Significance
Temporal lobe epilepsy is commonly drug resistant and is associated with dysregulated hippocampal gene expression. MicroRNAs are short noncoding RNAs which control protein levels by binding target mRNAs via Argonaute proteins. We sequenced Argonaute-bound microRNAs from the hippocampus of three rodent epilepsy models, identifying common and unique functioning microRNAs at each stage of epileptogenesis. We designed oligonucleotide inhibitors against six microRNAs shared among models in chronic epilepsy and show three of these protected against acute and spontaneous seizures in a mouse model. We demonstrate that normal brain physiology is not obviously disrupted by these treatments and used a multiomics approach to identify a common mechanistic pathway for the therapeutic protective effects. Overall, these studies reveal potential treatments for drug-resistant epilepsy.
Keywords: antisense oligonucleotide, biomarker, epigenetic, epilepsy, noncoding RNA
Abstract
Temporal lobe epilepsy is the most common drug-resistant form of epilepsy in adults. The reorganization of neural networks and the gene expression landscape underlying pathophysiologic network behavior in brain structures such as the hippocampus has been suggested to be controlled, in part, by microRNAs. To systematically assess their significance, we sequenced Argonaute-loaded microRNAs to define functionally engaged microRNAs in the hippocampus of three different animal models in two species and at six time points between the initial precipitating insult through to the establishment of chronic epilepsy. We then selected commonly up-regulated microRNAs for a functional in vivo therapeutic screen using oligonucleotide inhibitors. Argonaute sequencing generated 1.44 billion small RNA reads of which up to 82% were microRNAs, with over 400 unique microRNAs detected per model. Approximately half of the detected microRNAs were dysregulated in each epilepsy model. We prioritized commonly up-regulated microRNAs that were fully conserved in humans and designed custom antisense oligonucleotides for these candidate targets. Antiseizure phenotypes were observed upon knockdown of miR-10a-5p, miR-21a-5p, and miR-142a-5p and electrophysiological analyses indicated broad safety of this approach. Combined inhibition of these three microRNAs reduced spontaneous seizures in epileptic mice. Proteomic data, RNA sequencing, and pathway analysis on predicted and validated targets of these microRNAs implicated derepressed TGF-β signaling as a shared seizure-modifying mechanism. Correspondingly, inhibition of TGF-β signaling occluded the antiseizure effects of the antagomirs. Together, these results identify shared, dysregulated, and functionally active microRNAs during the pathogenesis of epilepsy which represent therapeutic antiseizure targets.
Temporal lobe epilepsy (TLE) is characterized by seizures arising from or involving the hippocampus and is the most common focal epilepsy syndrome in adults (1). TLE is frequently refractory to pharmacotherapy, often necessitating surgical resection of involved brain structures (2). The most common pathological finding within the removed hippocampus is select neuron loss and gliosis (3). Resected tissue from TLE patients also features neuroinflammation and remodeling of neuronal networks at both micro- and macroscopic scale (4, 5). Recent sequencing and array-based profiling of protein-coding transcripts and systems biology approaches have generated deep insights into the molecular pathophysiology and helped identify novel classes of molecule for therapeutic targeting (6–9).
MicroRNAs (miRNAs) are critical for shaping the gene expression landscape in the brain (10). They are short noncoding RNAs that primarily function posttranscriptionally, conferring precision to cellular protein fluctuations (11, 12). Biogenesis of miRNAs involves nuclear processing of a primary transcript followed by terminal loop processing in the cytoplasm, resulting in a miRNA duplex from which one strand is selected by an Argonaute (Ago) protein (13). Argonaute-2 (Ago2) is critically important for miRNA function, enriched in the hippocampus and, uniquely among Ago proteins, can directly cleave target RNAs (14). After miRNA loading and the formation of a RNA-induced silencing complex (RISC), potential mRNA targets are selected through imperfect base pairing between miRNA and mRNA (15). Upon identifying regions of sufficient complementarity, typically 7- to 8-nt matches between the miRNA and the 3′-untranslated region of the target mRNA, the RISC recruits further proteins, leading to translational repression or mRNA decay (16). Individual miRNAs often have multiple targets, increasing the scope for influencing several pathways or enhanced regulation of single pathways by multiple miRNAs, which may be an advantage for the treatment of TLE (11, 12).
Spatiotemporal changes to miRNA expression have been reported in the hippocampus following epileptogenic brain injuries and these persist in established epilepsy (17, 18). In parallel, in vivo deployment of oligonucleotide miRNA inhibitors (antagomirs) has demonstrated functional roles for a few miRNAs in seizure control and epileptogenesis (19, 20). It remains unknown how many more miRNAs may be suitable targets in epilepsy. Recent efforts have identified miRNAs dysregulated in TLE (21–23) but no study to date has focused on quantifying the amounts of functional Ago2-loaded miRNAs that are shared between TLE models. This is important since the specific enrichment for Ago2-loaded miRNAs provides greater coverage of the miRNA landscape and better predicts the regulatory potential of miRNAs (24).
Here, we performed small RNA sequencing (RNA-seq) of Ago2-loaded miRNAs from three different animal models across all phases of epilepsy development in two rodent species. Based on this resource, we deployed an in vivo antagomir screen and identified several antiseizure and neuroprotective phenotypes among miRNAs that were up-regulated across the three models. Pathway analysis combined with RNA sequencing and proteomic analysis implicated derepressed TGF-β signaling as a potential overlapping mechanism between the targeted miRNAs. In agreement with this, inhibition of TGF-β signaling blocked the antiseizure effects of the antagomirs. Our systems-level approach identifies an extensive class of miRNAs that may prove targetable for the treatment of seizures in human TLE.
Results
Ago2 Sequencing Provides a Quantitative Catalog of Functionally Engaged miRNAs in Experimental TLE.
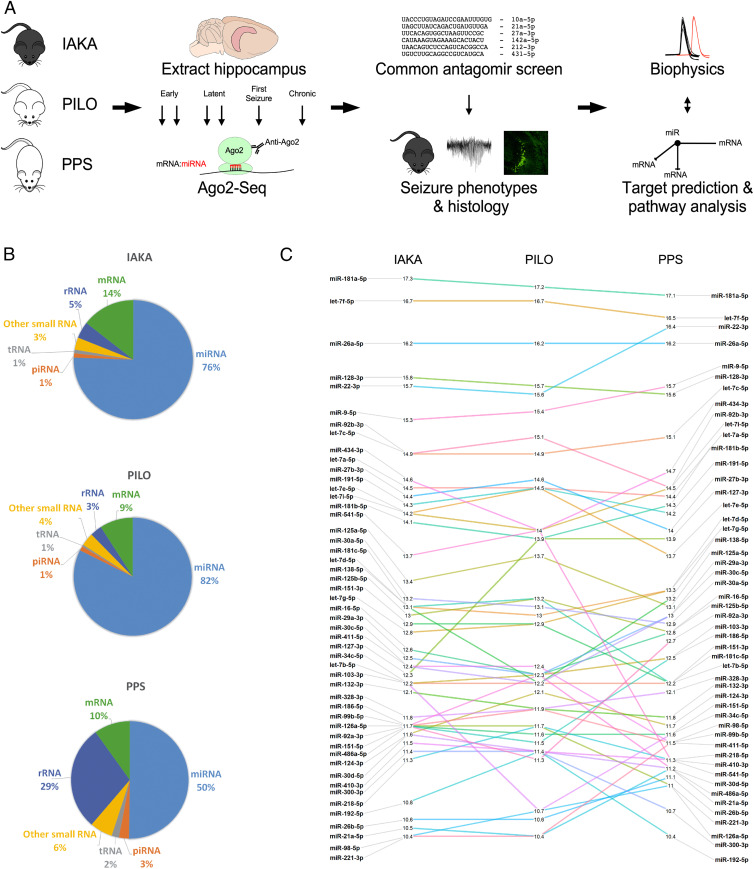
To identify shared functionally engaged miRNAs at each phase of epilepsy development, we performed Ago2 immunoprecipitation followed by small RNA sequencing on hippocampal samples from three different rodent models of TLE in two species under continuous electroencephalogram (EEG) monitoring, sampling tissue at six time points (intraamygdala kainic acid [IAKA]: n = 18 treated and 18 vehicle control; pilocarpine [PILO]: n = 18 treated and 18 vehicle control; perforant path stimulation [PPS]: n = 21 treated and 3 nonstimulated control, total n = 96; Fig. 1A, SI Appendix, Table S1, and Methods). This generated 1.44 billion small RNA reads of which up to 82% were miRNAs, with over 400 unique miRNAs detected per model (Fig. 1B and SI Appendix, Table S2). There was exceptionally high concordance for levels among the most abundant miRNAs, with sequencing reads often differing by as little as 1% across the three models (Fig. 1C).
Fig. 1.
Experimental design and small RNA sequencing. (A) Schematic showing the full study design. Step 1: Three rodent models of epilepsy were generated: IAKA (intraamygdala kainic acid-induced status epilepticus in C57BL/6 mice), PILO (pilocarpine-induced status epilepticus in NMRI mice), and PPS (perforant pathway stimulation-induced hippocampal lesioning in Sprague-Dawley rats). Step 2: Hippocampi were extracted at six (mice) or seven (rat) time points and processed for Ago2 immunoprecipitation and small RNA sequencing (Ago2-seq). Step 3: miRNAs with consistent up-regulation in all three models were selected for an antagomir-based screen for antiseizure phenotypes and neuroprotection. Step 4: Pathway modeling and biophysical analyses were used to investigate the function of the miRNAs. (B) The read mapping distribution for the three rodent models. Note, the majority of small RNA reads mapped to miRNAs. (C) Expression of the top 50 miRNAs between the three models showing highly similar expression levels.
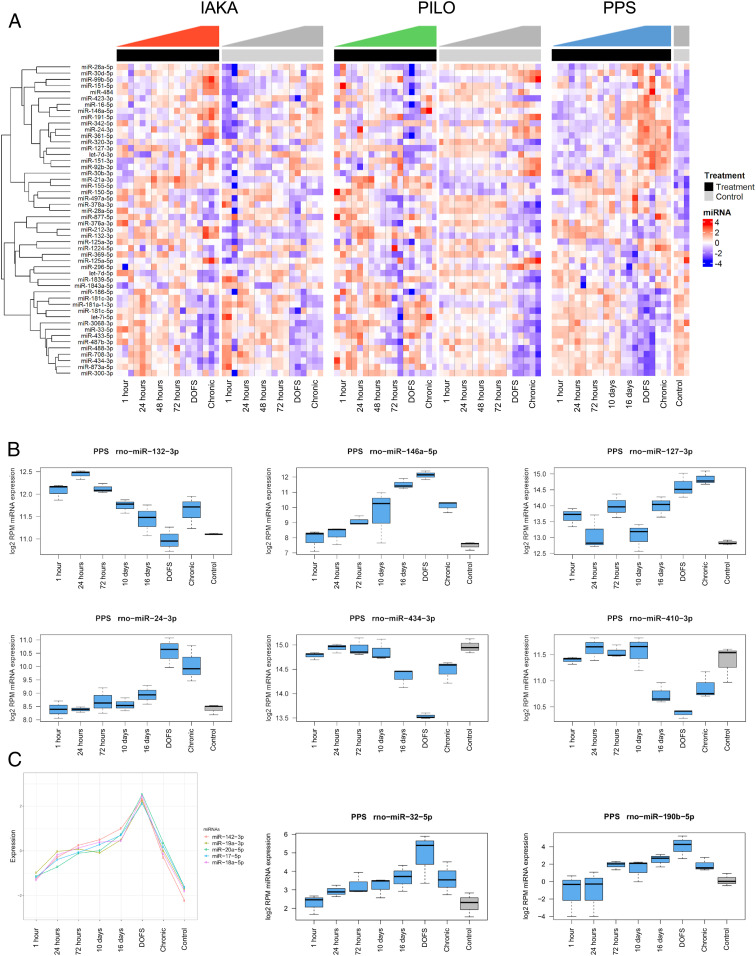
Induction of epilepsy led to significant changes in the abundance of approximately half of the detected miRNAs in the hippocampus in each model (Fig. 2A and Dataset S1). Expression changes showed disease stage-specific differences for individual miRNAs, including up- and down-regulation shortly after the epileptogenic insult, on the day of first spontaneous seizure and chronic epilepsy, indicating that all phases of epilepsy development are associated with specific miRNA changes (Fig. 2 A–C). For miRNAs that originate from the same primary transcript, expression levels of miRNAs within the cluster often followed similar patterns (Fig. 2C).
Fig. 2.
Extensive dysregulation of Ago2-loaded miRNAs across all phases of epilepsy development. (A) The 50 most significantly differentially expressed miRNAs are shown as a heatmap covering all samples from IAKA, PILO, and PPS models. Top annotation shows epileptic animals as black and control animals as gray. Shown are z scores of log2-transformed RPM values. (B) Examples of individual miRNA expression responses from the PPS model. Shown are miR-132 and miR-146 and potential novel epilepsy-associated miRNAs, miR-127, -24, -434, and -410. (C) Clustering analysis shows that miRNAs from the miR-17∼92 cluster peak at the day of first spontaneous seizure (DOFS). miR-142-3p also peaks at DOFS, though not transcribed from the miR-17∼92 cluster. Also shown are miR-32-5p and miR-190b-5p, both peaking at DOFS.
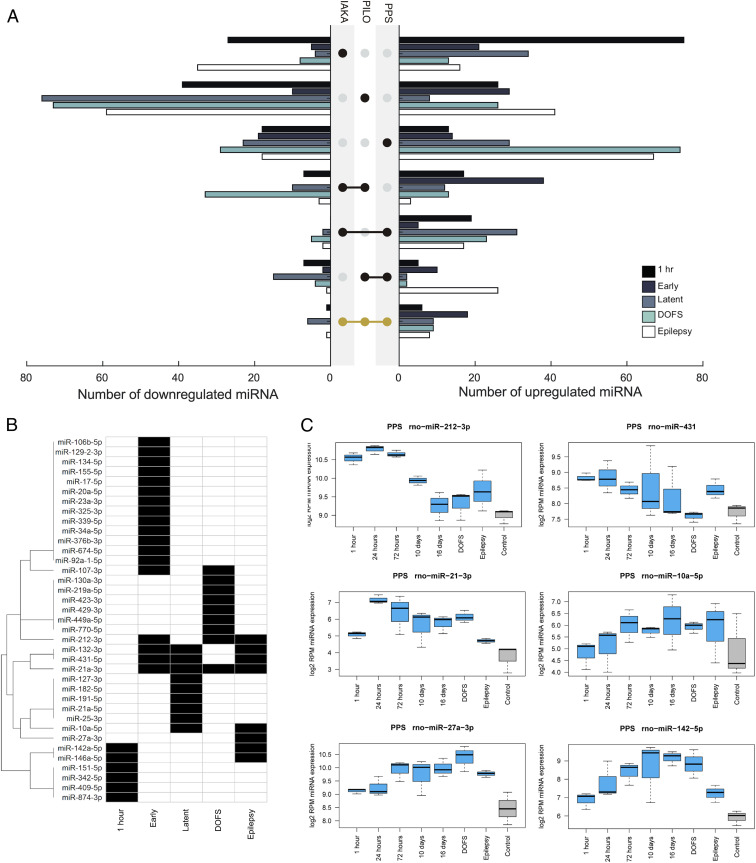
Next, we identified shared dysregulated (differential expression >25%) miRNAs across the three models, excluding low-abundance (<10 reads per million [RPM]) miRNAs for all time points analyzed (Fig. 3A). Within the chronic epilepsy phase, the period most relevant to how a miRNA-based therapeutic might be used clinically (i.e., treating patients with preexisting, refractory epilepsy) (17), we found eight up- and one down-regulated miRNAs common to all three models (Fig. 3 A–C). This included miR-132-3p and miR-146a-5p, for which there are already significant functional data linking them to epilepsy (25–28), and six miRNAs (miR-10a-5p, miR-21a-3p, miR-27a-3p, miR-142a-5p, miR-212-3p, and miR-431-5p) for which there are limited or no functional in vivo data linking to epilepsy (Fig. 3C).
Fig. 3.
Identification of common-to-all model miRNAs. (A) Graphs show the overlap of up-and down-regulated miRNAs between the three models at various phases of epilepsy development. (B) The miRNAs with consistent up-regulation in all three models, common-to-all miRNAs, are further highlighted. (C) Examples of the expression data from the PPS model for the common-to-all miRNAs up-regulated in chronic epilepsy (excluding miR-146a-5p and miR-132-3p).
In Vivo Antagomir Screening Identifies Three Antiseizure Phenotypes.
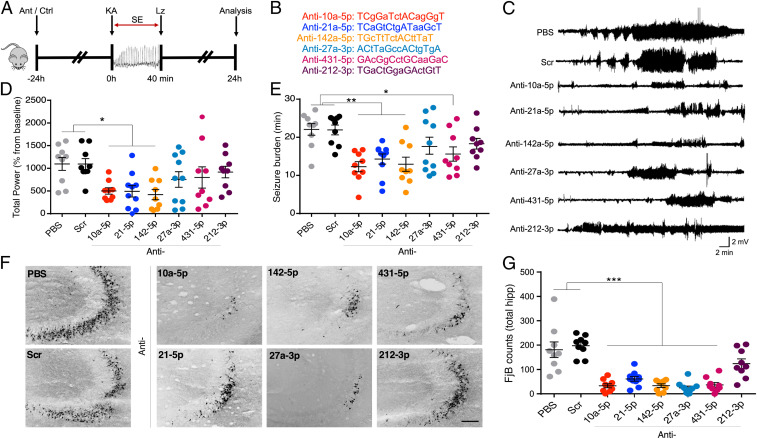
We hypothesized that the up-regulated miRNAs shared in the chronic epilepsy phase across the three models would be enriched for regulators of brain excitability, and we selected those which were fully conserved in humans. To test this, we designed custom locked nucleic acid (LNA)-modified oligonucleotide miRNA inhibitors (antagomirs) and used these to assess seizure responses after in vivo knockdown of miRNAs (Fig. 4 A and B). We excluded miR-132-3p and miR-146a-5p to prioritize miRNAs not previously linked to epilepsy and excluded miR-21a-3p because it is not fully conserved in humans, therefore limiting translational potential. Instead, we selected the fully conserved miR-21a-5p, which also satisfied basal expression criteria and up-regulation (at least 15%) in all three models. Mice received an intracerebroventricular injection of one of six targeting antagomirs (Fig. 4B), a scrambled antagomir (Scr) or vehicle (phosphate buffered saline [PBS]) 24 h before induction of status epilepticus (SE) by an intraamygdala microinjection of kainic acid (Fig. 4A). This procedure ensures an optimal miRNA knockdown at the time of testing seizure responses (29). EEG recordings were used to assess seizure severity and brains were later processed to quantify irreversible hippocampal damage (29).
Fig. 4.
Seizure phenotype screening of antagomirs. (A) Schematic shows the experimental design. Briefly, mice were equipped for EEG recordings and underwent intracerebroventricular injection of one of six antagomirs targeting miR-10a-5p, miR-21a-5p, miR-142a-5p, miR-27a-3p, -5p, miR-431-5p, miR-212-3p, or controls (PBS or scrambled antagomir). After 24 h, SE was induced (IAKA model) followed by lorazepam (Lz) to reduce mortality and morbidity. Hippocampal neuronal death was assessed at 24 h after SE. (B) Custom-made antagomir sequences. Capital letters represent LNAs and small letters, DNA phosphorothioates. (C) Representative traces show EEG recordings over time (starting from IAKA injection) for each group. (D) Graph shows EEG total power during SE as a percentage of each animal’s own baseline. Mice pretreated with antagomirs for miR-10a-5p, miR-21a-5p, or miR-142a-5p displayed reduced seizure severity when compared to PBS or scramble controls. (E) Graph shows seizure burden (time in ictal activity) for each group. (F) Representative photomicrographs and (G) graph from the dorsal ipsilateral hippocampus of mice 24 h after status epilepticus, stained using the irreversible damage marker Fluoro-jade B (FjB). (Scale bar, 100 μm). All error bars shown as mean ± SEM. n = 9 to 10/group; *P < 0.05, **P < 0.01, ***P < 0.001 compared either to PBS or Scr by one-way ANOVA with Bonferroni post hoc test.
Seizure severity, as determined by analysis of EEG total power (29), was significantly reduced during SE in mice preinjected with antagomirs against miR-10a-5p, miR-21a-5p, and miR-142a-5p (Fig. 4 C and D). Seizure burden, determined by measuring only ictal epileptiform activity (29), was significantly reduced by the same antagomirs and was also significant for antimiR-431-5p (Fig. 4E). Analysis of the brains from mice killed 24 h after SE revealed significant neuroprotection for five of the six antagomirs (those targeting miRNAs -10a-5p, -21a-5p, -27a-5p, -142a-5p, and -431-5p), relative to controls (Fig. 4 F and G). These results suggest that a high proportion of the shared miRNAs up-regulated in the chronic phase of experimental epilepsy may be maladaptive, contributing to enhanced network excitability and neuronal damage. Consequently, their targeting may offer approaches to control seizures.
Knockdown of miR-10a-5p, -21a-5p, and -142a-5p Has Limited Biophysical and Functional Effects in Naïve Rat Brains.
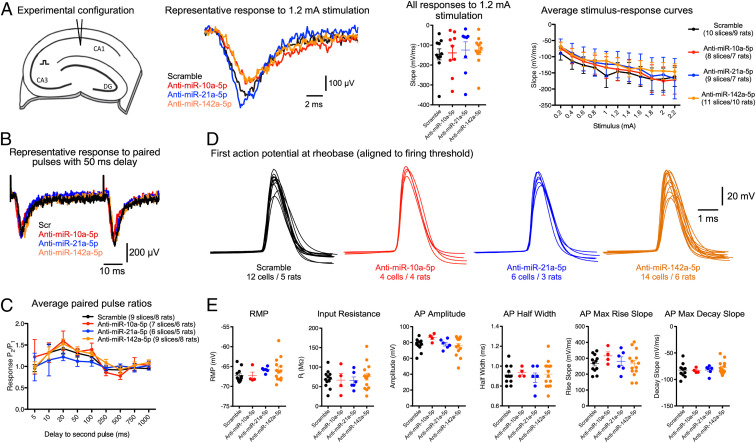
Current antiseizure drugs are associated with side effects arising from nonspecific dampening of brain excitability (1, 2). Focusing on the miRNAs which showed the most robust antiseizure phenotypes when targeted (miR-10a-5p, miR-21a-5p, and miR-142a-5p), we next assessed whether their knockdown had any adverse effects. We conducted a range of electrophysiological assessments, originally developed for antagomir-injected rats (30). We prepared ex vivo brain slices from the rats 2 to 4 d after antagomir injection, to coincide with maximal miRNA knockdown (29). Multiple electrophysiological measurements were unchanged in antagomir-injected animals, including population synaptic response to Schaffer collateral stimulation (Fig. 5A), paired pulse facilitation (Fig. 5 B and C), and action potential properties of pyramidal neurons (Fig. 5 D and E). Taken together, these studies indicate that the antiseizure antagomirs have seemingly limited and specific effects on hippocampal properties in naïve rodents.
Fig. 5.
Antagomir effects on hippocampal biophysics in naïve animals. (A) Stimulus–response curves. We stimulated the Schaffer collateral pathway in the hippocampus and recorded the population synaptic response in CA1 stratum radiatum (Left schematic). Robust responses were observed in all treatment groups (Middle Left) and no significant differences in excitability were seen between groups (Right; mixed ANOVA, P > 0.05). (B) Paired-pulse facilitation. We used the same electrode configuration as A but this time delivered two pulses (generating a response that was 30% of the maximum) at varying intervals. Robust facilitation was seen in all groups (B, representative raw data for 50-ms stimulation interval) with no clear differences between groups (C, mixed ANOVA, P > 0.05). (D) Single cell biophysics. We made current clamp recordings from CA1 pyramidal neurons and injected a current step protocol (−100 to +400 pA, 25-pA increments, 100-ms step duration). Raw data show the threshold action potential from each neuron. Total numbers of rats and cells used for each group are listed. (E) There were no differences in any passive properties (resting membrane potential [RMP] and input resistance scatterplots) or in properties of the threshold action potentials (all one-way ANOVA with Bonferroni post hoc test, P > 0.05).
Combinatorial miRNA Inhibition Reduces Seizures in Experimental TLE.
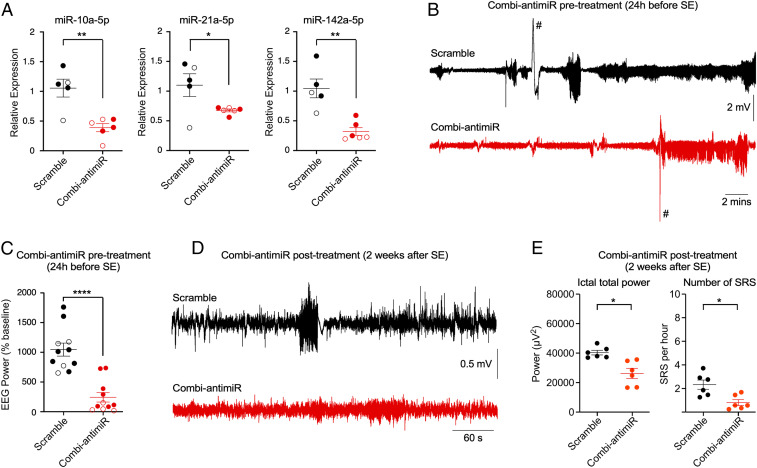
Next, we investigated whether targeting the identified miRNAs could affect spontaneous recurrent seizures. For this we combined the three most effective antagomirs (targeting miR-10a-5p, miR-21a-5p, and miR-142a-5p) into a single antagomir mixture (termed “combi-antimiR”). We confirmed the combi-antimiR mediated effective silencing of the three miRNA targets, comparable to the individual antagomirs and with no obvious sex difference (Fig. 6A). When preinjected with combi-antimiR, mice displayed reduced seizure severity during SE in the IAKA model, in line with the results of pretreatments targeting the three miRNAs individually (Fig. 6 B and C and SI Appendix, Fig. S1 A and B). Next, additional mice were subjected to IAKA and then monitored over the next 2 wk for spontaneous recurrent seizures (SRSs) (29, 31). After establishing equivalent baseline rates of SRSs (SI Appendix, Fig. S2), mice were randomly assigned to scramble or combi-antimiR posttreatments and SRS monitored over the next week. Combi-antimiR reduced both the occurrence and severity of seizures when administered as a posttreatment in already-epileptic mice, 2 wk after SE (Fig. 6 D and E and SI Appendix, Fig. S2C).
Fig. 6.
Combi-antimiR suppresses seizures in established epilepsy. (A) Relative levels of miRNAs measured 24 h after injection of combi-antimiR, confirming effective inhibition of all three targeted miRNAs (closed circles, male mice; open circles, female mice, pooled for statistics; all t test *P < 0.05, **P < 0.01; n = 5 mice [scramble] and 6 mice [combi-antimiR]). (B) Representative EEGs during SE induced by IAKA in mice pretreated 24 h previously with scramble or combi-antimiR (#, EEG artifacts [excluded from analysis]). (C) Scatterplot showing total EEG power during SE for all mice pretreated with combi-antimiR (closed circles, male mice; open circles, female mice, pooled for statistics; t test ****P < 0.0001, n = 11 mice per group). (D) Representative EEGs showing typical spontaneous recurrent seizure (SRS) in epileptic mice posttreated 2 wk after SE with either scramble or combi-antimiR. (E) Scatterplots show posttreatment of epileptic mice with combi-antimiR reduced EEG power during ictal periods and the number of SRS per hour averaged during the recording period compared to scramble (all t test, α-corrected to 0.025 for multiple testing, ictal power posttreatment P = 0.0061, number of SRS posttreatment, P = 0.011).
Target and Pathway Analysis Combined with RNA-Seq and Proteomics Reveal a Role for the TGF-β Signaling Pathway in the Antiseizure Effects of Combi-antimiR.
Finally, we sought to identify potential mechanisms underlying the antiseizure effects of the antagomirs and combi-antimiR and focused on identifying convergent pathways for miR-10a-5p, miR-21a-5p, and miR-142a-5p. The putative mRNA targets of the three miRNAs were identified using both predicted (miRDiP) (32) and experimentally validated [miRTarBase (33) and TarBase (34)] datasets. To reduce the risk of false positives, we applied strict miRNA–target interaction (MTI) filtering conditions based on miRDIP-assigned confidence levels and type of experimental validation (Methods). The estimated MTIs for each miRNA, along with brain expression information for each putative target, are listed in Dataset S2. We also performed in situ hybridization for miR-10a-5p, miR-21a-5p, and miR-142a-5p and this suggested neuronal as well as glial expression (SI Appendix, Fig. S3). While each miRNA had many unique targets, the three seizure-related miRNAs (miR-10a-5p, miR-21a-5p, and miR-142a-5p) shared 59 mRNA targets (Fig. 7A and SI Appendix, Table S3). Nineteen of these were not targeted by miR-27a-3p or miR-431, indicating that these targets could be specific to the observed antiseizure effects (Fig. 7A and SI Appendix, Table S3). Moreover, 48 mRNAs targeted by >1 of these miRNAs (of a total 525) have previously been associated with epilepsy, including GABA receptor, sodium, and potassium channel subunits (SI Appendix, Table S4).
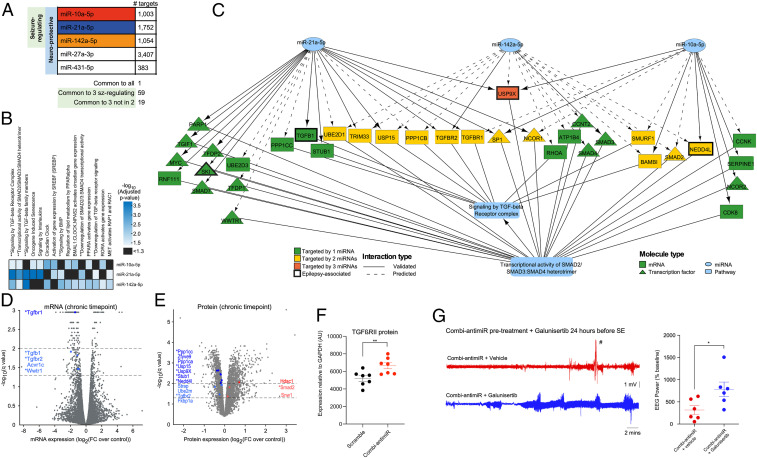
Fig. 7.
Target identification and pathway enrichment analysis identified TGF-β signaling as a potential convergent mechanism of the seizure-modifying miRNAs. (A) Number of mRNAs targeted by each miRNA. One mRNA (thyroid hormone receptor beta) is targeted by all 5 miRNAs. A total of 59 mRNAs are targeted by the 3 seizure-modifying miRNAs, 19 of which are not targeted by either miR-27a-3p or miR-431 (SI Appendix, Table S3). All targets are listed in Dataset S2. (B) Significantly enriched Reactome pathways for each of the seizure-modifying miRNAs. ** indicates pathways associated with TGF-β signaling. (C) Wiring diagram depicting mRNA targets of the 3 seizure-modifying miRNAs that are involved in the Reactome pathways: signaling by TGF-beta receptor complex and transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer, illustrating the convergence of diverse miRNA targets at the pathway level. (D) mRNA expression levels (normalized to control) of rat hippocampi isolated at the chronic time point of the PPS model. mRNAs above the dashed lines (drawn at −log10[q value] = 1.3 and 2.0) were considered statistically significant. Fold changes are shown on the x axis with significantly dysregulated mRNAs involved in the TGF-β signaling pathways highlighted in blue (all down-regulated). * denotes mRNAs which are targeted by miR-10a-5p, miR-21-5p, and/or miR-142-5p, as depicted in C. (E) Protein expression levels (normalized to control) of rat hippocampi isolated at the chronic time point of the PPS model. Proteins above the dashed lines (drawn at −log10[q value] = 1.3 and 2.0) are considered statistically significant. Fold changes are shown on the x axis with proteins involved in the TGF-β signaling pathways are highlighted in blue (down-regulation) and red (up-regulation). * denotes proteins which are targeted by miR-10a-5p, miR-21-5p, and/or miR-142-5p, as depicted in C. (F) Graph showing semiquantification of Western blot analysis of mouse brains taken 24 h after IAKA-induced SE shows that pretreament with combi-antimiR derepresses TGFβRII expression. Brain tissue samples were from mice in Fig. 6B, where combi-antimiR pretreatment reduced SE. (G) Coadministration of the TGF-β pathway inhibitor galunisertib occludes the antiseizure effect of combi-antimiR. (Left) Raw EEG traces show SE induced by IAKA in mice pretreated with combi-antimiR and galunisertib (blue trace) or vehicle control (red trace). (Right) TGF-β pathway inhibition with galunisertib blocks the antiseizure effects of combi-antimiR pretreatment (#, EEG artifact [excluded from analysis], n = 6 mice per group, t test *P < 0.05).
We next performed Reactome pathway enrichment analysis on the predicted targets for each of the miRNAs, using targets expressed in the hippocampus, and found that 15 pathways were enriched for targets of more than one seizure-modifying miRNA (Fig. 7B). Notably, six of these pathways are associated with TGF-β signaling, including the two pathways enriched in targets of all three miRNAs (“R-HSA-170834: Signaling by TGF-beta receptor complex” and its daughter pathway “R-HSA-2173793: Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer”). On further investigation, we noted that 35/73 genes involved in these two pathways are targeted by at least one of the seizure-modifying miRNAs, including TGF-β receptor type 2 (TGFβRII) (Fig. 7C).
To corroborate these systems-level predictions, we combined RNA-sequencing and mass spectrometry proteomic analyses on hippocampi isolated at the chronic time point (PPS model). This identified significant changes in the expression of multiple components involved in TGF-β signaling. Interestingly, all significantly dysregulated TGF-β-associated mRNAs were down-regulated (Fig. 7D and Dataset S3), and the main proteomic changes observed were down-regulated in the range of 0.7 to 0.9 fold change (FC) (Fig. 7E and Dataset S4). This is consistent with the actions of miRNAs to fine tune expression levels of targets in the same pathway. The majority of the significantly down-regulated mRNAs and proteins in the TGF-β signaling pathway are targeted by one or more of the three identified miRNAs as depicted in Fig. 7C. This led to a testable prediction that derepression of TGF-β signaling may be a convergent mechanism of the targeted miRNAs. We therefore measured levels of pathway components in mice that had previously received either scramble or the combi-antimiR 24 h before SE (Fig. 6B). Western blotting revealed TGFβRII protein levels were higher in mice treated with the combi-antimiR compared to scrambled controls (Fig. 7F and SI Appendix, Fig. S1C). To extend this, we coinjected the TGF-β pathway inhibitor galunisertib (2 mg/kg intraperitoneally [i.p.]), or vehicle control, with combi-antimiR before SE. Seizure severity was increased in combi-antimiR-treated mice coadministered galuninisertib compared to vehicle (Fig. 7G), providing additional evidence for a role of TGF-β signaling in the therapeutic effects of the antagomirs.
Discussion
The existence of conserved miRNA signatures in the development and maintenance of a seizure-prone state would provide important mechanistic insights and guide prioritization of miRNAs for therapeutic targeting. Here we undertook a coordinated effort, sequencing Ago2-bound miRNA to more accurately predict the regulatory potential of a given miRNA than by measuring overall miRNA levels in a sample (24), covering three different models, two species, and all stages from the initial precipitating insult to establishment of spontaneous recurrent seizures. The dataset contains robust statistics and fold change for individual miRNAs at each time point to illustrate expression variance and cross-model and cross-species comparisons. We found high concordance between the models and species in expression of known brain-enriched miRNAs, including miR-128-5p (35) and members of the let-7 family (36) whereas no reads were detected for nonbrain miRNAs such as miR-122-3p (liver specific) and miR-208b-3p (heart specific) (37, 38). The dataset features expected changes to neuronal activity-regulated miRNAs, including miR-132-3p (25) and miRNAs that regulate cellular responses to tissue injury, such as apoptosis-associated miR-34a-5p (39, 40). Together, the results offer important advances over previous work which focused on predetermined miRNAs (e.g., microarray based), lacked quantitative information on relative abundance, and lacked functional relevance (non-Ago2-loaded miRNAs) (21, 25, 41–45). The Ago2-sequencing (Ago2-seq) data provided in the current study are also an important companion to other databases on miRNA–epilepsy associations (46). The data complement, as well as reveal distinct profiles from Ago2-seq analysis of neural precursors (47) and should interest researchers working on disease mechanisms for which there is shared pathophysiology, such as traumatic brain injury (48).
By employing a multimodel sequencing approach, we were able to demonstrate that there are shared miRNAs dysregulated at all phases in the development of epilepsy, up to and including the period of active chronic epilepsy. Most of the miRNA changes fell within a 1.5- to 3-fold range although some, including miR-142a-5p, displayed much larger fold changes. There was no apparent species or model-specific bias, and numbers of shared miRNAs at the different stages of epilepsy development were quite similar, ranging from 6 to 18 among up-regulated miRNAs. We detected previously reported changes to miRNAs functionally linked to experimental epilepsy, including miR-22-3p (49), miR-129-5p (50), miR-134-5p (25), miR-146a-5p (27), and miR-324-5p (51). This indicates that Ago2-seq identifies robust miRNAs for targeting, a means to cross-compare between species and model, and a way to better prioritize miRNAs for functional assessment. A number of the miRNAs reported to be dysregulated in human TLE (52–54) were also differentially expressed in the chronic epilepsy state. This underscores the clinical relevance and translatability of our findings. It also invites additional predictions about human-dysregulated miRNAs which might be tested for function in animal models. The results extend evidence of a common miRNA signature in experimental epileptogenesis (23), contrasting conclusions from certain metaanalyses (22). Moreover, we report higher numbers of miRNAs and more differentially expressed miRNAs across these animal models than any previous epilepsy profiling study (21, 25, 41–45), indicating that miRNA dysregulation may impact on gene expression even more extensively than previously thought (17).
The potential for a miRNA-based therapeutic is gaining traction for disease modification in epilepsy (5, 17). LNA-based oligonucleotides as used here are particularly relevant for clinical translation as this backbone chemistry has been used in human trials of a miRNA-based therapy for hepatitis C (55). Here we show that robust antiseizure and neuroprotective effects can be achieved by targeting multiple miRNAs commonly up-regulated at the stage of chronic epilepsy. Notably, this included miRNAs for which there was no prior knowledge of a functional link to epilepsy. From these, we selected miRNAs which were fully conserved in humans, enhancing the translational potential of our findings. Our unbiased screen for antiseizure phenotypes identified five antagomirs that protect the brain against prolonged seizures, of which those targeting miR-10a-5p, miR-21a-5p, and miR-142a-5p had the most robust effects. Additionally, a combi-antimiR targeting all three miRNAs reduced spontaneous recurrent seizures when administered as a posttreatment to mice which had developed epilepsy. Taken together, this is a substantial addition to the number of miRNAs reported as potential targets for seizure control (20). It also suggests that many of the up-regulated miRNAs in the chronic epilepsy phase may be suppressing targets that would otherwise oppose hyperexcitability. While the antiseizure effects of targeting miR-10a-5p, miR-142a-5p, and the neuroprotection associated with inhibition of miR-431-5p have not previously been described, a recent study also found that targeting miR-21-5p could suppress seizures (56). Notably, our biophysical analyses of the electrophysiological properties of hippocampus from antagomir-treated rodents showed no obvious impairments. Together, these findings suggest broad safety and suitability to enter preclinical development.
The regulatory potential of miRNAs is enhanced where there is convergence upon a small number of targets or pathways (11, 12). An important effort in the present study was to combine mRNA targets of all miRNAs (experimentally validated and predicted interactions) to build superior pathways, building in a high-confidence threshold for target predictions. We found that the three seizure-regulating miRNAs converge on the TGF-β signaling pathway. Analyses at the mRNA and protein levels corroborated this pathway-level effect. Treatment with combi-antimiR derepressed TGFβRII, a validated target of both miR-21 (57) and miR-142 (58), while inhibition of TGF-β signaling occluded the antiseizure effect of combi-antimiR. These results implicate TGF-β signaling as an underlying therapeutic target mechanism. This is consistent with evidence that genetic defects in the TGF-β pathway can cause epilepsy in humans (59), although excessive TGF-β signaling has been implicated in epileptogenesis (60). Other overlapping targets, including ion channels and several of the genes targeted by two or all three of the seizure-regulating miRNAs, have previously been implicated in epilepsy.
There are some limitations and assumptions to consider in the present study. Some of the Ago2-bound miRNA pool may not be actively engaged with mRNA targets (61), Ago isoforms besides Ago2 may be important (14) and small RNA sequencing may over- or underestimate the abundance of certain miRNA species (62). We restricted our functional studies to targeting up-regulated miRNAs using antisense oligonucleotides. This has broad safety relative to overexpressing miRNAs which could saturate the RNA interference pathway (63), but down-regulated miRNAs may also represent therapeutic targets (23). Notably, target RNA-directed miRNA degradation (TDMD) has been reported, whereby miRNAs themselves are degraded upon binding a target site of high complementarity (64, 65). However, this has rarely been seen with endogenous target RNAs and we did not find evidence that the levels of the three miRNAs of main focus were influenced by TDMD (SI Appendix, Fig. S4). Another mechanism by which miRNA function can be reduced is via circular RNA (circRNA), which binds specific miRNAs rendering the bound miRNAs inactive, and studies have found circRNA levels to be altered in epilepsy (66). Adjustment of criteria for selecting miRNAs could yield additional miRNAs for functional studies. Indeed, several potentially new epilepsy-associated miRNAs not identified in multimodel or metaanalyses of miRNAs (21–23) showed significant regulation in two of the models here, including highly expressed miRNAs (thus likely to be functionally significant) such as miR-410-3p and miR-434-3p (down-regulated) and miR-24-3p and miR-127-3p (up-regulated). Primate or human-specific miRNAs would not have been detected by our screen and may represent additional therapeutic targets. Finally, genetic variation in miRNA coding regions could affect expression. However, the common-to-all miRNAs here do not originate from genomic regions known to have chromosomal mutations in epilepsy (67, 68). Large-scale DNA sequencing is warranted to test the impact of genetic variation on miRNA function in epilepsy covering coding genomic regions, miRNA processing genes, and miRNA target sites on mRNAs.
Conclusions
The present study generated a unique resource to explore the expression and dysregulation of miRNAs across multiple animal models of epilepsy and throughout the course of the disease. This systematic approach to discovery revealed a greater than previously anticipated, temporally specific dysregulation of miRNAs in epilepsy and showed this to be a rich source of seizure-regulatory miRNA targets. We identified multiple additional miRNA targets for seizure control, likely mediated through derepression of TGF-β signaling. Together, these results reinforce and extend the evidence that miRNAs are a major class of regulatory element in epilepsy with therapeutic potential for seizure control.
Methods
Animal Models of Epilepsy.
All animal experiments were performed in accordance with the European Communities Council Directive (2010/63/EU). All animals were housed in on-site barrier-controlled facilities having a 12-h light–dark cycle with ad libitum access to food and water.
Procedures in rats were approved by the local regulation authority (Philipps University Marburg, Germany: Regierungspraesidium Giessen, 73/2013), or according to the Animals (Scientific Procedures) Act 1986 (United Kingdom). Male Sprague-Dawley rats (325 to 350 g [Charles River] or 200 to 300 g [Harlan]) were used in all studies. Epilepsy was induced using PPS in rats, as described (69) and detailed in SI Appendix, Supplementary Methods. Rats were killed under deep anesthesia (xylazine + ketamine) by transcardial perfusion with ice-cold 0.9% NaCl solution at the following time points: 1 h, 24 h, 72 h, 10 d, and 16 d after PPS (epileptogenesis); within 1 d of first spontaneous seizure (early epilepsy); or 1 mo after first spontaneous seizure (chronic epilepsy). Control rats were killed on day 17 after surgery (corresponding to day 10 after PPS). Hippocampi were removed and snap frozen at −80 °C.
Procedures for inducing epilepsy using the IAKA technique in mice were approved by the Research Ethics Committee of the Royal College of Surgeons in Ireland (REC-842), under license from the Health Products Regulatory Authority (AE19127/001), Dublin, Ireland. Adult male C57BL/6 mice (20 to 25 g [Harlan]) were used, as described (70) and detailed in SI Appendix, Supplementary Methods. Mice were killed at 1 h, 24 h, 48 h, 72 h, day of first spontaneous seizure (typically 3 to 5 d after SE), or at 2 wk (chronic epilepsy). At the time of killing, mice were deeply anesthetized with phenobarbital and transcardially perfused with ice-cold PBS to remove blood contaminants. Hippocampi were frozen and stored at −80 °C.
Procedures for inducing epilepsy using the PILO model in mice were approved by the University of Verona Research Ethics Committee under license from the Italian Ministry of Health (27/2014-PR). Adult male NMRI mice (Harlan) were used, as detailed in SI Appendix, Supplementary Methods. Mice were killed at 1 h, 24 h, 48 h, 72 h, day of first spontaneous seizure (typically 1 to 2 w after SE), or at 4 wk (chronic epilepsy).
Immunoprecipitation of Ago2, RNA Extraction, and Sequencing (Ago2-Seq).
Frozen hippocampi were thawed on ice. Tissue was homogenized in 200 µL of immunoprecipitation (IP) buffer (300 mM NaCl, 5 mM MgCl2, 0.1% Nonidet P-40, 50 mM Tris⋅HCl pH 7.5, protease and RNase inhibitors). The homogenate was centrifuged at 16,000 × g for 15 min at 4 °C to pellet nuclei and membranes. Supernatant (considered total cell lysate) was transferred to a clean Eppendorf tube. Bradford assay was performed to quantify protein content of total cell lysate. The lysate was precleared by adding 10 µL of 50% Protein A/G beads (Santa Cruz Biotechnology) to 400 µg of protein lysate, final volume was adjusted to 1 mL using IP buffer, and lysate was incubated rotating for 1 h at 4 °C, then centrifuged at 13,000 × g for 5 min at 4 °C to pellet the beads, and supernatant was transferred to a new Eppendorf tube. A total of 5 µg (5 µL of AGO-2, Cell Signaling Cat. No. 2897) antibody was added to precleared cell lysate, vortexed, and incubated rotating overnight at 4 °C. A total of 20 µL of 50% A/G agarose beads was added to lysate antibody solution and incubated, rotating for 2 h at 4 °C, then centrifuged at 16,000 × g for 15 min at 4 °C, and supernatant removed. The pellet was washed twice with 500 µL IP buffer by gently resuspending pellet, centrifuging at 16,000 × g for 1 min at 4 °C, and removing supernatant. TRIzol RNA purification was performed after which pelleted RNA was dissolved in 12 µL dH20 and heated to 60 °C for 10 min. Purified RNA was stored at −80 °C until small RNA library preparation. A total of 5 µL of purified RNA was prepared using TruSeq small RNA library preparation kit (Illumina), for rat samples, using standard procedure and 12 PCR cycles and for mouse, using half the amount of primers and reagents and 15 PCR cycles. Pippin prep (Sage Science) was used to size fractionate libraries to the 140-bp to 160-bp size range. Library size and purity was validated on a Bioanalyzer 2100 (Agilent) using a high-sensitivity DNA chip, and the concentration was quantified using a KAPA Library Quantification Kit. Prepared libraries were pooled as required and sequenced on a NextSeq500 (Illumina) at Exiqon.
Analysis of Small RNA Sequencing Data.
FASTX-Toolkit was used to quality-filter reads and cutadapt was used to remove adaptor sequences. Filtered reads were mapped using Bowtie to a list of datasets. First, reads were mapped to miRNAs from miRBase v21 allowing zero mismatches, but allowing for nontemplated 3′ A and T bases. Reads not mapping to miRNAs were mapped against other relevant small RNA datasets: piRNA, tRNA, snRNA, snoRNA, and Y RNA allowing one mismatch. The remaining unmapped reads were mapped to mRNA and rRNA datasets. miRNAs were normalized as reads per million miRNA mapping reads (RPM). Statistical significance was calculated by one-way ANOVA with false discovery rate (FDR) (Benjamini–Hochberg). Common-to-all miRNAs were defined as having mean basal expression above 10 RPM, while exhibiting same-directional expression change of 25% or higher in all three models at key time points, as detailed above for each model.
Systematic Antagomir Screening.
Antagomir screening was performed in the IAKA mouse model according to previously described techniques (71) and detailed in SI Appendix, Supplementary Methods. An additional subset of adult female C57BL/6 mice (Harlan, weight matched to males [20 to 25 g], coordinates for IAKA verified by ink injection) were used in some experiments. For pretreatment studies, antagomirs or combi-antimiR (SI Appendix, Supplementary Methods) were administered 24 h before SE induction by IAKA. A subset of mice were given the TGFβRI/II inhibitor galunisertib (Tocris, Cat. No. 6956, 2 mg/kg via i.p. injection; vehicle, 10% dimethyl sulfoxide in PBS). A first galunisertib injection was given at the same time as combi-antimiR, 24 h before SE, and a second injection of galunisertib alone was given 30 min before SE. Twenty-four hours after SE, mice were transcardially perfused and brains removed for histopathological analysis of hippocampal damage or molecular analysis. Seizure-induced neuronal damage was analyzed on 12-µm coronal sections at the level of medial hippocampus (AP = −1.70 mm) using Fluoro-Jade B (FJB) (Millipore Ireland B.V.) as described (70). For posttreatment study, combi-antimiR was given 14 d after induction of SE by IAKA and mice were perfused 21 d after SE.
RNA Extraction and RT-qPCR to Assess Antagomir-Mediated miRNA Knockdown.
Hippocampi were homogenized in 750 µL of TRIzol and centrifuged at 12,000 × g for 10 min at 4 °C. Phase separation was performed by adding 200 µL of chloroform to each sample and vigorously mixing for 15 s before incubating at RT. Samples were centrifuged at 15,600 × g for 15 min at 4 °C. The upper phase was removed and 450 µL of isopropanol was added and samples were stored at −20 °C overnight. Samples were centrifuged at maximum speed for 30 min at 4 °C. A total of 750 µL of 75% cold ethanol was used to wash the pellet. Samples were centrifuged at 13,300 × g for 5 min and the ethanol was removed. This step was repeated twice. The pellets were left to dry for 1 h and resuspended in 25 µL of RNase free H20. Samples were incubated for 10 min at 60 °C with 60 × g of agitation. Samples were stored at −80 °. A total of 500 ng was reverse transcribed using stem-loop Multiplex primer pools (Applied Biosystems). We used reverse-transcriptase-specific primers for the hsa-miR-10a (Applied Biosystems miRNA assay ID 000387), hsa-miR-21a (Applied Biosystems miRNA assay ID 000397), and hsa-miR-142a (Applied Biosystems miRNA assay ID 002248) and real-time quantitative PCR was carried out on a 7900HT Fast Realtime System (Applied Biosystems) using TaqMan miRNA assays (Applied Biosystems). U19 (Applied Biosystems miRNA assay ID 001003) was used for normalization. A relative fold change in expression of the target gene transcript was determined using the comparative cycle threshold method (2-∆∆CT).
In Vitro Assay of Effects of Anti-miR-10a-5p, Anti-miR-21a-5p, and Anti-miR-142a-5p.
Stereotaxic injection for each miRNA knockdown was performed on adult male Sprague–Dawley rats (weight range 270 to 380 g) as described previously (30) and detailed in SI Appendix, Supplementary Methods. Ex vivo brain slices were prepared between 2 and 4 d after surgery, to coincide with the maximal miRNA silencing effect (30). All slice electrophysiology was performed using a membrane chamber (30) perfused with oxygenated recording artificial cerebrospinal fluid (ACSF), heated to 34 °C, at a rate of 16 mL min−1. Electrophysiological data were filtered at 10 kHz using a Multiclamp 700B amplifier (Molecular Devices), digitized at 10 kHz with a Power1401 (Cambridge Electronic Design), and recorded using Signal software (Cambridge Electronic Design). For extracellular recordings, we stimulated the Schaffer Collateral pathway with a concentric bipolar stimulating electrode (FHC, CBARC75) and recorded the response in CA1 stratum radiatum using a thin-walled borosilicate glass microelectrode (∼5 MΩ) filled with recording ACSF (in mM: 125 NaCl, 10 glucose, 26 NaHCO3, 1.25 NaH2PO4.H2O, 3 KCl, 2 CaCl2, 1 MgCl2). Patch clamp recordings used ∼5-MΩ glass microelectrodes filled with intracellular solution (in mM: 135 K-gluconate, 4 KCl, 10 Hepes, 4 Mg-ATP, 0.3 Na-GTP, 10 Na2-phosphocreatine; pH 7.3; 290 mOsm). After at least 5 min, neurons were injected with a series of hyperpolarizing and depolarizing current steps (100-ms current injections with 1 s between steps; −100 to +400 pA in 25-pA increments). The first action potential elicited by a depolarizing step was selected for analysis. All recordings were made with bridge balance compensated, access resistance <20 MΩ, and rejected if action potentials did not overshoot 0 mV.
Bioinformatics: mRNA Target Identification, miRNA Target Interaction (MTI) Prioritization, and Pathway Enrichment Analysis.
Predicted MTIs were downloaded from miRDiP V4.1, a database that integrates 30 prediction algorithms and calculates an MTI confidence score based on statistical inference (32). Experimentally validated MTIs were downloaded from miRTarBase V7 (33) and TarBase V7.0 (34). To ensure interoperability, all miRNA names were translated to miRBase V22 using miRNAmeConverter (72) and mRNA names were converted to official gene symbols using Ensembl V91. Nonhuman MTIs were excluded to constrain analyses to putative translatable mechanisms. Strict filtering criteria were devised to prioritize MTIs and reduce the risk of false-positive MTI identification; predicted MTIs were retained only if their miRDIP-assigned confidence levels were “very high,” while experimentally validated MTIs were excluded if the only form of validation was high-throughput CLiP experiments performed prior to 2013, as these are considered less reliable due to poor antibody specificity and RNA contamination (73). Baseline mRNA tissue expression information were from the GTEx-EBI Expression Atlas (74) and transcription factor information from TRRUST V2.0 [https://www.grnpedia.org/trrust/], ENCODE (75), and ChEA (76).
mRNAs implicated in epilepsy were identified using an in-house database collating information from CARPEDB [http://carpedb.ua.edu], epiGAD (77), Wang et al. (78), and curated epilepsy genes from the Comparative Toxicogenomics Database (CTD) (79).
Pathway analysis was performed on Reactome pathways containing 10 to 500 genes by applying the cumulative hypergeometric distribution for P value comparison (80). Pathways with corrected P values of <0.05 (Benjamini–Hochberg) were considered significantly enriched.
RNA Sequencing (mRNA).
RNA was rRNA depleted using the Ribo-Zero Magnetic Kit (human/mouse/rat; Illumina). Sequencing libraries were generated using the ScriptSeq v2 kit (Illumina), quality controlled on the 2100 Bioanalyzer (Agilent). Sequencing was done on an Illumina HiSeq sequencer.
Sequencing data were quality controlled and adaptor trimmed using Trim Galore. Filtered data were mapped to the rat genome Rn6 using TopHat2. Cufflinks was used to assemble transcripts from the mapped reads guided by gene annotation from Ensembl Release 87. Cuffmerge, cuffquant, and cuffdiff (part of the Cufflinks software package) were used to quantify transcript abundance in each sample and to perform differential expression analysis of genes and gene isoforms. P values were corrected for multiple testing using Benjamini–Hochberg correction.
Proteomic Analysis.
Hippocampi for proteomic analysis were taken from rats at the chronic time point of the PPS model and processed as detailed in SI Appendix, Supplementary Methods. Differential expression was evaluated using a combined limma and rank product test to obtain q values for each protein. Volcano plots were created by plotting log2 average ratios against −log10 q values obtained in the above-mentioned limma and rank product test.
Statistics.
For Ago2-seq, statistical significance was calculated by one-way ANOVA with FDR (Benjamini–Hochberg). In vivo seizure screening used one-way ANOVA with Bonferroni post hoc correction. Ex vivo slice experiments used unpaired t test, one-way ANOVA, or mixed ANOVA, as appropriate. The statistical approaches for pathway analysis, mRNA sequencing, and proteomics are detailed above.
Availability of Data and Materials.
The sequencing data have been deposited to the Gene Expression Omnibus (GEO) under accession no. GSE137473 (81). The proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD019098 (82). Custom analysis codes are available on https://github.com/g-morris/Ago2Seq. All data generated or analyzed during this study are included in this published article (and its SI Appendix files). For questions on the MTI analysis, contact jprehn@rcsi.ie.
Supplementary Material
Acknowledgments
We thank Lisa Anne Byrne for support with ethics and Anne Færch Nielsen, for careful editing and suggestions on the manuscript. We thank Nora Kalabrezi and Christian Siebert for help with EEG evaluation in the PPS model. This publication has emanated from research conducted with the financial support of the European Union’s “Seventh Framework” Programme (FP7) under Grant Agreement 602130 (EpimiRNA). Additionally, this publication has emanated from research supported in part by a research grant from Science Foundation Ireland (SFI) under Grant 16/RC/3948 and cofunded under the European Regional Development Fund and by FutureNeuro industry partners. Additional support from SFI was through grants SFI/13/IA/1891 and 12/COEN/18. G.M. and J.P.H. are supported by Marie Skłodowska-Curie Actions Individual Fellowships (G.M.: “EpimiRTherapy,” H2020-MSCA-IF-2018 840262; J.P.H.: “AstroMiRimage,” H2020-MSCA-IF-2017 798644). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Competing interest statement: D.C.H. reports US Patent No. US 9,803,200 B2 “Inhibition of microRNA-134 for the treatment of seizure-related disorders and neurologic injuries.”
This article is a PNAS Direct Submission.
Data deposition: The sequencing data reported in this paper have been deposited to the gene expression omnibus (GEO) under accession no. GSE137473. The proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD019098. Custom analysis codes are available in Github at https://github.com/g-morris/Ago2Seq.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919313117/-/DCSupplemental.
References
- 1.Schuele S. U., Lüders H. O., Intractable epilepsy: Management and therapeutic alternatives. Lancet Neurol. 7, 514–524 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Kwan P., Schachter S. C., Brodie M. J., Drug-resistant epilepsy. N. Engl. J. Med. 365, 919–926 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Blumcke I.et al.; EEBB Consortium , Histopathological findings in brain tissue obtained during epilepsy surgery. N. Engl. J. Med. 377, 1648–1656 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Pitkänen A., Lukasiuk K., Mechanisms of epileptogenesis and potential treatment targets. Lancet Neurol. 10, 173–186 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Devinsky O.et al., Epilepsy. Nat. Rev. Dis. Primers 4, 18024 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Gorter J. A.et al., Potential new antiepileptogenic targets indicated by microarray analysis in a rat model for temporal lobe epilepsy. J. Neurosci. 26, 11083–11110 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McClelland S.et al., The transcription factor NRSF contributes to epileptogenesis by selective repression of a subset of target genes. eLife 3, e01267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson M. R.et al., Systems genetics identifies Sestrin 3 as a regulator of a proconvulsant gene network in human epileptic hippocampus. Nat. Commun. 6, 6031 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srivastava P. K.et al., A systems-level framework for drug discovery identifies Csf1R as an anti-epileptic drug target. Nat. Commun. 9, 3561 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosik K. S., The neuronal microRNA system. Nat. Rev. Neurosci. 7, 911–920 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Schmiedel J. M.et al., Gene expression. MicroRNA control of protein expression noise. Science 348, 128–132 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Bartel D. P., Metazoan MicroRNAs. Cell 173, 20–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ha M., Kim V. N., Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15, 509–524 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Czech B., Hannon G. J., Small RNA sorting: Matchmaking for argonautes. Nat. Rev. Genet. 12, 19–31 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandradoss S. D., Schirle N. T., Szczepaniak M., MacRae I. J., Joo C., A dynamic search process underlies microRNA targeting. Cell 162, 96–107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gebert L. F. R., MacRae I. J., Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 20, 21–37 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henshall D. C.et al., MicroRNAs in epilepsy: Pathophysiology and clinical utility. Lancet Neurol. 15, 1368–1376 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Brennan G. P., Henshall D. C., microRNAs in the pathophysiology of epilepsy. Neurosci. Lett. 667, 47–52 (2018). [DOI] [PubMed] [Google Scholar]
- 19.van Rooij E., Kauppinen S., Development of microRNA therapeutics is coming of age. EMBO Mol. Med. 6, 851–864 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henshall D. C., Manipulating microRNAs in murine models: Targeting the multi-targeting in epilepsy. Epilepsy Curr. 17, 43–47 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kretschmann A.et al., Different microRNA profiles in chronic epilepsy versus acute seizure mouse models. J. Mol. Neurosci. 55, 466–479 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srivastava P. K.et al., Meta-analysis of microRNAs dysregulated in the hippocampal dentate gyrus of animal models of epilepsy. eNeuro 4, ENEURO.0152-17.2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cava C., Manna I., Gambardella A., Bertoli G., Castiglioni I., Potential role of miRNAs as theranostic biomarkers of epilepsy. Mol. Ther. Nucleic Acids 13, 275–290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flores O., Kennedy E. M., Skalsky R. L., Cullen B. R., Differential RISC association of endogenous human microRNAs predicts their inhibitory potential. Nucleic Acids Res. 42, 4629–4639 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jimenez-Mateos E. M.et al., miRNA Expression profile after status epilepticus and hippocampal neuroprotection by targeting miR-132. Am. J. Pathol. 179, 2519–2532 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y., Guo J., Wang Q., Chen Y., MicroRNA-132 silencing decreases the spontaneous recurrent seizures. Int. J. Clin. Exp. Med. 7, 1639–1649 (2014). [PMC free article] [PubMed] [Google Scholar]
- 27.Iori V.et al., Blockade of the IL-1R1/TLR4 pathway mediates disease-modification therapeutic effects in a model of acquired epilepsy. Neurobiol. Dis. 99, 12–23 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Tao H.et al., Intranasal delivery of miR-146a mimics delayed seizure onset in the lithium-pilocarpine mouse model. Mediators Inflamm. 2017, 6512620 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jimenez-Mateos E. M.et al., Silencing microRNA-134 produces neuroprotective and prolonged seizure-suppressive effects. Nat. Med. 18, 1087–1094 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris G., Brennan G. P., Reschke C. R., Henshall D. C., Schorge S., Spared CA1 pyramidal neuron function and hippocampal performance following antisense knockdown of microRNA-134. Epilepsia 59, 1518–1526 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vangoor V. R.et al., Antagonizing increased miR-135a levels at the chronic stage of experimental TLE reduces spontaneous recurrent seizures. J. Neurosci. 39, 5064–5079 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tokar T.et al., mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 46, D360–D370 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chou C. H.et al., miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 46, D296–D302 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karagkouni D.et al., DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 46, D239–D245 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan C. L.et al., MicroRNA-128 governs neuronal excitability and motor behavior in mice. Science 342, 1254–1258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shinohara Y.et al., miRNA profiling of bilateral rat hippocampal CA3 by deep sequencing. Biochem. Biophys. Res. Commun. 409, 293–298 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Sempere L. F.et al., Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5, R13 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ludwig N.et al., Distribution of miRNA expression across human tissues. Nucleic Acids Res. 44, 3865–3877 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang T. C.et al., Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell 26, 745–752 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sano T.et al., MicroRNA-34a upregulation during seizure-induced neuronal death. Cell Death Dis. 3, e287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu K.et al., Expression profile of microRNAs in rat hippocampus following lithium-pilocarpine-induced status epilepticus. Neurosci. Lett. 488, 252–257 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Bot A. M., Dębski K. J., Lukasiuk K., Alterations in miRNA levels in the dentate gyrus in epileptic rats. PLoS One 8, e76051 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gorter J. A.et al., Hippocampal subregion-specific microRNA expression during epileptogenesis in experimental temporal lobe epilepsy. Neurobiol. Dis. 62, 508–520 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Li M. M.et al., Genome-wide microRNA expression profiles in hippocampus of rats with chronic temporal lobe epilepsy. Sci. Rep. 4, 4734 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roncon P.et al., MicroRNA profiles in hippocampal granule cells and plasma of rats with pilocarpine-induced epilepsy–Comparison with human epileptic samples. Sci. Rep. 5, 14143 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mooney C., Becker B. A., Raoof R., Henshall D. C., EpimiRBase: A comprehensive database of microRNA-epilepsy associations. Bioinformatics 32, 1436–1438 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Liu X. S.et al., Identification of miRNomes associated with adult neurogenesis after stroke using Argonaute 2-based RNA sequencing. RNA Biol. 14, 488–499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liou A. K., Clark R. S., Henshall D. C., Yin X. M., Chen J., To die or not to die for neurons in ischemia, traumatic brain injury and epilepsy: A review on the stress-activated signaling pathways and apoptotic pathways. Prog. Neurobiol. 69, 103–142 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Jimenez-Mateos E. M.et al., microRNA targeting of the P2X7 purinoceptor opposes a contralateral epileptogenic focus in the hippocampus. Sci. Rep. 5, 17486 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rajman M.et al., A microRNA-129-5p/Rbfox crosstalk coordinates homeostatic downscaling of excitatory synapses. EMBO J. 36, 1770–1787 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gross C.et al., MicroRNA-mediated downregulation of the potassium channel Kv4.2 contributes to seizure onset. Cell Rep. 17, 37–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bencurova P.et al., MicroRNA and mesial temporal lobe epilepsy with hippocampal sclerosis: Whole miRNome profiling of human hippocampus. Epilepsia 58, 1782–1793 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Miller-Delaney S. F.et al., Differential DNA methylation profiles of coding and non-coding genes define hippocampal sclerosis in human temporal lobe epilepsy. Brain 138, 616–631 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kan A. A.et al., Genome-wide microRNA profiling of human temporal lobe epilepsy identifies modulators of the immune response. Cell. Mol. Life Sci. 69, 3127–3145 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Janssen H. L.et al., Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 368, 1685–1694 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Tang C.et al., Targeting of microRNA-21-5p protects against seizure damage in a kainic acid-induced status epilepticus model via PTEN-mTOR. Epilepsy Res. 144, 34–42 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Papagiannakopoulos T., Shapiro A., Kosik K. S., MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 68, 8164–8172 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Ma Z.et al., MicroRNA regulatory pathway analysis identifies miR-142-5p as a negative regulator of TGF-β pathway via targeting SMAD3. Oncotarget 7, 71504–71513 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kotlarz D.et al., Human TGF-β1 deficiency causes severe inflammatory bowel disease and encephalopathy. Nat. Genet. 50, 344–348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ivens S.et al., TGF-beta receptor-mediated albumin uptake into astrocytes is involved in neocortical epileptogenesis. Brain 130, 535–547 (2007). [DOI] [PubMed] [Google Scholar]
- 61.La Rocca G.et al., In vivo, Argonaute-bound microRNAs exist predominantly in a reservoir of low molecular weight complexes not associated with mRNA. Proc. Natl. Acad. Sci. U.S.A. 112, 767–772 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leshkowitz D., Horn-Saban S., Parmet Y., Feldmesser E., Differences in microRNA detection levels are technology and sequence dependent. RNA 19, 527–538 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Gestel M. A.et al., shRNA-induced saturation of the microRNA pathway in the rat brain. Gene Ther. 21, 205–211 (2014). [DOI] [PubMed] [Google Scholar]
- 64.de la Mata M.et al., Potent degradation of neuronal miRNAs induced by highly complementary targets. EMBO Rep. 16, 500–511 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bitetti A.et al., MicroRNA degradation by a conserved target RNA regulates animal behavior. Nat. Struct. Mol. Biol. 25, 244–251 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Gong G. H.et al., Comprehensive circular RNA profiling reveals the regulatory role of the CircRNA-0067835/miR-155 pathway in temporal lobe epilepsy. Cell. Physiol. Biochem. 51, 1399–1409 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Epi4K consortium; Epilepsy Phenome/Genome Project , Ultra-rare genetic variation in common epilepsies: A case-control sequencing study. Lancet Neurol. 16, 135–143 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Leu C., Coppola A., Sisodiya S. M., Progress from genome-wide association studies and copy number variant studies in epilepsy. Curr. Opin. Neurol. 29, 158–167 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Norwood B. A.et al., Classic hippocampal sclerosis and hippocampal-onset epilepsy produced by a single “cryptic” episode of focal hippocampal excitation in awake rats. J. Comp. Neurol. 518, 3381–3407 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mouri G.et al., Unilateral hippocampal CA3-predominant damage and short latency epileptogenesis after intra-amygdala microinjection of kainic acid in mice. Brain Res. 1213, 140–151 (2008). [DOI] [PubMed] [Google Scholar]
- 71.Reschke C. R.et al., Potent anti-seizure effects of locked nucleic acid antagomirs targeting miR-134 in multiple mouse and rat models of epilepsy. Mol. Ther. Nucleic Acids 6, 45–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haunsberger S. J., Connolly N. M., Prehn J. H., miRNAmeConverter: An R/bioconductor package for translating mature miRNA names to different miRBase versions. Bioinformatics 33, 592–593 (2017). [DOI] [PubMed] [Google Scholar]
- 73.Moore M. J.et al., Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat. Protoc. 9, 263–293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.European Molecular Biology Laboratory , RNA-seq from 53 human tissue samples from the Genotype-Tissue Expression (GTEx) Project. Expression Atlas. https://www.ebi.ac.uk/gxa/experiments/E-MTAB-5214/Results. Accessed 5 April 2018.
- 75.Ma'ayan Laboratory of Computational Systems Biology , ENCODE transcription factor targets. Harmonizome. http://amp.pharm.mssm.edu/Harmonizome/dataset/ENCODE+Transcription+Factor+Targets. Accessed 13 October 2018.
- 76.Ma'ayan Laboratory of Computational Systems Biology , CHEA transcription factor targets. Harmonizome. http://amp.pharm.mssm.edu/Harmonizome/dataset/CHEA+Transcription+Factor+Targets. Accessed 13 October 2018.
- 77.Tan N. C., Berkovic S. F., The epilepsy genetic association database (epiGAD): Analysis of 165 genetic association studies, 1996-2008. Epilepsia 51, 686–689 (2010). [DOI] [PubMed] [Google Scholar]
- 78.Wang J.et al., Epilepsy-associated genes. Seizure 44, 11–20 (2017). [DOI] [PubMed] [Google Scholar]
- 79.Davis A. P.et al., The comparative toxicogenomics database: Update 2019. Nucleic Acids Res. 47, D948–D954 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boyle E. I.et al., GO:TermFinder—Open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20, 3710–3715 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Veno M., Kjems J., Henshall D. C., A systems approach delivers a functional microRNA catalog and expanded targets for seizure suppression in temporal lobe epilepsy. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137473. Deposited 16 September 2019.
- 82.Andersen J. S., A systems approach delivers a functional microRNA catalog and expanded targets for seizure suppression in temporal lobe epilepsy. PRIDE. https://www.ebi.ac.uk/pride/archive/projects/PXD019098. Deposited 12 May 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data have been deposited to the Gene Expression Omnibus (GEO) under accession no. GSE137473 (81). The proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD019098 (82). Custom analysis codes are available on https://github.com/g-morris/Ago2Seq. All data generated or analyzed during this study are included in this published article (and its SI Appendix files). For questions on the MTI analysis, contact jprehn@rcsi.ie.