Significance
Random errors and genetic lesions give rise to messenger RNAs encoding nonfunctional or toxic protein variants. An evolutionarily conserved cellular surveillance mechanism recognizes such aberrant transcripts and subjects them to destruction, a process known as nonsense-mediated decay (NMD). The UPF1 protein is an essential component of this NMD machinery. In a genetic screen aimed at identifying regulators of T cell development in zebrafish, we discovered a upf1 mutation that causes widespread changes in the patterns of pre-mRNA splicing. Among the mRNA splicing regulators whose transcripts were affected by the mutation, the noncanonical splicing factor rpl10a was identified as a determinant of T cell development, exemplifying the advantages of examining NMD deficiency in the context of the whole vertebrate organism.
Keywords: NMD, thymus, evolution, ribosomal protein
Abstract
The transcriptome of eukaryotic cells is constantly monitored for errors to avoid the production of undesired protein variants. The evolutionarily conserved nonsense-mediated mRNA decay (NMD) pathway degrades aberrant mRNAs, but also functions in the regulation of transcript abundance in response to changed physiological states. Here, we describe a zebrafish mutant of upf1, encoding the central component of the NMD machinery. Fish homozygous for the upf1t20450 allele (Y163X) survive until day 10 after fertilization, presenting with impaired T cell development as one of the most conspicuous features of the mutant phenotype. Analysis of differentially expressed genes identified dysregulation of the pre-mRNA splicing pathway, accompanied by perturbed autoregulation of canonical splicing activators (SRSF) and repressors (HNRNP). In upf1-deficient mutants, NMD-susceptible transcripts of ribosomal proteins that are known for their role as noncanonical splicing regulators were greatly increased, most notably, rpl10a. When the levels of NMD-susceptible rpl10a transcripts were artificially increased in zebrafish larvae, T cell development was significantly impaired, suggesting that perturbed autoregulation of rpl10a splicing contributes to failing T cell development in upf1 deficiency. Our results identify an extraribosomal tissue-specific function to rpl10a in the immune system, and thus exemplify the advantages of the zebrafish model to study the effects of upf1-deficiency in the context of a vertebrate organism.
The transcriptome of eukaryotic cells is constantly monitored for errors to avoid the production of undesired protein variants. Apart from random errors that occur during transcription, genetic lesions often give rise to premature termination codons, potentially leading to the formation of toxic proteins (1). Nonsense-mediated mRNA decay (NMD) is an evolutionarily conserved translation-dependent surveillance mechanism monitoring the integrity of mRNAs. NMD leads to degradation of aberrant mRNAs, that is, transcripts that harbor premature termination codons (PTCs), but it also functions in the regulation of transcript abundance in response to changed physiological states during development and differentiation (reviewed in refs. 2–4). In addition, a recently described transcriptional adaptation mechanism, which sets in when mutant alleles are transcribed and subjected to NMD, results in up-regulation of the transcription of genes related to mutant genes to mitigate the deleterious effects of genetic lesions (5).
The NMD pathway also shapes the landscape of transcriptomes in all eukaryotes, from yeast to plants to humans (6–15), via direct and indirect means. An important aspect of NMD-mediated regulation is its role in shaping the auto- and cross-regulatory networks of splicing regulators. This was demonstrated, for example, for ribosomal gene rpl12 in Caenorhabditis elegans and L ribosomal protein (RPL)3 and RPL12 in human (16), providing an important hint at unexpected RPLs (see below). NMD-mediated regulation also affects the expression levels of genes encoding core factors of the spliceosome, such as PRPF18 (17), and is key to determining the levels of several members of the SR family of splicing factors, such as SRSF2 (8, 17–19).
UPF1 is the central effector of the NMD pathway (20). Together with UPF2 and UPF3, this group of factors is conserved in all eukaroytes (21). UPF1 is a multidomain protein, and its ATPase and helicase activities are essential for its function, as are C-terminal domains that are the sites of key regulatory phosphorylation events (reviewed in refs. 2–4). Here, we describe the discovery and characterization of a recessive allele of upf1 in zebrafish, opening up unprecedented opportunities to examine the role of upf1 during embryonic and early larval development in a time-resolved manner, enabling the simultaneous study of the effects of upf1-deficiency in many tissue types. The upf1 allele was identified in large forward genetic screens aimed at identifying the genetic basis of T cell development in zebrafish (22). Our results provide evidence for tissue-specific differences in the requirement of NMD activity, describe the changes of the transcriptome landscape, focus on the consequences of upf1 deficiency on auto- and cross-regulation of splicing factors, such as SRSF and hnRNP, and finally uncover a previously unappreciated role for the autoregulatory regulation of rpl10a in T cell development.
Results and Discussion
Identification of a upf1 Mutant Allele Associated with Impaired T Cell Development.
An ENU (N-ethyl N-nitrosourea) mutagenesis screen was conducted aimed at identifying recessive point mutations that affect thymus and T cell development in zebrafish (22). By 5 d postfertilization (dpf) the mutants were identified by whole-mount RNA in situ hybridization (WISH) using rag1 as a marker for immature T cells, rearranging their T cell receptor gene loci in the thymus (23, 24). Compared to wild-type fish, the number of cells expressing rag 1 in the thymus is essentially absent in about 25% of fish in the HJ028 line (Fig. 1A). Meiotic recombination mapping localized the mutation to a critical interval of 0.4 cM (corresponding to about 230 kb) on chromosome 2, about 0.14 cM away from the closest informative genetic marker HJ028_25 (Fig. 1B). upf1 (ENSDARG00000016302) was the only gene located within the critical interval with a deleterious mutation; the upf1t20450 allele features a cytidine-to-adenine transversion, converting Y163 to a stop codon (Fig. 1C). UPF1 is an ∼120-kDa RNA-dependent ATPase and ATP-dependent RNA helicase and the key component in the NMD pathway (25). The predicted truncated mutant protein lacks most of the cysteine- and histidine-rich zinc-finger domain, and the Stalk, RecA1, and RecA2 domains (Fig. 1D). However, at present, we cannot exclude the possibility that the utilization of methionine codons downstream of the premature stop codon support the synthesis of N-terminally truncated proteins (SI Appendix, Fig. S1) that would be subject to SMG1-mediated phosphorylation of serine and threonine residues clustered at the C terminus of the protein that underlies the mRNA decay function of UPF1 (26–29). Hence, further work is required to determine whether the upf1t20450 allele is hypomorphic or amorphic in nature.
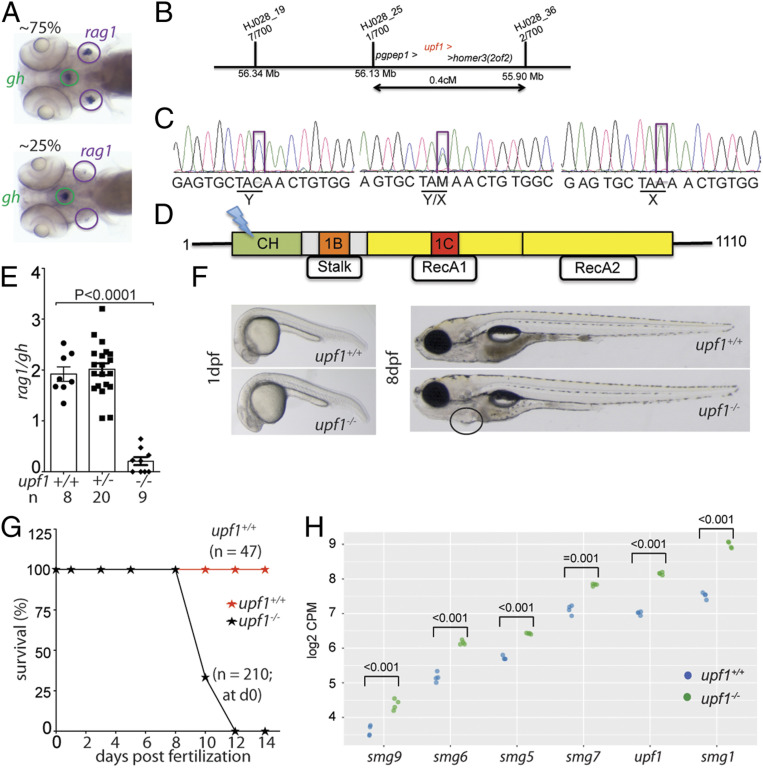
Fig. 1.
Characterization of a upf1 mutant allele. (A) In line HJ028, crosses of mutant carriers generate about 25% of fish with small numbers of rag1-expressing cells in the thymic rudiment, whereas the other siblings appear phenotypically normal. The expression of gh in cells of the hypophysis serves as an internal control. Note that some upf1 mutant fish lack detectable expression of rag1 (compare with quantitative analysis in E). (B) Schematic of the critical region for the HJ028 mutation on chromosome 2, as determined by linkage mapping with polymorphic markers and genome sequencing; the three genes located in the interval are indicated; the numbers of recombinants for each of the polymorphic markers observed in 700 meioses are indicated. (C) Sanger sequencing traces of the three genotypes in the HJ028 line. The C > A transversion is indicated (position 2:56,181,617; Zv11). (D) Schematic of the upf1 protein domains; cysteine- and histidine-rich zinc-finger domain (CH), amino acids 121 to 272; 1B/Stalk: amino acids 325 to 426; 1C: amino acids 567 to 620; RecA1: amino acids 449 to 700; RecA2: amino acids 701 to 918. The stop codon is introduced at amino acid position 163 (Y163X) in the cysteine- and histidine-rich zinc-finger domain. (E) Thymopoietic capacity of upf1 mutants as measured by RNA in situ hybridization using probes specific for rag1 (expressed in developing thymocytes) and gh (expressed in a subset of cells in the hypophysis). The latter probe serves as an internal control in two ways: Technical, to monitor the hybridization process as such; and biological, to ascertain the tissue specificity of the observed genetic effects. Each data point represents one fish; mean ± SEM. (F) Macroscopic appearance of upf1 mtants and their wild-type siblings at two time points. The cardiac region of the mutant is highlighted; the edema manifests itself as a ventral bulge, which is absent in wild-type siblings. (Magnification: 10×.) (G) Impaired survival of upf1 mutants. The survival rate of upf1 mutants was determined by analyzing aliquots of a clutch of initial 210 fish generated by a cross of heterozygous parents; the survival rate of upf1 wild-type fish was determined from a cross of wild-type fish of the same background. (H) Up-regulation of genes encoding components of the NMD pathway in upf1 mutants. Gene-level expression is shown as log2 counts per million (CPM); the levels of significance of differences between the two datasets for each gene are indicated.
Quantitative assessment of rag1 expression was determined using the thymopoietic index (30); at 5 dpf, the phenotype of wild-type fish and heterozygous siblings are indistinguishable, whereas mutant fish have a thymopoietic index of ∼10% compared to that of wild-type fish (Fig. 1E). upf1 morphants, generated by injection of an antisense oligonucleotide targeting the initiation codon, also lacked rag1 expression at 4 dpf, resulting in a low thymopoietic index (SI Appendix, Fig. S1). Interestingly, at early time points of development, lack of upf1 function primarily affects T cell development; development of the gh-expressing cells in the hypophysis is unaffected (Fig. 1A and SI Appendix, Fig. S1), as are cells in the neural crest, the pharyngeal ectoderm, and the endoderm, as indicated by normal patterns of expression of dlx2, gcm2, and foxn1 (SI Appendix, Fig. S1) tissue-specific marker genes. In contrast, cells expressing the lymphocyte marker ikzf1 are absent in the thymic rudiment (SI Appendix, Fig. S1), indicating that upf1 deficiency is associated with loss of developing thymocytes rather than misregulation of rag1 expression. Collectively, these results indicate that, compared to other cell types in the developing zebrafish larva, developing T cells are uniquely sensitive to perturbation of the NMD pathway.
The upf1 mutants exhibit normal overall morphology until 8 dpf, when the body size begins to become recognizably smaller and a pericardial edema becomes apparent (Fig. 1F and SI Appendix, Fig. S1); the mutants do not survive beyond day 12 (Fig. 1G); a similar phenotype was also described in a upf1 mutant line generated by CRISPR/Cas9 mutagenesis (5). In studies of upf1 morphants using translation block morpholino oligonucleotides a much earlier onset of developmental retardation and death was observed (31, 32), whereas our translation block morpholino oligonucleotide resulted in a milder phenotype; of note, our upf1 morphants are more severely affected than the embryos homozygous for the upf1t20450 allele. The reasons for these differences are unclear but may be related to different efficiencies of morpholinos or the residual activity of the upf1t20450 allele.
The NMD pathway is characterized by an autoregulatory negative feedback mechanism regulating the expression of core NMD factors; for example, in human cells knockdown of UPF1 significantly up-regulates the expression of other core NMD factors, such as UPF2, UPF3B, SMG1, SMG5, SMG6, and SMG7; moreover, knockdown of UPF3B up-regulates UPF1 mRNA levels (33). A similar pattern of compensatory up-regulation of core NMD factors upf1, smg1, smg5, smg6, smg7, and smg9 was associated with the zebrafish upf1 mutation (Fig. 1H). A survey of the mRNA sequences of upf1, smg1, smg6, smg7, and smg9 revealed the presence of NMD-inducing characteristics, either long 3′UTR sequences or uORFs or both (33). Collectively, these data are compatible with the known evolutionarily conserved autoregulatory structure of the NMD pathway (34) and strongly suggest that the upf1t20450 allele encodes a functionally impaired upf1 protein
Collectively, the phenotype of fish homozygous for the upf1t20450 allele establish the upf1 mutants as a unique animal model with which to examine the effect of an impaired NMD pathway on gene expression in vivo.
Lack of upf1 Impairs the NMD Pathway in Zebrafish.
Next, we systematically examined the effect of upf1 deficiency on the expression patterns of ∼600 zebrafish genes that are known to give rise to both transcripts with and transcripts without features relevant for NMD regulation (genome assembly GRCz10, available at www.ensembl.org/Danio_rerio/Info/Index?db = core) (Dataset S1). Excluding transcripts annotated as processed transcripts or nonprotein coding transcripts, a total of 663 endogenous mRNA transcripts arising from 603 genes exhibit PTC resulting from various kinds of alternative pre-mRNA splicing events in wild-type fish; in 646 of 663 transcripts, the PTCs are situated at >50 nucleotides upstream of the last exon–exon junction, which is considered to be an NMD-inducing feature (2–4) (Dataset S1). The transcriptome of upf1 mutants was determined at 5 dpf, when little morphological changes are yet apparent. Applying our stringent criteria of analysis (Materials and Methods), higher expression levels were observed for 119 (119 of 646 = 0.184) of PTC-containing transcripts, arising from 114 genes (114 of 603 = 0.19); in contrast, a reduction of expression levels was observed for only two transcripts (arising from two genes) (Fig. 2 A and B and Dataset S2). Collectively, these observations suggest that the impaired activity of the NMD pathway in upf1 mutants stabilizes at least about 20% of endogenous PTC-containing mRNA transcripts, leading to significant shifts in the ratio of expression levels for NMD versus non-NMD transcripts for these genes (Fig. 2 C and D). Of note, the introduction of a premature stop codon in exon 4 of the upf1 gene in the HJ028 line does not alter the splicing patterns of mutant transcripts (SI Appendix, Fig. S1).
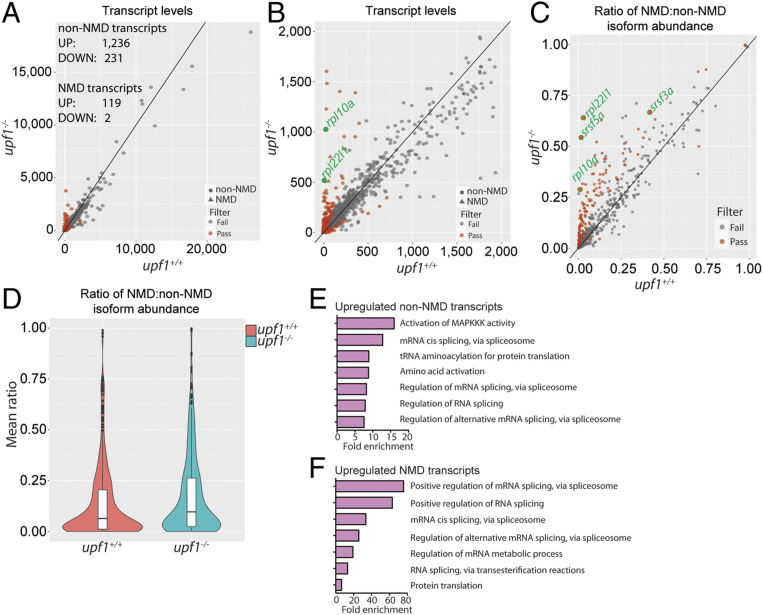
Fig. 2.
Characterization of the transcriptome of upf1 mutants at 5 dpf. (A) Differential transcript-level expression analysis of wild-type and mutant fish (Datasets S2 and S3). Expression levels are depicted as TPM. (B) Enlarged representation of data in A, focusing on genes with low expression levels. The two most highly expressed NMD transcripts (rpl22l1 and rpl10a) are indicated by green circles. (C) Increased NMD ratios in upf1 mutant fish; four selected genes are highlighted by green circles. For every gene, TPM counts were separately summed for NMD and non-NMD transcripts; the ratio was determined as division of NMD counts by the sum of NMD and non-NMD counts. (D) Violin plot of NMD ratios (P = 4.37 × 10−11). (E) GO term analysis of up-regulated non-NMD transcripts, indicating the enrichment of major categories. (F) GO term analysis of up-regulated NMD transcripts.
In addition to the effect on NMD-susceptible transcripts, upf1 deficiency impacts the expression levels of transcripts lacking PTCs. A total of 1,239 transcripts were found to be up-regulated, and 228 transcripts were found to be down-regulated (Fig. 2 A and B and Dataset S3); 519 up-regulated transcripts (42%) and 80 down-regulated transcripts (35%) were found to contain potential NMD-inducing features, such as long 3′UTRs, introns in the 3′UTR, and upstream ORFs (Dataset S3), suggesting the possibility that they also might be stabilized after the incapacitation of the NMD pathway. However, in contrast to the situation of premature stop codons, the NMD-inducing capacity of these alternative features is less well documented. Upf1 may affect the levels of certain transcripts even in the absence of known NMD-inducing features; alternatively, indirect effects may result for instance from increased or reduced levels of transcripts encoding transcriptional regulators. Further work is required to distinguish between these and other possibilities. The expression levels of many genes known to be involved in T cell differentiation, such as rag1, ccr9a, ccr9b, lck, and zap70 are down-regulated, reflecting the low number of lymphocytes in upf1 mutants; in contrast, expression levels of genes characteristic of the nonhematopoietic components of the immune system (for example, foxn1 and dlx2) were not changed in the mutants, confirming that—at least for the 5-dpf time point—the upf1 mutation primarily affects developing lymphocytes.
Biological Pathways Affected by upf1 Deficiency.
A Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (https://www.genome.jp/kegg/pathway.html) was conducted for the set of genes that are differentially expressed in upf1 mutants. Genes giving rise to dysregulated mRNAs lacking PTCs belong to three pathways (Fig. 2E): Spliceosome (KEGG dre03040; P = 1.5E-10), aminoacyl-tRNA biosynthesis (KEGG dre00970; P = 7.9E-05), and p53 signaling (KEGG dre04115; 3.7E-04). Among dysregulated NMD-susceptible transcripts, the most significantly enriched pathway is that of spliceosome (KEGG dre03040; P = 1.48E-10) (Fig. 2F). The moderate enrichment of ribosome biogenesis (pathway dre03010; P = 0.026) (Fig. 2F) is driven by overexpression of rpl10a and rpl22l1 genes (Fig. 2 B and C) that are also known for their extraribosomal function in RNA splicing (35, 36) (see below). Collectively, the pathway enrichment analysis points to alterations in pre-mRNA splicing as a major outcome of upf1 deficiency.
Alterations of Pre-mRNA Processing in upf1 Mutants.
The most prominent dysregulated KEGG signatures in upf1 mutants are associated with the regulation of pre-mRNA processing (Fig. 2 E and F), including a large number of genes encoding known regulators of alternative splicing. Indeed, the transcript levels of no fewer than 13 srsf and 16 hnrnp genes are affected by upf1-deficiency (Fig. 3A). Of note, this gene list encompasses several paralogous gene pairs—for example, srsf2a and srsf2b—indicating that they are subject to similar regulatory principles. This coregulation is unlikely to be related to the recently described process of transcriptional adaptation via up-regulation of closely related genes, since it is critically dependent on the activity of upf1 (5). Rather, the frequent coregulation of paralogous or closely related genes may be the result of pervasive cross-regulation, which example has been demonstrated for SRSF3 and SRSF5 (37), hnRNPD and hnRNPDL (38), hnRNPL and hnRNPLL (39), RBM5 and RMB10 (40), and RPL22 and RPL22L1 (35).
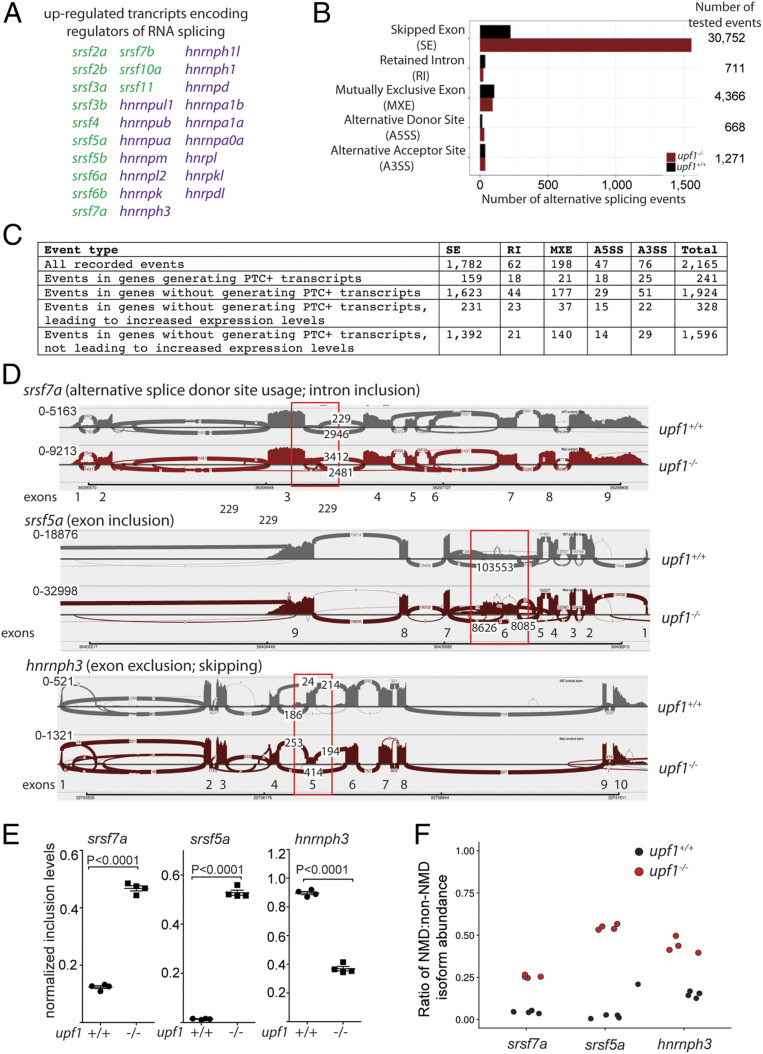
Fig. 3.
Altered landscape of pre-mRNA splicing in upf1 mutants. (A) Examples of genes encoding splice activators (srsf) and repressors (hnrnp), the transcripts of which are dysregulated in upf1 mutants as determined by transcript-level RNA-seq quantitation. (B) Summary of alternative splicing analysis. (C) Stratification of alternative splicing events. (D) Sashimi plots for three different genes indicating three different types of alternative splicing events; the affected exons are boxed. The NMD-transcript identifiers are as follows: srsf7a, ENSDART00000147803.3 (alternative donor site usage leading to inclusion of parts of intron sequences); srsf5a, ENSDART00000149400.2 (exon inclusion); hnrnph3, ENSDART00000108951.4 (exon exclusion/skipping). (E) Quantification of alternative splicing events resulting in generation of a PTC in the affected transcripts of srsf7a (inclusion of intron sequences), srsf5a (inclusion of exon), and hnrnph3 (exon inclusion equivalent to lower inclusion values) in upf1 mutant fish. (F) Higher levels of NMD transcripts of srsf7a, srsf5a, and hnrnph3 as a result of changes in alternative splicing events increase NMD/non-NMD transcript ratios. Black circles, upf1+/+; red circles, upf1−/−.
We found that well over 2,000 alternative splicing events distinguish wild-type and upf1-mutant fish (Fig. 3 B and C). About 11% (241 of 2,165) of alternative splicing events result in the introduction of a PTC; in the majority of these cases, this occurs by exon inclusion or exclusion (Fig. 3C). Of the 1,924 events affecting alternative splicing events without introducing a PTC, about 17% lead to increased expression levels, suggesting that they may also subject to NMD regulation in the wild-type background (Fig. 3C). Hence, the majority of alternative splicing events (∼75%; 1,596 of 2,165) may be the indirect consequence of altered activities of splice regulators, rather than a direct outcome of NMD inactivation. upf1 deficiency often results in the introduction of a PTC into the transcripts of genes encoding splicing activators. This occurs either through use of alternative splice sites (here exemplified by srsf7a) or through the inclusion of so-called “poison” exons (here exemplified by srsf5a) (Fig. 3 D and E); for splicing repressors, exon exclusion appears to be a major regulatory pathway, here exemplified by hnrnph3 (Fig. 3 D and E). In all of these cases (3), the changes in alternative splicing associated with upf1 deficiency (Fig. 3 D and E) lead to changes in the ratio of NMD and non-NMD transcripts (Fig. 3F).
In order to gain insight into the degree of evolutionary conservation of NMD-transcript stabilization, we compared our findings to a dataset obtained by the analysis of mouse liver and bone marrow-derived macrophages lacking Upf2 (41). The intersect between mouse and zebrafish genes included several genes encoding regulators of pre-mRNA processing whose transcripts are subject to NMD-mediated regulation (Dataset S4); these findings support the notion of an evolutionarily conserved and NMD-regulated balance between unproductive and productive transcript isoforms of splice factors (19).
Synthetic Genetic Interaction between upf1 and tnpo3 Mutant Alleles.
The evolutionarily conserved (19, 42) autoregulatory process determining the intracellular levels of srsf, hnrnp, and other splicing modulator proteins affords cells with a versatile mechanism to dynamically respond to changing physiological requirements. In zebrafish, deficiency of TNPO3, which is known to function as a transporter for a number of splice regulators—including SRSF proteins (43)—causes failure of intrathymic T cell development (24). However, when compared to wild-type fish, the transcriptome of tnpo3-deficient animals reveals a mere 178 differentially expressed transcripts (Fig. 4A), including modest changes for only three NMD-susceptible transcript isoforms: cpb1, tra2a, and srsf1b (Dataset S5). The ratios of NMD and non-NMD transcript versions of these genes remain practically unchanged (Datasets S5 and S6), indicating that tnpo3-deficiency has little effect on the balance between NMD and non-NMD transcripts (Fig. 4B), unlike the situation in upf1 mutants, where the ratios are drastically changed (Datasets S2 and S3). In contrast, a large number of alternative splicing events are seen (Fig. 4C), in line with previous analyses (24), and compatible with the notion that dysregulated intracellular transport of splice regulators directly alters the splicing landscape.
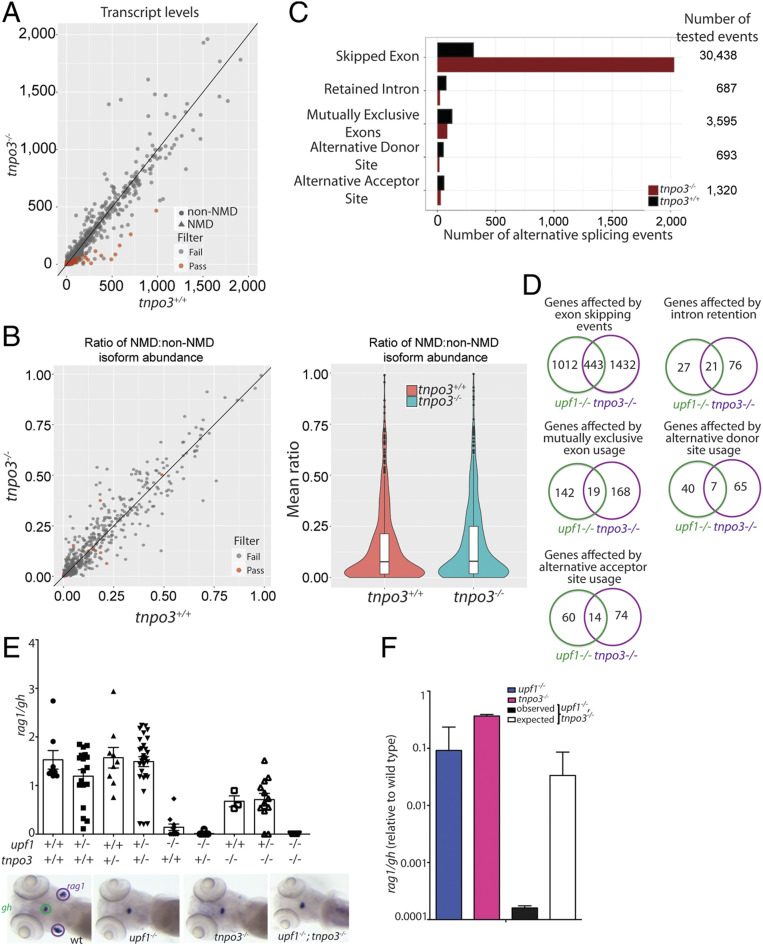
Fig. 4.
Characterization of the transcriptome of tnpo3 mutants at 5 dpf. (A) Differential transcript-level expression analysis of wild-type and mutant fish. Expression levels are depicted as TPM. (B) Minor effects on NMD ratios (Left); violin plot of NMD ratios (Right) (P = not significant). (C) Summary of differential splicing analysis. (D) Comparison of alternative splicing events between upf1 and tnpo3 mutants. The numbers of unique events are indicated for each class. (E) Synthetic interaction of upf1 and tnpo3 mutations. The thymopoietic capacities (expressed as rag1/gh ratios) of the nine genotypes resulting from an intercross of double heterozygous upf1+/−;tnpo3+/− parents are indicated. Each data point represents one fish; mean ± SEM. Representative WISH results for the key genotypes are shown (Lower). (Magnification: 10×.) (F) Comparison of phenotypes of the two single mutants (upf1−/− and tnpo3−/−, respectively), and the upf1−/−;tnpo3−/− double mutant; the latter phenotype is compared to the expected phenotype (multiplicative model). The thymopoietic capacity observed in the double mutant is significantly lower than expected from the combination of the single mutants, calculated under the assumption of no genetic interaction, indicative of synthetic genetic interaction.
Next, we determined the degree of similarity between alternative splicing events found in upf1 and tnpo3 mutants. The results indicate that although considerable overlap exists among the gene sets affected by the various types of alternative splicing, the majority of affected genes are unique to the respective mutants (Fig. 4D); for example, of the 1,785 genes affected by alternative splicing in upf1 mutants, 504 (28%) are shared with the 2,319 genes affected in tnpo3 mutants (22%). The overlapping gene sets are most likely associated with the similar morphological changes that are the result of the pervasive changes in alternative splicing patterns. The distribution of alternative splicing events across the different categories reveals little difference between upf1 and tnpo3 mutants, with the exception of more frequent intron retention in tnpo3 mutants (P = 0.013; χ2 test), whereas all other event types are equally frequent (Fig. 4D). Collectively, judging from the only partially overlapping changes in the splicing landscape, the common phenotypic features of upf1 and tnpo3 deficiencies may arise from largely distinct molecular perturbations. We used genetic interaction analysis to formally test this prediction. As expected from the transcriptome analysis, no rescue of the upf1-mediated loss of T cell development was observed in the additional absence of tnpo3 (Fig. 4E); rather, the phenotype of the double-mutant fish is worse than expected from the combination of the two single mutants (Fig. 4E). This outcome is indicative of synthetic lethality (44), indicating that upf1 and tnpo3 deficiencies impact T cell development (presumably by changes in pre-mRNA splicing) in different ways.
Synthetic Genetic Interaction between upf1 and lsm8 and upf1 and snapc3 Mutant Alleles.
We sought to substantiate this conclusion that upf1 and tnpo3 function in largely separate pathways to modulate the splicing landscape by analysis of genetic interactions between the upf1 mutation and mutations in genes encoding two components of the spliceosomal pathway, lsm8 and snapc3 (24). In previous work (24), we established that tnpo3, lsm8, snapc3, and gemin5 form a genetic network underlying mRNA splicing. gemin5 and tnpo3 mutations were found to interact in alleviating fashion, suggesting a partially overlapping mechanism of action; in contrast, lsm8 and gemin5 mutations were found to interact in synthetic fashion, indicating that they largely operate in parallel pathways (24). The phenotypes of upf1;lsm8 and upf1;snapc3 double mutants are compatible with the presence of synthetic interactions (SI Appendix, Fig. S2). The observation that both tnpo3 and lsm8 (and also snpac3) mutations interact in synthetic fashion with the upf1 mutation indicates that upf1 regulates the mRNA splicing process in distinct ways, reinforcing the view that the pathways affected by upf1 deficiency and the canonical pathways of pre-mRNA splicing are distinct.
upf1 and rag1 Mutations Do Not Interact Genetically.
Previous work has shown that developing mouse thymocytes overwhelmed with PTC-containing mRNAs transcribed from nonfunctionally rearranged Tcrb alleles die by apoptosis (45, 46). The up-regulation of the p53 pathway in upf1 mutants suggested that this phenomenon may contribute to their T cell phenotype. However, using upf1;rag1 double mutants, we found that this is not the case. In rag1-deficient animals, VDJ recombination of antigen receptor genes does not occur; hence, potentially nonproductive tcrb transcripts are not produced. Yet, thymocyte development is not rescued in upf1;rag1 double mutants (SI Appendix, Fig. S3). In fact, since the phenotype of the double mutant is not different from the expected value under the assumption of genetic neutrality (44), we conclude that the cellular pathways of upf1 and rag1 are distinct. These findings suggest that failure to eliminate nonproductive tcrb transcripts cannot be the only mechanism (if it does at all) contributing to the demise of developing thymocytes in upf1 mutants.
Extraribosomal Function of rpl10a in T Cell Development.
Our results show that NMD-susceptible transcripts of rpl10a and rpl22l1 are dramatically up-regulated in upf1 mutants (Fig. 2 B and C). Interestingly, both proteins are known for their extraribosomal function in regulation of pre-mRNA splicing (35, 36). It is known that RPL22 and its paralog RPL22L1 cross-regulate each other (35), and both have distinct functions in the hematopoietic system. RPL22 is known for its role in T cell development in zebrafish (47) and in mice (48, 49); lack of RPL22 leads to induction of endoplasmic reticulum stress in αβ T cells, and subsequently to activation of the p53 pathway (49). In zebrafish, RPL22L1 is known to be required for the emergence of hematopoietic stem cells in the aorta–gonad–mesonephros region (47), eventually causing the loss of T cell precursors. In upf1-deficient embryos, the majority of transcripts of rpl22l1 are of NMD-inducing nature, suggesting that the expression levels of productive mRNA are greatly diminished; hence, this particular function of RPL22L1 may contribute to the severe reduction of T cell development in upf1 mutants.
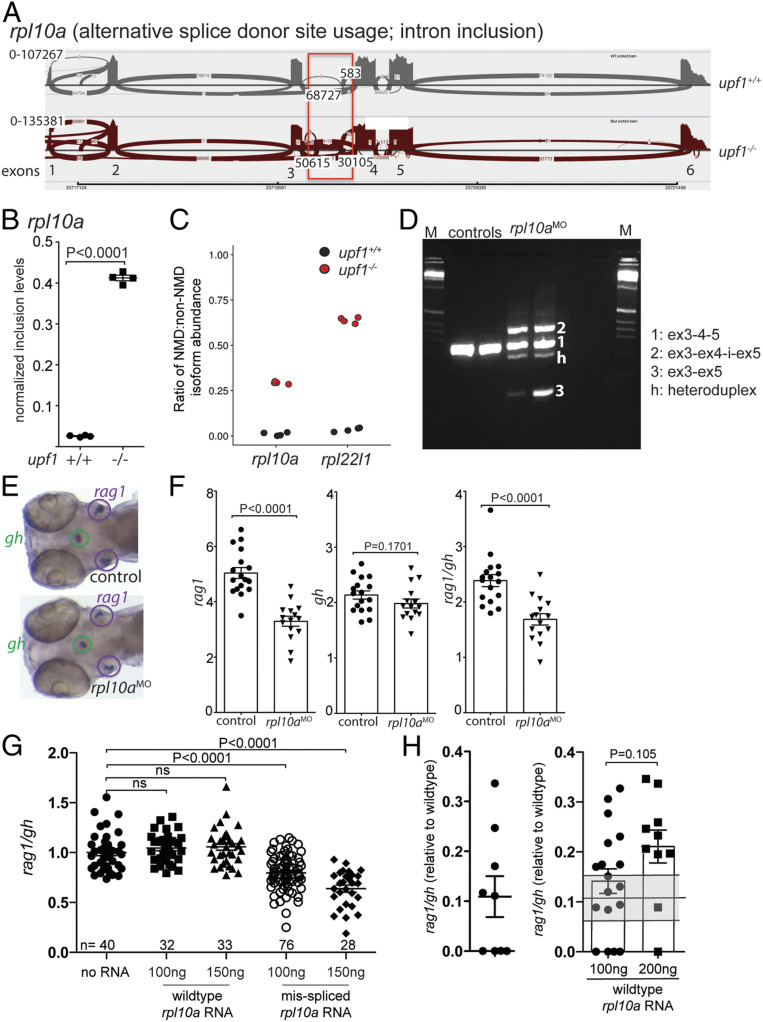
RPL10a also has extraribosomal functions, and in autoregulatory fashion determines the balance between its own productive and unproductive isoforms of mRNAs (36), although it may have additional targets. We therefore considered the possibility that the balance between productive and nonproductive isoforms of rpl10a mRNAs might affect T cell development. In upf1 mutants, a nonproductive mRNA of rpl10a is generated by the use of an alternative splice donor site downstream of exon 3 (Fig. 5A), leading to a drastic change in the ratio of NMD to non-NMD transcripts (Fig. 5B), similar in magnitude to that observed for rpl22l1 (Fig. 5C). In order to test the functional relevance of this aberration in rpl10a transcript composition, we aimed at artificially increasing the levels of NMD-susceptible transcript. Since the sequence context of the end of exon 3 was found to be unfavorable for sequence-specific interference, we instead targeted the splice donor site of exon 4 to generate a nonproductive transcript isoform (Fig. 5D). Two major types of aberrant splice products arising from exon skipping and intron retention were obtained. In one form, exon 4 is excluded from the transcript, leading to a PTC in exon 5; a second variation retains the intron between exons 4 and 5, leading to PTC in the intronic segment (Fig. 5D). Although these aberrant transcripts are nonfunctional, they are not identical in structure to the nonproductive version induced in upf1 mutants. Nonetheless, intrathymic T cell development is selectively impaired (Fig. 5 E and F); of note, rpl10a exhibits a certain degree of cell-type specificity, since growth hormone gene-expressing cells in the hypophysis were not affected (Fig. 5 E and F).
Fig. 5.
Requirement of rpl10a for T cell development. (A) Increased alternative splice donor site usage downstream of exon 3 (boxed) of the rpla10a gene in upf1 mutants. The transcript identifier for the NMD transcript is ENSDART00000138427.3. (B) Inclusion levels of intronic sequences in wild-type and upf1 mutant fish. (C) Increased NMD/non-NMD ratios for rpl10a and rpl22l1 in upf1 mutants. Black circles, upf1+/+; red circles, upf1−/−. (D) Transcript structure in rpl10a morphants generated by blocking the splice donor site of exon 4 with a site-specific antisense oligonucleotide. The composition of cDNAs as determined by direct sequencing of amplicons is indicated. (E) Representative WISH result for rpl10a morphants in D. Each data point represents one fish; mean ± SEM. (Magnification: 15×.) (F) The thymopoietic capacity of rpl10a morphants was calculated from rag1 (Left), gh (Center), and expressed as rag1/gh ratios (Right). (G) Detrimental effect of misspliced rpl10a mRNA on T cell development in zebrafish larvae. After injection of the indicated amounts of the two relevant RNA isoforms (compare with A), the thymopoietic index was determined at 5 dpf. (H) Alleviation of impaired T cell development in upf1 mutant larvae after injection of wild-type rpl10a mRNA as determined at 5 dpf. The thymopoietic indices are shown for uninjected upf1 mutants (data from Fig. 1E) (Left) and upf1 mutants injected with two different amounts of RNA (Right). Each data point represents one fish; mean ± SEM.
The experiment described above cannot differentiate between the effects of reduction of productive mRNAs and presence of nonproductive isoforms. To distinguish these possibilities, we injected the upf1 mutant associated misspliced rpl10a mRNA into wild-type embryos. We observed a statistically significant reduction of T cell development in embryos ectopically expressing the misspliced version (Fig. 5G), suggesting that its presence, rather than the reduction of normally spliced mRNA, is detrimental to T cell development. The common denominator of induction of nonproductive isoforms and the overexpression of misspliced mRNAs is a change in the ratio of productive to nonproductive transcripts. To test the hypothesis that altered ratios of NMD/non-NMD transcripts of rpl10a affect T cell development, we injected correctly spliced productive mRNAs into upf1 mutant embryos. Full rescue was not achieved (Fig. 5H); however, a trend (P = 0.01) toward increased T cell output was observed. The moderate reduction of T cell development after injection of misspliced rpl10a mRNA into wild-type fish, and the statistically nonsignificant rescue after injection of normally spliced rpl10a mRNA into upf1 mutant embryos indicate that rpl10a is only one of a number of factors (such as rpl22 and rpl22l1) that—in combination—mediate the effect of the upf1 mutation on larval T cell development. Thus, our findings not only support the notion that T cell development is regulated by a several members of the RPL-class of proteins, but also highlight the complexity of the downstream effects that are induced in upf1 mutants.
Intrigued by the possible connection between RPL22 and the p53 pathway (likely also relevant for other RPL-type proteins, such as RPL10a), we examined the physiological relevance of p53 activation under conditions of upf1 deficiency (SI Appendix, Fig. S4) by generating upf1;p53 double-mutant fish. Impaired T cell development in upf1 mutants was rescued in the p53 mutant background (SI Appendix, Fig. S4), suggesting that the outcomes of upf1 and p53 mutations are functionally connected. Indeed, positive (or alleviating) genetic interaction is often associated with activities of genes in the same pathway (44). Interestingly, inactivation of p53 also rescued at least some of the other, albeit less drastic abnormalities (when compared to developing T cells) in our upf1 mutants, here exemplified by the normalization of the size of the developing eye (SI Appendix, Fig. S4).
Collectively, our results implicate the upf1–rpl–p53 axis in T cell development, and suggest that the extraribosomal activities of the RPL-class of proteins are more widespread than previously assumed.
Conclusions.
Using an in vivo model of upf1 deficiency, we found that developing thymocytes in zebrafish embryos are uniquely sensitive to perturbations in the mRNA surveillance pathway, whereas other cell types and tissues appear to be less sensitive to the loss of upf1 activity, as clearly demonstrated by the grossly normal appearance of 5-dpf embryos. Differential sensitivity toward impaired mRNA surveillance during development and differentiation of various cell types and in cancer was previously documented (reviewed in refs. 2–4). We anticipate that further in vivo studies on the physiological consequences of failing NMD will be greatly facilitated by the availability of upf1-deficient zebrafish, exploiting the unique features of the zebrafish model, such as high fecundity, optical transparency, and the availability of a large arsenal of transgenic reporter lines.
The changes in the transcriptome of upf1 mutants observed here are best explained by a pervasive increase in the levels of NMD-susceptible transcripts predominantly of genes encoding splicing activators and repressors, markedly shifting their ratios of NMD-susceptible non-NMD transcripts. Since many splicing factors exploit evolutionarily conserved sequence features for autoregulation of their transcript levels, and in other cases also engage in cross-regulation of paralogous and highly related genes, the landscape of alternative splicing events drastically changed in upf1 mutants. Despite these considerable changes in the mutant transcriptome, it was possible to identify a new regulator of T cell development, rpl10a. Therefore, it appears that the extraribosomal functions of certain members of the RPL-class of proteins are particularly important for thymocyte development.
Materials and Methods
Animals.
The zebrafish (Danio rerio) strains wild-type in Kalkutta (WIK), AB, and Tübingen (TÜ) are maintained in the animal facility of the Max Planck Institute of Immunobiology and Epigenetics, Freiburg, Germany. The tnpo3, lsm8, and snapc3 mutant lines have been described previously (24), as have been the mutants for rag1 (50) and p53 (p53M214K) (51). All animal experiments were performed in accordance with relevant guidelines and regulations, approved by the review committee of the Max Planck Institute of Immunobiology and Epigenetics and the Regierungspräsidium Freiburg, Germany (license Az 35-9185.81/G-12/85). The upf1 mutant was isolated in the Tübingen 2000 screen (24).
Mutant Isolation.
Procedures for linkage analysis of recessive mutations, and whole-genome sequencing were described previously (24). The three polymorphic markers were amplified using the following primer pairs. HJ028_19L: 5′- TCTGAGCACCAGTTTATTAGAG; HJ028_19L: 5′- ACAGATTCCGTCTGTCCCTAA; HJ028_25L: 5′- CTTCGGTCTCTGTGTGACTG; HJ028_25R: 5′- GTGTGGGTTTGTGTATAGGAG; HJ028_36L: 5′- GTGTCCGCACTTCTTCTTGC; HJ028_36L: 5′- AAAGCAATCTACCCCACTGAC. The positions of markers on chromosome 2 relative to the zebrafish genome sequence (Zv11) are indicated. The upf1 mutants used for subsequent analyses were derived from fish that were outcrossed 7 to 10 times.
Morphants.
upf1 morphants were generated by injection of an antisense morpholino oligonucleotide (5′-GTACGCCTCCACACTCATCTTTATA; concentration in the injection buffer, 0.1 mM) to block translation of both maternal and zygotic mRNAs (“ATG morpholino”); rpl10a morphants were generated by injection of an antisense morpholino oligonucleotide (5′- CGGTTTTTAAACTCACCCAGCTTCT; concentration in the injection buffer, 0.1 mM) to block the use of the splice donor site of exon 4 (the overall procedure is described in ref. 24). Oligonucleotides were purchased from Gene Tools. A volume of 2 nL (∼16 ng) was injected per embryo. Morphants were analyzed at 5 dpf by RNA in situ hybridization using a combination of rag1- and gh-specific probes, as described previously (24). To determine transcript structure in rpl10a morphants, nested RT-PCR was performed using the following primers: First round, forward primer rpl10a_F1 5′-CGTGGAACTCCAGATCAGCT, reverse primer rpl10a_R1 5′-TCACCTCATCCACCTTGGTG; second round, forward primer rpl10a_F2 5′-ACTACGACCCTCAGAAGGAC, reverse primer rpl10a_R2 5′-CCAGGATACGAGGAATCTGC. The structure of cDNAs was deduced from the sequences of the amplicons.
Normal and misspliced forms of rpl10a mRNA were generated from cloned templates by in vitro transcription using T3 RNA polymerase (mMessage mMachine Kit; ThermoFisher Scientfic) and prepared for injection according to the procedure described in ref. 24.
RNA In Situ Hybridization.
The procedure and all genes-specific probes used here have been described previously (52). The thymopoietic capacity is expressed as the rag1/gh expression ratio as described previously (30).
Western Blot Analysis.
Protein lysates were resolved in 7.5% miniProtean TGX gels (Bio-Rad) and transferred to PVDF membranes (Bio-Rad). For detection of upf1 proteins, a 1:500 dilution of a polyclonal goat anti-upf1 antiserum (A300-036A; Bethyl) was used as a primary antibody, and a 1:2,000 diluted rabbit anti–goat-HRP antiserum (P0449; Agilent) as secondary antibody; as loading control, a 1:1,000 dilution of rabbit polyclonal antibody raised against β-tubulin (Ab209866; Abcam) was used as primary antibody, and a 1:2,000 diluted goat anti–rabbit-HRP antiserum (P0448; DAKO/Agilent) as secondary antibody. Immune complexes were detected using the ECL Prime Western Blotting Detection Reagent (RN 2232; Amersham).
RNA Extraction, cDNA Synthesis, RNA Sequencing.
These procedures were carried out as previously described (24). Four biological replicates were analyzed per genotype. RNA-sequencing (RNA-seq) data are deposited in National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) (53) and are accessible through GEO: GSE136669.
Computational Analysis of RNA-Seq Data.
Read alignment and detection of differential splicing events.
Reads were aligned to the GRCz10 genome assembly using STAR-2.5.2b with the following settings: Annotated splice junctions were based on Ensembl release 83 GTF file, maximum intron size was set to 11,000. Bams obtained from reads corresponding to the same sample were merged with samtools v1.3.1. Detection of differential splicing events was performed using rMATS v.3.2.5 (54) on merged bam files. The following parameters were specified: Read length (75 nucleotides), read type (paired), cutoff splicing difference (0.0001), analysis type (unpaired), and library type (fr-firststrand). Mutant bam files were submitted as group 1 (-b1), and wild-type as group 2 (-b2). Alternative splicing events supported by reads spanning splice junctions were reported. Filtering of rMATS result tables was done in R. For each class of alternative splicing events, entries were filtered for false-discovery rate < 0.01 and absolute inclusion level difference of 0.05. Events were annotated with mean inclusion levels in the wild-type and in the mutant groups.
Sashimi plots generation.
Sashimi plots were generated in IGV v.2.3.93 (55) using bam files output by rMATS prior to calling of differential splicing events.
Gene-level expression quantitation.
The number of reads per annotated gene (based on Ensembl 83 zebrafish gene annotation) were counted with featureCounts version v1.5.0-p1 (56). Fragments were only counted if both ends aligned on the same chromosome (-C and -B options) with a minimum mapping quality of 10 and in agreement with the strand orientation of the annotated gene (library type 2). Only primary alignments were counted. Gene-level expression for selected list of genes was plotted as a heatmap or a dotplot using the shiny app “RNA-seq Inspector” (https://github.com/maxplanck-ie/bulkRNAseq_MPI_shiny_app). Briefly, feature counts input was transformed to log2 counts per million using R package limma v3.36.5, and plots for selected genes were generated using gplots 3.0.1 (heatmap) or ggplot2 v. 3.2.0 (dotplot).
Transcript-level expression quantitation.
Reads corresponding to the same samples were merged, separately for each pair mate. Transcript isoform-wise expression was quantified using salmon v0.7.2 (57) on merged reads and the following parameters: Library type ISR, maxOcc 20, maxReadOcc 20, coverage 0.8, useVBOpt, numBootstraps 30. A reference fasta file was generated by running tophat-2.0.13 gtf_to_fasta on GRCz10 genome fasta and ensemble release 83 gtf file. Fmd-type salmon index was generated. Differential transcript isoform expression analysis was performed in R 3.3.1 using R packages wasabi (v0.2) and sleuth (v. 0.28.1). Results were filtered for q value < 0.05 and absolute log2 fold-change ≥ 1. Transcripts were annotated with gene id, gene symbol, and biotype using R package biomaRt (v2.35.12) corresponding to ensemble release 88. Transcripts per million (TPM) counts were aggregated by summing overall NMD and nonNMD transcript isoforms per gene, separately. The NMD ratio was calculated as ratio of counts coming from NMD transcripts to the sum of counts coming from all transcripts per gene. Changes in NMD ratio between mutant and wild-type fish groups were evaluated using R package limma on logit-transformed ratios. False-discovery rate threshold of 0.05 was applied to filter the results. The detailed R code underlying the calculations can be found in the “Supplementary code” document (https://github.com/katsikora/Lawir2019_SupplementaryCodeAndData) deposited in GitHub; the code is formatted in a way to facilitate understanding and interpretation by a lay R user. This analysis starts with sleuth (differential transcript expression) and salmon (transcript quantitation), as well as rMATS (differential splicing) output tables and walks the reader through the computational procedures underlying the paper’s figures. The vignette uses R 3.5.1 to reproduce manuscript figures.
Gene Ontology term enrichment analysis.
Unique gene symbols corresponding to non-NMD or NMD transcripts up-regulated in upf1 mutant fish were analyzed on geneontology.org/. Fold enrichments of identified gene ontology (GO) terms were plotted.
Statistical Analysis.
Two-tailed t tests were used to determine the significance levels of the differences between the means of two independent samples, considering equal or unequal variances as determined by the F-test. For multiple tests, the Bonferroni correction was applied. The χ2 test of association was used to compare frequencies of categorical variables.
Data Availability.
The sequences reported in this paper have been deposited in the GEO database (accession no. GSE136669). The detailed R code underlying the calculations can be found in the GitHub “Supplementary code” document (https://github.com/katsikora/Lawir2019_SupplementaryCodeAndData). The upf1 line is available via Material Transfer Agreement.
Supplementary Material
Acknowledgments
The mutant screens were conducted in collaboration with the Tübingen 2000 Screen Consortium and the Freiburg Screening Group. We thank E. Trompouki for discussion and provision of the p53 mutant strain; and the Deep Sequencing Unit of the Max-Planck Institute of Immunobiology and Epigenetics for next-generation sequencing. This work was supported by the Max Planck Society, and the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013), European Research Council Grant 323126.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE136669). The detailed R code underlying the calculations can be found in the GitHub “Supplementary code” document, https://github.com/katsikora/Lawir2019_SupplementaryCodeAndData.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917812117/-/DCSupplemental.
References
- 1.Thein S. L.et al., Molecular basis for dominantly inherited inclusion body beta-thalassemia. Proc. Natl. Acad. Sci. U.S.A. 87, 3924–3928 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurosaki T., Popp M. W., Maquat L. E., Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 20, 406–420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lykke-Andersen S., Jensen T. H., Nonsense-mediated mRNA decay: An intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 16, 665–677 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Anczuków O., Krainer A. R., Splicing-factor alterations in cancers. RNA 22, 1285–1301 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Brolosy M. A.et al., Genetic compensation triggered by mutant mRNA degradation. Nature 568, 193–197 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan Q.et al., Impact of nonsense-mediated mRNA decay on the global expression profile of budding yeast. PLoS Genet. 2, e203 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramani A. K.et al., High resolution transcriptome maps for wild-type and nonsense-mediated decay-defective Caenorhabditis elegans. Genome Biol. 10, R101 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tani H.et al., Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol. 9, 1370–1379 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rehwinkel J., Letunic I., Raes J., Bork P., Izaurralde E., Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA 11, 1530–1544 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurihara Y.et al., Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 2453–2458 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendell J. T., Sharifi N. A., Meyers J. L., Martinez-Murillo F., Dietz H. C., Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 36, 1073–1078 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Maquat L. E., Gong C., Gene expression networks: Competing mRNA decay pathways in mammalian cells. Biochem. Soc. Trans. 37, 1287–1292 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan Q.et al., Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev. 20, 153–158 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruno I. G.et al., Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol. Cell 42, 500–510 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green R. E.et al., Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics 19 (suppl. 1), i118–i121 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Cuccurese M., Russo G., Russo A., Pietropaolo C., Alternative splicing and nonsense-mediated mRNA decay regulate mammalian ribosomal gene expression. Nucleic Acids Res. 33, 5965–5977 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saltzman A. L.et al., Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol. Cell. Biol. 28, 4320–4330 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sureau A., Gattoni R., Dooghe Y., Stévenin J., Soret J., SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 20, 1785–1796 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lareau L. F., Inada M., Green R. E., Wengrod J. C., Brenner S. E., Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446, 926–929 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Leeds P., Peltz S. W., Jacobson A., Culbertson M. R., The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 5, 2303–2314 (1991). [DOI] [PubMed] [Google Scholar]
- 21.Kervestin S., Jacobson A., NMD: A multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 13, 700–712 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boehm T., Bleul C. C., Schorpp M., Genetic dissection of thymus development in mouse and zebrafish. Immunol. Rev. 195, 15–27 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Schorpp M.et al., Genetic dissection of thymus development. Curr. Top. Microbiol. Immunol. 251, 119–124 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Iwanami N.et al., Forward genetic screens in zebrafish identify pre-mRNA-processing pathways regulating early T cell development. Cell Rep. 17, 2259–2270 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim Y. K., Maquat L. E., UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond. RNA 25, 407–422 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arias-Palomo E.et al., The nonsense-mediated mRNA decay SMG-1 kinase is regulated by large-scale conformational changes controlled by SMG-8. Genes Dev. 25, 153–164 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamashita A.et al., SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 23, 1091–1105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deniaud A.et al., A network of SMG-8, SMG-9 and SMG-1 C-terminal insertion domain regulates UPF1 substrate recruitment and phosphorylation. Nucleic Acids Res. 43, 7600–7611 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melero R.et al., Structures of SMG1-UPFs complexes: SMG1 contributes to regulate UPF2-dependent activation of UPF1 in NMD. Structure 22, 1105–1119 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Lawir D.-F., Iwanami N., Schorpp M., Boehm T., A missense mutation in zbtb17 blocks the earliest steps of T cell differentiation in zebrafish. Sci. Rep. 7, 44145 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wittkopp N.et al., Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol. Cell. Biol. 29, 3517–3528 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anastasaki C., Longman D., Capper A., Patton E. E., Cáceres J. F., Dhx34 and Nbas function in the NMD pathway and are required for embryonic development in zebrafish. Nucleic Acids Res. 39, 3686–3694 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang L.et al., RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol. Cell 43, 950–961 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yepiskoposyan H., Aeschimann F., Nilsson D., Okoniewski M., Mühlemann O., Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA 17, 2108–2118 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y.et al., Ribosomal proteins Rpl22 and Rpl22l1 control morphogenesis by regulating pre-mRNA splicing. Cell Rep. 18, 545–556 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takei S., Togo-Ohno M., Suzuki Y., Kuroyanagi H., Evolutionarily conserved autoregulation of alternative pre-mRNA splicing by ribosomal protein L10a. Nucleic Acids Res. 44, 5585–5596 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ankö M. L., Morales L., Henry I., Beyer A., Neugebauer K. M., Global analysis reveals SRp20- and SRp75-specific mRNPs in cycling and neural cells. Nat. Struct. Mol. Biol. 17, 962–970 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Kemmerer K., Fischer S., Weigand J. E., Auto- and cross-regulation of the hnRNPs D and DL. RNA 24, 324–331 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rossbach O.et al., Auto- and cross-regulation of the hnRNP L proteins by alternative splicing. Mol. Cell. Biol. 29, 1442–1451 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Y.et al., Autoregulation of RBM10 and cross-regulation of RBM10/RBM5 via alternative splicing-coupled nonsense-mediated decay. Nucleic Acids Res. 45, 8524–8540 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weischenfeldt J.et al., Mammalian tissues defective in nonsense-mediated mRNA decay display highly aberrant splicing patterns. Genome Biol. 13, R35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lareau L. F., Brenner S. E., Regulation of splicing factors by alternative splicing and NMD is conserved between kingdoms yet evolutionarily flexible. Mol. Biol. Evol. 32, 1072–1079 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maertens G. N.et al., Structural basis for nuclear import of splicing factors by human Transportin 3. Proc. Natl. Acad. Sci. U.S.A. 111, 2728–2733 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baryshnikova A., Costanzo M., Myers C. L., Andrews B., Boone C., Genetic interaction networks: Toward an understanding of heritability. Annu. Rev. Genomics Hum. Genet. 14, 111–133 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Frischmeyer-Guerrerio P. A.et al., Perturbation of thymocyte development in nonsense-mediated decay (NMD)-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 108, 10638–10643 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weischenfeldt J.et al., NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 22, 1381–1396 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y.et al., Control of hematopoietic stem cell emergence by antagonistic functions of ribosomal protein paralogs. Dev. Cell 24, 411–425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson S. J.et al., Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity 26, 759–772 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Solanki N. R.et al., Rpl22 loss selectively impairs αβ T cell development by dysregulating endoplasmic reticulum stress signaling. J. Immunol. 197, 2280–2289 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wienholds E., Schulte-Merker S., Walderich B., Plasterk R. H., Target-selected inactivation of the zebrafish rag1 gene. Science 297, 99–102 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Berghmans S.et al., tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl. Acad. Sci. U.S.A. 102, 407–412 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schorpp M.et al., Conserved functions of Ikaros in vertebrate lymphocyte development: Genetic evidence for distinct larval and adult phases of T cell development and two lineages of B cells in zebrafish. J. Immunol. 177, 2463–2476 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Edgar R., Domrachev M., Lash A. E., Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen S.et al., rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc. Natl. Acad. Sci. U.S.A. 111, E5593–E5601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson J. T.et al., Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao Y., Smyth G. K., Shi W., The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47, e47 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences reported in this paper have been deposited in the GEO database (accession no. GSE136669). The detailed R code underlying the calculations can be found in the GitHub “Supplementary code” document (https://github.com/katsikora/Lawir2019_SupplementaryCodeAndData). The upf1 line is available via Material Transfer Agreement.