis. Orig. Life Evol. Biosph. 37, 423–428 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Maracci C., Rodnina M. V., Review: Translational GTPases. Biopolymers 105, 463–475 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pereira S. F., Gonzalez R. L. Jr., Dworkin J., Protein synthesis during cellular quiescence is inhibited by phosphorylation of a translational elongation factor. Proc. Natl. Acad. Sci. U.S.A. 112, E3274–E3281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamel E., Cashel M., Guanine nucleotides in protein synthesis. Utilization of pppGpp and dGTP by initiation factor 2 and elongation factor Tu. Arch. Biochem. Biophys. 162, 293–300 (1974). [DOI] [PubMed] [Google Scholar]
- 8.Gaca A. O., Colomer-Winter C., Lemos J. A., Many means to a common end: The intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J. Bacteriol. 197, 1146–1156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steinchen W., Bange G., The magic dance of the alarmones (p)ppGpp. Mol. Microbiol. 101, 531–544 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Artsimovitch I.et al., Structural basis for transcription regulation by alarmone ppGpp. Cell 117, 299–310 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Ross W.et al., ppGpp binding to a site at the RNAP-DksA interface accounts for its dramatic effects on transcription initiation during the stringent response. Mol. Cell 62, 811–823 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J. D., Sanders G. M., Grossman A. D., Nutritional control of elongation of DNA replication by (p)ppGpp. Cell 128, 865–875 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kriel A.et al., Direct regulation of GTP homeostasis by (p)ppGpp: A critical component of viability and stress resistance. Mol. Cell 48, 231–241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corrigan R. M., Bellows L. E., Wood A., Gründling A., ppGpp negatively impacts ribosome assembly affecting growth and antimicrobial tolerance in Gram-positive bacteria. Proc. Natl. Acad. Sci. U.S.A. 113, E1710–E1719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez-Vazquez P., Dewey C. N., Kitten N., Ross W., Gourse R. L., Genome-wide effects on Escherichia coli transcription from ppGpp binding to its two sites on RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 116, 8310–8319 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barker M. M., Gaal T., Josaitis C. A., Gourse R. L., Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305, 673–688 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Gaal T., Bartlett M. S., Ross W., Turnbough C. L. Jr., Gourse R. L., Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278, 2092–2097 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Paul B. J.et al., DksA: A critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118, 311–322 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Laffler T., Gallant J. A., Stringent control of protein synthesis in E. coli. Cell 3, 47–49 (1974). [DOI] [PubMed] [Google Scholar]
- 20.O’Farrell P. H., The suppression of defective translation by ppGpp and its role in the stringent response. Cell 14, 545–557 (1978). [DOI] [PubMed] [Google Scholar]
- 21.Svitil A. L., Cashel M., Zyskind J. W., Guanosine tetraphosphate inhibits protein synthesis in vivo. A possible protective mechanism for starvation stress in Escherichia coli. J. Biol. Chem. 268, 2307–2311 (1993). [PubMed] [Google Scholar]
- 22.Lindahl L., Post L., Nomura M., DNA-dependent in vitro synthesis of fibosomal proteins, protein elongation factors, and RNA polymerase subunit alpha: Inhibition by ppGpp. Cell 9, 439–448 (1976). [DOI] [PubMed] [Google Scholar]
- 23.Haseltine W. A., Block R., Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc. Natl. Acad. Sci. U.S.A. 70, 1564–1568 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown A., Fernández I. S., Gordiyenko Y., Ramakrishnan V., Ribosome-dependent activation of stringent control. Nature 534, 277–280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arenz S.et al., The stringent factor RelA adopts an open conformation on the ribosome to stimulate ppGpp synthesis. Nucleic Acids Res. 44, 6471–6481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loveland A. B.et al., Ribosome•RelA structures reveal the mechanism of stringent response activation. eLife 5, e17029 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner E. G., Kurland C. G., Translational accuracy enhanced in vitro by (p)ppGpp. Mol. Gen. Genet. 180, 139–145 (1980). [DOI] [PubMed] [Google Scholar]
- 28.Pausch P.et al., Structural basis for (p)ppGpp-mediated inhibition of the GTPase RbgA. J. Biol. Chem. 293, 19699–19709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rojas A. M., Ehrenberg M., Andersson S. G., Kurland C. G., ppGpp inhibition of elongation factors Tu, G and Ts during polypeptide synthesis. Mol. Gen. Genet. 197, 36–45 (1984). [DOI] [PubMed] [Google Scholar]
- 30.Milon P.et al., The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc. Natl. Acad. Sci. U.S.A. 103, 13962–13967 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitkevich V. A.et al., Thermodynamic characterization of ppGpp binding to EF-G or IF2 and of initiator tRNA binding to free IF2 in the presence of GDP, GTP, or ppGpp. J. Mol. Biol. 402, 838–846 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Persky N. S., Ferullo D. J., Cooper D. L., Moore H. R., Lovett S. T., The ObgE/CgtA GTPase influences the stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 73, 253–266 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J., Xu Y., Stoleru D., Salic A., Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc. Natl. Acad. Sci. U.S.A. 109, 413–418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murata M., Ohno S., Kumano M., Yamane K., Ohki R., Multidrug resistant phenotype of Bacillus subtilis spontaneous mutants isolated in the presence of puromycin and lincomycin. Can. J. Microbiol. 49, 71–77 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Kriel A.et al., GTP dysregulation in Bacillus subtilis cells lacking (p)ppGpp results in phenotypic amino acid auxotrophy and failure to adapt to nutrient downshift and regulate biosynthesis genes. J. Bacteriol. 196, 189–201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Potrykus K., Murphy H., Philippe N., Cashel M., ppGpp is the major source of growth rate control in E. coli. Environ. Microbiol. 13, 563–575 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray H. D., Schneider D. A., Gourse R. L., Control of rRNA expression by small molecules is dynamic and nonredundant. Mol. Cell 12, 125–134 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Boes N., Schreiber K., Schobert M., SpoT-triggered stringent response controls usp gene expression in Pseudomonas aeruginosa. J. Bacteriol. 190, 7189–7199 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nanamiya H.et al., Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol. Microbiol. 67, 291–304 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Srivatsan A.et al., High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 4, e1000139 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tagami K.et al., Expression of a small (p)ppGpp synthetase, YwaC, in the (p)ppGpp(0) mutant of Bacillus subtilis triggers YvyD-dependent dimerization of ribosome. MicrobiologyOpen 1, 115–134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krásný L., Gourse R. L., An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 23, 4473–4483 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mirouze N., Prepiak P., Dubnau D., Fluctuations in spo0A transcription control rare developmental transitions in Bacillus subtilis. PLoS Genet. 7, e1002048 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burgos H. L., O’Connor K., Sanchez-Vazquez P., Gourse R. L., Roles of transcriptional and translational control mechanisms in regulation of ribosomal protein synthesis in Escherichia coli. J. Bacteriol. 199, e00407-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimizu Y., Kuruma Y., Kanamori T., Ueda T., The PURE system for protein production. Methods Mol. Biol. 1118, 275–284 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Buckstein M. H., He J., Rubin H., Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J. Bacteriol. 190, 718–726 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friesen J. D., Fiil N., Accumulation of guanosine tetraphosphate in T7 bacteriophage-infected Escherichia coli. J. Bacteriol. 113, 697–703 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Legault L., Jeantet C., Gros F., Inhibition of in vitro protein synthesis by ppGpp. FEBS Lett. 27, 71–75 (1972). [DOI] [PubMed] [Google Scholar]
- 49.Buglino J., Shen V., Hakimian P., Lima C. D., Structural and biochemical analysis of the Obg GTP binding protein. Structure 10, 1581–1592 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Feng B.et al., Structural and functional insights into the mode of action of a universally conserved Obg GTPase. PLoS Biol. 12, e1001866 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y., Zborníková E., Rejman D., Gerdes K., Novel (p)ppGpp binding and metabolizing proteins of Escherichia coli. MBio 9, e02188-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wienk H.et al., Structural dynamics of bacterial translation initiation factor IF2. J. Biol. Chem. 287, 10922–10932 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roelofs K. G., Wang J., Sintim H. O., Lee V. T., Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc. Natl. Acad. Sci. U.S.A. 108, 15528–15533 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simonetti A.et al., Structure of the 30S translation initiation complex. Nature 455, 416–420 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Pavlov M. Y., Zorzet A., Andersson D. I., Ehrenberg M., Activation of initiation factor 2 by ligands and mutations for rapid docking of ribosomal subunits. EMBO J. 30, 289–301 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J., Caban K., Gonzalez R. L. Jr., Ribosomal initiation complex-driven changes in the stability and dynamics of initiation factor 2 regulate the fidelity of translation initiation. J. Mol. Biol. 427, 1819–1834 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caban K., Pavlov M., Ehrenberg M., Gonzalez R. L. Jr., A conformational switch in initiation factor 2 controls the fidelity of translation initiation in bacteria. Nat. Commun. 8, 1475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murphy M. C., Rasnik I., Cheng W., Lohman T. M., Ha T., Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys. J. 86, 2530–2537 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaca A. O.et al., Basal levels of (p)ppGpp in Enterococcus faecalis: The magic beyond the stringent response. MBio 4, e00646-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Puszynska A. M., O’Shea E. K., ppGpp controls global gene expression in light and in darkness in S. elongatus. Cell Rep. 21, 3155–3165 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Stapels D. A. C.et al., Salmonella persisters undermine host immune defenses during antibiotic treatment. Science 362, 1156–1160 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Schofield W. B., Zimmermann-Kogadeeva M., Zimmermann M., Barry N. A., Goodman A. L., The stringent response determines the ability of a commensal bacterium to survive starvation and to persist in the gut. Cell Host Microbe 24, 120–132.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hauryliuk V., Atkinson G. C., Murakami K. S., Tenson T., Gerdes K., Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar V.et al., Structure of BipA in GTP form bound to the ratcheted ribosome. Proc. Natl. Acad. Sci. U.S.A. 112, 10944–10949 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang B.et al., Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat. Chem. Biol. 15, 141–150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Julián P.et al., The Cryo-EM structure of a complete 30S translation initiation complex from Escherichia coli. PLoS Biol. 9, e1001095 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Libby E. A., Reuveni S., Dworkin J., Multisite phosphorylation drives phenotypic variation in (p)ppGpp synthetase-dependent antibiotic tolerance. Nat. Commun. 10, 5133 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dubnau D., Losick R., Bistability in bacteria. Mol. Microbiol. 61, 564–572 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Ferrell J. E. Jr., Self-perpetuating states in signal transduction: Positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 14, 140–148 (2002). [DOI] [PubMed] [Google Scholar]
- 70.Steinchen W.et al., Structural and mechanistic divergence of the small (p)ppGpp synthetases RelP and RelQ. Sci. Rep. 8, 2195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fei J.et al., A highly purified, fluorescently labeled in vitro translation system for single-molecule studies of protein synthesis. Methods Enzymol. 472, 221–259 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data for the paper are contained in the main text or SI Appendix. All plasmids and strains used will be made available upon request.