Significance
Haptoglobin (Hp) is one of the most abundant plasma proteins; it binds with high affinity to hemoglobin (Hb). Thereby Hp protects against the toxic effects of the heme when Hb leaks into plasma from damaged red blood cells. Therefore, serum Hp is an important antioxidant and a clinically important protein, often used as a protein biomarker. Here, we address in detail what proteoforms and proteoform assemblies co-occur in serum, and show how they differ in individuals with distinct genotypes. Our data, obtained by a range of state-of-the-art analytical methods, reveal in unprecedented detail how these hundreds of Hp proteoforms influence the scavenging of Hb through several distinctive molecular features of Hp genotypes.
Keywords: haptoglobin, genetic polymorphism, hybrid mass spectrometry, protein glycosylation, proteolytic processing
Abstract
The serum haptoglobin protein (Hp) scavenges toxic hemoglobin (Hb) leaked into the bloodstream from erythrocytes. In humans, there are two frequently occurring allelic forms of Hp, resulting in three genotypes: Homozygous Hp 1-1 and Hp 2-2, and heterozygous Hp 2-1. The Hp genetic polymorphism has an intriguing effect on the quaternary structure of Hp. The simplest form, Hp 1-1, forms dimers consisting of two α1β units, connected by disulfide bridges. Hp 2-1 forms mixtures of linear (α1)2(α2)n-2(β)n oligomers (n > 1) while Hp 2-2 occurs in cyclic (α2)n(β)n oligomers (n > 2). Different Hp genotypes bind Hb with different affinities, with Hp 2-2 being the weakest binder. This behavior has a significant influence on Hp’s antioxidant capacity, with potentially distinctive personalized clinical consequences. Although Hp has been studied extensively in the past, the finest molecular details of the observed differences in interactions between Hp and Hb are not yet fully understood. Here, we determined the full proteoform profiles and proteoform assemblies of all three most common genetic Hp variants. We combined several state-of-the-art analytical methods, including various forms of chromatography, mass photometry, and different tiers of mass spectrometry, to reveal how the tens to hundreds distinct proteoforms and their assemblies influence Hp’s capacity for Hb binding. We extend the current knowledge by showing that Hb binding does not just depend on the donor’s genotype, but is also affected by variations in Hp oligomerization, glycosylation, and proteolytic processing of the Hp α-chain.
Around 1900 Karl Landsteiner discovered the ABO blood group system, which resulted in the description of the first human genetic polymorphism (1). Identification of the genetic polymorphism of the histo-blood group ABO system glycosyltransferases, which control blood group antigen expression, has proven to be of essential importance, not only in transfusion medicine but also in the pathophysiology of hemolytic diseases of newborn babies and organ transplantation (2). Subsequently, genetic polymorphisms have been described for many other proteins (3), including several highly abundant plasma proteins, such as antitrypsin (4, 5), fetuin (6), and haptoglobin (Hp) (7, 8). Hp represents a particularly interesting case. It exists in two allelic forms in the human population, Hp1 and Hp2 (9–11) (SI Appendix, Fig. S1). Based on the most recent findings on the evolutionary origins of the Hp polymorphism, Hp1 likely arose from recurrent deletions in Hp2 (12). Hp is among the 10 most abundant proteins in serum (13, 14) with a significant function to clear toxic hemoglobin (Hb) (15, 16) through high-affinity binding to the macrophage scavenger receptor CD163 (7), but various other roles have been proposed as a potent antioxidant (12, 17) and in the immune response (18). Notably, one of the most striking effects of the Hp polymorphism resides in the structural heterogeneity represented by the existence of Hp in different oligomeric states, depending on the Hp genotype (19, 20). From a structural point of view, the Hp 1-1 homozygote allele type represents the simplest form, exhibiting a tetrachain structure composed of two α1- (molecular mass ∼9.2 kDa) and two β- (molecular mass ∼35 kDa) chains linked by disulfide bonds (21). The β-chain (containing a serine protease domain, SP) is common to all Hp genotypes, while the α-chain (containing the complement component control protein domain, CCP) has two (genetic) variants, α1 and α2. The α2-chain (molecular mass ∼15.9 kDa) has a high degree of sequence identity with α1, albeit with a repeated CCP domain (22). Importantly, the α1-chain possesses two interlinking Cys residues, one at the C terminus connecting with a β-chain (αβ unit), and the other one at the N terminus linking another αβ unit that forms the above-mentioned tetra-chain structure in Hp 1-1 (2 × α1β). The α2-chain contains a repeated CCP domain with three extra cysteines and therefore can interact with an additional αβ unit. This feature has a crucial effect on the quaternary structures of the other two Hp genotypes, the 2-1 heterozygote and the 2-2 homozygote. Structural studies have confirmed that Hp 2-1 forms linear oligomers of α2β units, in which α1β always forms the terminal units of the Hp protein oligomer. Hp 2-2 builds cyclic structures consisting of multiple α2β units (23).
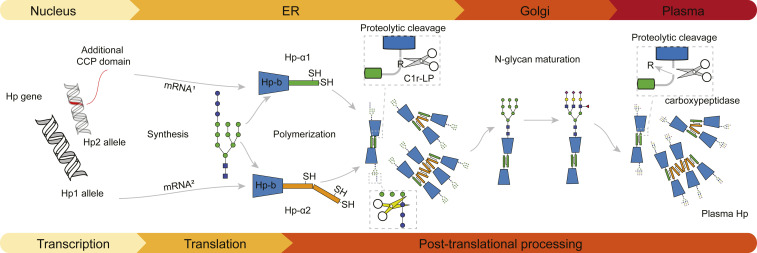
The complex structural heterogeneity of Hp implies that also its biosynthesis may stand out compared to other common serum proteins. Indeed, Hp synthesis is unusual as its proform is cleaved already in the endoplasmic reticulum (ER) before it enters the Golgi (24, 25), where most of the secretory proteins are typically cleaved (Fig. 1). Hp is synthesized predominantly in hepatocytes as a single 45-kDa polypeptide, which rapidly forms dimers through disulfide bond formation (26). Shortly after, the α-chain of Hp is proteolytically cleaved C-terminal to the Arg102 residue (Arg161 in Hp2), which is subsequently removed by a so far unknown plasma carboxypeptidase (27), resulting in the final tetrameric form, Hp 1-1. The initial cleavage after Arg102/161 is thought to be mediated by the C1 complement C1r-like protein (C1r-LP) that resides in the ER (28). A comparison of the amino acid sequences in proximity to the cleavage site in proforms of Hp, C1s, and C1r shows a remarkable similarity. This sequence resemblance pinpoints an evolutionary relationship of Hp to these serine proteases involved in activation of the complement system (29). The biological relevance of this proteolytic cleavage of the Hp proform was demonstrated to be essential for Hb binding (30, 31).
Fig. 1.
Model depicting the maturation pathway of Hp 2-1 oligomers. At the transcription level, the two Hp alleles lead to two distinctive mRNAs and thus two protein backbones. Initial glycosylation and oligomer formation occur in the ER, generating disulfide-bound oligomers. Furthermore, the glycans are maturing in the Golgi. Posttranslational elimination of Arg102/161 occurs in plasma. Eventually, the final mixture of mature oligomeric Hp assemblies is ready to capture Hb. See also SI Appendix, Fig. S1 for annotated amino acid sequences originating from the distinct allotypes Hp1 and Hp2.
Next to the diversity originating from distinct genotypes and processing, further variety in Hp stems from posttranslational modifications (PTMs), primarily glycosylation (32–34). Protein glycosylation is one of the most complex PTMs and dramatically influences the structural and functional properties of proteins. Hp possesses four N-glycosylation sites on the β-chain, located at Asn184, Asn207, Asn211, and Asn241 occupied by complex type N-glycans with a varying number of antennas, which may be fucosylated and sialylated (35, 36). In healthy individuals, the glycosylation sites, Asn184, Asn207, and Asn241, carry predominantly sialylated biantennary N-glycans, while Asn211 is primarily occupied by sialylated triantennary complex N-glycan. Fucosylation is primarily located on the antennas. Core fucosylation was observed only in trace amounts (37) and predominantly in serum of people with some cases of rare cancers (38). The functional role of these glycans is not yet fully understood. The vast majority of glycoproteomic studies dealing with Hp glycosylation have been focused on the analysis of aberrances and changes related to a particular physiological state, such as cancer (35, 36, 38), and discovery of novel biomarkers. Two of these studies investigated glycosylation differences among patients with liver cirrhosis and hepatocellular carcinoma (35, 36). Interestingly, both studies revealed discrepancies in Hp glycosylation patterns, specifically in the structural microheterogeneity of the identified sites. Such differences in glycosylation patterns between individuals can be caused by a mixture of genetic and environmental factors, as demonstrated in several glycomic and genomic studies (39, 40). Although the significance of Hp genotypes has already been reflected in clinical practice (41), mainly regarding the difficulty in the precise determination of Hp levels in individuals with different genotypes, it has not yet been addressed how such genetic differences may affect the glycosylation patterns and protein structure. We recently contributed to this discussion by demonstrating with MS that a genetic polymorphism substantially influences the proteoform profile of another plasma protein, human fetuin (42).
The in-tandem assembled CCP domains in the α2-chains of Hp2 have several crucial functional consequences. The best-established function of Hp is its binding to free Hb leaked from red blood cells (32). This heme detoxifying process in plasma mitigates oxidative stress and recycles iron within the body. Although still controversial (43), it has been proposed that the stoichiometry of the binding between Hp and Hb is identical for all forms of Hp and equals 1:1. That is, one αβ unit of Hp binds one αβ unit of Hb (22, 44, 45). In theory, 1 mole of the homozygote Hp 1-1 contains fewer binding sites (fewer αβ units) for Hb than the more complex oligomeric structures occurring in the heterozygote 2-1 and homozygote 2-2 genotypes. From that point of view, it is somewhat surprising that Hp 1-1 has been found to exhibit superior binding to Hb when compared to Hp 2-1 and 2-2 (17, 44). A molecular basis for the observed tight binding of Hp 1-1 to Hb (46) has been revealed by the X-ray structural analysis of a porcine Hp–Hb complex (21). However, an explanation for the apparent lower affinity of Hp 2-1 and 2-2 to Hb is currently missing. In a compensatory manner, it has been observed that protein assemblies of Hb with highly oligomeric Hp 2-2 exhibit higher functional affinity for the CD163 hemoglobin scavenger receptor than do complexes of Hb with dimeric Hp 1-1 (7).
The extraordinary ability of Hp to bind Hb is often used to isolate Hp from plasma. Recently, it has been reported that a small fraction of native Hp binds somewhat weaker to Hb compared to the tightly bound Hp fraction. This weakly bound Hp exhibited an altered glycosylation pattern (47), with relatively more branching complex glycans, somewhat less decorated with α2-6 linked sialic acid residues. The glycosylation effect was further investigated by Wu et al. (48, 49), who combined native MS and glycoproteomics to examine the impact of Hp glycosylation microheterogeneity on interactions with Hb and lectins. The authors revealed that N-glycan branching attenuates Hp–Hb binding affinity, and in contrast, fucosylation stabilizes the Hp–Hb binding. Overall, these studies indicate that N-glycosylation might fine-tune Hp and Hb interactions, although they focus exclusively on Hp 1-1. Here, we also applied Hp–Hb-binding assays to investigate properties of Hp oligomers isolated from single donors of all three of the most common genotypes and complement the acquired data with an in-depth analysis of co-occurring Hp proteoforms and proteoform assemblies. Our data, obtained by interferometric scattering microscopy (mass photometry) (50), revealed apparent differences in the Hb binding among different Hp oligomers originating from the three Hp genotypes. In-depth characterization of the Hp proteoform profiles by combining native separation methods such as gel electrophoresis, size-exclusion chromatography (SEC), ion-exchange chromatography (IEX), and state-of-the-art integrative MS approaches (51, 52) revealed hundreds of different proteoforms and proteoform assemblies. The data showed distinctive glycosylation profiles for different genotypes and oligomeric states. Additionally, we discovered a trend in the proteolytic processing of Hp that correlates well with the observed differences in Hb binding among the distinct Hp oligomers. Overall, our findings unravel a wealth of molecular details that fine-tune Hp–Hb interactions, laying the ground for a full explanation of proteogenotype-specific binding of Hp to Hb.
Results
The binding of Hp to dimeric oxygenated Hb is one of the strongest known noncovalent interactions occurring in human plasma with a reported equilibrium dissociation constant (Kd) of around 10−15 M (53). The effect of this interaction, formation of Hp–Hb complexes, has been studied for decades and, therefore, it may be surprising that several molecular details regulating these interactions remain elusive. A likely explanation lies in the sheer structural diversity of Hp originating from the coexistence of genetic variants and the presence of multiple oligomers, all decorated with a plethora of heterogeneous PTMs. Thus, the Hp–Hb complex is represented not just by a single configuration but by an entire class of different conformations and oligomeric states composed of a varying number of Hp monomers, also highly dependent on the Hp genotype. Human Hb is typically present in red blood cells as a hetero-tetramer Hb(αβ)2. However, this tetramer dissociates into two αβ heterodimers after entering plasma, where Hp captures Hb very efficiently (54). In the remainder, we refer to this binding Hp (αβ) unit as an Hp monomer. Although each of us needs Hp to scavenge Hb, this is done in a very personal manner, depending on each individual’s genotype. Therefore, here we set out to investigate the occurrence and properties of all Hp proteoforms with respect to their genotype. We obtained Hp from homozygote donors carrying either the Hp 1-1 genes or the Hp 2-2 genes, and Hp from a single heterozygote donor carrying Hp 2-1. For some of the experiments requiring more material, we used a commercially available Hp mixed type (termed Hp M), acquired from multiple donors, which turned out to be mostly Hp 2-1, with just traces of other Hp genotypes. We clearly state in the text when this Hp M sample was used.
Mass Photometry Reveals Genotype-Specific Oligomerization Status of Hp and Stoichiometry of Hp–Hb Complexes.
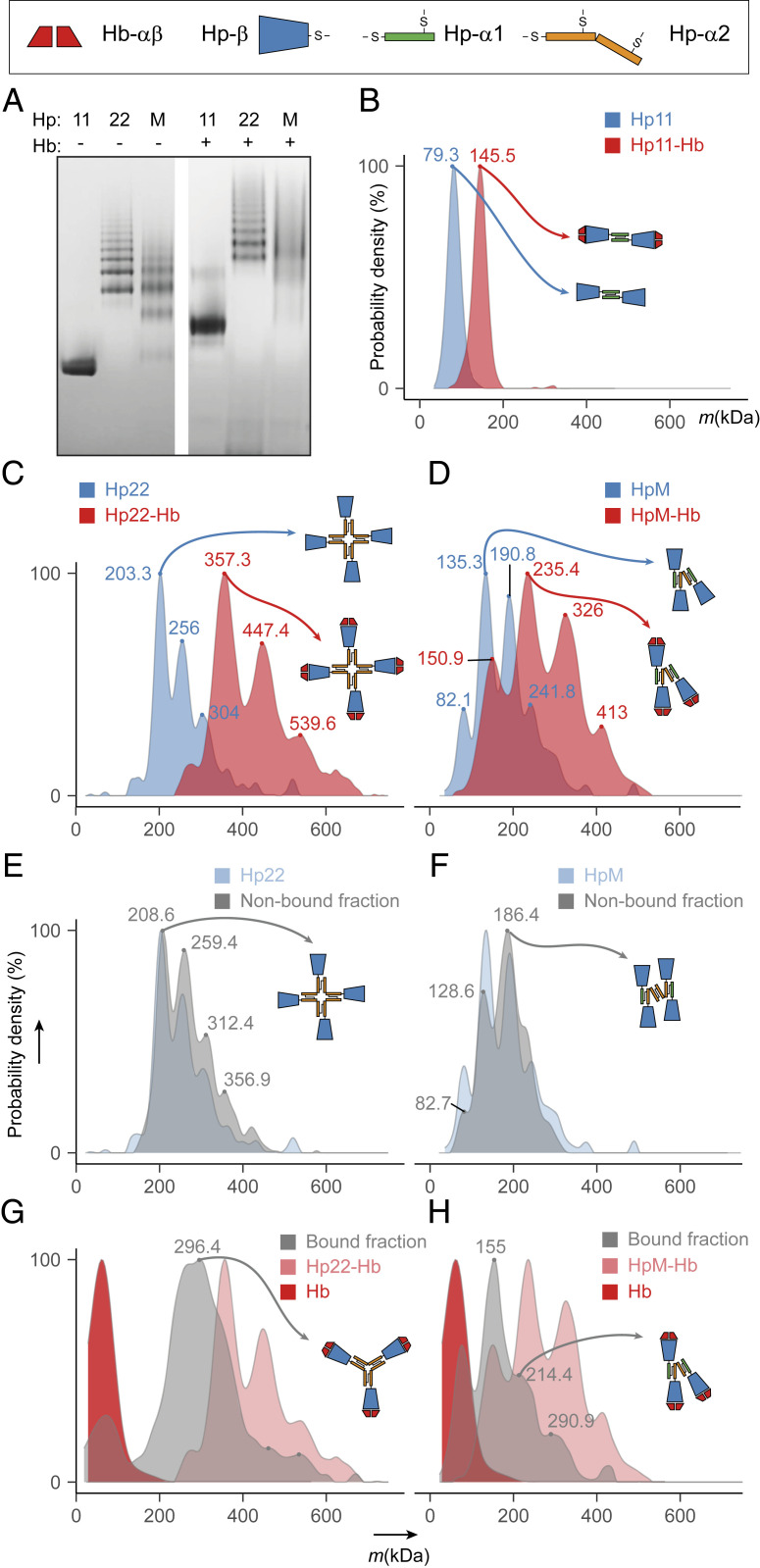
Most of the fundamental questions regarding Hp center around its primary function in blood, capturing Hb through the formation of tight Hp–Hb complexes. First, we investigated whether there are any differences in binding stoichiometry among the Hp–Hb complexes formed in the serum of donors of the three major genotypes of Hp: 1-1, 2-2, and 2-1. All three genetic variants of Hp were first separated and visualized by blue native-gel electrophoresis (Fig. 2A). While Hp 1-1 appears as a single band, Hp 2-2 and Hp 2-1 (here represented by Hp M) were detected as series of resolved bands corresponding to the different Hp oligomers. Our native-gel results resemble well previously reported data, showing similar distributions of the resolved bands of the distinct Hp oligomers (23). Incubating these Hp samples in molar excess of Hb compared to the monomer of Hp resulted in native gels with a clear mass shift of all bands toward higher masses, suggesting the extensive formation of Hp–Hb complexes. However, sample complexity and insufficient resolution did not allow us to estimate relative quantities and to determine binding stoichiometries of Hp oligomers accurately.
Fig. 2.
Hp oligomerization and Hb binding for different Hp genotypes. (A) Blue native-gel analysis of Hp 1-1, Hp 2-2, and Hp 2-1, represented by Hp M, alone (Left) and upon incubation with Hb (Right). (B) Mass photometry profiles obtained for Hp 1-1 (blue) and Hp (1, 1)-Hb (red) with the determined average molecular mass indicated on the peak apex. (C and D) Mass photometry profiles of Hp 2-2 and Hp (2, 2)-Hb (C) and of Hp M and Hp(M)-Hb (D). The blue profiles represent Hp alone and the red profiles correspond to Hb-bound Hp. (E and F) Constructed overlay of mass photometry profiles obtained for Hp 2-2 (E) or Hp M (F) and their respective Hb-unbound fractions (gray) demonstrates the relative depletion of larger oligomers in the Hb-unbound fraction. (G and H) Constructed overlay of mass photometry profiles obtained for Hp (2, 2)-Hb (G) or Hp(M)-Hb (H) and their respective Hb-bound fractions demonstrate enrichment of smaller oligomers. A mass profile of Hb alone is depicted in dark red. See also SI Appendix, Fig. S2 for a more detailed overview of all Hp oligomer mass measurements by using mass photometry.
Next, we used the recently introduced mass photometry technology, which has great potential in characterizing protein–protein interactions in solution, including the quantitative detection and analysis of binding stoichiometries (50). We applied mass photometry to quantify the relative abundance of each detected Hp oligomer and assess their binding stoichiometries with Hb. Fig. 2 B–D display the quantitative distributions of resolved Hp oligomers before and after incubation with Hb. In all cases, we observed a homogeneous shift of all peaks to higher masses in the mass photometry profiles after incubating Hp with Hb (see also SI Appendix, Fig. S2A). Therefore, we conclude that the binding stoichiometry is the same for all Hp monomers in each of the abundant Hp oligomers, independent of genotype; for example, a dimer of Hp 1-1 binds two dimers of Hb(αβ), and a pentamer of Hp 2-1 binds five dimers of Hb(αβ), one per monomer of Hp (SI Appendix, Fig. S2B). This observation is in good agreement with previous assessments of these stoichiometries (22, 44, 45). Additionally, we determined that the desialylation of the Hp samples by using neuraminidase does not affect the stoichiometry of Hb binding. Although our data indicate that sialylation does not affect Hp–Hb binding substantially, the sialic acid linkages on the N-glycans of Hp can be functionally relevant, for example in the uptake of Hp via the asialoglycoprotein receptor (55). The molecular masses of all Hp assemblies recorded by mass photometry were in good agreement with theoretically expected masses with an average mass error of approximately ±4.1% (SI Appendix, Fig. S2 C and D and Table S1). This error is higher compared to mass photometry measurements of nonglycosylated proteins, for which typically a 2% mass accuracy and 1-kDa mass precision have been determined (50). Because of a multitude of coexisting glycoproteoforms with subtle mass differences, the mass distribution of any Hp oligomer spans more than 4 kDa. Therefore, we expect that the heterogeneity of Hp broadens the mass distribution in our mass photometric measurements, which subsequently influences the determination of the mass accuracy and precision.
Hb-Binding Assays Reveal Differences in Hb Binding among Different Hp Oligomers.
Although the binding affinity between Hp and Hb is known to be particularly high, some minor variations in the binding strength within and between the Hp genotypes have been reported (8, 43, 48). To investigate such variations, we combined mass photometry with a well-described Hb-binding assay, in which we fractionated all three Hp samples by using an Hb-agarose gel column, following the protocol of Wu et al. (48), who performed this earlier, albeit for Hp 1-1 alone. Subsequently, we obtained mass photometry profiles for Hb-bound and -unbound fractions and compared them to the mass profiles of the original unfractionated Hp (Fig. 2 E–H). Notably, we observed in both Hp 2-2 and 2-1 (represented by Hp M) clear changes in the mass distribution of Hp oligomers. In the unbound Hp 2-2 and 2-1 fraction, the relative abundance of the higher–molecular-mass Hp oligomers was increased compared to their original profiles (Fig. 2 E and F). Concurrently, the Hp-bound fraction was relatively enriched for lower–molecular-mass Hp oligomers in both genotypes (Fig. 2 G and H). These differences in the mass profiles suggest that lower-mass Hp oligomers have a higher affinity to Hb compared with higher-mass Hp oligomers. To further illustrate this point, the Hp 2-2 trimer, which comprises less than 10% of the total abundance in the original unfractionated Hp 2-2 mass profile, is nearly five-times enriched (47%) in the Hb-bound fraction, suggesting it has a higher affinity to Hb compared to the higher–molecular-mass Hp 2-2 oligomers (SI Appendix, Fig. S2E). All quantitative data, including fractional abundances of each Hp oligomer in all analyzed samples, are listed in SI Appendix, Tables S1 and S2. The previous findings that Hp 1-1 has a higher affinity for Hb than Hp 2-2 (8, 43) may, thus, stem from the fact that Hp 1-1 forms exclusively dimers while Hp 2-2 forms higher Hp oligomers. Notably, this claim is surrounded by some controversy (56, 57).
Characterizing Hp Proteoforms and Proteoform Assemblies by Integrative MS.
The distinct mass photometry profiles obtained for native Hp oligomers in Hb-bound/unbound fractions demonstrated that lower–molecular-mass Hp complexes have a higher affinity to Hb compared to higher–molecular-mass Hp complexes. Up to now, we lack a clear explanation of this phenomenon. The X-ray structural models of the porcine Hp (1, 1)-Hb complex (21) and human Hp–Hb complex with an Hp–Hb receptor from Trypanosoma brucei brucei (58) provide the best insights so far into the molecular basis of the strong binding of Hp to Hb. Unfortunately, high-resolution structures of Hp oligomers and their complexes with Hb are not yet available. Moreover, it has been hypothesized that the microheterogeneity at the level of PTMs fine-tunes Hp–Hb complexation (48), which is more challenging to address with the current high-resolution structural biology methods that average over many particles. Distinctively, mass spectrometry enables charting of the wealth of proteoforms (59) and their assemblies by detecting subtle mass deviations, which stem from distinctive PTM patterns or sequence variations (60, 61). Here, we combined native and intact denaturing MS to characterize Hp oligomers from all three genotype variants.
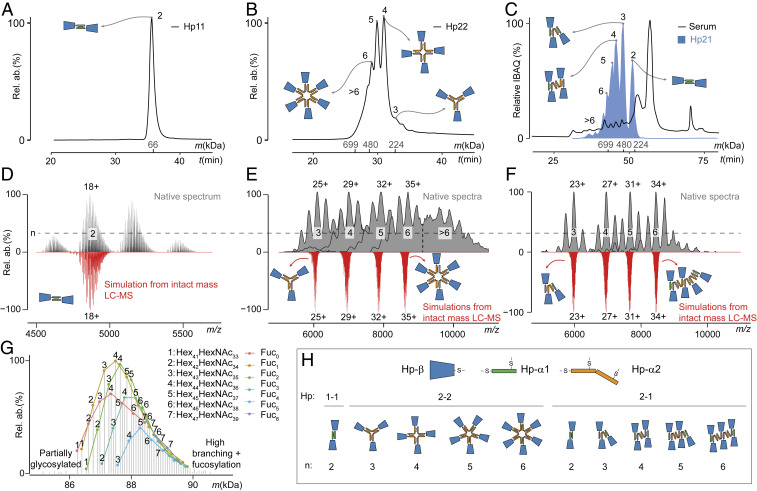
To mass-analyze in depth each Hp oligomer, we first fractionated them by SEC (Fig. 3 A–C). While Hp 1-1 and Hp 2-2 samples were obtained commercially as purified proteins (Fig. 3 A and B) from genotyped donors, for Hp 2-1 we purified Hp oligomers directly from the serum of one individual (Fig. 3C). In the latter case, to separate the Hp 2-1 from copurified serum proteins, Hp-containing SEC fractions were further cleaned up by IEX. Before MS analysis, each fraction was treated with neuraminidase to remove sialic acid moieties, which are present in high abundance on the various N-glycans of Hp molecules. We observed that for reducing the complexity in the native MS spectra, desialylation was essential and helped us to discern between double fucosylation and sialylation since they have similar masses (i.e., 291.1 and 292.1 Da, respectively).
Fig. 3.
Characterization of SEC separated Hp oligomers by high-resolution native MS, and in silico reconstruction based on denatured intact protein LC-MS data. (A and B) SEC chromatograms of Hp 1-1 and Hp 2-2, respectively, with the apparent molecular mass and oligomerization status annotated. (C) SEC profile of serum from a donor with Hp 2-1 genotype (black line). The abundance profile of Hp proteins (based on iBAQ [intensity-based absolute quantification] proteomics values) is displayed in blue. (D–F) High-resolution native MS spectra of desialylated Hp 1-1 (D) and SEC separated oligomers of desialylated Hp 2-2 (E) and Hp 2-1 (F) (black) and corresponding in silico-generated spectra only for the most abundant charge states (red). (G) Annotation of peaks in the native MS spectrum of Hp 1-1 with integers corresponding to the base glycan composition and color denoting the number of terminal fucoses carried by Hp 1-1 (SI Appendix, Table S5). See also SI Appendix, Fig. S3 for annotations of the proteoforms of Hp 2-1 oligomers. (H) Cartoons of the complexes observed for Hp 1-1, Hp 2-2, and Hp 2-1 in the most abundant SEC fractions (A–C).
Even after desialylation, the native MS spectra of Hp revealed highly complex proteoform profiles for all measured samples, which could be prevalently attributed to the microheterogeneity of N-glycosylation. In addition to the accurate mass assignment of each detected Hp proteoform, high-resolution native MS spectra can confirm the overall composition of protein complexes (62). However, direct annotation of the complicated MS profiles of Hp turned out to be very challenging due to isobaric species or species with minute mass differences, which were present in these heavily glycosylated samples. One way to untangle such complex native MS spectra is by simulating them based on data from either peptide LC-MS (63) or intact protein mass measurements, where distinct denatured and reduced polypeptide chains are separated by LC and measured by MS (51). Here, we used the intact mass denaturing LC-MS analysis of Hp fractions to simulate and annotate the corresponding native MS spectra. In this approach, masses and abundances of the distinct Hp–β- and Hp–α-chains were determined and used for in silico construction of masses for Hp oligomers (SI Appendix, Tables S3 and S4). The upper panels of Fig. 3 D–F display the original native MS spectra of the most abundant Hp oligomers from the Hp 1-1, 2-2, and 2-1 genotypes, respectively, while the lower panels display the in silico constructed native-like proteoform profiles for the most abundant charge state of each oligomer (see Material and Methods for details and Fig. 3Hfor cartoon representation of Hp oligomers).
The high correlation of the in silico reconstructed mass profiles with the native spectra (e.g., r = 0.96 for the Hp 1-1 dimer and r = 0.92 for the Hp 2-1 trimer) supports that this hybrid method is suitable for the comprehensive annotation of complex native MS spectra of heavily glycosylated Hp oligomers. An example of such a fully annotated native MS spectrum can be seen in Fig. 3G, where we visualize different glycoproteoform series for the Hp 1-1 variant. Our data for Hp 1-1 agrees with the annotation provided by Wu et al. (48). In addition, this approach allowed us to reduce ambiguity and accurately determine the oligomeric state and composition of Hp oligomers in the SEC fractions of Hp 2-1. For example, without in silico-generated spectra, it would be challenging to determine that the Hp trimer in the SEC fraction 3 (Fig. 3C) had the (α1)2α2β3 composition ruling out alternative possibilities (e.g., α1(α2)2β3), which can also fit into the broad mass profiles observed. Simulation of the mass profiles for the Hp 2-1 trimer (α1)2α2β3, tetramer (α1)2(α2)2β4, and pentamer (α1)2(α2)3β5 are provided in the SI Appendix, Fig. S3. In summary, this hybrid MS approach allowed us to determine the oligomeric state of Hp complexes observed in each SEC fraction and provided a comprehensive list of tens to hundreds of Hp proteoforms with their PTM compositions (SI Appendix, Tables S5–S8). A more extensive description of the data processing pipeline used for the identification and simulation of the Hp native mass spectra is provided in SI Appendix, Fig. S4.
Posttranslational Processing of Hp Correlates with Its Higher-Order Structure and Hb Binding.
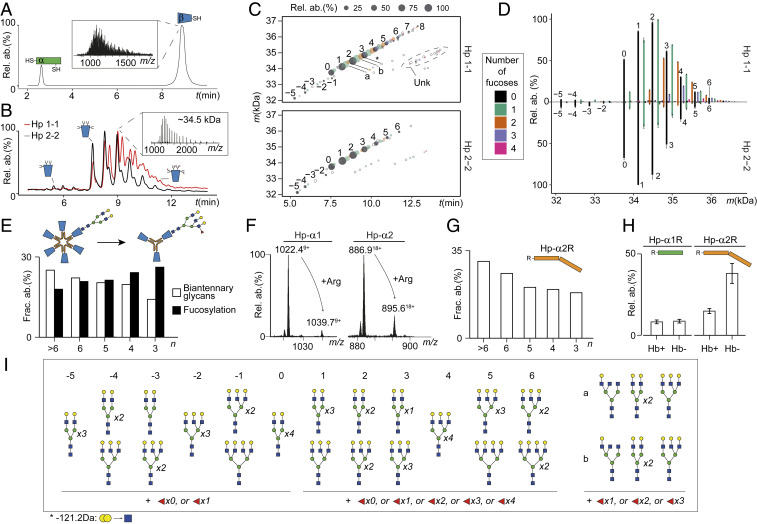
The Hp monomer is composed of an α- and β-chain linked by a disulfide bridge, which after reduction results into an α1 or α2 chain exhibiting molecular masses of 9.2 kDa and 15.9 kDa, respectively (depending on the Hp genotype), and a β-chain with a molecular mass of 35 kDa (including the averaged mass of the N-glycans). Principally, all of these Hp chains and their proteoforms can be chromatographically separated and analyzed by MS. Initially, we used reverse-phase LC-MS that provided an excellent separation of the α- and β-chains (Fig. 4A). However, the reverse-phase chromatographic mechanism, based on hydrophobic interactions, did not allow for sufficient separation of the complex glycoproteoforms of the β-chain (Fig. 4 A, Inset). To further investigate glycosylation differences, we performed hydrophilic interaction chromatography (HILIC) using an amide-based stationary phase (64). By injecting desialylated and reduced Hp 1-1 and Hp 2-2 onto this HILIC system, we attained decent separation of Hp-β glycoproteoforms based on their distinct glycosylation patterns (Fig. 4B). The high resolving power achieved by HILIC-LC-MS allowed us to detect distinct proteoforms per scan (Fig. 4 B, Inset), enabling accurate mass determination. Intriguingly, the recorded HILIC-LC-MS data revealed significant differences in glycosylation patterns between the β-chains originating from Hp 1-1 and Hp 2-2. To further interpret the HILIC-LC-MS data, we performed mass matching of observed mass features to the theoretical list of Hp-β glycoproteoforms (see Materials and Methods for details). For this, we assumed the presence of only complex N-glycans on all four glycosylation sites in Hp-β, as previously reported in detail (37, 48).
Fig. 4.
Sequence processing and N-glycosylation fine-tune Hp oligomerization and Hb binding. (A) Separation and detection of intact Hp 1-1 α- and β-proteoforms with reverse-phase LC-MS. The nonresolved glycoforms of Hp-β are shown in the Inset. (B) HILIC-LC-MS of Hp 1-1 and Hp 2-2 shows good separation of the Hp-β glycoforms, enabling accurate mass determination of each separated proteoform (Inset). (C) Proteoform elution maps obtained by HILIC-LC-MS of Hp 1-1 and Hp 2-2 depicting qualitatively and quantitatively all mass features detected in the samples as a function of retention time. The color and size of a dot reflect the number of fucoses carried by the glycoform and its relative abundance in the sample, respectively. Filled dots represent mass features detected in triplicate, empty circles represent mass features detected in only one LC-MS run. (D) Mirror plot of average abundances of Hp-β glycoforms in Hp 1-1 and Hp 2-2 based on the three technical replicates. Error bars represent SEM. The color scheme is the same as in C. (E) Levels of fucosylation (filled bar) and biantennary glycans (empty bar), as observed in Hp 2-2 oligomers. Illustratively, a schematic representation of a biantennary glycan attached to a higher Hp oligomer and a branched and fucosylated glycan on a smaller oligomer of Hp 2-2 is depicted above. (F) Mass (m/z) profiles of Hp-α proteoforms detected with and without Arg for Hp 1-1 and Hp 2-2. (G) Abundance levels of the noncleaved Hp-α2R form in distinct Hp 2-2 oligomers. (H) Abundance levels of noncleaved Hp-α1R and Hp-α2R in Hb-bound (Hb+) and Hb-unbound (Hb−) fractions following the above mentioned Hb-binding assays of Hp 1-1 and Hp 2-2 (see main text) reveal that the processing is crucial for tight Hb binding. (I) Overview of the primary glycan compositions for glycoforms of Hp-β depicted by the integer and lowercase Roman letters in C and D. Number of fucoses detected in this study is indicated underneath the glycan structures.
Our analysis supported that the majority of Hp proteoforms contain complex N-glycans on all of their four potential N-glycosylation sites with a pronounced level of macro and microheterogeneity (Fig. 4C). By fragmenting the most abundant glycoproteoforms using top-down electron transfer dissociation (ETD)-MS/MS we observed that branching and fucosylation occur predominantly on sites Asn207/Asn211, while Asn184 and Asn241 primarily harbor complex biantennary N-glycans (SI Appendix, Fig. S5). These results are nicely in agreement with reported data obtained by a more detailed bottom-up glycopeptide analyses (35, 36). We also detected Hp-β glycoforms with only three N-glycosylation sites occupied, which were somewhat more abundant in the Hp 2-2 samples, although in all cases they were significantly less abundant (2.15 ± 0.27% of total Hp-β proteoforms for Hp 1-1 and 5.50 ± 0.94% for Hp 2-2) compared to fully occupied forms. Top-down ETD-MS/MS of this partially glycosylated Hp-β revealed that the putative Asn241 glycosylation site does not harbor any glycans. For some of the highly hydrophilic, albeit very low abundant, glycoform masses (Fig. 4C, labeled as unk), we were not able to match their determined by us glycosylation compositions (e.g., H26HN15F1) to any of the N-glycan combinations reported for Hp; therefore, we chose to exclude them from further analysis.
Overall, we observed that the Hp1-β contains more branched complex N-glycans with a substantially higher level of fucosylation compared to the Hp2-β chain (Fig. 4D). These observed apparent differences between the two genetic variants motivated us to further investigate the glycosylation patterns as a function of the Hp oligomeric state, which is the primary distinctive feature between Hp 1-1 and Hp 2-2. For this, we performed HILIC-LC-MS analysis of several of the above-mentioned Hp 2-2 SEC fractions (Fig. 3B), which correspond to different Hp oligomers. As branching and fucosylation appeared to be different between Hp 1-1 and Hp 2-2, we specifically focused on these two features in the glycosylation patterns of the distinct Hp 2-2 oligomers. Interestingly, the data revealed that with the decreasing size of the Hp oligomer, both branching and fucosylation were increasing (Fig. 4E). The same trend was observed for oligomers of Hp 2-1 (SI Appendix, Fig. S6A). These trends of decreasing complexity in glycosylation with increasing oligomeric state correlate with the mass photometry results for Hb-bound and -unbound fractions of Hp 2-2 and Hp 2-1, wherein the binding affinity of Hp to Hb is decreasing with the increasing size of the Hp complex. Recently, fucosylation and branching of N-glycans were shown to enhance the binding affinity of Hp 1-1 to Hb (48). Here, we hypothesize that glycosylation not only mediates Hb binding within the single oligomeric state of Hp 1-1 but, concurrently, plays an essential role in fine-tuning Hb binding in all of the co-occurring distinctive oligomeric states of Hp 2-2 and Hp 2-1.
In addition to the glycosylation patterns of Hp-β being dependent on the oligomeric state, we discovered a trend in the processing of Hp-α, which also correlates with Hb binding. Hp-α is formed when the proform of Hp is cleaved after Arg102 or Arg161, depending on the genotype, in the ER before entering the Golgi (11). Following this cleavage, a newly formed C-terminal Arg on the Hp–α-chain is thought to be removed by an unknown carboxypeptidase while in circulation (27) (Fig. 1). Our data revealed that for serum Hp this clipping reaction is not complete, as evidenced by detecting in the LC-MS data two distinct Hp-α variants, with and without Arg residue, for both Hp-α1 and Hp-α2 (Fig. 4F). The fraction of nonclipped Hp-α (Hp-αR) follows a similar trend as observed for the glycosylation patterns of Hp-β in Hp 2-2 oligomers. Higher Hp oligomers have a significantly higher level of Hp-αR (∼30% of total Hp-α) compared to smaller size Hp 2-2 oligomers (∼20%) or Hp 1-1 (∼13%) (Fig. 4G). Similar behavior of the nonclipped Hp-αR form was observed for oligomers of the Hp 2-1 genotype (SI Appendix, Fig. S6 B and C). Moreover, we observed that the Hp containing the Arg is about two times more abundant in the Hb-unbound (Hb-) fractions of Hp 2-2 compared to the Hb-bound (Hb+) fraction (Fig. 4H). For the Hp 1-1 genotype, there was no difference between Hb-bound and Hb-unbound fractions, with overall lower levels of Hp-αR forms in Hp 1-1 compared to Hp 2-2.
Overall, our data highlight that various features are influencing the binding efficacy of Hp to Hb. This fine-tuning of binding is at the molecular level affected by the cumulative effect of many factors, including the genotype, the heterogeneous glycosylation pattern, and the Hp-α posttranslational proteolytic processing.
Discussion
Since the discovery of Hp in human plasma (65), more than 3,000 articles have been published wherein Hp was mentioned in the abstract, title, or keywords. Such immense interest is likely driven by the vital function of Hp in protecting tissues against Hb-derived heme toxicity (15). Under normal physiological conditions, free intravascular Hb is bound by Hp and rapidly cleared from circulation through high-affinity binding to the macrophage scavenger receptor CD163 (7). However, in the case of severe and acute conditions such as sepsis, thrombosis, malaria, or sickle cell anemia, the heme-group released from free Hb can trigger vascular and organ pathologies that may lead to severe complications (66). Due to its detoxifying qualities, Hp is often considered as the major therapeutic candidate for critically ill patients with excessive intravascular hemolysis (30, 67). The primary bottleneck for expanding the therapeutic use of Hp is in its sheer molecular complexity, mainly originating from the distinctive conformational landscape observed for the different Hp genotypes. Multiple studies have linked the Hp genotype as a risk factor for vascular complications of diabetes (68, 69), which can be partly explained by the differential antioxidant capacity observed for the different Hp genotypes (44). Hp antioxidant function has been found to correlate with the size of Hp oligomers and their ability to neutralize the oxidative effects of Hb. However, these functional assays largely ignore factors that drive differences in Hp–Hb binding at the more-detailed molecular level. For all of these reasons, a detailed characterization of Hp structure is critical for the future application of Hp as a potential biomarker and as a therapeutic agent.
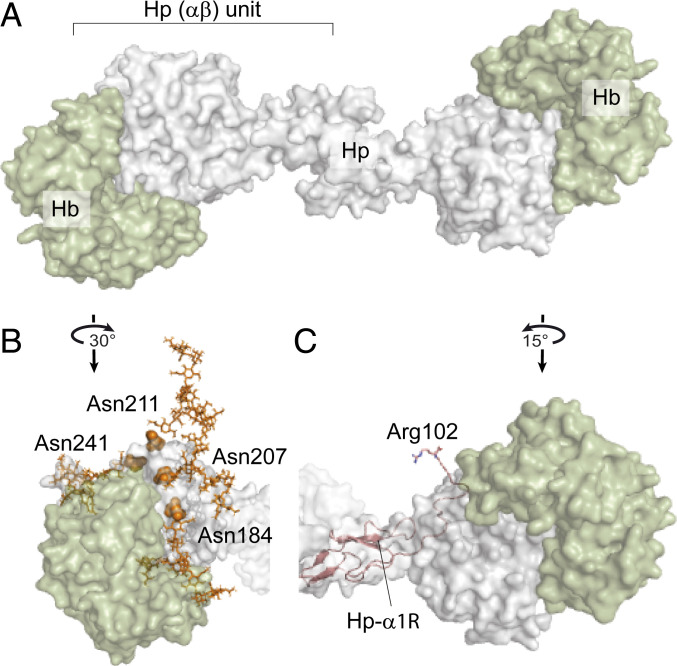
For a long time it was a great challenge to determine a crystal structure of an Hp–Hb complex and define the structural basis for Hp-mediated recognition of Hb. Eventually, a high-resolution structural model of porcine haptoglobin provided a first necessary explanation for the very tight binding between Hp and Hb (21), wherein primarily the Hp serine protease domains form extensive interactions with both the α- and β-chains of Hb. In this work, we investigated the implications of the human genetic polymorphism on the molecular diversity of Hp, which affects not only Hp oligomerization but also Hp glycosylation and Hp-α proteolytic processing. Initially, mass photometry measurements of Hp from different genotypes before and after fractionation using an Hb-affinity agarose column demonstrated that higher–molecular-mass Hp oligomers bind Hb weaker than lower–molecular-mass oligomers. To put these data into context, we characterized the proteoforms of Hp oligomers from the three major Hp genotypes, 1-1, 2-1, and 2-2 by using an integrative mass spectrometry approach combining native MS and intact mass denaturing HILIC-LC-MS. By further investigating Hp proteoforms between and within genotypes, we observed a clear correlation between changes in glycosylation and the size of Hp oligomer. Notably, the level of fucosylation and glycan branching is decreasing with the increasing size of the Hp oligomer in both Hp 2-1 and Hp 2-2. Considering that higher Hp oligomers are weaker binders of Hb, it is tempting to conclude that both extended fucosylation and branching enhance the binding of Hp to Hb, which however, would partly contradict previous data from Wu et al. (48), where Hp 1-1 with a higher branching showed lower affinity to Hb. However, the two observations are rather complementary since in the study of Wu et al. the effect of glycosylation on Hp–Hb binding was described only for the Hp 1-1 genotype. We extend this observation by exploring global glycosylation differences among the three most common Hp genotypes and reveal a relation between the glycosylation pattern and the quaternary structure of Hp. The direct effect of these glycans on the Hp–Hb interaction is difficult to assess; nonetheless, based on the structural model, all of the glycosylation sites are proximal to the Hp–Hb interaction interface (Fig. 5 A and B), which strongly suggests their involvement.
Fig. 5.
Assessing the effect of Hp glycosylation and proteolytic processing on Hp–Hb binding based on the structure of the human Hp–Hb–CD163 complex (PDB ID code 4WJG). (A) Structure of Hp 1-1 (gray) fully saturated with two Hb (αβ) dimers (green). (B) Structure of one Hp (αβ) unit bound to Hb (αβ) highlighting the Asn residues of Hp-β that carry biantennary N-glycans displayed in orange balls and sticks, respectively, revealing their proximity to the interaction site. (C) Cleaved Hp-α1R chain fitted into the available structural model of Hp 1-1 (shown as a pink cartoon) reveals that the C-terminal Arg102 is in the proximity of the Hb-α molecule (green).
In addition to our observation of distinctive glycosylation patterns between different Hp oligomers, we detected that proteolytic processing of the Hp–α-chain is also dependent on the oligomeric state of Hp. Prohaptoglobin is cleaved after the Arg102/161 residue in the sequence motif PVQR by the complement C1r-LP (11). Interestingly, this is in striking resemblance with the first enzymatic event in the classic pathway of complement activation, where the autoactivation of C1r involves the cleavage after the Arg446 located in the same sequence motif (PVQR) (70). The newly created C-terminal Arg is directly involved in the interaction observed in the C1r enzyme–product complex (71). While the functionality of C1r highlights the great importance of the C-terminal Arg following autoactivation, the function of the C-terminal Arg in the Hp–Hb complex is so far unknown. Interestingly, this Arg was reported to be partially removed in plasma by a hitherto unknown carboxypeptidase (27). Our data support this finding as we detected variants with and without Arg at the C terminus of the Hp–α-chain (Fig. 4F). Remarkably, we observed that the levels of the form still possessing the C-terminal Arg are not equally distributed among the Hp oligomers but show a continuous trend, similar to the glycosylation of Hp-β. Higher Hp oligomers, which have weaker binding to Hb, exhibit relatively higher levels of the Hp-αR isoform compared to lower Hp oligomers. We corroborate that the presence of Arg102/161 correlates with the binding affinity of Hp to Hb by detecting depleted levels of this isoform in the fraction of Hp oligomers that were enriched by Hb-affinity agarose column. Unfortunately, the available structural models of Hp–Hb complexes do not cover the C-terminal region of the Hp–α-chain; therefore, we can only speculate about the potential role of this Arg processing. Notably, by modeling the complete Hp-αR isoform and placing it within the available Hp–Hb structure, we found that the C-terminal region of Hp-α is flexible and can exist in close proximity to Hp–Hb interaction interface. This indicates that the C-terminal Arg may electrostatically influence Hb binding due to its positive charge (Fig. 5C).
While we clearly show that the oligomeric state of Hp, its glycosylation status, and proteolytic processing all correlate with the strength of Hp–Hb binding, the exact contribution of each of these factors is challenging to determine. Based on the current knowledge about Hp biosynthesis, oligomerization occurs in the ER and, subsequently, the N-glycans are sequentially processed in the Golgi (24, 26). The various structural conformations of Hp must define an action space for glycan processing enzymes in the Golgi; for example, less branched (more compact) glycans are placed on higher Hp oligomers, which may endure more steric hindrance in the space between the monomers. At the same time, our data indicate that the higher-order Hp structure somehow defines the rate of Arg clipping, potentially through steric effects that may influence the accessibility of the Arg residue for enzymatic removal. However, in this case, the causality is less obvious, as the hitherto carboxypeptidase that performs the clipping is still unknown. In conclusion, as shown here in molecular detail, the scavenging of hemoglobin by haptoglobin is influenced by multiple factors, whereby, very likely, their cumulative effect fine-tunes this crucial Hp–Hb interaction.
Materials and Methods
Chemicals and Materials.
Hp 1-1, 2-2, and mixed type were purchased from Athens Research and Technology. Human Hb was acquired from Sigma-Aldrich. Individual Hp multimers from Hp 2-2 were purified by SEC. Hp 2-1 oligomers were purified from blood serum of a single donor. Additional chemicals and materials are described in SI Appendix, Supplementary Methods.
Blue Native Gel of Hp Samples from Different Genotypes.
A system for native gel electrophoresis NativePAGE Novex Bis-Tris was obtained from Thermo Fisher Scientific. Aliquots of 5 µg of Hp protein samples (Hp1-1, Hp 2-2, and Hp mixed type) and their variants incubated with two times molar excess of Hb (αβ) unit to Hp (αβ) unit were loaded on the top of a precast polyacrylamide minidiscontinuous gel (4–16%), 10-well, 1 mm. The whole procedure was performed according to the vendor’s instructions.
SEC Separation of Hp Oligomers.
An Agilent 1290 Infinity HPLC system (Agilent Technologies) consisting of a vacuum degasser, binary pump, refrigerated autosampler with a 100-µL injector loop, thermostated two-column compartment, auto collection fraction module, and multiwavelength detector, was used in this study. The dual-column set-up, comprising a tandem Yarra 4000-Yarra 3000 (SEC-4000, 300 × 7.8 mm i.d., 3 µm, 500 Å; SEC-3000, 300 × 7.8 mm i.d., 3 µm, 290 Å) two-stage set-up. Both columns were obtained from Phenomenex. The column compartment was cooled to 17 °C while the other bays were chilled to 4 °C to minimize sample degradation. The mobile phase buffer consisted of 150 mM AMAC in water, which was filtered using a 0.22-µm disposable membrane cartridge (Millipore) before use. Typically, 200 µg in the case of commercially obtained Hp 1-1 and Hp 2-2 or 100 µL of serum (Hp 2-1) (∼7 mg of total protein) were injected per run. Elution was achieved using isocratic flow within 60 min and the flow rate was set to 350 µL min−1. The chromatograms were monitored at 280 nm and peak based fractions collected using an automated fraction collector.
Bottom-Up LC-MS/MS Analysis of Hp 2-1 SEC Fractions.
We used bottom-up LC-MS/MS analysis to determine SEC elution profile of Hp 2-1 oligomers. This information was further used to collect only the SEC fractions containing Hp assemblies that were later cleaned up by IEX and analyzed by hybrid MS. In total, we collected 99 SEC fractions, which were digested by trypsin and analyzed by bottom-up proteomics approach, basically as previously described (72). See the details in SI Appendix, Supplementary Methods.
Dual-Column IEX Separation of Hp 2-1 SEC Fractions.
Collected SEC fractions of Hp 2-1 purified from serum of one individual were further cleaned up from interfering serum proteins. The same chromatographic system was used as in the case of SEC with some minor adjustments, which will be described. Autosampler was equipped with a 2,500-µL injection loop instead of 100 µL. The dual-column set-up was comprised of a tandem IEX WAX-CAT (PolyWAX LP, 200 × 2.1 mm i.d., 5 µm, 1,000 Å; PolyCAT A, 50 × 2.1 mm i.d., 5 µm, 1,000 Å), which was previously described (42, 63, 73). Both columns were obtained from PolyLC Inc. The mobile phase consisted of Buffer A (100 mM AMAC in water) and Buffer B (2.5 M AMAC in water), which was filtered using a 0.22-µm disposable membrane cartridge (Millipore) before use. Injections were 2,500 µL of collected SEC fraction containing specific Hp oligomer per run. Elution was achieved using a multistep gradient, consisting of six transitions with increasing proportions of Buffer B: (step 1; equilibration) 0%B, 0 to 6 min; (step 2; salt gradient) 0 to 20%B, 6 to 11 min; (step 3; salt gradient) 20 to 36%B, 11 to 24 min; (step 4; high salt rinse) 36 to 100%B, 24 to 28 min; (step 5; high salt wash) 100%B, 28 to 32 min; (step 6; restoration) 100 to 0%B. The flow rate was set to 800 µL/min.
Hb-Affinity Purification for Hp.
For Hb-affinity purification for Hp, 100 µg of Hb in 200 µL of PBS was added and immobilized to NHS-activated agarose resin (Pierce NHS-activated agarose spin columns, Thermo Scientific) following the vendor’s instructions. Next, 200 µL of settled Hb-agarose gel was incubated with 200 µg of either Hp 1-1, Hp 2-2, or Hp M at a final volume of 400 µL for 2 h with end-over-end mixing at room temperature. The mixture of Hp and Hb-agarose slurry was washed with 2 mL PBS. The flow-through and washes were pooled as Hb-unbound Hp (Hb− fraction). The Hp bound to Hb (Hb+ fraction) was eluted from Hb-agarose gel with 2 mL of elution buffer (Pierce IgG Elution Buffer, Thermo Scientific). The Hb− and Hb+ Hp fractions were concentrated and buffer exchanged into 150 mM ammonium acetate through several dilution and concentration cycles using ultrafiltration spin columns with 10-kDa cutoff (Vivaspin 500 µL; Sartorius Stedim Biotech).
Preparation of Hp and Hp–Hb Samples for Mass Photometry Measurements.
Hp and Hb stock solutions were prepared in 150 mM ammonium acetate solution (pH 7.6) at a concentration of 1 mg/mL and 2 mg/mL, respectively. Hp and Hb were mixed and incubated at room temperature for 30 min with a two times molar excess of Hb (αβ) unit compared to Hp (αβ) unit.
Mass Photometry.
Mass photometry data were collected on a Refeyn OneMP instrument. The instrument was calibrated with a native marker protein standard mixture (NativeMark Unstained Protein Standard, Thermo Scientific), which contains proteins in the range from 20 to 1,200 kDa. The following masses were used to generate a standard calibration curve: 66, 146, 480, and 1,048 kDa. Borosilicate coverslips were extensively cleaned with Milli-Q water and isopropanol prior to the measurements. Typically, 10 µL of Hp was applied to 10 µL buffer on a coverslip resulting in a final concentration 2 to 5 nM. Mass profiles of following samples were acquired: Hp 1-1, Hp 2-2, Hp mixed type, Hp (1, 1)-Hb, Hp (2, 2)-Hb, Hp(M)-Hb, Hp (1, 1) Hb+ fraction, Hp (1, 1) Hb− fraction, Hp (2, 2) Hb+ fraction, Hp (2, 2) Hb− fraction, Hp(M) Hb+ fraction, Hp(M) Hb− fraction. Movies were acquired by using AcquireMP software for 60 to 100 s with a frame rate of 100 Hz. From 1,000 to 4,000 particle landing events were detected per movie. We used for threshold 1 and threshold 2 parameters 1.50 and 0.25, respectively. All data were processed in DiscoverMP software. Masses of Hp oligomers were estimated by fitting a Gaussian distribution into mass histograms and taking the value at the mode of the distribution. Probability density functions were extracted and overlayed in R to generate Fig. 2 B–H.
Native MS Analysis Using a QE-UHMR Mass Spectrometer.
Samples were introduced into a modified Q Exactive Orbitrap instrument with Ultra High Mass Range detection capability (QE-UHMR; Thermo Fisher Scientific) via gold-coated borosilicate capillaries prepared in-house (74). The following MS parameters were used. Capillary voltage: 1,100 to 1,300 V in positive ion mode; source temperature: 250 °C; S-lens RF level: 200; in-source trapping desolvation voltage: −100 V; higher-energy collisional dissociation energy: 1 V; automatic gain control mode: Fixed; noise level parameter: 3.64; resolution: 35,000 (at m/z 200); injection time: 50 ms. Ion transfer optics and voltage gradients throughout the instrument were manually tuned to achieve optimal transmission of each Hp multimer. Nitrogen was used in the higher-energy collisional dissociation cell with trapping gas pressure setting set to 3. The instrument was calibrated in the range of up to 11,000 m/z using a CsI solution. Acquisition of spectra was performed by averaging 1,000 µscans in the time domain and subsequently recording 10 scans (10 µscans each). Spectra were viewed in Excalibur QualBrowser software (Thermo Fisher Scientific).
The Intact Mass LC-MS Analysis.
Chromatographic separation of intact protein samples was performed on a Thermo Scientific Vanquish Flex UHPLC equipped either with MAbPac column (2.1 mm × 100 mm) for reversed-phase separation of Hp chains or with ACQUITY UPLC Glycoprotein BEH Amide column (300 Å, 1.7-µm particles, 2.1 mm × 150 mm) for HILIC LC-MS of Hp-β glycoproteoforms. Next 2 to 5 µg of Hp was loaded on either column heated to 80 °C. In the case of reverse phase, LC-MS runtime was set to 12 min with flow rate of 250 µL/min. Gradient elution was performed using mobile phases A (water/0.1% formic acid) and B (ACN/0.1% formic acid): 20 to 40% B for 7 min, and 40 to 60% B over additional 1 min. For HILIC, the runtime was set to 25 min with a flow rate of 250 µL/min. The gradient was created by mobile phases A (water/0.1% trifluoroacetic acid) and B (ACN/0.1% trifluoroacetic acid): 85 to 67% B for 0.5 min, 67 to 60% B for 17 min, and 60 to 0% B over 1 min. Trifluoroacetic acid was used in HILIC for ion-pairing of amine groups of Hp proteins to reduce the unspecific binding of Hp-β glycoproteoforms. All intact mass LC-MS experiments were performed on a Thermo Fisher Scientific Orbitrap Fusion Lumos Tribrid mass spectrometer. LC-MS data were collected with an instrument set to the Intact Protein Mode and low-pressure setting. The Orbitrap resolution parameter was set to 7,500 (at 200 m/z) corresponding to a 16-ms transient signal. Full MS scans were acquired for the range of 900 to 3,000 m/z with the automatic gain control target set to 3e6. The maximum injection time was defined at 50 ms with 5 µscans recorded. Source-induced desolvation (SID) voltage was set to 15 V.
Identification of Hp Glycoproteoforms and Their Assemblies.
Raw native spectra were deconvoluted with UniDec in order to obtain zero-charge mass distributions (75). Raw intact mass LC-MS data were deconvoluted using sliding window deconvolution and ReSpect algorithm (Protein Deconvolution 4.0, Thermo Fisher Scientific). Average masses of common monosaccharides were used for the PTM assignments, including hexose (Hex; 162.14 Da), N-acetylhexosamine (HexNAc; 203.20 Da), and deoxyhexose (Fuc; 146.14 Da). The theoretical average masses of the Hp amino acid chains were calculated by extracting protein sequences from the UniProt database (https://www.uniprot.org/uniprot/) (SI Appendix, Fig. S1). The possible glycosylation compositions were calculated by ranging the number of monosaccharides in the following boundaries: 15:28 for Hex, 12:24 for HexNAc, and 0:8 for Fuc, as per the Hp–β-chain. Average masses of native proteoform assemblies determined with native MS and denatured and reduced Hp glycoproteoforms determined by intact mass HILIC-LC-MS were matched against the generated list of theoretical glycoproteoforms with 100-ppm tolerance. Glycans of matched glycoproteoforms were deduced based on known biosynthetic pathways and verified based on the N-glycans reported for Hp-β (35, 48). All additional data analysis and visualization were performed in R supplemented with the ggplot2 package (76, 77).
In Silico Reconstruction of Native Mass Profiles of Purified Hp Multimers.
First, the masses of theoretical Hp–β- and Hp–α-chains were assigned the abundances extracted from intact mass LC-MS analysis of denatured and reduced Hp 1-1, and purified multimers of Hp 2-2 and Hp 2-1. Next, the glycoproteoforms of distinct Hp multimers were calculated for each Hp genotype as sum of permuted chain masses in accordance with the composition formulas: (α1)2(β)2 for Hp 1-1, (α1)2(α2)n-2(β)n for Hp 2-1 (2 < n < 6), and (α2)n(β)n for Hp 2-2 (2 < n < 7). Then, for each native glycoproteoform of Hp multimer, we calculated relative abundance as the product of multiplying normalized relative abundances of involved Hp–α- and Hp–β-chains obtained in intact mass LC-MS experiments. Finally, mass profiles were generated by binning the data points into mass ranges and kernel smoothing the resulting binned data. Pearson correlation was used to estimate similarities between experimental and in silico reconstructed mass profiles. The overview of deconvoluted native MS profiles and the in silico reconstructed masses of Hp proteoforms can be interactively viewed at htmlpreview.github.io/?https://github.com/stamara6/Haptoglobin/blob/master/hp_oligomers.html. The underlying code and used datasets are available at https://github.com/stamara6/Haptoglobin.
Structural Modeling of Hp-α1R and N-Glycans on Hp-β.
For comparative modeling of the complete Hp-α1R chain, we used web portal Phyre2 (78), which provides tools to predict and analyze protein structure. The monomer of Hp from PDB ID code 4WJG was selected as a template for the structural remodeling of the soluble Hp to its putative form. N-glycans were modeled using Glycam online prediction service (glycam.org/), which uses AMBER for energy minimization. The resulting models were eventually processed and visualized using PyMOL Molecular Graphic System.
Data Availability.
The raw and processed data used in this work have been uploaded to Figshare (https://doi.org/10.6084/m9.figshare.12098592.v1).
Supplementary Material
Acknowledgments
We acknowledge support from the Netherlands Organization for Scientific Research (NWO), funding the Netherlands Proteomics Centre through the X-omics Road Map program (Project 184.034.019). V.F. and A.J.R.H. acknowledge further support by the NWO TOP-Punt Grant 718.015.003, NWO Satin Grant 731.017.202, and the European Union Horizon 2020 program INFRAIA project Epic-XS (Project 823839).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The raw and processed data used in this work have been uploaded to Figshare (https://doi.org/10.6084/m9.figshare.12098592.v1).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002483117/-/DCSupplemental.
References
- 1.Crow J. F., Felix Bernstein and the first human marker locus. Genetics 133, 4–7 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avent N. D., Large-scale blood group genotyping: Clinical implications. Br. J. Haematol. 144, 3–13 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Delanghe J., Speeckaert M., De Buyzere M. L., Langlois M., Torck M., Human plasma protein polymorphisms and the persistence of cultural diversity. Proc. Natl. Acad. Sci. U.S.A. 109, E2914 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chappell S.et al., A polymorphism of the alpha1-antitrypsin gene represents a risk factor for liver disease. Hepatology 47, 127–132 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Topic A., Juranic Z., Jelic S., Magazinovic I. G., Polymorphism of alpha-1-antitrypsin in hematological malignancies. Genet. Mol. Biol. 32, 716–719 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox D. W., Andrews B. J., Wills D. E., Genetic polymorphism of alpha 2HS-glycoprotein. Am. J. Hum. Genet. 38, 699–706 (1986). [PMC free article] [PubMed] [Google Scholar]
- 7.Kristiansen M.et al., Identification of the haemoglobin scavenger receptor. Nature 409, 198–201 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Langlois M. R., Delanghe J. R., Biological and clinical significance of haptoglobin polymorphism in humans. Clin. Chem. 42, 1589–1600 (1996). [PubMed] [Google Scholar]
- 9.Maeda N., Yang F., Barnett D. R., Bowman B. H., Smithies O., Duplication within the haptoglobin Hp2 gene. Nature 309, 131–135 (1984). [DOI] [PubMed] [Google Scholar]
- 10.Raugei G.et al., Sequence of human haptoglobin cDNA: Evidence that the alpha and beta subunits are coded by the same mRNA. Nucleic Acids Res. 11, 5811–5819 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang F., Brune J. L., Baldwin W. D., Barnett D. R., Bowman B. H., Identification and characterization of human haptoglobin cDNA. Proc. Natl. Acad. Sci. U.S.A. 80, 5875–5879 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boettger L. M.et al., Recurring exon deletions in the HP (haptoglobin) gene contribute to lower blood cholesterol levels. Nat. Genet. 48, 359–366 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martosella J., Zolotarjova N., Multi-component immunoaffinity subtraction and reversed-phase chromatography of human serum. Methods. Mol. Biol. 425, 27–39 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Xiao M.et al., Analysis of the whole serum proteome using an integrated 2D LC-MS/MS system. Anal. Methods 6, 7157–7160 (2014). [Google Scholar]
- 15.Alayash A. I., Andersen C. B. F., Moestrup S. K., Bülow L., Haptoglobin: The hemoglobin detoxifier in plasma. Trends Biotechnol. 31, 2–3 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Allison A. C., Rees W. A., The binding of haemoglobin by plasma proteins (haptoglobins); its bearing on the renal threshold for haemoglobin and the aetiology of haemoglobinuria. BMJ 2, 1137–1143 (1957). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tseng C. F., Lin C. C., Huang H. Y., Liu H. C., Mao S. J. T., Antioxidant role of human haptoglobin. Proteomics 4, 2221–2228 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Huntoon K. M.et al., The acute phase protein haptoglobin regulates host immunity. J. Leukoc. Biol. 84, 170–181 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smithies O., Zone electrophoresis in starch gels: Group variations in the serum proteins of normal human adults. Biochem. J. 61, 629–641 (1955). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smithies O., Walker N. F., Notation for serum-protein groups and the genes controlling their inheritance. Nature 178, 694–695 (1956). [DOI] [PubMed] [Google Scholar]
- 21.Andersen C. B. F.et al., Structure of the haptoglobin-haemoglobin complex. Nature 489, 456–459 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Bowman B. H., Kurosky A., Haptoglobin: The evolutionary product of duplication, unequal crossing over, and point mutation. Adv. Hum. Genet. 12, 189–261 (1982). [DOI] [PubMed] [Google Scholar]
- 23.Wejman J. C., Hovsepian D., Wall J. S., Hainfeld J. F., Greer J., Structure and assembly of haptoglobin polymers by electron microscopy. J. Mol. Biol. 174, 343–368 (1984). [DOI] [PubMed] [Google Scholar]
- 24.Hanley J. M., Haugen T. H., Heath E. C., Biosynthesis and processing of rat haptoglobin. J. Biol. Chem. 258, 7858–7869 (1983). [PubMed] [Google Scholar]
- 25.Wassler M., Fries E., Proteolytic cleavage of haptoglobin occurs in a subcompartment of the endoplasmic reticulum: Evidence from membrane fusion in vitro. J. Cell Biol. 123, 285–291 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haugen T. H., Hanley J. M., Heath E. C., Haptoglobin. A novel mode of biosynthesis of a liver secretory glycoprotein. J. Biol. Chem. 256, 1055–1057 (1981). [PubMed] [Google Scholar]
- 27.Mikkat S., Koy C., Ulbrich M., Ringel B., Glocker M. O., Mass spectrometric protein structure characterization reveals cause of migration differences of haptoglobin α chains in two-dimensional gel electrophoresis. Proteomics 4, 3921–3932 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Wicher K. B., Fries E., Prohaptoglobin is proteolytically cleaved in the endoplasmic reticulum by the complement C1r-like protein. Proc. Natl. Acad. Sci. U.S.A. 101, 14390–14395 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wicher K. B., Fries E., Haptoglobin, a hemoglobin-binding plasma protein, is present in bony fish and mammals but not in frog and chicken. Proc. Natl. Acad. Sci. U.S.A. 103, 4168–4173 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaer C. A.et al., Phenotype-specific recombinant haptoglobin polymers co-expressed with C1r-like protein as optimized hemoglobin-binding therapeutics. BMC Biotechnol. 18, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinderyckx M., Jacobs P., Bollen A., Secretion of glycosylated human recombinant haptoglobin in baculovirus-infected insect cells. Mol. Biol. Rep. 13, 225–232 (1988). [DOI] [PubMed] [Google Scholar]
- 32.Alayash A. I., Haptoglobin: Old protein with new functions. Clin. Chim. Acta 412, 493–498 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Dobryszycka W., Biological functions of haptoglobin—New pieces to an old puzzle. Eur. J. Clin. Chem. Clin. Biochem. 35, 647–654 (1997). [PubMed] [Google Scholar]
- 34.Suttapitugsakul S., Sun F., Wu R., Recent advances in glycoproteomic analysis by mass spectrometry. Anal. Chem. 92, 267–291 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S., Jiang K., Sun C., Lu H., Liu Y., Quantitative analysis of site-specific N-glycans on sera haptoglobin β chain in liver diseases. Acta Biochim. Biophys. Sin. (Shanghai) 45, 1021–1029 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Pompach P.et al., Site-specific glycoforms of haptoglobin in liver cirrhosis and hepatocellular carcinoma. Mol. Cell. Proteomics 12, 1281–1293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pompach P.et al., Protein and site specificity of fucosylation in liver-secreted glycoproteins. J. Proteome Res. 13, 5561–5569 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang S., Shang S., Li W., Qin X., Liu Y., Insights on N-glycosylation of human haptoglobin and its association with cancers. Glycobiology 26, 684–692 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Lauc G.et al., Genomics meets glycomics-the first GWAS study of human N-Glycome identifies HNF1α as a master regulator of plasma protein fucosylation. PLoS Genet. 6, e1001256 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klarić L.et al., Glycosylation of immunoglobulin G is regulated by a large network of genes pleiotropic with inflammatory diseases. Sci. Adv. 6, eaax0301 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng T.-M.et al., Immunochemical property of human haptoglobin phenotypes: Determination of plasma haptoglobin using type-matched standards. Clin. Biochem. 40, 1045–1056 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Lin Y.-H., Zhu J., Meijer S., Franc V., Heck A. J. R., Glycoproteogenomics: A frequent gene polymorphism affects the glycosylation pattern of the human serum fetuin/α-2-HS-glycoprotein. Mol. Cell. Proteomics 18, 1479–1490 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Javid J., The effect of haptoglobin polymer size on hemoglobin binding capacity. Vox Sang. 10, 320–325 (1965). [DOI] [PubMed] [Google Scholar]
- 44.Melamed-Frank M.et al., Structure-function analysis of the antioxidant properties of haptoglobin. Blood 98, 3693–3698 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Hamaguchi H., Purification and some properties of the three common genetic types of haptoglovins and the hemoglobin-haptoglobin complexes. Am. J. Hum. Genet. 21, 440–456 (1969). [PMC free article] [PubMed] [Google Scholar]
- 46.Nielsen M. J.et al., Haptoglobin-related protein is a high-affinity hemoglobin-binding plasma protein. Blood 108, 2846–2849 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Spagnuolo M. S., Cigliano L., Maresca B., Pugliese C. R., Abrescia P., Identification of plasma haptoglobin forms which loosely bind hemoglobin. Biol. Chem. 392, 371–376 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Wu D., Struwe W. B., Harvey D. J., Ferguson M. A. J., Robinson C. V., N-glycan microheterogeneity regulates interactions of plasma proteins. Proc. Natl. Acad. Sci. U.S.A. 115, 8763–8768 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu D., Li J., Struwe W. B., Robinson C. V., Probing N-glycoprotein microheterogeneity by lectin affinity purification-mass spectrometry analysis. Chem. Sci. 10, 5146–5155 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young G.et al., Quantitative mass imaging of single biological macromolecules. Science 360, 423–427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamara S., Hoek M., Scheltema R. A., Leney A. C., Heck A. J. R., A colorful pallet of B-phycoerythrin proteoforms exposed by a multimodal mass spectrometry approach. Chem 5, 1302–1317 (2019). [Google Scholar]
- 52.Yang Y.et al., Hybrid mass spectrometry approaches in glycoprotein analysis and their usage in scoring biosimilarity. Nat. Commun. 7, 13397 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hwang P. K., Greer J., Interaction between hemoglobin subunits in the hemoglobin. Haptoglobin complex. J. Biol. Chem. 255, 3038–3041 (1980). [PubMed] [Google Scholar]
- 54.Chiancone E., Dissociation of hemoglobin into subunits. II. Human oxyhemoglobin: Gel filtration studies. J. Biol. Chem. 243, 1212–1219 (1968). [PubMed] [Google Scholar]
- 55.Rensen P. C.et al., Determination of the upper size limit for uptake and processing of ligands by the asialoglycoprotein receptor on hepatocytes in vitro and in vivo. J. Biol. Chem. 276, 37577–37584 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Okazaki T., Yanagisawa Y., Nagai T., Determination of intermediates of hemoglobin-haptoglobin complex of haptoglobin polymers by “Crossed hemoglobin electrophoresis”. Anal. Biochem. 239, 123–129 (1996). [DOI] [PubMed] [Google Scholar]
- 57.Okazaki T., Nagai T., Difference in hemoglobin-binding ability of polymers among haptoglobin phenotypes. Clin. Chem. 43, 2012–2013 (1997). [PubMed] [Google Scholar]
- 58.Stødkilde K., Torvund-Jensen M., Moestrup S. K., Andersen C. B. F., Structural basis for trypanosomal haem acquisition and susceptibility to the host innate immune system. Nat. Commun. 5, 5487 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Smith L. M., Kelleher N. L.; Consortium for Top Down Proteomics , Proteoform: A single term describing protein complexity. Nat. Methods 10, 186–187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Čaval T., Tian W., Yang Z., Clausen H., Heck A. J. R., Direct quality control of glycoengineered erythropoietin variants. Nat. Commun. 9, 3342 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toby T. K., Fornelli L., Kelleher N. L., Progress in top-down proteomics and the analysis of proteoforms. Annu. Rev. Anal. Chem. (Palo Alto, Calif.) 9, 499–519 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leney A. C., Heck A. J. R., Native mass spectrometry: What is in the name? J. Am. Soc. Mass Spectrom. 28, 5–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Franc V., Zhu J., Heck A. J. R., Comprehensive proteoform characterization of plasma complement component C8αβγ by hybrid mass spectrometry approaches. J. Am. Soc. Mass Spectrom. 29, 1099–1110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buszewski B., Noga S., Hydrophilic interaction liquid chromatography (HILIC)—A powerful separation technique. Anal. Bioanal. Chem. 402, 231–247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Polonovski M., Existence dans le plasma sanguim d’une substance activant l’action peroxydasique de l’hemoglobine. C. R. Soc. Biol. 129, 457–460 (1938). [Google Scholar]
- 66.Schaer D. J., Buehler P. W., Alayash A. I., Belcher J. D., Vercellotti G. M., Hemolysis and free hemoglobin revisited: Exploring hemoglobin and hemin scavengers as a novel class of therapeutic proteins. Blood 121, 1276–1284 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schaer D. J., Buehler P. W., Cell-free hemoglobin and its scavenger proteins: New disease models leading the way to targeted therapies. Cold Spring Harb. Perspect. Med. 3, 1–18 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Levy A. P.et al., Haptoglobin phenotype and vascular complications in patients with diabetes. N. Engl. J. Med. 343, 969–970 (2000). [DOI] [PubMed] [Google Scholar]
- 69.Roguin A.et al., Haptoglobin phenotype as a predictor of restenosis after percutaneous transluminal coronary angioplasty. Am. J. Cardiol. 87, 330–332, A9 (2001). [DOI] [PubMed] [Google Scholar]
- 70.Budayova-Spano M.et al., Monomeric structures of the zymogen and active catalytic domain of complement protease c1r: Further insights into the c1 activation mechanism. Structure 10, 1509–1519 (2002). [DOI] [PubMed] [Google Scholar]
- 71.Kardos J.et al., Revisiting the mechanism of the autoactivation of the complement protease C1r in the C1 complex: Structure of the active catalytic region of C1r. Mol. Immunol. 45, 1752–1760 (2008). [DOI] [PubMed] [Google Scholar]
- 72.van de Waterbeemd M.et al., Dissecting ribosomal particles throughout the kingdoms of life using advanced hybrid mass spectrometry methods. Nat. Commun. 9, 2493 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Havugimana P. C., Wong P., Emili A., Improved proteomic discovery by sample pre-fractionation using dual-column ion-exchange high performance liquid chromatography. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 847, 54–61 (2007). [DOI] [PubMed] [Google Scholar]
- 74.van de Waterbeemd M.et al., High-fidelity mass analysis unveils heterogeneity in intact ribosomal particles. Nat. Methods 14, 283–286 (2017). [DOI] [PubMed] [Google Scholar]
- 75.Marty M. T.et al., Bayesian deconvolution of mass and ion mobility spectra: From binary interactions to polydisperse ensembles. Anal. Chem. 87, 4370–4376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wickham H., ggplot2: Elegant Graphics for Data Analysis, (Springer, New York, 2009). [Google Scholar]
- 77.R Core Team , R: A Language and Environment for Statistical Computing, (R Foundation for Statistical Computing, Vienna, 2012). [Google Scholar]
- 78.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. E., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw and processed data used in this work have been uploaded to Figshare (https://doi.org/10.6084/m9.figshare.12098592.v1).