Significance
Aging is associated with a myriad of changes at all levels of organization of an organism. However, how it affects protein synthesis, a major metabolic activity of the cell, is unknown. We discovered deregulation of protein synthesis and ribosome biogenesis machinery specifically at the translational level with age. Moreover, ribosomes were depleted in the vicinity of start codons and increased near stop codons. These findings reveal systematic, multilevel deregulation in gene expression and protein synthesis, showing how this major cellular process declines with age.
Keywords: aging, proteostasis, ribosome profiling, transcriptome, 5′-TOP
Abstract
Protein synthesis represents a major metabolic activity of the cell. However, how it is affected by aging and how this in turn impacts cell function remains largely unexplored. To address this question, herein we characterized age-related changes in both the transcriptome and translatome of mouse tissues over the entire life span. We showed that the transcriptome changes govern those in the translatome and are associated with altered expression of genes involved in inflammation, extracellular matrix, and lipid metabolism. We also identified genes that may serve as candidate biomarkers of aging. At the translational level, we uncovered sustained down-regulation of a set of 5′-terminal oligopyrimidine (5′-TOP) transcripts encoding protein synthesis and ribosome biogenesis machinery and regulated by the mTOR pathway. For many of them, ribosome occupancy dropped twofold or even more. Moreover, with age, ribosome coverage gradually decreased in the vicinity of start codons and increased near stop codons, revealing complex age-related changes in the translation process. Taken together, our results reveal systematic and multidimensional deregulation of protein synthesis, showing how this major cellular process declines with age.
Aging is associated with a gradual decline of organismal function and fitness, which in turn is tightly linked with changes in the proteome. The balance between protein synthesis and degradation, proteostasis, and proper protein quality control is required to maintain cell homeostasis (1–3). Among other processes, molecular damage accumulating in cells with age influences the proteome, the endpoint of gene expression (4). Additionally, with advancing age, protein damage manifests itself in the form of posttranslational modifications such as oxidation and glycation, impairing function; damaged proteins are also prone to form toxic oligomers and insoluble aggregates. In fact, disruption of proteostasis is a well-known cause of aging-associated diseases. However, this age-related dysfunction is also believed to influence organisms in a systemic and chronic way, decreasing their stress resistance and the ability to clear misfolded proteins (1). Indeed, compared to their closest relatives, long-lived species exhibit an increased proteome stability (5) and resistance to protein oxidation (6). Protein turnover and quality control include several distinct but tightly connected biological processes and can be conditionally divided into the phases of protein synthesis, folding, activity, posttranslation regulation, and degradation. Among them, only folding and degradation are relatively well understood in the context of aging, and their impairment indeed explains some aging-related pathologies (1). On the other hand, protein synthesis changes with age remain notably less explored (7, 8).
In the second half of the 20th century, several studies studying various species showed that the overall rate of protein synthesis, activity, concentration of elongation and initiation factors, and tRNA aminoacylation levels decrease with age (9). Recent studies in mammals are also in agreement with the idea of age-related decline in overall translation rate. In particular, it was shown that the levels of total mRNA, as well as the expression of RNA polymerase I, eIF2Bε, and eEF2, decrease with age in rat tissues (10). Increased promoter methylation in ribosomal RNA genes and decreased ribosomal RNA concentration during aging were also reported (11). In addition, down-regulation of translation with age was confirmed in vivo in the sheep (12) as well as in replicatively aged yeast (13). Recently, analyses of liver and brain of 6- and 24-mo-old rats revealed age-related translatome changes in mammals (14).
The role of protein synthesis in aging is further supported by indirect evidence. The decreased rate of protein synthesis generally leads to increased life span of animals (reviewed in ref. 7). For instance, knockout or knockdown of several translation machinery components in worms significantly increased average life span and accelerated the effects of life-extending mutations (15). In addition, overexpression of translation initiation repressor 4E-BP1 increased life span (16) and mediated life span extension effects of dietary restriction in fruit flies (17), whereas inhibition of biogenesis of the 60S ribosomal subunit prolonged life span of yeast (18). Notably, most of the interventions known to extend life span are associated with suppression of metabolism (e.g., caloric restriction) or inhibition of nutrient signaling (e.g., rapamycin), regulating protein synthesis and biosynthesis of translation machinery components (19). At least in part, this can be explained by the reduced load on the protein quality control machinery and decreased energy use (20). Moreover, increased fidelity of translation and decreased protein turnover rate were recently found to be associated with longevity; e.g., the naked mole rat, an animal with extremely long life span compared to its rodent relatives, possesses a highly accurate translation apparatus (21–23).
Despite a growing number of studies on protein synthesis alterations with age in mammals, previous research did not address translatome changes with the temporal and quantitative resolution sufficient to reveal principles of protein synthesis alteration with age at the whole-transcriptome scale. In this study, we took advantage of a combination of ribosome profiling (Ribo-seq) (24) and RNA-seq to characterize age-related changes in protein synthesis throughout the entire life span, focusing on liver and kidney of mice. This approach supported the identification of functional groups of genes exhibiting age-related changes in transcription and/or translation. Interestingly, dozens of transcripts encoding ribosome biogenesis and protein synthesis machinery components were specifically down-regulated with age at the translational level, consistent with the decline in protein synthesis with age. Ribo-seq analyses also revealed a transcriptome-wide redistribution of ribosome coverage from the beginning to the end of mRNA coding regions as well as other features associated with complex and multifactorial deregulation of protein synthesis with age.
Results
Gene Expression Changes in Mouse Liver and Kidney Reflect Age-Related Dysfunction.
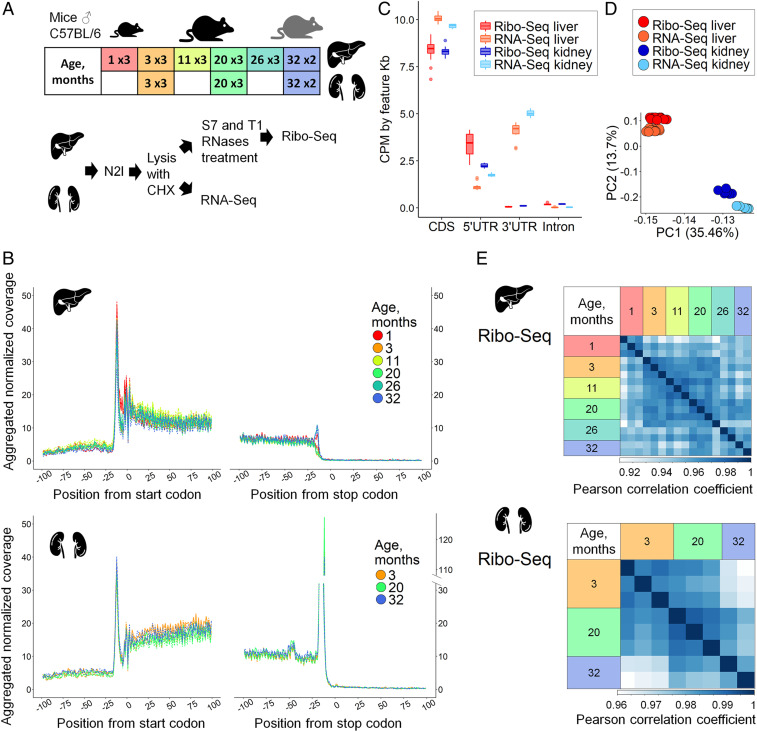
To characterize age-related changes in protein synthesis, we subjected mouse tissue samples to both transcriptome sequencing and ribosome profiling (Dataset S1), as this approach allows to separate the contribution of transcription and translation processes. Liver samples were collected from male mice representing six age groups (1, 3, 11, 20, 26, and 32 mo old), and kidney samples from three age groups (3, 20, and 32 mo old) (Fig. 1A). This broad range of ages was chosen to cover the entire adult mouse life span, from the final stages of development (1 mo) to the very advanced ages (32 mo). For each age group, there were three biological replicates (three mice), except for the 32-mo mice (two replicates).
Fig. 1.
Ribo-seq and RNA-seq of aging mouse liver and kidney. (A) Overview of experimental design. Mouse livers representing six age groups (1-, 3-, 11-, 20-, 26-, and 32-mo-old) and kidneys representing three age groups (3-, 20-, and 32-mo-old) were used. For each age, three biological replicates were prepared (three C57BL/6 male mice), except for the 32-mo group (two mice). Ribo-seq and RNA-seq libraries were prepared from the same cytoplasmic cell lysate. (B) Metagene profiles of ribosomal footprint 5′ ends in 200-nt windows centered at start and stop codons built for 2,920 and 4,566 transcripts for liver and kidney, respectively. For each transcript, raw Ribo-seq coverage was normalized to the sum of transcript coverage divided by its length. Normalized transcript coverage in the window was then aggregated for all selected transcripts. (C) Distribution of Ribo-seq and RNA-seq coverage in different gene regions. (D) Principal-component analysis (PCA) of 8,562 genes in Ribo-seq and RNA-seq datasets of mouse liver and kidney. (E) Heatmaps of Pearson correlation coefficients for replicates of mouse liver and kidney analyzed by Ribo-seq. For PCA and calculation of Pearson correlation coefficients, and further in the study, Ribo-seq was analyzed together with the RNA-seq dataset, but separately for organs. In total, the number of genes covered in each sample was 8,992 in liver and 11,461 in kidney.
In our Ribo-seq protocol, we applied the combination of T1 and S7 RNases, because it most efficiently converts polysomes to monosomes in mouse tissues while retaining ribosome integrity (SI Appendix, Fig. S1A) (25). The resulting footprints had the mean length of 28 nt (SI Appendix, Fig. S1B), displayed clear triplet periodicity (Fig. 1B), and showed a higher CDS and 5′-untranslated region (5′-UTR) coverage compared to 3′-UTRs and introns (Fig. 1C). In both organs, there were pronounced peaks at start codons, while the peak at stop codons was more pronounced in kidney, in concordance with previous observations (26, 27). Principal-component analysis (PCA) has shown that 61% of variance can be explained by the first three principal components (PCs): PC1 and PC2 attributed to the sequenced tissue and PC3 to the type of experiment (Ribo- or RNA-seq). Sample separation by age was achieved by PC4 with 3% of the explained variance (Fig. 1D and SI Appendix, Fig. S1C). Expression profiles were highly reproducible across the replicates (Fig. 1E and SI Appendix, Fig. S1D), especially within each age group, in both liver and kidney, reflecting the fact that gene expression profiles between individual mice within an age group are more similar than those observed across different ages. Samples from 1-mo-old mice formed a separate group with high correlation across the replicates in both Ribo-seq and RNA-seq data, whereas for the older mice we detected a slight increase in gene expression variance with age (Fig. 1E and SI Appendix, Fig. S1D), in agreement with previous reports (28).
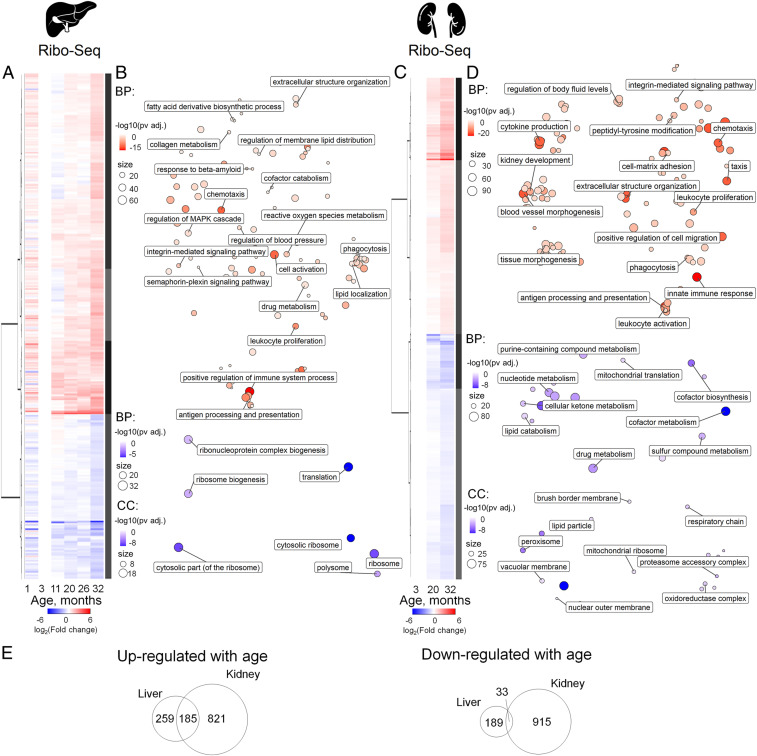
We further assessed Ribo-seq–based temporal patterns of gene expression in mouse liver (Fig. 2 A and B) and kidney (Fig. 2 C and D). Here, Ribo-seq was not normalized to transcriptome coverage, therefore reflecting gene expression with respect to both transcription and translation changes. As liver from 1-mo-old mice differed from that of other ages (Fig. 1E), we used 3-mo-old mice as the reference and selected genes that were differentially expressed at least in one comparison (SI Appendix, Fig. S3 and Dataset S2). Most genes followed a U-shaped pattern of expression changes with the maximum or minimum at 3 mo (Fig. 2A), which served as a turning point (29, 30) and may reflect a transition from development to the adult state. Indeed, genes up-regulated in the 1-mo-old liver (compared to 3-mo samples) were associated with urogenital and vascular system development, chromatin assembly, extracellular matrix organization, and amino acid and nucleotide metabolism (Dataset S3).
Fig. 2.
Expression profiling of aging mouse liver and kidney by Ribo-seq. (A and C) Heatmap of age-related gene expression changes in liver (A) and kidney (C). For each age, differential expression, in comparison to that of 3-mo-old mice, was calculated. Data from three mice per age were analyzed except for the 32-mo-old samples (two mice). For DE genes with adjusted P value less than 0.05, at least in one age compared to 3 mo old, log2(Fold change) values were clustered and presented on a heatmap (Dataset S2). The number of DE genes is summarized in SI Appendix, Fig. S3. (B and D) GO BP (biological process) and GO CC (cellular compartment) functional enrichment of genes up-regulated (red) or down-regulated (blue) with age in liver (B) and kidney (D) (Dataset S3). (E) Comparison of gene sets differentially expressed in liver and kidney. Venn diagrams show genes up- or down-regulated with age according to Ribo-seq data.
Functional patterns of Ribo-seq changes (Fig. 2), as well as changes in the expression of particular genes (SI Appendix, Fig. S2), were somewhat similar in liver and kidney, especially for genes up-regulated with age. We observed a robust increase in the expression of inflammation and immune system genes, reflecting “inflammaging”—chronic inflammation that progresses with age (reviewed in ref. 31). Common markers of senescent cells—lysosomal proteins (32)—were up-regulated in both tissues. Five genes encoding inflammatory and lysosomal proteins (Ctss, C1qa, C1qb, C1qc, and Laptm5) were up-regulated in both liver and kidney in our study as well as several other aging transcriptome and translatome datasets in different organs and species (14, 33, 34), suggesting that they may be considered to be reliable biomarkers of aging.
In contrast to inflammation, mitochondrial function is known to decline with age (reviewed in ref. 35). Indeed, in kidney, we observed a gradually decreased expression of nuclear genes coding for mitochondrial proteins, although it was less pronounced in liver (yet many genes followed the pattern, e.g., Uqcc2, Fxn, Mrps16, Mrpl9, Mrpl30, Mrpl54, Mterf4, Atp5k, Aadat, Mtch2). Expression of genes participating in redox homeostasis was changed in both liver and kidney, consistent with the role of oxidative stress in aging (reviewed in ref. 36). Some of these genes were down-regulated in kidney and liver (Pex16, Sod1), whereas other genes were up-regulated (Gpx3, Gsta2: in both liver and kidney; Gstt3, Sod3: in liver; Gstt1: in kidney). Other functional groups of genes with increased expression in both organs were related to regulation of blood pressure and precursors of amyloid proteins (Prnp: in liver and kidney; App, Aplp2: in liver).
Genes down-regulated in kidney included those participating in the response to glucocorticoid hormones, cofactor biosynthesis, and lipid metabolism. Notably, 21 genes encoding peroxisomal components showed decreased expression with age in kidney, in agreement with association of age-related alterations in lipid metabolism with renal disorders (37).
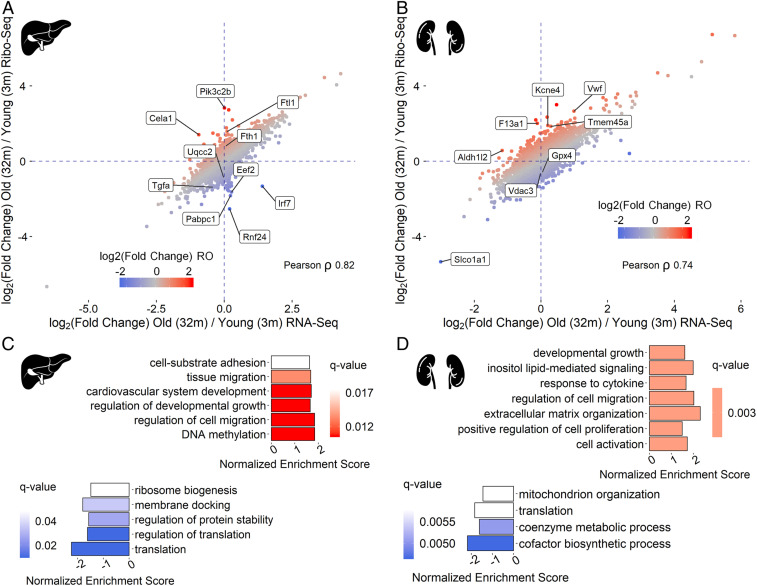
The majority of age-related changes in the translatome were correlated with those in the transcriptome in both organs, suggesting that the primary changes of gene expression with age occur at the transcriptional level (Fig. 3 A and B and SI Appendix, Fig. S3). Therefore, we searched for putative transcription factors (TFs) regulating genes up- or down-regulated with increased age (Table 1 and Dataset S4). This analysis revealed RelA (an NF-κB p65 subunit) and Spi-B (lymphocyte-specific transcription activator), which may activate transcription of their targets with aging in both liver and kidney. Most of their targets up-regulated with age participate in inflammatory response and immune processes. In addition, 55 out of 217 RelA targets up-regulated in kidney were found to be shared with another transcription factor, Jun (an AP-1 transcription factor subunit), which in cooperation with NF-κB promotes cell survival (38). In turn, shared targets of Jun and Smad3 were also enriched for genes with an age-related increase in expression, reflecting the response to proapoptotic cytokine TGFβ (39). Additional factors that can participate in transcriptional activation of immunity-related genes with age were MafB in liver and Irf3 and Stat5a in kidney. Targets of peroxisome proliferator-activated receptor α, PPARα (alone or together with retinoic acid receptor RXRα), were enriched for up-regulated lipid metabolism genes in kidney. The PPARα/RXRα dimer activates transcription of genes participating in fatty acid oxidation and catabolism (40). Moreover, activation of PPARα represses NF-κB function, and the reduction in PPARα levels is associated with aging (41). Ppara itself is among the Foxa3 transcription factor targets, which was enriched in kidney genes up-regulated with age. For down-regulated genes, we found two potential transcription factors involved in kidney, Arid1a and Hnf4a, regulating expression of genes responsible for lipid and cofactor metabolism.
Fig. 3.
Age-dependent changes in ribosome occupancy of functional gene groups. (A and B) Comparison of transcriptional (RNA-seq) and translational output (Ribo-seq) for liver (A) and kidney (B) of 32-mo-old mice (two replicates) vs. 3-mo-old-mice (three replicates) (Dataset S2). (C and D) Functional groups of genes with age-related changes in RO presented as the GSEA results. RO linear changes with age (from 3- to 32-mo-old mice) were estimated with edgeR (14 mice in total). Genes were sorted according to their signed P values [−log10(P value)*sign(log2(Fold change))], in liver (C) and kidney (D). GO BP terms with q value less than 0.25 are shown (Dataset S3).
Table 1.
Transcription factors with binding sites enriched in the promoters of genes up- or down-regulated with age
| Gene symbol | P value, adjusted* | Odds ratio | No. of targets |
| Potential regulators up-regulated with age in liver | |||
| Spib | 9.17E-03 | 3.29 | 19 |
| Rela | 1.25E-02 | 1.57 | 109 |
| Mafb | 4.96E-02 | 1.84 | 42 |
| Potential regulators up-regulated with age in kidney | |||
| Irf3 | 1.87E-06 | 2.43 | 66 |
| Msx1 | 4.67E-04 | 1.43 | 261 |
| Rela | 7.64E-04 | 1.44 | 217 |
| Spib | 1.97E-02 | 2.29 | 28 |
| Ppara | 2.34E-02 | 1.49 | 101 |
| Cebpa | 4.41E-02 | 1.27 | 254 |
| Foxa3 | 4.41E-02 | 1.86 | 36 |
| Jarid2 | 4.41E-02 | 1.23 | 535 |
| Stat5a | 4.64E-02 | 1.30 | 194 |
| Potential regulators down-regulated with age in kidney | |||
| Hnf4a | 1.55E-02 | 1.45 | 156 |
| Arid1a | 1.55E-02 | 1.35 | 244 |
| Interactions of two transcription factors | |||
| Potential regulators up-regulated with age in kidney | |||
| Rela + Jun | 1.10E-02 | 1.82 | 55 |
| Ppara + Rxra | 3.08E-02 | 1.56 | 77 |
| Ppara + Gata4 | 3.08E-02 | 2.93 | 16 |
| Msx1 + Tbp | 3.20E-02 | 1.92 | 35 |
| Jun + Smad3 | 3.20E-02 | 1.71 | 49 |
| Stat5a + Esr1 | 3.20E-02 | 1.50 | 83 |
P values of right-sided Fisher’s exact test were adjusted with the Benjamini–Hochberg correction.
Aging Affects Ribosome Occupancy of Specific Groups of Transcripts.
Many genes down-regulated with age in liver Ribo-seq data were related to protein synthesis, noncoding RNA metabolism, and ribosome biogenesis. Among them were the genes encoding numerous ribosomal proteins, translation factors, proteins involved in large (Nol9, Nsa2) and small (Utp14a, Tsr1) ribosome subunits biogenesis, RNA polymerase I components (Cd3eap), nuclear import and export proteins (Nmd3, Ipo4, Sdad1), RNA helicases (Ddx1, Ddx17, Ddx21), noncoding RNA processing nucleases (Elac2), aminoacyl-tRNA synthetases (Mars, Qars, Farsb), components of rRNA pseudouridylation complex (e.g., Nhp2), and RNA-binding proteins with various functions (Cirbp, Aimp1, Pa2g4, Rtraf). In kidney, various genes related to protein synthesis also showed a decreased expression (e.g., Eif3h, Eif4g3, Eif5, Eif5a, Dars, Sars, Ddx3x, Cirbp, Rtraf). This functional group was not observed in previous age-related transcriptome studies, suggesting down-regulation specifically at the posttranscriptional level. To investigate this further, we analyzed contributions of transcription and translation by comparing age-related changes detected in RNA-seq and Ribo-seq data (Fig. 3 and SI Appendix, Fig. S3).
Together, Ribo-seq and RNA-seq allow decomposition of the contribution of transcription and translation to gene expression changes by analyzing ribosome occupancy (RO) (number of ribosome footprints normalized to transcript abundance) of particular transcripts. RO is a proxy of translation efficiency, since in most cases the more ribosomes translate the mRNA, the more products are made. Thus, we first compared age-dependent changes in gene expression identified by Ribo- and RNA-seq in each organ (Fig. 3 A and B). Even though age-related changes in gene expression were mostly driven by transcriptome changes, there were also distinct outliers, and almost all of them were differentially expressed at the translational and not the transcriptional level.
To identify functional groups of genes whose translation is age-dependent, we performed gene set enrichment analysis (GSEA). The linear RO changes from 3- to 32-mo-old mice were estimated considering the data from all respective age groups. The lists of genes were sorted according to the signed P values. In both organs, up-regulated genes represented the GO terms related to inflammation, development, and differentiation of different cell types, whereas the GO terms encompassing down-regulated genes in kidney included mitochondrial components related to the electron transport chain, mitochondrial translation, and membrane (Fig. 3 C and D and Dataset S3). Although a decline in mitochondrial function is a well-documented phenomenon in aging, translational control of this process has not yet been reported. Intriguingly, many nucleus-encoded mRNAs related to mitochondria activity and biogenesis are regulated at the translational level by mTOR pathway through 4E-BP–dependent translational regulation (42). Thus, several functional groups of genes indeed exhibited translational regulation affected by aging, although there were no individual genes passing false discovery rate-adjusted P value. However, by examining the top and bottom of the ranked gene list, we found genes encoding functionally related products. Among the genes with the RO decreased with age, a striking example was IFN regulatory transcription factor 7 (Irf7), known to be specifically repressed through 4E-BPs (43). Its RO was approximately six times lower in old compared to young samples. The most prominent GO terms enriched in down-regulated genes in liver were related to protein biosynthesis (Fig. 3 A and C). In contrast, mRNAs encoding ferritin subunits Fth1 and Ftl1, known to be translationally regulated by an iron-responsive element in their 5′-UTRs (44), were both up-regulated in liver. This may reflect the well-known disturbances in iron metabolism and storage associated with aging (45), or be a consequence of the aging-related inflammation, as these mRNAs are translationally up-regulated upon inflammation response (46).
The “translation” GO term included genes that were also down-regulated in kidney, although with lower significance. It could be that the observed changes in RO are linked to changes in transcript isoform abundance. To clarify this, we analyzed changes in transcript isoform abundance with age (Dataset S5) and compared them with RO changes of the corresponding genes. However, we detected neither significant switches of major transcript isoforms with age nor association between isoform abundance and RO changes (SI Appendix, Figs. S4 and S5).
Translation of 5′-Terminal Oligopyrimidine Transcripts Encoding Components of Protein Synthesis Machinery Is Repressed with Age.
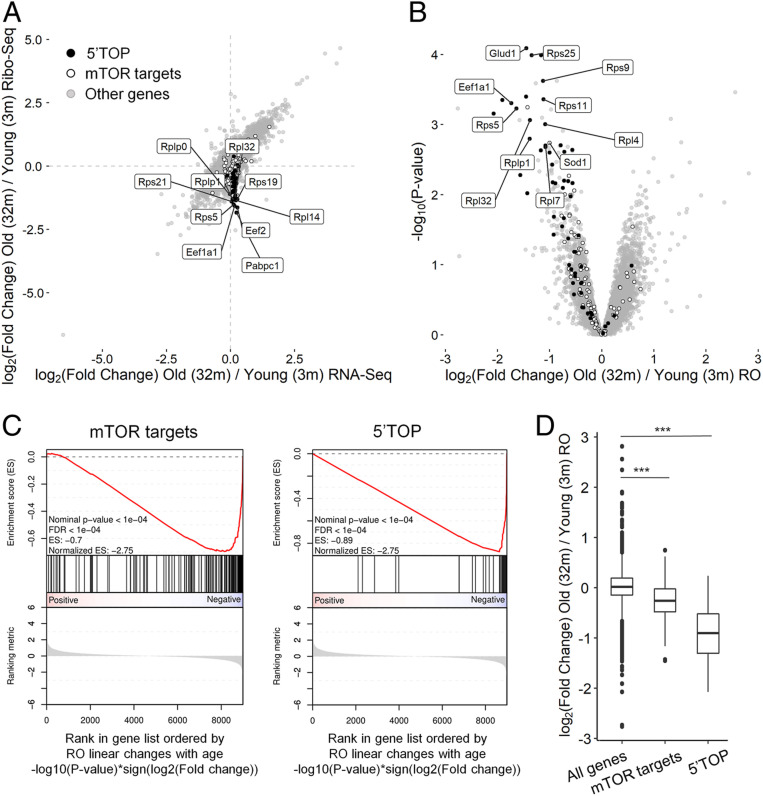
Translation of the mRNAs encoding the components of protein synthesis machinery is known to be regulated through the mTOR pathway. Specifically, many of mTOR mRNA targets carry the 5′-terminal oligopyrimidine (5′-TOP) regulatory sequence (47), making them more sensitive to mTOR inhibition than the rest of the transcriptome (48). We analyzed the age-related RO changes of these transcripts (Fig. 4). In liver, translation of mTOR targets was down-regulated in old animals (32- vs. to 3-mo-old mice) and among them the 5′-TOP transcripts were the most affected (Fig. 4A and B and SI Appendix, Fig. S8A). For example, genes encoding ribosomal proteins Rps5, Rps11, Rps25, Rps21, elongation factor Eef2, and Pabpc1 were among those with the strongest RO decrease in liver (Fig. 4A and B and SI Appendix, Fig. S7). Most of these genes had a shared pattern of RO change with age: Ribo-seq footprints count increased from 1 to 3 mo and decreased gradually from 3 mo to the oldest age, whereas RNA-seq counts were more or less stable in different age groups (SI Appendix, Fig. S7). Decrease of RO was also detected if linear RO change with age was supplied to GSEA (Fig. 4C) and Mann–Whitney U test (Fig. 4D). The significant, although weaker decrease in RO of 5′-TOP transcripts and mTOR targets was also observed when the 32-mo-old mice were excluded from the analysis (SI Appendix, Fig. S8 B and C), suggesting that the 5′-TOP genes and mTOR targets tend to exhibit lower ROs earlier in life, with the most prominent decrease at the older age.
Fig. 4.
Decreased ribosome occupancy of transcripts encoding ribosomal and other translation-related proteins with age in liver. (A) Comparison of transcriptome (RNA-seq) and translation output (Ribo-seq) log2(Fold change) between 32-mo-old mice (two mice) and 3-mo-old-mice (three mice). (B) The volcano plot shows the log2(Fold change) of the RO between 32-mo-old mice (two mice) and 3-mo-old-mice (three mice). (C) GSEA of age-related changes in RO of 41 5′-TOP and 160 mTOR-sensitive genes (59) in liver. RO linear changes with age (from 3- to 32-mo-old mice) were estimated with edgeR (14 mice in total). Genes were sorted according to their signed P values [−log10(P value)*sign(log2(Fold change))]. (D) Box plot showing distribution of mTOR regulated and 5′-TOP genes ROs. Statistical significance was calculated with Mann–Whitney U test.
Thus, the data suggest that the observed decline in the translation of transcripts encoding components of the protein synthesis apparatus can be explained by down-regulation of mTOR with age. Interestingly, we found that the abundance of mTOR mRNA itself was negatively associated with age according to our Ribo-seq and RNA-seq data (Dataset S2), suggesting that during aging mTOR kinase can be regulated at the transcriptional level.
In kidney, a similar but weaker pattern of changes was observed for transcripts encoding translation-related components (SI Appendix, Fig. S6). Although Ribo-seq counts slightly decreased from 3 to 32 mo, RNA-seq did not change or even increased for most of these genes, thus compensating for the observed effect (SI Appendix, Fig. S7). This is probably why the translation GO term was not detected in the overall differential expression profile in kidney (Fig. 2A). Nevertheless, even with a similar pattern of change in translation-related transcripts and RO, the repertoire of these transcripts was not identical for liver and kidney (e.g., compare Fig. 4 A and B and SI Appendix, Fig. S6 A and B and Dataset S6). We also observed down-regulation of 5′-TOP transcript translation with age and a decrease in mTOR transcript levels (Dataset S2), while in this case we did not find a pronounced decrease in RO of mTOR-sensitive transcripts (SI Appendix, Fig. S6C).
Redistribution of Ribosome Coverage toward the 3′ End of Coding Sequences with Age.
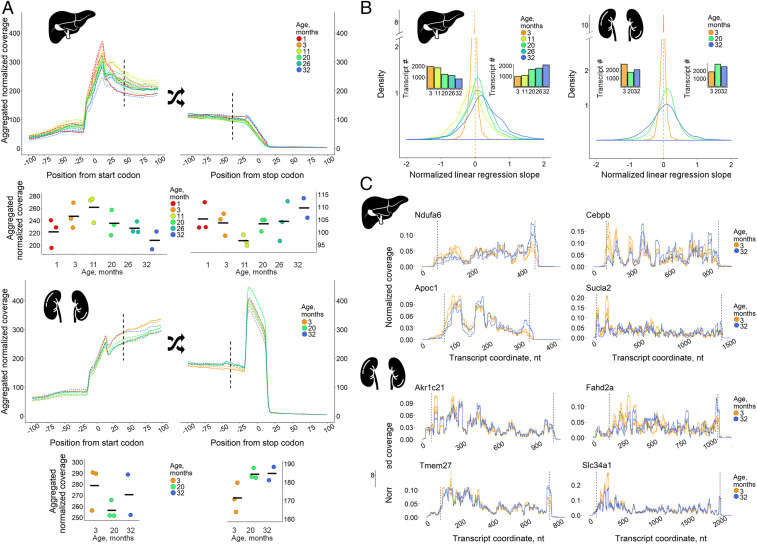
To analyze possible age-related changes in ribosomal distribution along the transcripts, we constructed metagene profiles of ribosomal coverage in 100-nt windows surrounding the start and stop codons of reliably expressed transcripts in liver and kidney (Fig. 5A). Strikingly, we observed an age-dependent decrease in ribosomal footprint density at the 5′ end of coding regions and an increase at its 3′ proximal part, in both organs. In liver, there was a clear turning point at 11 mo. A change in ribosome distribution of a limited subset of mRNAs may strongly contribute to the observed pattern, but it may also be explained by minor changes in many mRNAs. To distinguish between these possibilities, we analyzed the positional profiles of ribosome footprints along individual transcripts, relative to the 3-mo reference point. We split each transcript into a fixed set of segments (Materials and Methods) and fitted a linear model of footprint coverage using a relative transcript coordinate as the predictor variable (Fig. 5B and SI Appendix, Fig. S9 and Supplementary Methods, section 5). For each age, we found a significant shift of the distribution of linear regression slopes across transcript (compared to the 3-mo old mice using the sign test [SI Appendix, Fig. S9]), whereas only a small number of transcripts showed ribosome coverage increases in the 5′ proximal part. The effect gradually became more pronounced for samples from 11- to 32-mo-old mice (Fig. 5B). Thus, changes in ribosomal coverage along the transcript reflect a global tendency of translation deregulation with aging. These age-dependent changes are illustrated in Fig. 5C by showing representative transcripts. The effect was most pronounced in the vicinity of start and stop codons, but also was detectable along whole transcripts (SI Appendix, Fig. S9). Overall, this redistribution pattern uncovers yet another layer of age-related translation deregulation.
Fig. 5.
Gradual age-related rearrangement of ribosome footprints toward the 3′ end of coding sequence. (A) Metagene profiles of ribosomal coverage in the vicinity of start and stop codons (200-nt windows) of 2,920 and 4,566 transcripts for liver and kidney, respectively. Metagene coverage values at +42-nt position from start codons and at −42-nt position from stop codons are presented on separate graphs below the main graphs (coverage values in replicates, mean shown as horizontal lines). (B) Distribution of linear regression slopes for ribosome footprint profiles normalized to mean coverage at 3 mo and smoothed with the relative transcript coordinate as the predictor variable (SI Appendix, Supplementary Methods, section 5). Bar plots depict the number of transcripts with negative (Left) and positive (Right) slopes. Data from three mice per age were analyzed except for the 32-mo-old samples (two mice). (C) Representative transcripts exhibiting increased ribosome footprint coverage in liver and kidney with age. The dashed lines denote start and stop codons.
Discussion
Alteration of protein synthesis with age has been a contentious issue for some time (reviewed in ref. 7). Although the overall protein synthesis in mammals is thought to decrease with age, mechanistic details remained elusive. In this work, we applied Ribo-seq and RNA-seq to analyze age-related changes in the translatomes of mouse liver and kidney over the entire life span. This allowed us to characterize functional groups of genes, whose expression is altered with age at either transcription or translation levels, and to uncover candidate genes that may serve as markers of aging in examined tissues. In addition, we identified specific translational deregulation of 5′-TOP transcripts encoding components of protein synthesis machinery during aging. We also discovered that the positional profile of ribosomes along the transcript differs between young and old animals, reflecting a further layer of age-related changes in translation as animals age.
The gene expression changes that we observed (Fig. 2) were generally consistent with the previously described transcriptomic patterns during aging (e.g., refs. 33 and 34), pointing to the major processes altered with age at the level of gene expression, such as inflammation, regulation of blood pressure, lipid and glucocorticoid biosynthesis, proteasomal protein degradation, mitochondrial activity, and oxidative stress. A strong correlation between Ribo-seq and RNA-seq profiles as well as concordance with data reported in previous studies indicate that age-related changes in gene expression are manifested predominantly at the transcriptional level, at least in liver and kidney (Fig. 3). Expression of the majority of genes differentially expressed over the life span followed a U-shaped curve with the turning point around 3 mo of age. In previous studies, such a pattern was not observed, probably because age-related gene expression changes were examined either only at two time points (14, 33) or they did not include young animals (1 mo old) (14, 33, 34). However, the U-shaped, “reversal” pattern of gene expression was previously reported for human (29, 30) and rat (49) brain, with the turning points at ∼3.5 and ∼20 y for humans and between 6 and 12 mo for rats.
Apart from age-related changes in gene expression at the transcriptome level, we found a number of transcripts affected specifically at the level of translation. These include transcripts coding for mitochondrial proteins, immunity-related transcription factor Irf7, clinically relevant hemostasis factors F13a1 and Vwf, ferroptosis-related peroxidase Gpx4, and two ferritin subunits, Fth1 and Ftl1. Additionally, genes from the GO terms related to development and cell differentiation were enriched among the genes with RO up-regulated with age in both organs. On other hand, we did not find an association between isoform abundance and RO changes (SI Appendix, Figs. S4 and S5 and Dataset S5), suggesting that alternative splicing is unlikely to be the source of the observed RO changes.
Most importantly, our analysis revealed a particular subset of transcripts, the 5′-TOP mRNAs encoding multiple components of protein synthesis machinery, whose translation efficiency is gradually decreased with age (Fig. 4). This group of transcripts is specifically regulated by the mTOR/4E-BP axis (47), a signaling pathway known to be associated with aging, life span control, and longevity (for review, see ref. 7). Interestingly, we also observed down-regulation of mTOR mRNA abundance with age, suggesting the existence of a transcriptional component of mTOR regulation during aging (Dataset S2). Although ribosome profiling provides expression data for protein-coding genes only, it is likely that rRNA and tRNA synthesis are also compromised during aging, as their expression is controlled by the mTOR pathway as well (50).
Thus, we showed that translation of mRNAs encoding protein synthesis machinery components is decreased with age in both liver and kidney. This pattern well correlates with the previously observed decline in overall protein synthesis with age (for review, see ref. 7). However, in contrast to rapidly proliferating cancer cells, where the mTOR-dependent transcripts constitute a major fraction of polysome-associated mRNAs, in our data, collected from terminally differentiated cells of mouse organs, 5′-TOP transcripts were not enriched among the highly translated transcripts. Therefore, their decreased association with polysomes is not supposed to significantly contribute to the overall decline in protein synthesis in old animals, but rather affects indirectly by altering translation machinery abundance and composition. Of note, the observed decrease in the overall expression of translation machinery components is most notable in liver, whereas in kidney changes in the translation rate of these transcripts are substantially compensated for at the transcriptional level (SI Appendix, Fig. S6). We also cannot exclude a contribution of age-related changes in tissue composition, as whole-tissue lysates were used for experimental analyses.
The described translational down-regulation of transcripts encoding components of protein synthesis machinery was not detected in the pairwise comparison of gene expression in liver and brain of young and old rats, performed by an earlier ribosome profiling study, presumably because changes in the synthesis of translation machinery components are more prominent at older ages (14). However, the findings are consistent with the translatome changes during yeast replicative aging (13). Interestingly, as the yeast transcripts encoding translation-related components do not exhibit 5′-TOP motifs (47) and thus are unlikely to be translationally regulated by TOR in the manner similar to the mammalian transcripts. In this case, the reduction of both overall translation and ribosome protein synthesis is achieved by distinct mechanisms, i.e., activation of the GCN2/eIF2α regulatory pathway and elevated mRNA recruitment to P bodies in aging cells (13). Interestingly, the GCN2/eIF2α axis was shown to regulate translation of 5′-TOP mRNAs also in mammals (50), so its possible link with the observed phenomenon is worthy of further investigation.
Another important observation in our study concerns changes in the metagene profile of ribosome coverage along coding regions (Fig. 5), pointing to a systemic alteration of translation with age. We suggest that these changes reflect the decrease in overall translation initiation efficiency caused by the observed down-regulation of the mTOR pathway (Fig. 4), as well as a possible decline in translation termination or ribosome recycling rate. Our model is based on the idea of the differential elongation speed in different parts of the coding sequence (51, 52). It is known that slow codons are distributed within transcripts in a nonrandom fashion and are particularly enriched in the region following the initiation site, while the distal parts of the coding regions have a much lower content of slow codons (51, 52). As a result, the first ∼30 to 50 codons are usually translated more slowly, whereas the last ∼50 codons are the fastest (51). Thus, under conditions when translation initiation is inhibited, ribosome density along the coding sequence should be redistributed from the 5′ proximal to the distal part of the transcript, while inhibition of termination and/or of ribosome recycling should increase the density in its 3′ proximal part. It should be noted, however, that elevated elongation speed and perhaps other factors could also lead to a similar pattern of metagene profile changes with age (53).
In summary, our results revealed previously unknown modes of translation deregulation with age. The decrease in translation rate may reflect an attempt of the cell to cope with the accumulation of damaged proteins or compensate for the deficit of energy with age (7). For many model organisms from yeast to primates, pharmacological, dietary, and genetic interventions reducing protein synthesis rate and inhibiting mTOR signaling have been shown to significantly increase life span (7, 8, 54). However, our study clearly shows that younger tissues are actually characterized by more active protein synthesis and elevated translation of mTOR-dependent mRNAs. Thus, returning the cell to a younger state should include renewal and reactivation of protein synthesis machinery, accompanied by simultaneous reinforcement of the cellular proteostasis network. As revealed by our study, age-related translation deregulation has many faces, together contributing to dysfunction in this most important cellular process during aging.
Materials and Methods
Tissue Collection and Lysis.
Tissue samples were collected from male C57BL/6 mice of indicated ages from the National Institute on Aging Aged Rodent Colony, as described in ref. 55. For each age, three biological replicates were prepared (three mice), except for the 32-mo group (two mice). Liver and kidney samples were sliced and frozen in liquid N2 and stored at −80 °C. Tissue samples (∼55 and ∼75 mg for liver and kidney, respectively) were used for subsequent analyses. Tissues were thawed and mechanically disrupted in the lysis buffer (20 mM Tris⋅HCl, pH 7.5, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, 1% Triton X-100) supplemented with 0.1 mg/mL cycloheximide (CHX) as described previously (4). Flash-freezing and rapid thawing followed by disruption in the presence of high concentrations of CHX allowed to minimize artifacts associated with the CHX treatment (56). We note that no CHX-induced artifacts were reported for animal tissue samples (57). After homogenization and centrifugation, 250 µL of lysate were supplied with 20 units of SUPERase-In RNase inhibitor and taken for RNA-seq library preparation, and the 500 µL of lysate was brought to 1 mL with lysis buffer and taken for Ribo-seq library preparation.
Ribosome Profiling Sequencing Library Preparation.
Ribosome profiling libraries were prepared as described previously (4) with the modification in RNase digestion as indicated below. RNA digestion of lysates was performed for 1 h with the mixture of 2,000 units of RNase T1 (Epicentre) and 300 units of RNase S7 (Roche). After 30 min of incubation, 0.8 mg of heparin was added to inhibit all RNases except for RNase T1. After digestion, lysates were supplied with 80 units of SUPERase-In RNase inhibitor.
Transcriptome Library Preparation and Sequencing.
Total RNA was isolated from 250 µL of lysate with 750 µL of TRIzol LS Reagent and treated with RQ1 RNase-Free DNase (1 unit for 1 µg of total RNA) for 30 min at 37 °C with subsequent water-saturated acidic phenol extraction and precipitation with ethanol (with the addition of 1/100 volume of glycogen RNA grade). Five hundred nanograms of DNase I-treated total RNA was depleted of ribosomal RNA with NEBNext rRNA Depletion Kit (Human/Mouse/Rat) (#E6310) and used for transcriptome library preparation with NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (#E7760). Both ribosome profiling and transcriptome libraries were sequenced on the Illumina NextSeq 500/550 system (Faculty of Bioengineering and Bioinformatics, Lomonosov Moscow State University, and Institute for Information Transmission Problems, Russian Academy of Sciences).
Ribo-Seq and RNA-Seq Sequencing, Data Processing, and Bioinformatic Analyses.
The bioinformatic analysis of the Ribo-seq and RNA-seq sequencing data are described in detail in SI Appendix, Supplementary Methods. Briefly, the Ribo-seq and RNA-seq reads were aligned to the mouse transcriptome and genome assemblies (mm10, GRCm38.p5) using GENCODE M13 annotation. For the differential expression analysis, RNA-seq and Ribo-seq read counts were relative log expression-normalized separately for kidney and liver and transformed to cpm (counts per million). Genes with less than 1 cpm at least in one sample were excluded. Differential expression was analyzed with the generalized linear model of the edgeR package (58). Paired age comparisons and a model describing linear changes of gene expression with age were used in the study. To analyze ribosomal coverage changes and construct metagene profiles, the reads were aligned to the transcriptome. The used transcripts with less than 2.5 transcripts per kilobase million (tpm) at least in one sample were excluded. To construct the metagene profiles for each transcript, the coverage profile was normalized to the respective (5′-end or full-length footprint) Ribo-seq whole-transcript average coverage, and then, the normalized values were summed up for all of the transcripts. The dynamics of age-related changes in ribosomal coverage of protein-coding regions were assessed as follows. The coverage profile for each transcript was processed to obtain a list of nonoverlapping segments with stable footprint coverage, and normalized to the mean coverage at 3 mo. A linear regression analysis for the normalized ribosome footprint coverage was conducted with the relative transcript coordinate as a predictor variable (see SI Appendix, Supplementary Methods, section 5, for details).
Ethics Approval.
Experiments were carried out according to the protocols approved by the Institutional Animal Care and Use Committee of the Brigham and Women’s Hospital.
Data Availability.
All raw and processed sequencing data generated in this study have been deposited in the Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE123981).
Supplementary Material
Acknowledgments
We are grateful to Maria D. Logacheva, Faculty of Bioengineering and Bioinformatics, Lomonosov Moscow State University, and Institute for Information Transmission Problems, Russian Academy of Sciences, for Illumina sequencing and valuable comments. We also thank Irina A. Eliseeva, Pavel V. Baranov, Dmitry E. Andreev, and Nadezhda E. Makarova for discussion, and Ekaterina A. Sakharova, Alexander Tyshkovskiy, and Philipp O. Gusev for assistance with bioinformatics analyses. The work was funded by Russian Federation Grant 14.W03.31.0012 and NIH Grants DK117149 and AG047745, and the bioinformatics analyses were supported by Russian Science Foundation Grant 18-14-00291.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: All raw and processed sequencing data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE123981).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001788117/-/DCSupplemental.
References
- 1.Hipp M. S., Kasturi P., Hartl F. U., The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 20, 421–435 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Steffen K. K., Dillin A., A ribosomal perspective on proteostasis and aging. Cell Metab. 23, 1004–1012 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Morimoto R. I., Cuervo A. M., Proteostasis and the aging proteome in health and disease. J. Gerontol. A Biol. Sci. Med. Sci. 69 (suppl. 1), S33–S38 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerashchenko M. V., Peterfi Z., Gladyshev V. N., Organ-specific translation elongation rates measured by in vivo ribosome profiling. bioRxiv:279257 (8 March 2018).
- 5.Treaster S. B., et al., Superior proteome stability in the longest lived animal. Age 36, 9597 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pérez V. I., et al., Protein stability and resistance to oxidative stress are determinants of longevity in the longest-living rodent, the naked mole-rat. Proc. Natl. Acad. Sci. U.S.A. 106, 3059–3064 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anisimova A. S., Alexandrov A. I., Makarova N. E., Gladyshev V. N., Dmitriev S. E., Protein synthesis and quality control in aging. Aging 10, 4269–4288 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonskikh Y., Polacek N., Alterations of the translation apparatus during aging and stress response. Mech. Ageing Dev. 168, 30–36 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Van Remmen H., Ward W. F., Sabia R. V., Richardson A., Gene Expression and Protein Degradation (Wiley, 1995). [Google Scholar]
- 10.Mobley C. B., et al., Aging in rats differentially affects markers of transcriptional and translational capacity in soleus and plantaris muscle. Front. Physiol. 8, 518 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Aquila P., et al., Methylation of the ribosomal RNA gene promoter is associated with aging and age-related decline. Aging Cell 16, 966–975 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connors M. T., Poppi D. P., Cant J. P., Protein elongation rates in tissues of growing and adult sheep. J. Anim. Sci. 86, 2288–2295 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Hu Z., et al., Ssd1 and Gcn2 suppress global translation efficiency in replicatively aged yeast while their activation extends lifespan. eLife 7, e35551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ori A., et al., Integrated transcriptome and proteome analyses reveal organ-specific proteome deterioration in old rats. Cell Syst. 1, 224–237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curran S. P., Ruvkun G., Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 3, e56 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teleman A. A., Chen Y. W., Cohen S. M., 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes Dev. 19, 1844–1848 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zid B. M., et al., 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell 139, 149–160 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steffen K. K., et al., Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell 133, 292–302 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennedy B. K., Lamming D. W., The mechanistic target of rapamycin: The grand ConducTOR of metabolism and aging. Cell 23, 990–1003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hipkiss A. R., On why decreasing protein synthesis can increase lifespan. Mech. Ageing Dev. 128, 412–414 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Azpurua J., et al., Naked mole-rat has increased translational fidelity compared with the mouse, as well as a unique 28S ribosomal RNA cleavage. Proc. Natl. Acad. Sci. U.S.A. 110, 17350–17355 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ke Z., et al., Translation fidelity coevolves with longevity. Aging Cell 16, 988–993 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swovick K., et al., Cross-species comparison of proteome turnover kinetics. Mol. Cell. Proteomics 17, 580–591 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ingolia N. T., Ghaemmaghami S., Newman J. R. S., Weissman J. S., Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerashchenko M. V., Gladyshev V. N., Ribonuclease selection for ribosome profiling. Nucleic Acids Res. 45, e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingolia N. T. T., Lareau L. F. F., Weissman J. S. S., Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castelo-Szekely V., Arpat A. B., Janich P., Gatfield D., Translational contributions to tissue specificity in rhythmic and constitutive gene expression. Genome Biol. 18, 116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Somel M., Khaitovich P., Bahn S., Pääbo S., Lachmann M., Gene expression becomes heterogeneous with age. Curr. Biol. 16, R359–R360 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Colantuoni C., et al., Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 478, 519–523 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dönertaş H. M., et al., Gene expression reversal toward pre-adult levels in the aging human brain and age-related loss of cellular identity. Sci. Rep. 7, 5894 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franceschi C., Garagnani P., Parini P., Giuliani C., Santoro A., Inflammaging: A new immune-metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 14, 576–590 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Lee B. Y., et al., Senescence-associated β-galactosidase is lysosomal β-galactosidase. Aging Cell 5, 187–195 (2006). [DOI] [PubMed] [Google Scholar]
- 33.de Magalhães J. P., Curado J., Church G. M., Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 25, 875–881 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White R. R., et al., Comprehensive transcriptional landscape of aging mouse liver. BMC Genomics 16, 899 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun N., Youle R. J., Finkel T., The mitochondrial basis of aging. Mol. Cell 61, 654–666 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Labunskyy V. M., Gladyshev V. N., Role of reactive oxygen species-mediated signaling in aging. Antioxid. Redox Signal. 19, 1362–1372 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang T., Liebman S. E., Lucia M. S., Li J., Levi M., Role of altered renal lipid metabolism and the sterol regulatory element binding proteins in the pathogenesis of age-related renal disease. Kidney Int. 68, 2608–2620 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Baud V., Karin M., Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 11, 372–377 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y., Feng X.-H., Derynck R., Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature 394, 909–913 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Lefebvre P., Chinetti G., Fruchart J.-C., Staels B., Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J. Clin. Invest. 116, 571–580 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erol A., The functions of PPARs in aging and longevity. PPAR Res. 2007, 39654 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morita M., et al., mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 18, 698–711 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Colina R., et al., Translational control of the innate immune response through IRF-7. Nature 452, 323–328 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Muckenthaler M., Gray N. K., Hentze M. W., IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Mol. Cell 2, 383–388 (1998). [DOI] [PubMed] [Google Scholar]
- 45.Xu J., Jia Z., Knutson M. D., Leeuwenburgh C., Impaired iron status in aging research. Int. J. Mol. Sci. 13, 2368–2386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell C. H., Solgonick R. M., Linder M. C., Translational regulation of ferritin synthesis in rat spleen: Effects of iron and inflammation. Biochem. Biophys. Res. Commun. 160, 453–459 (1989). [DOI] [PubMed] [Google Scholar]
- 47.Meyuhas O., Kahan T., The race to decipher the top secrets of TOP mRNAs. Biochim. Biophys. Acta 1849, 801–811 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Philippe L., Vasseur J.-J., Debart F., Thoreen C. C., La-related protein 1 (LARP1) repression of TOP mRNA translation is mediated through its cap-binding domain and controlled by an adjacent regulatory region. Nucleic Acids Res. 46, 1457–1469 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wood S. H., Craig T., Li Y., Merry B., de Magalhães J. P., Whole transcriptome sequencing of the aging rat brain reveals dynamic RNA changes in the dark matter of the genome. Age 35, 763–776 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li B. B., et al., Targeted profiling of RNA translation reveals mTOR-4EBP1/2-independent translation regulation of mRNAs encoding ribosomal proteins. Proc. Natl. Acad. Sci. U.S.A. 115, E9325–E9332 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tuller T., et al., An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141, 344–354 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Tuller T., Zur H., Multiple roles of the coding sequence 5′ end in gene expression regulation. Nucleic Acids Res. 43, 13–28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kasari V., Margus T., Atkinson G. C., Johansson M. J. O., Hauryliuk V., Ribosome profiling analysis of eEF3-depleted Saccharomyces cerevisiae. Sci. Rep. 9, 3037 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kennedy B. K., Kaeberlein M., Hot topics in aging research: Protein translation, 2009. Aging Cell 8, 617–623 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petkovich D. A., et al., Using DNA methylation profiling to evaluate biological age and longevity interventions. Cell Metab. 25, 954–960.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gerashchenko M. V., Gladyshev V. N., Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 42, e134 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharma P., Nilges B. S., Wu J., Leidel S. A., The translation inhibitor cycloheximide affects ribosome profiling data in a species-specific manner. bioRxiv:746255 (24 August 2019).
- 58.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thoreen C. C., et al., A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and processed sequencing data generated in this study have been deposited in the Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE123981).